Abstract
Land plant organellar genomes encode a small number of genes, many of which are essential for respiration and photosynthesis. Organellar gene expression is characterized by a multitude of RNA processing events that lead to stable, translatable transcripts. RNA binding proteins (RBPs), have been shown to generate and protect transcript termini and eventually induce the accumulation of short RNA footprints. We applied knowledge of such RBP-derived footprints to develop software (sRNA miner) that enables identification of RBP footprints, or other clusters of small RNAs, in organelles. We used this tool to determine mitochondrial and chloroplast cosRNAs (clustered organellar sRNAs) in Arabidopsis. We found that in mitochondria, cosRNAs coincide with transcript 3′-ends, but are largely absent from 5′-ends. In chloroplasts this bias is absent, suggesting a different mode of 5′ processing, possibly owing to different sets of RNases. Furthermore, we identified a large number of cosRNAs that represent silenced insertions of mitochondrial DNA in the nuclear genome of Arabidopsis. Steady-state RNA analyses demonstrate that cosRNAs display differential accumulation during development. Finally, we demonstrate that the chloroplast RBP PPR10 associates in vivo with its cognate cosRNA. A hypothetical role of cosRNAs as competitors of mRNAs for PPR proteins is discussed.
INTRODUCTION
RNA metabolism in land plant mitochondria and chloroplasts depends on nuclear-encoded RNA binding proteins (RBPs). These RBPs are involved in RNA editing, splicing and post-transcriptional end processing of organellar RNAs. All of these steps are required for proper mitochondrial respiration and chloroplast photosynthesis (1,2).
The genomes of mitochondria and chloroplasts encode gene expression machinery components adjacent to genes encoding proteins for respiration and photosynthesis, respectively. Transcription initiation and termination in these organelles is relaxed, allowing the vast majority of their chromosomes to be transcribed. This includes genomic regions with no known function, including antisense strands of genes, which give rise to transcripts that are rapidly degraded in wild-type plants (3–5). Degradation in plant organelles is carried out by a number of nuclear-encoded RNases (6,7). A reduction in the level of such RNases, for example the exonucleases polynucleotide phosphorylase (PNPase) or RNase II (RNR1), leads to an accumulation of presumably non-functional RNAs (4). Moreover, many mRNAs are not correctly processed at their 3′-end in PNPase-deficient and RNR1-deficient plants (3,8–9). This suggests that organellar RNases are non-specific safeguards against the accumulation of aberrant, unwanted transcripts. How then do functional mRNAs and other stably accumulating RNA species escape degradation by organellar exonucleases?
Recent work on selected chloroplast RBPs suggests an emerging model for RNA stabilization in plant organelles (10). According to this model, helical-hairpin-repeat proteins bind tightly to specific sequences in untranslated regions (UTRs) of mRNAs and thus act as roadblocks against both 5′-to-3′ and 3′-to-5′ exonucleases (11). In cases where an RBP binds to an internal UTR of a polycistronic transcript, two different products of degradation are found: one corresponds to the upstream cistron, the other corresponds to the downstream cistron (10). These two mRNAs overlap in a short area of only a few dozen nucleotides that represents the binding site of the RBP. This model suggests that rare endonucleolytic events in the upstream or downstream cistron create exonuclease-sensitive transcript ends that are degraded up to the position of the RBP. In cases where exonucleolytic degradation proceeds toward the RBP from both sides, only the RNA directly protected by the protein survives as an RBP footprint.
At present, this model is supported mostly by data from studies on chloroplast RNA metabolism. Several processed 5′- and 3′-ends of chloroplast mRNAs overlap with RBP binding sites (10,12–14). However, in only a few cases have biochemical and/or genetic links been established between the presence of a small RNA (sRNA) and a chloroplast RBP. The founding example is the pentatricopeptide repeat (PPR) protein PPR10, which is responsible for protection of the mRNAs atpH and rpl33, and leads to accumulation of sRNAs that correspond to its binding sites (10,14–15). Similarly, the PPR protein HCF152 protects an sRNA fragment of the psbH-petB intergenic spacer and is required both for upstream and downstream mRNA protection (13,16–17). CRP1 (chloroplast RNA processing 1) protects petB and petD mRNAs, and leaves behind a footprint of 29 nucleotides (nt) (14). HCF107, a half-a-tetratricopeptide (HAT) repeat protein, protects psbB and psbH mRNAs against exonucleolytic decay, also generating sRNAs (14,18). This function is evolutionarily maintained in the Chlamydomonas ortholog, MBB1 (12). All proteins involved in generating sRNAs belong to the helical-hairpin-repeat superfamily of proteins. Members of this family are capable of binding RNAs with high specificity and high affinity. The genomic potential for further RBP-sRNA pairs is large. Most prominent in this regard is the PPR protein family, which is represented in land plants by several hundred members.
PPR proteins are involved in all organellar RNA processing steps, both in plastids and mitochondria (11). Only a subset of PPR proteins is believed to function as road-blocks against exonucleolytic degradation and thus eventually cause sRNA footprints. In vitro, PPR proteins can bind to sRNA fragments corresponding to their minimal binding site (15). Thus, sRNAs and cognate mRNAs could be competitors in vivo for their PPR protein ligands, highlighting a potential regulatory role of sRNA fragments. Such a regulatory role could be reflected in differential accumulation of sRNA in response to various signals. However, at present, no expression studies for small organellar RNAs are available.
How many PPR proteins and how many other helical-hairpin-repeat proteins will ultimately serve to end-protect organellar RNAs is unclear at present. However, recent transcriptome-wide studies might provide a clue about the number of sRNAs in chloroplasts. These studies have identified a large number of sRNAs in chloroplast UTR regions of Chlamydomonas, barley, maize, rice and Arabidopsis, pointing to a dominant and evolutionarily conserved role of protein-mediated protection of plastid transcripts (12–14). In mitochondria, similar global investigations are lacking, but recently, the PPR protein MTSF1 was shown to define the processed 3′-end of the mitochondrial mRNA, nad4. In an mtsf1 mutant, nad4 mRNA and a corresponding sRNA in the nad4 UTR do not accumulate, likely owing to degradation from the 3′-end (19). A systematic and exhaustive analysis of sRNAs in mitochondria is needed as a prerequisite for defining RBP-sRNA pairs and thus understanding RNA stabilization and, potentially, RNA regulation. To date, however, the tools needed to rapidly and automatically mine short RNA-Seq datasets for RBP footprints have been lacking.
We established a bioinformatics pipeline termed sRNA miner to identify potential RBP footprints in plant RNA-Seq datasets. The clusters of reads identified by this software are representing such RBP footprints and are named here clustered organellar sRNAs (cosRNAs). Using this pipeline, we re-analyzed available sRNA-Seq datasets to identify cosRNAs that likely originate from mitochondrial and chloroplast genomes and possibly represent footprints of organelle-targeted RBPs. Similar to chloroplasts, mitochondria also contain a large number of cosRNAs, suggesting that RBPs help to stabilize transcripts and protect them against exonucleolytic decay. Unlike the case in chloroplasts, we found that cosRNAs accumulate at the majority of 3′-ends of mitochondrial mRNAs, but only at a minor fraction of processed 5′-ends. The latter appear to be generated differently in mitochondria.
MATERIALS AND METHODS
Analysis of sRNA sequencing data
The sRNAs from Arabidopsis thaliana ecotype Columbia-0 analyzed here were extracted from previously established RNA sequencing libraries dedicated to nuclear siRNA analysis (20). Sequences were obtained from the sequence read archive at NCBI: SRA035939. We performed adapter and quality trimming using cutadapt (21) and mapped sRNA sequences to the Arabidopsis nuclear and organellar genomes (NCBI:JF729201, TAIR10; 22) using Bowtie (version 1.1.1) (23) with the following parameters –a --best --strata –v 2. Using these settings, Bowtie reports all alignments with the highest score and a maximum of two mismatches. Read mappings were processed using SAMtools (24), and coverage graphs were extracted using BEDTools (25). Coverage graphs were visualized using the Integrated Genome Browser (26). The 5′ sharpness of cosRNAs that are positioned at 3′-ends of mitochondrial and plastid transcripts was calculated as the number of nucleotides required to decrease coverage from 80 to 10%. The 3′ sharpness of cosRNAs positioned at 5′-ends of mRNAs was calculated in an analogous manner.
Identification of cosRNAs using sRNA miner
sRNA miner is implemented in R/Bioconductor (27), and is freely available at GitHub repository (https://github.com/MolGen/sRNAminer). The algorithm calculates alignment end density at every genome position in a strand- and end-type-specific manner from a mapping file in bam format. Maxima in a sliding 31-nt window are retained for the detection of dominant 5′- and 3′-ends. The algorithm tests for sharp ends at these maxima by dividing the number of ends in a window of ±1 nt of the maximum by the number of ends in a window of ±15 nt. The output of the software—cosRNAs detected—depends on a user-defined threshold, set at 0.75 for the dataset used in this study. The software also allows filtering by the number of read ends at the positions of maxima. This parameter can be adjusted to allow cosRNAs expressed at low levels to be detected and to adjust for sequencing depth. The threshold for the dataset described in this study was set to 40 read ends. The software allows the detection of cosRNAs with both sharp 5′- and 3′-ends; however, less stringent settings, which require only one end to exceed the specified threshold, are also supported. This less stringent setting was applied for the detection of small organellar RNAs in this work. The corresponding 3′-end of a cosRNA for a sharp 5′-end is defined as the 3′-end maxima in a window of +15 to +50 from the obtained 5′-end. The 5′-ends for cosRNAs identified by a sharp 3′-end were detected in a similar manner.
sRNA mappings and identified cosRNAs are available at https://www.molgen.hu-berlin.de/projects-jbrowse-athaliana.php.
sRNA gel blot
RNA was extracted using TRIzol reagent (Thermo Scientific) and enrichment of low molecular weight RNA was carried out as described (28). sRNA gel blot hybridization was carried out as described previously (12). Sequences of DNA oligonucleotides used as probes are listed in Supplementary Table S3.
Rapid amplification of cDNA ends (RACE)
Col-0 total RNA (2 μg) was ligated with 10 pmol of an adapter oligonucleotide (Supplementary Table S3) using T4 RNA ligase I (NEB). After phenol–chloroform extraction of RNA, cDNA was synthesized using Protoscript II reverse transcriptase (NEB) and an adapter-specific primer (Supplementary Table S3). Polymerase chain reaction (PCR) amplification was performed using primers given in Supplementary Table S3. PCR products were gel purified, ligated into the pJet1.2/blunt vector (Thermo Scientific) and sequenced.
RNase protection assay
Intact chloroplasts from 10-day-old maize seedlings were isolated as previously described (29). Preparation of stromal fractions, co-immunoprecipitation and isolation of nucleic acid were performed as described previously (30) using 5 μl of affinity-purified antibodies against PPR10 and PPR4 (10,31). RNase protection assays were performed as described previously (13). Oligonucleotides used to generate the template for in vitro transcription are listed in Supplementary Table S3.
Statistical analysis
Values are expressed as means ± standard deviation (SD). The significance of differences in 5′- or 3′-end sharpness between mitochondrial and plastid cosRNAs (number of nucleotides needed to decrease sRNA coverage at the 5′- or 3′-end from 80 to 10%), were determined using two-tailed, unpaired Student's t-tests.
RESULTS AND DISCUSSION
Most 24-nt sRNAs that map to the mitochondrial genome map equally well to the nuclear genome
To identify novel, small organellar RNAs in Arabidopsis that potentially represent footprints of RBPs, we aligned sRNA sequences from a previously published library to both nuclear and organellar genomes (20). The library contains RNAs size-selected (15–50 nt) from total RNA, and thus includes sequences derived from the two DNA-containing organelles. A total of 18 939 882 reads aligned with the chloroplast genome and 1 492 111 reads aligned with the mitochondrial genome, corresponding to 20.4 and 1.6% of all reads, respectively.
Typical nuclear-encoded sRNAs include miRNAs, siRNAs and derivatives that are in a size range of 21–24 nt (32). Indeed, an investigation of read lengths of sRNAs mapped to the nuclear genome revealed a peak in this size range (Supplementary Figure S1A). Chloroplast-mapped reads did not show this distribution (Supplementary Figure S1B). Instead, the length distribution showed a single peak at 22 nt, representing 4.5 million reads. Of these, 3.9 million reads corresponded to a single genomic location upstream of the ndhB gene, a location that coincides with the 5′-end of ndhB mRNA (33). The sequence of this cosRNA is not present in the nuclear or mitochondrial genome, suggesting that it is a genuine chloroplast product. The abundance of reads for this ndhB cosRNA exceeds read numbers of known abundant miRNAs like miR159a, for which 360 000 reads were found in the library investigated. It is also the most abundant cosRNA in our previous analysis (13). This cosRNA likely represents the footprint of the PPR protein CRR2, known to act on the 5′-UTR of ndhB to serve as an mRNA stabilizer (33).
In mitochondria, the distribution of mapped reads showed a bias toward sequences with a read length of 24 nt (Supplementary Figure S1C). This bias is specific to sRNAs that map to the mitochondrial genome and was not detected in mappings to the plastid genome (Supplementary Figure S1B). A total of 24-nt sRNAs, the most abundant class of sRNAs in plants, mostly represent siRNAs associated with heterochromatic regions in the nuclear genome (32). About 45% of sRNAs that we mapped to the mitochondrial genome mapped equally well to the nuclear genome. The sequenced Arabidopsis nuclear genome (22) contains a recent insertion of large parts of the mitochondrial genome in the centromeric region of chromosome 2, a so-called numt (nuclear mitochondrial DNA; 34,35). In fact, this same centromeric region was previously reported to be associated mainly with sRNAs 24 nt in length (36,37). We therefore conclude that 24-nt sRNAs that map to the mitochondrial genome are largely nuclear siRNAs involved in heterochromatin maintenance of mitochondrial DNA insertions in chromosome 2. Other numts are likely to add to the pool of 24-nt long siRNAs with mitochondrial sequences. In fact, since organellar gene transfer is an ongoing and frequent process (38,39), any short RNA read could potentially map to a recent numt/nupt. Future efforts to sequence sRNAs isolated from purified mitochondria will help to unequivocally assign reads to this genome.
sRNA miner—a versatile tool for extracting clusters of organellar sRNAs from sRNA mappings
Our previous analysis of small organellar RNAs was purely based on read abundance (13). Here, we developed a bioinformatics pipeline to detect small organellar RNAs based on key characteristics, with the foremost being that they are short (<50 nt) and have sharp ends. The pipeline thus allows the identification of defined clusters of sRNAs even in regions of higher background, resulting from random decay of abundant RNA species. Such random decay products dominate the mapping in some regions. We implemented software that uses these criteria to identify cosRNAs in RNA-Seq datasets starting from a mapped read file in bam format. The software, termed sRNA miner, allows for adjustment of the number of reads and sharpness of 5′- and 3′-ends, which proved instrumental in the detection process.
cosRNAs are located preferentially in intergenic regions in both chloroplasts and mitochondria
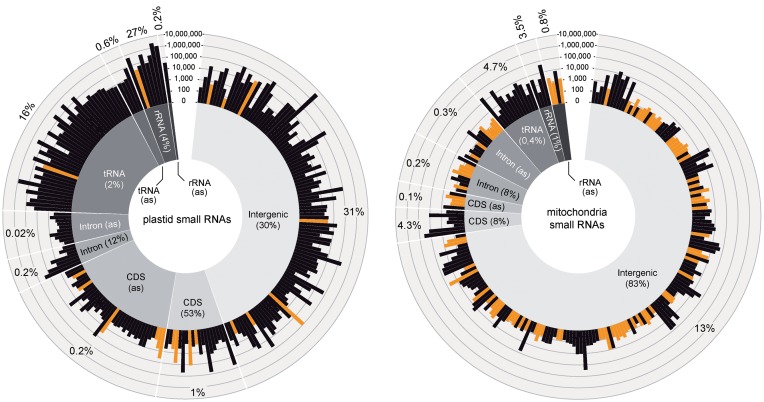
Using sRNA miner with the settings described in ‘Materials and Methods’ section, we identified a total of 229 cosRNAs in chloroplast genomes and investigated whether these cosRNAs were located in non-coding or coding regions, or in tRNA or rRNA genes (Figure 1). The largest group of cosRNAs was found within non-coding regions, consistent with their presumptive biogenesis through RBPs binding to UTR sequences.
Figure 1.
Genomic location of cosRNAs identified using sRNA miner. cosRNAs identified using our sRNA miner algorithm were ascribed to organellar chromosome locations using the gene annotations from TAIR10 for the nuclear and the chloroplast chromosomes (22) and from (NCBI: JF729201) for the mitochondrial chromosome. Overlaps of cosRNAs with tRNA, rRNA or mRNA annotations were identified using BEDTools intersect in a strand-specific manner requiring a 50% overlap with the annotations (25). The abundance of individual cosRNAs is shown in a log10 scale and is also available in Supplementary Table S1. The percentage of reads found in the cosRNAs of the different categories with respect to all reads mapping to the respective organelle genome are indicated in the outer circle. The percentage of genome space occupied by the different categories is indicated in the inner circle. cosRNAs with a length of 24 nt, which likely represent nuclear siRNAs are indicated in orange (grey in print version). The graphs were produced using Circos (40).
A considerable number of cosRNAs were also found in protein-coding regions, in both sense and antisense orientations. The chloroplast genome is relatively gene-dense. As a result, cosRNAs found inside coding regions could represent footprints of RBPs that bind the UTR sequence of an up- or downstream gene. We previously showed that a cosRNA in psbC overlaps with a processed transcript end of the downstream psbD (13). A similar situation is shown in Figure 3B, where a cosRNA in rpoB is located just 70 nt upstream of rpoC1. cosRNAs identified in intergenic regions generally show higher accumulation levels than cosRNAs positioned antisense to coding regions: sense cosRNAs have a median read number of 1454 versus 290 reads for antisense cosRNAs (based on all mitochondrial and chloroplast intergenic and coding sequence (CDS) antisense cosRNAs shown in Supplementary Table S1, excluding the 24-nt long cosRNAs). If the cosRNAs antisense to chloroplast genes are indeed footprints of RBPs, then what is the reason for this binding? A large number of antisense transcripts accumulate in plastids (4) and antisense RNA can influence the processing of chloroplast RNAs (41–43). RBPs may be involved in stabilization of such antisense RNAs, but whether this is functionally relevant is unclear. Alternatively, these antisense RNAs may represent off-targets of RBPs, which could lead to the accumulation of non-functional antisense transcripts.
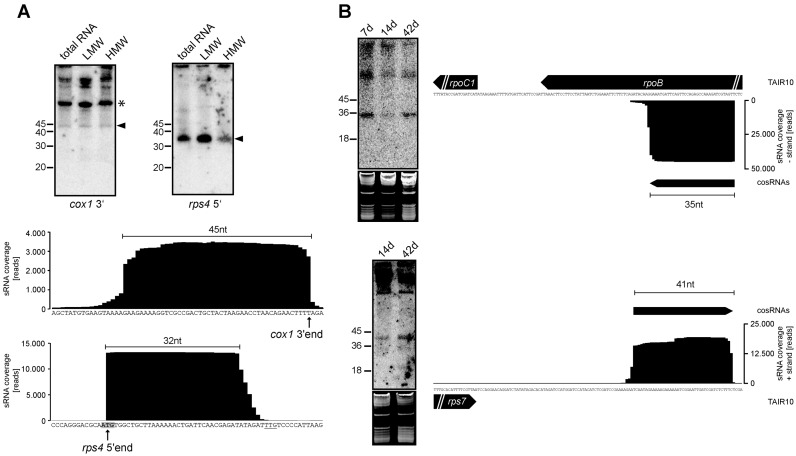
Figure 3.
Novel, ‘long’ organellar sRNAs detected in mitochondria and plastids. (A) Two mitochondrial cosRNAs, cox1 3′ and rps4 5′, detected by RNA gel blot analysis (upper panel). Total RNA (10 μg), low-molecular-weight (LMW) RNA and high-molecular-weight (HMW) RNA were run on a denaturing polyacrylamide gel and detected using 32P-end-labeled oligonucleotide probes after transfer to a nylon membrane. Hybridization signals of the expected size are marked by arrowheads. The asterisk marks a potential 5′-extended form of the cosRNA. sRNA coverage for the respective genome regions is shown below the RNA gel blot images. The cox1 3′-end and rps4 5′-end (47), are indicated by arrows. The annotated rps4 start codon is shown in bold and shaded. A potential alternate start codon at a conserved position is underlined. (B) sRNA coverage for two newly identified cosRNAs (indicated by arrows) upstream of rpoC1 and downstream of rps7 is shown on the left. The cosRNAs were detected by RNA gel blot analysis (show on right). Total RNA (10 μg) from 7-, 14- and 42-day-old plants was separated on denaturing polyacrylamide gels, and cosRNAs were detected as described in (A). Ethidium bromide staining of the gel before transfer, shown below the radiographs, served as a control for loading differences.
Two other classes of abundant cosRNAs found here are fragments of tRNAs and rRNAs. Such fragments have been described previously, but whether they are functional remains to be resolved (44). In case of tRNAs, most fragments identified are sense-oriented (50 of 57) and 42 of 57 share either the 5′- or 3′- end with their respective tRNA, suggesting that they are degradation products of mature tRNAs. These cosRNAs are heterogeneous in size and a majority terminates within the anticodon stem-loop of tRNAs, where endonucleolytic degradation is known to start. A further subgroup of cosRNAs is found immediately downstream of tRNAs, likely generated by the activity of RNase Z, an endonuclease that generates tRNA 3′ ends (45). Surprisingly, we also detected fragments that are antisense to tRNA genes and are thus not simple degradation products of abundant tRNAs (Figure 1). It is possible that RNAs antisense to tRNAs also fold into stable nuclease-resistant structures and therefore are captured in the library analyzed here.
A similar distribution of cosRNAs into different categories was seen for mitochondria. Again, of the 222 cosRNAs identified, the largest fraction was found in non-coding regions. In addition, a considerable number of cosRNAs were also found in coding regions, with several abundant cosRNAs in a sense orientation and few abundant cosRNAs in an antisense orientation. In contrast to the plastid sRNAs, mitochondria exhibit a bias toward sRNAs 24 nt in length (Figure 1). As discussed above, many of these likely represent abundant siRNAs of nuclear origin.
In total, 77% of the reads that map to the chloroplast genome are found within the detected cosRNAs. In mitochondria, 27% of the reads are located in cosRNAs. The cosRNAs identified for the two organelles could serve as an excellent database for the computational prediction of RNA targets of PPR proteins using the recently described PPR code (46). The sRNA mappings and identified cosRNAs can be viewed online at https://www.molgen.hu-berlin.de/projects-jbrowse-athaliana.php.
Mitochondrial cosRNAs overlap with previously mapped 3′-ends of transcripts
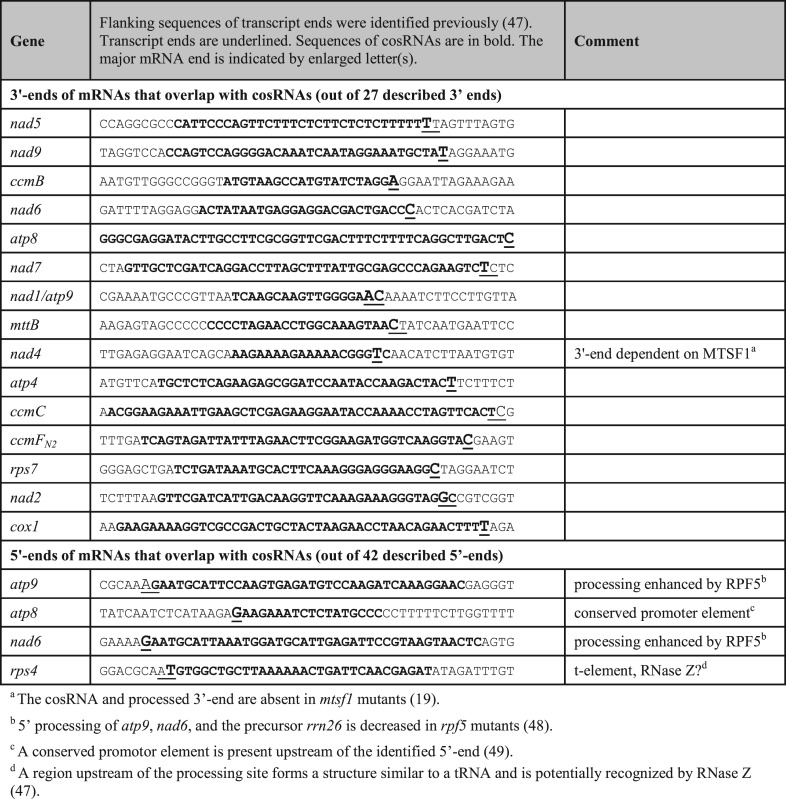
Plastid cosRNAs have been shown to overlap with transcript ends that are created post-transcriptionally, including both 5′- and 3′-ends of mRNAs (13,14). For mitochondria, we observed a strong overlap of cosRNAs with previously mapped 3′-ends of mitochondrial transcripts (47). Of the 27 described 3′-ends of mitochondrial transcripts, 15 were associated with cosRNAs (Table 1). We also observed high coverage of reads for an additional four mRNA 3′ termini (ccmFC, cob, rps4 and cox3), but the algorithm did not detect these as cosRNAs because 5′- and 3′-ends were not well defined.
Table 1. cosRNAs coinciding with mitochondrial transcript ends.
|
It was recently shown that the PPR protein MTSF1 is indispensable for the stabilization and correct 3′-end processing of nad4 mRNA. In vitro, MTSF1 binds to the last 20 nt of the 3′-UTR of nad4 mRNA. A cosRNA that is also present in our dataset (Table 1) is missing in the corresponding mutants, indicating that the cosRNA is the footprint of MTSF1 (19). Similar to MTSF1, other RBPs could stabilize mRNAs that accumulate cosRNAs at their 3′-ends (Table 1). The large number of cosRNAs at 3′-ends points to a dominant role of protein-mediated protection of 3′-ends in plant mitochondria. Such proteins will be nuclear-encoded and are interesting candidates for the post-transcriptional adjustment of mRNA levels. Little is known about the mechanisms of mitochondrial gene regulation in plants. However, it is clear that steady state levels of mRNAs do not correspond well with their respective transcription rates (50), arguing for regulation at later steps of gene expression. In chloroplasts of the green alga Chlamydomonas reinhardtii, RBPs like MCA1 can be rate limiting for transcript accumulation (51). There will also likely be rate-limiting factors among the multitude of RBPs predicted here to mediate mitochondrial transcript stabilization in higher plants.
Another asset of the cosRNA analysis is that it allows for the prediction of additional transcript ends in mitochondria. A case in point is the cosRNA in the intergenic region of rpl2 and mttB (M216 in Supplementary Table S1). This cosRNA predicts the existence of a monocistronic form of rpl2. To address this, we mapped transcript ends of rpl2 by rapid amplification of cDNA ends. Indeed, 3′-ends of rpl2 overlap with the identified cosRNA (Supplementary Figure S2). Moreover, recent RNA gel blot analyses showed that two spliced rpl2 transcript forms exist in Arabidopsis (52), with the shorter one possibly being monocistronic. This example demonstrates that in general, mitochondrial cosRNAs in intergenic regions are predictive of transcript ends, potentially uncovering novel functional transcription units.
cosRNAs coincide with 5′-ends of mRNAs in chloroplasts, but rarely do so in mitochondria
It was previously shown that chloroplast cosRNAs overlap with 5′-ends of mRNAs (13,14). Mitochondrial transcripts usually harbor multiple distinct processed 5′-ends (47). In our analysis, only a minority of mitochondrial 5′-ends overlapped with cosRNAs (Table 1, bottom)—a striking contrast to the overlap we observed at 3′-ends. Even though PPR proteins have been implicated in the generation of certain 5′ termini in mitochondrial transcripts (53–55), none of these PPR-dependent 5′-termini overlapped with a cosRNA. This suggests that this specific subset of PPR proteins may fulfill its function in 5′ processing by only transiently attaching to its RNA target, and thus fail to produce a footprint, a role quite different from that of its PPR10-like relatives in the chloroplast.
Among the cosRNAs that co-localize with 5′-ends is one that protects rps4 mRNA. This is the second-most abundant of all cosRNAs that map to the mitochondrial genome (Supplementary Table S1). It has been proposed that this end is generated by RNase Z, which is suggested to recognize a tRNA-like element immediately upstream of the rps4 5′-end (47). This processed 5′-end of rps4 mRNA is located at position +2 in the coding sequence. Therefore, mRNAs with this particular 5′-end lack a complete start codon (47); however, it is possible that an alternative start codon is used for rps4. Indeed, the start codon in non-brassicaceae is located 13 codons downstream of the annotated Arabidopsis start codon (56). In Arabidopsis, a UUG codon encoding leucine is found at this position (Figure 1). UUG codons have been identified as alternate start codons in both bacteria and mitochondria (57,58); thus, the true start codon for Arabidopsis rps4 is likely the UUG codon at position +39 of the processed end. This finding emphasizes once more that cosRNAs are useful tools for defining the physical boundaries of RNAs within plant organellar transcriptomes.
Defined 3′-ends versus relaxed 5′-ends of mitochondrial cosRNAs positioned at mRNA 3′-ends
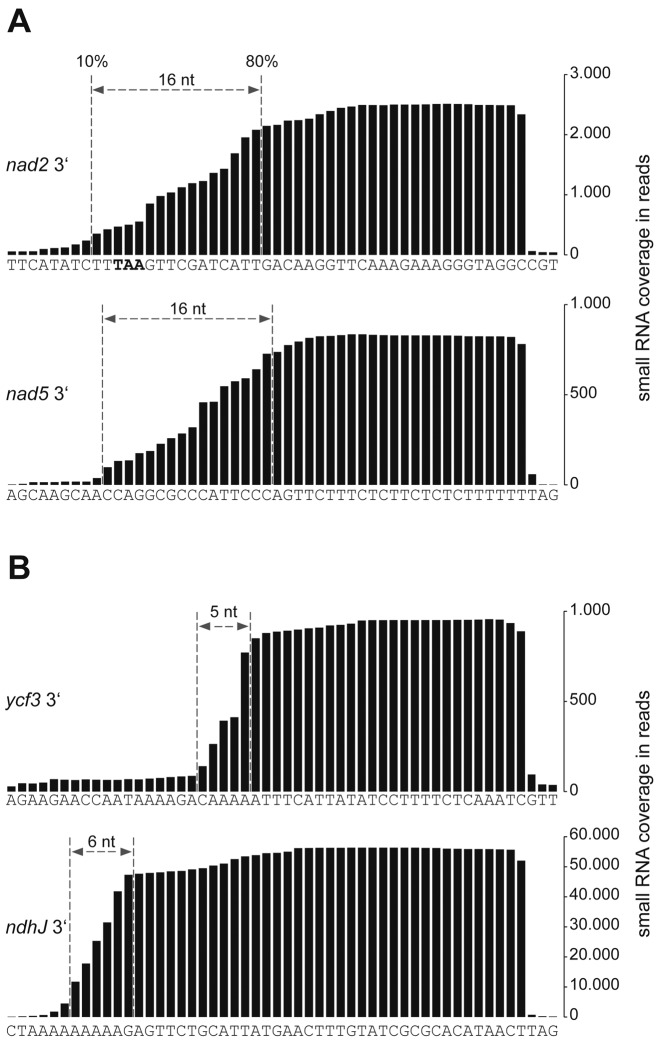
A hallmark of chloroplast cosRNAs is the sharpness of their termini (13), which are generated by the activity of exonucleases (15,16). This can be visualized by displaying the RNA sequencing coverage for individual cosRNAs, as shown in Figure 2 for two exemplary cosRNAs of each organelle located at 3′-ends of mRNAs known to accumulate in organelles. All four cosRNAs have sharper 3′-ends than 5′-ends. In addition, the 5′-ends of chloroplast cosRNAs appear much sharper than those of mitochondria. To quantify the ‘sharpness’ of cosRNA 5′-ends, we determined the number of nucleotides needed to decrease cosRNA coverage at the 5′-end from 80 to 10% (Figure 2). We applied this to all 16 mitochondrial cosRNAs that coincide with described 3′-termini of mRNAs (Supplementary Figure S4), including the two mitochondrial cosRNAs shown in Figure 2. For comparison with chloroplasts, we included all cosRNAs that overlap described 3′-ends of plastid transcripts (Supplementary Figure S4).
Figure 2.
5′-ends of mitochondrial cosRNAs at transcript 3′-ends are loosely defined. Comparison of selected cosRNAs from mitochondria (A) and plastids (B) associated with 3′-ends of mRNAs. cosRNA coverage is plotted against genome position. The decrease in cosRNA coverage at the 5′-end was measured as the number of nucleotides needed to decrease maximal coverage from 80 to 10% (dashed lines). See text for comparisons of additional cosRNAs.
The nine cosRNAs from plastids on average needed 4 ± 8 nt to decrease coverage from 80 to 10%. In contrast, mitochondrial cosRNAs required an average of 14 ± 6 nt, a highly significant difference (P = 0.007; two-tailed unpaired Student's t-test). Thus, cosRNAs in mitochondria display a different, less stringent maturation of their 5′-ends than chloroplast cosRNAs. We performed an analogous analysis of 3′ ‘sharpness’ of cosRNAs positioned at mRNA 5′-ends (Supplementary Figure S5) and could not detected a significant difference.
Sharpening the 5′-ends of multiple RNAs requires a non-specific 5′-to-3′ exonuclease activity. An RNase J with such activity has been detected experimentally in plastids, where it contributes to degradation of spurious RNAs in chloroplasts (5). No such activity has been found in mitochondria, and we assume that mitochondrial endonucleases from RNase H and RNase P families (6) generate the observed loose 5′-ends of cosRNAs. The absence of a 5′-to-3′ exonuclease activity in mitochondria might explain the low abundance of cosRNAs in the 5′-UTR of mitochondrial RNAs, since no protection by RBPs is needed. Furthermore, the absence of a 5′-to-3′ exonuclease activity might explain the dramatic phenotype of mitochondrial PNPase mutants compared to chloroplast PNPase mutants (8,59). If PNPase is the only, or only major, exonuclease in mitochondria, its loss could lead to over-accumulation of transcripts that are detrimental for gene expression, for example because of base-pairings with mRNAs. In chloroplasts, the loss of PNPase can be partially compensated by RNase J, an alternative that is lacking in mitochondria.
In sum, the lack of defined 5′-ends of mitochondrial cosRNAs is indicative of the absence of a major 5′-to-3′ exonuclease in plant mitochondria.
cosRNAs longer than 30 nt accumulate in mitochondria and chloroplasts
Accumulation of chloroplast cosRNAs has been demonstrated in several organisms by high-throughput RNA sequencing (12–14,44), and we have added further to this analysis by performing RNA gel blot hybridizations (Supplementary Figure S3). In the case of mitochondrial cosRNAs, only the MTSF1 footprint has been experimentally verified. Thus, to confirm the data from the RNA-Seq analysis, we analyzed selected mitochondrial cosRNAs by RNA gel blot hybridization. A cosRNA that overlaps with the processed 5′-end of rps4 was detected at the expected size of ∼35 nt (Figure 3A). The presence of another cosRNA with a size of 45 nt located at the 3′-end of cox1 mRNA also confirms the RNA-Seq data.
The dataset we used for our analysis contains cosRNAs in the range of 15–50 nt. This enabled us to detect longer cosRNAs that escaped our earlier analysis (13). Our current sRNA miner analysis revealed several chloroplast and mitochondrial cosRNAs between 28 and 50 nt, (Supplementary Table S1). The two mitochondrial cosRNAs shown in Figure 3A are cases in point. We also tested two longer plastid cosRNAs, upstream of rpoC1 in the rpoB gene and downstream of the rps7 open reading frame. Signals of the predicted size were found in both cases (Figure 3B). The latter cosRNA overlapped with transcript ends we reported previously for rps7, but no cosRNA was identified in this earlier study owing to the size limitation of the library analyzed (13). How can such rather extended RNAs be generated? The prevailing model for the interaction of PPR proteins with RNAs posits that each repeat associates with one nucleotide (11). The PPR tracts described in Arabidopsis range from 2 to 26 repeats, with an average of 12 (60). Taking into account that nucleotides adjacent to binding sites will also not be fully accessible to nucleases for steric reasons (9,15,18), we have to assume a maximum of about 30 nt for the footprint of a single PPR protein. This size limit is breached by cosRNAs shown in Figure 3 and additional cosRNAs shown in Supplementary Table S1. It is possible that nucleotides or stretches of nucleotides are looped out of the binding site, but remain protected (61). Such loops are in fact expected since steric constraints due to the superhelical nature of PPR proteins impose a physical size limit for consecutively bound nucleotides. This size limit may be in the range of only 9 nt (46). An alternative explanation for long footprints would be that several PPR proteins, or PPR proteins together with other RBPs, act as a larger complex to protect RNA. An example of this is a chloroplast RRM (RNA recognition motif) protein, which supports, but is not essential for, the accumulation of a footprint in the 3′-UTR of chloroplast ndhF mRNA (30). Also, recent reports that PPRs involved in editing interact with RRM-proteins and other factors suggest that larger protein complexes that assemble around a PPR may increase footprint size (62,63).
We also considered the alternative explanation that these large cosRNAs are accumulating due to stable secondary structures that survive nuclease attack. Therefore, we calculated the free energy of all mitochondrial and chloroplast cosRNAs (Supplementary Table S1). Only three cosRNAs matched free energy values known from stem-loop structure known to be biologically relevant and resistant to nuclease activity (64,65). This is preliminary evidence that the cosRNAs are not simply stabilized due to extensive RNA structure. Still, despite displaying typical characteristics of RBP-footprints, it cannot be excluded at present that such longer fragments are unprotected mRNA fragments that are generated by precise endonucleolytic action or transcription termination. It will be interesting to see whether even larger footprints can be retrieved using RNA sequencing libraries designed to capture RNA fragments above the 50-nt limit that defines the library used in the current study.
Chloroplast cosRNAs show development-dependent accumulation
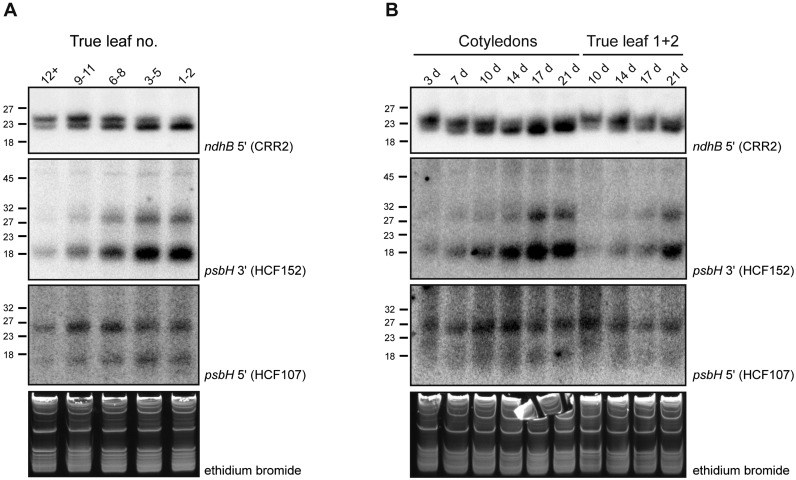
Many regulatory short RNAs show specific expression profiles, best exemplified by miRNAs in plants and many other organisms (66). cosRNAs generated by end-protective PPR proteins have to be considered degradation products without specific function. However, they could have a secondary role as regulators. Functions in trans, similar to miRNAs or bacterial sRNAs, or a role as a sink for the cognate RBP have been proposed (13). Experimental support for either of these functions is currently lacking. As an initial step toward a functional study of cosRNAs, we analyzed their accumulation during plant development by RNA gel blot analysis. First, different leaves were harvested from 32-day-old plants. Leaves from these plants were sampled according to their time of emergence and analyzed for the accumulation of cosRNAs (Figure 4A). Second, we harvested either the cotyledons or the first and second true leaves from plants of different age after germination and tested them using the same procedure (Figure 4B). For this analysis, we selected three cosRNAs for which the cognate RBP is known. The PPR proteins CRR2 and HCF152 are required for the accumulation of ndhB 5′ cosRNA and the cosRNA downstream of psbH, respectively (unpublished data and 13). The HAT-protein HCF107 is linked to a cosRNA in the psbH 5′-UTR (18).
Figure 4.
Developmental and age-dependent accumulation of plastid cosRNAs. (A) True leaves of 32-day-old Arabidopsis plants were harvested according to their time of emergence and separated into five samples. Leaves 1 and 2 represent the first emerging leaves. Total RNA (10 μg) was separated on denaturing polyacrylamide gels, and RNA gel blots were performed using 32P-end-labeled oligonucleotide probes antisense to three cosRNAs representing footprints of the RNA binding proteins CRR2, HCF152 and HCF107. Ethidium bromide staining of the gel before transfer served as a control for equal loading. Sizes of marker oligonucleotides are indicated. (B) Analysis of cosRNAs during leaf aging in cotyledons and first true leaves. The day of harvest after sowing (d) is indicated for each sample. Oligoprobes used are the same as in (A).
All three cosRNAs showed differential accumulation in different-aged leaves. The CRR2 footprint consisted of two isoforms that differ by only two nucleotides at their 3′-end. The two isoforms show a reciprocal pattern of leaf-age-dependent accumulation: the larger cosRNA accumulated early and declined during leaf aging, whereas the smaller one increased with increasing leaf age (Figure 4A and B). This suggests that there is a slow conversion of the large ndhB 5′ sRNA species into the small sRNA species, likely reflecting a slow nibbling away of the terminal nucleotides. For the psbH 3′ sRNA, accumulation increased in aging tissue and was most pronounced in the oldest leaves (leaf 1 + 2) of the 32-day-old plant (Figure 4A and B) as well as 21-day-old cotyledons (Figure 4B). This trend of accumulation over time is consistent with continuous expression and decay of psbH mRNA, leading to a constant influx of sRNA. Whether these sRNAs are free or predominantly bound by their cognate PPR proteins is unclear at present. In any case, this would make for a formidable diversion of PPR proteins from their true target, the mRNAs.
One intriguing observation is that the two cosRNAs flanking psbH showed different patterns of accumulation in leaf development. Whereas psbH 3′ increased over time, psbH 5′ remained rather constant, with a slight peak accumulation in leaves no. 6–11 and in 10–14-day-old cotyledons (Figure 4A and B). Thus, the accumulation pattern during development of at least one of the two cosRNA species does not passively follow the accumulation of its cognate psbH mRNA. A possible explanation for the differences in accumulation would be differences in the degradation of the upstream psbT or downstream petB mRNAs. Alternatively, the stabilizing trans-factors HCF107 and HCF152 may exhibit differential availability during development, i.e. they could limit cosRNA accumulation depending on their own expression levels. If the amount of protective PPR proteins is limiting for cosRNA accumulation, cosRNA amounts would parallel PPR amounts. Of course, this would only be true if PPR proteins are able to associate with free cosRNAs, as they do with their target mRNAs.
PPR proteins associate with cosRNAs in vivo
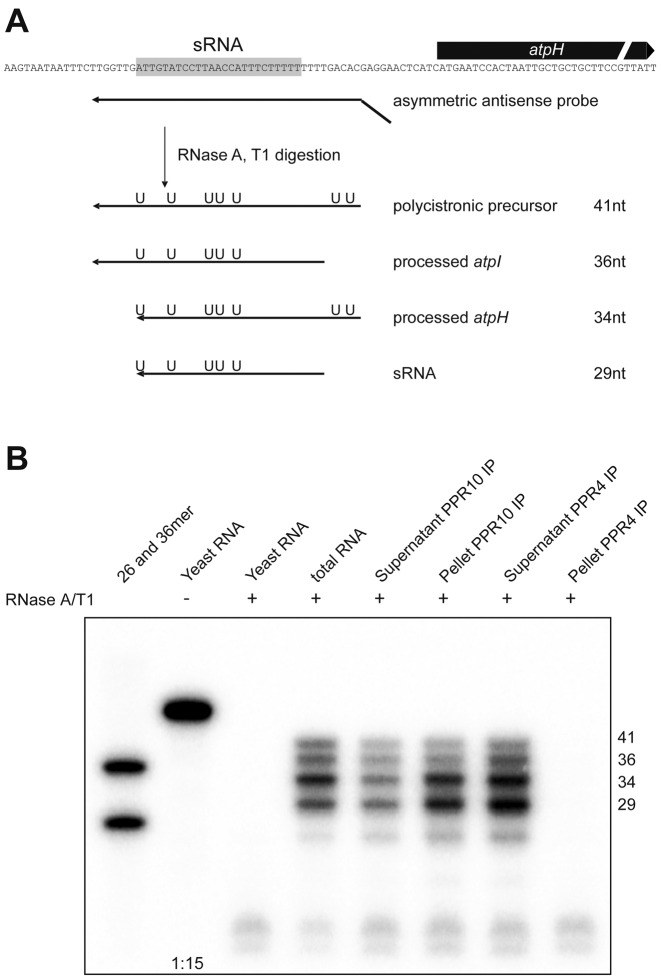
An important question for understanding PPR proteins as well as cosRNAs is whether PPR proteins actually associate with cosRNAs in vivo. If so, cosRNAs could act as competitors for the mRNA targets of PPRs, and thus potentially have regulatory functions. At least in vitro, the PPR protein PPR10 from maize can bind to the cosRNA atpH (15). However, whether the concentration of target RNAs in such electrophoretic mobility shift assay studies is comparable to in vivo concentrations is questionable. Also, such an analysis does not represent the competitive situation that a PPR protein faces in vivo, where it could attach to polycistronic mRNAs, processed mRNAs or cosRNAs. To investigate the specific RNAs associated with maize PPR10, we coupled immunoprecipitation of PPR10–RNA complexes with RNase protection analyses. We investigated maize PPR10 since this protein is well understood in terms of its binding sites and footprint, has been studied in vitro and, most importantly, an antibody is available that works in immunoprecipitations (10). We used a radiolabeled probe for the RNase protection assay designed such that degradation leads to four different protected RNA fragments (Figure 5A). The protected fragments represent the polycistronic precursor, the two processed transcripts and the cosRNA.
Figure 5.
The RNA binding protein PPR10 associates with its footprint in vivo. (A) Schematic representation of the probe and protected fragments in the RNase protection assay used to identify RNAs associated with PPR10 in vivo. The sequence of the cosRNA upstream of atpH is highlighted (10). The probe used is shown as an arrow indicating the strand. A short artificial sequence at the 5′-end of the probe is not aligned with the chloroplast genome sequence. Protected fragments originating from hybridization with the different possible RNA species are shown with the position of radiolabeled uridines indicated. (B) RNase protection assay using total RNA from the first leaf of 10-day-old maize seedlings and RNAs co-precipitated with PPR10 from maize stroma. Total RNA (1 μg) and RNA isolated from supernatants were used. The same partial volumes as for supernatants were also used for pellet fractions. RNAs were hybridized at 42°C with a 32P-labeled antisense RNA, and non-hybridized regions of the probe were digested with a mixture of RNase A and RNase T1. Two end-labeled RNA oligonucleotides are included as size markers. Hybridization with yeast RNA controlled for probe integrity during the experiment (−RNase, 1:15 dilution) and self-protection of the probe (+RNase). Immunoprecipitation was performed using specific antibodies for PPR10 (10) and PPR4 (31), with the latter representing a non-specific control that precipitates an unrelated PPR protein.
We used total RNA as a control to visualize the ratio of the four different transcripts potentially bound to PPR10. In this sample, the strongest signal was found for the fragment representing the processed atpH RNA, followed by the fragment for the cosRNA (Figure 5, right). The probe is body-labeled; therefore, the shorter the degradation product, the less radioactivity is incorporated. This leads to an underestimation of the abundance of the fragment representing the cosRNA relative to longer fragments. Still, it is clear from this analysis that the cosRNA signal and signals for the other transcripts are within the same intensity range, which is supported by quantifications of the autoradiograph after correction for the numbers of radiolabeled U's in the protected fragments (Supplementary Figure S6). Thus, the cosRNA is not rare relative to its precursors. Importantly, the relative abundance of the cosRNA was also similar, if not increased in comparison to the precursor RNAs in the PPR10 immunoprecipitation pellet. By contrast, no atpH signal was observed in the control PPR4 immunoprecipitation pellet, demonstrating the specificity of the interaction of PPR10 with atpH mRNA.
This analysis demonstrates that PPR proteins can bind to cosRNAs in vivo. cosRNAs constitute a substantial amount of the total population of RNA ligands of PPR10. It follows that a considerable fraction of all PPR10 proteins present in the chloroplast is absorbed by cosRNAs and is thus not available to stabilize mRNAs. This finding has obvious implications for the function of PPR proteins involved in RNA stabilization. The plant must provide sufficient PPR protein to stabilize the processed mRNAs, but is also required to express additional PPR proteins to account for titration by cosRNAs. An important parameter in these considerations is the on/off rate of the PPR–RNA complexes. We assume that the association with cosRNA and mRNAs follows similar biophysical properties, but evidence for this is currently lacking. Alternatively, a higher affinity of the PPR protein for mRNAs than cosRNAs could ensure that all mRNAs are covered and cosRNAs remain as secondary targets. Such a preference, however, is not consistent with the finding that PPR proteins prefer linear, unstructured targets (11), which is a more likely conformation for a short-footprint RNA than for a long, convoluted mRNA.
The PPR10 footprint does not appear to be exceptional in any respect, and is of intermediate abundance compared with other cosRNAs identified here (Supplementary Table S1). Furthermore, many, if not most PPR proteins are expected to bind to several RNAs (11) and are thus expected to cause multiple footprints of related, but not identical sequence. Some of these targets will be biologically meaningful, while others could be off-targets. Thus, off-targets like the abundant antisense RNAs in chloroplasts or fragments of rRNAs and tRNAs with sequence homology to PPR binding sites would further contribute to titrating a PPR. The efficiency of titration would depend on the on/off-rate of a PPR protein–RNA pair. In sum, on the basis of these findings and considerations, we hypothesize here that cosRNAs could act as competitors with mRNAs for PPR proteins and related proteins. A future test of this hypothesis would be to overexpress the PPR10 footprint or other cosRNAs using a transplastomic approach and monitor the effect on cognate mRNA abundance. cosRNAs in plant organelles are just emerging as potential players in organellar gene regulation.
AVAILABILITY
sRNA miner is implemented in R/Bioconductor (27), and is freely available at GitHub repository (https://github.com/MolGen/sRNAminer) for non-commercial use.
Supplementary Material
Acknowledgments
The gift of PPR10 antisera by Alice Barkan is gratefully acknowledged.
Footnotes
Present address: Sandra Gusewski, Department of Genetics, Friedrich Schiller University Jena, Philosophenweg 12, 07743 Jena, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [SCHM1698/5-1 to C.S.L.]. Funding for open access charge: Deutsche Forschungsgemeinschaft [SCHM1698/5-1 to C.S.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Barkan A. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 2011;155:1520–1532. doi: 10.1104/pp.110.171231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammani K., Giege P. RNA metabolism in plant mitochondria. Trends Plant. Sci. 2014;19:380–389. doi: 10.1016/j.tplants.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Holec S., Lange H., Kuhn K., Alioua M., Borner T., Gagliardi D. Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol. Cell. Biol. 2006;26:2869–2876. doi: 10.1128/MCB.26.7.2869-2876.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hotto A.M., Schmitz R.J., Fei Z., Ecker J.R., Stern D.B. Unexpected diversity of chloroplast noncoding RNAs as revealed by deep sequencing of the arabidopsis transcriptome. G3. 2011;1:559–570. doi: 10.1534/g3.111.000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharwood R.E., Halpert M., Luro S., Schuster G., Stern D.B. Chloroplast RNase J compensates for inefficient transcription termination by removal of antisense RNA. RNA. 2011;17:2165–2176. doi: 10.1261/rna.028043.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoppel R., Meurer J. The cutting crew - ribonucleases are key players in the control of plastid gene expression. J. Exp. Bot. 2012;63:1663–1673. doi: 10.1093/jxb/err401. [DOI] [PubMed] [Google Scholar]
- 7.Germain A., Hotto A.M., Barkan A., Stern D.B. RNA processing and decay in plastids. Wiley Interdiscip. Rev. RNA. 2013;4:295–316. doi: 10.1002/wrna.1161. [DOI] [PubMed] [Google Scholar]
- 8.Perrin R., Meyer E.H., Zaepfel M., Kim Y.J., Mache R., Grienenberger J.M., Gualberto J.M., Gagliardi D. Two exoribonucleases act sequentially to process mature 3′-ends of atp9 mRNAs in Arabidopsis mitochondria. J. Biol. Chem. 2004;279:25440–25446. doi: 10.1074/jbc.M401182200. [DOI] [PubMed] [Google Scholar]
- 9.Germain A., Kim S.H., Gutierrez R., Stern D.B. Ribonuclease II preserves chloroplast RNA homeostasis by increasing mRNA decay rates, and cooperates with polynucleotide phosphorylase in 3′ end maturation. Plant J. 2012;72:960–971. doi: 10.1111/tpj.12006. [DOI] [PubMed] [Google Scholar]
- 10.Pfalz J., Bayraktar O.A., Prikryl J., Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009;28:2042–2052. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barkan A., Small I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014;65:415–442. doi: 10.1146/annurev-arplant-050213-040159. [DOI] [PubMed] [Google Scholar]
- 12.Loizeau K., Qu Y., Depp S., Fiechter V., Ruwe H., Lefebvre-Legendre L., Schmitz-Linneweber C., Goldschmidt-Clermont M. Small RNAs reveal two target sites of the RNA-maturation factor Mbb1 in the chloroplast of Chlamydomonas. Nucleic Acids Res. 2014;42:3286–3297. doi: 10.1093/nar/gkt1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruwe H., Schmitz-Linneweber C. Short non-coding RNA fragments accumulating in chloroplasts: footprints of RNA binding proteins? Nucleic Acids Res. 2012;40:3106–3116. doi: 10.1093/nar/gkr1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhelyazkova P., Hammani K., Rojas M., Voelker R., Vargas-Suarez M., Borner T., Barkan A. Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res. 2012;40:3092–3105. doi: 10.1093/nar/gkr1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prikryl J., Rojas M., Schuster G., Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. U.S.A. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luro S., Germain A., Sharwood R.E., Stern D.B. RNase J participates in a pentatricopeptide repeat protein-mediated 5′ end maturation of chloroplast mRNAs. Nucleic Acids Res. 2013;41:9141–9151. doi: 10.1093/nar/gkt640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meierhoff K., Felder S., Nakamura T., Bechtold N., Schuster G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15:1480–1495. doi: 10.1105/tpc.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammani K., Cook W.B., Barkan A. RNA binding and RNA remodeling activities of the half-a-tetratricopeptide (HAT) protein HCF107 underlie its effects on gene expression. Proc. Natl. Acad. Sci. U.S.A. 2012;109:5651–5656. doi: 10.1073/pnas.1200318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haili N., Arnal N., Quadrado M., Amiar S., Tcherkez G., Dahan J., Briozzo P., Colas des Francs-Small C., Vrielynck N., Mireau H. The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Res. 2013;41:6650–6663. doi: 10.1093/nar/gkt337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitz R.J., Schultz M.D., Lewsey M.G., O'Malley R.C., Urich M.A., Libiger O., Schork N.J., Ecker J.R. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 22.Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M., et al. The arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicol J.W., Helt G.A., Blanchard S.G., Jr, Raja A., Loraine A.E. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu C., Meyers B.C., Green P.J. Construction of small RNA cDNA libraries for deep sequencing. Methods. 2007;43:110–117. doi: 10.1016/j.ymeth.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 29.Voelker R., Barkan A. Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J. 1995;14:3905–3914. doi: 10.1002/j.1460-2075.1995.tb00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kupsch C., Ruwe H., Gusewski S., Tillich M., Small I., Schmitz-Linneweber C. Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps. Plant Cell. 2012;24:4266–4280. doi: 10.1105/tpc.112.103002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitz-Linneweber C., Williams-Carrier R.E., Williams-Voelker P.M., Kroeger T.S., Vichas A., Barkan A. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell. 2006;18:2650–2663. doi: 10.1105/tpc.106.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 33.Hashimoto M., Endo T., Peltier G., Tasaka M., Shikanai T. A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 2003;36:541–549. doi: 10.1046/j.1365-313x.2003.01900.x. [DOI] [PubMed] [Google Scholar]
- 34.Michalovova M., Vyskot B., Kejnovsky E. Analysis of plastid and mitochondrial DNA insertions in the nucleus (NUPTs and NUMTs) of six plant species: size, relative age and chromosomal localization. Heredity. 2013;111:314–320. doi: 10.1038/hdy.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin X., Kaul S., Rounsley S., Shea T.P., Benito M.I., Town C.D., Fujii C.Y., Mason T., Bowman C.L., Barnstead M., et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- 36.Kasschau K.D., Fahlgren N., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Huang C.Y., Ayliffe M.A., Timmis J.N. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature. 2003;422:72–76. doi: 10.1038/nature01435. [DOI] [PubMed] [Google Scholar]
- 39.Stegemann S., Hartmann S., Ruf S., Bock R. High-frequency gene transfer from the chloroplast genome to the nucleus. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8828–8833. doi: 10.1073/pnas.1430924100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hegeman C.E., Halter C.P., Owens T.G., Hanson M.R. Expression of complementary RNA from chloroplast transgenes affects editing efficiency of transgene and endogenous chloroplast transcripts. Nucleic Acids Res. 2005;33:1454–1464. doi: 10.1093/nar/gki286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hotto A.M., Huston Z.E., Stern D.B. Overexpression of a natural chloroplast-encoded antisense RNA in tobacco destabilizes 5S rRNA and retards plant growth. BMC Plant Biol. 2010;10:213. doi: 10.1186/1471-2229-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharwood R.E., Hotto A.M., Bollenbach T.J., Stern D.B. Overaccumulation of the chloroplast antisense RNA AS5 is correlated with decreased abundance of 5S rRNA in vivo and inefficient 5S rRNA maturation in vitro. RNA. 2011;17:230–243. doi: 10.1261/rna.2336611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L., Yu X., Wang H., Lu Y.Z., de Ruiter M., Prins M., He Y.K. A novel class of heat-responsive small RNAs derived from the chloroplast genome of Chinese cabbage (Brassica rapa) BMC Genomics. 2011;12:289. doi: 10.1186/1471-2164-12-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogel A., Schilling O., Späth B., Marchfelder A. The tRNase Z family of proteins: physiological functions, substrate specificity and structural properties. Biol. Chem. 2005;386:1253–1264. doi: 10.1515/BC.2005.142. [DOI] [PubMed] [Google Scholar]
- 46.Barkan A., Rojas M., Fujii S., Yap A., Chong Y.S., Bond C.S., Small I. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8:e1002910. doi: 10.1371/journal.pgen.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forner J., Weber B., Thuss S., Wildum S., Binder S. Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res. 2007;35:3676–3692. doi: 10.1093/nar/gkm270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hauler A., Jonietz C., Stoll B., Stoll K., Braun H.P., Binder S. RNA Processing Factor 5 is required for efficient 5′ cleavage at a processing site conserved in RNAs of three different mitochondrial genes in Arabidopsis thaliana. Plant J. 2013;74:593–604. doi: 10.1111/tpj.12143. [DOI] [PubMed] [Google Scholar]
- 49.Kuhn K., Weihe A., Borner T. Multiple promoters are a common feature of mitochondrial genes in Arabidopsis. Nucleic Acids Res. 2005;33:337–346. doi: 10.1093/nar/gki179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giege P., Hoffmann M., Binder S., Brennicke A. RNA degradation buffers asymmetries of transcription in Arabidopsis mitochondria. EMBO Rep. 2000;1:164–170. doi: 10.1093/embo-reports/kvd024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raynaud C., Loiselay C., Wostrikoff K., Kuras R., Girard-Bascou J., Wollman F.-A., Choquet Y. Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc. Natl. Acad. Sci. U.S.A. 2007;104:9093–9098. doi: 10.1073/pnas.0703162104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Francs-Small C.C., Kroeger T., Zmudjak M., Ostersetzer-Biran O., Rahimi N., Small I., Barkan A. A PORR domain protein required for rpl2 and ccmF(C) intron splicing and for the biogenesis of c-type cytochromes in Arabidopsis mitochondria. Plant J. 2012;69:996–1005. doi: 10.1111/j.1365-313X.2011.04849.x. [DOI] [PubMed] [Google Scholar]
- 53.Holzle A., Jonietz C., Torjek O., Altmann T., Binder S., Forner J. A RESTORER OF FERTILITY-like PPR gene is required for 5′-end processing of the nad4 mRNA in mitochondria of Arabidopsis thaliana. Plant J. 2011;65:737–744. doi: 10.1111/j.1365-313X.2010.04460.x. [DOI] [PubMed] [Google Scholar]
- 54.Jonietz C., Forner J., Hildebrandt T., Binder S. RNA PROCESSING FACTOR3 is crucial for the accumulation of mature ccmC transcripts in mitochondria of Arabidopsis accession Columbia. Plant Physiol. 2011;157:1430–1439. doi: 10.1104/pp.111.181552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jonietz C., Forner J., Holzle A., Thuss S., Binder S. RNA PROCESSING FACTOR2 is required for 5′ end processing of nad9 and cox3 mRNAs in mitochondria of Arabidopsis thaliana. Plant Cell. 2010;22:443–453. doi: 10.1105/tpc.109.066944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Regina T.M.R., Lopez L., Picardi E., Quagliariello C. Striking differences in RNA editing requirements to express the rps4 gene in magnolia and sunflower mitochondria. Gene. 2002;286:33–41. doi: 10.1016/s0378-1119(01)00802-2. [DOI] [PubMed] [Google Scholar]
- 57.Blattner F.R., Plunkett G., Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 58.Okimoto R., Macfarlane J.L., Wolstenholme D.R. Evidence for the frequent use of TTG as the translation initiation codon of mitochondrial protein genes in the nematodes, Ascaris suum and Caenorhabditis elegans. Nucleic Acids Res. 1990;18:6113–6118. doi: 10.1093/nar/18.20.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marchive C., Yehudai-Resheff S., Germain A., Fei Z., Jiang X., Judkins J., Wu H., Fernie A.R., Fait A., Stern D.B. Abnormal physiological and molecular mutant phenotypes link chloroplast polynucleotide phosphorylase to the phosphorus deprivation response in Arabidopsis. Plant Physiol. 2009;151:905–924. doi: 10.1104/pp.109.145144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lurin C., Andres C., Aubourg S., Bellaoui M., Bitton F., Bruyere C., Caboche M., Debast C., Gualberto J., Hoffmann B., et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmitz-Linneweber C., Williams-Carrier R., Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast Pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi X., Hanson M.R., Bentolila S. Two RNA recognition motif-containing proteins are plant mitochondrial editing factors. Nucleic Acids Res. 2015;43:3814–3825. doi: 10.1093/nar/gkv245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takenaka M., Verbitskiy D., Zehrmann A., Härtel B., Bayer-Császár E., Glass F., Brennicke A. RNA editing in plant mitochondria—connecting RNA target sequences and acting proteins. Mitochondrion. 2014;19 Pt B:191–197. doi: 10.1016/j.mito.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 64.Stern D.B., Gruissem W. Control of plastid gene expression: 3′ inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987;51:1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- 65.Jiao H.S., Hicks A., Simpson C., Stern D.B. Short dispersed repeats in the Chlamydomonas chloroplast genome are collocated with sites for mRNA 3′ end formation. Curr. Genet. 2004;45:311–322. doi: 10.1007/s00294-004-0487-7. [DOI] [PubMed] [Google Scholar]
- 66.Li C., Zhang B. MicroRNAs in control of plant development. J. Cell. Physiol. 2016;231:303–313. doi: 10.1002/jcp.25125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.