Abstract
The transcriptional response of Saccharomyces cerevisiae to cell wall stress is mainly mediated by the cell wall integrity (CWI) pathway through the MAPK Slt2 and the transcription factor Rlm1. Once activated, Rlm1 interacts with the chromatin remodeling SWI/SNF complex which locally alters nucleosome positioning at the target promoters. Here we show that the SAGA complex plays along with the SWI/SNF complex an important role for eliciting both early induction and sustained gene expression upon stress. Gcn5 co-regulates together with Swi3 the majority of the CWI transcriptional program, except for a group of genes which are only dependent on the SWI/SNF complex. SAGA subunits are recruited to the promoter of CWI-responsive genes in a Slt2, Rlm1 and SWI/SNF-dependent manner. However, Gcn5 mediates acetylation and nucleosome eviction only at the promoters of the SAGA-dependent genes. This process is not essential for pre-initiation transcriptional complex assembly but rather increase the extent of the remodeling mediated by SWI/SNF. As a consequence, H3 eviction and Rlm1 recruitment is completely blocked in a swi3Δ gcn5Δ double mutant. Therefore, SAGA complex, through its histone acetylase activity, cooperates with the SWI/SNF complex for the mandatory nucleosome displacement required for full gene expression through the CWI pathway.
INTRODUCTION
Adverse environmental conditions elicit in yeast cellular stress responses mediated by specific signaling pathways, most of them regulated through mitogen-activated protein kinase (MAPK) cascades. The activation of the MAPK induces changes in the global transcriptional pattern, generating efficient adaptive responses that guarantee cell survival. In Saccharomyces cerevisiae, stressful conditions damaging the cell wall, an essential structure for viability, trigger the cell wall integrity (CWI) pathway coordinated by the MAPK Slt2 (reviewed in (1)). CWI is sensed through cell membrane proteins, mainly Mid2 and Wsc1. Under activation conditions, these sensors interact with the guanine nucleotide exchange factor Rom2, activating the small GTPase Rho1, which then interacts and activates Pkc1. The main role of activated Pkc1 is to trigger a MAPK module comprising the MAPKKK Bck1, the redundant MAPKKs Mkk1/Mkk2 and the MAPK Slt2/Mpk1. Phosphorylation of the MAPK Slt2 leads to the activation of the transcription factors SCB-binding factor complex: Swi4/Swi6 (SBF) (2) and Rlm1 (3). Slt2 regulates the Rlm1 transcriptional activation function by phosphorylation of Ser427 and Thr439 (4), being this transcription factor responsible for most of the transcriptional output of the CWI pathway upon cell wall stress (5,6). Additionally, a non catalytic mechanism by which Slt2 and its pseudokinase Mlp1 activate SBF to regulate the transcription of a small subset of CWI-responsive genes has been described (7,8).
The high complexity of DNA packaged into chromatin points the removal of nucleosomes at the promoter as an important step in transcriptional activation. Recently, we have characterized the essential role of the SWI/SNF adenosine triphosphate (ATP)-dependent chromatin remodeling complex in developing adequate transcriptional responses under cell wall stress conditions. Upon activation of the MAPK Slt2, SWI/SNF is recruited to the CWI-responsive gene promoters to locally alters nucleosome positioning, allowing further Rlm1 binding at previously occluded Rlm1-binding sites and accessibility of the RNA Pol II to DNA (9).
Transcription initiation is therefore a complicated process that requires the coordinated activities of not only one co-activator but a large number of factors to ensure an appropriate regulation. In addition to ATP chromatin remodeling machinery, the activity of histone modifier complexes, as the acetyltransferase SAGA complex, is also required. SAGA (Spt-Ada-Gcn5-acetyltransferase) is a multi-subunit complex (18–20 proteins) that is conserved from yeast to mammals (10) and has two known enzymatic modules that mediate acetylation (the histone acetyltransferase (HAT) Gcn5 together with Ada2, Ada3 and Sgf29) (11–13) and deubiquitination (the ubiquitin-specific protease Ubp8, Sgf11, Sgf73 and Sus1) (14–18) of histones as well as non-histone substrates. H3 acetylation by Gcn5 (primarily at K14 andK9 (19)) has long been positively correlated with transcription (20,21), indeed, Gcn5 is generally recruited to the promoter of active genes (22). Acetylation of histones neutralizes the positive charge of the histone tails and decreases their affinity for the negatively charged DNA affecting higher order packing of chromatin that would otherwise be inhibitory for regulatory factors recruitment and transcription (23,24). This modification also create specific binding surfaces for bromodomain-containing proteins, providing evidences that acetyl-lysines in histones function to recruit or stabilize machinery involved in DNA-mediated processes (25–27). However, other SAGA subunits, as Spt3 and Spt8, enhance pre-initiation complex assembly by delivering TBP (TATA-binding protein) to promoters, a mechanism that goes beyond HAT activity (28–30) and that has been related to the expression of highly regulated genes that are induced under stress conditions (31,32).
In the context of stress, the role of the yeast SAGA complex has been largely studied in the upregulation of gene expression mediated by heat shock (31,33,34), acute glucose limitation (35) and DNA-damage (36–38). Related to MAPK-signaling pathways, SAGA also participates in the response to osmotic stress through the MAPK Hog1, being targeted to the osmoresponsive genes to mediate RNA Pol II recruitment and gene expression, independently of its HAT activity (39,40). In contrast, under oxidative conditions, Gcn5 is recruited to promoters by the MAPK Atf1 in Schizosaccharomyces pombe to mediate the histone acetylation necessary for Pol II progression along genes, although Pol II recruitment is not affected by the lack of Gcn5 during the oxidative response (41). Accordingly, the expression of specific groups of genes is more sensitive to the deletion of subunits within one module versus another, making different the participation of SAGA complex in the regulation of each stress response. Moreover, cooperation between different chromatin modifying complexes seems to be necessary for regulating chromatin structure and proper transcriptional activation, without a concerted mechanism of activation for the different environmental conditions (42).
Here, we show that the SAGA complex plays along with the chromatin remodeling SWI/SNF complex a critical role in orchestrating the transcriptional responses regulated by Rlm1 and the MAPK Slt2 in response to cell wall stress. Gcn5 co-regulates, together with Swi3, the majority of the transcriptional response induced upon stress. Furthermore, the SAGA complex enters on cell wall responsive gene promoters under cell wall stress to mediate H3 acetylation cooperating with the SWI/SNF complex for eliciting nucleosome reorganization and gene expression in response to cell wall stress through the CWI pathway.
MATERIALS AND METHODS
Yeast strains, plasmids and growth conditions
Experiments were performed with the S. cerevisiae wild-type (WT) strain BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) and mutant derivatives provided by Euroscarf (Frankfurt, Germany). The tagged strains in BY4741 background WT SPT20-13MYC::HIS3, WT GCN5-13MYC::HIS3 and the corresponding rlm1Δ SPT20-13MYC::HIS3, slt2Δ SPT20-13MYC::HIS3 and snf2Δ SPT20-13MYC::HIS3 mutants were obtained using the one-step polymerase chain reaction (PCR)-mediated technique for gene modification (43). The fragment including the 13Myc-His3 was amplified by PCR using primers designed as described and the pFA6a-13Myc-His3MX6 plasmid as template (43). Primer sequences are available upon request. The resulting fragment was integrated by homologous recombination into the SPT20 and GCN5 locus. Correct integration was confirmed with a PCR-based strategy. The strain SJ01 (WT RLM1-3HA::LEU2) was described in (9). Mutant gcn5Δ RLM1-3HA::HIS3 was obtained as described above but using the pFA6a-3 HA-His3MX6 plasmid as template. The double mutant swi3Δ gcn5Δ (swi3::KanR, gcn5::HIS3) was obtained by sporulation and tetrad analyses using standard yeast genetic techniques after generation of the corresponding diploid heterozygous disruptant. Strains YJS6, YJS7 and YKH100 (31) were kindly provided by B. Franklin Pugh (Pennsylvania State University, PA, USA). The plasmid YEp352-RLM1HA expressing the fusion protein Rlm1-HA under the control of the native Rlm1 promoter was previously described (9). Plasmid pRS416-RLM1HA was constructed by digestion of plasmid YEp352-RLM1HA with PvuII and SacI and cloning of the derived fragment containing RLM1HA and its promoter into the vector pRS416 using SmaI and SacI sites.
Routinely, cells were grown overnight in liquid YEPD (1% yeast extract, 2% peptone and 2% glucose) or synthetic defined (SD) (0.17% yeast nitrogen base, 0.5% ammonium sulphate, 2% glucose, supplemented with the required amino acids) medium in the case of cells bearing plasmids, at 220 rpm and 24°C to an optical density of 0.8–1 (A600). The culture was refreshed to 0.2 (A600) in YEPD, grown for an additional 2 h 30 min and then divided into two parts. One part continued growing under the same conditions (non-treated culture), while the other one was supplemented with Congo red (CR) (30 μg/ml; Merck, Darmstadt, Germany). Cells were collected at the indicated times and processed depending on the experimental approach.
Phenotypic analyses
Yeast cells were grown at 24°C in YEPD to mid-log phase and sensitivity assays to the cell wall perturbing agent CR were performed as previously described (44).
Microarray experiments
Total RNA isolation and purification was carried out as detailed elsewhere (6). Genome-wide transcriptional profiles were obtained using AffymetrixGeneChip® Yeast Genome 2.0 arrays. cDNA synthesis and chip hybridization, image analysis, data processing and statistical analysis were basically carried out as described (45). The files generated from the scanning (.CEL) were converted to gene expression signals using RMA algorithm included in Affymetrix® Expression Console™ (Affymetrix®). For each experimental condition, three microarray experiments corresponding to three independent biological samples were processed and analyzed. Fold changes between experimental conditions under comparison were calculated as a ratio between the mean of the gene expression signals. The genes were considered to be up- or down regulated when their expression ratio under the conditions tested was ≥2 or ≤0.5, respectively. Statistical analysis was performed with the Student t-test. Those values with a P-value < 0.05 were considered as significant and the corresponding genes considered for further analysis.
To determine whether the gene induction observed in the WT strain after CR treatment (30 μg/ml, 3 h) was significantly reduced in gcn5Δ mutant we used the relationship between the response of the mutant (mutant ratio, +/− CR treatment) versus the one of the WT strain (WT ratio, +/− CR treatment). Thus, an arbitrary value of mutant ratio/WT ratio of 0.65 was considered the threshold for defining a significant reduction in gene induction (46). In any case, genes whose mutant ratio was <1.6 were deemed as not being upregulated. The same threshold for the relationship between mutant/WT versus WT ratio was used to determine genes whose basal expression levels was dependent on GCN5. Clustering analysis was performed using the MeV MultiExperiment Viewer, version 4.9 (47).
The microarray data described here follow the minimum information about a microarray experiment recommendations and have been deposited at the NCBI gene expression and hybridization array data repository with accession number GSE71433.
Quantitative RT-PCR assays
Total RNA isolation and purification was carried out as detailed elsewhere (6). Real-time quantitative RT-PCR (RT-qPCR) assays were performed as previously detailed with several modifications (6). In this case, real-time PCR was performed using an 7900 HT Fast Real-Time PCR System instrument (Applied Biosystems) in a final volume of 10.7 μl containing 4.5 μl of a 100-fold dilution of the RT reaction, 5 μl of the Power SYBR Green PCR Master Mix (Roche) and 0.6 μl of each 5 μM primer stock (Sigma). Basic analysis was performed using the SDS 2.3 and RQ-Manager 1.2 softwares (Applied Biosystems). For quantification, the abundance of each gene was determined relative to the standard transcript of ACT1 for input cDNA normalization, and the final data on relative gene expression between the conditions tested were calculated following the 2−ΔΔCt method (48). Primer sequences are listed in Supplementary Table S1.
Western blotting assays
The procedures used for immunoblot analyses, including cell collection and lysis, collection of proteins, fractionation by sodium dodecyl sulphate-polyacrylamide gel electrophoresis, and transfer to nitrocellulose membranes, have been described previously (44).
Chromatin immunoprecipitation assays
ChIP was performed as described elsewhere (9,49). The antibodies used in these experiments were: monoclonal anti-Myc (9E10, Covance), polyclonal anti-HA (ab9110, Abcam), monoclonal anti-Pol II (8WG16, Covance), polyclonal anti-Snf2 antibody kindly provided by Joseph C. Reese (Pennsylnania State University, USA), polyclonal anti-H3 (ab1791, Abcam) and polyclonal anti-acetyl-histone H3 (06-599, Millipore). The immunoprecipitated DNA was quantified by qPCR using primers that amplified the following regions (locations are indicated by the distance from the respective ATG initiation codon): MLP1-BOX1 (RLM1 binding domain at the promoter), −453/−313; MLP1-POLII (promoter/ORF), −143/+56; MLP1-ORF2 (ORF), +555/+687; MLP1-ORF3 (3′ end ORF), +1210/+1359; YLR194C-BOX (RLM1 binding domains at the promoter), −254/−123; YLR194C-POLII (promoter/ORF), −130/+60; YLR194C-ORF2 (ORF), +359/+520; YLR194C-ORF3 (3′ end ORF), +666/+805; YPL088W-BOX1 (RLM1 binding domain at the promoter), −573/−423; YPL088W-ORF2 (ORF), +562/+712; YPL088W-ORF3 (3′ end ORF), +911/+1064; and VMA8 (promoter), −321/−191. Primer sequences are listed in Supplementary Table S1. The fold enrichment (FE) at specific DNA regions was calculated using the Comparative Ct Method (50) and the promoter region of the VMA8 gene, whose expression does not vary upon cell wall stress, as a control sequence. Thus, the Ct of the input sample was subtracted from the Ct of the immunoprecipitated sample to calculate the ΔCt, both in the control sequence (ΔCtcont) and in the target DNA (ΔCtexp) for each condition. Finally, FE was calculated by using the following formula: FE = 2–[ΔCtexp-ΔCtcont]. Data represent the mean and standard deviation of at least three independent experiments.
RESULTS
Deletion of SAGA histone acetyltransferase module subunits causes Congo red hypersensitivity
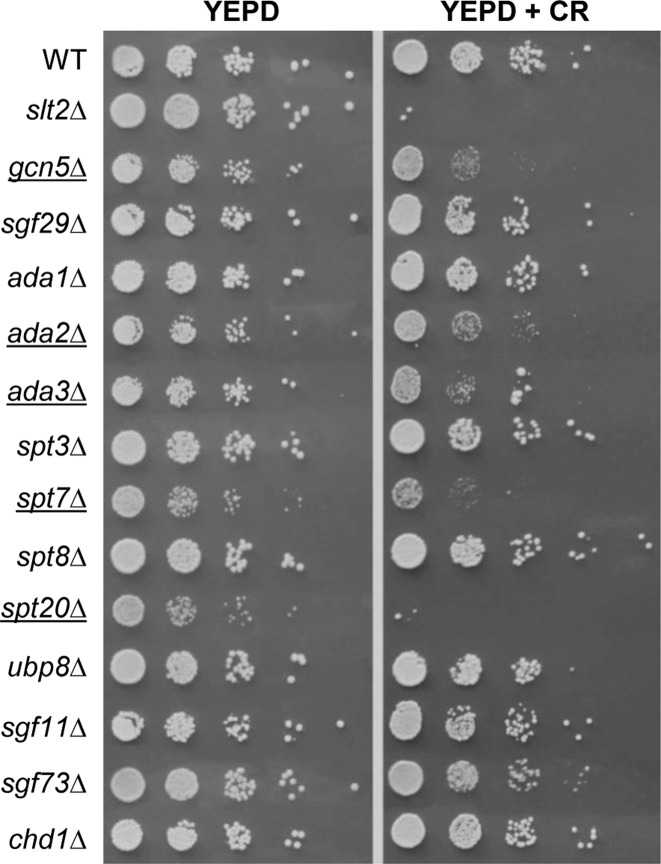
We had previously developed a large-scale screening using the whole collection of haploid deletion strains transformed with the CWI reporter system pSJ05 (pMLP1-NAT1) to identify genes required for the transcriptional response triggered by cell wall stress through the CWI pathway (9). In addition to the SWI/SNF remodeling complex, whose functional relevance for the CWI transcriptional response was described, several subunits of the SAGA complex were also identified as defective in the induction of the cell wall stress responsive gene MLP1/KDX1 (9), although its precise role in transcription regulation was not established. To further characterize the functional role of this complex in transcription through the CWI pathway, we first analyzed the importance of the SAGA complex in the maintaining of the CWI. We performed a phenotypic analysis of a full set of viable saga mutants growing in the presence of the cell wall-interfering compound Congo red (CR, 100 μg/ml). As shown in Figure 1, mutants in subunits related to the acetyltransferase activity as Gcn5, Ada2 and Ada3, as well as structural components, Spt7 and Spt20, were hypersensitive to Congo red. However, deletion of those genes related with ubiquitin modification (UBP8, SGF11 and SGF73) and TBP recruitment (SPT3 and SPT8) did not have any effect on yeast cell growth relative to the WT strain. These phenotypes suggest that the disruption of the histone modifying function of the complex results in the strongest defect in the maintenance of a stable cell wall.
Figure 1.
Deletion of genes that encode several SAGA subunits renders cells sensitive to cell wall stress. The indicated strains were spotted on YEPD plates without or with 100 μg/ml CR, and plates were incubated for 48 h at 30°C. Mutants underlined correspond to strains showing hypersensitivity to this compound.
SAGA complex is required for the transcriptional response mediated by the CWI pathway
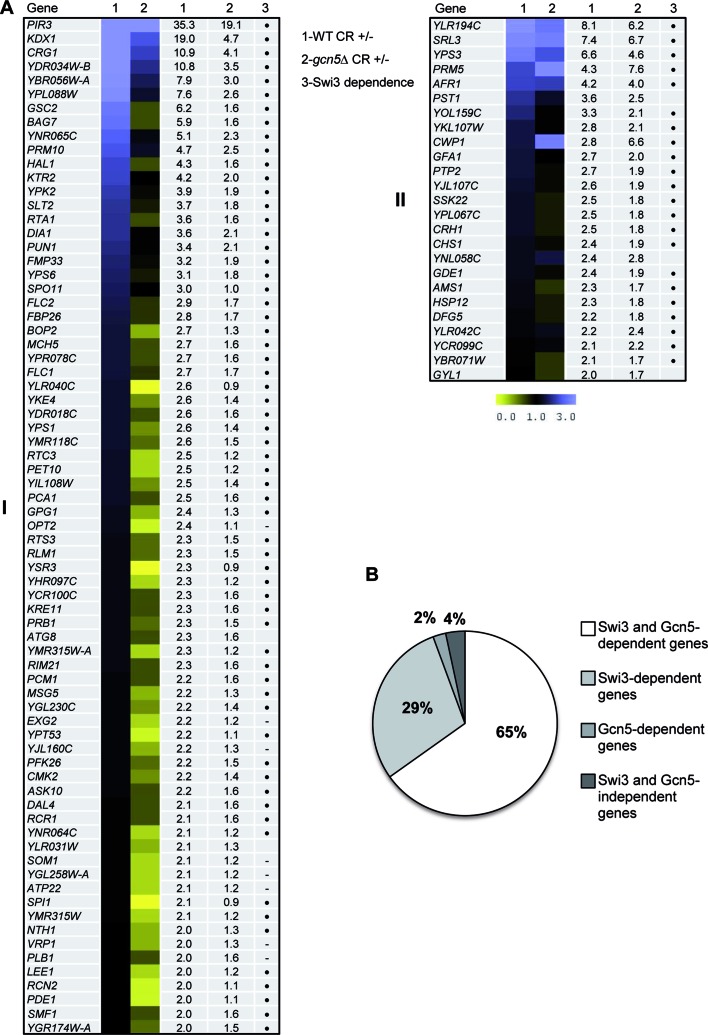
In order to establish a role for the HAT SAGA module in the transcriptional response dependent on the CWI pathway, genome-wide expression profiles of WT and gcn5Δ mutant strains under basal and cell wall stress (CR, 3 h) conditions were characterized. Microarray analyses revealed that 98 genes were induced with ratios of transcript levels ≥2 upon CR treatment in a WT strain. Comparison of the transcriptional profiles of both strains revealed that upregulation of gene expression was severely affected in the gcn5Δ mutant. As a result, 73 out of the 98 genes of the WT induction response (74.4%) depended on Gcn5 (Figure 2A), confirming an essential role of SAGA complex in CWI-mediated gene expression. Moreover, comparison of the transcriptional profiles of gcn5Δ cells generated in this work to those of a swi3Δ mutant upon CR stress (9), revealed that the majority of those genes dependent on Gcn5 were also dependent on Swi3 (Figure 2A and B), further suggesting a cooperative function of both complexes in the regulation of gene expression responses through the CWI pathway.
Figure 2.
Genome-wide expression profiles of wild-type and gcn5Δ strains after CR treatment. (A) The heat map obtained by the MeV 4.9 software shows gene expression ratios comparing the transcriptional response to CR (3 h) (treated versus untreated) in WT and gcn5Δ strains (columns 1 and 2), respectively. The degree of color saturation represents the expression log2 ratio value, as indicated by the scale bar. Numeric ratio values are also included in additional columns. Those genes upregulated in the WT strain by CR treatment were grouped based on the dependence on Gcn5 (see ‘Materials and Methods’ section for details): (I) genes whose induction was dependent on Gcn5 and (II) genes induced by cell wall stress independently on Gcn5. Swi3 dependence for all genes according to previously published data (9) is also indicated with a dot (hyphen represents no data available). (B) Schematic representation of the comparison of the transcriptional profiles of gcn5Δ and swi3Δ (9) mutants after CR treatment respect to the WT strain. Data are represented as the % of genes dependent on Swi3, Gcn5 or both.
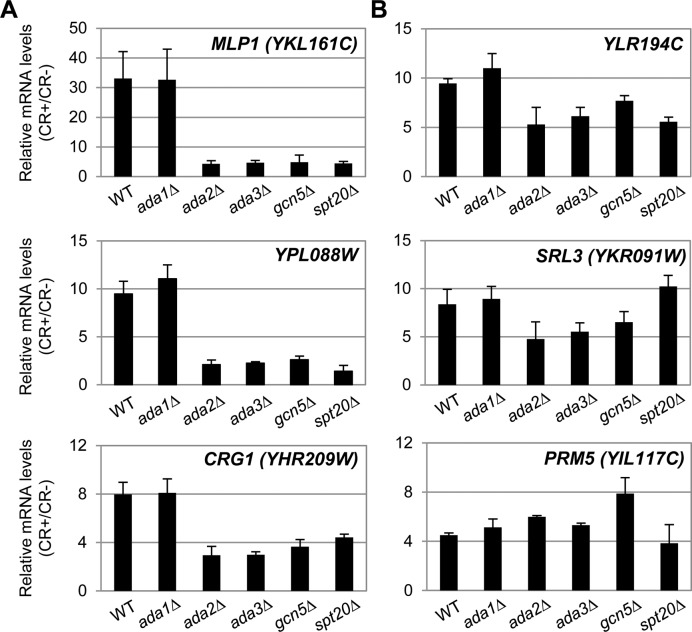
These results were further confirmed by RT-qPCR. The transcriptional activation of several of the genes identified above, including both Gcn5 dependent and not-dependent genes was analyzed not only in a gcn5Δ mutant but also in ada1Δ, ada2Δ, ada3Δ and spt20Δ strains. As shown in Figure 3 and in agreement with the genome-wide analysis, induction of MLP1, YPL088W and CRG1 was severely impaired in ada2Δ, ada3Δ, spt20Δ and gcn5Δ strains. However, the induction of YLR194C and SRL3 was affected to a lesser extent whereas induction of PRM5 was not altered. It is worth to mention here that in the case of PRM5, and also for AFR1 and CWP1, all of them included in the group of Gcn5 not-dependent genes, their basal expression in the absence of stress is highly affected by deletion of GCN5. Additionally, the absence of ADA1 did not affect the transcriptional output of any of these genes (Figure 3). Therefore, there was a good correlation between phenotypes and transcriptional profiles.
Figure 3.
Gene expression analysis of CWI-responsive genes in WT and mutants in several subunits of the SAGA complex. mRNA levels of several selected CWI-responsive genes, dependent (A) and not-dependent (B) on Gcn5 based on the gcn5Δ genome-wide transcriptional profile, were analyzed by RT-qPCR in a WT strain and in the indicated saga mutants after 3 h of CR treatment. Values represent the ratio between CR-treated and non-treated cells. Data correspond to the mean and standard deviation of at least three independent experiments.
To rule out possible signaling alterations of the CWI pathway as a consequence of SAGA deletion, we characterized the activation of the MAPK Slt2 in a selection of saga mutants. Importantly, these mutants showed similar Slt2 phosphorylation levels than the WT (Supplementary Figure S1A), indicating that the absence of SAGA does not affect the functionality of the CWI pathway. In a previous work, we had observed that Rlm1 protein levels were significantly reduced in a swi3Δ strain with respect to the WT (9). This is likely due to the lack of positive transcriptional feedback mechanisms mediated by the transcription factor Rlm1 on itself and on SLT2 (51). Both SLT2 and RLM1 are induced as consequence of cell wall stress in an Rlm1-dependent manner (6). Since SWI/SNF complex is necessary for chromatin remodeling at CWI-responsive genes (9), this feedback mechanism should not be operative in swi/snf mutants and probably nor in saga mutants. For this reason we analyzed both Slt2 and Rlm1 protein levels in different saga mutants, in the absence or presence of stress. As expected, Slt2 and Rlm1 protein levels were also affected in the ada2Δ, ada3Δ, gcn5Δ, spt7Δ and spt20Δ mutants but not in ada1Δ, spt3Δ and spt8Δ (Supplementary Figure S1B and C). Equalizing the amount of Rlm1 protein in a gcn5Δ strain, by overexpression of Rlm1, did not rescue the levels of expression of GCN5-dependent genes like MLP1, CRG1, BAG7 and YPL088W upon stress (Supplementary Figure S2), further demonstrating the importance of the SAGA complex in cell wall stress-mediated transcription of this group of genes. For this reason, oncoming experiments were carried out in cells expressing RLM1 under the control of its own promoter from an episomal plasmid to bypass the effect of low Rlm1 levels.
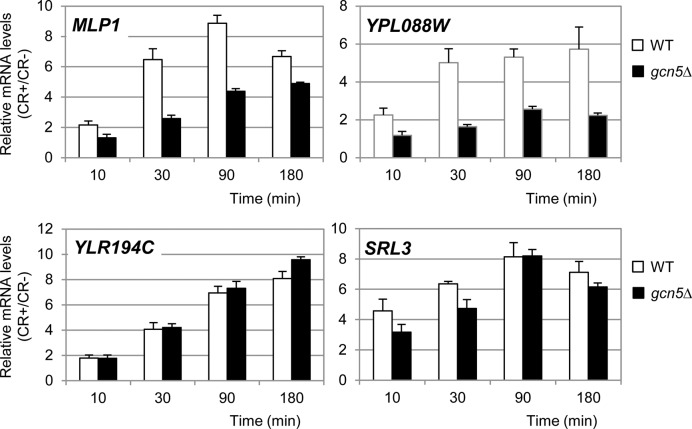
Trying to understand the temporal regulation of CWI-responsive genes by the SAGA complex and to figure out whether SAGA was required to turning on gene expression or to maintain a more sustained expression over time, we first followed gene expression kinetics of MLP1, YPL088W, YLR194C and SRL3 at different times of cell wall stress in WT and gcn5Δ cells. As shown in Figure 4, for MLP1 and YPL088W, the expression both at early or late post stress times was dependent on Gcn5, whereas the temporal pattern of gene expression of YLR194C and SRL3 in response to cell wall stress did not vary in a gcn5Δ mutant compared to the WT (Figure 4). Our previous studies showed that transcriptional induction of the four genes (MLP1, YPL088W, YLR194C and SRL3) by cell wall stress was highly dependent on SWI/SNF (9). Therefore, MLP1 and YPL088W are SWI/SNF and SAGA-dependent genes, whereas YLR194C and SRL3 are SWI/SNF-dependent but SAGA-independent genes. Our results indicate that SAGA complex is required for both early induction and sustained expression of genes regulated by cell wall stress, as it was previously described for the SWI/SNF complex.
Figure 4.
Gene expression time course of CWI-responsive genes in WT and gcn5Δ strains. mRNA levels of Gcn5-dependent genes (MLP1 and YPL088W) and Gcn5 not-dependent genes (YLR194C and SRL3) were analyzed by RT-qPCR in WT and gcn5Δ strains upon CR treatment. Values represent the ratio between CR-treated and non-treated cells. Data represent the mean and standard deviation of at least three independent experiments.
SAGA complex binds to DNA in response to cell wall stress, and this requires Slt2, Rlm1 and SWI/SNF complex
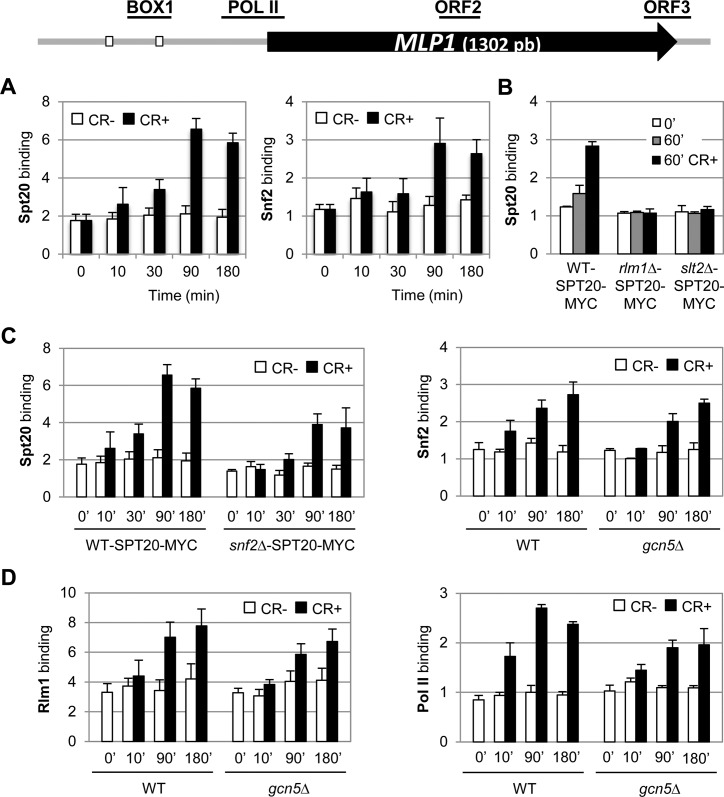
The mechanisms by which Rlm1 induce the transcription of CWI-responsive genes under cell wall stress is still not completely understood. Upon cell wall stress, Slt2 phosphorylates the transcription factor Rlm1 that physically interacts with SWI/SNF to direct its association with target promoters to finally mediate the chromatin remodeling necessary for developing adequate transcriptional responses (9). However, the role of SAGA complex in this context has yet to be defined. Using chromatin immunoprecipitation (ChIP) analysis, we first followed the binding kinetics of SAGA complex to MLP1 (SWI/SNF- and SAGA-dependent gene) in the presence or absence of cell wall stress at different times of treatment. Chromatin from a WT strain expressing a functional myc epitope-tagged Spt20 was immunoprecipitated with anti-Myc antibody and analyzed by qPCR to check occupation of the promoter of the MLP1 gene. As shown in Figure 5A (left panel), cell wall stress induced Spt20 recruitment after 10 min of Congo red treatment, rising maximum levels after 90 min of stress in the region of the Rlm1 box 1 where the binding of Rlm1 and SWI/SNF is also more pronounced (9). A similar recruitment pattern was also observed for Gcn5 at MLP1 promoter and for Spt20 at the promoter of YPL088W, the other SWI/SNF- and SAGA-dependent gene (Supplementary Figure S3A). However, SAGA subunit Spt20 was not found through the coding region of MLP1 (Supplemental Figure S3B). We further explored the recruitment of the SAGA complex to the MLP1 promoter in the absence of Rlm1 or Slt2. As shown in Figure 5B and similarly to Snf2 entry (9), Spt20 recruitment observed in a WT strain as a consequence of cell wall stress was completely blocked in slt2Δ and rlm1Δ mutants, indicating the requirement of an active CWI pathway for SAGA recruitment.
Figure 5.
SAGA complex binds to MLP1 gene after CR treatment in a Slt2, Rlm1 and SWI/SNF-dependent manner and its absence slightly affects the recruitment of Snf2, Rlm1 and Pol II. (A) The recruitment of Spt20 and Snf2 was analyzed by ChIP in a WT strain expressing Spt20-myc in the absence and presence of CR (CR 30 μg/ml, 3 h) at the indicated times. A schematic representation of the MLP1 gene is depicted above, where Rlm1 binding sites are denoted by the small white boxes and the regions analyzed for ChIP experiments are shown as horizontal lines. (B) Recruitment of Spt20 was determined by ChIP in WT, rlm1Δ and slt2Δ strains expressing Spt20-myc, in the absence and presence of CR at the indicated time. (C) Spt20 binding was determined by ChIP in WT and snf2Δ strains expressing the Spt20-myc tagged protein (left panel). Snf2 binding was analyzed by ChIP in WT and gcn5Δ strains exposed to cell wall stress at the indicated times (right panel). (D) Rlm1-HA (left panel) and RNA Pol II (right panel) recruitment were analyzed by ChIP experiments in WT and gcn5Δ strains exposed to cell wall stress. The regions analyzed were MLP1-BOX1 and MLP1-POL II (only for RNA Pol II). Data represent the mean and standard deviation of at least three independent experiments.
ChIP assays revealed that SWI/SNF is also recruited into the MLP1 promoter after 10 min of stress, rising maximum levels after 90 min of CR addition (Figure 5A, right panel). Thus, the temporal kinetics of Snf2 entry into the MLP1 promoter, as a consequence of the stress, was almost identical to that of the SAGA complex, in agreement with the requirement of both complexes for the early induction and sustained expression of genes regulated by cell wall stress.
To establish a separate or interdependent role of SWI/SNF and SAGA complexes on initial recruitment of each other, the kinetics of Spt20 and Snf2 recruitment to the MLP1 promoter in a WT strain were compared to those of snf2Δ and gcn5Δ strains, respectively. As shown in Figure 5C (left panel), binding of Spt20 to the SAGA-dependent gene MLP1 clearly diminished in cells lacking Snf2 along the CR treatment, further suggesting that recruitment of SAGA subunits to the CWI promoters require the presence of the SWI/SNF complex. In contrast, recruitment of Snf2 did not almost require Gcn5 (Figure 5C, right panel).
Entry of the transcriptional machinery to CWI-responsive genes is slightly affected in a gcn5Δ mutant
To analyze whether SAGA complex was affecting recruitment and assembly of other elements of the pre-initiation transcriptional complex (PIC) upon cell wall stress, we measured what effect does loss of Gcn5 have on recruitment of the transcription factor Rlm1 as well as Pol II in a time-dependent manner. As shown in Figure 5D (left panel), Rlm1 recruitment to the MLP1 promoter slightly decreased through the time course and particularly at 90 and 180 min of CR treatment in the absence of Gcn5. Moreover, initial recruitment (10 min post stress) and later occupation (90 and 180 min post stress) of the MLP1 promoter by Pol II was also partially affected (Figure 5D, right panel). Taken together, these results suggest that Gcn5 is not essential for PIC assembly, although facilitate this process.
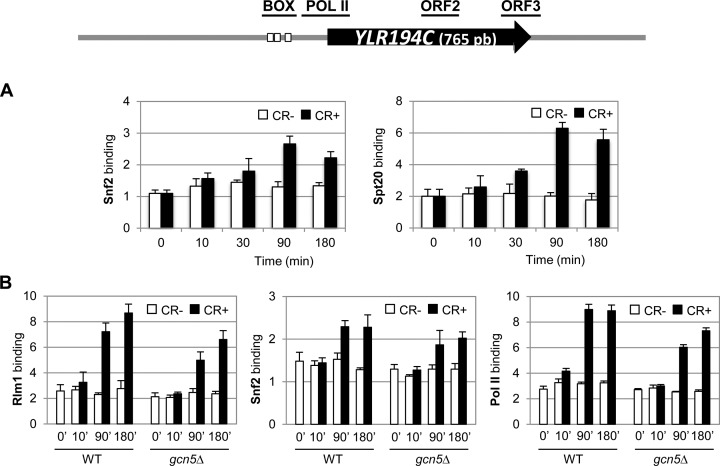
What about genes like YLR194C and SRL3, whose induction is SWI/SNF dependent but SAGA independent? As expected for its dependence on SWI/SNF complex, Snf2 concentrates into YLR194C promoters as consequence of the stress following a similar kinetics to that of MLP1 (Figure 6A, left panel). However, unexpectedly Spt20 was also bound to the promoter region of YLR194C, following a similar temporal profile to that of the SAGA-dependent gene MLP1 (Figure 6A, right panel). Moreover, recruitment of Rlm1, Snf2 and Pol II to the YLR194C promoter was also slightly affected in a gcn5Δ strain (Figure 6B).
Figure 6.
Recruitment of SWI/SNF and SAGA complexes to YLR194C and effect of GCN5 deletion on Rlm1, Snf2 and Pol II binding. (A) Snf2 (left panel) and Spt20 (right panel) binding were determined by ChIP in a WT strain expressing the Spt20-myc tagged protein during CR treatment. A schematic representation of the YLR194C gene is depicted above, where Rlm1 binding sites are denoted by the small white boxes and the regions analyzed for ChIP experiments are shown as horizontal lines. (B) Rlm1-HA (left panel), Snf2 (central panel) and RNA Pol II (right panel) recruitments were analyzed by ChIP experiments in WT and gcn5Δ strains exposed to cell wall stress. YLR194C-BOX region was analyzed in all cases, except for RNA Pol II whose entry was analyzed at YLR194C-POL II region. Data represent the mean and standard deviation of at least three independent experiments.
H3 acetylation by SAGA complex is necessary for gene induction during cell wall stress
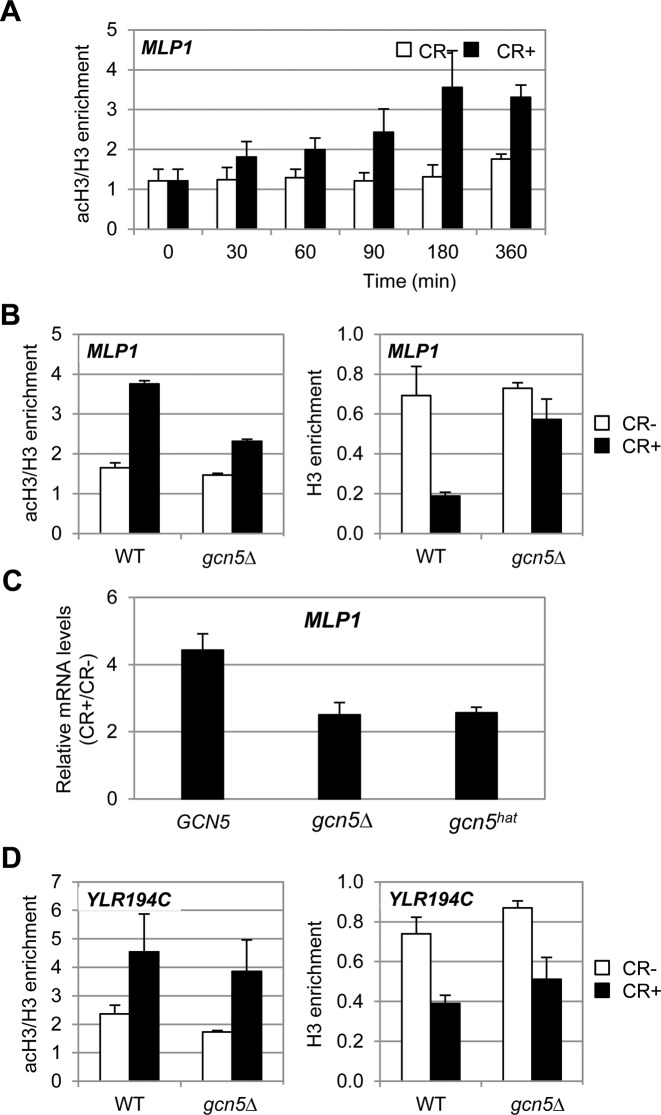
To verify whether the effect of SAGA complex over gene transcription mediated by cell wall stress was due to its HAT activity, we first performed ChIP experiments to study the level of histone acetylation at the MLP1 promoter in a WT strain before and after CR treatment. Since there is a displacement of H3 under these circumstances (9), levels of acetylated H3 were determined necessarily as the ratio of acetylated H3 per total histone H3. Chromatin was isolated at the indicated times and double immunoprecipitation was performed, one using the anti-H3 antibody and the other one with an anti-acetyl H3 antibody, to detect levels of total H3 and diacetylated H3 K9/K14, respectively. As shown in Figure 7A, the ratio of histone H3 acetylation significantly increased upon stress, reaching a maximum value after 180 min. Moreover, this process was highly dependent on Gcn5 (Figure 7B, left panel).
Figure 7.
Gcn5 promotes histone H3 acetylation and H3 eviction at MLP1 but not at YLR194C gene. (A) H3 acetylation was determined by ChIP analysis in a WT strain treated with CR for the indicated time points at MLP1-BOX1 region, using antibodies specific for diacetylated H3 (K9 and K14) (acH3) and against total H3 (H3). Values are expressed as the ratio of acetylated H3 versus total H3 enrichment. (B) H3 acetylation (left panel) and H3 eviction (right panel) was determined at MLP1-BOX1 region in WT and gcn5Δ strains after 3 h of CR addition. (C) mRNA levels of MLP1 were analyzed by RT-qPCR in YJS6 (GCN5), YKH100 (gcn5Δ) and YJS7 (gcn5hat) strains after 3 h of CR addition. Values represent the ratio between CR-treated and non-treated cells. (D) H3 acetylation (left panel) and H3 enrichment (right panel) were analyzed in YLR194C-BOX region as in (B). Data represent the mean and standard deviation of at least three independent experiments.
We have previously shown a decrease in H3 occupancy at different regions of MLP1 promoter, with a peak of H3 eviction after 3 h of CR treatment (9). As a next step, we wanted to know if acetylation mediated by Gcn5 was affecting chromatin remodeling at MLP1 promoter upon stress. We measured H3 displacement at the MLP1 promoter after CR treatment, both in WT and gcn5Δ strains. The results in Figure 7B (right panel) show a substantial reduction in H3 eviction in the gcn5Δ respect to the WT strain, suggesting a dependence on Gcn5 for chromatin remodeling at the promoter of this gene. Moreover, the levels of MLP1 mRNA upon cell wall stress were recovered in a gcn5Δ strain by expression of a WT copy of GCN5 but not by a gcn5hat allele (possessing the KQL HAT mutation) (Figure 7C), indicating that the HAT activity of Gcn5 is required for an adequate MLP1 transcriptional response.
Interestingly, in contrast to SAGA-dependent genes, lack of Gcn5 did not affect H3 eviction at SAGA-independent genes, as shown in Figure 7D (right panel) for the YLR194C promoter. Moreover, the increase in acetylated H3 levels at this promoter in a WT strain upon stress was independent on Gcn5 (Figure 7D, left panel). All these results indicate that Gcn5 mediates H3 acetylation and nucleosome eviction at the promoters of SAGA-dependent genes but not at SAGA-independent genes. Therefore, although SAGA complex is binding to the promoters of the latter genes upon stress, H3 acetylation and chromatin remodeling is not dependent on this complex.
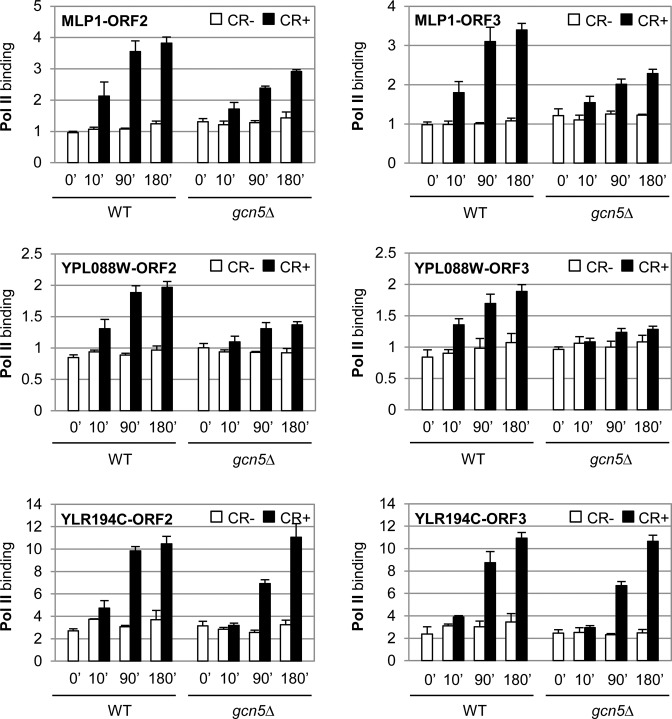
Since we had only found slight differences on Rlm1 and Pol II assembly in a gcn5Δ strain respect to the WT and these differences were found in both, SAGA-dependent and -independent genes (Figures 5 and 6), all these results suggest that chromatin remodeling mediated by Gcn5 is important to control binding of other PIC elements or post recruitment mechanisms necessary to regulate gene expression. In agreement, binding of Pol II at the coding regions of MLP1 and particularly YPL088W, upon cell wall stress, was clearly impaired in cells lacking Gcn5 during the whole time-course (Figure 8). In contrast, Pol II association with the coding regions of YLR194C was very similar in WT and gcn5Δ cells (Figure 8).
Figure 8.
Deletion of GCN5 affects Pol II progression at MLP1 and YPL088W but not at YLR194C. Pol II association was measured by ChIP at ORF2 and ORF3 coding regions of the indicated genes in a WT and gcn5Δ strains at the indicated times after CR addition. Data represent the mean and standard deviation of at least three independent experiments.
SWI/SNF and SAGA complex cooperate for eliciting gene expression in response to cell wall stress
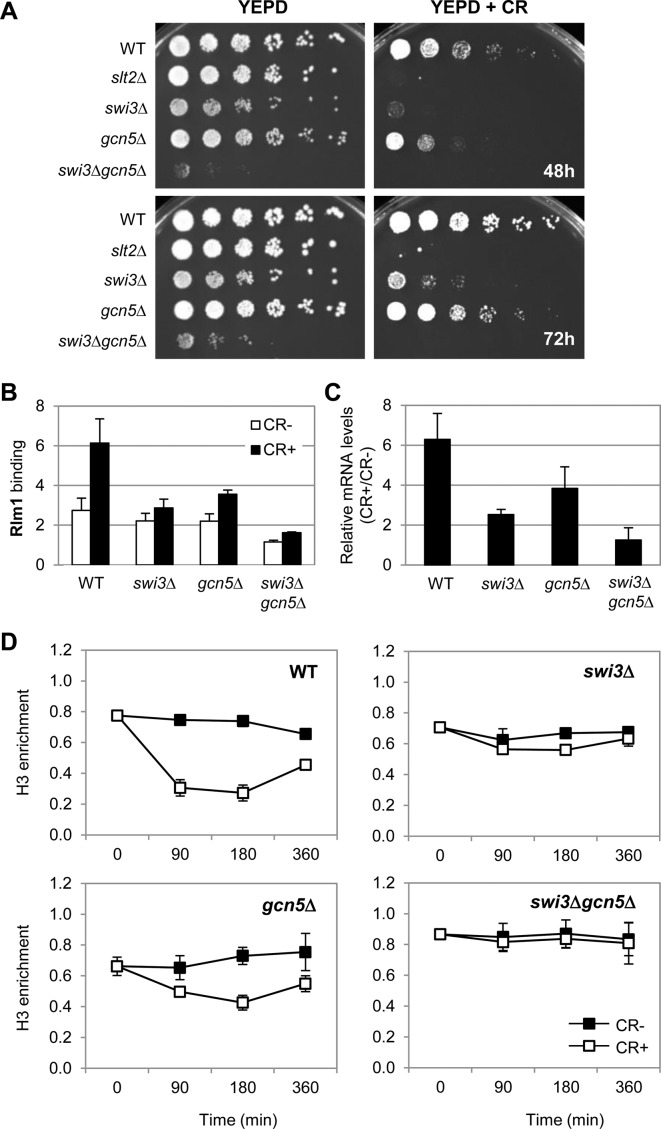
All the results shown above, together with the fact that the SWI/SNF chromatin remodeling complex function is very important for the transcription mediated by the yeast CWI pathway, prompted us to investigate a potential cooperation between SWI/SNF and SAGA complexes in response to cell wall stress. To this end, we constructed a swi3Δ gcn5Δ double mutant strain lacking one of the most important subunits of each complex. As shown in Figure 9A, this mutant is viable but it exhibits a severe cell growth phenotype as deduced from its growth in YEPD compared to the WT and single swi3Δ and gcn5Δ strains. Deletion of GCN5 rendered cells hypersensitive to CR, but less hypersensitive than SWI3 deletion. Moreover, similarly to the strain lacking Slt2 (MAPK of the pathway), the swi3Δ gcn5Δ double mutant was unable to growth at the same concentration of CR. These results suggest a cooperative functional role for Swi3 and Gcn5 to overcome cell wall damage.
Figure 9.
SWI/SNF and SAGA complexes cooperate to displace H3 histone at MLP1 and regulate gene transcription in response to cell wall stress. (A) The indicated strains were spotted on YEPD plates without or with 100 μg/ml of CR and plates were incubated for 48 and 72 h at 30°C. (B) Rlm1 enrichment at MLP1 promoter (MLP1-BOX1) was analyzed by ChIP in the indicated strains exposed to cell wall stress (CR 30 μg/ml, 3 h). (C) mRNA levels of MLP1 were analyzed by RT-qPCR in the same strains and growth conditions indicated in (A). Values represent the ratio between CR-treated and non-treated cells. Data represent the mean and standard deviation of at least three independent experiments. (D) The kinetics of histone H3 binding to MLP1 promoter (MLP1-BOX1) was determined by ChIP in WT, swi3Δ, gcn5Δ and swi3Δ gcn5Δ strains under CR treatment at the indicated times. ChIP assays were performed using an antibody against total H3. Data represent the mean and standard deviation of at least three independent experiments.
To better understand this cooperation, we characterized the binding of Rlm1 at MLP1 promoter (Gcn5- and Swi3-dependent gene) by ChIP assays in the corresponding single and double mutants in the presence or absence of cell wall stress. As shown in Figure 9B, Rlm1 binding decreased in both single strains being more affected in a swi3Δ than in gcn5Δ, whereas it was completely blocked in the swi3Δ gcn5Δ double mutant both under basal and stress conditions (3 h of CR treatment). Moreover, the induction of MLP1 expression by cell wall stress was totally abolished in the absence of both complex subunits, showing an additive effect respect to the alteration of MLP1 mRNA levels in the single mutant strains (Figure 9C).
To further support cooperation between these complexes for chromatin remodeling, we evaluated histone H3 eviction upon cell wall stress either in the absence of SAGA, SWI/SNF or both complexes. The kinetics of H3 enrichment at MLP1 promoter in WT, gcn5Δ, swi3Δ and swi3Δ gcn5Δ strains under basal growth conditions and in the presence of cell wall stress are shown in Figure 9D. Looking at H3 levels in the absence of stress, similar values of H3 enrichment (around 0.7-fold) were found in WT, gcn5Δ and swi3Δ strains, whereas the double mutant showed higher levels, suggesting that simultaneous abrogation of Swi3 and Gcn5 mediated activities conduct to a more static chromatin in this strain. High levels of H3 eviction were detected in a WT strain upon stress. This eviction was reduced in the gcn5Δ strain, more reduced in a swi3Δ strain and completely lost in a swi3Δ gcn5Δ double mutant strain (Figure 9D), demonstrating the cooperative effect of SWI/SNF and SAGA complexes in chromatin remodeling upon cell wall stress.
DISCUSSION
Upon cell wall stress, S. cerevisiae elicits an adaptive transcriptional response to overcome damage in this essential structure. This response is mediated by a MAPK signaling cascade coordinated by the MAPK Slt2. The activation of Slt2 triggers direct phosphorylation of the transcription factor Rlm1 that is recruited to CWI-responsive genes upon stress. Assembly of a preinitiation complex near the transcriptional start site of CWI-responsive genes, leading to Pol II recruitment, requires the participation of protein complexes that remodel and modify chromatin. Thus, once Rlm1 is activated, interacts with the chromatin remodeling complex SWI/SNF to direct its association with the specific binding sites at the promoters of the genes transcriptionally induced upon CWI pathway activation. The activity of the SWI/SNF complex is necessary to displace nucleosomes positioned at the occluded Rlm1-binding sites and surrounding regions to permit Rlm1 and Pol II assembly (9). In addition, we have established here an additional role for the acetyltransferase SAGA complex to elicit adequate transcriptional responses upon cell wall stress. Genome-wide gene expression analysis revealed that a high percentage of the CWI transcriptional response is dependent on Gcn5. The majority of these genes, i.e MLP1 and YPL088W, are also dependent on the SWI/SNF complex for their induction. Both complexes are required not only for early induction but also for sustained expression of these genes upon cell wall stress. Additionally, we identified a group of genes, represented by YLR194C and SRL3, whose induction is SWI/SNF dependent but SAGA independent.
Under cell wall stress, we show here that entry of SAGA complex into MLP1 promoter was dependent on Slt2 and Rlm1. In this context, SAGA could be recruited to target promoters by Rlm1, as deduced by the stress-induced interaction between Gcn5 and Ada2 with the MAPK Slt2 in an Rlm1-dependent manner (52). Consistent with this, we have detected a major enrichment of SAGA subunits within the Rlm1-binding sites at the promoter regions of the activated genes. As suggested for Rlm1, which would be able to target SWI/SNF and SAGA complexes, other transcriptional activators as Gcn4, Swi5 or Gal4 were also found to recruit a large number of co-activators (53–56). However, it is important to note that the order of events in the recruitment of these different factors is not universal, but rather depends on the transcription factor itself, the nature of promoter and the chromatin structure in which it resides and also the cellular demand for gene products, making necessary to study specific genes at specific times (reviewed in (57)).
To delineate a separate or interdependent role of SWI/SNF and SAGA complexes in CWI-mediated gene expression, we have characterized the interdependence on recruitment of each other as well as the temporal kinetics of entry for both complexes upon cell wall stress. SAGA entry depends on SWI/SNF complex whereas Gcn5 is almost not required for Snf2 recruitment. In consequence, it is possible that SWI/SNF complex could be recruited by Rlm1 before SAGA, being SWI/SNF recruitment in this case necessary for the later entry of SAGA. However, it is also possible that a simultaneous entry of both complexes occurs. In this case, the dependency on the SWI/SNF for SAGA recruitment and not the opposite could be explained by a more important role in chromatin remodeling for the former complex. The fact that no temporal differences in the recruitment of both complexes were observed would be in agreement with a simultaneous recruitment, although subtle differences in the times of entry, not observable with our approach, could also exist. Parallel recruitments of different co-activators have been observed in the transcriptional initiation of stress-inducible genes, probably to support the rapid upregulation required during adverse conditions. For example, simultaneous recruitment of SAGA and SWI/SNF has also been observed at osmotic stress-inducible genes (39) and at the glucose-repressed SUC1 gene (35).
We have previously demonstrated that the SWI/SNF complex mediates nucleosome eviction at the promoter region of CWI-responsive genes to regulate gene expression. How is SAGA complex affecting the upregulation of cell wall stress genes? SAGA and TFIID are the two major complexes that deliver TBP to promoter in yeast. Genome-wide studies reveal that SAGA regulates highly inducible stress responsive genes, whereas TFIID regulates housekeeping genes. At some promoters, the contribution of both complexes is more or less equivalent (31). SAGA mediates this process through Spt3 and Spt8 subunits although we found that deletion of both subunits does not render cell wall stress hypersensitivity. Thus, our results suggest that SAGA is probably not the factor that delivers TBP to MLP1 or at least, not exclusively as in other stress-responsive genes. Phenotypic assays prompted us to study in detail the HAT activity of SAGA complex due to the observation that deletion of ADA2, ADA3 and GCN5 genes caused hypersensitivity to CR and serious impairment of the transcriptional reprogramming under cell wall stress.
Here we show that SAGA acetylates histone H3 at the promoters of CWI-responsive SAGA-dependent genes like MLP1. HAT activity of Gcn5 is essential, cooperatively with the SWI/SNF complex, to mediate the chromatin-remodeling activity necessary for regulation of gene expression through the CWI pathway. Acetylation of histones can mediate different actions. On one hand, this modification can create a target for proteins containing bromodomains, including Gcn5 and Spt7 of SAGA complex (58,59) and Snf2 of SWI/SNF complex. Thus, the interaction between SAGA and chromatin is stabilized by its own HAT activity (60), and also, acetylation of lysine residues by SAGA stabilizes SWI/SNF binding in vitro to promoter nucleosomes (60–62). However, a role for the Snf2 bromodomain mediating the HAT-facilitated SWI/SNF association in response to cell wall stress is unlikely since the recruitment of Snf2 is almost not affected in gcn5Δ strain. On the other hand, the acetylated nucleosomes by SAGA are more easily displaced by the SWI/SNF complex, increasing the rate of gene expression (63,64). Interestingly, the SAGA complex is recruited not only to the promoters of SAGA-dependent genes but also to SAGA-independent genes. However, Gcn5 mediates acetylation and nucleosome eviction at the promoters of the former genes, like MLP1, whereas in the latter ones (i.e YLR194C), H3 acetylation and chromatin remodeling is not dependent on this complex.
Pol II, Snf2 and Rlm1 recruitment to the promoters of CWI genes upon stress is only slightly dependent on Gcn5, suggesting that acetylation by Gcn5 is not essential for nucleosome eviction necessary for PIC assembly, but rather increase the extent of the remodeling mediated by SWI/SNF. In addition, the effect on chromatin remodeling mediated by Gcn5 at the promoters of MLP1 and YPL088W genes, also affects Pol II progression whereas this progression is almost not affected in genes not dependent on SAGA. In S. pombe, upon oxidative stress conditions, Gcn5 facilitates Pol II progression rather than its recruitment to nucleosome-depleted stress promoters (41). However, in that case, Gcn5 mediates the eviction of nucleosomes downstream of the promoters, whereas we have shown here that Gcn5 is not recruited to the coding regions of CWI-responsive genes. Our results suggest that the remodeling mediated by SAGA at the MLP1 and YPL088W gene promoters would be required for the recruitment of additional elements necessary for Pol II progression and effective production of mRNA transcripts.
As deduced from the results shown here and those previously published (9), SWI/SNF would have a more important effect on chromatin remodeling at CWI promoters. However, cooperation between both complexes, SWI/SNF and SAGA, is necessary to obtain a complete nucleosome remodeling of those genes transcriptionally induced in response to cell wall stress, as deduced by the analysis of the MLP1 promoter in the swi3Δ gcn5Δ double mutant. Indeed, the loss of both proteins, in addition to block chromatin remodeling and completely abrogates Rlm1 binding and gene expression in the corresponding single mutants, it leads to an increased hypersensitivity to cell wall stress. Therefore, upon cell wall stress, the MAPK Slt2 mediates through Rlm1, complete nucleosome rearrangements at CWI-responsive genes by selective targeting and cooperative action of SAGA and SWI/SNF complexes.
Supplementary Material
Acknowledgments
We want to thank Pilar García-Broncano for her help at the initial stages of this work, Sonia Díez-Muñiz for technical assistance, B. Franklin Pugh for strains and plasmids provided, Joseph C. Reese for the anti-Snf2 antibody and Craig Peterson for fruitful discussions. We are also in debt with Jesús García-Cantalejo and Pedro Botías at the Genomics Unit (Genomics and Proteomics Center, UCM) for their help in genome-wide expression analysis and with Miriam Sansó and Elena Hidalgo for their advices about H3 acetylation experiments. All members of our research group (UCM-920640: Yeast Functional Genomics) at the Department of Microbiology II (UCM) are also acknowledged for their support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Economía (MINECO) [BIO2010-22146, BIO2013-48136-P to J.A]; Comunidad de Madrid [S2010/BDM-2414 to J.A]. Funding for open access charge: Ministerio de Economía (MINECO, Spain) [BIO2013-48136-P to J.A].
Conflict of interest statement. None declared.
REFERENCES
- 1.Levin D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics. 2011;189:1145–1175. doi: 10.1534/genetics.111.128264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breeden L.L. Periodic transcription: a cycle within a cycle. Curr. Biol. 2003;13:R31–R38. doi: 10.1016/s0960-9822(02)01386-6. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe Y., Irie K., Matsumoto K. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell Biol. 1995;15:5740–5749. doi: 10.1128/mcb.15.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung U.S., Sobering A.K., Romeo M.J., Levin D.E. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 2002;46:781–789. doi: 10.1046/j.1365-2958.2002.03198.x. [DOI] [PubMed] [Google Scholar]
- 5.Jung U.S., Levin D.E. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 1999;34:1049–1057. doi: 10.1046/j.1365-2958.1999.01667.x. [DOI] [PubMed] [Google Scholar]
- 6.García R., Bermejo C., Grau C., Pérez R., Rodríguez-Peña J.M., Francois J., Nombela C., Arroyo J. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 2004;279:15183–15195. doi: 10.1074/jbc.M312954200. [DOI] [PubMed] [Google Scholar]
- 7.Kim K.Y., Levin D.E. Transcriptional reporters for genes activated by cell wall stress through a non-catalytic mechanism involving Mpk1 and SBF. Yeast. 2010;27:541–548. doi: 10.1002/yea.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim K.Y., Truman A.W., Levin D.E. Yeast Mpk1 mitogen-activated protein kinase activates transcription through Swi4/Swi6 by a noncatalytic mechanism that requires upstream signal. Mol. Cell Biol. 2008;28:2579–2589. doi: 10.1128/MCB.01795-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanz A.B., García R., Rodríguez-Peña J.M., Díez-Muñiz S., Nombela C., Peterson C.L., Arroyo J. Chromatin remodeling by the SWI/SNF complex is essential for transcription mediated by the yeast cell wall integrity MAPK pathway. Mol. Biol. Cell. 2012;23:2805–2817. doi: 10.1091/mbc.E12-04-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee K.K., Workman J.L. Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 11.Balasubramanian R., Pray-Grant M.G., Selleck W., Grant P.A., Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- 12.Grant P.A., Eberharter A., John S., Cook R.G., Turner B.M., Workman J.L. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 13.Sterner D.E., Grant P.A., Roberts S.M., Duggan L.J., Belotserkovskaya R., Pacella L.A., Winston F., Workman J.L., Berger S.L. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henry K.W., Wyce A., Lo W.S., Duggan L.J., Emre N.C., Kao C.F., Pillus L., Shilatifard A., Osley M.A., Berger S.L. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingvarsdottir K., Krogan N.J., Emre N.C., Wyce A., Thompson N.J., Emili A., Hughes T.R., Greenblatt J.F., Berger S.L. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol. Cell. Biol. 2005;25:1162–1172. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee K.K., Florens L., Swanson S.K., Washburn M.P., Workman J.L. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol. Cell. Biol. 2005;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee K.K., Swanson S.K., Florens L., Washburn M.P., Workman J.L. Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik(SALSA) HAT complexes. Epigenetics Chromatin. 2009;2:2. doi: 10.1186/1756-8935-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohler A., Pascual-García P., Llopis A., Zapater M., Posas F., Hurt E., Rodríguez-Navarro S. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol. Biol. Cell. 2006;17:4228–4236. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo Y.M., Andrews A.J. Quantitating the specificity and selectivity of Gcn5-mediated acetylation of histone H3. PLoS One. 2013;8:e54896. doi: 10.1371/journal.pone.0054896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 21.Liu C.L., Kaplan T., Kim M., Buratowski S., Schreiber S.L., Friedman N., Rando O.J. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robert F., Pokholok D.K., Hannett N.M., Rinaldi N.J., Chandy M., Rolfe A., Workman J.L., Gifford D.K., Young R.A. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 24.Horn P.J., Peterson C.L. Molecular biology. Chromatin higher order folding: wrapping up transcription. Science. 2002;297:1824–1827. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- 25.Yang X.J. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays. 2004;26:1076–1087. doi: 10.1002/bies.20104. [DOI] [PubMed] [Google Scholar]
- 26.Zeng L., Zhou M.M. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 27.Winston F., Allis C.D. The bromodomain: a chromatin-targeting module? Nat. Struct. Biol. 1999;6:601–604. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- 28.Mohibullah N., Hahn S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 2008;22:2994–3006. doi: 10.1101/gad.1724408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhaumik S.R., Green M.R. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 2002;22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dudley A.M., Rougeulle C., Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huisinga K.L., Pugh B.F. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 32.Lee T.I., Causton H.C., Holstege F.C., Shen W.C., Hannett N., Jennings E.G., Winston F., Green M.R., Young R.A. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- 33.Kremer S.B., Gross D.S. SAGA and Rpd3 chromatin modification complexes dynamically regulate heat shock gene structure and expression. J. Biol. Chem. 2009;284:32914–32931. doi: 10.1074/jbc.M109.058610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zanton S.J., Pugh B.F. Changes in genomewide occupancy of core transcriptional regulators during heat stress. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16843–16848. doi: 10.1073/pnas.0404988101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geng F., Laurent B.C. Roles of SWI/SNF and HATs throughout the dynamic transcription of a yeast glucose-repressible gene. EMBO J. 2004;23:127–137. doi: 10.1038/sj.emboj.7600035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma V.M., Li B., Reese J.C. SWI/SNF-dependent chromatin remodeling of RNR3 requires TAF(II)s and the general transcription machinery. Genes Dev. 2003;17:502–515. doi: 10.1101/gad.1039503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh S., Pugh B.F. Sequential recruitment of SAGA and TFIID in a genomic response to DNA damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 2011;31:190–202. doi: 10.1128/MCB.00317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H., Kruk J.A., Reese J.C. Dissection of coactivator requirement at RNR3 reveals unexpected contributions from TFIID and SAGA. J. Biol. Chem. 2008;283:27360–27368. doi: 10.1074/jbc.M803831200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Proft M., Struhl K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell. 2002;9:1307–1317. doi: 10.1016/s1097-2765(02)00557-9. [DOI] [PubMed] [Google Scholar]
- 40.Zapater M., Sohrmann M., Peter M., Posas F., de Nadal E. Selective requirement for SAGA in Hog1-mediated gene expression depending on the severity of the external osmostress conditions. Mol. Cell. Biol. 2007;27:3900–3910. doi: 10.1128/MCB.00089-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sansó M., Vargas-Pérez I., Quintales L., Antequera F., Ayte J., Hidalgo E. Gcn5 facilitates Pol II progression, rather than recruitment to nucleosome-depleted stress promoters, in Schizosaccharomyces pombe. Nucleic Acids Res. 2011;39:6369–6379. doi: 10.1093/nar/gkr255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uffenbeck S.R., Krebs J.E. The role of chromatin structure in regulating stress-induced transcription in Saccharomyces cerevisiae. Biochem. Cell Biol. 2006;84:477–489. doi: 10.1139/o06-079. [DOI] [PubMed] [Google Scholar]
- 43.Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 44.Bermejo C., Rodríguez E., García R., Rodríguez-Peña J.M., Rodríguez de la Concepcion M.L., Rivas C., Arias P., Nombela C., Posas F., Arroyo J. The sequential activation of the yeast HOG and SLT2 pathways is required for cell survival to cell wall stress. Mol. Biol. Cell. 2008;19:1113–1124. doi: 10.1091/mbc.E07-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arroyo J., Hutzler J., Bermejo C., Ragni E., García-Cantalejo J., Botias P., Piberger H., Schott A., Sanz A.B., Strahl S. Functional and genomic analyses of blocked protein O-mannosylation in baker's yeast. Mol. Microbiol. 2011;79:1529–1546. doi: 10.1111/j.1365-2958.2011.07537.x. [DOI] [PubMed] [Google Scholar]
- 46.García R., Rodríguez-Peña J.M., Bermejo C., Nombela C., Arroyo J. The high osmotic response and cell wall integrity pathways cooperate to regulate transcriptional responses to zymolyase-induced cell wall stress in Saccharomyces cerevisiae. J. Biol. Chem. 2009;284:10901–10911. doi: 10.1074/jbc.M808693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saeed A.I., Bhagabati N.K., Braisted J.C., Liang W., Sharov V., Howe E.A., Li J., Thiagarajan M., White J.A., Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 48.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 49.Papamichos-Chronakis M., Peterson C.L. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat. Struct. Mol. Biol. 2008;15:338–345. [Google Scholar]
- 50.Aparicio O., Geisberg J.V., Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr. Protoc. Cell Biol. 2004;23:17.7.1–17.7.23. doi: 10.1002/0471143030.cb1707s23. [DOI] [PubMed] [Google Scholar]
- 51.García R., Sanz A.B., Rodríguez-Peña J.M., Nombela C., Arroyo J. Rlm1 mediates a positive autoregulatory transcriptional feedback essential for Slt2 MAPK dependent gene expression. J. Cell Sci. 2016;129:1649–1660. doi: 10.1242/jcs.180190. [DOI] [PubMed] [Google Scholar]
- 52.Kim K.Y., Levin D.E. Mpk1 MAPK association with the Paf1 complex blocks Sen1-mediated premature transcription termination. Cell. 2011;144:745–756. doi: 10.1016/j.cell.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swanson M.J., Qiu H., Sumibcay L., Krueger A., Kim S.J., Natarajan K., Yoon S., Hinnebusch A.G. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 2003;23:2800–2820. doi: 10.1128/MCB.23.8.2800-2820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bryant G.O., Ptashne M. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell. 2003;11:1301–1309. doi: 10.1016/s1097-2765(03)00144-8. [DOI] [PubMed] [Google Scholar]
- 55.Burns L.G., Peterson C.L. The yeast SWI-SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol. Cell Biol. 1997;17:4811–4819. doi: 10.1128/mcb.17.8.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neely K.E., Hassan A.H., Wallberg A.E., Steger D.J., Cairns B.R., Wright A.P., Workman J.L. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- 57.Venters B.J., Pugh B.F. How eukaryotic genes are transcribed. Crit. Rev. Biochem. Mol. Biol. 2009;44:117–141. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brownell J.E., Zhou J., Ranalli T., Kobayashi R., Edmondson D.G., Roth S.Y., Allis C.D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 59.Gansheroff L.J., Dollard C., Tan P., Winston F. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics. 1995;139:523–536. doi: 10.1093/genetics/139.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hassan A.H., Prochasson P., Neely K.E., Galasinski S.C., Chandy M., Carrozza M.J., Workman J.L. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 61.Mitra D., Parnell E.J., Landon J.W., Yu Y., Stillman D.J. SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment. Mol. Cell. Biol. 2006;26:4095–4110. doi: 10.1128/MCB.01849-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hassan A.H., Neely K.E., Workman J.L. Histone acetyltransferase complexes stabilize SWI/SNF binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- 63.Chandy M., Gutierrez J.L., Prochasson P., Workman J.L. SWI/SNF displaces SAGA-acetylated nucleosomes. Eukaryot. Cell. 2006;5:1738–1747. doi: 10.1128/EC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barbaric S., Walker J., Schmid A., Svejstrup J.Q., Horz W. Increasing the rate of chromatin remodeling and gene activation-a novel role for the histone acetyltransferase Gcn5. EMBO J. 2001;20:4944–4951. doi: 10.1093/emboj/20.17.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.