Abstract
Although RNA-Seq data provide unprecedented isoform-level expression information, detection of alternative isoform regulation (AIR) remains difficult, particularly when working with an incomplete transcript annotation. We introduce JunctionSeq, a new method that builds on the statistical techniques used by the well-established DEXSeq package to detect differential usage of both exonic regions and splice junctions. In particular, JunctionSeq is capable of detecting differential usage of novel splice junctions without the need for an additional isoform assembly step, greatly improving performance when the available transcript annotation is flawed or incomplete. JunctionSeq also provides a powerful and streamlined visualization toolset that allows bioinformaticians to quickly and intuitively interpret their results. We tested our method on publicly available data from several experiments performed on the rat pineal gland and Toxoplasma gondii, successfully detecting known and previously validated AIR genes in 19 out of 19 gene-level hypothesis tests. Due to its ability to query novel splice sites, JunctionSeq is still able to detect these differences even when all alternative isoforms for these genes were not included in the transcript annotation. JunctionSeq thus provides a powerful method for detecting alternative isoform regulation even with low-quality annotations. An implementation of JunctionSeq is available as an R/Bioconductor package.
INTRODUCTION
In 2015 alone, hundreds of research papers have reported differential gene expression (DGE) based on RNA-Seq data (1–10). In general, RNA-Seq studies focus primarily on detecting gene-wide effects, in which entire genes are upregulated or downregulated depending on some experimental or biological condition. Although the statistical methodologies have advanced considerably, these studies generally follow the same basic design principles as previous microarray-based expression research.
However, RNA-Seq data provide more than simple measurements of gene-level expression. In theory, RNA-Seq can be used to study more complex regulatory phenomena at the isoform level, even when the isoforms in question are unannotated. Numerous tools have been developed to detect alternative isoform regulation (AIR; also known as differential transcript usage or DTU) (11–19); however, only a few of these tools have seen serious application outside their respective methodology papers. Many RNA-Seq studies do not even attempt the detection of AIR (1–10), with a few notable exceptions (14,20,21).
Detecting alternative isoform regulation is inherently difficult in RNA-Seq, as sequencer reads are often one or more orders of magnitude shorter than the transcripts themselves. While there are several utilities that attempt to de-convolute read data into isoform abundances, the accuracy and robustness of these methods is difficult to establish (22,23). Isoform expression estimates seem to vary considerably between different tools, and generally depend on the quality and completeness of the transcript assembly (24,25). Most of the newest and most popular tools do not assess or model unannotated isoforms (including eXpress (26), RSEM (27) or Kallisto (unpublished)), and the presence of such isoforms can substantially alter the estimated abundances of the known isoforms belonging to a gene. Thus: accurate analysis of differential isoform regulation may be very difficult when the available transcript annotation is incomplete.
In addition, almost all existing AIR analysis tools share one common shortcoming: the results are difficult to interpret. This is not a trivial issue: unlike simple DGE, instances of AIR cannot be adequately characterized by a single fold change and P-value. Alternative isoform regulation is a broad and diverse class of phenomena that can involve alternative splice sites, alternative promoters, nucleosome occupancy, cassette exons, alternative donors/acceptors, long non-coding RNAs, alternative polyadenylation, gene-level differential expression, or any number of these factors in combination. A gene may be composed of dozens of distinct isoforms, each controlled by its own set of regulatory mechanisms. As a consequence, the raw results of alternative isoform regulation analyses are often counterintuitive and resistant to interpretation.
The interpretation of these results is critical: in order to be considered credible by the community, any detected instances of AIR will generally require validation by secondary methods (such as qRT-PCR or SMRT sequencing). Such validation is often costly and time consuming. Detecting the mere presence of an effect is insufficient for these purposes: the investigator must also be able to identify which specific isoforms are being differentially used and assess the strength, direction and credibility of the effect. Furthermore, since hundreds of AIR genes may be detected, this interpretation process must be streamlined, scalable, and intuitive.
The DEXSeq software package tests for differential usage of exonic regions as a proxy for alternative isoform regulation, and provides a powerful suite of visualization tools (18,28). However, DEXSeq is only effective at detecting AIR when it results in changes in the expression of the annotated exonic regions. This method has two major weaknesses: firstly, it does not query all forms of alternative isoform regulation, as not all forms of AIR necessarily produce differentials in the exon counts (for two illustrative examples of hypothetical scenarios in which AIR does not result in large exon count differences, see Supplemental Figures S14 and S15). Secondly, this method is strongly dependent on the reference annotation, and cannot directly identify differences in novel exons and splice junctions. Additionally, DEXSeq output plots often obscure vital information, particularly for genes with a large number of isoforms and splice variants.
Here, we introduce JunctionSeq, a new method and associated Bioconductor package that builds upon the popular and well-established DEXSeq methodology in order to detect differential usage of both exons and known or novel splice junctions. Unlike most similar tools, JunctionSeq can reliably detect alternative isoform regulation even when the alternatively regulated isoforms themselves are not annotated. JunctionSeq also provides improved visualization tools that produce readable and informative expression profiles across all potential genes of interest, and across the genome as a whole.
MATERIALS AND METHODS
Differential usage of exons and junctions
Alternative isoform regulation is a biological phenomenon in which specific isoforms belonging to a multi-isoform gene are differentially regulated relative to one another with respect to some biological condition (18). Equivalently, it can be defined as a difference in the isoform fractions between different biological conditions (29,30).
Estimating the true isoform abundance of overlapping multi-kb transcripts using hundred-base-pair reads is an inherently difficult and error-prone task, particularly when some of the isoforms are not known a priori. As a consequence, it is difficult to detect AIR directly. Rather than attempting to directly detect AIR using estimates of isoform abundances, we instead attempt to detect differentials in quantities that are directly observable: the read counts for exonic regions and splice junction loci.
‘Differential usage’ (DU) of exons and junctions is an observed phenomenon in which individual exons or splice junctions display expression that is inconsistent with that of the gene as a whole. This can sometimes be counterintuitive: if a gene is differentially expressed, an individual sub-component that displays constant expression across all samples might be considered ‘differentially used’, as its expression is not consistent with that of the gene. Differential usage of exons and junctions thus serves as a proxy for the detection of AIR.
Testing for differential usage of splice junctions has a number of benefits. Firstly: all major aligners designed for RNA-Seq will align across novel (unannotated) splice junctions (31), and thus we can include novel splicing variants when they splice to/from known genes. This allows us to indirectly query for differential regulation in unknown isoforms, improving performance on sparsely annotated genomes. Furthermore, some forms of AIR do not necessarily result in observable differences in the exon-level counts. An intron retention, for example, will alter splice junction counts but not the counts of the flanking exons. As a result, our method substantially broadens the variety of regulatory phenomena that can be effectively detected. See Supplemental Figures S14 and S15 for two scenarios in which exon counts alone cannot be used to adequately characterize cases of alternative isoform regulation.
Statistical methodology
Like the DEXSeq Bioconductor package, we first partition each gene into a set of mutually non-overlapping exonic regions, and then use the read (or read-pair) counts for each exonic region to estimate the relative expression of each exon for each experimental condition (18). Unlike the DEXSeq package, we also calculate counts for each splice junction belonging to each gene, including novel splice junctions that are within the gene's span that surpass a user-specified normalized mean coverage threshold (we recommend 1–3 reads per sample). We use the DESeq2 package along with a set of specialized multivariate generalized linear models (GLM) to individually test for differential usage of each exonic region and splice junction (28). It should be noted that the arbitrary threshold used to ‘detect’ novel junctions is only used to determine whether such junctions will be assigned unique identifiers and be included in the count tables. JunctionSeq then uses the ‘automatic independent filtering’ method proposed and implemented by DESeq2 to determine which features should be filtered prior to hypothesis testing. If desired, the initial filtering threshold can be set to 0 to include all observed junctions at this initial step.
Previous studies have done similar analyses simply by plugging splice junction counts into DEXSeq (32), however, we found this method to be inadequate as it did not account for the numerous differences in the distribution and structure of the splice junction count data. A number of modifications to the basic DEXSeq methodology were found to be necessary.
To begin with: for most datasets DEXSeq will double- or triple-counts reads in each hypothesis test, as it uses the sum of all exonic regions as a proxy for estimating gene-level expression. This would be even more pronounced in JunctionSeq, and while it would not technically invalidate the hypothesis tests it can bias the fold-change estimates by over-weighting variant-dense regions, producing confusing artifacts under certain conditions. Thus, we use gene-level counts as the basis for our estimates of gene-wide expression rather than the sum of all exonic regions. This means that in the JunctionSeq framework no read or read-pair is counted more than once in any given statistical model. See section 2.1.1 in the supplement for an in-depth, illustrated explanation of the two counting methods. These altered count vectors were applied to both the hypothesis test and the effect estimation steps. For similar reasons, our size factor estimation is carried out using the gene-level counts rather than the exon/junction counts.
In addition, we found that splice junctions and exonic regions generally followed different dispersion trends from one another (see supplemental methods, Supplemental Table S4). This is not surprising, given the various biological and technical differences between the two count types. To account for this difference, JunctionSeq (by default) fits separate dispersion trends for exonic regions and for splice junctions. As in DEXSeq, the final dispersion estimates used for hypothesis testing are calculating by estimating the maximum a priori (MAP) dispersions for each exon and junction, which ‘shrinks’ each feature-specific dispersion estimate towards its respective fitted dispersion estimate (28).
In our RSEM simulations analyses (see Results), we found that (like almost all differential isoform usage tools) JunctionSeq appears to suffer from inflated false discovery rates (29). The precise cause of this issue is unclear, but after investigating this in detail (see results) we found that the false discovery rates can be greatly reduced by combining two options available in JunctionSeq which restrict hypothesis testing to splice junctions only and deactivate the maximum a posteriori dispersion estimation. Under this mode, the dispersions are instead calculated by taking the simple maximum of the fitted and unshared dispersions. This combination of options is referred to as the ‘SJ+noMAP’ mode in Figures 1 and 2.
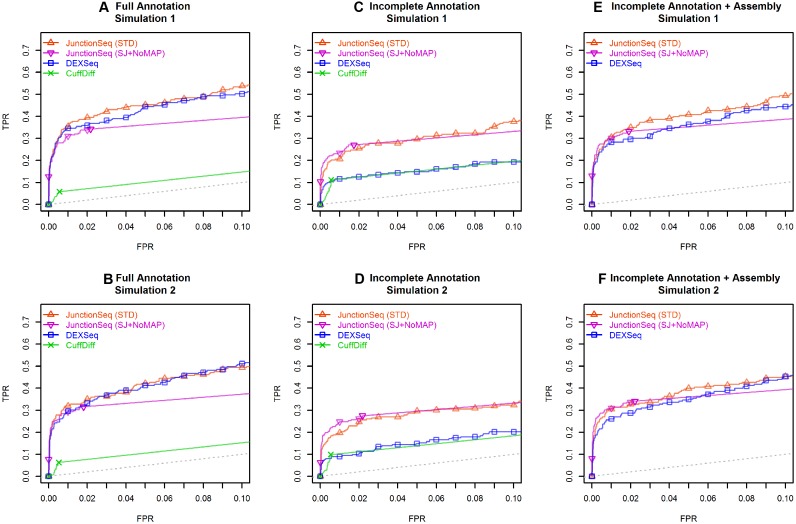
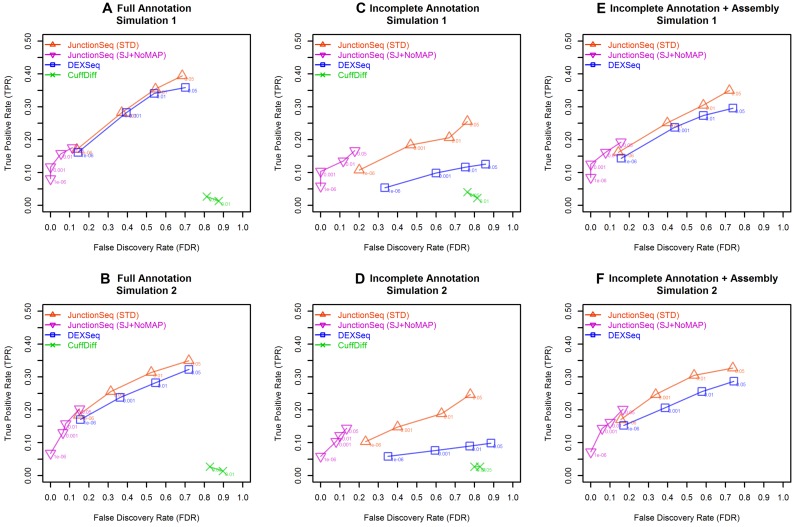
Figure 1.
ROC curves for JunctionSeq, DEXSeq, and CuffDiff for the two simulated datasets. This plot indicates how well each tool discriminates AIR genes from non-AIR genes. Plots A–C and D–F show the results for simulated datasets 1 and 2, respectively. The y-axis is the true positive rate (TPR, # true AIR genes detected / total # AIR genes), and the x-axis indicates false positive rate (FPR, # non-AIR genes detected AIR / total # of non-AIR genes). The ROC curve indicates the TPR/FPR over all possible adjusted P-value thresholds. Plots (A) and (B) show the results using the full annotation. (B) and (C) show the results using the incomplete annotation, and (E) and (F) show the results for DEXSeq and JunctionSeq using the incomplete annotation along with CuffLinks-assembled splice junctions and exonic regions. Two lines are drawn for JunctionSeq, displaying the results for a standard JunctionSeq run with the standard options (red), and for a secondary analysis with more conservative settings (only query splice junctions, do not use the maximum a posteriori dispersion estimates). Note that when provided with the complete annotation, both JunctionSeq and DEXSeq are able to discriminate AIR genes with approximately the same efficacy. However, when provided with the reduced annotation, with or without a CuffLinks assembly, DEXSeq has weaker discrimination than JunctionSeq. CuffDiff demonstrates low discrimination in all tests. The full-range ROC curves are available in the online supplement (see Supplemental Figure S20).
For a complete description of the JunctionSeq methodology, see the supplemental methods online.
The interpretation problem
Most existing AIR utilities provide little-to-no functionality to assist the end-user in the interpretation of the results. Some tools provide basic analysis-wide summary plots (12,18,33) and/or expression profile plots for individual samples (12,14,34–38), but very few provide methods for directly comparing gene expression profiles between multi-sample experimental groups. Many tools provide little information to the user beyond a text file of raw test statistics (11,13,15–17).
The DEXSeq visualization toolset, while unparalleled in its class, was found to be insufficient for our purposes (18). DEXSeq generates a number of gene profile plots that show read/read-pair coverage across each exonic region, plotted above a representation of the isoform annotation (see Figure 4b). However, genes vary widely in the number of exons and isoforms they possess, and as a result these plots vary widely in the complexity of the data they present. Consequently: regardless of the specific graphical settings, DEXSeq-generated plots often suffer from ‘over-plotting’, in which data are concealed by being drawn less than a pixel apart.
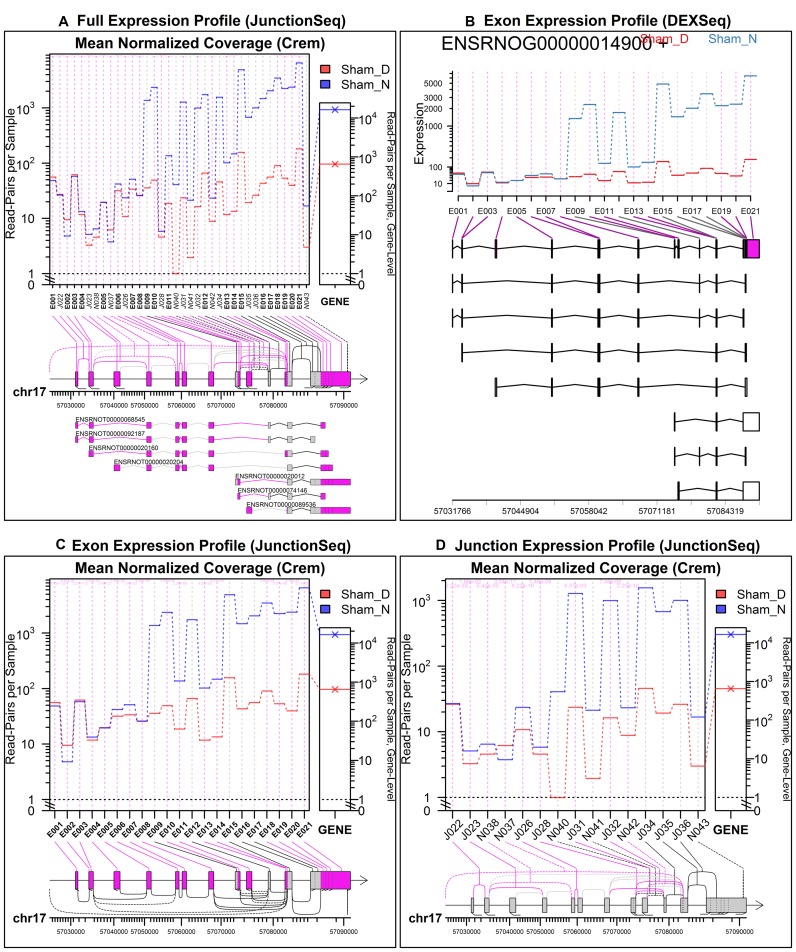
Figure 4.
Day/Night gene profile plots for the Crem gene in the rat pineal gland, sham-surgery group. Plots (A), (C) and (D) were produced by JunctionSeq, and (B) was produced by an equivalent analysis using DEXSeq. The full standard JunctionSeq gene profile plot (A) includes both exon and splice junction information. The equivalent DEXSeq plot (B) only displays exon information. Optionally, JunctionSeq can produce similar exon profile plots (C), or plots displaying only splice-junction information (D). Beneath the plotting regions in each figure a gene diagram displays the features’ positions on the genomic scale (note that small features are expanded for readability in the JunctionSeq versions). Novel junction loci are drawn using dashed lines. In the upper plots (A and B), all known transcripts are displayed beneath the main plotting area. Similar plots are available online for the control day/night comparison, as well as the two treated-vs-untreated comparisons (see Supplementary Figures S9–S11)
JunctionSeq implements a number of refinements designed to streamline and improve this process. Many parameters are automatically adjusted for each figure to improve readability, including adjustments to the feature label size and orientation, figure aspect ratio, relative size of the left and right panels, y-axis scaling, figure margins, and label positioning (see Figures 3, 4 and 6). Other improvements were added to make the plots more informative, including the nonlinear expansion of small features, highlighting of significant features, nested splice junction diagrams, and the inclusion of a gene-level expression plot. The various plots can either be viewed manually or browsed using a set of automatically-generated html pages, designed for easy navigation between genes and between experiments.
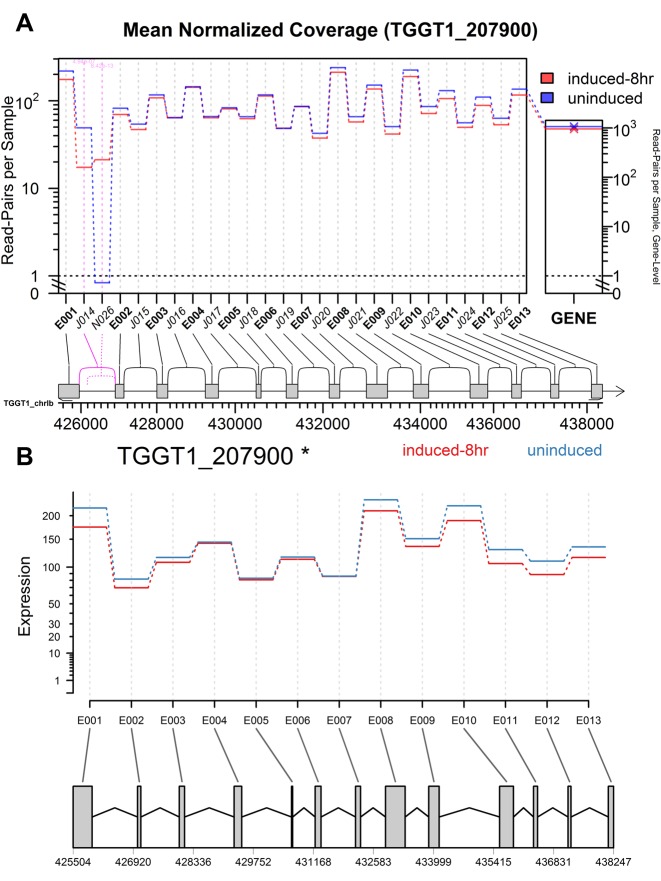
Figure 3.
Gene profile plots from (A) JunctionSeq and (B) DEXSeq for the TGGT1_207900 gene, in the 8-hour-induced vs un-induced Toxoplasma gondii experiment. The large central plotting panel of (A) and (B) displays the estimates for the mean normalized read counts for each exon or splice junction for the 8-hour-induced (red) or uninduced (blue) sample groups. The narrow panel on the right in (A) displays the gene-level mean normalized read counts. In each plot a gene diagram is drawn beneath the main plotting panels, showing the location and layout of the gene. Statistically significant (P-adjust < 0.01) exons or junctions are drawn with pink, and features that had counts that were too low to test are drawn in light gray (or they would be, if there were any such features). Known splice junctions are drawn with solid lines and unannotated splice junctions are drawn with dashed lines. Note in the JunctionSeq plot the first two splice junctions are strongly and significantly differentially used (in opposing directions). This effect was confirmed in a previous study via qRT-PCR (40). Also note that differential usage is not apparent in the DEXSeq plot, as the differentially used features are unannotated. Similar plots for the other two Toxoplasma gondii experiments can be found online, and show similar results (see Supplementary Figures S1 and S2).
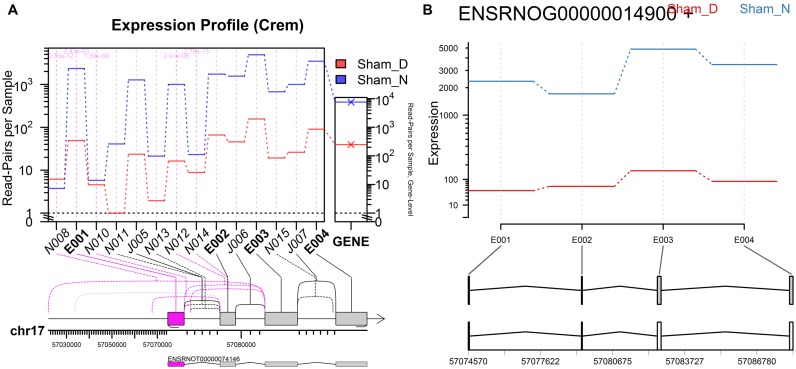
Figure 6.
Day/Night gene profile plots for the Crem gene, created by JunctionSeq (A) and DEXSeq (B), both using an incomplete transcript annotation. These plots are equivalent to Figure 4 (A) and (B), except that all the transcripts except one (transcript ENSRNOT00000074146) were removed from the annotation prior to analysis. Without the a priori knowledge of the missing transcripts, DEXSeq cannot reliably detect differential usage. Note that the ‘novel’ junction N010 is actually known junction J028 from Figure 4. Similarly, N014 is J032 and N015 is J035. The other novel junctions are not present even in the full annotation. It should be noted that exon E001 shows borderline statistical significance in the DEXSeq plot (P-adjust = 0.016).
While these features might seem cosmetic, they vastly improve the utility and scalability of this tool and allow investigators to quickly examine a large number of potential genes of interest in order to identify, characterize, and assess interesting biological phenomena.
The JunctionSeq analysis pipeline also generates genome-wide browser tracks suitable for use with IGV or the UCSC genome browser (See Figure 5). These tracks allow investigators to interactively browse expression profiles, splice junction counts, and statistically significant features across the entire genome, all alongside the numerous publicly available annotation tracks (38,39).
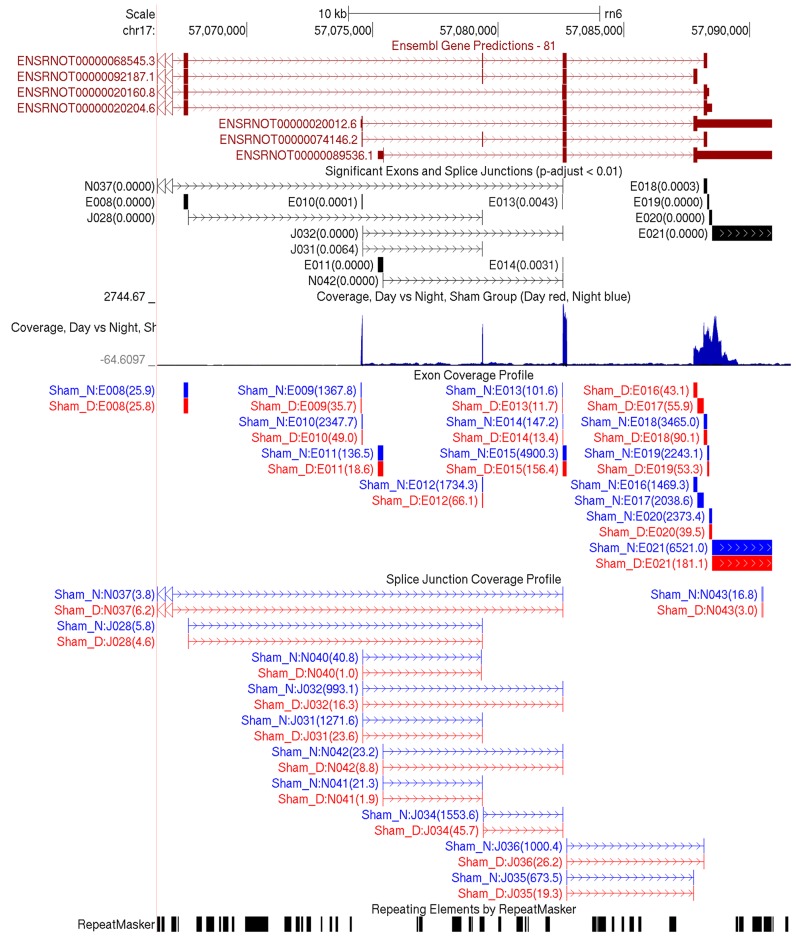
Figure 5.
Genome-wide Browser tracks produced in the QoRTs/JunctionSeq pipeline. The above screenshot displays much of the same information found in Figure 4, except using the UCSC genome browser. The top track displays the ensembl gene annotation. The second track displays the statistically significant features, with the adjusted P-value included in parentheses. The next track is a ‘wiggle’ track that displays coverage over both the forward and reverse strand (above and below the x-axis, respectively), in red and blue for day and night, respectively (overlap is colored black). The next two tracks display all exons and splice junctions, respectively, that were tested for DU by JunctionSeq. The day/night normalized mean expression values from Figure 4 are included in parentheses. The final track is from RepeatMasker, and displays regions with repeating or low-complexity elements. Using these tracks together can be vital for the purposes of interpretation and validation.
RESULTS
To demonstrate the strengths of our new method compared with other similar methods, we applied JunctionSeq, DEXSeq and CuffDiff to several datasets, including a simulated dataset generated via RSEM (27) as well as two publicly available (real) datasets with known and previously-validated AIR genes.
The simulated dataset included four separate analyses, two containing 250 simulated AIR genes and two null comparisons with no AIR genes. The first real dataset was in Toxoplasma gondii and included 3 analyses; the second was in rat pineal glands and included four analyses. Both real datasets included known and validated AIR genes, one gene in the Toxoplasma gondii dataset and four genes in the rat pineal gland dataset. Thus, there were a total of 19 gene-level hypothesis tests in which we expected to detect differential usage, acting as positive controls in our analysis.
JunctionSeq consistently detected differential usage in AIR genes across all experiments, even when the alternative isoforms were not included in the transcript annotation. When the annotation was incomplete, JunctionSeq substantially outperformed both DEXSeq and CuffDiff.
Test dataset 1: RSEM simulations
Our simulation methodology was loosely based on a recent review paper of several alternative isoform regulation methods (29). Briefly: twelve samples were simulated on the human transcriptome, six cases and six controls. Expression levels were assigned randomly at the gene level based on the coverage distribution curve of the rat pineal gland. Then 500 randomly selected genes were assigned gene-level differential expression and 250 genes were assigned alternative isoform regulation across a spectrum of effect sizes. To simulate the case in which a complete transcript annotation is not available, we generated a ‘reduced’ annotation in which the annotation for each of the 250 AIR genes were cut down to only the most highly expressed respective transcripts. For more information, see the supplemental methods. It should be noted that these simulated datasets had fewer and more modest case/control differences compared with the simulations in (29).
Four analyses were run for each analysis tool. First: two ‘DU’ tests (in which there was known differential usage) composed of three cases and three controls compared to one another. Then two 3 vs 3 ‘Null’ analyses were performed, comparing cases to cases and controls to controls.
We found that JunctionSeq detected far more genes in the ‘DU’ analyses than in the ‘Null’ analyses, both with and without the complete annotation (see Supplemental Figures S16 and S17). JunctionSeq also displayed strong discrimination between AIR and non-AIR genes (see Figure 1). Additional plots produced by the simulations analysis are available in the online supplement (see Supplemental Figures S16–S24). Interestingly, we found that the majority of the false discoveries were caused by exonic regions, not splice junctions (see Supplemental Figures S16 and S17).
Excess false discovery rates
A recent publication (29) reported that practically all AIR/DTU detection tools (including DEXSeq) return inflated P-values when run on RSEM simulation data. The cause of this phenomenon is unclear, but we have replicated these findings in our own simulations using different simulation parameters (see Figure 2). Although the JunctionSeq false discovery rate (FDR) was higher than the reported levels, it was as good as or better than the DEXSeq and CuffLinks results, particularly when running on the incomplete transcript annotation (see Figure 2C and D). Note that the false discovery rates in these simulated datasets are substantially worse than the real detection rates in the DCN and SCGX rat pineal experiments (see results for test dataset 3), possibly suggesting that the RSEM simulation methodology exaggerates this issue.
Figure 2.
True/False detection rates for JunctionSeq, DEXSeq, and CuffDiff for the two simulated datasets, at various adjusted-P-value cutoffs. This plot is analogous to Figure 2 in a recently published paper that compared various AIR detection tools (29). The y-axis indicates the true positive rate (TPR: # true AIR genes detected / total # of AIR genes), and the x-axis indicates the false discovery rate (FDR: # genes detected that are NOT truly AIR/total # of significant genes). Labelled points are placed at four selected adjusted P-value thresholds (P-adjust < 0.05, 0.01, 0.001 and 1e−6). Note that JunctionSeq is consistently superior to DEXSeq, particularly when the transcript annotation is incomplete. In all cases and cutoffs, CuffDiff performs worse than both JunctionSeq and DEXSeq.
A disproportionate number of the false discoveries were the result of exonic regions rather than splice junctions (see Supplemental Figures S16 and S17). As a result, the inflated false discovery rates seem to be less pronounced when JunctionSeq is restricted to testing splice junctions only (see Supplemental Figures S21–S23). The false discovery rates were also reduced by deactivating the maximum a posteriori (MAP) estimation of the final dispersion, and instead using a simple maximum of the unshared and fitted dispersion estimates (see Supplemental Figure S24). Combining both optional alternatives reduces the FDR even further; this combination of options is referred to as ‘SJ+noMAP’ in Figures 1 and 2. Using these options, JunctionSeq produces much more conservative adjusted-P-values which are much closer to the true FDR (see Figure 2), while still maintaining high-end discrimination that is comparable to the standard JunctionSeq method (see Figure 1; Supplementary Figures S18 and S24). It should be noted that previous versions of DEXSeq did not use the MAP dispersions, but this is no longer a supported option in the current version.
JunctionSeq offers these options (and numerous others) to end-users to apply as they see fit.
Test dataset 2: Toxoplasma gondii and TgSR3
Our first real test dataset originated from a previous study in which alternative splicing was detected and validated in Toxoplasma gondii between control samples and samples in which overexpression of the TgSR3 gene was induced (40). There were four sample groups of 3 biological replicates each: untreated; induced, 4 h; induced, 8 h and induced, 24 h. The dataset is available from the NCBI short read archive (SRA), accession number PRJNA252680.
In the original study, numerous genes were found to display differential splicing between the induced and untreated sample groups. One particular gene, TGGT1_207900, was found to display strong differential splicing across an unannotated 5′ variant in all three comparisons. This effect was detected using DEXSeq via a CuffLinks assembly, and was subsequently confirmed via qRT-PCR. In order to demonstrate JunctionSeq's ability to detect differential usage of novel variants, we performed the same analysis using JunctionSeq, but without the benefit of the CuffLinks assembly step.
Summary plots for these analyses are available online (see Supplemental Figures S3 and S4).
Detection of AIR without CuffLinks assembly
Even without a complete transcript assembly, JunctionSeq detected differential usage of the previously-validated novel splice variant in TGGT1_207900 in all three experiments, with adjusted P-values of 0.00023, 8.5 × 10−13, and 0.0098 for the untreated versus 4-h, 8-h and 24-h experiments, respectively. The gene profile plots clearly displayed the same form of differential splicing found in the original experiment (see Figure 3 and Supplemental Figures S1 and S2) (40).
This demonstrates that JunctionSeq can accurately detect differential usage in novel splicing variants, and does not require a complete and comprehensive transcript annotation in order to detect alternative isoform regulation.
Even when run using the much more conservative splice-junctions-only/no-MAP-dispersion mode, JunctionSeq still detects significant differential usage in two of the three experiments (see Supplemental Figure S27).
Test dataset 3: circadian Rhythms in the rat pineal gland
The rat pineal gland is known to display strong and consistent differential expression resulting from neural stimulation across hundreds or thousands of genes (41,42). Most if not all of these changes are believed to be controlled via neural innervation of the pineal gland by the SCG, using the neurotransmitter norepinephrine (NE) and the second messenger cyclic AMP (cAMP) (43–50).
Several genes have already been found in the literature to exhibit neurally-controlled alternative isoform usage in the rat pineal gland: Crem (51–53), Pde4b (54), Atp7b (55) and Slc15a1 (formerly known as Pept1) (56,57). It should be noted that all of these genes were discovered in previous studies using different datasets, and all are validated and well-established in the literature.
We performed four comparisons in which we expected to detect differential splicing in genes that are neurally controlled by norepinephrine and cAMP: two in vivo analyses comparing night and day conditions in no-surgery (Ctrl) and sham-surgery (Sham) rats, as well as two in vitro analyses comparing pineal glands in organ culture that had been treated with norepinephrine (NE) or dibutyryl cyclic AMP (DBcAMP, an analog of the second messenger, cyclic AMP), each versus an untreated control set (CN). Given that the four known-AIR genes are neurally controlled, we expect to detect differential usage in all four genes across all four comparisons.
In addition, we performed similar analyses one two datasets in which neural stimulation of the pineal gland was eliminated via decentralization (DCN) or removal (SCGX) of the superior cervical ganglia (SCG). We expected that most of the night/day differences would be eliminated in these analyses, providing an upper bound for the false discovery rate.
Summary plots for these analyses are available online (see Supplementary Figures S5 and S6).
Detection of known AIR genes
For the four known AIR genes, we found strong genome-wide statistical significance for all 16 gene-level hypothesis tests (see Table 2). The genes Crem, Pde4b and Atp7b were detected by JunctionSeq at an extremely high significance level in all analyses (P-adjust < 1e−8 for all three genes and all four comparisons), and the gene Slc15a1 was detected at a moderately high significance level in all analyses (P-adjust < 0.01). See Figure 4a for an example plot displaying the JunctionSeq results for the Crem gene in the sham-surgery group.
Table 2. JunctionSeq gene-level adjusted P-values for four known-AIR genes in the rat pineal gland, both with and without a complete isoform annotation.
| Gene Symbol | Full Annotation | Incomplete Annotation (1 ‘known’ isoform) | ||||||
|---|---|---|---|---|---|---|---|---|
| Ctrl day/night | Sham day/night | CN versus NE | CN versus DBcAMP | Ctrl day/night | Sham day/night | CN versus NE | CN versus DBcAMP | |
| Atp7b | <1e−8 | <1e−8 | <1e−8 | <1e−8 | <1e−8 | <1e−8 | <1e−8 | <1e−8 |
| Crem | <1e−8 | <1e−8 | <1e−8 | <1e−8 | <1e−8 | <1e−8 | <1e−8 | <1e−8 |
| Pde4b | <1e−8 | <1e−8 | <1e−8 | <1e−8 | <1e−8 | <1e−8 | <1e−8 | 2.8e−7 |
| Slc15a1 | <1e−8 | <1e−8 | 0.0034 | 0.0015 | <1e−8* | <1e−8* | 0.0034* | 0.0016* |
The left four columns display the results from a normal analysis, the right four columns display the results from an analysis in which all but one isoform was removed from the annotation for each gene, simulating a scenario in which the gene is poorly studied and the annotation incomplete. *Note: since the Slc15a1 gene actually only has one known transcript, the ‘full’ and ‘incomplete’ annotation analyses for this gene are equivalent, differing only slightly due to minor analysis-wide differences in the dispersion estimation and multiplicity correction.
Using the much more conservative splice-junctions-only/no-MAP-dispersion mode the JunctionSeq results are more modest, with 13 out of 16 tests showing statistically significant differential usage (P-adjust < 0.01; see Supplemental Table S2).
Differential usage of novel variants
To demonstrate JunctionSeq's ability to detect differential usage of novel splice junctions even with an incomplete transcript assembly, we performed a second set of analyses with a reduced annotation. For each of the three known AIR genes that had multiple annotated transcripts (Crem, Pde4b and Atp7b), we manually removed all but one transcript from the ensembl annotation GTF and then re-ran the analyses. This was intended to simulate the scenario in which AIR occurs in poorly-annotated genes. The gene Slc15a1 only has one transcript in the current annotation, and thus the annotation was left unchanged.
Even with only one annotated transcript, JunctionSeq was still able to detect differential usage of ‘novel’ splice sites for all four genes across all four comparisons (P-adjust < 0.01, see the right half of Table 2). See Figure 6A for an example plot displaying the incomplete-annotation JunctionSeq results for the Crem gene in the sham-surgery group.
Using the much more conservative splice-junctions-only/no-MAP-dispersion mode, 12 out of 16 tests still show statistically significant differential usage (P-adjust < 0.01; see Supplemental Table S2).
Replicability and consistency:
In addition to confirming known AIR genes and providing a strong positive control for JunctionSeq, we can further use these analyses to demonstrate the reliability and replicability of our methods by examining the overlap between the four comparisons.
While these experiments are not direct replications, isoforms whose regulation is controlled specifically by neural innervation of the pineal gland through the SCG (via norepinephrine and cyclic AMP) should theoretically exhibit similar expression regulation across all four experiments.
In each comparison hundreds of genes were found to display statistically significant differential exon or splice-junction usage (at P-adjust < 0.01), and 42 of these genes displayed differential usage in all four analyses (see Table 1 and Supplemental Figure S7). The strong concordance between the four experiments spanning very different (but biologically related) experimental conditions demonstrates that JunctionSeq produces consistent and replicable results.
Table 1. JunctionSeq results.
| Adjusted P-value threshold | In vivo | In vitro | No stimulus (in vivo) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ctrl day/night | Sham day/night | Overlap, in vivo | CN versus NE | CN versus DBcAMP | Overlap, in vitro | Overlap, All four | SCGX Day/Night | DCN day/night | Overlap, no stimulus | |
| 0.01 | 447 | 320 | 168 | 144 | 195 | 90 | 42 | 38 | 9 | 2 |
| 0.001 | 300 | 202 | 116 | 89 | 127 | 61 | 28 | 24 | 5 | 0 |
| 0.0001 | 227 | 151 | 94 | 67 | 91 | 48 | 18 | 20 | 2 | 0 |
| 0.00001 | 182 | 119 | 79 | 51 | 74 | 38 | 15 | 14 | 2 | 0 |
| 0.000001 | 151 | 98 | 61 | 43 | 65 | 34 | 14 | 11 | 2 | 0 |
The numbers of genes found to exhibit significant differential exon or splice junction usage for the four rat pineal gland analyses at various P-value thresholds.
False discovery rate
In the SCGX and DCN experiments, we found that the night/day differences were greatly reduced, particularly in the DCN experiment. In the SCGX group only 38 genes were found with statistically significant differential usage at the P-adjust < 0.01 level, and in the DCN group only nine genes were found (see Table 1). The majority of these detected genes showed only moderate-to-weak statistical significance and small fold-changes (see Table 1, Supplemental Figure S5).
Not all of these detected genes are necessarily false discoveries: a small number of genes are known to display night/day differential regulation in the pineal gland in SCGX rats due to circulating catecholamines (42,58,59). These circulating catecholamines would theoretically be blocked from stimulating the pineal gland in the DCN rats (42).
Although these analyses are not perfect negative controls, they do provide a rough upper bound to the false discovery rate. The fact that the DCN analysis found so few statistically significant effects demonstrates that false discoveries are relatively rare, and furthermore, the fact that none of the detected genes replicated in both SCGX and DCN experiments at high significance levels (P-adjust < 0.001) suggests that high significance thresholds and proper replication should effectively eliminate false discoveries.
Even fewer genes appear statistically significant in the DCN and SCGX analyses using the splice-junctions-only/no-MAP-dispersion options, further reinforcing the evidence that these options make JunctionSeq less prone to false discovery, albeit at the cost of statistical power (see Supplementary Table S3).
Comparison with existing tools
Comparisons between differential isoform regulation tools are difficult, as many are actually designed to detect subtly distinct phenomena. As a consequence: even if both tools perform with perfect accuracy they may still return different results. CuffDiff, for example, performs several separate tests of transcript switching and alternative promoter usage for each gene. Other transcript-alignment-based tools like eXpress (26), RSEM (27) or Kallisto/Sleuth (unpublished, preprint available at http://arxiv.org/abs/1505.02710) only detect overall differential expression of individual transcripts and do not attempt to detect differential usage of transcripts relative to one another. Thus, results from these tools would likely consist predominantly of differentially expressed genes, and would not specifically target differential splicing. Furthermore, most such tools are strongly annotation-dependent and do not attempt to assess novel splice variants.
The obvious comparison, however, is with the DEXSeq software tool (18). We found that when all affected isoforms are known, JunctionSeq and DEXSeq seem to perform with similar efficacy. However, when unannotated isoforms are involved, JunctionSeq demonstrates clear superiority due to its ability to query unannotated splice junctions.
With or without the complete annotation, both JunctionSeq and DEXSeq outperformed CuffDiff, which failed to detect any differentials in the rat pineal gland and in the simulations data produced higher false discovery rates and much lower true positive rates.
Toxoplasma gondii analyses
Without a CuffLinks assembly, DEXSeq was unable to detect any differential usage in the validated gene (TGGT1_207900) in any of the three Toxoplasma gondii analyses (see Figure 3b and Supplemental Figures S1b and S2b). JunctionSeq, however, detects the differential usage of the alternative start site in all three analyses, even without the CuffLinks assembly (see Figure 3a and Supplemental Figures S1a and S2a).
Rat pineal gland analyses
In the rat pineal gland data, JunctionSeq and DEXSeq seemed to perform similarly when the full transcript annotation was used (see Table 1, Supplemental Table S1, and Supplemental Figures S7–S11). Across all experiments JunctionSeq detected at least as many statistically significant genes in each experiment individually and found more genes that overlapped between all four analyses.
For the four known-AIR genes, DEXSeq and JunctionSeq returned very similar results when the full annotation was used, although JunctionSeq reported slightly weaker significance for the gene Slc15a1 (see Tables 2 and 3).
Table 3. DEXSeq gene-level adjusted P-values for four known-AIR genes in the rat pineal gland, both with and without a complete isoform annotation.
| Gene symbol | Full annotation | Incomplete annotation (1 ‘known’ isoform) | ||||||
|---|---|---|---|---|---|---|---|---|
| Ctrl Day/Night | Sham Day/Night | CN vs NE | CN vs DBcAMP | Ctrl Day/Night | Sham Day/Night | CN vs NE | CN vs DBcAMP | |
| Atp7b | <1e-8 | <1e-8 | <1e-8 | <1e-8 | <1e-8 | <1e-8 | <1e-8 | <1e-8 |
| Crem | <1e-8 | <1e-8 | <1e-8 | <1e-8 | 1.2e-5 | 0.0357 | 6.5e-6 | 9.3e-5 |
| Pde4b | <1e-8 | <1e-8 | <1e-8 | <1e-8 | 1.2e-4 | 0.0041 | 1.0 | 1.0 |
| Slc15a1 | <1e-8 | <1e-8 | 3.3e-5 | 2.7e-5 | <1e-8* | <1e-8* | 3.2e-5* | 3.0e-5* |
See Table 2. Note that without the complete annotation, several tests do not show significant differential usage or have much less significant P-values.
When the transcript annotation was incomplete, DEXSeq fails to detect differQ2QQential usage in 3 of the 16 tests, one for Crem and two for Pde4b (at P-adjust < 0.01, see Table 3). JunctionSeq, on the other hand, still reports differential usage in all 16 tests (see Table 2). Furthermore, although the other five DEXSeq tests for the genes Crem and Pde4b are still statistically significant at P-adjust < 0.01, all of the reported P-values are several orders of magnitude weaker than those found in either the corresponding JunctionSeq analyses or the corresponding full-annotation DEXSeq analyses.
Even with the complete annotation, CuffDiff failed to detect any isoform switching or differential promoter usage in any of the four known-AIR genes across any of the four analyses (see Table 4).
Table 4. CuffDiff results for the four known AIR genes in the rat pineal gland, using the full annotation.
| Output file | Gene symbol | # Tests for gene | Experiments (full annotation) | |||
|---|---|---|---|---|---|---|
| Ctrl day/night | Sham day/night | CN versus NE | CN versus DBcAMP | |||
| Splicing.diff | Atp7b | 4 | 1 | 1 | 1 | 1 |
| Crem | 9 | 0.99991 | 0.99991 | 0.99991 | 0.9999 | |
| Pde4b | 30 | 0.99991 | 0.99991 | 0.99991 | 0.1444 | |
| Slc15a1 | 3 | 1 | 1 | 1 | 1 | |
| Promoters.diff | Atp7b | 1 | 0.13110 | 1 | 1 | 1 |
| Crem | 1 | 0.99991 | 0.99991 | 0.99991 | 0.99991 | |
| Pde4b | 1 | 1 | 0.99991 | 1 | 1 | |
| Slc15a1 | 1 | 1 | 1 | 1 | 1 | |
| cds.diff | Atp7b | 1 | 1 | 1 | 1 | 1 |
| Crem | 1 | 0.99808 | 0.99862 | 0.99988 | 0.99896 | |
| Pde4b | 1 | 0.99808 | 0.99862 | 0.99988 | 0.99896 | |
| Slc15a1 | 1 | 1 | 1 | 1 | 1 | |
The CuffDiff analysis design differs somewhat from that of DEXSeq or JunctionSeq. CuffDiff peforms separate analyses testing for isoform switching (splicing.diff), differences in CDS expression (cds.diff), and differential promoter usage (promoters.diff), producing several tests for each gene. The results for the four known-AIR genes are shown below. When an analysis file contained multiple tests for a given gene, the most significant adjusted P-value is shown. (Note: CuffDiff was not tested on the incomplete annotation set.)
Simulated datasets
We ran JunctionSeq, DEXSeq, and the full CuffLinks/CuffDiff pipeline using both the full and incomplete annotations. For JunctionSeq and DEXSeq we also ran an analysis using the set of exons and splice junctions discovered in the CuffLinks assembly on the incomplete annotation. We found that when the annotation was complete, JunctionSeq and DEXSeq once again seemed to perform with approximately equal effectiveness (see Figures 1 and 2, Supplemental Figure S20). However, when the annotation was incomplete, JunctionSeq provided clearly superior performance in both AIR-gene discrimination (see Figure 1) and control of the false discovery rate (Figure 2). Both the AIR-gene discrimination and FDR control of CuffDiff was substantially worse than either method, with or without the complete annotation.
Even when CuffLinks is used to recover some of the absent exonic regions and splice junctions in the incomplete annotation, JunctionSeq still visibly outperforms DEXSeq in both simulated datasets.
Additional efficacy metrics and summary plots comparing the three tools under various options and conditions are available in the online supplement (see Supplemental Figures S16–S24).
It is important to note that since AIR/DTU is actually a broad category of related phenomena, the particular results from any given simulation analysis will not necessarily generalize to all actual datasets. The relative efficacy of these methods will depend strongly on numerous factors, including the annotation, genome build, tissue type, read length, and organism. In addition: efficacy may vary depending on the nature of the actual biological phenomena that are occurring in each particular experiment. For example: in one previous study (29), CuffDiff displayed a much lower FDR, most likely due to the presence of a very large number of highly expressed genes with extremely strong differential usage.
The results of all three test datasets lead us to the same conclusion: when the transcript annotation is complete and comprehensive, DEXSeq and JunctionSeq produce similar results. However, when novel isoforms are involved JunctionSeq provides a clear improvement over other methods.
Other advantages of JunctionSeq
The improved visualization tools provided by JunctionSeq further increase its utility. In general, simply detecting the presence of AIR is insufficient; the investigator must also be able to determine precisely which isoforms or splice variants are responsible for the apparent differences. In many cases, DEXSeq detects differential usage in the same genes as JunctionSeq, however, even when manually examining the DEXSeq plots it is often impossible to identify the specific splice variants that are being differentially expressed, particularly when the relevant exons or splice junctions are unannotated.
For example, in the Crem gene there are several clusters of small exonic regions (E009–E010, E013–E015, E017–E021, see Figure 4) that are completely indistinguishable in the DEXSeq gene/transcript diagram due to ‘over-plotting’, in which such features are plotted less than a pixel apart (see bottom of Figure 4b). These same features, however, can be easily identified and matched to their corresponding isoforms in the JunctionSeq plot, due to the nonlinear expansion of small features (see bottom of Figure 4A and C). Other visualization tools, like the IGV browser views and ‘sashimi’ plots, often suffer from similar issues (see Supplemental Figures S25 and S26).
Similarly, when novel isoforms are involved, it is often impossible to identify the relevant splicing variants in the DEXSeq plots, even when statistical significance is detected in the gene. This is because DEXSeq will often detect the indirect effects of alternative isoform usage, but the causal variants themselves will remain obscured.
For example, in the incomplete-annotation analysis shown in Figure 6b, the first exon (E001) actually displays borderline statistical significance in the DEXSeq analysis (P-adjust = 0.016), due to the fact that this exon is not present in the (unobserved) alternative isoforms. However, even if this is considered significant it is impossible to identify the actual variants responsible for this effect, as they are not directly observable in the DEXSeq plots. The JunctionSeq plots, on the other hand, clearly show the source of the differential usage in the various ‘novel’ splice junctions, most of which lead to the upstream alternative promoter site.
If desired, JunctionSeq can (optionally) run pure exon-based analyses, reducing the number of comparisons (see Supplemental Figures S21–S23). One of the major strengths of JunctionSeq is that it queries a broader array of regulatory phenomena, however, this comes at the cost of additional comparisons and potentially reducing power. Running a purely exon-based analysis may provide superior results when working with a well-characterized tissue on a comprehensively annotated genome. In fact, with a certain set of options (documented in the user manual), JunctionSeq will precisely reproduce a standard DEXSeq analysis while still providing the user with the enhanced visualization tools of JunctionSeq. To demonstrate the advantages of the JunctionSeq plotting engine we plotted identical analyses run by JunctionSeq and DEXSeq for a large and complex human gene using simulated data (see Supplemental Figures S12 and S13).
Example interpretation
For the purposes of demonstration we will examine a well-known AIR gene, Crem, in the rat pineal gland dataset. The mechanism behind the circadian alternative isoform regulation of the Crem gene is already well understood, and the patterns of expression of this gene's various isoforms are well-characterized (51,52,60). Briefly, an internal promoter is greatly upregulated at night, resulting in large quantities of a number of small transcripts collectively known as ICER. ICER is known to play a major role in the melatonin synthesis pathway (61).
By default, JunctionSeq automatically generates gene profile plots for every gene that contains one or more differentially used exon or splice junction. Figure 4 (A–D) displays a few of the available plots in the sham-surgery night/day experiment.
As seen in the small rightmost panel of each JunctionSeq plot (i.e. the narrow panels labelled ‘GENE’), the Crem gene as a whole appears to display strong upregulation at night (∼15 000 versus ∼650 read-pairs per sample). Looking at the gene profile plots we can see that this is not uniform across the gene: some of the exonic regions and splice junctions display strong upregulation at night while others do not. Exonic regions E009 through E021 all display strong differentials (>8×, see Figure 4C), but exonic regions E001 through E008 display consistently low counts at both day and night. The splice junction plot (see Figure 4D) shows similar results for the splice junction coverage.
It may seem counterintuitive that the constant-expression exons (E001–E008) are marked as statistically significant. This is because JunctionSeq (like DEXSeq) tests for differential usage, not differential expression. The expression of each sub-feature is compared with the expression of the gene as a whole (see the rightmost panel of each JunctionSeq plot in Figure 4). Since the gene as a whole has strong differential expression, exonic regions and splice junctions that do not display such differentials are considered ‘differentially used’ relative to the gene.
Using the genome browser tracks produced in the QoRTs/JunctionSeq pipeline (Figure 5), we can examine the read coverage across the genome and over all known and novel splice junctions. These can be examined alongside external annotation tracks such as the RepeatMasker track or the UCSC-maintained EST and mRNA databases. Visual examination alongside these tracks can be critical, as it can determine whether novel splice variants have been previously detected, or if apparent difference might be the result of alignment artifacts or flaws in the annotation.
Taken together, these visualizations lead towards a clear and obvious hypothesis: the full-length isoforms of the Crem gene display constant low-level expression at day and night, whereas the isoforms originating in the internal (‘ICER’) promoter are greatly upregulated at night. This ‘hypothesis’ matches the known behavior and function of this gene in the literature (51,52,60,61).
Similar plots are available online for the Crem gene in the other three rat pineal gland experiments (see Supplementary Figure S9–S11).
DISCUSSION
JunctionSeq offers a powerful, flexible, statistically robust and efficient solution for the identification, characterization, and interpretation of differential isoform regulation. The underlying methodology has a strong theoretical basis and is built upon established statistical methods that are already widely accepted by the community. It includes a number of powerful improvements that allow it to query a broader class of regulatory phenomena, including the differential regulation of novel splicing variants in the absence of an accurate and comprehensive transcript annotation.
This is a notable addition to the community, as DEXSeq cannot consistently detect differential transcript usage in novel transcripts, and since many popular tools such as eXpress (26), RSEM (27) or Kallisto (unpublished, preprint available at http://arxiv.org/abs/1505.02710) cannot assess novel variants at all. Furthermore, many transcript quantification tools seem to perform poorly when used with an incomplete transcript annotation (24). Although JunctionSeq may not necessarily provide uniform superiority over existing methods when the annotation is comprehensive, it provides a valuable tool for researchers studying esoteric tissues and/or less-common species.
Another major advantage of the JunctionSeq software toolset is its suite of powerful automated visualization and interpretation tools, which allow investigators to quickly and intuitively examine hundreds of genes. This assists investigators in identifying and characterizing genes of interest for further validation and study.
The JunctionSeq R package
We implemented the described method in a new Bioconductor package, JunctionSeq, written entirely in the R statistical programming language.
The JunctionSeq analysis pipeline requires the QoRTs quality-control/data-processing software package (62) in order to generate the raw gene, exon, and splice junction counts. QoRTs is also used to create the multi-sample normalized-mean ‘wiggle’ tracks for use with IGV or the UCSC genome browser.
The JunctionSeq package is extensively documented and includes a comprehensive walkthrough and example dataset, with line-by-line instructions describing the complete analysis pipeline. JunctionSeq will be included in Bioconductor release 3.3 (http://bioconductor.org/packages/JunctionSeq/), and is available now along with additional online help and documentation at the JunctionSeq GitHub page: http://hartleys.github.io/JunctionSeq/.
ACCESSION NUMBERS
The datasets used in the application sections are available from the NCBI short read archive (SRA), with accession numbers PRJNA267246 and PRJNA252680 for the Rattus norvegicus (42) and the Toxoplasma gondii (40) datasets, respectively.
Supplementary Material
Acknowledgments
We would like to thank Steven L. Coon and David C. Klein from the Eunice Kennedy Shriver National Institute of Child Health and Human Development for their input and assistance, particularly in generating and conceiving the rat pineal experiments.
We would also like to thank John Didion and Peter Chines from the Medical Genomics and Metabolic Genetics Branch at the National Human Genome Research Institute for their assistance in the testing and development of this software.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Human Genome Research Institute; National Institutes of Health. Funding for open access charge: Comparative Genomics Analysis Unit, Cancer Genomics and Comarative Genomics Branch, National Human Genome Research Institute, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nurnberg S.T., Cheng K., Raiesdana A., Kundu R., Miller C.L., Kim J.B., Arora K., Carcamo-Oribe I., Xiong Y., Tellakula N. Coronary artery disease associated transcription factor TCF21 regulates smooth muscle precursor cells that contribute to the fibrous cap. PLoS genetics. 2015;11:e1005155. doi: 10.1371/journal.pgen.1005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu J., Shao Z., Li D., Xie H., Kim W., Huang J., Taylor J.E., Pinello L., Glass K., Jaffe J.D. Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Mol. Cell. 2015;57:304–316. doi: 10.1016/j.molcel.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagenblast E., Soto M., Gutiérrez-Ángel S., Hartl C.A., Gable A.L., Maceli A.R., Erard N., Williams A.M., Kim S.Y., Dickopf S. A model of breast cancer heterogeneity reveals vascular mimicry as a driver of metastasis. Nature. 2015;520:358–362. doi: 10.1038/nature14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boj S.F., Hwang C.-I., Baker L.A., Chio I.I.C., Engle D.D., Corbo V., Jager M., Ponz-Sarvise M., Tiriac H., Spector M.S. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber D., Heisig J., Kneitz S., Wolf E., Eilers M., Gessler M. Mechanisms of epigenetic and cell-type specific regulation of Hey target genes in ES cells and cardiomyocytes. J. Mol. Cell. Cardiol. 2015;79:79–88. doi: 10.1016/j.yjmcc.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Bettigole S.E., Lis R., Adoro S., Lee A.-H., Spencer L.A., Weller P.F., Glimcher L.H. The transcription factor XBP1 is selectively required for eosinophil differentiation. Nat. Immunol. 2015;16:829–837. doi: 10.1038/ni.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai T., Jen H.-I., Kang H., Klisch T.J., Zoghbi H.Y., Groves A.K. Characterization of the Transcriptome of Nascent Hair Cells and Identification of Direct Targets of the Atoh1 Transcription Factor. J. Neurosci. 2015;35:5870–5883. doi: 10.1523/JNEUROSCI.5083-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park S.-M., Gönen M., Vu L., Minuesa G., Tivnan P., Barlowe T.S., Taggart J., Lu Y., Deering R.P., Hacohen N. Musashi2 sustains the mixed-lineage leukemia–driven stem cell regulatory program. J. Clin. Invest. 2015;125:1286–1298. doi: 10.1172/JCI78440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S., Kim C., Lee J.M., LEE H.A., Fei Z., Wang L., Apel K. Blocking the QB‐binding site of photosystem II by tenuazonic acid, a non–host‐specific toxin of Alternaria alternata, activates singlet oxygen‐mediated and EXECUTER‐dependent signalling in Arabidopsis. Plant Cell Environ. 2015;38:1069–1080. doi: 10.1111/pce.12462. [DOI] [PubMed] [Google Scholar]
- 10.Wong E.S., Thybert D., Schmitt B.M., Stefflova K., Odom D.T., Flicek P. Decoupling of evolutionary changes in transcription factor binding and gene expression in mammals. Genome Res. 2015;25:167–178. doi: 10.1101/gr.177840.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katz Y., Wang E.T., Airoldi E.M., Burge C.B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu J., Akerman M., Sun S., McCombie W.R., Krainer A.R., Zhang M.Q. SpliceTrap: a method to quantify alternative splicing under single cellular conditions. Bioinformatics. 2011;27:3010–3016. doi: 10.1093/bioinformatics/btr508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffith M., Griffith O.L., Mwenifumbo J., Goya R., Morrissy A.S., Morin R.D., Corbett R., Tang M.J., Hou Y.C., Pugh T.J., et al. Alternative expression analysis by RNA sequencing. Nat. Methods. 2010;7:843–847. doi: 10.1038/nmeth.1503. [DOI] [PubMed] [Google Scholar]
- 15.Shi Y., Jiang H. rSeqDiff: detecting differential isoform expression from RNA-Seq data using hierarchical likelihood ratio test. PLoS One. 2013;8:e79448. doi: 10.1371/journal.pone.0079448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh D., Orellana C.F., Hu Y., Jones C.D., Liu Y., Chiang D.Y., Liu J., Prins J.F. FDM: a graph-based statistical method to detect differential transcription using RNA-seq data. Bioinformatics. 2011;27:2633–2640. doi: 10.1093/bioinformatics/btr458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y., Huang Y., Du Y., Orellana C.F., Singh D., Johnson A.R., Monroy A., Kuan P.F., Hammond S.M., Makowski L., et al. DiffSplice: the genome-wide detection of differential splicing events with RNA-seq. Nucleic Acids Res. 2013;41:e39. doi: 10.1093/nar/gks1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anders S., Reyes A., Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alamancos G.P., Agirre E., Eyras E. Methods to study splicing from high-throughput RNA Sequencing data. Spliceosomal Pre-mRNA Splicing: Methods Protoc. 2014:357–397. doi: 10.1007/978-1-62703-980-2_26. [DOI] [PubMed] [Google Scholar]
- 20.Colla S., Ong D.S.T., Ogoti Y., Marchesini M., Mistry N.A., Clise-Dwyer K., Ang S.A., Storti P., Viale A., Giuliani N. Telomere dysfunction drives aberrant hematopoietic differentiation and myelodysplastic syndrome. Cancer Cell. 2015;27:644–657. doi: 10.1016/j.ccell.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandramohan R., Wu P.Y., Phan J.H., Wang M.D. Benchmarking RNA-Seq quantification tools. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2013;2013:647–650. doi: 10.1109/EMBC.2013.6609583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z.H., Jhaveri D.J., Marshall V.M., Bauer D.C., Edson J., Narayanan R.K., Robinson G.J., Lundberg A.E., Bartlett P.F., Wray N.R., et al. A comparative study of techniques for differential expression analysis on RNA-Seq data. PLoS One. 2014;9:e103207. doi: 10.1371/journal.pone.0103207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanitz A., Gypas F., Gruber A.J., Gruber A.R., Martin G., Zavolan M. Comparative assessment of methods for the computational inference of transcript isoform abundance from RNA-seq data. Genome Biol. 2015;16:1–26. doi: 10.1186/s13059-015-0702-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rehrauer H., Opitz L., Tan G., Sieverling L., Schlapbach R. Blind spots of quantitative RNA-seq: the limits for assessing abundance, differential expression, and isoform switching. BMC Bioinformatics. 2013;14:370. doi: 10.1186/1471-2105-14-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts A., Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soneson C., Matthes K.L., Nowicka M., Law C.W., Robinson M.D. Isoform prefiltering improves performance of count-based methods for analysis of differential transcript usage. Genome Biol. 2016;17:1. doi: 10.1186/s13059-015-0862-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engstrom P.G., Steijger T., Sipos B., Grant G.R., Kahles A., Ratsch G., Goldman N., Hubbard T.J., Harrow J., Guigo R., et al. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat. Methods. 2013;10:1185–1191. doi: 10.1038/nmeth.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Rao X., Mattox W.W., Amos C.I., Liu B. RNA-Seq analysis of differential splice junction usage and intron retentions by DEXSeq. PLoS One. 2015;10:e0136653. doi: 10.1371/journal.pone.0136653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goff L.A., Trapnell C., Kelley D. R Package Version 2.2. 2012. CummeRbund: visualization and exploration of Cufflinks high-throughput sequencing data. [Google Scholar]
- 34.Liu Q., Chen C., Shen E., Zhao F., Sun Z., Wu J. Detection, annotation and visualization of alternative splicing from RNA-Seq data with SplicingViewer. Genomics. 2012;99:178–182. doi: 10.1016/j.ygeno.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Ryan M.C., Cleland J., Kim R., Wong W.C., Weinstein J.N. SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts. Bioinformatics. 2012;28:2385–2387. doi: 10.1093/bioinformatics/bts452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aschoff M., Hotz-Wagenblatt A., Glatting K.H., Fischer M., Eils R., Konig R. SplicingCompass: differential splicing detection using RNA-seq data. Bioinformatics. 2013;29:1141–1148. doi: 10.1093/bioinformatics/btt101. [DOI] [PubMed] [Google Scholar]
- 37.Rogers M.F., Thomas J., Reddy A.S., Ben-Hur A. SpliceGrapher: detecting patterns of alternative splicing from RNA-Seq data in the context of gene models and EST data. Genome Biol. 2012;13:R4. doi: 10.1186/gb-2012-13-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeoh L.M., Goodman C.D., Hall N.E., van Dooren G.G., McFadden G.I., Ralph S.A. A serine–arginine-rich (SR) splicing factor modulates alternative splicing of over a thousand genes in Toxoplasma gondii. Nucleic Acids Res. 2015;43:4661–4675. doi: 10.1093/nar/gkv311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailey M.J., Coon S.L., Carter D.A., Humphries A., Kim J.S., Shi Q., Gaildrat P., Morin F., Ganguly S., Hogenesch J.B., et al. Night/day changes in pineal expression of >600 genes: central role of adrenergic/cAMP signaling. J. Biol. Chem. 2009;284:7606–7622. doi: 10.1074/jbc.M808394200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartley S.W., Coon S.L., Savastano L.E., Mullikin J.C., Program N.C.S., Fu C., Klein D.C. Neurotranscriptomics: the effects of neonatal stimulus deprivation on the rat pineal transcriptome. PLoS One. 2015;10:e0137548. doi: 10.1371/journal.pone.0137548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein D.C., Weller J.L., Moore R.Y. Melatonin metabolism: neural regulation of pineal serotonin: acetyl coenzyme A N-acetyltransferase activity. Proc. Natl. Aad. Sci. U.SA. 1971;68:3107–3110. doi: 10.1073/pnas.68.12.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bowers C.W., Dahm L.M., Zigmond R.E. The number and distribution of sympathetic neurons that innervate the rat pineal gland. Neuroscience. 1984;13:87–96. doi: 10.1016/0306-4522(84)90261-6. [DOI] [PubMed] [Google Scholar]
- 45.Bowers C.W., Zigmond R.E. The influence of the frequency and pattern of sympathetic nerve activity on serotonin N-acetyltransferase in the rat pineal gland. J. Physiol. 1982;330:279–296. doi: 10.1113/jphysiol.1982.sp014341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lingappa J.R., Zigmond R.E. A histochemical study of the adrenergic innervation of the rat pineal gland: evidence for overlap of the innervation from the two superior cervical ganglia and for sprouting following unilateral denervation. Neuroscience. 1987;21:893–902. doi: 10.1016/0306-4522(87)90045-5. [DOI] [PubMed] [Google Scholar]
- 47.Maronde E., Pfeffer M., von Gall C., Dehghani F., Schomerus C., Wicht H., Kroeber S., Olcese J., Stehle J.H., Korf H.W. Signal transduction in the rodent pineal organ. From the membrane to the nucleus. Adv. Exp. Med. Biol. 1999;460:109–131. doi: 10.1007/0-306-46814-x_14. [DOI] [PubMed] [Google Scholar]
- 48.Roseboom P.H., Klein D.C. Norepinephrine stimulation of pineal cyclic AMP response element-binding protein phosphorylation: primary role of a beta-adrenergic receptor/cyclic AMP mechanism. Mol. Pharmacol. 1995;47:439–449. [PubMed] [Google Scholar]
- 49.Maronde E., Schomerus C., Stehle J.H., Korf H.W. Control of CREB phosphorylation and its role for induction of melatonin synthesis in rat pinealocytes. Bio. Cell. 1997;89:505–511. doi: 10.1016/s0248-4900(98)80006-3. [DOI] [PubMed] [Google Scholar]
- 50.Tamotsu S., Schomerus C., Stehle J.H., Roseboom P.H., Korf H.W. Norepinephrine-induced phosphorylation of the transcription factor CREB in isolated rat pinealocytes: an immunocytochemical study. Cell and tissue research. 1995;282:219–226. doi: 10.1007/BF00319113. [DOI] [PubMed] [Google Scholar]
- 51.Foulkes N.S., Borjigin J., Snyder S.H., Sassone-Corsi P. Transcriptional control of circadian hormone synthesis via the CREM feedback loop. Proc. Natl. Acad. Sci. U.S.A. 1996;93:14140–14145. doi: 10.1073/pnas.93.24.14140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korf H.-W., Schomerus C., Maronde E., Stehle J. Signal transduction molecules in the rat pineal organ: Ca2+, pCREB, and ICER. Naturwissenschaften. 1996;83:535–543. doi: 10.1007/BF01141978. [DOI] [PubMed] [Google Scholar]
- 53.Schwartz W.J., Aronin N., Sassone-Corsi P. Photoinducible and rhythmic ICER-CREM immunoreactivity in the rat suprachiasmatic nucleus. Neurosci. Lett. 2005;385:87–91. doi: 10.1016/j.neulet.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 54.Kim J.-S., Bailey M.J., Ho A.K., Møller M., Gaildrat P., Klein D.C. Daily rhythm in pineal phosphodiesterase (PDE) activity reflects adrenergic/3′, 5′-cyclic adenosine 5′-monophosphate induction of the PDE4B2 variant. Endocrinology. 2007;148:1475–1485. doi: 10.1210/en.2006-1420. [DOI] [PubMed] [Google Scholar]
- 55.Borjigin J., Payne A.S., Deng J., Li X., Wang M.M., Ovodenko B., Gitlin J.D., Snyder S.H. A novel pineal night-specific ATPase encoded by the Wilson disease gene. J. Neurosci. 1999;19:1018–1026. doi: 10.1523/JNEUROSCI.19-03-01018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaildrat P., Møller M., Mukda S., Humphries A., Carter D.A., Ganapathy V., Klein D.C. A novel pineal-specific product of the oligopeptide transporter PepT1 gene circadian expression mediated by cAMP activation of an intronic promoter. J. Biol. Chem. 2005;280:16851–16860. doi: 10.1074/jbc.M414587200. [DOI] [PubMed] [Google Scholar]
- 57.Maronde E., Stehle J.H. The mammalian pineal gland: known facts, unknown facets. Trends Endocrinol. Metab. 2007;18:142–149. doi: 10.1016/j.tem.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 58.Sugden D., Klein D.C. Regulation of rat pineal hydroxyindole‐O‐methyltransferase in neonatal and adult rats. J. Neurochem. 1983;40:1647–1653. doi: 10.1111/j.1471-4159.1983.tb08138.x. [DOI] [PubMed] [Google Scholar]
- 59.Jimenez J., Osuna C., Reiter R.J., Rubio A., Guerrero J.M. Adrenalectomy or superior cervical ganglionectomy modifies the nocturnal increase in rat pineal type II thyroxine 5′-deiodinase. Chronobiol. Int. 1993;10:87–93. doi: 10.1080/07420529309059696. [DOI] [PubMed] [Google Scholar]
- 60.Stehle J.H., Foulkes N.S., Pevet P., Sassone-Corsi P. Developmental maturation of pineal gland function: synchronized CREM inducibility and adrenergic stimulation. Mol. Endocrinol. 1995;9:706–716. doi: 10.1210/mend.9.6.8592516. [DOI] [PubMed] [Google Scholar]
- 61.Foulkes N.S., Borjigin J., Snyder S.H. Rhythmic transcription: the molecular basis of circadian melatonin synthesis. Trends Neurosci. 1997;20:487–492. doi: 10.1016/s0166-2236(97)01109-0. [DOI] [PubMed] [Google Scholar]
- 62.Hartley S.W., Mullikin J.C. QoRTs: a comprehensive toolset for quality control and data processing of RNA-Seq experiments. BMC Bioinformatics. 2015;16:224. doi: 10.1186/s12859-015-0670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.