Abstract
According to recent models, as yet poorly studied architectural proteins appear to be required for local regulation of enhancer–promoter interactions, as well as for global chromosome organization. Transcription factors ZIPIC, Pita and Zw5 belong to the class of chromatin insulator proteins and preferentially bind to promoters near the TSS and extensively colocalize with cohesin and condensin complexes. ZIPIC, Pita and Zw5 are structurally similar in containing the N-terminal zinc finger-associated domain (ZAD) and different numbers of C2H2-type zinc fingers at the C-terminus. Here we have shown that the ZAD domains of ZIPIC, Pita and Zw5 form homodimers. In Drosophila transgenic lines, these proteins are able to support long-distance interaction between GAL4 activator and the reporter gene promoter. However, no functional interaction between binding sites for different proteins has been revealed, suggesting that such interactions are highly specific. ZIPIC facilitates long-distance stimulation of the reporter gene by GAL4 activator in yeast model system. Many of the genomic binding sites of ZIPIC, Pita and Zw5 are located at the boundaries of topologically associated domains (TADs). Thus, ZAD-containing zinc-finger proteins can be attributed to the class of architectural proteins.
INTRODUCTION
In recent years, considerable progress has been made in understanding chromosome organization (for reviews, see (1–3)). High-resolution chromosome conformation capture techniques have provided evidence that chromosomes in the genomes of human, mouse and Drosophila are partitioned into a series of discrete topologically associating domains (TADs) (4–7). Their characteristic feature is that regulatory elements within a TAD display extensive long-range interactions with each other but interact far less frequently with regulatory elements located outside their domain. TADs themselves are often organized hierarchically and include smaller domains (sub-TADs) interspaced with short boundary elements or longer spacing regions (inter-TADs) that contain active chromatin and constitutively transcribed (housekeeping) genes. Partitioning of the mammalian and Drosophila genomes into TADs appears to be largely cell-lineage independent and evolutionary conserved (5,8,9).
However, despite progress in the study of chromosomal architecture, we still do not have a clear mechanistic picture of how long-range interactions between distant regulatory regions are established and maintained through the cell cycle. In the past few years, the concept has arisen that there is a special class of architectural proteins that are responsible not only for global chromosome architecture but also for the local regulation of enhancer–promoter interactions (1,6,10–12). Architectural proteins include molecules differing in structure and functions, but the mechanisms and protein domains involved in long-distance interactions are not well understood. The question is still unresolved as to how architectural (insulator) proteins can organize specific interactions between distantly located sites.
Many transcription factors involved in insulator activity have been attributed to the category of architectural proteins. Insulators in the Drosophila and vertebrate genomes have been identified based on their ability to disrupt the communication between an enhancer and a promoter when inserted between them (11,13–17). The growing amount of data show that insulator proteins fulfill an architectural function in mediating inter- and intrachromosomal interactions and in contacting regulatory elements such as promoters or enhancers (18). In mammals, cohesin and insulator protein CTCF are often found at TAD boundaries and play a major role in long-range contact formation (8,19). In addition, a condensin complex and transcription factors like TFIIIC and ZFP143 have been detected at these sites (20,21).
The bulk of information about potential transcription factors involved in long-range interactions has been obtained in studies of Drosophila insulators. The Drosophila genome contains many sequences with an insulator function (15,22,23). As shown in transgenic lines, pairing of two identical insulators can support distant activation of a promoter by an enhancer or yeast GAL4 activator (24–28). The relative orientation of two identical insulators defines the mode of loop formation that either allows or blocks enhancer (GAL4)–promoter interaction (26,27,29). Supposedly, this orientation-dependent interaction is accounted for by at least two insulator-bound proteins that are involved in specific protein–protein interactions. It has also been found that two identical insulators can support interactions between regulatory elements located in transgenes inserted at distances of up to several megabases from each other (30–34). The most striking example is the insulator named Homie that is located between the TER94 promoter and regulatory region of the eve gene (35). The presence of Homie in a transgene as far as 3.3 Mb away from the endogenous copy facilitates long-range communication between endogenous eve enhancers located near Homie and a promoter placed on the transgene (35,36). These facts suggest that proteins bound to insulators can support very specific distant interactions through the cell cycle.
The first insulators to be identified in Drosophila were scs and scs’ located at the boundaries of two heat shock 70 genes (37,38). One protein, Zw5, binds to scs and partially accounts for its insulator properties (39,40). Four reiterated binding sites for Zw5 can function as an effective insulator (39). The Zw5 protein contains C2H2-type zinc fingers (ZF) at the C-terminus and, by this criterion, belongs to the largest group of transcription factors in higher metazoans (41–43). A C2H2-ZF domain can specify a wide range of three or four base pair targets, and tandem arrays of these domains bind contiguous DNA sequences, giving the C2H2-ZF proteins the ability to recognize an incredibly diverse set of sequence motifs (44–46). At the N-terminus, Zw5 proved to contain a zinc finger-associated domain (ZAD) that is almost exclusively found in association with zinc finger proteins (ZFP) (39,47,48). Two new transcription factors described recently, Pita and ZIPIC, function as insulator proteins and have a ZAD domain at the N-termini (49). In addition, both these factors interact with the 190-kDa centrosome-associated protein, named CP190, which has been suggested to have a global role in the function of chromatin insulators (50–52).
The ZAD domain is characteristic of the subfamily of mostly clustered Drosophila ZFP genes and appears to be restricted to the dipteran and closely related insect genomes (48,53,54). The ZAD domain is characterized by a conserved constellation of four cysteines, which form a zinc-coordinated fold. Data on the crystal structure of the ZAD of the Grauzone protein provide evidence that two ZAD molecules interact in a head-to-tail mode to form a dimer (55). The ability to form a homodimer was also shown for the ZAD domain of the Serendipity-δ (56) and Weckle (57) proteins. It has been suggested that ZAD domains of other proteins are also able to self-associate.
Here, we tested the ZAD domains of three known insulator proteins—Zw5, Pita, and ZIPIC—for the ability to form homo- or heterodimers. The results showed that, these domains were capable only of homodimerization. Moreover, ZIPIC and Pita each could support long-range interaction between the yeast GAL4 activator and the white promoter in transgenic Drosophila lines, as was previously found for the Zw5 protein (27). However, we did not observe any functional interactions between different ZAD proteins. These results suggest the ZAD domains account for highly specific interactions between the insulator proteins.
MATERIALS AND METHODS
ChIP-Seq
Embryo collection and ChIP were performed as described (58). In particular, wild-type Oregon-R embryos were collected at 0–12 h and fixed with 1.7% formaldehyde. Chromatin was precipitated with rabbit anti-Pita, anti-ZIPIC, anti-Zw5 antibodies. To construct the libraries, DNA was processed as described in the TruSeq RNA Sample Preparation Guide (Illumina) v. 2 from the end repair stage. This protocol has been chosen because of a very low amount of input DNA (<10ng). Amplified libraries were quantified using fluorometry with Qubit (Invitrogen, United States) and real-time PCR and diluted to a final concentration of 10 pM. Diluted libraries were clustered on a single-read flowcell using cBot instrument and sequenced in 51 cycles using a HiSeq2000 sequencer with TruSeq SBS Kit v3-HS (Illumina, United States). We sequenced the input sample as well as by one sample for Pita, ZIPIC, Zw5 and preimmune chromatin immunoprecipitates. Individual ChIP-Seq data were extensively validated by specific qPCR (Supplementary Figure S1). Raw and processed data were deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE76997.
Computational processing of ChIP-Seq data
Raw reads were processed in the following way: adapters were trimmed using cutadapt (59) and subsequent sliding window-based adaptive trimming with sickle (https://github.com/najoshi/sickle); reads shorter than 20 bps were discarded. The remaining reads were mapped with Bowtie (60) (the optimal alignments with ‘–best –strata –tryhard’ parameters) to the BDGP r5 / UCSC dm3 Drosophila melanogaster genome assembly, and only uniquely mapped (‘–m 1’) reads were kept. Reads overlapping with ENCODE blacklist regions (https://sites.google.com/site/anshulkundaje/projects/blacklists) (61) were discarded.
Peak calling was done using MACS2 (62) (https://github.com/taoliu/MACS) against preimmune and input control data. Peak sets obtained with the preimmune control were smaller and were utilized for motif discovery with diChIPMunk (63,64). Resulting dinucleotide position weight matrices were used for motif finding in peaks obtained with the input control. To detect peaks with strong motif occurrences, we estimated a P-value for the best motif hit in each peak. To estimate the P-values, Drosophila genome dinucleotide composition was used as a background model for PERFECTOS-APE (65). For each ChIP-Seq peak, the motif statistical significance (S) was estimated as the probability that a random sequence of the same length had at least one motif hit scoring no less than the best hit observed in the ChIP-Seq peak (66):
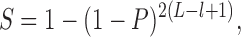 |
where P is the best hit P-value, L is the length of a particular peak, l is the length of the motif; the motif hits (including hits on both DNA strands and overlapping hits) were considered independent. All peaks with S < 0.05 (>1.3 after –log10 transformation) were considered carrying strong motif hits.
Peak annotation was performed in R with ChIPpeakAnno (67), biomaRt (68) and GenomicRanges (69). During peak annotation, we sequentially checked if a peak overlapped with a promoter/upstream/intron/exon/intergenic segments and assigned the peak into the first appropriate category (e.g. a peak annotated as promoter-related could also partly overlap the upstream and/or intron/exon segments). Promoter segments were considered as [–100; TSS], and upstream segments, as [–1000; –100] from the TSS.
Antibodies
Antibodies against ZIPIC [aa 84–257] and Pita [aa 99–302 and 550–683] were described in (49). Antibodies against Zw5 [aa 98–302] were raised in rabbits and purified from the sera by ammonium sulfate fractionation followed by affinity purification on CNBr-activated Sepharose (GE Healthcare, USA) according to standard protocols. Anti-FLAG M2 and anti-HA antibodies were from Sigma (Unites States), and anti-GST antibody was from Pierce (United States).
Plasmid construction
To express ZAD domains in S2 cells, protein-coding sequences were cloned in frame with 3×FLAG or 3×HA, excised, and subcloned into the pAc5.1 plasmid (Life Technologies).
Plasmids for yeast two-hybrid assay were prepared for full-sized proteins and ZAD domains alone as C-termini fused with DNA-binding or activation domain of GAL4 in corresponding pGBT9 and pGAD424 vectors from Clontech. We also used a modification of standard pGBT9 vector in which ZAD domain was cloned at the N-terminus of GAL4 DNA-binding domain. In such a system, the ZAD domain was separated from the GAL4 DNA-binding domain with the (GluAlaAlaAlaLys)4 linker.
For in vitro experiments, each ZAD domain was cloned into two plasmids, pAc28-TRX and pMAL-C5X (New England Biolabs). The pAc28-TRX vector was produced by cloning thioredoxin from pET32a (Novagen) into pAc28 (70). Thus, we obtained a combination of pACYC and pET28a(+) (Novagen) containing the p15A replication origin, Kanamycin-resistance gene, and pET28a(+) MCS. Fragments taken for cloning were as follows: Grauzone, aa 1–84; Zw5, aa 1–103; ZIPIC, aa 3–81; and Pita, aa 1–109.
The constructs for testing long-range interactions in transgenic flies were made as described previously (71).
Generation and analysis of transgenic lines
The transgenic construct and P25.7wc plasmid were injected into yacw1118 preblastoderm embryos (72). The resultant flies were crossed with yacw1118 flies, and the transgenic progeny were identified by their eye color under a Stemi 2000 stereomicroscope (Carl Zeiss, Germany). The transformed lines were tested for transposon integrity and copy number by RT-PCR. Only single-copy transformants were included in the study.
To obtain transgenic flies with insertion in 86Fb, the DNA of reporter constructs was injected into preblastoderm embryos of y1 genotype (73). The emerging adults were crossed with the y ac w1118 flies, and the progeny carrying the transgene in the 86Fb region were identified by pigmented bristles and eyes.
The lines with DNA fragment excisions were obtained by crossing the transposon-bearing flies with Cre (yw; Cyo, P[w+,cre]/Sco;+) recombinase-expressing lines. The Cre recombinase induces 100% excisions in the next generation. All excisions were confirmed by PCR analysis. Details of the crosses and primers used for genetic analysis and the excision of functional elements are available upon request.
Protein expression and purification
Escherichia coli BL21 cells were transformed with plasmids encoding a protein fused with an affinity purification tag. Protein expression was induced with 1 mM IPTG. After induction, the cells were incubated in the presence of 0.2 mM ZnCl2 at 18°C overnight and disrupted by sonication in buffer A (for purification with 6×His tag: 50 mM HEPES–KOH, pH 7.6, with 500 mM NaCl, 5 mM MgCl2, 0.1 mM ZnCl2, 20 mM imidazole, 5% glycerol, 0.1% NP-40, and 5 mM β-mercaptoethanol; for purification with MBP tag: 20 mM HEPES–KOH, pH 7.6, with 150 mM NaCl, 5 mM MgCl2, 0.1 mM ZnCl2, 10% glycerol, 0.1% NP-40, and 1 mM DTT) containing 1 mM PMSF and Calbiochem Complete Protease Inhibitor Cocktail VII (5 μL/ml). The cell lysate was centrifuged, and the supernatant was incubated with 10 μl of Co-IDA-Agarose (Biontex) for purification with 6×His tag or with Amylose Resin (New England Biolabs) for purification with MBP tag (20 min at room temperature), washed with four portions of buffer A (5 min each at room temperature), and eluted with elution buffer (for purification with 6×His tag: 50 mM HEPES-KOH, pH 7.6, with 500 mM NaCl, 0.1 mM ZnCl2, 250 mM imidazole and 5 mM β-mercaptoethanol; for purification with MBP tag: 20 mM Tris–HCl, pH 7.5, with 50 mM NaCl, 0.1 mM ZnCl2, 10 mM maltose and 1 mM DTT) (20 min at room temperature).
Protein chemical crosslinking
ZAD domains fused with thioredoxin and thioredoxin alone (as a negative control) were taken for crosslinking. Protein concentration was adjusted to 10 μM. Crosslinking was carried out in 50 mM HEPES–KOH, pH 7.6, with 500 mM NaCl, 0.1 mM ZnCl2, 250 mM imidazole, 5 mM β-mercaptoethanol, and 0.02 or 0.1% glutaraldehyde (with additional 0.05% for thioredoxin) or without it (10 min at room temperature). Crosslinking was quenched with 200 mM Tris. The crosslinked samples were resolved by SDS-PAGE and stained with silver.
Pull-down assays
For pull-down assays, ZAD domains fused with 6×His-thioredoxin and MBP were coexpressed in Escherichia coli BL21 cells. For this purpose, we transformed E. coli BL21 with two plasmids and used media containing ampicillin and kanamycin.
After induction of expression, each probe was divided into two portions: one used for purification with 6×His tag, and the other, with MBP tag. After elution, the samples were analyzed by SDS-PAGE with Coomassie staining.
Co-immunoprecipitation assay
S2 cells grown in SFX medium were cotransfected by 3×FLAG- and 3×HA-fused plasmids with Cellfectin (Life Technologies) as recommended by the manufacturer. After transfection, the cells were incubated for 48 h and then collected by centrifugation at 700 g for 5 min, washed once with 1×PBS, and resuspended in 20 packed cell volumes of hypotonic lysis buffer (20 mM Tris–HCl, pH 7.4, with 10 mM KCl, 10 mM MgCl2, 2 mM EDTA, 10% glycerol, 1% Triton X-100, 1 mM DTT, and Calbiochem Complete Protease Inhibitor Cocktail V). After incubation on ice for 10 min, the cells were sonicated (2 × 15 s on ice at 20% output), NaCl was added to a final concentration of 420 mM, and incubation on ice continued for 60 min, with periodic mixing. Sonication was repeated as above to reduce viscosity, cell debris was pelleted by centrifugation at 10 000 g for 30 min at 4°C, and the supernatant was collected for immunoprecipitation with anti-FLAG- and anti-HA-conjugated Sepharose (Sigma) equilibrated in incubation buffer-150 (20 mM Tris–HCl, pH 7.4, with 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 10% glycerol and 0.1% NP-40). Mouse IgG conjugated to Protein G Agarose beads (by incubating in the same buffer on a rotary shaker at 4°C for 1 h) was used as negative control. The protein extract (50 μg protein) was adjusted to a volume of 500 μl with buffer-150, mixed with antibody-conjugated beads (30 μl), and incubated on a rotary shaker overnight at 4°C. The beads were then washed with two portions of buffer-150, three portions of incubation buffer-400 (with 400 mM NaCl), and one portion of incubation buffer-150, resuspended in SDS-PAGE loading buffer, boiled, and analyzed by western blotting. Proteins were detected using the ECL Plus Western Blotting substrate (Pierce).
Electrophoretic mobility shift assay (EMSA)
Aliquots of purified recombinant proteins (10–15 μg) were incubated with a radioactively labeled DNA fragments of artificial Zw5-binding sites (100 cps) in the presence of nonspecific binding competitor poly(dI-dC). Incubation was performed in PBS (pH 8.0) containing 5 mM MgCl2, 0.1 mM ZnSO4, 1 mM DTT, 0.1% NP-40 and 10% glycerol at room temperature for 30 min. The mixtures were then resolved by nondenaturing 5% PAGE in 0.5×TBE buffer at 5 V/cm.
Yeast two-hybrid assay
Yeast two-hybrid assay was carried out using yeast strain pJ69-4A, with plasmids and protocols from Clontech. For growth assays, plasmids were transformed into yeast strain pJ69-4A by the lithium acetate method, as described by the manufacturer, and plated on media without tryptophan and leucine. After 2 days of growth at 30°C, the cells were plated on selective media without tryptophan, leucine, histidine, and adenine, and their growth was compared after 2–3 days. Each assay was repeated three times.
RESULTS
Genomic colocalization of Zw5, Pita, and ZIPIC in Drosophila embryos
In previous studies, binding sites for Pita and ZIPIC were identified in the genome of S2 cells (49); in case of Zw5, only ChIP-on-chip data were available (74). To compare the genome-wide distribution of these ZAD-containing insulator proteins, we performed ChIP experiments with 0- to 12-h embryos, with subsequent sequencing by Illumina's massively parallel sequencing technology. ChIP-Seq peak calling against input control data revealed 1845/575/695 peaks for Pita/ZIPIC/Zw5 respectively (Supplementary Table S1).
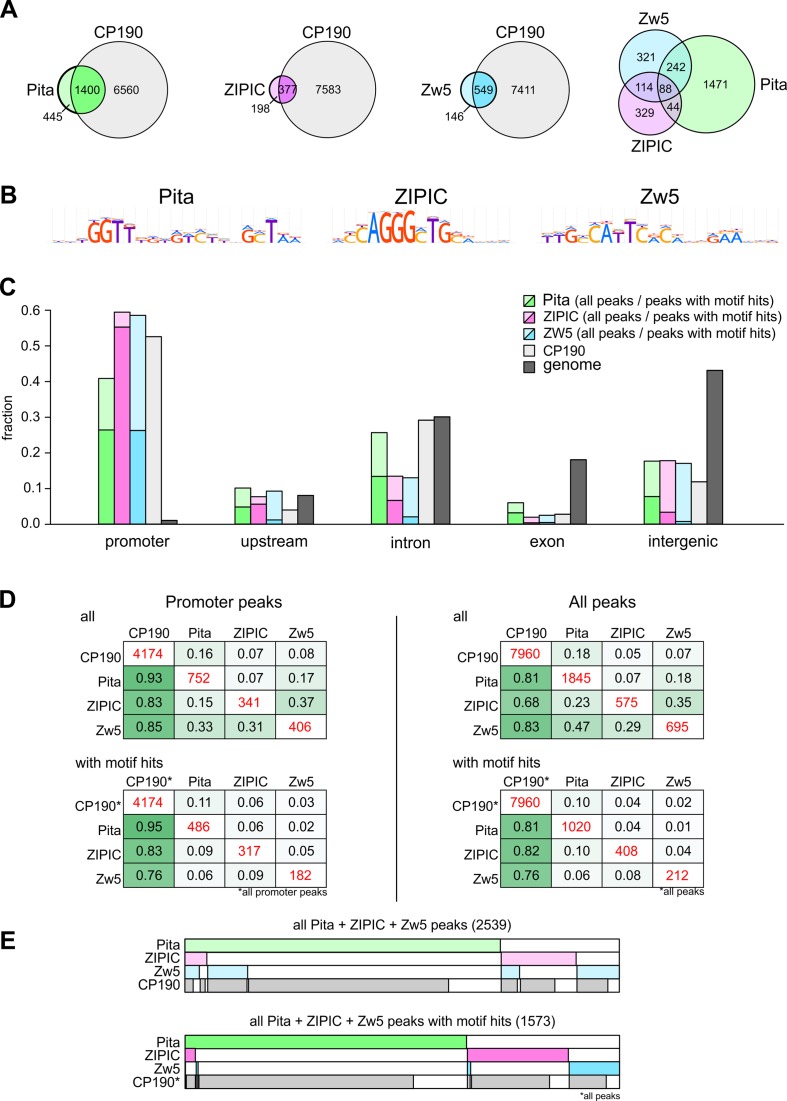
More than 75% of Pita peaks, 65% of ZIPIC peaks and 79% of Zw5 peaks overlapped CP190 binding regions (75) (Figure 1A). It is noteworthy that no direct interaction between Zw5 and CP190 was revealed in a yeast two-hybrid assay. The overlap between the peak sets of Pita/ZIPIC/Zw5 themselves was notably less exhibited (10–20%, see Figure 1A). According to the current model, insulator proteins organize chromatin architecture in cooperation with condensin and cohesin complexes. We tested colocalization of Pita/ZIPIC/ZW5 with CapH2 (condensin complex) (76) and Rad-21/Nipped-B (cohesin complex) (77) and observed notable overlapping of these subunits with all three insulator proteins under study. Approximately half of Pita/ZIPIC/Zw5 peaks colocalized with CapH2/Rad-21/Nipped-B (Supplementary Figure S2A). As expected, the subunits of condensin and cohesin complexes were found at many other genomic sites that showed no colocalization with the Pita/ZIPIC/Zw5 peaks (e.g. see Supplementary Figure S2B).
Figure 1.
Genome-wide study of Pita, ZIPIC, and Zw5 binding sites. (A) Venn diagrams of Pita, ZIPIC, Zw5 and CP190; the number of overlapping ChIP-Seq peaks is shown. (B) Sequence logos for binding motifs of Pita, ZIPIC and Zw5 obtained from ChIP-Seq data. (C) Proportions of peak sets overlapping different genomic regions for Pita, ZIPIC, Zw5 and CP190 ChIP-Seq data. The proportion of the total genome length covered by different region types is shown for comparison. The deeper colored areas show peaks with high motif occurrence (see Methods). (D) Pairwise comparisons of ChIP-Seq data sets (all peaks and promoter-overlapping subsets). In each matrix, cells at the diagonal show the total number of peak calls (in red), the other cells show the proportion of peaks of each particular data set (rows) that overlap peaks of another data set (columns). Matrices at the bottom show the same information on peaks with high motif occurrence for Pita, ZIPIC and Zw5, respectively. (E) Binary heat maps of Pita, ZIPIC and Zw5 peaks classified into groups on the basis of their overlap with CP190 peaks and with each other.
Sequence motifs for Pita and ZIPIC were previously identified in S2 cells (49). We performed de novo motif search by ChIP-Seq in embryos, where the motifs proved to have the same DNA pattern as in S2 cells (Figure 1B). Interestingly, about 55% of Pita peaks and 71% of ZIPIC peaks were characterized by a high occurrence of the motif (Figure 1D), while the respective proportions of such peaks in S2 cells were only 40 and 37% (49).
Only one binding site for Zw5 was previously described in the SCS insulator (39). Our search for the motif for Zw5 based on embryo ChIP-Seq data resulted in identification of a 17-bp pattern (Figure 1B), and the SCS binding site fitted to the identified consensus sequence. Nearly 31% of Zw5 peaks showed high motif occurrence (Figure 1D), and Zw5 binding to the consensus sequence was successfully confirmed by EMSA (Supplementary Figure S3).
Next, we assessed genomic annotation of ChIP-Seq peaks. The peaks were enriched in the promoter regions in close proximity to TSS (from –100 bp to TSS) (as in the case of CP190), and depleted in intergenic and coding regions (Figure 1C). In particular, most of peaks with high motif occurrence were found in promoter regions: there were 65% of Pita peaks, 93% of ZIPIC peaks, and 45% of Zw5 peaks (Figure 1C and D). The occurrence of motifs in peaks associated with intronic and intergenic regions was rare (Figure 1C), suggesting that such peaks were probably a result of some trans interactions. Furthermore, promoter-associated Pita, ZIPIC and Zw5 peaks showed a very high correlation with CP190 (compare ‘Promoter peaks’ and ‘All peaks’ panels in Figure 1D), while pairwise overlapping between Pita, ZIPIC and Zw5 peaks was observed in a relatively small number of cases (133 overlaps for Pita–ZIPIC, 238 overlaps for Pita–Zw5 and 112 overlaps for ZIPIC–Zw5) (Figure 1D and E). Only 89 overlaps were found for the peaks of all three proteins (Figure 1A and E). Among motif-containing peaks, only two were with such a ternary combination, but there were 40 peaks with Pita–ZIPIC, 10 peaks with Pita–Zw5, and 15 peaks with ZIPIC–Zw5 combinations. This allowed us to conclude that these proteins are mainly responsible for chromatin organization of different regulatory regions, primarily at the promoters.
Hi-C data available for S2, Kc167, Bg3 and OSC cells (9) allowed us to analyze how Pita/ZIPIC/Zw5 peaks were located relative to the TAD/inter-TAD and, specifically, TAD boundaries as the key points of long-range interactions. We found peaks for Pita, ZIPIC, and Zw5 both at TADs and inter-TADs, with the latter case demonstrating 1.5- to 2-fold enrichment of insulator proteins binding (binomial test, P << 0.05; Supplementary Table S2). Then TAD boundaries were taken to be at ±20 kb and ±10 kb from TAD start/end locations. We found that about 50% (for ±20 kb) and 25–30% (for ±10 kb) of peaks for Pita, ZIPIC and Zw5 proteins were anchored within these boundaries (Supplementary Table S3). These amounts were 1.5–2 times greater than expected, taking into account the total length of the TAD boundaries (binomial test, P << 0.05; Supplementary Table S3). As for the TAD boundaries themselves, Pita was found at almost 60% of them (40% for ±10 kb window); while ZIPIC and Zw5 occupied a lower proportion of the boundaries (∼30% for ±20 kb window and 20% for ±10 kb window) (Supplementary Table S3). In many cases, we found the TADs being flanked by binding sites for Pita, ZIPIC, or Zw5 (Supplementary Table S4); i.e. there were multiple TADs with both boundaries containing ChIP-Seq peaks. Two regions of the high resolution Hi-C map (76) were randomly selected and overlaid with Pita/ZIPIC/Zw5 peaks (Supplementary Figure S4). It can be seen that many sites for one or several insulator proteins flank TADs or sub-TADs or are located in genomic regions corresponding to rare contacts between different TADs. These observations suggest that Pita, ZIPIC and Zw5 can be classified as architectural proteins that support long-range interactions between genomic regions.
ZAD domains effectively form homodimers, but not heterodimers
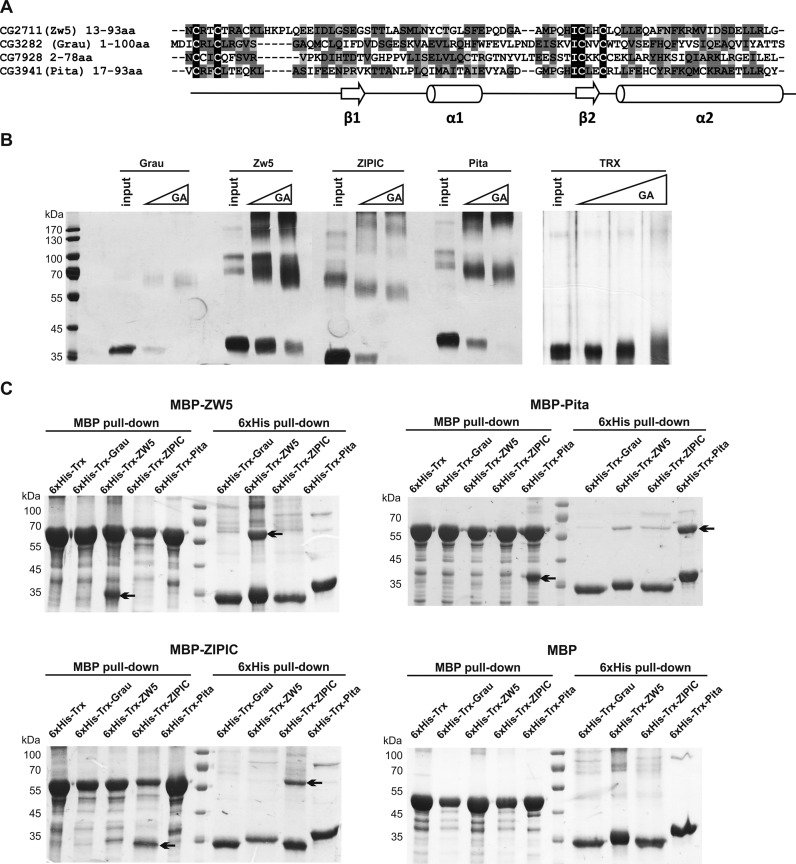
According to the crystal structure, two ZAD domains of Grau interact in a head-to-tail mode to form a dimer (55). The ZAD domain of Grau has only 5.8% identical and 72.1% similar positions in amino acid sequence alignment with the ZAD domains of insulator proteins Zw5, ZIPIC and Pita (Figure 2A). Pairwise comparisons of Zw5, ZIPIC and Pita showed a limited degree of similarity, as in comparisons with Grau. We used chemical crosslinking by glutaraldehyde to confirm that the ZAD domains of these proteins are also able to homodimerize, with the ZAD domain of Grau serving as a positive control and thioredoxin as a negative control (Figure 2B). The crosslinked ZAD domains of Grau, Pita, ZIPIC and Zw5 migrated as dimers. The fractions of monomeric (non-crosslinked) species could appear due to inefficient intermolecular crosslinking explained by the lack of neighboring lysines. Thus, the ZAD domains of these four proteins are able to homodimerize in vitro.
Figure 2.
Testing for homo- and heterodimerization of the ZAD domains in vitro. (A) Sequence comparison of ZAD domains of Grau, Pita, ZIPIC and Zw5. (B) Testing for homodimerization of Grau, Pita, ZIPIC, and Zw5 ZAD domains by chemical crosslinking with glutaraldehyde. (C) The results of MBP and 6×His pull-down assay showing that the ZAD domains of proteins expressed in vitro formed only homodimers. For MBP pull-down, beads with bound MBP–Pita, MBP–ZIPIC, MBP–Zw5 or MBP alone were incubated with 6×His-Trx-Pita, 6×His-Trx-ZIPIC, 6×His-Trx-Zw5, 6×His-Trx-Grau or 6×His-Trx. For 6×His pull-down, beads with bound 6×His-Trx-Pita, 6×His-Trx-ZIPIC, 6×His-Trx-Zw5, 6×His-Trx-Grau, were incubated with MBP–Pita, MBP–ZIPIC, MBP–Zw5 or MBP alone. The precipitated proteins were resolved by SDS-PAGE and stained with Coomassie. Grau was used as positive control for homodimer formation, while MBP and 6×His-Trx were used as negative control. Arrows indicate the precipitated prey protein after pull-down with bait protein.
We experienced some problems while testing in vitro interactions between ZAD domains expressed in bacteria. Firstly, ZAD domains from the same protein but with different tags (6×His-Thioredoxin or MBP) showed no interaction when expressed separately and mixed after purification. This could be explained by the stability of their dimers, which could not dissociate and reassociate with another domain during the experiment. Extension of the incubation time up to 7 days and changes in the buffer composition produced no effect except increasing the level of nonspecific interactions. Then we carried out a coexpression assay in which both differently tagged ZAD domains were expressed in the same E. coli cells, using compatible vectors. In this assay, the effective formation of homodimers was observed. Although homodimerization obviously prevailed over heterodimerization, the frequency of supposedly nonspecific heterodimeric interactions was relatively high. A probable explanation is that protein concentrations in the cells were high enough to exceed the threshold of heterodimer assembly, even though it is much higher than the threshold sufficient for homodimerization (Figure 2C). This assumption is supported by the results of yeast two-hybrid and immunoprecipitation assays, which deal with small protein concentrations within the cells.
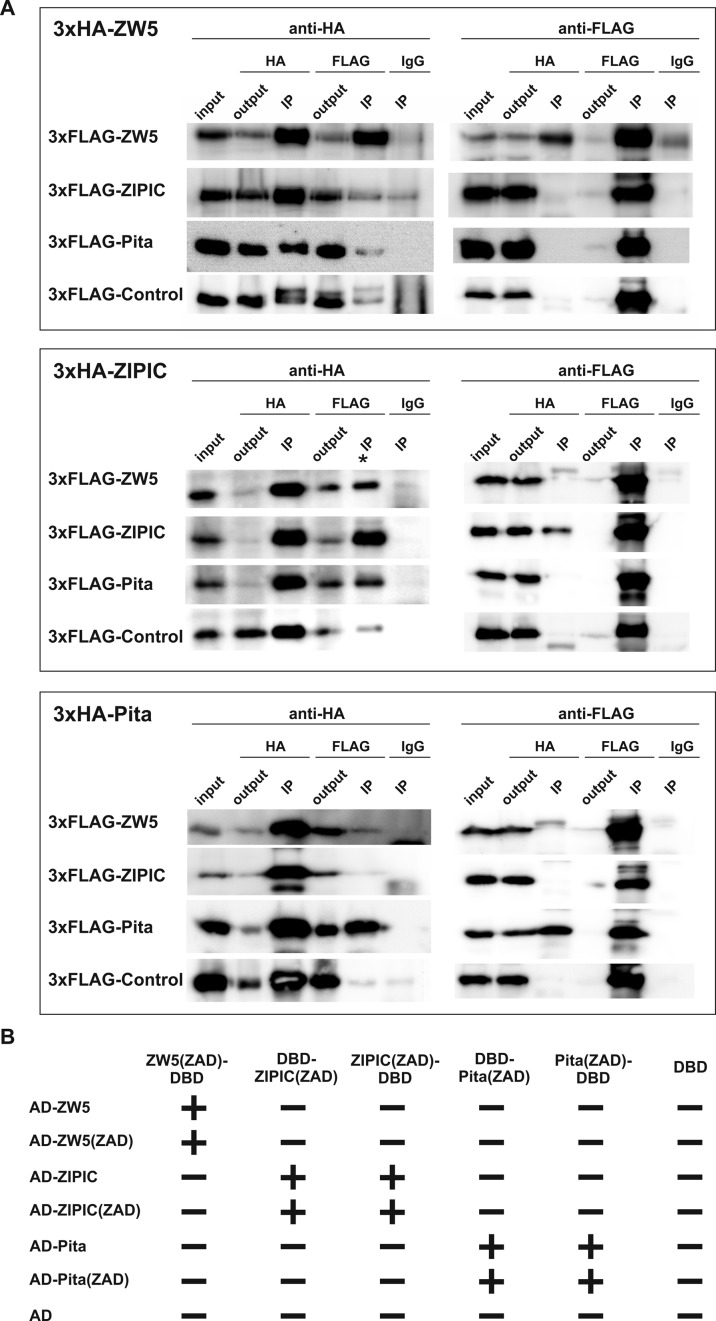
To further confirm the observed property of ZAD domains, we analyzed co-immunoprecipitation of 3×FLAG- and 3×HA-targeted ZAD domains in transfected S2 cells (Figure 3A). Each ZAD domain was fused with either 3×FLAG or 3×HA epitope and transfected in all combinations into S2 cells. The C-terminus of dCTCF fused with 3×FLAG was used as a negative control for interaction with the ZAD domains. After immunoprecipitation with HA-Sepharose, only the bands corresponding to homodimers of ZAD domains were detectable, whereas in the reverse experiment with FLAG-Sepharose we observed strong homodimer bands and weak, probably nonspecific bands for some heterodimer combinations.
Figure 3.
Testing for homo- and heterodimerization of the ZAD domains in vivo. (A) Co-immunoprecipitation of ZAD domains fused with 3×FLAG or 3×HA and expressed in S2 cells. Protein extracts from Drosophila S2 cells cotransfected with 3×FLAG- and 3×HA-fused plasmids were immunoprecipitated with antibodies against 3×FLAG or 3×HA (using nonspecific IgG as a negative control), and the immunoprecipitates (IP) were analyzed by Western blotting for the presence of HA- or FLAG-tagged proteins. ‘Input’ refers to samples of the initial protein extract; ‘output,’ to the supernatant after the removal of immunoprecipitate (IP). Nonspecific IP is indicated with an asterisk. (B) Testing Pita, Zw5 and ZIPIC for interaction in the yeast two-hybrid assay. Fragments of Pita, ZIPIC and Zw5 corresponding to the ZAD domains were fused to the GAL4 DNA-binding domain at the N- and C-termini and tested for interaction with Pita, ZIPIC and Zw5 (full-size or only the ZAD domain) fused to the GAL4 activating domain. All fragments were tested for the absence of interaction with the GAL4 activating and GAL4 DNA-binding domains alone. The results are summarized in columns where the plus and minus signs indicate a strong interaction or the absence of interaction, respectively.
Finally, we tested the ZAD domains in the yeast two-hybrid system (Figure 3B). The sequences encoding the ZAD domains were fused in frame to the yeast GAL4 DNA-binding domains (BD) and activation domains (AD). Since steric hindrance can interfere with transcriptional activation in the two-hybrid system, the ZAD domains were placed at both the N-terminus (ZAD-AD and ZAD-BD) and the C-terminus (AD-ZAD and BD-ZAD) of the fusion protein. The results provided additional evidence that the ZAD domains of the test proteins were mainly homodimerized: in all experimental variants, the interaction was observed only when the same ZAD domain was fused to the DNA-binding and activation domains of GAL4.
Pita and ZIPIC are able to support long-range interactions in Drosophila transgenic lines
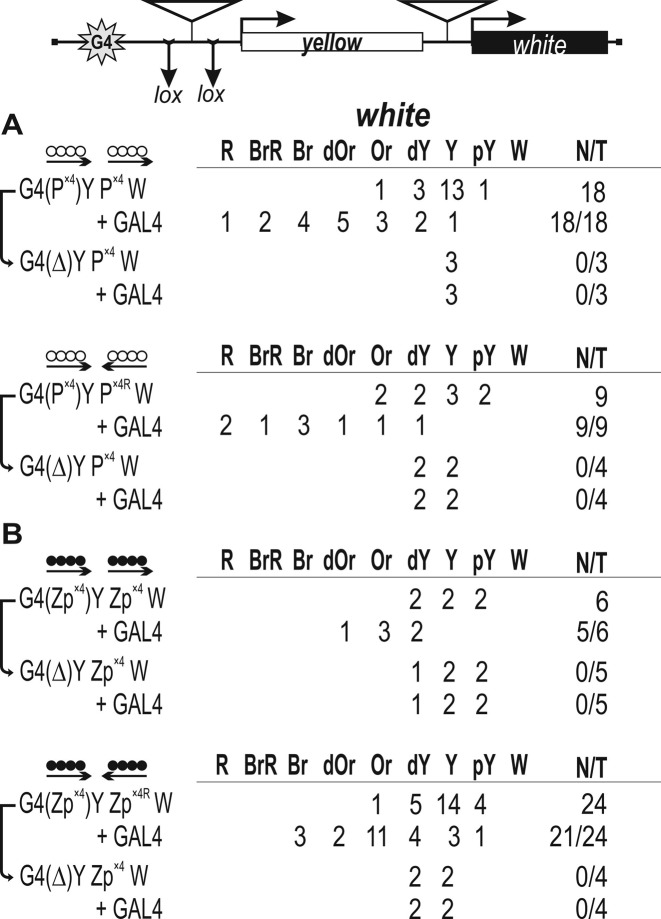
To find out whether the binding regions of ZAD-containing proteins can support long-range interactions, we used the GAL4/white assay (Supplementary Figure S5), which is based on the finding that the yeast GAL4 activator bound to sites located upstream of the yellow gene fails to stimulate the white promoter placed downstream of the yellow 3′ end (26). In the test constructs (Figure 4), ten GAL4 binding sites (G4) were inserted at –893 relative to the yellow transcription start site. As a result, the distance between the white gene and G4 was almost 5 kb. As shown previously (27), eight binding sites for Zw5 from the SCS insulator can efficiently support the long-range interaction in the GAL4/white assay.
Figure 4.
Testing for the functional interaction between DNA fragments containing binding sites for (A) Pita (white circles) or (B) ZIPIC (black circles) in the GAL4/white model system. At the top is a reductive scheme of transgenic construct used to examine the functional interaction between the binding sites for the test proteins. The GAL4 binding sites (indicated as G4) are at a distance of 5 kb from the white gene. The yellow gene is inserted between the GAL4 binding sites and the white promoter. ‘+GAL4’ indicates that eye phenotypes in transgenic lines were examined after induction of GAL4 expression. The yellow and white genes are shown as boxes, with arrows indicating the direction of their transcription. The inverted triangles indicate sites that used for insertion of tested DNA fragments. Downward arrows indicate target sites for Cre recombinase (lox). The same sites in construct names are denoted by parentheses. The superscript index ‘R’ indicates that the corresponding element is inserted in the reverse orientation in the construct. The ‘white’ column shows the numbers of transgenic lines with different levels of white expression. The wild-type white expression determined the bright red eye color (R); in the absence of white expression, the eyes were white (W). Intermediate levels of pigmentation, with the eye color ranging from pale yellow (pY) through yellow (Y), dark yellow (dY), orange (Or), dark orange (dOr) and brown (Br) to brownish red (BrR), reflect the increasing levels of white expression. N is the number of lines in which flies acquired a new white (w) phenotype upon deletion (Δ) of the specified DNA fragment; T is the total number of lines examined for each particular construct.
The question arose as to whether four binding sites for either Pita or ZIPIC could also support this interaction, taking into account our data (49) that Pita proved to be recruited to a DNA fragment containing four Pita binding sites arranged in the same orientation (P×4). The P×4 fragment flanked by LOX sites was inserted near the GAL4 sites. The second P×4 fragment was inserted in either direct (P×4) or reverse orientation (P×4R) upstream of the white promoter (Figure 4A). The white promoter usually accounts for the basal expression level, with eye pigmentation ranging from pale yellow to dark yellow (Supplementary Figure S6). To express the GAL4 protein, we used a transgenic line carrying the GAL4 gene under control of the ubiquitous tubulin promoter (26). These experiments showed that the pair of Pita binding sites provided for stimulation by GAL4 in both orientations relative to each other (Figure 4A). To demonstrate that white stimulation by GAL4 was supported by the Pita binding sites, we deleted the DNA fragments located near GAL4 binding sites by crossing with a line expressing the Cre recombinase. As a result, GAL4 lost the ability to stimulate white expression in all the lines tested. Thus, Pita binding sites can support long-range stimulation of the white promoter by GAL4 in an orientation-independent manner. Similar results were obtained with ZIPIC binding sites: they supported the long-range interaction regardless of their orientation relative to each other (Figure 4B).
We then tested whether Pita and ZIPIC binding sites could support the long-range interaction between GAL4 and the white promoter (Figure 5A). The P×4 fragment flanked by LOX sites was inserted near the GAL4 sites, and the ZIPIC fragment was inserted in either direct (Zp×4) or reverse orientation (Zp×4R) upstream of the white promoter. GAL4 failed to stimulate white transcription in any of the transgenic lines, showing that there was no effective hetero-interaction between Pita and ZIPIC. In similar tests for the interaction between binding sites for Zw5 and Pita (Figure 5B), these sites also failed to support white activation by GAL4. Thus, we observed no functional interactions between binding sites for different proteins.
Figure 5.
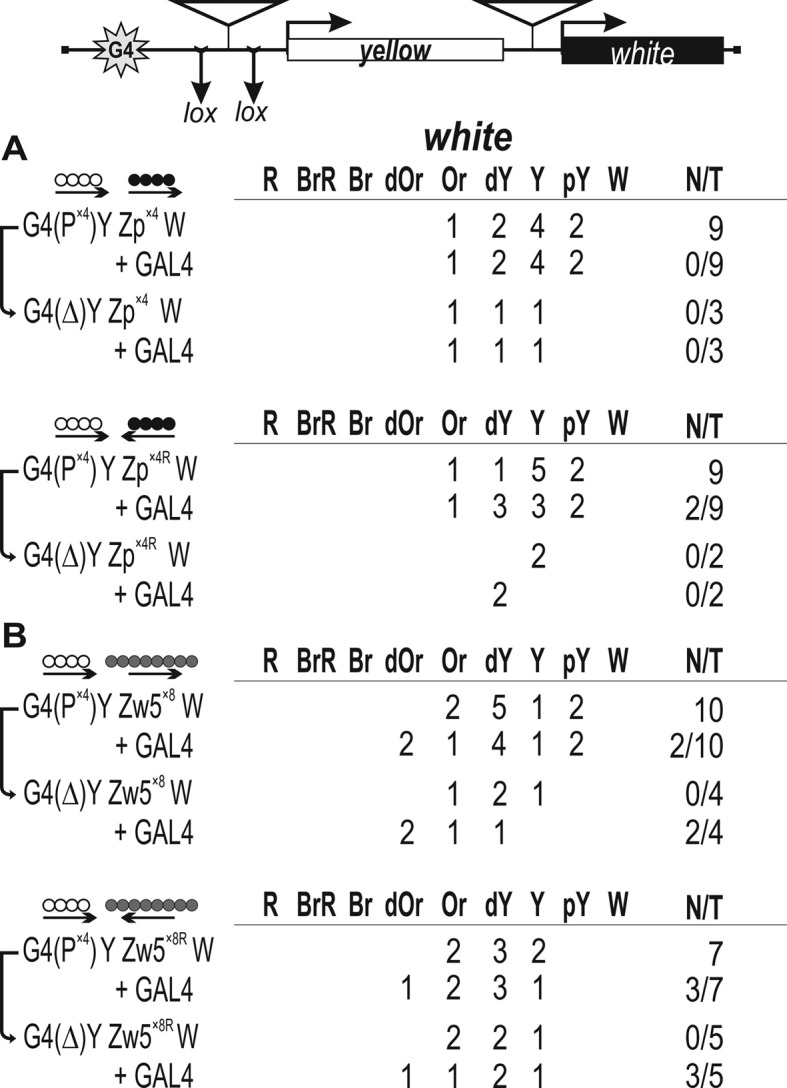
Testing for the functional interaction between DNA fragments containing (A) binding sites for Pita and ZIPIC or (B) binding sites for Pita and Zw5 (gray circles) in the GAL4/white model system. At the top is a reductive scheme of transgenic construct used to examine the functional interaction between the binding sites for the test proteins. In some transgenic lines, weak activation of the white reporter by GAL4 was observed. However, deletion of the binding sites inserted between lox sites in transgenic lines did not abolish GAL4-mediated activation, suggesting the absence of interaction between binding sites for different proteins. Other designations are as in Figure 4.
In addition, we performed an analysis for the functional interaction between binding sites for Pita, ZIPIC, and Zw5 using a phiC31-based integration system (73). All possible combinations between Pita, ZIPIC, and Zw5 were inserted in the same genomic region 86Fb (Supplementary Figure S7). A functional interaction was observed between binding sites for the same protein: Pita–Pita, ZIPIC–ZIPIC, and Zw5–Zw5; however, no white stimulation by GAL4 took place in variants with binding sites for different ZAD proteins, confirming the absence of interaction between them.
ZIPIC can support long-range stimulation of reporter gene by GAL4 in yeast
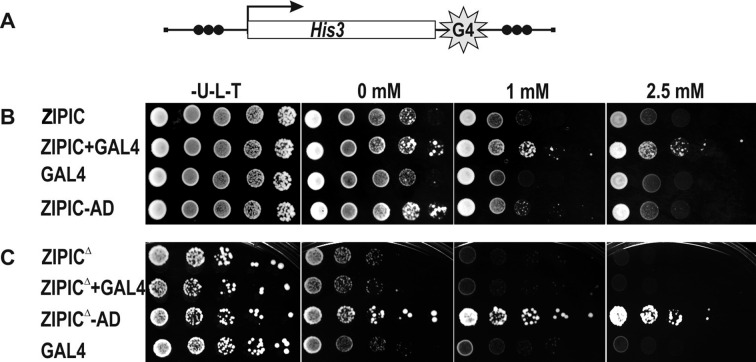
Next, we examined whether ZIPIC could support long-range interactions between regulatory elements in yeast, where such interactions are rare. For example, the GAL4 activator can stimulate yeast promoters only when its binding sites are located in relatively close proximity to the promoter, at a distance of no more than a few hundred base pairs (78–80). To test whether heterologous ZAD-containing protein can facilitate long-range interaction between GAL4 and a promoter, we used the his gene under control of its own promoter (Figure 6A). Ten GAL4 binding sites were inserted on the 3′ side of the his reporter, and a DNA fragment containing three ZIPIC binding sites was inserted upstream of the his promoter and downstream of the GAL4 sites. This construct was integrated in the yeast genome using homologous recombination. To demonstrate that ZIPIC can bind to its sites in yeast, we transformed the resulting yeast line with ZIPIC fused with the GAL4 binding domain. As a result, strong activation of the his promoter by GAL4 was observed, confirming the binding of ZIPIC to its sites (Figure 6B). Next, the yeast strain was transformed with expression vectors for either GAL4 and ZIPIC or GAL4 alone. The results showed that GAL4 failed to stimulate the promoter when the binding sites were located on the 3′ side of the gene. At the same time, coexpression of ZIPIC and GAL4 induced strong reporter activation. Hence, we conclude that GAL4 is recruited to the his promoter due to the interaction between ZIPIC proteins bound to the corresponding sites downstream and upstream of the his gene.
Figure 6.
ZIPIC mediates long-distance activation of a promoter by GAL4 in yeast. (A) Structure of the his gene reporter system to monitor ZIPIC activity in yeast. Ten GAL4 binding sites (G4) were inserted at the 3′ end of the his gene. The DNA fragments containing three ZIPIC sites (indicated as black circles) were inserted in reverse orientation upstream of the his promoter and downstream of the GAL4 binding sites. (B) Expression of ZIPIC promotes transcriptional activation at large distances. The reporter construct depicted in (A) was integrated into the genome. Expression of the his reporter was spot-assayed by measuring colony formation on plates containing the 3-amino-1,2,4-triazole (3-AT): the higher the concentration of 3-AT, the higher the level of HIS3 protein required for growth (97). (C) Mutant ZIPIC protein lacking the ZAD domain fails to support long-distance activation by GAL4.
To test for role of ZAD domain in long-range interactions, we prepared mutant ZIPIC devoid of this domain (ZIPICΔ) and transformed the yeast line with ZIPICΔ fused with the GAL4 activation domain (Figure 6C). As a result, activation of the reporter was observed, suggesting that ZIPICΔ bound to sites in the promoter region of the his gene. Coexpression of GAL4 and ZIPICΔ or expression of GAL4 alone did not stimulate the his promoter. Therefore, the ZAD domain is required for the ability of ZIPIC to facilitate distant GAL4 activation.
To demonstrate that the interaction between ZIPIC binding sites is required for long-range GAL4-mediated stimulation, we made a construct that contained ZIPIC binding sites only near the his promoter (Supplementary Figure S8). In this case, coexpression of GAL4 and ZIPIC did not stimulate the expression of the reporter gene. Taken together, these results suggest that ZIPIC can support long-range interactions in yeast and that the ZAD domain is required for this activity.
DISCUSSION
The zinc-finger associated domain (ZAD) family is the largest transcription factor family in dipteran insects, which consists of 91 members (47,48). This accounts for ∼10% of all Drosophila transcription factors. The ZAD domain is evolutionarily conserved within arthropod genomes and is represented by only one member in vertebrates (47,48). Most of ZAD-containing proteins in Drosophila are expressed in the female germline and embryos, suggesting that proteins containing these motifs play important roles in oocyte development and during embryogenesis (47,48,81). However, only a small part of ZAD transcription factors have been characterized with regard to their transcriptional regulatory activity in D. melanogaster. They include Grau (82,83), Sry-δ (84,85), Zw5 (39), Pita (86,87), Hang (88), Weckle (57), Pad (89), M1BP (90) and Trem (91).
The Pita, ZIPIC, and Zw5 proteins have been recently shown to have properties similar to those of the well-described insulator protein Su(Hw) (39,49). They are strongly expressed at all Drosophila life stages, especially in embryos, which is indicative of their key role in gene regulation. This conclusion is confirmed by the lethal phenotype of null mutations in the pita and zw5 genes (39,87). Pita, ZIPIC and Zw5 bind mainly to promoter regions near the TSS. Therefore, these proteins may be involved in the generation of an open chromatin structure in the promoter area and the recruitment of complexes regulating transcription. The Pita, ZIPIC and Zw5 binding is also enriched at the TAD boundaries and in inter-TAD regions. The proteins are extensively colocalized with subunits of cohesin and condensin complexes, as previously shown for mammalian architectural protein CTCF (92). On the basis of these facts, the Pita, ZIPIC and Zw5 proteins can be classified as architectural.
The ability to form homodimers has been shown for the ZAD domains of three proteins, Grau, Sry-δ and Wek (55–57). Here, similar results were obtained for three more proteins, suggesting that this ability is a common property of the ZAD domains. At the same time, the domains tested in this study did not form heterodimers. However, they had a low level of homology to each other, which leaves the possibility that ZAD domains with a relatively high level of homology are capable of heterodimerization.
The pairs of binding sites for ZAD proteins Pita, ZIPIC (examined here) and Zw5 (27) can support long-range interactions between the GAL4 activator and the white promoter in transgenic lines. Similar results were previously obtained for the Su(Hw) and CTCF insulator proteins (27,29,71). There are two lines of evidence for the critical role of the ZAD domain in organizing such interactions. First, ZIPIC can support long-range activation of the reporter gene by GAL4, with the ZAD domain being required for this activity. Since Drosophila insulator proteins have no homologs in the yeast genome, ZIPIC apparently facilitates this interaction in yeast without help of partners. The importance of ZAD domain for long-range interactions is also supported by the inability of different ZAD proteins to facilitate such an interaction in the model system. Interestingly, although no functional interaction is observed between binding sites for ZIPIC and Pita, both these proteins interact with CP190 (49), the protein that appears to be important for long-range interactions between insulator proteins (50–52). Based on these results we suggest that homodimerization of the ZAD domain is important for organizing the long-range interaction between binding sites for the corresponding protein.
Chromatin looping between different types of regulatory elements (promoters, enhancers, silencers, and insulators) is widely observed and appears to be a general mechanism for establishing long-range functional interactions in the genomes of higher eukaryotes (6,19,93–96). The results of studies on Drosophila insulator elements allow us to conclude that all tested Drosophila insulators contain binding sites for more than one insulator protein that can be involved in specific long-range interactions (11,27,29,49,71).
It has been suggested that regulatory elements (insulator, promoter, enhancer) containing similar combinations of binding sites for insulator/architectural proteins can be involved in supporting specific long-range interactions in chromatin. It seems likely that architectural proteins can contribute to the initial organization of specific interactions between remote regulatory elements, while cohesin and condensin complexes support such interaction during the cell cycle. The large family of ZAD transcription factors that includes 91 members (47,48,53) may have a key role in organization of Drosophila chromatin architecture. Further studies are required to understand the functional role and properties of this poorly studied group of transcription factors.
Supplementary Material
Acknowledgments
We are grateful to V.J. Makeev for supervising of bioinformatics analysis and N.A. Gorgolyuk for his help in preparing the manuscript and to Farhod Hasanov and Aleksander Parshikov for fly injection. This study was performed using the equipment of the IGB RAS facilities supported by the Ministry of Science and Education of the Russian Federation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Science Foundation [14-24-00166 to P.G.]. Funding for open access charge: Russian Science Foundation [14-24-00166].
Conflict of interest statement. None declared.
REFERENCES
- 1.Gibcus J.H., Dekker J. The hierarchy of the 3D genome. Mol. Cell. 2013;49:773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Laat W., Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- 3.Sexton T., Cavalli G. The role of chromosome domains in shaping the functional genome. Cell. 2015;160:1049–1059. doi: 10.1016/j.cell.2015.02.040. [DOI] [PubMed] [Google Scholar]
- 4.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghavi-Helm Y., Klein F.A., Pakozdi T., Ciglar L., Noordermeer D., Huber W., Furlong E.E. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512:96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- 7.Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Vietri Rudan M., Barrington C., Henderson S., Ernst C., Odom D.T., Tanay A., Hadjur S. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulianov S.V., Khrameeva E.E., Gavrilov A.A., Flyamer I.M., Kos P., Mikhaleva E.A., Penin A.A., Logacheva M.D., Imakaev M.V., Chertovich A., et al. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res. 2016;26:70–84. doi: 10.1101/gr.196006.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maksimenko O., Golovnin A., Georgiev P. Enhancer-promoter communication is regulated by insulator pairing in a Drosophila model bigenic locus. Mol. Cell. Biol. 2008;28:5469–5477. doi: 10.1128/MCB.00461-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyrchanova O., Georgiev P. Chromatin insulators and long-distance interactions in Drosophila. FEBS Lett. 2014;588:8–14. doi: 10.1016/j.febslet.2013.10.039. [DOI] [PubMed] [Google Scholar]
- 12.Bouwman B.A., de Laat W. Architectural hallmarks of the pluripotent genome. FEBS Lett. 2015;589:2905–2913. doi: 10.1016/j.febslet.2015.04.055. [DOI] [PubMed] [Google Scholar]
- 13.Raab J.R., Kamakaka R.T. Insulators and promoters: closer than we think. Nat. Rev. Genet. 2010;11:439–446. doi: 10.1038/nrg2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghirlando R., Giles K., Gowher H., Xiao T., Xu Z., Yao H., Felsenfeld G. Chromatin domains, insulators, and the regulation of gene expression. Biochim. Biophys. Acta. 2012;1819:644–651. doi: 10.1016/j.bbagrm.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herold M., Bartkuhn M., Renkawitz R. CTCF: insights into insulator function during development. Development. 2012;139:1045–1057. doi: 10.1242/dev.065268. [DOI] [PubMed] [Google Scholar]
- 16.Matzat L.H., Lei E.P. Surviving an identity crisis: A revised view of chromatin insulators in the genomics era. Biochim. Biophys. Acta. 2013;1839:203–214. doi: 10.1016/j.bbagrm.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chetverina D., Aoki T., Erokhin M., Georgiev P., Schedl P. Making connections: Insulators organize eukaryotic chromosomes into independent cis-regulatory networks. BioEssays. 2014;36:163–172. doi: 10.1002/bies.201300125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maksimenko O., Georgiev P. Mechanisms and proteins involved in long-distance interactions. Front. Genet. 2014;5:28. doi: 10.3389/fgene.2014.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Bortle K., Nichols M.H., Li L., Ong C.T., Takenaka N., Qin Z.S., Corces V.G. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014;15:R82. doi: 10.1186/gb-2014-15-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey S.D., Zhang X., Desai K., Aid M., Corradin O., Cowper-Sal Lari R., Akhtar-Zaidi B., Scacheri P.C., Haibe-Kains B., Lupien M. ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters. Nat. Commun. 2015;2:6186. doi: 10.1038/ncomms7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Negre N., Brown C.D., Shah P.K., Kheradpour P., Morrison C.A., Henikoff J.G., Feng X., Ahmad K., Russell S., White R.A., et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Negre N., Brown C.D., Ma L., Bristow C.A., Miller S.W., Wagner U., Kheradpour P., Eaton M.L., Loriaux P., Sealfon R., et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai H.N., Shen P. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science. 2001;291:493–495. doi: 10.1126/science.291.5503.493. [DOI] [PubMed] [Google Scholar]
- 25.Muravyova E., Golovnin A., Gracheva E., Parshikov A., Belenkaya T., Pirrotta V., Georgiev P. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science. 2001;291:495–498. doi: 10.1126/science.291.5503.495. [DOI] [PubMed] [Google Scholar]
- 26.Kyrchanova O., Toshchakov S., Parshikov A., Georgiev P. Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: effects of insulator pairing on enhancer-promoter communication. Mol. Cell. Biol. 2007;27:3035–3043. doi: 10.1128/MCB.02203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kyrchanova O., Chetverina D., Maksimenko O., Kullyev A., Georgiev P. Orientation-dependent interaction between Drosophila insulators is a property of this class of regulatory elements. Nucleic Acids Res. 2008;36:7019–7028. doi: 10.1093/nar/gkn781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyrchanova O., Toshchakov S., Podstreshnaya Y., Parshikov A., Georgiev P. Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the Drosophila Abd-B gene. Mol. Cell. Biol. 2008;28:4188–4195. doi: 10.1128/MCB.00229-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kyrchanova O., Maksimenko O., Stakhov V., Ivlieva T., Parshikov A., Studitsky V.M., Georgiev P. Effective blocking of the white enhancer requires cooperation between two main mechanisms suggested for the insulator function. PLoS Genet. 2013;9:e1003606. doi: 10.1371/journal.pgen.1003606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sigrist C.J., Pirrotta V. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics. 1997;147:209–221. doi: 10.1093/genetics/147.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller M., Hagstrom K., Gyurkovics H., Pirrotta V., Schedl P. The mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics. 1999;153:1333–1356. doi: 10.1093/genetics/153.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kravchenko E., Savitskaya E., Kravchuk O., Parshikov A., Georgiev P., Savitsky M. Pairing between gypsy insulators facilitates the enhancer action in trans throughout the Drosophila genome. Mol. Cell. Biol. 2005;25:9283–9291. doi: 10.1128/MCB.25.21.9283-9291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vazquez J., Muller M., Pirrotta V., Sedat J.W. The Mcp element mediates stable long-range chromosome-chromosome interactions in Drosophila. Mol. Biol. Cell. 2006;17:2158–2165. doi: 10.1091/mbc.E06-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H.B., Muller M., Bahechar I.A., Kyrchanova O., Ohno K., Georgiev P., Pirrotta V. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol. Cell. Biol. 2011;31:616–625. doi: 10.1128/MCB.00849-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujioka M., Wu X., Jaynes J.B. A chromatin insulator mediates transgene homing and very long-range enhancer-promoter communication. Development. 2009;136:3077–3087. doi: 10.1242/dev.036467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujioka M., Sun G., Jaynes J.B. The Drosophila eve insulator Homie promotes eve expression and protects the adjacent gene from repression by polycomb spreading. PLoS Genet. 2013;9:e1003883. doi: 10.1371/journal.pgen.1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kellum R., Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kellum R., Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 39.Gaszner M., Vazquez J., Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 1999;13:2098–2107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blanton J., Gaszner M., Schedl P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 2003;17:664–675. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nowick K., Stubbs L. Lineage-specific transcription factors and the evolution of gene regulatory networks. Brief. Funct. Genomics. 2010;9:65–78. doi: 10.1093/bfgp/elp056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Razin S.V., Borunova V.V., Maksimenko O.G., Kantidze O.L. Cys2His2 zinc finger protein family: classification, functions, and major members. Biochem. Biokhimiia. 2012;77:217–226. doi: 10.1134/S0006297912030017. [DOI] [PubMed] [Google Scholar]
- 43.Najafabadi H.S., Mnaimneh S., Schmitges F.W., Garton M., Lam K.N., Yang A., Albu M., Weirauch M.T., Radovani E., Kim P.M., et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 2015;33:555–562. doi: 10.1038/nbt.3128. [DOI] [PubMed] [Google Scholar]
- 44.Iuchi S. Three classes of C2H2 zinc finger proteins. Cell. Mol. Life Sci: CMLS. 2001;58:625–635. doi: 10.1007/PL00000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta A., Christensen R.G., Bell H.A., Goodwin M., Patel R.Y., Pandey M., Enuameh M.S., Rayla A.L., Zhu C., Thibodeau-Beganny S., et al. An improved predictive recognition model for Cys(2)-His(2) zinc finger proteins. Nucleic Acids Res. 2014;42:4800–4812. doi: 10.1093/nar/gku132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Persikov A.V., Wetzel J.L., Rowland E.F., Oakes B.L., Xu D.J., Singh M., Noyes M.B. A systematic survey of the Cys2His2 zinc finger DNA-binding landscape. Nucleic Acids Res. 2015;43:1965–1984. doi: 10.1093/nar/gku1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chung H.R., Lohr U., Jackle H. Lineage-specific expansion of the zinc finger associated domain ZAD. Mol. Biol. Evol. 2007;24:1934–1943. doi: 10.1093/molbev/msm121. [DOI] [PubMed] [Google Scholar]
- 48.Chung H.R., Schafer U., Jackle H., Bohm S. Genomic expansion and clustering of ZAD-containing C2H2 zinc-finger genes in Drosophila. EMBO Rep. 2002;3:1158–1162. doi: 10.1093/embo-reports/kvf243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maksimenko O., Bartkuhn M., Stakhov V., Herold M., Zolotarev N., Jox T., Buxa M.K., Kirsch R., Bonchuk A., Fedotova A., et al. Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin. Genome Res. 2015;25:89–99. doi: 10.1101/gr.174169.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartkuhn M., Straub T., Herold M., Herrmann M., Rathke C., Saumweber H., Gilfillan G.D., Becker P.B., Renkawitz R. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009;28:877–888. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahanger S.H., Shouche Y.S., Mishra R.K. Functional sub-division of the Drosophila genome via chromatin looping: the emerging importance of CP190. Nucleus. 2013;4:115–122. doi: 10.4161/nucl.23389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vogelmann J., Le Gall A., Dejardin S., Allemand F., Gamot A., Labesse G., Cuvier O., Negre N., Cohen-Gonsaud M., Margeat E., et al. Chromatin insulator factors involved in long-range DNA interactions and their role in the folding of the Drosophila genome. PLoS Genet. 2014;10:e1004544. doi: 10.1371/journal.pgen.1004544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lespinet O., Wolf Y.I., Koonin E.V., Aravind L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002;12:1048–1059. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krystel J., Ayyanathan K. Global analysis of target genes of 21 members of the ZAD transcription factor family in Drosophila melanogaster. Gene. 2013;512:373–382. doi: 10.1016/j.gene.2012.09.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jauch R., Bourenkov G.P., Chung H.R., Urlaub H., Reidt U., Jackle H., Wahl M.C. The zinc finger-associated domain of the Drosophila transcription factor grauzone is a novel zinc-coordinating protein-protein interaction module. Structure. 2003;11:1393–1402. doi: 10.1016/j.str.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 56.Payre F., Buono P., Vanzo N., Vincent A. Two types of zinc fingers are required for dimerization of the serendipity delta transcriptional activator. Mol. Cell. Biol. 1997;17:3137–3145. doi: 10.1128/mcb.17.6.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen L.Y., Wang J.C., Hyvert Y., Lin H.P., Perrimon N., Imler J.L., Hsu J.C. Weckle is a zinc finger adaptor of the toll pathway in dorsoventral patterning of the Drosophila embryo. Curr. Biol: CB. 2006;16:1183–1193. doi: 10.1016/j.cub.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 58.Ghavi-Helm Y., Furlong E.E. Analyzing transcription factor occupancy during embryo development using ChIP-seq. Methods Mol. Biol. 2012;786:229–245. doi: 10.1007/978-1-61779-292-2_14. [DOI] [PubMed] [Google Scholar]
- 59.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 60.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kulakovskiy I.V., Boeva V.A., Favorov A.V., Makeev V.J. Deep and wide digging for binding motifs in ChIP-Seq data. Bioinformatics. 2010;26:2622–2623. doi: 10.1093/bioinformatics/btq488. [DOI] [PubMed] [Google Scholar]
- 64.Kulakovskiy I., Levitsky V., Oshchepkov D., Bryzgalov L., Vorontsov I., Makeev V. From binding motifs in ChIP-Seq data to improved models of transcription factor binding sites. J. Bioinformatics Comput. Biol. 2013;11:1340004. doi: 10.1142/S0219720013400040. [DOI] [PubMed] [Google Scholar]
- 65.Vorontsov I.E., K I.V., Khimulya G., Nikolaeva D.D., Makeev V.J. PERFECTOS-APE – predicting regulatory functional effect of SNPs by approximate P-value estimation. Proceedings of the International Conference on Bioinformatics Models Methods and Algorithms. 2015:102–108. [Google Scholar]
- 66.Kulakovskiy I.V., Vorontsov I.E., Yevshin I.S., Soboleva A.V., Kasianov A.S., Ashoor H., Ba-Alawi W., Bajic V.B., Medvedeva Y.A., Kolpakov F.A., et al. HOCOMOCO: expansion and enhancement of the collection of transcription factor binding sites models. Nucleic Acids Res. 2016;44:D116–D125. doi: 10.1093/nar/gkv1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu L.J., Gazin C., Lawson N.D., Pages H., Lin S.M., Lapointe D.S., Green M.R. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Durinck S., Spellman P.T., Birney E., Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kholod N., Mustelin T. Novel vectors for co-expression of two proteins in E. coli. BioTechniques. 2001;31:322–328. doi: 10.2144/01312st03. [DOI] [PubMed] [Google Scholar]
- 71.Kyrchanova O., Ivlieva T., Toshchakov S., Parshikov A., Maksimenko O., Georgiev P. Selective interactions of boundaries with upstream region of Abd-B promoter in Drosophila bithorax complex and role of dCTCF in this process. Nucleic Acids Res. 2011;39:3042–3052. doi: 10.1093/nar/gkq1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karess R.E., Rubin G.M. Analysis of P transposable element functions in Drosophila. Cell. 1984;38:135–146. doi: 10.1016/0092-8674(84)90534-8. [DOI] [PubMed] [Google Scholar]
- 73.Bischof J., Maeda R.K., Hediger M., Karch F., Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schwartz Y.B., Linder-Basso D., Kharchenko P.V., Tolstorukov M.Y., Kim M., Li H.B., Gorchakov A.A., Minoda A., Shanower G., Alekseyenko A.A., et al. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 2012;22:2188–2198. doi: 10.1101/gr.138156.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Celniker S.E., Dillon L.A., Gerstein M.B., Gunsalus K.C., Henikoff S., Karpen G.H., Kellis M., Lai E.C., Lieb J.D., MacAlpine D.M., et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li L., Lyu X., Hou C., Takenaka N., Nguyen H.Q., Ong C.T., Cubenas-Potts C., Hu M., Lei E.P., Bosco G., et al. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol. Cell. 2015;58:216–231. doi: 10.1016/j.molcel.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schaaf C.A., Misulovin Z., Gause M., Koenig A., Gohara D.W., Watson A., Dorsett D. Cohesin and polycomb proteins functionally interact to control transcription at silenced and active genes. PLoS Genet. 2013;9:e1003560. doi: 10.1371/journal.pgen.1003560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guarente L., Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the ‘TATA box’. Proc. Natl. Acad. Sci. U.S.A. 1984;81:7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Struhl K. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc. Natl. Acad. Sci. U.S.A. 1984;81:7865–7869. doi: 10.1073/pnas.81.24.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Petrascheck M., Escher D., Mahmoudi T., Verrijzer C.P., Schaffner W., Barberis A. DNA looping induced by a transcriptional enhancer in vivo. Nucleic Acids Res. 2005;33:3743–3750. doi: 10.1093/nar/gki689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tomancak P., Berman B.P., Beaton A., Weiszmann R., Kwan E., Hartenstein V., Celniker S.E., Rubin G.M. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2007;8:R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Harms E., Chu T., Henrion G., Strickland S. The only function of Grauzone required for Drosophila oocyte meiosis is transcriptional activation of the cortex gene. Genetics. 2000;155:1831–1839. doi: 10.1093/genetics/155.4.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen B., Harms E., Chu T., Henrion G., Strickland S. Completion of meiosis in Drosophila oocytes requires transcriptional control by grauzone, a new zinc finger protein. Development. 2000;127:1243–1251. doi: 10.1242/dev.127.6.1243. [DOI] [PubMed] [Google Scholar]
- 84.Payre F., Crozatier M., Vincent A. Direct control of transcription of the Drosophila morphogen bicoid by the serendipity delta zinc finger protein, as revealed by in vivo analysis of a finger swap. Genes Dev. 1994;8:2718–2728. doi: 10.1101/gad.8.22.2718. [DOI] [PubMed] [Google Scholar]
- 85.Payre F., Noselli S., Lefrere V., Vincent A. The closely related Drosophila sry beta and sry delta zinc finger proteins show differential embryonic expression and distinct patterns of binding sites on polytene chromosomes. Development. 1990;110:141–149. doi: 10.1242/dev.110.1.141. [DOI] [PubMed] [Google Scholar]
- 86.Laundrie B., Peterson J.S., Baum J.S., Chang J.C., Fileppo D., Thompson S.R., McCall K. Germline cell death is inhibited by P-element insertions disrupting the dcp-1/pita nested gene pair in Drosophila. Genetics. 2003;165:1881–1888. doi: 10.1093/genetics/165.4.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Page A.R., Kovacs A., Deak P., Torok T., Kiss I., Dario P., Bastos C., Batista P., Gomes R., Ohkura H., et al. Spotted-dick, a zinc-finger protein of Drosophila required for expression of Orc4 and S phase. EMBO J. 2005;24:4304–4315. doi: 10.1038/sj.emboj.7600890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Scholz H., Franz M., Heberlein U. The hangover gene defines a stress pathway required for ethanol tolerance development. Nature. 2005;436:845–847. doi: 10.1038/nature03864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gibert J.M., Marcellini S., David J.R., Schlotterer C., Simpson P. A major bristle QTL from a selected population of Drosophila uncovers the zinc-finger transcription factor poils-au-dos, a repressor of achaete-scute. Dev. Biol. 2005;288:194–205. doi: 10.1016/j.ydbio.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 90.Li J., Gilmour D.S. Distinct mechanisms of transcriptional pausing orchestrated by GAGA factor and M1BP, a novel transcription factor. EMBO J. 2013;32:1829–1841. doi: 10.1038/emboj.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lake C.M., Nielsen R.J., Hawley R.S. The Drosophila zinc finger protein trade embargo is required for double strand break formation in meiosis. PLoS Genet. 2011;7:e1002005. doi: 10.1371/journal.pgen.1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ali T., Renkawitz R., Bartkuhn M. Insulators and domains of gene expression. Curr. Opin. Genet. Dev. 2016;37:17–26. doi: 10.1016/j.gde.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 93.Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S., et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heidari N., Phanstiel D.H., He C., Grubert F., Jahanbani F., Kasowski M., Zhang M.Q., Snyder M.P. Genome-wide map of regulatory interactions in the human genome. Genome Res. 2014;24:1905–1917. doi: 10.1101/gr.176586.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bothma J.P., Garcia H.G., Ng S., Perry M.W., Gregor T., Levine M. Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. eLife. 2015;4 doi: 10.7554/eLife.07956. doi:10.7554/eLife.07956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ing-Simmons E., Seitan V.C., Faure A.J., Flicek P., Carroll T., Dekker J., Fisher A.G., Lenhard B., Merkenschlager M. Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Res. 2015;25:504–513. doi: 10.1101/gr.184986.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Alexandre C., Grueneberg D. A., Gilman M. Z. Studying heterologous transcription factors in yeast. Methods Companion Methods Enzymol. 1993;5:147–155. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.