Abstract
The intolerance of DNA polymerase δ (Polδ) to incorrect base pairing contributes to its extremely high accuracy during replication, but is believed to inhibit translesion synthesis (TLS). However, chicken DT40 cells lacking the POLD3 subunit of Polδ are deficient in TLS. Previous genetic and biochemical analysis showed that POLD3 may promote lesion bypass by Polδ itself independently of the translesion polymerase Polζ of which POLD3 is also a subunit. To test this hypothesis, we have inactivated Polδ proofreading in pold3 cells. This significantly restored TLS in pold3 mutants, enhancing dA incorporation opposite abasic sites. Purified proofreading-deficient human Polδ holoenzyme performs TLS of abasic sites in vitro much more efficiently than the wild type enzyme, with over 90% of TLS events resulting in dA incorporation. Furthermore, proofreading deficiency enhances the capability of Polδ to continue DNA synthesis over UV lesions both in vivo and in vitro. These data support Polδ contributing to TLS in vivo and suggest that the mutagenesis resulting from loss of Polδ proofreading activity may in part be explained by enhanced lesion bypass.
INTRODUCTION
Polδ synthesizes DNA with remarkably high fidelity making only a single error per 106 nucleotides synthesised in vivo (1). It achieves this accuracy in two ways. In common with other replicative polymerases, it is able to discriminate very accurately between correct and incorrect base pairs at the polymerase active site (2,3). Second, incorrect nucleotides can be removed by the proofreading nuclease domain of Polδ further increasing overall accuracy by 10- to 60-fold (4). The enzymatic properties of its active site also inhibit Polδ from bypassing many DNA lesions in vitro (5). In addition, effective TLS by Polδ will be countered by elimination of nucleotides inserted opposite the damaged base by the proofreading exonuclease activity of the enzyme.
Thus, a prevalent model is that Polδ and Polϵ are unable to bypass DNA lesions, leading to arrest of DNA synthesis with the resulting replication blocks being released by specialized translesion DNA polymerases, such as Polη and Polζ, enzymes that have less spatially constrained active sites and that can thus accommodate the distorted base pairing created by damaged bases (6). While these characteristics allow TLS polymerases to bypass lesions, when coupled with the enzymes’ lack of proofreading activity, their deployment results in a reduction in fidelity of several orders of magnitude compared with Polδ and Polϵ (reviewed in (1)).
Polδ consists of four subunits: POLD1/p125, POLD2/p50, POLD3/p66, and POLD4/p12 (7). Although the Polδ holoenzyme is capable of bypass of some lesions in vitro (5,8–11), direct evidence for participation of Polδ in TLS in vivo is lacking. The POLD1 subunit contains both the DNA polymerase and 3′ to 5′ proofreading exonuclease domains. Genetic and biochemical studies in budding yeast have indicated that POLD3, a subunit that is not essential for cellular proliferation (12), contributes to TLS as an integral component of Polζ (13–16). POLD3 is also a subunit of both Polδ and Polζ in mammalian cells and it has been proposed that it contributes to TLS through its interaction with Polζ (15–18). However, we recently showed that POLD3 contributes to TLS even in the absence of Polζ. POLD3−/− (pold3) chicken DT40 cells, but not cells lacking POLζ−/− (polζ), are deficient in maintenance of replication fork progression along a UV-damaged template, and exhibit an altered pattern of abasic site bypass in the immunoglobulin light chain gene (19). Further, we demonstrated that human POLD3 facilitates abasic site bypass by Polδ in vitro by promoting extension from the nucleotide inserted opposite the lesion (19).
We advanced a model suggesting that POLD3 may alter the balance between nucleotide incorporation and proofreading by Polδ, increasing the probability that it could complete TLS. A prediction of this idea is that introducing a proofreading mutation into Polδ should bypass the requirement for the POLD3 subunit and at least partially restore TLS to the pold3 cell line. In this study we test this hypothesis in vivo and in vitro. We show that inactivation of the proofreading activity of one allele of POLD1 does indeed restore TLS past UV damage and abasic sites in pold3 cells but not cells deficient in Polζ. Moreover, expression of proofreading-deficient POLD1 substantially changes the spectrum of mutagenesis arising from TLS past UV damage and abasic sites in POLD3+ cells. These observations provide direct evidence that Polδ makes a substantial contribution to TLS in vivo and suggests that at least some of the mutagenesis in the absence of the proofreading activity of Polδ, as observed for instance in a subset of cancers, is the result of more proficient lesion bypass by the enzyme.
MATERIALS AND METHODS
Cell lines
The generation of pold3, polζ and polη DT40 single and combination mutants has been described previously (19–22).
Knock-in of Pold1exo− mutation
A pold1exo-mutation knock-in construct was generated from the genomic sequence covering the POLD1 gene isolated from a genomic library. POLD1 genomic sequence was isolated from a genomic library by hybridization and a 2.6 kb PstI fragment containing exons10 and 11 cloned into pBlueScript SK. A conserved residue in exonuclease domain, 402Asp (encoded in exon 10) was mutated to Ala using following primers. 5′-CAGAACTTCGCCCTGCCCTAC-3′ and 5′-GTAGGGCAGGGCGAAGTTCTG-3′. This mutation has been previously shown to completely eliminate the exonuclease activity of Polδ in vitro (11). The mutation concurrently disrupts a recognition site for a restriction enzyme, TaqI. A HisD selection-marker gene flanked by loxP sequences was inserted into the NdeI site in intron 10 to generate a pold1exo-mutation knock-in construct. Wild type and pold3 cells were transfected with pold1exo−HisD. The 0.1 kb fragment of cDNA covering exon 10 was used as a probe for Southern blot analysis to screen gene-targeting events as previously described (23). The HisD selection-marker gene was removed by the transient expression of CRE recombinase. Knock-in of the mutation was confirmed by digestion of the RT-PCR products with TaqI. Efficiency of targeting was 31.2% (5/16) and all targeted clones carried the pold1exo-mutation.
Sensitivity of cells to genotoxic agents to evaluate DNA repair
Sensitivity of cells to MMS and H2O2 was measured as a fraction of living cells after proliferation in liquid culture for 48 h. For exposure of cells to MMS or H2O2, 1 × 106 cells were treated for 1 h at 39.5°C in 1 ml of PBS containing 1% FCS and MMS or complete medium containing H2O2. 1 × 104 cells were seeded into 24-well plates with 1 ml of medium per well. Plates were incubated at 39.5°C for 48 h. Cell survival was determined using the CellTiter-Glo (Promega). Briefly, 100 μl CellTiter-Glo solution was mixed with 100 μl of cell culture from each well in 96-well plate. After 5 min, luminescence was measured by Fluoroskan Ascent (Thermo).
AID overexpression by retrovirus infection and analysis of Ig Vλ diversification
AID overexpression by retrovirus infection was carried out as described previously (24,25). The efficiency of infection was about 70%, as assayed by GFP expression. Twenty four hours after retrovirus infection, limiting dilution was performed to isolate single colonies. Genomic DNA was extracted at 14 days after limiting dilution from at least three independent colonies. The rearranged Vλ segments were PCR amplified using primers 5′-CAGGAGCTCGCGGGGCCGTCACTGATTGCCG-3′, forward in the Vλ leader intron, and 5′-GCGCAAGCTTCCCCAGCCTGCCGCCAAGTCCAAG-3′, reverse in the JCλ intron. To minimize PCR-introduced mutation, a high-fidelity polymerase, Prime Star (Takara) was used for amplification. The PCR products were cloned into TOPO Zeroblunt vector (Invitrogen) and sequenced with the M13 forward (−20) primer. Sequence alignment with DNASIS-MAC v3.3 (HITACHI) allowed identification of changes from the consensus sequence of each clone. Mutations were classified as described previously (24,25).
Dynamic molecular combing and immunofluorescent detection
Asynchronously growing DT40 cells were sequentially labelled for 15 min with 25 μM IdU and for 15 min with 25 μM CIdU. UV treated cells were irradiated at 20 J/m2 just before the CldU treatment. At the end of the labelling period (30 min), cells were placed in ice cold 1× PBS (1 volume of cells for 2 volumes of 1× PBS) and centrifuged at 250 g for 5 min at 4°C, washed in ice-cold PBS, and resuspended in PBS to a final concentration of 1 × 106 cells/ml. Three microliters of the cell suspension was spotted onto clean glass Superfrost slides and lysed with 7 μl of 0.5% SDS in 200 mM Tris–HCl (pH 5.5) and 50 mM EDTA (5 min, at room temperature). Slides were tilted at 15° to horizontal, allowing the DNA to run slowly down the slide. Slides were then air dried and fixed in 3:1 methanol/acetic acid, and stored at 4°C before immunolabelling. IdU, CldU, DNA revelations and analysis were performed as described (26), with minor modifications: the DNA was denatured for 30 min in 2.5 N HCl, and CldU was detected using rat anti BrdU (ABD Serotec, Raleigh, NC, USA) at 1/750. A stretching factor of 2.6 for conversion from μm to kb was applied, as previously described for the method in (27). Slides were mounted in 10% 1× PBS and 90% glycerol, kept at −20°C and imaged using a Nikon C1-si confocal microscope.
PiggyBlock assay
To insert a SalI restriction enzyme site into the original PiggyBlock plasmid (28), we inserted duplex oligonucleotides made of 5′-AATTGGAAGACCCGTCGACCA-3′ and 5′-TATGGTCGACGGGTCTTCC-3′ into the MfeI/NdeI sites (piggyBlock-SalI). A 30-nucleotide oligonucleotide, CTCGTCAGCATC(TT)CATCATACAGTCAGTG carrying CPD on (TT), and a 16-nucleotide oligonucleotide, TCGAGCGACACTGGAT, was annealed with complementary 46-nucleotide oligonucleotide, AATTCACTGACTGTATGATGGCGATGCTGACGAGATCCAGTGTCGC. To make piggyBlock-op plasmid, CTCGTCAGCATC(TT)CATCATACAGTCAGTG and TCGAGCGACACTGGAT were annealed with AATTCACTGACTGTATGATG(TT)GATGCTGACGAGATCCAGTGTCGC. The resultant duplex fragment carrying a single CPD lesion was ligated with the piggyBlock-SalI plasmid digested with MfeI/SalI, and ligated plasmid was gel purified (Qiagen), as previously described (28). Ten ng of the ligated plasmid together with 1 μg of the transposase expression vector was transfected into DT40 cells using the Neon transfection system (Invitrogen, CA, USA) with settings, 1350 V, 10 m sec, and three pulses. Transfected cells were subjected to limiting dilution immediately after transfection. Puromycin was added at 30 h after transfection. Genomic DNAs from individual puromycin resistant clones were purified, and were PCR amplified using primers (ACTGATTTTGAACTATAACGACCGCGTGAG) and (ACTAGTGAGACGTGCTACTTCCATTTGTCA) to examine DNA sequences at the CPD lesion. If a single puromycin resistant clone contained two different sequences, we counted as two independent DNA synthesis events. We obtained sequences from xpa, polη/polζ/xpa and polη/polζ/xpa/pold1exo− cells, respectively. We analyzed them following the method described previously (28).
Protein purification and primer extension assays
The human Polδ holoenzyme, with N-terminal His-tagged p50, was expressed using a baculovirus vector (pBacPAK9, Clontech, Palo Alto, CA, USA) in insect cells (High Five, Life Technologies, Palo Alto, CA), as described previously (29). To inactivate proofreading exonuclease activity, p125 Asp 402 was replaced by Ala. For primer-extension analysis, DNA synthesis was carried out with 0.06 pmol 32P-labeled primer. For examining abasic site bypass, 17 mer primer (AGCTATGACCATGATTA) annealed with a 49-mer template oligo DNA (AGCTACCATGCCTGCACGAATXAAGCAATTCGTAATCATGGTCATAGCT), where X can be an abasic site were used. For examining CPD baypass, 16 mer primer (CACTGACTGTATGATG) annealed with 30 mer oligo DNA (CTCGTCAGCATC(TT)CATCATACAGTCAGTG), where (TT) can be CPD were used as illustrated in (11). The assay was carried out in a reaction mixture (5 μl) containing 30 mM HEPES–NaOH (pH 7.4), 7 mM MgCl2, 8 mM NaCl, 0.5 mM dithiothreitol, and 10 μM each dNTP in the presence of 15 nM of primer/template complex, 2 nM of Polδ and 50 nM of PCNA for 15 min at 37°C. At the end of the reaction, the products were denatured with formamide and loaded onto 15.6% polyacrylamide gels containing 7 M urea in TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA). After electrophoresis, radioactivity was measured with a Fuji Image analyzer, FLA2500 (Fujifilm, Tokyo, Japan).
RESULTS
Expression of proofreading-deficient Polδ rescues the DNA damage hypersensitivity of pold3 cells
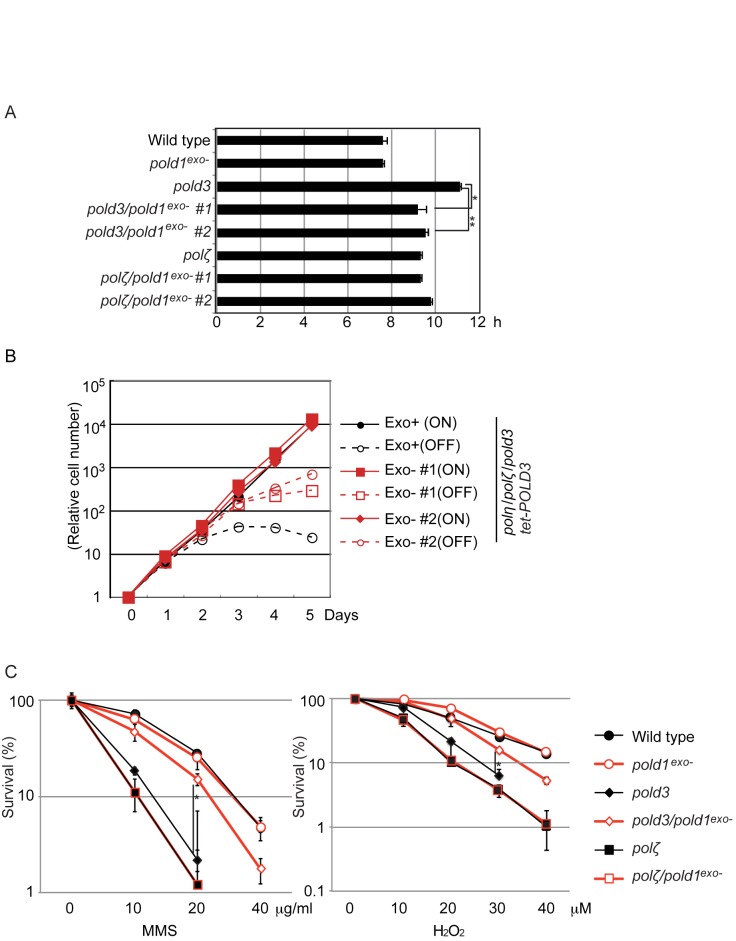
To test the hypothesis that expression of proofreading-defective Polδ would suppress the TLS defect of pold3 cells, we inserted a point mutation (D402A) into one of the two allelic POLD1 loci in wild type cells as well as in pold3 and polζ mutant DT40 cells. This generated pold1exo−, pold3/pold1exo− and polζ/pold1exo− cells (Supplementary Figure S1A–D). As expected, pold1exo− cells were viable and grew normally (Figure 1A). Interestingly, expression of proofreading deficient Polδ partially normalized the slow proliferation of pold3 cells, but not that of polζ cells (Figure 1A).
Figure 1.
pold1exo- mutation restores mutant phenotype of pold3 cells. (A) pold1exo- mutation significantly suppresses the growth defect of pold3 cells. The doubling time for the indicated genotypes is indicated. Error bars represent standard deviations (SD) from three independent assays. Statistical significance was determined by a Student's t-test and P-value was calculated. (*) P < 0.01, (**) P < 0.001. (B) Expression of POLD1exo− reverses the synthetic lethality of polη/polζ/pold3 cells. Growth curves of the indicated cells are shown after addition of doxycycline at time zero. The tet-POLD3 transcription was active without doxycyclin (ON) and inhibited upon addition of doxycyclin (OFF). (C) Expression of POLD1exo− reverses sensitivities of pold3 cells to MMS and H2O2. Indicated cells were exposed to MMS or H2O2. The dose of the genotoxic agent is displayed on the x-axis on a linear scale, while the percentage fraction of surviving cells is displayed on the y-axis on a logarithmic scale. Error bars show the SD for three independent assays. Statistical significance was determined by a Student's t-test and P-value was calculated. (*) P < 0.01.
We have previously shown that cells lacking both POLD3 and two major TLS polymerases, Polη and Polζ, are inviable (19). We inserted the pold1exo− mutation into polη/polζ/pold3 cells, in which viability was supported by expression of a POLD3 transgene under the control of the doxycycline-repressible promoter (tet-POLD3 transgene). The resulting polη/polζ/pold3/tet-POLD3 cells stopped proliferating on the third day after addition of doxycycline, as previously reported (19) (Figure 1B). However, the pold1exo− mutation significantly improved the viability of the triple mutant, polη/polζ/pold3 cells after addition of doxycycline, but not polη/polζ cells (Figure 1B).
We next asked whether expression of proofreading-deficient Polδ enhanced the tolerance of pold3 and polζ cells to DNA damaging agents. pold3/pold1exo− cells displayed significantly increased tolerance to the alkylating agent methyl methane sulfonate (MMS) and the oxidising agent H2O2, compared with pold3 cells (Figure 1C). In contrast, polζ and polζ/pold1exo− cells exhibited indistinguishable sensitivity to MMS and H2O2. Thus, Polδ proofreading significantly contributes to the DNA damage sensitivity of the pold3 mutant cells. The reversion of the mutant phenotype associated with pold3 but not polζ by the pold1exo− mutation suggests that the proofreading activity counteracts TLS by Polδ but not TLS by Polζ.
Expression of proofreading-deficient Polδ affects TLS past abasic sites in Ig V gene
To test the role of Polδ in TLS in vivo, we examined the diversification of the immunoglobulin (Ig) Vλ region in the DT40 B cell line during in vitro passage. DT40 cells constitutively diversify their Ig VJλ segment through two mechanisms, TLS dependent hypermutation and gene conversion from upstream pseudo-Vλ segments (30,31). Hypermutation at C/G basepairs in this locus is caused by TLS across abasic sites (32,33). Thus, the nucleotide sequence analysis of Ig V diversification during clonal expansion of cells provides the opportunity for measuring the rate of TLS as well as identifying the nucleotides inserted opposite to abasic sites (20).
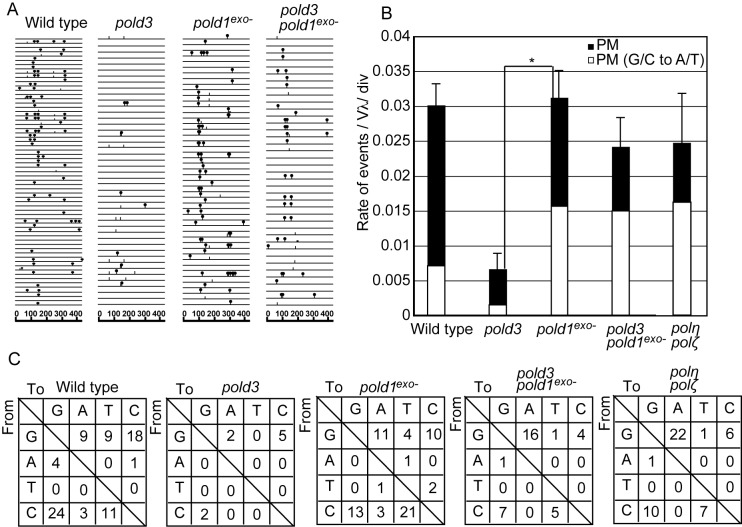
We overexpressed AID to enhance Ig V diversification. The resultant AID overexpressing cells were subcloned and cultured for two weeks. Then we subjected PCR-amplified VJλ segment to nucleotide sequence analysis (Figure 2A). pold3 cells exhibited a significant decrease in the rate of TLS dependent hypermutation as reported previously (19) (Figure 2B). Remarkably, TLS dependent hypermutation was restored in pold3/pold1exo− cells to a nearly wild-type level (Figure 2B). Thus, loss of proofreading exonuclease activity of Polδ bypasses requirement of POLD3 to execute TLS past abasic site (15,16). Interestingly, the restoration was associated with an increase in the proportion of G/C to A/T transitions (Chi-square test, P = 0.0050, Figure 2B and C). Polδ preferentially incorporates dA opposite abasic sites (A-rule (34)) while Rev1 preferentially incorporates dC (C-rule (35)). Importantly, the presence of a proofreading deficient allele of Polδ also increases the proportion of dA incorporation opposite C even when POLD3 is present (Chi-square test, P = 0.0068, Figure 2C and C). The reversal of the abasic site TLS defect of the pold3 mutant and marked bias towards G/C to A/T transitions (Figure 2B and C) induced by expression of proofreading-deficient Polδ supports the idea that Polδ can perform TLS past abasic sites in vivo and that this is facilitated by the POLD3 subunit.
Figure 2.
Expression of the proof reading exonuclease deficient Polδ substantially changes the mutation spectrum of the Ig Vλ hypermutation. (A) Ig Vλ segments isolated from indicated cells, clonally expanded for two weeks. Horizontal lines represent the rearranged Ig Vλ (450 bp), with hypermutation (lollipop shapes), single-nucleotide substitutions that could be the result of hypermutation or gene conversion (vertical bars). At least three cellular clones were expanded for two weeks and analyzed for each data set. (B) The rates of TLS-dependent hypermutation (PM) are indicated with standard error. White bars represent the rate of G/C to A/T mutations, TLS following A-rule. Data from polη/polζ cells are taken from (19) for comparison. Statistical significance was determined by a Fisher’s exact test and P-value was calculated. (*) P < 0.05. (C) Pattern of point mutation in wild type, pold3, pold1exo− and pold3/pold1exo- cells. Tables showing the pattern of mutation in each line. Some of data for wild type, pold3 and polη/polζ cells are from (19).
Expression of proofreading-deficient Polδ rescues the attenuated replication fork progression of pold3 cells after UV irradiation
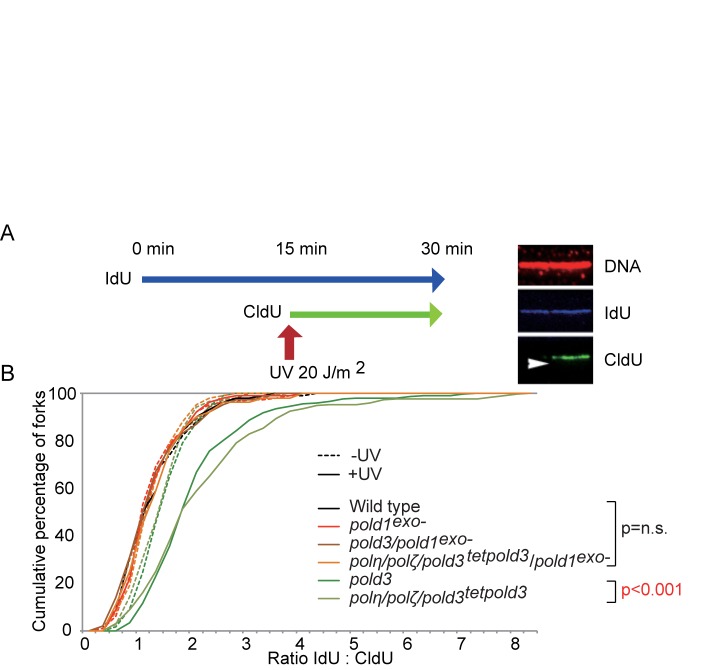
Previous work in DT40 cells has revealed temporally separated modes of lesion bypass. One mechanism is responsible for timely filling of postreplicative gaps at UV-damage sites, while another operates at or very close to stalled replication forks and maintains normal fork progression on UV-damaged DNA (36). pold3 cells are deficient only in the latter mode of damage bypass (19), which can be assessed by DNA molecular combing (Figure 3A). To examine replication fork progression after UV irradiation, we labeled nascent strands with IdU for 15 min, irradiated the cells with UV, and then continued labeling the nascent strands with CldU for a further 15 min (Figure 3A). After DNA combing, we detected the tracts of CldU and IdU with immunofluorescence and calculated the ratio between them to compare the total DNA synthesized before and after UV exposure on a fork-by-fork basis. Counterstaining the fibers for DNA allowed us to distinguish fork stalls from broken DNA. We plotted the data as a cumulative percentage of forks at each ratio (Figure 3B).
Figure 3.
Expression of POLD1exo− restores defective replication fork progression past UV damaged DNA in pold3 cells. (A) Schematic for DNA fiber labelling. DT40 cells were labeled sequentially with IdU and CldU with or without UV treatment after IdU labeling. The right hand panel shows an example of an ongoing fork. The arrowhead indicates the direction of replication. (B) The data for cells carrying the indicated genotypes was plotted as a cumulative percentage (y-axis) of forks at each ratio (x-axis). The transcription of tet-POLD3 was repressed by doxycycline for 1 day. The P-values of the Kolmogorov-Smirnov test for ratio distribution of each mutant for UV compared to wild type are indicated. n.s.: not significant. A part of data for pold3 and polη/polζ/pold3 cells were from (19).
pold3 and polη/polζ/pold3 cells exhibited a significant reduction in the DNA synthesized during labeling period after UV, as reported previously (19). Interestingly, inactivation of Polδ proofreading significantly restored the defective fork progression of pold3 cells after UV (Figure 3B), suggesting that the defective TLS of pold3 cells is suppressed by proofreading-deficiency of the Polδ catalytic subunit. Likewise, the inactivation of Polδ proofreading activity completely restored the UV-induced fork progression defect of polη/polζ/pold3 cells (Figure 3B), indicating that POLD3 is operating independently of Polη or Polζ in this context. We therefore conclude that Polδ proofreading-deficiency alleviates the in vivo TLS defect induced by loss of POLD3, which in turn does not depend on Polη or Polζ.
Expression of proofreading-deficient Polδ alters pattern of UV induced mutagenesis
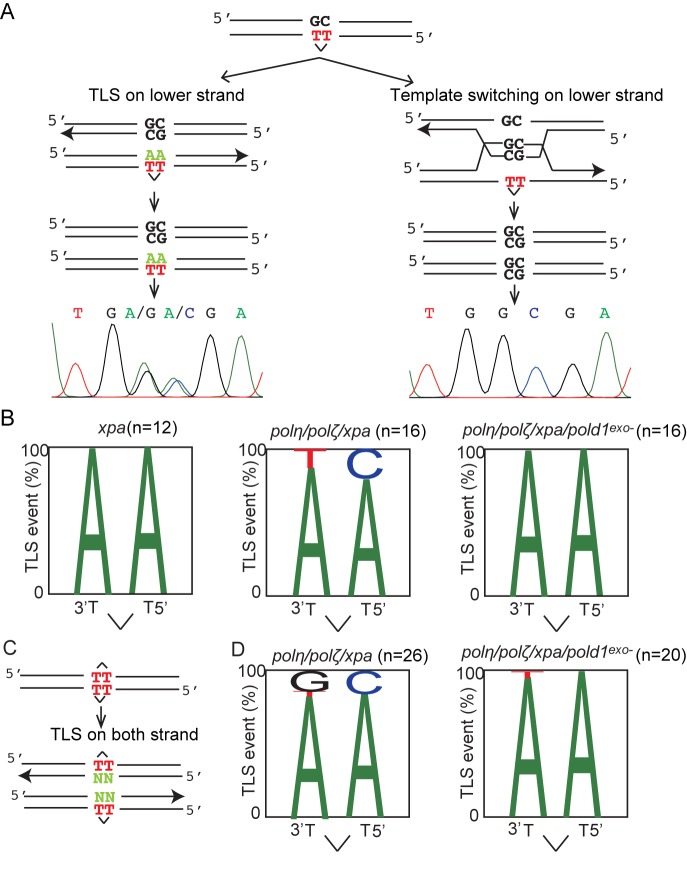
We next investigated whether expression of proofreading-deficient Polδ alters the pattern of TLS-induced mutagenesis at a UV damage (cyclobutane pyrimidine dimer (CPD)) integrated into chromosomal DNA. To this end, we inserted a CPD into the ‘piggyBlock’ transposon-based vector (28), transfected the CPD-carrying vector into the cells and picked individual clones having randomly integrated it (Figure 4A and Supplementary Figure S2). To avoid elimination of the integrated CPD by nucleotide excision repair and TLS by Polη and Polζ, we performed all experiments in polη/polζ/xpa background. We analyzed individual clones. These clones were mosaics as the cells within the clone inherit either the Watson or Crick strand of the parental integrant (Figure 4A and Supplementary Figure S2). Thus, in this assay, release of replication block at the CPD site by error-free template switching and by TLS can be distinguished as the CPD-containing TpT is placed opposite a GpC. Template switching would result in GpC at the CPD site, while TLS would insert ApA (accurate TLS) or other bases (inaccurate TLS) at the site (note, insertion of GpC opposite the T-T CPD would be unusual. (13,37)). Accordingly, TLS events are expected to give rise to a dual peak in the sequencing fluorogram (Figure 4A). Template switching, on the other hand, proceeds through the base opposite the lesion, and consequently, its signature consists of a single peak.
Figure 4.
Expression of POLD1exo− changes the mutation spectrum of TLS past CPD. (A) A CPD placed opposite GpC mismatch was randomly integrated into the genome using the PiggyBlock vector. TLS across the CPD results in a dual peak in the resulting cellular clone (left), while template switching results in a homogenous GC read (right). (B) The pattern of nucleotide incorporation opposite the CPD site. The percentage of each nucleotide incorporated at each position is indicated by the size of the letter of the nucleotide in the column. The total numbers of TLS events are shown. (C) A schematic representation of the opposed arrangement of CPD photoproducts in the piggyBlock plasmid (piggyBlock-op) and possible outcomes of DNA replication, only by TLS, over the lesion. (D) The pattern of nucleotide incorporation opposite the CPD in polη/polζ/xpa and polη/polζ/xpa/pold1exo− cells in piggyBlock-op plasmid.
Based on this principle, we determined the frequency of TLS relative to template switching (Supplementary Figure S3). We observed a decrease in the use of TLS from 35% to 5.8% following disruption of both POLη and POLζ in xpa cells. Further, the proportion of accurate TLS (i.e. incorporation of ApA) decreased from 100% in xpa cells to 85% in polη/polζ/xpa cells (Figure 4B). Thus, as expected, Polη and Polζ contribute significantly to accurate TLS past CPDs. Importantly, the expression of proofreading-deficient Polδ in polη/polζ/xpa increased the proportion of accurate TLS from 85% to 100% (Figure 4B). Thus, proofreading-deficient Polδ may contribute to the residual TLS past this UV lesion in cells lacking Polη and Polζ. To confirm this conclusion, we designed another piggyBlock vector, piggyBlock-op, in which the lesions are placed non-physiologically opposite each other, a configuration that forces bypass to be executed only by TLS (Figure 4C). Using this approach, we also observed an increase in the proportion of accurate TLS in polη/polζ/xpa/pold1exo− cells in comparison with polη/polζ/xpa cells (Figure 4D). These data support the idea that Polη and Polζ are the primary enzymes responsible for TLS past CPDs, but that Polδ can also contribute to bypass of this lesion, particularly in the absence of these canonical TLS enzymes.
Proofreading-deficiency causes a dramatic increase in the efficiency of TLS by purified human Polδ holoenzyme
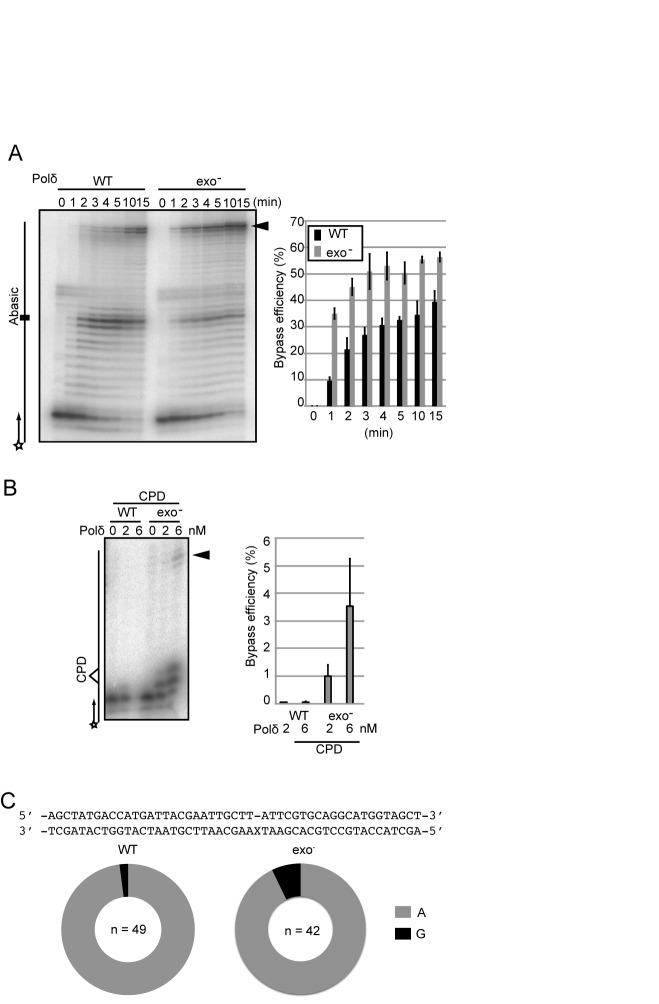
We next tested the capability of purified proofreading-deficient and proofreading-proficient Polδ to perform TLS. Using a physiological concentration (10 μM) of deoxynucleotides, both proofreading-deficient and proofreading-proficient Polδ exhibited comparable efficiency of DNA synthesis over intact template strands. We then analyzed TLS using three sets of primer and template strands, two containing an abasic site and the other one containing CPD (Supplementary Figure S4). We optimized the concentration of Polδ for the in vitro DNA synthesis analysis, and decided to use 2 and 6 nM (Supplementary Figure S5A). The efficiency of TLS was evaluated by measuring the amount of fully synthesized products as a function of time (Figure 5A, B; arrowhead). Proofreading-proficient Polδ generated more prominent bands corresponding to stalling at the abasic site, and one nucleotide before site than did the proofreading-deficient enzyme (Figure 5A and Supplementary Figure S5B), suggesting that proofreading-proficient Polδ repeats futile cycles of incorporation and proofreading. Consistent with our in vivo observations, proofreading-deficient Polδ performed TLS past an abasic site with a few times higher efficiency than proofreading-proficient Polδ (Figure 5A). To confirm which nucleotides are inserted opposite the abasic site, we purified the DNA synthesis products using a biotinylated primer, PCR amplified them and determined the nucleotide sequence. We did not detect any DNA slippage events in either case. The percentage of DNA synthesis products following the A-rule was 98% and 93% for proofreading-proficient and deficient Polδ, respectively (Figure 5C). This is consistent with the increase in the proportion of the A-rule mutations observed in pold1exo− cells in comparison with wild type cells (Figure 2B and C). Lastly, we measured TLS past a CPD site. We found that proofreading-deficient Polδ performed TLS with higher efficiency than proofreading-proficient Polδ (Figure 5B). These observations indicate that proofreading-deficiency significantly enhances the capacity for Polδ to perform TLS past abasic sites and CPDs in vitro.
Figure 5.
Inactivation of proofreading activity significantly increases the efficiency of Polδ (POLD3+) to perform TLS past abasic sites and UV damage. (A) DNA synthesis reactions were carried out with 2 nM of proofreading proficient (WT) or deficient (exo−) Polδ holoenzyme for the indicated duration. The histogram shows amounts of the fully extended product at the indicated time points. Error bars show the SD for three independent assays. (B) DNA synthesis reactions carried out with the indicated Polδ holoenzymes on template and primer strands, which are schematically shown on the left. Amount of the fully extended product was analyzed at 15 min. Error bars show the SD for three independent assays. (C) The pie charts indicate percentage of the nucleotides inserted opposite abasic site.
DISCUSSION
Testing the role of Polδ in TLS directly by inactivating the catalytic activity of the enzyme is not possible due to the essential role of Polδ in DNA replication. Several studies have shown that Polδ is capable of lesion bypass in vitro (5,8,10,11,38). However, it has remained unclear whether this replicative polymerase contributes significantly to translesion synthesis in vivo. Here we provide a number of lines of evidence that provide the strongest evidence to date that it does. First, the POLD1 proofreading deficiency is able to suppress the defect in translesion synthesis caused by loss of POLD3, but not Polζ (Figure 1C), which plays a critical role in completing TLS by extending DNA synthesis after other TLS polymerases have inserted nucleotides opposite damaged template bases (20,39). Thus, the data shown in Figure 1C indicate that when completion of TLS is inefficient due to the loss of Polζ, the POLD1 proofreading activity cannot substitute and rescue the damage sensitivity of polζ cells. While the proofreading activity of Polδ has been shown to be able to operate in trans for replication errors (40,41), these observations suggest that POLD1 proofreading acts preferentially on TLS nucleotide incorporation events mediated by POLD1 rather than by other TLS polymerases. Consistent with this, inactivation of POLD1 proofreading activity in vitro increases the efficiency of lesion bypass by the Polδ holoenzyme (Figure 5). Further, analysis of the pattern of abasic site bypass in the immunoglobulin light chain locus shows that inactivation of POLD1 proofreading significantly increases the frequency of dA incorporation consistent with POLD1 proofreading its own translesion synthesis nucleotide incorporations.
Given the ability of Polδ to perform error-prone TLS, an interesting question is whether Polδ can modulate its fidelity and proofreading activity when it encounters template damage. We have previously argued that POLD3 may facilitate the ability of Polδ to complete TLS by allowing extension from a base incorporated opposite a lesion (19). We suggested that in the absence of POLD3, Polδ may undergo futile cycles of incorporation and proofreading resulting in its stalling at a lesion. The observations we present here are consistent with this model, by showing that loss of proofreading facilitates completion of TLS by Polδ in vivo and in vitro. This ability of Polδ to alter its catalytic properties so as to carry out TLS, and therefore to act mutagenically, is at odds with its role as a replicative polymerase. It suggests that the catalytic site and proofreading need to be regulated when the enzyme encounters a lesion. How this ‘fidelity switch’ is regulated remains to be explored, but might be promoted by post-translational modification, for instance of POLD3 itself (42,43). Alternatively, given the proposed role of POLD3 as an anchor between POLD1 and PCNA (44), it may be due to a constrained ‘TLS mode’ interaction of the enzyme with the clamp. Another intriguing possibility is that the TLS ability of the replicative polymerases is regulated local modulation of dNTP concentration. TLS by replicative polymerases in eukaryotic cells has been invoked to explain the increased mutation frequency following exposure to 4-nitro-quinoline oxide (4-NQO) of S. cerevisiae in which all TLS polymerases had been deleted (45). DNA damage increases dNTP concentration and this increase promotes lesion bypass by Polϵ, which at normal physiological dNTP concentrations is unable to carry translesion synthesis (45). Since the proofreading exonuclease activity of replicative polymerases is also suppressed by elevated concentration of dNTP (19), it is possible that locally elevated dNTP concentrations in the vicinity of DNA damage allows the replicative polymerases to engage in damage bypass. Supporting of this view, inactivation of exonuclease activity of E.coli PolIII (Polδ homolog) enhances bypass replication (46,47).
Our data suggest that bypass by Polδ may be a relatively frequent event, at least at abasic sites and CPDs. Important questions for future studies concern the order in which Polδ and the canonical TLS polymerases are deployed in lesion bypass, and whether Polδ can contribute to TLS equally at leading and lagging strand obstacles. We previously showed that A-rule mutagenesis is also increased in polη/polζ cells in comparison with wild type cells, suggesting that Polδ serves as a backup for Polη–Polζ axis (20). Thus, if TLS polymerases fail to restore DNA replication, Polδ might attempt TLS as a last resort. Alternatively, since Polδ is likely to be the first enzyme to encounter a template strand lesion, TLS by Polδ opposite a weakly blocking lesion may well be the most pragmatic mechanism to ensure maintenance of processive replication. In this model, whether the classical TLS apparatus is deployed may depend on whether Polδ can complete the reaction in a reasonable time, or on other contextual cues surrounding the lesion (48). Thus, it will be interesting to determine whether Polδ is used for TLS on both leading and lagging strands, given its prominent role as the lagging strand replicase (49) and a possible interplay among yeast replicative DNA polymerases δ and ϵ (50).
In summary, to completely replicate the whole genomic DNA in a timely fashion, cells have evolved multiple mechanisms, including the firing of dormant replication origins, homologous recombination, and TLS polymerases. Our study provides an insight into a fourth mechanism, bypassing lesions directly with replicative DNA polymerases.
Supplementary Material
Acknowledgments
We acknowledge the Radioisotope Research Center in Kyoto University, Kyushu University and Tokyo Metropolitan University for support in the use of isotopes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
JSPS KAKENHI [25281021, 26116518 and 24114509 to K.H.]; JSPS KAKENHI [25650006 and 23221005 to S.T.; Medical Research Council [LMB, U105178808 to J.E.S.]; Flight Attendant Medical Research Institute, Florida, USA; Israel Science Foundation [684/12]; Mike and Valeria Rosenbloom through the Mike Rosenbloom Foundation (to Z.L.). Funding for open access charge: KAKENHI.
Conflict of interest statement. None declared.
REFERENCES
- 1.McCulloch S.D., Kunkel T.A. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doublie S., Tabor S., Long A.M., Richardson C.C., Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 3.Swan M.K., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat. Struct. Mol. Biol. 2009;16:979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fortune J.M., Pavlov Y.I., Welch C.M., Johansson E., Burgers P.M., Kunkel T.A. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 5.Schmitt M.W., Matsumoto Y., Loeb L.A. High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie. 2009;91:1163–1172. doi: 10.1016/j.biochi.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sale J.E. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb. Perspect. Biol. 2013;5:a012708. doi: 10.1101/cshperspect.a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Podust V.N., Chang L.S., Ott R., Dianov G.L., Fanning E. Reconstitution of human DNA polymerase delta using recombinant baculoviruses: the p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme. J. Biol. Chem. 2002;277:3894–3901. doi: 10.1074/jbc.M109684200. [DOI] [PubMed] [Google Scholar]
- 8.Choi J.Y., Lim S., Kim E.J., Jo A., Guengerich F.P. Translesion synthesis across abasic lesions by human B-family and Y-family DNA polymerases alpha, delta, eta, iota, kappa, and REV1. J. Mol. Biol. 2010;404:34–44. doi: 10.1016/j.jmb.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daube S.S., Tomer G., Livneh Z. Translesion replication by DNA polymerase delta depends on processivity accessory proteins and differs in specificity from DNA polymerase beta. Biochemistry. 2000;39:348–355. doi: 10.1021/bi9917784. [DOI] [PubMed] [Google Scholar]
- 10.Meng X., Zhou Y., Zhang S., Lee E.Y., Frick D.N., Lee M.Y. DNA damage alters DNA polymerase delta to a form that exhibits increased discrimination against modified template bases and mismatched primers. Nucleic Acids Res. 2009;37:647–657. doi: 10.1093/nar/gkn1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narita T., Tsurimoto T., Yamamoto J., Nishihara K., Ogawa K., Ohashi E., Evans T., Iwai S., Takeda S., Hirota K. Human replicative DNA polymerase delta can bypass T-T (6-4) ultraviolet photoproducts on template strands. Genes Cells. 2010;15:1228–1239. doi: 10.1111/j.1365-2443.2010.01457.x. [DOI] [PubMed] [Google Scholar]
- 12.Gerik K.J., Li X., Pautz A., Burgers P.M. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 1998;273:19747–19755. doi: 10.1074/jbc.273.31.19747. [DOI] [PubMed] [Google Scholar]
- 13.Gibbs P.E., McDonald J., Woodgate R., Lawrence C.W. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang M.E., de Calignon A., Nicolas A., Galibert F. POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase delta, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr. Genet. 2000;38:178–187. doi: 10.1007/s002940000149. [DOI] [PubMed] [Google Scholar]
- 15.Johnson R.E., Prakash L., Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase zeta. Proc. Natl. Acad. Sci. U.S.A. 2012;109:12455–12460. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarova A.V., Stodola J.L., Burgers P.M. A four-subunit DNA polymerase zeta complex containing Pol delta accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012;40:11618–11626. doi: 10.1093/nar/gks948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baranovskiy A.G., Lada A.G., Siebler H.M., Zhang Y., Pavlov Y.I., Tahirov T.H. DNA polymerase delta and zeta switch by sharing accessory subunits of DNA polymerase delta. J. Biol. Chem. 2012;287:17281–17287. doi: 10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Netz D.J., Stith C.M., Stumpfig M., Kopf G., Vogel D., Genau H.M., Stodola J.L., Lill R., Burgers P.M., Pierik A.J. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 2011;8:125–132. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirota K., Yoshikiyo K., Guikbaud G., Tsurimoto T., Murai J., Tsuda M., Phillips L., Narita T., Nishihara K., Kobayashi K., et al. The POLD3 subunit of DNA polymerase δ can promote translesion synthesis independently of DNA polymerase ζ. Nucleic Acids Res. 2015;43:1671–1683. doi: 10.1093/nar/gkv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirota K., Sonoda E., Kawamoto T., Motegi A., Masutani C., Hanaoka F., Szuts D., Iwai S., Sale J.E., Lehmann A., et al. Simultaneous disruption of two DNA polymerases, Poleta and Polzeta, in Avian DT40 cells unmasks the role of Poleta in cellular response to various DNA lesions. PLoS Genet. 2010;6:e1001151. doi: 10.1371/journal.pgen.1001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawamoto T., Araki K., Sonoda E., Yamashita Y.M., Harada K., Kikuchi K., Masutani C., Hanaoka F., Nozaki K., Hashimoto N., et al. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol. Cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 22.Sonoda E., Okada T., Zhao G.Y., Tateishi S., Araki K., Yamaizumi M., Yagi T., Verkaik N.S., van Gent D.C., Takata M., et al. Multiple roles of Rev3, the catalytic subunit of polzeta in maintaining genome stability in vertebrates. EMBO J. 2003;22:3188–3197. doi: 10.1093/emboj/cdg308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi K., Fujii T., Asada R., Ooka M., Hirota K. Development of a targeted flip-in system in avian DT40 cells. PLoS One. 2015;10:e0122006. doi: 10.1371/journal.pone.0122006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saberi A., Nakahara M., Sale J.E., Kikuchi K., Arakawa H., Buerstedde J.M., Yamamoto K., Takeda S., Sonoda E. The 9-1-1 DNA clamp is required for immunoglobulin gene conversion. Mol. Cell. Biol. 2008;28:6113–6122. doi: 10.1128/MCB.00156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shinkura R., Ito S., Begum N.A., Nagaoka H., Muramatsu M., Kinoshita K., Sakakibara Y., Hijikata H., Honjo T. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat. Immunol. 2004;5:707–712. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- 26.Guilbaud G., Rappailles A., Baker A., Chen C.L., Arneodo A., Goldar A., d'Aubenton-Carafa Y., Thermes C., Audit B., Hyrien O. Evidence for sequential and increasing activation of replication origins along replication timing gradients in the human genome. PLoS Comput. Biol. 2011;7:e1002322. doi: 10.1371/journal.pcbi.1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson D.A., Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen I.S., Bar C., Paz-Elizur T., Ainbinder E., Leopold K., de Wind N., Geacintov N., Livneh Z. DNA lesion identity drives choice of damage tolerance pathway in murine cell chromosomes. Nucleic Acids Res. 2015;43:1637–1645. doi: 10.1093/nar/gku1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shikata K., Ohta S., Yamada K., Obuse C., Yoshikawa H., Tsurimoto T. The human homologue of fission Yeast cdc27, p66, is a component of active human DNA polymerase delta. J. Biochem. 2001;129:699–708. doi: 10.1093/oxfordjournals.jbchem.a002909. [DOI] [PubMed] [Google Scholar]
- 30.Arakawa H., Moldovan G.L., Saribasak H., Saribasak N.N., Jentsch S., Buerstedde J.M. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 2006;4:e366. doi: 10.1371/journal.pbio.0040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Noia J., Neuberger M.S. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- 32.Ross A.L., Sale J.E. The catalytic activity of REV1 is employed during immunoglobulin gene diversification in DT40. Mol. Immunol. 2006;43:1587–1594. doi: 10.1016/j.molimm.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 33.Simpson L.J., Sale J.E. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 2003;22:1654–1664. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mozzherin D.J., Shibutani S., Tan C.K., Downey K.M., Fisher P.A. Proliferating cell nuclear antigen promotes DNA synthesis past template lesions by mammalian DNA polymerase delta. Proc. Natl. Acad. Sci. U.S.A. 1997;94:6126–6131. doi: 10.1073/pnas.94.12.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson J.R., Lawrence C.W., Hinkle D.C. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 36.Edmunds C.E., Simpson L.J., Sale J.E. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol. Cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 37.Szuts D., Marcus A.P., Himoto M., Iwai S., Sale J.E. REV1 restrains DNA polymerase zeta to ensure frame fidelity during translesion synthesis of UV photoproducts in vivo. Nucleic Acids Res. 2008;36:6767–6780. doi: 10.1093/nar/gkn651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acharya N., Johnson R.E., Pages V., Prakash L., Prakash S. Yeast Rev1 protein promotes complex formation of DNA polymerase zeta with Pol32 subunit of DNA polymerase delta. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9631–9636. doi: 10.1073/pnas.0902175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedberg E.C., Lehmann A.R., Fuchs R.P. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 40.Pavlov Y.I., Frahm C., Nick McElhinny S.A., Niimi A., Suzuki M., Kunkel T.A. Evidence that errors made by DNA polymerase alpha are corrected by DNA polymerase delta. Curr. Biol. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Flood C.L., Rodriguez G.P., Bao G., Shockley A.H., Kow Y.W., Crouse G.F. Replicative DNA polymerase delta but not epsilon proofreads errors in Cis and in Trans. PLoS Genet. 2015;11:e1005049. doi: 10.1371/journal.pgen.1005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bursomanno S., McGouran J.F., Kessler B.M., Hickson I.D., Liu Y. Regulation of SUMO2 Target Proteins by the Proteasome in Human Cells Exposed to Replication Stress. J. Proteome Res. 2015;14:1687–1699. doi: 10.1021/pr500997p. [DOI] [PubMed] [Google Scholar]
- 43.Liu G., Warbrick E. The p66 and p12 subunits of DNA polymerase delta are modified by ubiquitin and ubiquitin-like proteins. Biochem. Biophys. Res. Commun. 2006;349:360–366. doi: 10.1016/j.bbrc.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 44.Ducoux M., Urbach S., Baldacci G., Hubscher U., Koundrioukoff S., Christensen J., Hughes P. Mediation of proliferating cell nuclear antigen (PCNA)-dependent DNA replication through a conserved p21(Cip1)-like PCNA-binding motif present in the third subunit of human DNA polymerase delta. J. Biol. Chem. 2001;276:49258–49266. doi: 10.1074/jbc.M106990200. [DOI] [PubMed] [Google Scholar]
- 45.Sabouri N., Viberg J., Goyal D.K., Johansson E., Chabes A. Evidence for lesion bypass by yeast replicative DNA polymerases during DNA damage. Nucleic Acids Res. 2008;36:5660–5667. doi: 10.1093/nar/gkn555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fuchs R.P., Napolitano R.L. Inactivation of DNA proofreading obviates the need for SOS induction in frameshift mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13114–13119. doi: 10.1073/pnas.95.22.13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pages V., Janel-Bintz R., Fuchs R.P. Pol III proofreading activity prevents lesion bypass as evidenced by its molecular signature within E.coli cells. J. Mol. Biol. 2005;352:501–509. doi: 10.1016/j.jmb.2005.07.063. [DOI] [PubMed] [Google Scholar]
- 48.Sale J.E. Competition, collaboration and coordination–determining how cells bypass DNA damage. J. Cell Sci. 2012;125:1633–1643. doi: 10.1242/jcs.094748. [DOI] [PubMed] [Google Scholar]
- 49.Nick McElhinny S.A., Gordenin D.A., Stith C.M., Burgers P.M., Kunkel T.A. Division of labor at the eukaryotic replication fork. Mol. Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pavlov Y.I., Maki S., Maki H., Kunkel T.A. Evidence for interplay among yeast replicative DNA polymerases alpha, delta and epsilon from studies of exonuclease and polymerase active site mutations. BMC Biol. 2004;2:11. doi: 10.1186/1741-7007-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.