Abstract
Eukaryotic translation initiation is a highly regulated process involving multiple steps, from 43S pre-initiation complex (PIC) assembly, to ribosomal subunit joining. Subunit joining is controlled by the G-protein eukaryotic translation initiation factor 5B (eIF5B). Another protein, eIF1A, is involved in virtually all steps, including subunit joining. The intrinsically disordered eIF1A C-terminal tail (eIF1A-CTT) binds to eIF5B Domain-4 (eIF5B-D4). The ribosomal complex undergoes conformational rearrangements at every step of translation initiation; however, the underlying molecular mechanisms are poorly understood. Here we report three novel interactions involving eIF5B and eIF1A: (i) a second binding interface between eIF5B and eIF1A; (ii) a dynamic intramolecular interaction in eIF1A between the folded domain and eIF1A-CTT; and (iii) an intramolecular interaction between eIF5B-D3 and -D4. The intramolecular interactions within eIF1A and eIF5B interfere with one or both eIF5B/eIF1A contact interfaces, but are disrupted on the ribosome at different stages of translation initiation. Therefore, our results indicate that the interactions between eIF1A and eIF5B are being continuously rearranged during translation initiation. We present a model how the dynamic eIF1A/eIF5B interaction network can promote remodeling of the translation initiation complexes, and the roles in the process played by intrinsically disordered protein segments.
INTRODUCTION
Eukaryotic translation initiation is a multistep process involving ribosomes, mRNAs, tRNAs and a number of proteins called eukaryotic translation initiation factors (eIFs). The key stages of translation initiation are: (i) assembly of a 43S pre-initiation complex (43S PIC) on the small, 40S ribosomal subunit; (ii) binding of the 43S PIC to mRNA; (iii) scanning by the PIC along the mRNA in search of the start codon; (iv) start codon selection through basepairing of the initiator Met-tRNAi with mRNA to form the 48S PIC; and (v) ribosomal subunit joining, yielding the 80S initiation complex (80S IC). Start codon selection and ribosomal subunit joining are controlled by the G-proteins eIF2 and eIF5B, respectively, and require multiple proteins, including eIF1A, a protein with pleiotropic functions in virtually all stages of translation initiation (reviewed in (1–5)).
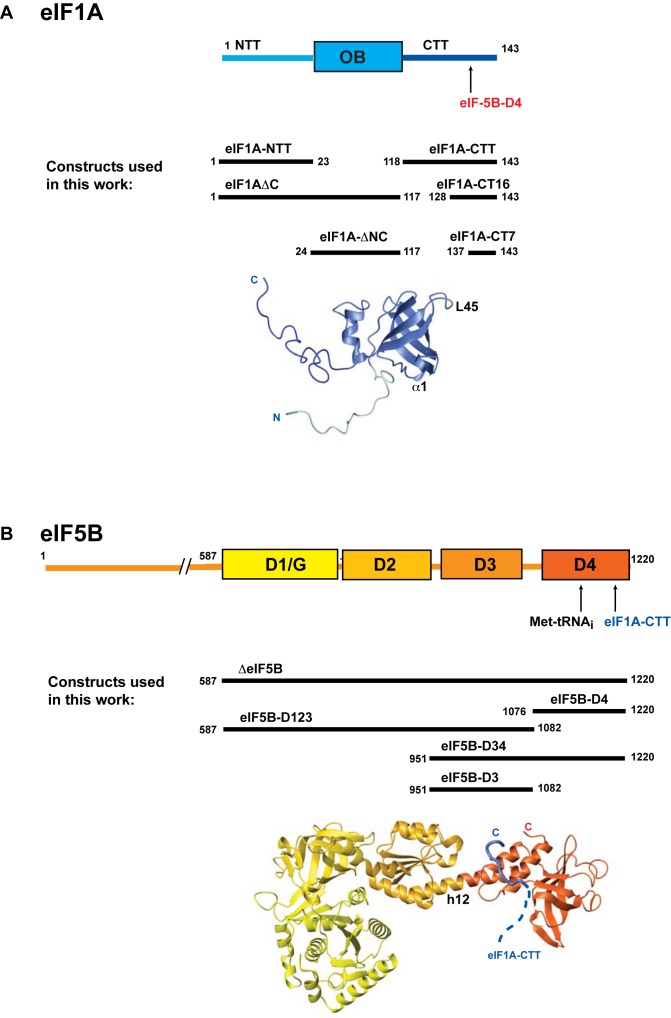
eIF1A has an oligonucleotide/oligosaccharide-binding (OB) fold domain and two flexible intrinsically disordered tails (6): a positively charged N-terminal tail (NTT) and a negatively charged C-terminal tail (CTT) (Figure 1A). eIF1A binds to the A-site on the 40S ribosomal subunit with its two tails extending into the P-site (7–10). Upon start codon recognition, eIF1A-NTT remains in the P-site while eIF1A-CTT is displaced (7,11). eIF5B contains four conserved domains in its C-terminal half (D1–D4) (Figure 1B), the first of which, D1 is the GTPase domain (12–14). The N-terminal region is less conserved and is not essential in vitro in mammalian or yeast systems, or in vivo in Saccharomyces cerevisiae (12,13). Upon start codon recognition, eIF5B displaces eIF2-GDP from Met-tRNAi (15) and promotes ribosomal subunit joining, together with eIF1A. Ribosomal subunit joining triggers GTP hydrolysis by eIF5B (13), followed by the coordinated release of eIF5B and eIF1A (16).
Figure 1.
Domain structure and interactions of eIF5B and eIF1A. (A) Top, domain structure of eIF1A. The binding site for eIF5B-D4 is labeled. Middle, constructs used in this work. Bottom, structure of human eIF1A (6). Helix α1 and loop L45, discussed in the text, are labeled. (B) Top, domain structure of eIF5B. The binding sites for eIF1A-CTT and Met-tRNAi are labeled. Middle, constructs used in this work. Bottom, structure of Saccharomyces cerevisiae eIF5B in complex with an eIF1A C-terminal peptide (34). Helix h12, discussed in the text, is labeled.
eIF1A-CTT binds to eIF5B-D4 (17,18), and the interaction is important for subunit joining and for release of eIF1A and eIF5B (16,19–20). The two proteins occupy adjacent sites on the ribosome, bringing eIF5B-D3 near the eIF1A-OB domain (7,21), but no interaction between these regions has been reported in eukaryotes. It has been reported, however, that bacterial IF1 and IF2 (the homologs of eIF1A and eIF5B, respectively), can be cross-linked to each other when part of the translation IC. The cross-linking was mapped to a segment encompassing IF2 domains D2 and D3 (22). It has been proposed, based on Cryo-EM data, that IF1 contacts the linker between IF2-D2 and D3 (23), whereas recent Cryo-EM reconstructions observe contacts between IF1 and IF2-D3 (24).
Intrinsically disordered proteins (IDPs) and regions (IDPRs) are unfolded and dynamic under native conditions. IDPs and IDPRs have been found to play important roles and have attracted increasing attention in recent years. IDPRs often contain binding sites for other proteins and ligands and can fold upon binding to their target. A less well known fact is that IDPRs can be involved in dynamic interactions while remaining unfolded and flexible (reviewed in (25–28)). We recently reported that in the bacteriophage T7 ssDNA-binding protein gp2.5, the negatively charged intrinsically disordered CTT dynamically contacts the DNA-binding surface and binds to the T7 DNA polymerase. Deletion of the tail increases the affinity for ssDNA and allows gp2.5 to also bind dsDNA (29). gp2.5 has an OB-fold domain with structure homology to the eIF1A OB domain. Sequence/structure-specific nucleic acid-binding proteins tend to have significant non-specific affinity for any nucleic acid. Thus, the intramolecular contacts in gp2.5 play a dual role: they reduce the non-specific, dsDNA-binding affinity, as well as coordinate ssDNA binding with binding of other proteins to the gp2.5 CTT (29). Similar to gp2.5, deleting the CTT of yeast eIF1A increases its affinity for the 40S ribosomal subunit (30), and eIF1A tends to self-associate at high protein concentrations at physiological salt, but not at high salt. These observations led to the hypotheses that the conserved intrinsically disordered eIF1A-CTT may contact the eIF1A ribosome-binding surface, and that these intramolecular interactions may modulate the interactions of eIF1A with other proteins.
Here we report that eIF1A-CTT does indeed dynamically contact the ribosome-binding surface of the eIF1A-OB domain. We also observed an intramolecular interaction between eIF5B-D3 and -D4. eIF5B-D3 binds weakly to eIF1A-OB, but only in the absence of eIF1A-CTT. The intramolecular interactions, both within eIF1A and within eIF5B, weaken the interaction between eIF1A-CTT and eIF5B-D4. Our results show that the affinity between eIF1A and eIF5B on the ribosome is much greater than that between the free proteins in solution. Since eIF1A and eIF5B bind to adjacent sites on the ribosome, their effective concentrations with respect to each other would also be increased, further stabilizing their interaction. We present a model for the interactions within and between eIF1A and eIF5B, their dynamic remodeling at various stages of translation initiation and their respective roles in the process.
MATERIALS AND METHODS
Protein expression and purification
All protein constructs are shown in Figure 1. The proteins were cloned in pET21a with an N-terminal GB1 tag, a His6-tag and a TEV protease cleavage site, and expressed in Bl21(DE3) cells. The eIF5B constructs were expressed at 20°C O/N and purified on a TALON Cell-Thru His-tag affinity column (Clontech) in buffer containing 20 mM Tris, pH 7.0, 300 mM KCl, 1 mM EDTA, 1 mM DTT and 0.1 mM AEBSF. The GB1 tag was cleaved using TEV and removed using IgG column. Ion exchange chromatography on a Uno Q or a Uno S column was used, where necessary, for additional purification. The eIF1A constructs were expressed at 37°C for 3 h, except eIF1AΔN and eIF1AΔNC, which were expressed at 20°C. The purification was the same as for eIF5B constructs, except 1 M NaCl was used for eIF1A, eIF1AΔN and eIF1AΔNC, and 150 mM KCl was used for eIF1A-CTT constructs. Fluorescein-labeled eIF1A-CT7 was chemically synthesized. For 15N-, 13C- and 2H-labeling, bacteria were grown on minimal medium supplemented with 15N-NH4Cl, 13C-glucose, and/or 2H2O, respectively.
NMR experiments
Nuclear magnetic resonance (NMR) experiments were performed in buffer containing 20 mM Tris, pH 7.0, 150 mM KCl, 1 mM EDTA, 1 mM DTT and 0.1 mM AEBSF, with 5% 2H2O, except where noted. NMR data were collected on a 500 MHz Bruker spectrometer (Boston University School of Medicine), an 800 MHz Bruker spectrometer (Brandeis University) and an 850 MHz Bruker spectrometer (Brown University), all equipped with cryoprobes.
Chemical shift perturbation assay
Backbone resonance assignments for eIF5B-D3 were obtained using standard triple-resonance experiments (reviewed in (31)) on 15N/13C-labeled samples at 500 mM NaCl. The backbone assignments for eIF1A and eIF5B-D4 have been published previously (6,17). 15N heteronuclear single-quantum coherence (HSQC) experiments on 15N-labeled proteins were used for NMR binding and deletion analysis for smaller proteins. Transverse relaxation optimized spectroscopy (TROSY)-HSQC on 15N/2H-labeled proteins was used for larger proteins and complexes. For binding experiments, a 15N-labeled protein sample was titrated with increasing concentrations of unlabeled binding partner, until saturation (where no further chemical shift changes are observed) or until the solubility limit of the unlabeled protein was reached. Chemical shift changes were calculated according to the formula δ = ((δH)2 + (δN/5)2)1/2 and affected residues were mapped on the surface of the protein. For NMR deletion analysis, the spectra of 15N-labeled full-length proteins were compared to the spectra of deletion mutants and analyzed as above.
Paramagnetic relaxation enhancement
Paramagnetic groups induce increased rates of relaxation (loss of signal) of nuclei located up to ∼25 Ǻ away, which can be converted into distance restraints for structure determination by NMR (32). For paramagnetic relaxation enhancement (PRE) experiments, WT proteins containing solvent-exposed cysteine side chains or engineered single-cysteine mutants were labeled with N-(1-Oxyl-2,2,6,6-tetramethyl-4-piperidinyl)maleimide (OPM) (Toronto Research Chemicals, Inc.) in degassed buffer in the absence of a reducing agent. OPM attaches to cysteine sulfhydryl groups through its maleimide group and the resulting bond is not reversible by reducing agents. 15N-labeled proteins were OPM-labeled for detection of intramolecular PRE effects. Unlabeled proteins were OPM-labeled and added to a 15N-labeled protein to detect intermolecular PRE effects in a complex. The spectra of the samples under oxidizing conditions, where OPM is paramagnetic, were compared to spectra under reducing conditions, where OPM is diamagnetic. Samples were reduced using 2 mM ascorbic acid. The intensities of each peak under oxidized and reducing conditions were quantified to identify nuclei that are near the paramagnetic center and experience faster relaxation. The intensity loss due to the PRE effect was converted into distance restraints, which were used for docking, as described (32).
Fluorescence anisotropy
Fluorescence anisotropy (FA) measurements were done on a QuantaMaster QM4 fluorescent spectrometer (PTI), equipped with polarizers and dual monochromators. eIF5B fragments were titrated into synthetic fluorescein-labeled eIF1A-CT7 peptide (Fl-eIF1A-CT7), to determine their KDs for the fluorescently labeled peptide. The increase in FA as a function of competitor concentration was recorded and used to fit the KD of the interaction. Competition assays were used to determine the KDs of unlabeled eIF1A and eIF1A fragments for eIF5B domains. Increasing concentrations of a competitor were added to a mixture of Fl-eIF1A-CT7 and the eIF5B fragment of interest, and the drop in FA as a function of competitor concentration was recorded and used to fit the KD of the interaction. The experiments were performed in a buffer containing 20 mM Tris–HCl, pH 7.0, 150 mM KCl, 1 mM EDTA, 1 mM DTT, 0.1 mM AEBSF, at 20°C. Data analysis was done in SigmaPlot.
RESULTS
Intramolecular interactions within eIF1A
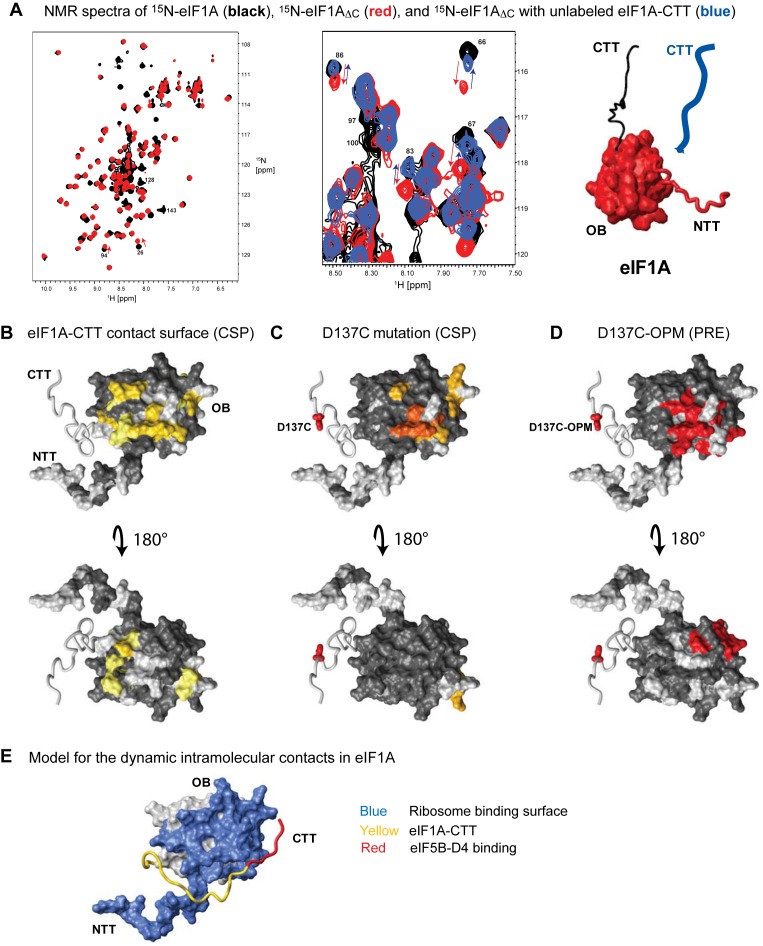
To test whether eIF1A-CTT and -OB interact, we used NMR deletion mapping, a variant of the chemical shift perturbation (CSP) assay, which is a sensitive method for mapping protein-protein and protein–ligand interactions. NMR chemical shifts are affected by changes in the environment at and around the contact interface upon binding, causing changes in the positions of the corresponding peaks in the NMR spectra (peak ‘movement’). In the CSP assay, NMR spectra of a labeled protein are recorded in the presence and absence of an unlabeled binding partner; the peaks affected by the interaction are identified; and the corresponding residues are mapped on the structure of the protein, to identify the surfaces affected by the interaction. In the NMR deletion analysis, the NMR spectra of a full-length protein and a deletion mutant are compared. Chemical shift changes between the two spectra are mapped on the protein surface in the same way as with the ‘traditional’ CSP assay to identify the contact interface of the deleted segment with the rest of the protein (reviewed in (31)). Since eIF1A-CTT and -NTT are intrinsically disordered (6), their deletion is not expected to affect the folding and stability of the OB domain of eIF1A (see Figure 1A for constructs used in this work). However, if they contact the eIF1A-OB at least transiently, their deletion would change the chemical environment at the surfaces they contact.
Comparison of the NMR spectra of 15N-labeled full-length eIF1A and eIF1AΔC, missing the entire 26-residue eIF1A-CTT (black and red, respectively in Figure 2A, left), shows that deletion of eIF1A-CTT strongly affects multiple peaks belonging to residues from the eIF1A-OB domain. Mapping the affected residues on the eIF1A surface (Figure 2B) demonstrates that eIF1A-CTT not only binds to the OB domain, but specifically contacts a distinct surface: around helix α1 and loop L45 (labeled in the structure in Figure 1A, the numbering of secondary structure elements in eIF1A is from (6)). Adding eIF1A-CTT to 15N-labeled eIF1AΔC (blue in Figure 2A, center) or eIF1AΔNC (missing both the NTT and the CTT, data not shown) affects most of the same surfaces as the respective deletion, showing that the interaction between eIF1A-CTT and -OB is strong enough to occur even when the tail is not covalently attached. In one region the chemical shift changes amount to reversing the changes due to the deletion of eIF1A-CTT. In another—the reversal is only partial. Surfaces in the vicinity of the site of the deletion are not affected by adding eIF1A-CTT, consistent with those effects being due to the covalent attachment of the CTT and not to productive interactions between the CTT and the OB domain. At saturating concentrations of eIF1A-CTT, the peaks corresponding to the affected residues in eIF1AΔC move to the same positions as in full-length eIF1A, but do not move past them (Figure 2A, center). Therefore, even though eIF1A-CTT is natively unfolded and mobile in full-length eIF1A, it spends most of the time in contact with eIF1AΔC at a distinct surface on the eIF1A-OB domain. For instance, if eIF1A-CTT spends half the time bound to the eIF1A-OB domain, the peak positions in full-length eIF1A would be intermediate between those in free eIF1AΔC and eIF1AΔC in the presence of saturating eIF1A-CTT concentrations.
Figure 2.
Intramolecular interactions between eIF1A-OB and -CTT observed by NMR. (A) Left, overlay of NMR spectra of 15N-eIF1A (black) and 15N-eIF1AΔC (red). Center, overlay of a portion of NMR spectra of 15N-eIF1A (black), 15N-eIF1AΔC (red) and 15N-eIF1AΔC with unlabeled eIF1A-CTT (blue). The arrows show peak movements upon eIF1A-CTT deletion (red) and upon addition of unlabeled eIF1A-CTT (blue). At saturation, binding of eIF1A-CTT reverses the changes caused by its deletion. Right, schematic of the eIF1A fragments used: the OB domain is in surface representation; the NTT is red wire; the CTT is black wire; and the added unlabeled CTT is blue. (B) eIF1A-OB domain surfaces affected by eIF1A-CTT deletion in the Chemical Shift Perturbation (CSP) assay shown in (A). In the top panel, eIF1A is in the same orientation as in Figure 1A. Affected residues are colored in yellow (smaller effects) and dark yellow (larger effects); eIF1A is in surface representation, except the CTT, which is shown as wire. Residues with no significant changes are colored in dark gray; residues that could not be analyzed are colored in light gray. (C) eIF1A-OB domain surfaces affected by the D137C mutation in CSP assay. Affected residues are colored orange (smaller effects) and dark orange (larger effects). The side-chain of D137 is shown as red sticks. (D) Paramagnetic Relaxation Enhancement (PRE) effects on eIF1A-OB residues in Oxytetramethyl Piperidinyl Maleimide (OPM) labeled eIF1AD137C. Affected residues are colored red. Effects on residues in the CTT proximal to D137 are not shown. (E) Model for the dynamic interaction between eIF1A-CTT and -OB. Surfaces contacting the 40S ribosomal subunit are colored blue; the CTT is colored yellow; the eIF5B-D4 binding segment of the CTT is colored red. Note that the CTT/OB contacts are dynamic and the CTT remains mobile.
In order to determine whether the 16 C-terminal residues in eIF1A (eIF1A-CT16) are sufficient to bind to the OB domain, we repeated the above experiments with eIF1A-CT16. Deletion of eIF1A-CT16 affects a distinct surface on eIF1A-OB, which is a subset of that affected by deletion of eIF1A-CTT (compare Figure 2B and Supplementary Figure S1A). Adding eIF1A-CT16 to 15N-labeled eIF1AΔC or eIF1AΔNC affects most of the same surfaces affected by the respective deletion (data not shown). Therefore, eIF1A-CT16 contacts the OB domain at a specific surface.
Position of eIF1A-CTT on the OB domain
To map more precisely the interaction between eIF1A-CTT and -OB, we introduced a point mutation, D137C, in eIF1A-CTT. In principle, a point mutation affects the environment in the close vicinity of the mutated residue, in much the same way as does ligand binding or deletion. The approach of combining NMR with ‘soft’ mutations, which change residues at an interface, without disrupting the interaction, has been successfully used by the Gierasch group to study intramolecular interactions within Hsp70 (33). The D137C mutation not only affects nearby residues in the CTT, but also affects a specific surface on the eIF1A OB domain (Figure 2C), helping to pinpoint the location of D137 binding on the eIF1A-OB domain. We then performed PRE experiments with oxytetramethyl piperidinyl maleimide (OPM) labeled eIF1AD137C. Paramagnetic probes cause increased relaxation (loss of signal) in nuclei at a distance of up to ∼20–25 Ǻ. Comparison of NMR spectra of paramagnetically labeled proteins with spectra where the probe is reduced (diamagnetic) provides long-range distance restraints that can be used for structure determination (32). Paramagnetic labeling of eIF1AD137C causes loss of signal in a specific surface on the eIF1A OB domain (Figure 2D). The observed PRE effects cover an area similar to, but wider than the surface, where the D137C mutation induced chemical shift changes (compare Figure 2C and D), consistent with the long-range nature of the PRE effects. The PRE effects were localized to a distinct surface, confirming that the interaction between eIF1A-CTT and -OB is specific, and that D137 in eIF1A-CTT does not spend significant amount of time at any other eIF1A-OB domain surfaces. Some residues were affected in the CSP experiment comparing WT eIF1A and eIF1AD137C (Figure 2C), but not in the PRE experiment (Figure 2D), indicating that the corresponding CSP effects were likely due to allosteric effects, rather than proximity to D137.
To further delineate the contacts between eIF1A-OB and -CTT, we used two cysteine mutants in the OB domain at the interface with the CTT: R65C in helix α1 and D85C in loop L45 (6). PRE experiments with paramagnetically labeled eIF1AR65C show that the proximal portion of eIF1A-CTT spends time in the vicinity of R65 (Supplementary Figure S1C). In contrast, similar experiments with the eIF1AD85C mutant show that only the distal portion of eIF1A-CTT (the extreme C-terminus) reaches the area around D85 (Supplementary Figure S1D). Therefore, the natively unfolded eIF1A-CTT dynamically contacts a specific surface of the folded domain of eIF1A (Figure 2E).
The PRE experiments with the R65C and D85C mutants also showed that the entire eIF1A-NTT spends some time in the vicinity of R65, but not D85 (compare Supplementary Figure S1C and D). Deletion of the 24-residue eIF1A-NTT affects a distinct surface on eIF1A-OB (Supplementary Figure S1B). However, unlike the case with eIF1A-CTT, no significant effects were observed when adding eIF1A-NTT to 15N-labeled eIF1AΔN (up to ∼200 μM, data not shown). Therefore, the eIF1A-NTT:OB interaction is too weak to observe when the NTT is not covalently attached to the rest of eIF1A, at least at the protein concentrations we could achieve.
eIF1A-CTT contacts the ribosome-binding surface of eIF1A-OB
Next, we asked whether the newly observed contacts of eIF1A-OB with the CTT and NTT can occur when eIF1A is bound to the 40S ribosomal subunit. A comparison of the binding interface between eIF1A-CTT and -OB with the structure of the eIF1A:40S complex (10) shows significant overlap between the eIF1A-OB surfaces that contact eIF1A-CTT and the 40S subunit (compare Figure 2B and E). Therefore, the dynamic interaction of eIF1A-CTT with the rest of eIF1A must be eliminated upon ribosome binding. This can explain why deletion of the CTT of S. cerevisiae eIF1A increases its affinity for the 40S subunit ∼4-fold (30). Similar analysis for eIF1A-NTT shows that the main NTT contact surface on the OB is compatible with the position of the eIF1A-NTT in 40S-bound eIF1A (compare Figure 2E and Supplementary Figure S1B). However, on the 40S ribosomal subunit, eIF1A-NTT would no longer be able to reach the vicinity of R65, which is buried at the interface with the ribosome (compare Figure 2E and Supplementary Figure S1C). Therefore, the transient contacts of the OB-proximal portion of eIF1A-NTT with the rest of eIF1A are maintained upon ribosome binding, whereas any additional contacts are lost.
The intramolecular interaction within eIF1A modulates eIF1A-CTT binding to eIF5B-D4
Comparison of NMR spectra of 15N-labeled eIF1A and eIF1A-CTT shows that virtually the entire eIF1A-CTT contacts the eIF1A-OB domain (data not shown). The eIF1A C-terminus is known to bind eIF5B-D4 (17,18). Therefore, the intramolecular interaction between eIF1A-OB and -CTT could interfere with eIF1A-CTT binding to eIF5B-D4. To test this hypothesis, we determined the affinities of a series of eIF1A constructs for eIF5B-D4 using FA with a fluorescein-labeled synthetic eIF1A-CT7 peptide (Fl-eIF1A-CT7). The affinity of Fl-eIF1A-CT7 for eIF5B-D4 was 12 μM. Competition FA assays using Fl-eIF1A-CT7 showed that the binding affinities of eIF1A-CT16 (12 μM) and of the full 26-residue eIF1A-CTT (12 μM) were similar (Figure 3A and B, Table 1). In contrast, the affinity of full-length eIF1A for eIF5B-D4 was 39 μM, much weaker than that of eIF1A-CTT (Figure 3B and Table 1). These results show that the intramolecular interaction within eIF1A interferes with eIF1A-CTT binding to eIF5B-D4, as expected based on the physical overlap between the two interfaces. Comparisons of NMR spectra of 15N-labeled eIF5B-D4 with eIF1A, eIF1A-CTT, and eIF1A-CT16 showed that all three eIF1A constructs contact the same surface on eIF5B-D4 (Supplementary Figure S2 and data not shown), fully consistent with our previous results (17). These findings are also in line with the recently published crystal structure of yeast eIF5B in complex with eIF1A, in which only the 11 C-terminal eIF1A residues are visible, while the rest of the protein is not (34). Since no significant chemical shift changes are observed on eIF1A-OB upon eIF5B-D4 binding ((17) and data not shown), while the eIF1A C-terminus is bound to eIF5B-D4, the interaction of eIF1A-OB with the rest of the CTT remains mostly unperturbed.
Figure 3.
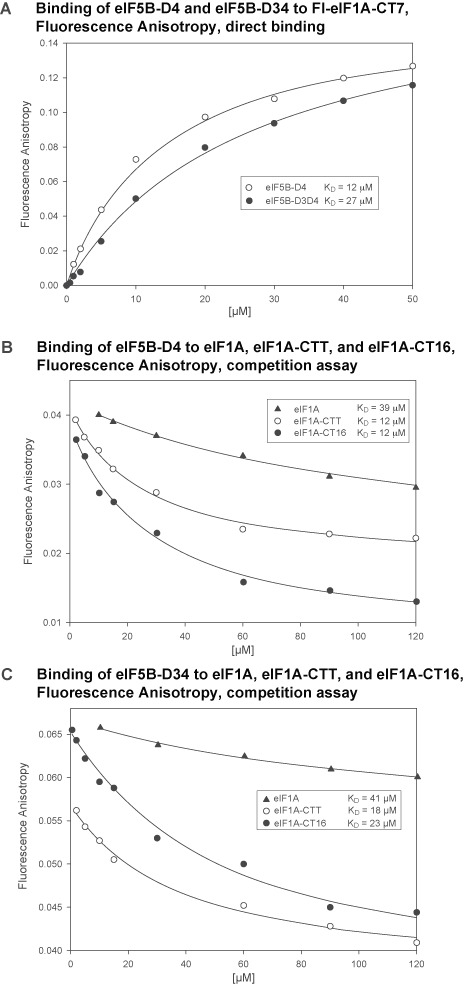
Binding affinities between eIF1A and eIF5B fragments determined by fluorescence anisotropy (FA). (A) Direct titration of fluorescein-labeled eIF1A-CT7 (Fl-eIF1A-CT7) with eIF5B-D4 and eIF5B-D34. (B) Competition assay with eIF1A, eIF1A-CTT and eIF1A-CT16 binding to eIF5B-D4. (C) Competition assay with eIF1A, eIF1A-CTT and eIF1A-CT16 binding to eIF5B-D34.
Table 1. Binding affinities between eIF1A and eIF5B constructs determined by Fluorescence Anisotropy (FA).
| eIF5B-D4 (μM) | eIF5B-D34 (μM) | |
|---|---|---|
| Fl-eIF1A-CT71 | 12 ± 1 | 27 ± 3 |
| eIF1A-CT162 | 12 ± 3 | 23 ± 6 |
| eIF1A-CTT2 | 12 ± 2 | 18 ± 5 |
| eIF1A2 | 39 ± 9 | 41 ± 9 |
1Direct binding FA assay to fluoresceine-labeled synthetic eIF1A-CT7 peptide (Fl-eIF1A-CT7).
2Competition FA assay.
eIF1A-OB interacts with eIF5B-D3
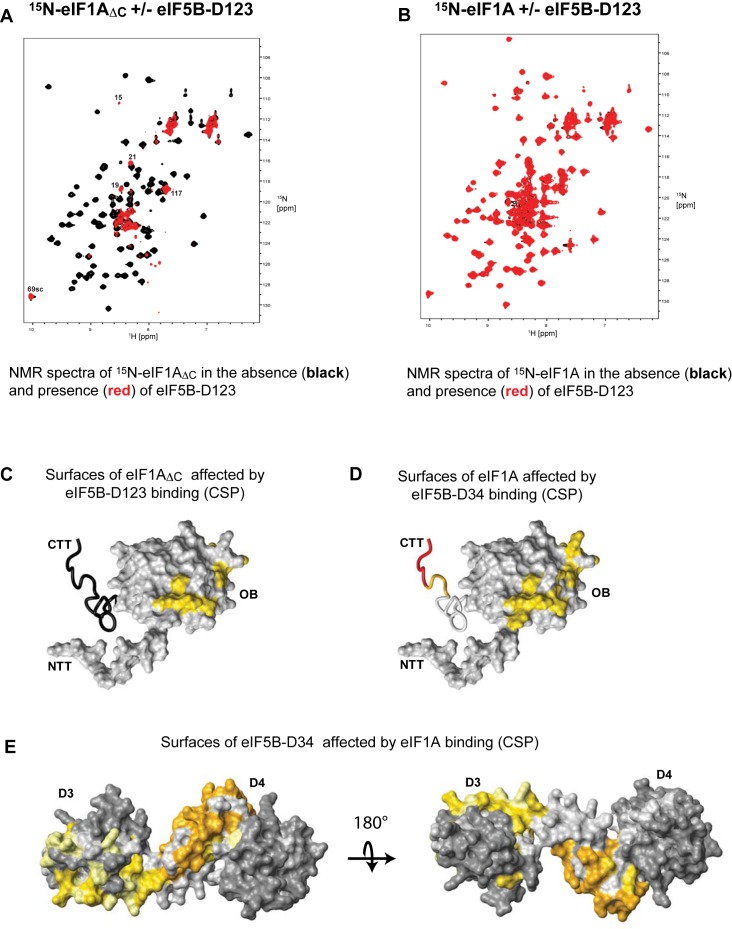
The eIF1A-OB domain has been proposed to bind to eIF5B-D2 and/or -D3 on the ribosome (see e.g. (17)), because of their proximity on the ribosome and because such an interaction has been reported between their bacterial homologs (22,24). However, the interaction has never been observed off the ribosome in either bacteria or eukaryotes. The intramolecular interactions shown in Figure 2 and Supplementary Figure S1 involve a surface on the eIF1A-OB domain around loop L45 (6), which faces in the direction of eIF5B in the translation IC, and is thus the most likely eIF5B contact surface on the OB domain. Therefore, we tested both full-length eIF1A and eIF1AΔC for binding to eIF5B-D123, encompassing domains D1, D2 and D3. eIF5B-D123 contains D2 and D3, but not D4 and thus cannot bind to eIF1A-CTT. We found that eIF5B-D123 binds to 15N-labeled eIF1AΔC, but not to full-length 15N-eIF1A (Figure 4A and B). Binding leads to severe broadening of peaks corresponding to the eIF1A-OB domain, but not the natively unfolded NTT. Mapping the binding surface on eIF1A-OB using TROSY and 15N/2H-eIF1AΔC shows that the affected surfaces on the eIF1A-OB (Figure 4C and Supplementary Figure S3A) overlap with those affected by eIF1A-CTT binding (compare with Figure 2B). Therefore, eIF1A-OB does indeed bind eIF5B, and eIF1A-CTT sterically prevents this interaction.
Figure 4.
eIF1A-OB binds to eIF5B-D3. (A) Overlay of NMR spectra of 15N-eIF1AΔC in the absence (black) and presence of eIF5B-D123 (red). Select peaks from eIF1A-NTT, as well as other peaks that remain visible in the presence of eIF5B-D123 are labeled. Loss of most peaks, except those belonging to residues in the eIF1A-NTT and other flexible regions, indicates formation of a large complex, in which the NTT remains dynamic. (B) Overlay of NMR spectra of 15N-eIF1A in the absence (black) and presence of eIF5B-D123 (red). No loss of signal or chemical shift changes are observed, indicating no significant binding. (C) Surfaces of 15N/2H-eIF1AΔC affected by eIF5B-D123 binding in CSP assay. eIF1A is in the same orientation as in Figure 1A, in surface representation, except for eIF1A-CTT (deleted), shown as black wire. Affected residues are painted yellow. (D) Surfaces of 15N/2H-eIF1A affected by eIF5B-D34 binding in CSP assay. Affected residues are painted from yellow (smaller effects) to red (larger effects). (E) Surfaces of 15N/2H-eIF5B-D34 affected by eIF1A binding in CSP assay. In the left panel, eIF5B-D34 is in the same orientation as in the eIF5B structure in Figure 1B. Affected residues are painted from yellow (smaller effects) to orange (larger effects). Residues with no significant effects are dark gray and residues that could not be analyzed are light gray.
To further map the eIF5B segment that binds to eIF1A-OB, we tested whether eIF5B-D34 can bind to eIF1AΔC. eIF5B-D34 binds to eIF1AΔC and causes similar effects to those observed with eIF5B-D123 (compare Supplementary Figure S3B with the inset of S3A), indicating that D3, the common segment between the two eIF5B constructs, is both necessary and sufficient for the interaction. Indeed, eIF5B-D3 binds to eIF1AΔC and causes similar effects to those observed with eIF5B-D123 and eIF5B-D34 (compare Supplementary Figure S3C with A and B). eIF5B-D4 does not interact with eIF1A-OB or eIF1A-NTT (data not shown), consistent with previous data (17). Therefore, eIF1A-OB can interact with eIF5B-D3, when not blocked by eIF1A-CTT. Since eIF1A-CTT is likely displaced from the OB domain upon binding to the 40S ribosomal subunit (see above), eIF5B-D3 and eIF1A-OB can interact when eIF5B and eIF1A are on the ribosome.
We then proceeded to test whether eIF1A-OB and eIF5B-D3 interact in the context of the eIF1A:eIF5B-D34 complex, where eIF1A-CTT interferes with the eIF1A-OB–eIF5B-D3 interaction. The rationale was that in the complex held together by binding of eIF1A-CTT to eIF5B-D4, eIF1A-OB and eIF5B-D3 are brought in proximity to each other, which could allow them to overcome, at least partially, the inhibitory effect of eIF1A-CTT. eIF5B-D34 binding to 15N-labeled eIF1A causes severe line-broadening in both the extreme eIF1A-C-terminus and the eIF1A-OB domain (Supplementary Figure S3D), as expected for formation of a large complex. The eIF1A-NTT and most of the eIF1A-CTT remain flexible. Comparison of TROSY-HSQC spectra of 15N/2H-labeled eIF1A in the presence and absence of eIF5B-D34 shows strong effects in the eIF1A-CTT, as well as weak effects at the eIF5B-D3 binding surface of eIF1A-OB (Figure 4D). Therefore, in the eIF1A:eIF5B-D34 complex, eIF5B-D3 and D4 both contact eIF1A. The chemical shift changes on the OB domain are much smaller than those caused by deletion of eIF1A-CTT; therefore, the intramolecular interaction between eIF1A-OB and -CTT is not significantly disturbed in the complex. The chemical shift changes in the L45 region were similar to those observed with eIF5B-D3 binding to eIF1AΔC (compare e.g. the 83 peak movement in Supplementary Figure S3C and D), consistent with direct contact between eIF5B-D3 and eIF1A-OB. In contrast, chemical shift changes in the α1 helix region were different from those observed with eIF5B-D3 binding to eIF1AΔC, instead moving slightly in the same direction as when eIF1A-CTT is deleted (compare e.g. the 67 peak movement in Figure 2A, Supplementary Figure S3C and D). This observation indicates slight destabilization of the intramolecular eIF1A-OB/-CTT contacts. Therefore, eIF5B-D4 binding appears to cause ‘fraying’ of the eIF1A-CTT away from the eIF1A-OB surface sufficient to allow eIF5B-D3 to contact the L45 region of eIF1A-OB.
To map the eIF1A contact surfaces on eIF5B, we obtained the NMR backbone resonance assignments of human eIF5B-D34 and used them in CSP assay. eIF1A binding to 15N/2H-labeled eIF5B-D34 shows strong effects in eIF5B-D4, which interacts with eIF1A-CTT, as well as weaker effects in eIF5B-D3 (Figure 4E, weaker effects are shown in yellow, and stronger effects—in orange).
eIF5B-D3 contacts eIF5B-D4 and interferes with binding to eIF1A-CTT
The above experiments mapping the interaction between eIF1A and eIF5B-D34 indicated that eIF1A can contact both eIF5B-D3 and -D4 simultaneously. It was therefore, interesting to determine the binding affinity between eIF1A and eIF5B-D34. This was done using competition FA assay, as described above for eIF5B-D4. Surprisingly, the fluorescently labeled peptide Fl-eIF1A-CT7, used for the competition FA assay, had lower affinity for eIF5B-D34 (27 μM) than for eIF5B-D4 (12 μM) (Figure 3A and Table 1). The affinities of eIF1A-CT16 (23 μM) and eIF1A-CT26 (18 μM) for eIF5B-D34 were also weaker than for eIF5B-D4. Therefore, the presence of D3 interferes with binding of D4 to eIF1A-CTT. This observation indicates that eIF5B-D3 and D4 may contact each other. The eIF1A affinity for eIF5B-D34 (41 μM)(Figure 3C and Table 1) was nearly identical to that for eIF5B-D4 (39 μM)(Figure 3B and Table 1). Since all eIF1A-CTT constructs have lower affinities for eIF5B-D34 than for eIF5B-D4, this indicates a modest stabilizing effect of the eIF1A-OB–eIF5B-D3 interaction, even in the presence of the inhibitory eIF1A-OB/CTT interaction.
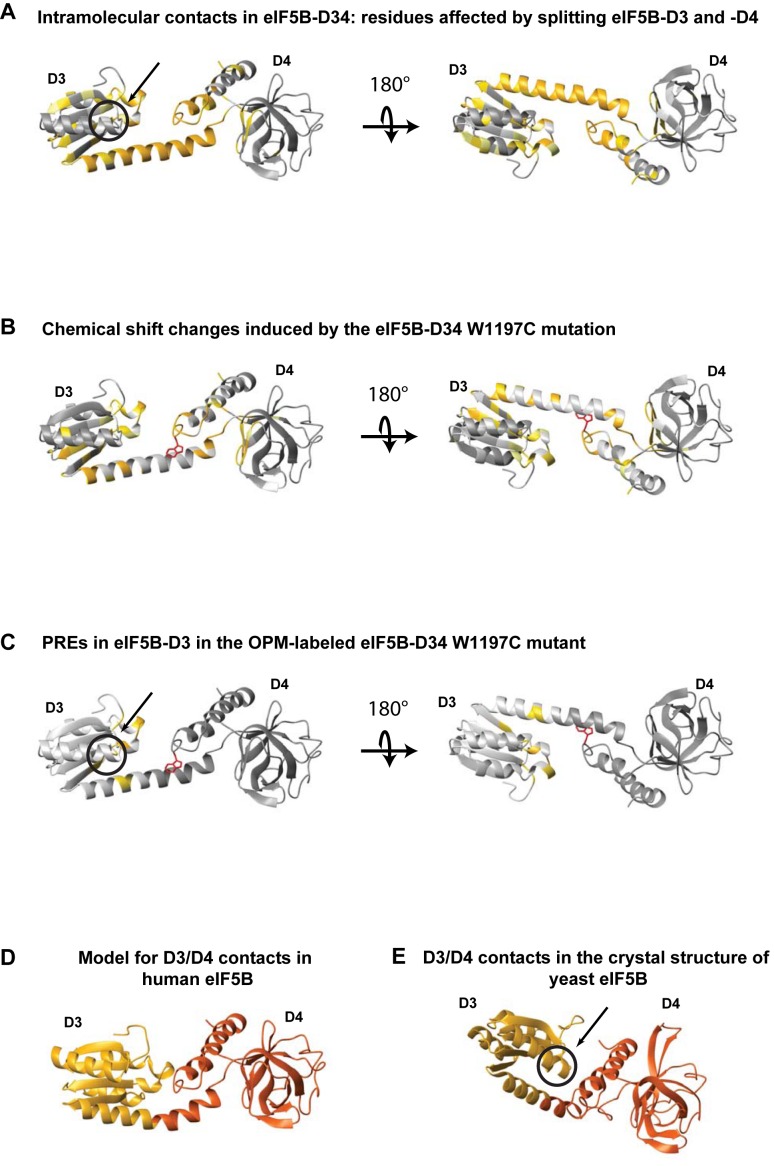
eIF5B-D3 and -D4 are connected by the long helix h12 (labeled in Figure 1B, the numbering of secondary structure elements in eIF5B is from (14)), which appears rigid in the structure of the archaeal eIF5B homolog aIF5B (14). However, recent reports show that D4 is in fact attached flexibly to the rest of eIF5B (34–37). Furthermore, one of several crystal forms of yeast eIF5B reported in (34) shows D3 and D4 in direct contact with each other. If such contact exists in solution, it could explain the inhibitory effect of D3 on the binding of D4 to eIF1A-CTT. To test whether D3 and D4 interact in solution, we used NMR deletion mapping, as described above for eIF1A. Comparison of the spectra of eIF5B-D34 to those of the individual domains shows that D3 and D4 do indeed contact each other. The affected surface on D4 (Figure 5A) partially overlaps with the eIF1A-CTT binding surface (Figure 4E), explaining the effect of the intramolecular interaction within eIF5B on the affinity for eIF1A. The effects on eIF5B-D3 are more extensive and include the entire helix h12 connecting D3 with D4, as well as regions of D3 that pack against it, indicating changes in conformation and/or dynamics in D3 between the eIF5B-D3 and eIF5B-D34 constructs. Therefore, it was difficult to unambiguously distinguish direct effects on D3 due to contacts with D4 from indirect effects mediated by conformational changes. Interestingly, whereas the eIF5B-D4 surfaces affected by the presence or absence of D3 are at least partially compatible with contacts observed in the recent yeast eIF5B crystal structure (34), the affected surfaces on D3 are clearly different from the surface that contacts D4 in that crystal structure (compare Figure 5A and E), indicating that the contact interface between human eIF5B and eIF1A is different from that observed in the yeast eIF5B/eIF1A crystal structure.
Figure 5.
Intramolecular contacts between eIF5B-D3 and -D4. (A) eIF5B-D3 and -D4 surfaces affected by deletion of eIF5B-D4 and -D3, respectively, in CSP assay. Affected residues are colored yellow (smaller effects) and orange (larger effects); residues with no significant effects are dark gray; and residues that could not be analyzed are light gray. The region corresponding to the contact surface in the structure of yeast eIF5B (34) (circled and marked with an arrow) is unaffected. (B) eIF5B-D34 surfaces affected by the W1197C mutation in CSP assay. Affected residues are colored yellow (smaller effects) and orange (stronger effects). The side-chain of W1197 is shown as red sticks. (C) PRE effects on eIF5B-D3 residues in OPM-labeled eIF5BW1197C. Affected residues are colored yellow. Effects on residues in D4 are not shown. (D) Model for the dynamic interaction between eIF5B-D3 and -D4. (E) D3/D4 contacts in the crystal structure of Saccharomyces cerevisiae eIF5B (34). The main contact surface on D3 is circled.
To further elucidate the interaction interface between eIF5B-D3 and -D4, we used a combination of site-directed mutagenesis and PRE analysis. To this end, we introduced the W1197C mutation in eIF5B-D34. The sidechain of W1197 in D4 packs against the long helix h12 that connects D3 with D4 and the NH chemical shifts of W1197 are strongly affected by the presence/absence of D3. W1197 is also at the periphery of the eIF1A-CTT binding surface of D4. The W1197C mutation in the two-domain eIF5B-D34 construct causes chemical shift changes not only in the surrounding region of D4 (as expected from its proximity), but also in D3. The regions of D3 affected by the W1197C mutation (Figure 5B) are essentially the same as those affected by deletion of D4 (Figure 5A), although the magnitude of the chemical shift changes is somewhat smaller. Therefore, the mutation appears to weaken the D3:D4 interaction. PRE experiments with paramagnetically labeled eIF5B-D34W1197C (Figure 5C) allowed us to differentiate between direct and indirect effects of D4 on D3 and determine the mutual orientation of the two domains (Figure 5D). The interdomain orientation of eIF5B-D34 was obtained using distance restraints from intramolecular PRE effects within eIF5B (Figure 5C). These results also confirmed that at least in human eIF5B, the D3/D4 contact interface is different from that observed in (34)(compare Figure 5D and E).
Comparison of the eIF5B-D3/D4 interface (Figure 5) with the eIF5B surfaces affected by eIF1A binding (Figure 4E) indicates that eIF1A binding has a modest effect on the D3/D4 interface. The magnitude and direction of the changes in the D4-binding surface of D3 suggest that eIF1A binding causes weakening of the intramolecular D3/D4 interaction, consistent with the D3/D4 interaction itself weakening eIF1A binding.
Structure of the dynamic eIF1A:eIF5B-D34 complex
To determine the precise mutual orientation of eIF1A and eIF5B-D34, we performed PRE experiments with three eIF1A cysteine mutants: D85C, in the OB domain, at the interface with eIF5B-D4; R65C, also in the OB domain, farther from eIF5B; and D137C, in the CTT, in close proximity to the D4-binding eIF1A C-terminus (Supplementary Figure S4AB, and data not shown). Weak to moderate PREs from OPM-labeled eIF1AD85C were observed in both D3 and D4 in the eIF1A:eIF5B-D34 complex (Supplementary Figure S4A). PRE effects from eIF1AD137C were observed only in D4 (Supplementary Figure S4B), as expected based on its position next to the D4-binding site. Only weak PRE effects were observed with eIF1A-R65C (data not shown), limited to a small subset of the residues affected by the D137C mutant. These results are consistent with D137 contacting the OB domain in the vicinity of R65 (see Figure 2D), as well as with eIF1A-CTT being located between the eIF1A-OB domain and eIF5B-D4 in the eIF1A:eIF5B complex.
We used our results, together with previously available structural data, to build a model for the structure of the dynamic eIF5B:eIF1A complex (Supplementary Figure S4C). The interaction of eIF5B-D4 with the eIF1A C-terminus was modeled after the crystal structure of the corresponding complex between yeast eIF5B and eIF1A-CTT (34), because our data were consistent with it. The orientation of the eIF1A-OB domain with respect to eIF5B-D34 was obtained using intermolecular PRE effects (Supplementary Figure S4A and B). Since our NMR data indicated that most of the intramolecular contacts within eIF1A and eIF5B-D34 remain intact in their complex, the two proteins were docked initially as rigid bodies. The only exception was the eIF1A C-terminus, whose interactions with the eIF1A-OB domain and eIF5B-D4 were not compatible with each other. Therefore, the C-terminal 11 residues of eIF1A were modeled bound to eIF5B-D4 and away from the OB domain. This eIF1A-CTT conformation also allows the OB domain to contact eIF5B-D3 in the complex, as indicated by our experimental data. In summary, in the eIF1A:eIF5B complex in solution, the dynamic intramolecular interactions remain largely intact, albeit slightly destabilized (meaning they may spend a little more time away from each other than in the free proteins), with the exception of the eIF1A C-terminus, which appears to fray away from the OB domain. This is reflected in the model in Supplementary Figure S4C, where the eIF1A and eIF5B-D34 conformations are the same as those of the free proteins (Figures 2E and 5D, respectively), except for the eIF1A C-terminus, which is in a new conformation bound to eIF5B-D4. While the eIF1A-CTT:eIF5B-D4 interaction is stable and involves folding of the eIF1A C-terminus upon binding, the other contacts (both intra- and intermolecular) in the eIF1A:eIF5B complex in solution remain dynamic. Therefore, like the eIF1A and eIF5B-D34 models in Figures 2E and 5D, the model for the eIF1A:eIF5B-D34 complex in Supplementary Figure S4C is only a snapshot of a dynamic complex with mobile parts and not a rigid structure.
In the model in Supplementary Figure S4C, the eIF1A-C-terminus comes in close proximity with D3, consistent with the observed modest inhibitory effect of D3 on the D4:eIF1A-CTT interaction (Table 1). Thus, the effect could be due to any of the following: unfavorable contacts between side chains, long-range repulsive electrostatic interactions or allosteric effect on the interface, because both interactions cause CSPs in overlapping sets of residues, which could involve changes in conformation or dynamics. The absence of steric clashes is consistent with the rather modest effect (∼2-fold), since a steric clash would likely have had a much greater effect.
The resulting eIF1A:eIF5B-D34 model represents the dynamic contacts between eIF1A and eIF5B in solution, but not on the ribosome. Upon binding to the 40S subunit, eIF1A-CTT will dissociate from the OB domain and eIF5B-D4 will also have to move together with eIF1A-CTT, as well as to avoid steric clashes with the 40S ribosomal subunit.
DISCUSSION
eIF1A and eIF5B are the only two universally conserved translation initiation factors. Their ribosomal positions, as well as many of their functions are also conserved between bacteria and eukaryotes. It has been widely accepted that these two proteins, together with the rest of the IC components, undergo conformational changes as the IC progresses through the stages of translation initiation (reviewed in (2–5)). Yet we only recently started to understand the nature of these changes and their roles.
In this work, we show the existence of dynamic intramolecular interactions within both eIF1A and eIF5B. Each of these interactions lowers the affinity of eIF5B–eIF1A binding. Both intramolecular interactions affect eIF1A-CTT binding to eIF5B-D4, while the intramolecular contacts within eIF1A also block the newly discovered eIF1A-OB–eIF5B-D3 binding. These observations point toward potential mechanisms for modulation of the eIF1A:eIF5B interaction, and through it, for remodeling the IC, provided that the inhibitory intramolecular interactions are disrupted at certain stages of translation initiation. Remarkably, we already know of stages when this is indeed the case: the eIF1A-OB/-CTT interaction must be disrupted when eIF1A binds to the ribosome (7), and the eIF5B-D3/-D4 interaction is disrupted at least upon ribosomal subunit joining (35,37).
When eIF1A binds to the small ribosomal subunit A-site, its NTT and CTT both extend into the P-site, where the NTT appears to bind to the ribosome, while the CTT remains mobile. Upon start codon recognition, eIF1A-CTT is displaced from the P-site (7,11). It is not known when exactly eIF5B is recruited to the 40S subunit; however, its role in helping displace eIF2-GDP from Met-tRNAi after start codon recognition (15), indicates that it has to be recruited at that stage or earlier. The eIF1A-CTT:eIF5B-D4 interaction is most likely established when eIF1A-CTT is ‘evicted’ from the P-site upon start codon recognition, and it is known to be important for both ribosomal subunit joining and coordinated release of eIF1A and eIF5B after subunit joining and GTP hydrolysis by eIF5B (16,19–20). If eIF5B is not already present in the PIC, the eIF1A-CTT:eIF5B-D4 interaction may also play a role in eIF5B recruitment. eIF1A binding to the ribosome uncovers the eIF5B-D3 binding surface on eIF1A-OB by displacing eIF1A-CTT, as we report here. The eIF1A-OB/eIF5B-D3 interface is similar to that observed by Cryo-EM between IF1 and IF2 in the 30S PIC (24). Therefore, eIF5B could bind to the PIC earlier than start codon recognition, via interactions with ribosomal protein rpS23 and eIF1A-OB. If eIF5B is already present before the scanning PIC has reached the start codon, this would be beneficial for the rate of translation initiation because otherwise the PIC would stay idle until eIF5B binds, which would not be instantaneous, considering the cellular levels of eIFs (38). Alternatively, if the scanning PIC does not (always) contain an eIF5B, that would allow for an additional level of regulation by the cellular eIF5B levels, whereby lower eIF5B concentrations would slow down the rate of translation initiation and potentially cause an increased frequency of leaky scanning as the 48S PIC dwells longer at the start codon before subunit joining takes place. Dependence of start codon selection in vitro on the eIF5B concentration has been observed experimentally (39,40) and the same has been shown for other initiation factors in vivo, e.g. eIF1 and eIF5 (41–43).
The KD of the eIF1A-CTT:eIF5B-D4 interaction is weakened ∼3-fold by the eIF1A-OB/-CTT contacts and another ∼1.5-fold by the eIF5B-D3/-D4 contacts (Table 1). Therefore, when the inhibitory intramolecular contacts are eliminated on the ribosome, the contribution of the eIF1A-CTT:eIF5B-D4 interaction for the overall eIF1A:eIF5B affinity increases by ∼4.5-fold. While we do not know the exact affinity of the second component, the eIF1A-OB:eIF5B-D3 interaction, it should have a KD < 1 mM, because we were able to observe it by NMR at protein concentrations of ∼200 uM. In solution, the intramolecular eIF1A-OB/-CTT contacts are largely unperturbed (Supplementary Figure S3D) and the contribution of the eIF1A-OB:eIF5B-D3 interaction appears to be only minimal (Table 1). In contrast, on the ribosome, the eIF1A-OB:eIF5B-D3 interaction will be unobstructed and could increase the overall affinity between eIF1A and eIF5B by order(s) of magnitude. The effect of this increase in absolute affinity is magnified by the increased effective concentration of the two proteins with respect to each other, when they are both bound to adjacent sites on the ribosome.
While there is no available structure of an 80S IC containing both eIF1A and eIF5B, eIF5B-D3 and eIF1A-OB likely no longer contact each other at that stage, judging from the position of D3 in the 80S IC (35,37), similar to the bacterial 70S IC (23,44). It is interesting to note that while bacterial IF2-D3 has different positions in 30S IC and 70S IC (23–24,44), it contacts rpS12 (the bacterial homolog of rpS23) in both of these complexes, but via distinct interfaces. The structures of the 80S IC (35,37) and the eIF5B-D3:eIF1A-OB interaction reported here indicate that the same is likely the case for eIF5B-D3 and rpS23.
The eIF1A interaction with eIF5B-D4 is mediated by eIF1A-CTT and the helical subdomain of D4. However, while the D4 helical subdomain is present in archaeal aIF5B, archaeal aIF1A lacks a CTT. Therefore, the D4 helical subdomain must play other role(s), at least in archaea, and likely also in eukaryotes. Binding to D3 is one such role, but may not be the only one. Remarkably, modeling eIF1A into the structure of the mammalian eIF5B/80S IC (37), places it in very close proximity, even touching eIF5B-D4 (Supplementary Figure S5A). A common feature shared between archaea and eukaryotes, but different from bacteria, is that a/eIF5B does not bring the initiator tRNA to the ribosome. Instead, a/eIF2 recruits the initiator tRNA and is displaced by a/eIF5B upon start codon recognition and GTP hydrolysis. Therefore, it seems logical to propose that the interaction of D4 with D3, and possibly with eIF1A-OB, is important for ribosomal recruitment and positioning of a/eIF5B, before it can displace a/eIF2 from the tRNA. The D3:D4 interaction is compatible with the D3:eIF1A-OB interaction and is thus likely present in the PIC upon eIF5B recruitment. As shown in Supplementary Figure S5A, D4 can contact eIF1A-OB in 80S ICs, while D3 no longer contacts D4 and eIF1A-OB (37). It should be noted, however, that we were unable to observe interaction between eIF1A-OB and eIF5B-D4 in solution (data not shown), indicating that if such an interaction does exist, it would be weak and depend on eIF1A and eIF5B being brought in proximity on the ribosome. It remains to be determined whether D4 contacts eIF1A-OB in 43S and 48S PICs, before and/or after eIF5B displaces eIF2-GDP and binds to Met-tRNAi; as well as whether D4 still contacts D3 after eIF5B binds to Met-tRNAi. The position of eIF2 in the 48S PICs (45) shows that eIF5B-D4 cannot occupy the same position as it does in 80S IC (35,37), because eIF2 would sterically prevent it. The overlap is more significant in the open 48S PIC (Supplementary Figure S5B) than in the closed (Supplementary Figure S5C), but is present in both. We suggest that, after eIF5B displaces eIF2-GDP, Met-tRNAi-bound D4 occupies a position similar to that in 80S ICs and can contact eIF1A-OB. At this stage D4 has likely lost contact with D3: for D4 to move toward Met-tRNAi, either its dynamic contacts with D3 would have to be disrupted, or the D3/eIF1A-OB contacts would have to be rearranged, which we find much less likely. Furthermore, eIF1A-CTT binding to D4 weakens the D3/D4 interaction, which would make it easier to disrupt the D3/D4 contacts upon Met-tRNAi binding. Testing this hypothesis directly would have to await solving the structure of the 48S PIC with Met-tRNAi bound to eIF5B. Modeling the eIF1A-CTT:eIF5B-D4 interaction in the context of the 80S IC (Supplementary Figure S6A) shows that the length of human eIF1A-CTT is sufficient to reach D4, consistent with the role of this interaction for subunit joining (19). It is interesting that, if this model is correct, the C-terminus of eIF1A reaches to almost the same position on eIF1A-OB as in free eIF1A, but approaching from the opposite direction (compare Supplementary Figure S6B and C). eIF1A-CTT is even longer in S. cerevisiae; however, it is six residues shorter in Schizosaccharomyces pombe, which appears to be the minimum, or near-minimum CTT length allowing eIF1A-CTT to bind eIF5B-D4 as shown in Supplementary Figure S6A.
A recently reported crystal structure of archaeal aIF5B from Aeropyrum pernix has an additional C-terminal helix that packs against the first two helices of the D4 helical subdomain, occupying the surface that in eukaryotic eIF5B-D4 binds to eIF1A-CTT (46). Interestingly, this C-terminal helix is not present in the structure of another archaeal aIF5B from Methanobacterium thermoautotrophicum (M. therm.). Instead, the C-terminus of M. therm. aIF5B is disordered (14). It is not clear whether the difference between these aIF5B structures reflects genuine differences between the two proteins or is an indication that the C-terminus of aIF5B can alternate between a disordered state and a helix packed against the other two helices in D4. Supplementary Figure S6 indicates that whether the aIF5B C-terminus is helical or disordered, it is likely to be in close proximity to aIF1A-OB and could mediate binding between aIF5B-D4 and aIF1A.
The D4 helical subdomain is not present in bacterial IF2; thus, it is possible that the D3:D4 interaction is also unique to eukaryotes and archaea. However, bacterial IF2-D3 and -D4 may also interact, including on the ribosome (47). The contact surface on D3 may be similar, while the one on D4 would obviously be different and would have to involve the OB fold of IF2-D4. The fact that h12, connecting D3 to D4, is shorter in IF2 than in eIF5B is also consistent with such an idea. eIF5B and IF2 have similar structures and must have similar overall dimensions, since they fit in similar cavities on the ribosome. Thus, the C-terminal helical subdomain in eIF5B-D4 fits in the space between D1/D2 and D4. The positions of D1/D2 and D4 on the ribosome are similar between eIF5B and IF2. However, the range of motion of eIF5B-D3 is restricted by the extra helices in D4.
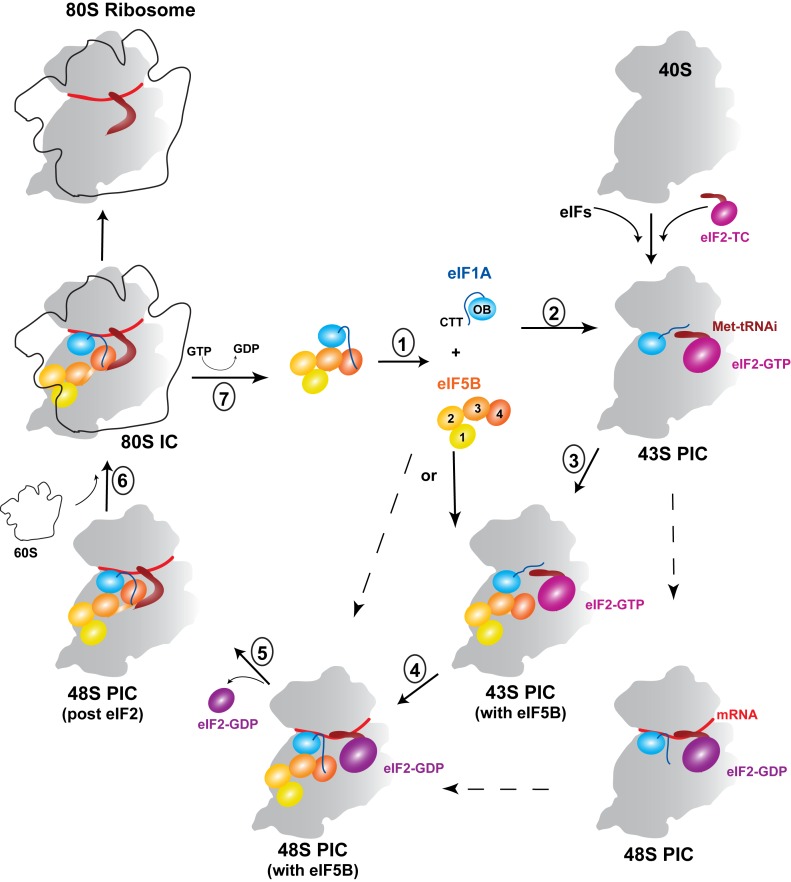
Model for the dynamic interactions of eIF1A and eIF5B on and off the ribosome
Based on our results and recent advances in the field, we built a model for the dynamics of eIF1A and eIF5B on and off the ribosome (Figure 6).
In solution, eIF1A and eIF5B are likely to exist in equilibrium between free species and the complex shown in Supplementary Figure S4C.
Upon eIF1A binding to the 40S subunit, eIF1A-CTT is displaced from the OB domain and is now located in the ribosomal P-site. While this would increase the eIF1A-CTT affinity for eIF5B-D4 (see Table 1), its location on the ribosome away from eIF5B may preclude such an interaction. The displacement of eIF1A-CTT from eIF1A-OB also exposes the surface on the OB domain that interacts with eIF5B-D3.
eIF5B is recruited to the PIC either before or after start codon recognition (see text). At this stage, eIF5B-D3 contacts both rpS23 and eIF1A-OB. While weak in isolation, the eIF5B-D3:eIF1A-OB interaction is likely to play a role in eIF5B recruitment and/or influence its orientation within the IC, since eIF1A and eIF5B are bound to adjacent sites on the 40S subunit, and their effective concentrations with respect to each other can be quite high.
Upon start codon recognition, eIF1A-CTT is ‘evicted’ from the P-site and is now able to access eIF5B-D4.
eIF5B-GTP displaces eIF2-GDP from the acceptor end of Met-tRNAi. At this stage, eIF5B-D4 is released from D3 and can contact eIF1A-OB. Disruption of the D3:D4 contacts would also allow the eIF5B-D4:eIF1A-CTT interaction to reach maximum affinity (see Table 1). The result is cooperative binding of eIF5B simultaneously to GTP, Met-tRNAi and eIF1A, in a conformation able to promote efficient ribosomal subunit joining.
Subunit joining causes D3 to move to the GTPase-activating center, as seen in Cryo-EM reconstructions (35,37), away from eIF1A-OB.
Upon subunit joining, eIF5B hydrolyzes GTP, which changes its conformation and lowers its affinity for the ribosome. eIF5B-GDP then dissociates from the 80S IC, while still bound tightly to eIF1A-CTT. Once off the ribosome, the intramolecular interactions in both eIF1A and eIF5B are restored, which weakens their complex.
Figure 6.
Model for the dynamic interactions of eIF1A and eIF5B on and off the ribosome. The steps are numbered as in the text (see the ‘Discussion’ section' for detailed descriptions). Alternative pathways are marked with dashed arrows. Only the relevant initiation factors are shown. The coloring of eIF5B is as in Figure 1B: from yellow (D1) to dark orange (D4); the N-terminal region of eIF5B is not shown. eIF1A-OB is light blue; eIF1A-CTT is blue; and eIF1A-NTT is omitted for clarity. eIF2-GTP is magenta; eIF2-GDP is purple. The small 40S ribosomal subunit is gray. The large 60S ribosomal subunit is shown as an outline. eIF5B may bind to the PIC either before or after start codon recognition. Ribosomal subunit joining induces GTP hydrolysis by eIF5B, which leads to release of eIF5B-GDP and eIF1A. eIF5B-GDP undergoes spontaneous nucleotide exchange to eIF5B-GTP (not shown). Conformations of eIF1A and eIF5B-D34 at individual steps: (i) in solution, the intramolecular interactions in both eIF1A and eIF5B-D34 are formed and their conformations should be dynamic and similar to those shown in Figure 2E (eIF1A) and Figure 5D (eIF5B-D34). (ii) Upon binding to the 40S subunit, eIF1A-NTT and -CTT extend into the P-site, where the NTT becomes at least partially folded, while the CTT remains disordered (see e.g. Figure 1B in ref. (10)). (iii) In the 43S PIC, the contacts between eIF5B-D3 and -D4 are retained. The interaction between eIF5B-D3 and eIF1A-OB should be similar to that between IF2-D3 and IF1 in the bacterial 30S IC (see e.g. Figure 2B in ref. (24)). (iv) Upon start codon recognition, the interactions within eIF5B-D34 and between eIF5B-D3 and eIF1A-OB likely remain unchanged, but eIF1A-CTT now binds eIF5B-D4, as shown in Figure 1B. (v) When eIF5B displaces eIF2 from the Met-tRNAi, the position of eIF5B-D3 and its contacts with eIF1A-OB remain the same. eIF5B-D4 moves away from -D3 and contacts Met-tRNAi, and possibly eIF1A-OB. Its position should be similar to that in the 80S IC (Supplementary Figure S6A). (vi) Upon subunit joining, eIF5B-D3 moves away from eIF1A-OB, while the position of eIF5B-D4 likely does not change much. The model for the mutual orientations of eIF1A and eIF5B in the 80S IC is shown in Figure 6A.
It is interesting to see how this model relates to the available experimental evidence and current models. The roles of the eIF1A-CTT:eIF5B-D4 interaction in both ribosomal subunit joining and coordinated release of eIF1A and eIF5B from the ribosome have been shown experimentally (16,19–20). Displacement of eIF1A-CTT from the OB domain upon binding to the 40S subunit is consistent with the modest increase in yeast eIF1A affinity for the ribosome upon deletion of the CTT (30).
In human, the eIF1A:eIF5B complex is expected to only form transiently in solution, and to have a limited role, because the KD of the interaction we determined for eIF1A:eIF5B-D34 is ∼40 μM (Table 1), likely greater than the eIF1A and eIF5B cellular concentrations. The situation may be different in S. cerevisiae, where the eIF1A:eIF5B interaction was shown by pull-down (12,18), a non-equilibrium assay that typically only detects tighter interactions. The eIF5B-binding motif at the C-terminus of S. cerevisiae eIF1A has an extra hydrophobic residue (LDIDDI), compared to that of human eIF1A (EDIDDI), which may account for higher binding affinity for eIF5B. The difference between human and yeast may not be just quantitative, however, because a 23-residue S. cerevisiae eIF1A C-terminal fragment bound tighter to eIF5B than a 14-residue fragment, and a deletion mutant lacking the last 14 residues (eIF1AΔ141-153), was still able to bind eIF5B, albeit very weakly (18). This is clearly not the case with human eIF1A and eIF5B (Table 1), which indicates that S. cerevisiae eIF1A and eIF5B may have greater affinity for each other in solution and may form a complex off the ribosome. Therefore, it would be interesting to know where and how tightly S. cerevisiae eIF1AΔ141-153 binds to eIF5B, as well as whether this interaction mediates yeast-specific functions. A greater affinity between yeast eIF1A and eIF5B, compared to their human counterparts, could, for instance, indicate a greater role of yeast eIF1A-CTT in recruiting eIF5B to the ribosome.
A ‘domain release mechanism’ was recently proposed, which postulates that GTP binding causes the eIF5B-D3 to disengage from D1 (the G-domain) and D2, rendering the D3/D4 segment of eIF5B flexible and able to sample different orientations and eventually become immobilized upon binding to Met-tRNAi (36). The weakening of the interaction of D3 with D1 and D2 was supported with experimental evidence. However, the eIF5B affinity for GTP increased only 2–3-fold upon deletion of D3/D4. Therefore, GTP binding should weaken the D1/D2 interaction with D3 by 2–3-fold. Such modest thermodynamic coupling is more likely to weaken the contacts with D3 than to completely abolish them. Therefore, it is difficult to distinguish whether GTP binding completely eliminates the interdomain interactions, as per the ‘domain release’ model, or merely weakens them. An advantage of the ‘weakened interactions’ alternative is that if D3 still contacts D1 in its GTP-bound state at least transiently, and if this contact is further weakened by eIF5B binding to the 40S subunit and/or Met-tRNAi, this would contribute to cooperative binding of GTP and the Met-tRNAi to eIF5B on the ribosome. Either variation of the ‘domain release’ model is compatible with the model shown in Figure 6.
The structure of the archaeal eIF5B homolog, aIF5B (14) showed a rigid chalice-like structure, with modest conformational changes induced by the addition of GTP. This led the authors to propose an ‘articulated lever’ model, whereby eIF5B-D3/D4 swing as a rigid body. While this model has shaped our views for over a decade, recent results from multiple groups clearly show that both IF2 and eIF5B are much more flexible than the aIF5B structure led us to believe. Not only is the movement of D1 and D2 relative to each other much more significant, but the D2-D3 and D3-D4 linkers act as flexible hinges, allowing the protein to sample multiple conformations (24,34,36,48). That said, there is consensus in the field that when bound to the initiator tRNA on the small ribosomal subunit, both IF2 and eIF5B adopt a discrete conformation, conducive for binding of the large ribosomal subunit, while flexible enough to undergo rearrangements upon subunit joining. It was also suggested that in eukaryotes, eIF1A-CTT could stabilize the eIF5B-Met-tRNAi conformation, serving as one of the legs of a tripod (34). However, our data show that only the extreme eIF1A C-terminus folds upon binding to eIF5B-D4, whereas the rest of the CTT remains disordered. There is also no indication that the eIF1A-CTT sequence has a propensity to form a rigid ‘leg-like’ structure on its own. Furthermore, the eIF1A-CTT length varies significantly among species, from as few as 20 residues in S. pombe (likely close to the minimum possible length, see above), to 35 in S. cerevisae, while the eIF1A-CTT length in mammals is 26 residues. Therefore, it seems more likely that the eIF1A-CTT role is to keep eIF5B-D4 near without fixing its position.
IDPRs are present in a large number of translation initiation factors and many of them are known to mediate important protein–protein and protein–RNA interactions. While binding-induced folding is well known, some dynamic interactions mediated by IDPRs do not lead to folding and the resulting ‘fuzzy’ complexes present a much greater conceptual challenge. Such dynamic interactions typically have a strong electrostatic component, because the strength of electrostatic interactions decreases with the first power of the distance between the charges (the strength of van der Waals interactions, for instance, decreases with the sixth power of the distance). Thus, in a disordered peptide, modest movements of the interacting charges away from the optimal distance may carry lower enthalpy loss, compared to the significant entropy loss associated with folding the peptide (reviewed in (25–28)). A notable example of IDPR undergoing binding-induced folding is the N-terminal portion of eIF4G, which tethers the PIC to the 5′-Cap by binding to eIF4E and to the 3′-polyA tail by binding to PABP. In both of these cases, the interaction induces folding of a portion of the IDPR (reviewed in (3–5)). eIF1A presents an interesting example of a combination of binding-induced folding, as well as dynamic interactions. eIF1A-NTT and -CTT are IDPRs (6), which as we show here (Figure 2 and Supplementary Figure S1), dynamically contact the folded OB domain, without becoming folded. The interaction involving eIF1A-CTT interferes with eIF1A binding to eIF5B (Figures 3 and 4) and likely also with nonspecific binding to cellular RNAs, but not with the high-affinity specific binding to the 40S ribosomal subunit. As discussed above, upon binding to the 40S, both the NTT and CTT end up in the ribosomal P site (step 2 in Figure 6), where they play important regulatory roles in scanning and start codon selection (7,11,42,49). The NTT undergoes binding-induced folding in the P-site, whereas the CTT remains dynamic while performing its role in the P-site (7,9–11). Upon start codon selection, the CTT is ejected from the P-site and binds to eIF5B-D4 (step 4 in Figure 6), which causes binding-induced folding of its extreme C-terminus (17,34). As described here (see also Supplementary Figure S3D), the rest of the CTT remains disordered, which results in eIF5B-D4 being tethered by eIF1A-CTT, while retaining its mobility.
Many questions remain about the roles eIF1A and eIF5B play in translation initiation. While there is ample experimental support for the main framework of the model presented here (Figure 6), it is not always known when certain interactions are formed and broken. In particular, it would be important to determine when and how eIF5B is recruited to the 40S ribosomal subunit, its position in 43S and 48S ICs; and what role, if any, the eIF5B N-terminal region plays in this process.
Supplementary Material
Acknowledgments
We thank Andrew Bogorad for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM095720 to A.M.; Shared Instrument Grant, 1S10-OD11941 to C. J. McKnight]; Bridgewater State University [FLRG grant to B.M].
Conflict of interest statement. None declared.
REFERENCES
- 1.Hinnebusch A.G. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 2011;75:434–467. doi: 10.1128/MMBR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinnebusch A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 3.Jackson R.J., Hellen C.U., Pestova T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marintchev A., Wagner G. Translation initiation: structures, mechanisms and evolution. Q. Rev. Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- 5.Sonenberg N., Hinnebusch A.G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battiste J.L., Pestova T.V., Hellen C.U., Wagner G. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell. 2000;5:109–119. doi: 10.1016/s1097-2765(00)80407-4. [DOI] [PubMed] [Google Scholar]
- 7.Yu Y., Marintchev A., Kolupaeva V.G., Unbehaun A., Veryasova T., Lai S.C., Hong P., Wagner G., Hellen C.U., Pestova T.V. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res. 2009;37:5167–5182. doi: 10.1093/nar/gkp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashem Y., des Georges A., Dhote V., Langlois R., Liao H.Y., Grassucci R.A., Hellen C.U., Pestova T.V., Frank J. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell. 2013;153:1108–1119. doi: 10.1016/j.cell.2013.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lomakin I.B., Steitz T.A. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature. 2013;500:307–311. doi: 10.1038/nature12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weisser M., Voigts-Hoffmann F., Rabl J., Leibundgut M., Ban N. The crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A. Nat. Struct. Mol. Biol. 2013;20:1015–1017. doi: 10.1038/nsmb.2622. [DOI] [PubMed] [Google Scholar]
- 11.Saini A.K., Nanda J.S., Lorsch J.R., Hinnebusch A.G. Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNA(i)(Met) binding to the ribosome. Genes Dev. 2010;24:97–110. doi: 10.1101/gad.1871910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi S.K., Olsen D.S., Roll-Mecak A., Martung A., Remo K.L., Burley S.K., Hinnebusch A.G., Dever T.E. Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol. Cell. Biol. 2000;20:7183–7191. doi: 10.1128/mcb.20.19.7183-7191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pestova T.V., Lomakin I.B., Lee J.H., Choi S.K., Dever T.E., Hellen C.U. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- 14.Roll-Mecak A., Cao C., Dever T.E., Burley S.K. X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell. 2000;103:781–792. doi: 10.1016/s0092-8674(00)00181-1. [DOI] [PubMed] [Google Scholar]
- 15.Pisarev A.V., Kolupaeva V.G., Pisareva V.P., Merrick W.C., Hellen C.U., Pestova T.V. Specific functional interactions of nucleotides at key -3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20:624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fringer J.M., Acker M.G., Fekete C.A., Lorsch J.R., Dever T.E. Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining. Mol. Cell. Biol. 2007;27:2384–2397. doi: 10.1128/MCB.02254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marintchev A., Kolupaeva V.G., Pestova T.V., Wagner G. Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: a new interaction between old partners. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1535–1540. doi: 10.1073/pnas.0437845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olsen D.S., Savner E.M., Mathew A., Zhang F., Krishnamoorthy T., Phan L., Hinnebusch A.G. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J. 2003;22:193–204. doi: 10.1093/emboj/cdg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acker M.G., Shin B.S., Dever T.E., Lorsch J.R. Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J. Biol. Chem. 2006;281:8469–8475. doi: 10.1074/jbc.M600210200. [DOI] [PubMed] [Google Scholar]
- 20.Acker M.G., Shin B.S., Nanda J.S., Saini A.K., Dever T.E., Lorsch J.R. Kinetic analysis of late steps of eukaryotic translation initiation. J. Mol. Biol. 2009;385:491–506. doi: 10.1016/j.jmb.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unbehaun A., Marintchev A., Lomakin I.B., Didenko T., Wagner G., Hellen C.U., Pestova T.V. Position of eukaryotic initiation factor eIF5B on the 80S ribosome mapped by directed hydroxyl radical probing. EMBO J. 2007;26:3109–3123. doi: 10.1038/sj.emboj.7601751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boileau G., Butler P., Hershey J.W., Traut R.R. Direct cross-links between initiation factors 1, 2, and 3 and ribosomal proteins promoted by 2-iminothiolane. Biochemistry. 1983;22:3162–3170. doi: 10.1021/bi00282a020. [DOI] [PubMed] [Google Scholar]
- 23.Allen G.S., Zavialov A., Gursky R., Ehrenberg M., Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 24.Simonetti A., Marzi S., Billas I.M., Tsai A., Fabbretti A., Myasnikov A.G., Roblin P., Vaiana A.C., Hazemann I., Eiler D., et al. Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor. Proc. Natl. Acad. Sci. U.S.A. 2013;110:15656–15661. doi: 10.1073/pnas.1309578110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uversky V.N. The most important thing is the tail: multitudinous functionalities of intrinsically disordered protein termini. FEBS Lett. 2013;587:1891–1901. doi: 10.1016/j.febslet.2013.04.042. [DOI] [PubMed] [Google Scholar]
- 26.Oldfield C.J., Dunker A.K. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 27.Chen T., Song J., Chan H.S. Theoretical perspectives on nonnative interactions and intrinsic disorder in protein folding and binding. Curr. Opin. Struct. Biol. 2015;30:32–42. doi: 10.1016/j.sbi.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Das R.K., Ruff K.M., Pappu R.V. Relating sequence encoded information to form and function of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2015;32:102–112. doi: 10.1016/j.sbi.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marintcheva B., Marintchev A., Wagner G., Richardson C.C. Acidic C-terminal tail of the ssDNA-binding protein of bacteriophage T7 and ssDNA compete for the same binding surface. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1855–1860. doi: 10.1073/pnas.0711919105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fekete C.A., Applefield D.J., Blakely S.A., Shirokikh N., Pestova T., Lorsch J.R., Hinnebusch A.G. The eIF1A C-terminal domain promotes initiation complex assembly, scanning and AUG selection in vivo. EMBO J. 2005;24:3588–3601. doi: 10.1038/sj.emboj.7600821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marintchev A., Frueh D., Wagner G. NMR methods for studying protein-protein interactions involved in translation initiation. Methods Enzymol. 2007;430:283–331. doi: 10.1016/S0076-6879(07)30012-8. [DOI] [PubMed] [Google Scholar]
- 32.Battiste J.L., Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 33.Zhuravleva A., Clerico E.M., Gierasch L.M. An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell. 2012;151:1296–1307. doi: 10.1016/j.cell.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng A., Yu J., Yamamoto R., Ose T., Tanaka I., Yao M. X-ray structures of eIF5B and the eIF5B-eIF1A complex: the conformational flexibility of eIF5B is restricted on the ribosome by interaction with eIF1A. Acta Crystallogr. D. Biol. Crystallogr. 2014;70:3090–3098. doi: 10.1107/S1399004714021476. [DOI] [PubMed] [Google Scholar]
- 35.Fernandez I.S., Bai X.C., Hussain T., Kelley A.C., Lorsch J.R., Ramakrishnan V., Scheres S.H. Molecular architecture of a eukaryotic translational initiation complex. Science. 2013;342:824. doi: 10.1126/science.1240585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuhle B., Ficner R. eIF5B employs a novel domain release mechanism to catalyze ribosomal subunit joining. EMBO J. 2014;33:1177–1191. doi: 10.1002/embj.201387344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto H., Unbehaun A., Loerke J., Behrmann E., Collier M., Burger J., Mielke T., Spahn C.M. Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA. Nat. Struct. Mol. Biol. 2014;21:721–727. doi: 10.1038/nsmb.2859. [DOI] [PubMed] [Google Scholar]
- 38.von der Haar T., McCarthy J.E. Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol. Microbiol. 2002;46:531–544. doi: 10.1046/j.1365-2958.2002.03172.x. [DOI] [PubMed] [Google Scholar]
- 39.Pestova T.V., de Breyne S., Pisarev A.V., Abaeva I.S., Hellen C.U. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J. 2008;27:1060–1072. doi: 10.1038/emboj.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pestova T.V., Kolupaeva V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanov I.P., Loughran G., Sachs M.S., Atkins J.F. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1) Proc. Natl. Acad. Sci. U.S.A. 2010;107:18056–18060. doi: 10.1073/pnas.1009269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin-Marcos P., Cheung Y.N., Hinnebusch A.G. Functional elements in initiation factors 1, 1A, and 2beta discriminate against poor AUG context and non-AUG start codons. Mol. Cell. Biol. 2011;31:4814–4831. doi: 10.1128/MCB.05819-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loughran G., Sachs M.S., Atkins J.F., Ivanov I.P. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res. 2012;40:2898–2906. doi: 10.1093/nar/gkr1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Myasnikov A.G., Marzi S., Simonetti A., Giuliodori A.M., Gualerzi C.O., Yusupova G., Yusupov M., Klaholz B.P. Conformational transition of initiation factor 2 from the GTP- to GDP-bound state visualized on the ribosome. Nat. Struct. Mol. Biol. 2005;12:1145–1149. doi: 10.1038/nsmb1012. [DOI] [PubMed] [Google Scholar]
- 45.Llacer J.L., Hussain T., Marler L., Aitken C.E., Thakur A., Lorsch J.R., Hinnebusch A.G., Ramakrishnan V. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol. Cell. 2015;59:399–412. doi: 10.1016/j.molcel.2015.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murakami R., Miyoshi T., Uchiumi T., Ito K. Crystal structure of translation initiation factor 5B from the crenarchaeon Aeropyrum pernix. Proteins. 84:712–717. doi: 10.1002/prot.25009. [DOI] [PubMed] [Google Scholar]
- 47.Simonetti A., Marzi S., Myasnikov A.G., Fabbretti A., Yusupov M., Gualerzi C.O., Klaholz B.P. Structure of the 30S translation initiation complex. Nature. 2008;455:416–420. doi: 10.1038/nature07192. [DOI] [PubMed] [Google Scholar]
- 48.Eiler D., Lin J., Simonetti A., Klaholz B.P., Steitz T.A. Initiation factor 2 crystal structure reveals a different domain organization from eukaryotic initiation factor 5B and mechanism among translational GTPases. Proc. Natl. Acad. Sci. U.S.A. 2013;110:15662–15667. doi: 10.1073/pnas.1309360110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nanda J.S., Saini A.K., Munoz A.M., Hinnebusch A.G., Lorsch J.R. Coordinated movements of eukaryotic translation initiation factors eIF1, eIF1A, and eIF5 trigger phosphate release from eIF2 in response to start codon recognition by the ribosomal preinitiation complex. J. Biol. Chem. 2013;288:5316–5329. doi: 10.1074/jbc.M112.440693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.