Abstract
Ribosome assembly is an essential and conserved cellular process in eukaryotes that requires numerous assembly factors. The six-subunit UTPB complex is an essential component of the 90S precursor of the small ribosomal subunit. Here, we analyzed the molecular architecture of UTPB using an integrative structural biology approach. We mapped the major interactions that associate each of six UTPB proteins. Crystallographic studies showed that Utp1, Utp21, Utp12 and Utp13 are evolutionarily related and form a dimer of dimers (Utp1–Utp21, Utp12–Utp13) through their homologous helical C-terminal domains. Molecular docking with crosslinking restraints showed that the WD domains of Utp12 and Utp13 are associated, as are the WD domains of Utp1, Utp21 and Utp18. Electron microscopy images of the entire UTPB complex revealed that it predominantly adopts elongated conformations and possesses internal flexibility. We also determined crystal structures of the WD domain of Utp18 and the HAT and deviant HAT domains of Utp6. A structural model of UTPB was derived based on these data.
INTRODUCTION
Ribosome assembly is an essential cellular process in every organism. The eukaryotic ribosome is composed of a 40S small subunit (SSU) and a 60S large subunit (LSU) and is assembled from four ribosomal RNAs (rRNAs) and ∼80 ribosomal proteins (r-proteins). Extensive studies in the yeast Saccharomyces cerevisiae have found ∼200 trans-acting proteins and many snoRNAs involved in ribosome assembly. Most of these ribosome assembly factors are conserved in other eukaryotes (1). These factors transiently associate with ribosomal subunits, forming various pre-ribosomal particles and function in the modification and processing of rRNA, r-protein assembly and structural remodeling and export of pre-ribosomal particles (2–5).
Except for 5S rRNA, which is transcribed separately, three of the four rRNAs are transcribed in the nucleolus as a single 35S precursor that additionally encodes external and internal spacer sequences (ETS and ITS). The spacers are removed through a series of nucleolytic cleavage steps in the context of pre-ribosomal particles. The pre-rRNA is co-transcriptionally packed into a large particle, called the 90S pre-ribosome or SSU processome (6–8). This particle likely corresponds to the ∼40 nm knob structure at the 5′-terminus of nascent pre-rRNA transcripts (8). Upon formation of 90S particles, the pre-rRNA is cleaved at the A0 and A1 sites of 5′ ETS and at the A2 site of ITS1. The 90S particle is then converted into a pre-40S particle that contains a 20S pre-rRNA (9). The pre-40S is exported to the cytoplasm and maturates into the SSU. The 3′ portion of the pre-rRNA encoding LSU rRNAs is packed into pre-60S particles and processed into 5.8S and 25S rRNA.
Biochemical studies have revealed that the 90S particle contains 35S pre-rRNA, U3 snoRNA, r-proteins and ∼50 stably associated assembly factors (6,7,10). The molecular mass of 90S has been estimated to be 5 MDa, almost four times that of the mature 40S ribosome, making it one of the most complex ribonucleoprotein (RNP) particles in cells. A few stable subcomplexes of 90S assembly factors, including UTPA, UTPB, UTPC and U3 snoRNP, have been purified (6,11,12). These subcomplexes appear to assemble into 90S as independent modules (13,14). The structure of 90S is poorly understood at present and is highly challenging to investigate. Structural analysis of its individual components and subcomplexes is an important step to understand this highly complicated RNP structure.
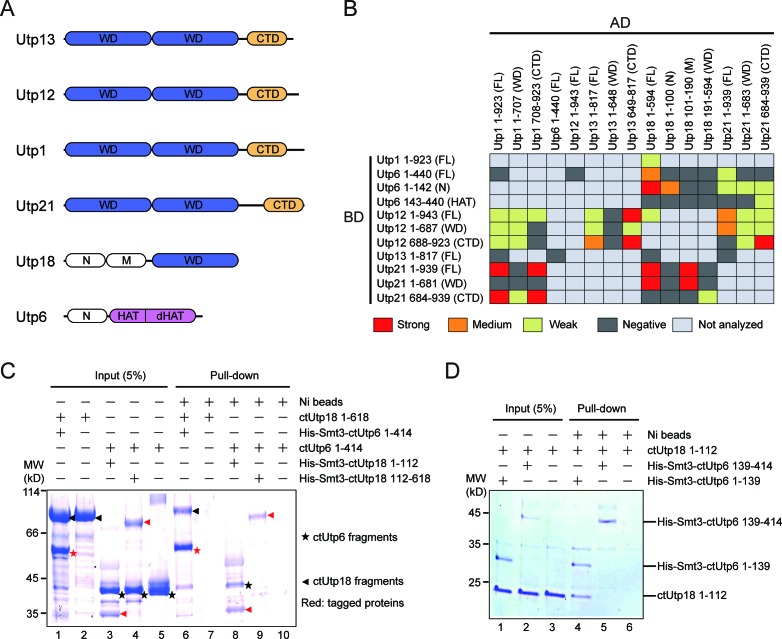
The UTPB complex is made up of six proteins (Utp1/Pwp2, Utp6, Utp12/Dip2, Utp13, Utp18 and Utp21) and has a total molecular mass of 525 kDa (6,11) (Figure 1A). The WD domain is one of the most common domains in eukaryotes and often mediates protein-protein interactions. Utp1, Utp12, Utp13 and Utp21 each consist of tandem WD domains at the N-terminus and 170–260 residues of unknown structure and function at the C-terminus. There is no apparent sequence homology among the C-terminal domains (CTDs) of the four proteins. Utp18 bears a single WD domain preceded by ∼250 residues at the N-terminus. Utp6 is predicted to contain three HAT (half-a-tetratricopeptide) repeats at the middle region (15,16). The HAT repeats fold into elongated helical structures (17,18) and likely mediate protein interactions. A short sequence in the Utp21 WD domain has been found to bind the Utp6 HAT domain (15).
Figure 1.
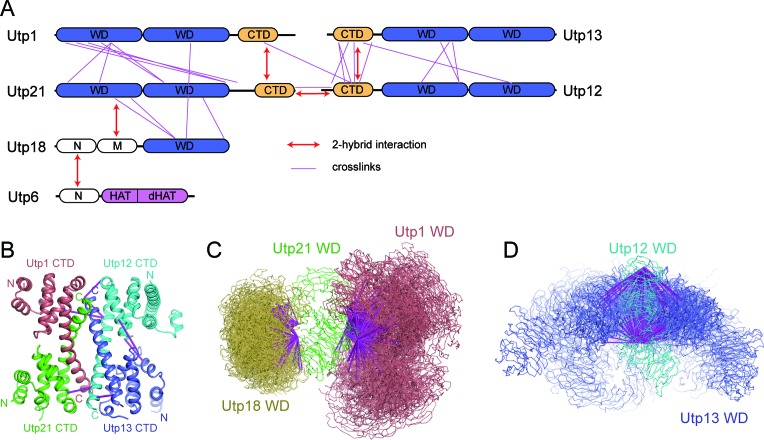
Domain–domain interaction of UTPB proteins. (A) Domain diagrams of UTPB proteins. Utp13, Utp12, Utp1 and Utp21 are composed of tandem WD domains and a C-terminal domain (CTD). Utp18 is composed of an N, M and WD domain. Utp6 is composed of an N, HAT and deviant HAT (dHAT) domain. (B) Two-hybrid interactions among UTPB protein fragments. The indicated Saccharomyces cerevisiae genes were fused to the activation domain (AD) and GAL4 DNA-binding domain (BD) and co-expressed in the AH109 strain. An interaction was identified as weak, medium or strong when the two-hybrid strain is able to grown on SC medium lacking Leu, Trp and His (low stringency), or the above medium additionally containing 5 mM 3- AT (medium stringency) or lacking Ade (high stringency). Negative interaction indicates no growth on the low stringency medium. FL, full-length. The previously published interactions between Utp21 and Utp18 are included for completeness (22). (C and D) Pull-down assay of ctUtp6 and ctUtp18. One protein contained a His-Smt3-tag and was pulled down with Ni Sepharose beads. The tagged proteins were marked with red symbol in C. The input (5%) and bound proteins were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie staining. The positions of molecular weight markers are indicated on the left.
UTPB is a critical component of 90S, yet its exact function is not known. Depletion of Utp1 blocks assembly of U3 snoRNA and also abolishes the A0, A1 and A2 cleavage of pre-rRNA (19). All six UTPB proteins are universally conserved in eukaryotes and some have been associated with human diseases. For example, mutations of the human UTP21 gene are the causative agents of some forms of primary open angle glaucoma (20) and heterozygous deletion of the UTP6 gene is a candidate modifier of neurofibromatosis type 1 (21).
The structure of UTPB has been investigated with two-hybrid assays, protein-fragment complementation assays, chemical crosslinking and mass spectrometry (CXMS), in vitro reconstitution and crystallography (15,22–25). High resolution structures have been determined only for the tandem WD domain of Utp21 (22). The structural organization of UTPB remains largely unknown. In this study, we present the first structural model of UTPB that integrates results from X-ray crystallography, homology modeling, protein interaction data, chemical crosslinking, molecular docking and electron microscopy (EM).
MATERIALS AND METHODS
DNA cloning
The genes for the UTPB proteins were polymerase chain reaction (PCR) amplified from S. cerevisiae and Chaetomium thermophilum genomic DNAs. For the C. thermophilum genes containing introns, the individual exon sequences were amplified and joined by PCR. The genes were cloned into plasmid pET28a without any tag, a modified pET28a with an N-terminal 6-histidine and Smt3 (His-Smt3) tag or pGEX-6p-1 with an N-terminal GST-tag. All constructs were confirmed by DNA sequencing.
Protein purification
The plasmids were transformed into Escherichia coli strains BL21(DE3)-RIL unless otherwise stated. After the cells were grown to an A600 of 1.0 at 37°C, the temperature was shifted to 22°C and isopropyl-β-D-thiogalactoside was added to a final concentration of 0.5 mM to induce protein expression. The cells were broken by a high-pressure cell disruptor (JNBIO).
His-Smt3-tagged ctUtp12 748–956 and GST-tagged ctUtp13 738–912 were co-expressed. The cells were harvested and broken in buffer A (25 mM Tris–HCl, pH 8.0, 50 mM imidazole, 500 mM NaCl). After clarification, the supernatant was loaded onto a HisTrap column (GE Healthcare) and the protein was eluted with buffer B (25 mM Tris–HCl, pH 8.0, 300 mM imidazole and 500 mM NaCl). The His-Smt3 tag of ctUtp12 was cleaved with Ulp1 overnight at 4°C. The sample was bound to glutathione Sepharose resin (GE Healthcare) in a PD-20 column (Bio-Rad), followed by washing with buffer A. The GST-tag of ctUtp13 was cleaved on column with PreScission proteinase overnight. The flow through was collected, concentrated and loaded onto a HiLoad 16/60 Superdex 200 column running in buffer C (10 mM Tris–HCl, pH 8.0, 200 mM NaCl). The peak fractions were pooled and concentrated with centrifugal filters.
To assemble and purify the CTD tetramer, untagged ctUtp1 701–849 and untagged ctUtp21 806–1049 were expressed separately in Rosetta(DE3). His-Smt3-tagged ctUtp12 769–931 and GST-tagged ctUtp13 738–889 were co-expressed in BL21(DE3)-RIL. These cells were mixed and disrupted. The CTD tetramer was purified in the same way as the CTD dimer of ctUtp12 and ctUtp13.
His-Smt3-tagged ctUtp6 81–414 was purified with HisTrap chromatography. After removal of the His-Smt3 tag with Ulp1, the protein was purified with gel filtration chromatography.
His-Smt3-tagged ctUtp18 197–618 was purified through HisTrap chromatography. After removal of the His-Smt3 tag with Ulp1, the sample was loaded on a heparin column and eluted with a NaCl gradient. The eluted protein was concentrated and subjected to gel filtration chromatography.
Proteins were labeled with selenomethionine (SeMet) as previously described (22). All SeMet-labeled proteins were purified similarly to native proteins.
Crystallization
Crystallization was performed with the vapor diffusion method using sitting drop for screening and hanging drop for optimization at 20°C. Normally, 1 μl of protein solution was mixed with 1 μl of reservoir solution.
Crystals of the complex of ctUtp12 748–956 and ctUtp13 738–912 (25 mg/ml protein in 10 mM Tris–HCl, pH 8.0, 200 mM NaCl) were obtained from the reservoir solution containing 0.1 M Bis–Tris–HCl, pH 6.5, 50% pentaerythritol, 0.35 M ammonium sulfate and 0.2 M sodium thiocyanate and were cryoprotected in the reservoir solution supplemented with 15% (v/v) ethanediol before flash freezing in liquid nitrogen. SeMet-labeled crystals were obtained under the same conditions.
The CTD tetramer complex was composed of ctUtp1 701–849, ctUtp21 806–1049, ctUtp12 769–931 and ctUtp13 738–889. Crystals of the CTD tetramer (11 mg/ml protein in 10 mM Tris–HCl, pH 8.0, 200 mM NaCl) were obtained from the reservoir solution containing 0.1 M HEPES-Na, pH 7.5, 45% w/v poly(acrylic acid sodium salt) 5100, 0.02 M magnesium chloride and 0.01 M spermidine and were cryoprotected in the reservoir solution supplemented with 15% (v/v) ethanediol. SeMet-labeled crystals were grown under the same conditions.
Crystals of ctUtp6 81–414 (20 mg/ml protein in 10 mM Tris–HCl, pH 8.0, 200 mM NaCl) were obtained from the reservoir solution containing 0.1 M HEPES-Na, pH 7.5, 10% 2-proponal, 20% PEG 4000 and 0.05 M sodium fluoride and were cryoprotected in the mother solution supplemented with 15% (v/v) glycerol. SeMet-labeled crystals were obtained under the same conditions.
Crystals of ctUtp18 197–618 (25 mg/ml protein in 10 mM Tris–HCl, pH 8.0, 200 mM NaCl) were obtained from the reservoir solution containing 0.1 M HEPES-Na, pH 7.5, 0.1 M sodium chloride, 1.75 M ammonium sulfate and 0.01 M calcium chloride and were cryoprotected in the mother solution supplemented with 15% (v/v) ethylene glycol. SeMet-labeled crystals were grown under the same conditions.
Structure determination
Diffraction data were collected for the SeMet-labeled crystals at the K-edge wavelength of selenium at beamline BL17U of the Shanghai Synchrotron Radiation Facility (SSRF). The data were processed by the HKL2000 package (26). The statistics for data processing and refinement are summarized in Supplementary Table S1. All structures were determined with the single-wavelength anomalous dispersion (SAD) method using autosol in PHENIX (27). Structural models were built in COOT (28) and refined against the SeMet datasets in PHENIX. Model building was guided by Se atom positions.
In the case of the CTD tetramer structure, the SAD phases were combined with the molecular replacement phases that were obtained using the CTD dimer structure of ctUtp12 and ctUtp13 as a search model. The phase combination significantly improved the electron density map. During model building and refinement, the SAD phases were also iteratively combined with the phases of partial models.
The crystal of the CTD complex of ctUtp12 and ctUtp13 contains two copies of the heterodimer in the asymmetric unit (ASU). One copy contains ctUtp12 residues 771–798, 825–885 and 893–932 and ctUtp13 residues 742–840 and 856–888. The second copy contains ctUtp12 residues 773–817, 825–839, 850–885 and 894–931 and ctUtp13 residues 767–838 and 858–888.
The structure of the CTD tetramer contains ctUtp1 residues 729–841, ctUtp21 residues 923–956 and 965–1046, ctUtp12 residues 769–931 and ctUtp13 residues 744–846 and 855–889. Many terminal residues and two internal loops (ctUtp21 957–964 and ctUtp13 847–854) were not modeled due to poor electron density. In particular, residues 806–922 at the N-terminus of the ctUtp21 CTD were included in crystallization, yet largely missing in the structure. An α-helix likely from this region was modeled as UNK residues.
The crystal of ctUtp6 81–414 contains four molecules in the ASU. The most complete model (chain A) contains residues 86–196 and 237–409. The structure of ctUtp18 197–618 includes ctUtp18 residues 254–458, 463–502 and 524–618, 6 sulfate ions and 97 water molecules.
Rigid body docking with crosslinking distance restrains
The scUtp21 WD domain structure was previously determined (22). The other WD domain structures were built with MODELLER (29). The structure of the scUtp18 WD domain was modeled on the ctUtp18 WD domain structure. The tandem WD domain structures of scUtp1, scUtp12 and scUtp13 were modeled on the tandem WD domain structure of scUtp21. Multiple sequence alignments were generated with MUSCLE (30) and manually adjusted.
The molecular docking was conducted in Xplor–NIH following the previously published protocol for docking protein complexes from intermolecular NOE distance restraints (31). The individual domains were treated as rigid bodies. The energy function contains a square-well quadratic term for inter-Cα distance restraints and a quartic van der Waals repulsion term. The energy term of gyration radius was not used. Distance restrains were set for the Cα atoms of crosslinked residue pairs with a lower bound of 3 Å and a upper bound of 24 Å (Supplementary Table S2). If a crosslinked residue was absent in the structural model, its nearest residue in the primary sequence present in the model was used and the upper bound of the distance restraint was increased by 4 Å per missing residue.
Docking was conducted separately for the scUtp12 and scUtp13 WD domain complex and the scUtp1, scUtp21 and scUtp18 WD domain complex. One hundred starting structures of randomly orientated domains were generated for each of two complexes and subjected to simulated annealing. The Utp12 and Utp13 WD complex was docked with three crosslinking distance restraints, yielding 80 structures of zero energy (no violation). The Utp1, Utp21 and Utp18 WD domain complex was docked with 6 restraints between Utp21 and Utp1 and 3 restraints between Utp21 and Utp18, yielding 91 structures of zero energy.
Yeast two-hybrid assay
The two-hybrid assay was conducted using the MATCH MAKER GAL4 two-hybrid system (Clontech) as previously described (22). The genes of interest were cloned into pGBKT7 and pGADT7 vectors. Two vectors expressing a bait and a prey protein were co-transformed into yeast strain AH109, which contains the HIS3 and ADE2 reporter genes under a GAL4 promoter. The cell was spotted on plates with Synthetic Complete (SC) medium lacking Leu, Trp and His (low stringency), or the above medium that additionally contained 5 mM 3-amino-1,2,4-triazole (3-AT) (medium stringency) or lacked Ade (high stringency). The plates were incubated for 3 days at 30°C and checked for cell growth.
Pull-down assay
His-Smt3 tagged proteins were purified with HisTrap chromatography. If needed, the His-Smt3 tag was removed by Ulp1 cleavage. The proteins were further purified through ion exchange chromatography. The purified proteins were mixed and bound to Ni Sepharose beads (GE Healthcare). The beads were washed three times with 25 mM Tris–HCl, pH 8.0, 500 mM NaCl and 40 mM imidazole, followed by elution with buffer B. The samples were mixed with an equal volume of 2× sodium dodecyl sulphate loading buffer and boiled at 95°C for 5 min before loading onto sodium dodecyl sulphate-polyacrylamide gel electrophoresis gels. The gel was stained with Coomassie blue.
Electron microscopy
The six proteins of UTPB were co-expressed and co-purified from insect cells as previously described (24). For EM analysis, UTPB was crosslinked with the GraFix method (32). Briefly, two mother solutions of 10 and 30% glycerol were prepared in buffer 50 mM HEPES-Na, pH 8.0, 100 mM NaCl. The high-density solution contained additional 0.2% glutaraldehyde. Other concentrations of glycerol were formed by mixing the two mother solutions in proper ratios. Ultracentrifugation tubes were iteratively layered with 1 ml of glycerol solution of decreasing concentrations (30, 23.3, 16.7 and 10%) and frozen in liquid nitrogen after addition of every new layer. The tubes were placed at 4°C overnight to allow formation of 10–30% linear gradients. Then, 100 μl of UTPB (∼1 mg/ml) was layered on the gradients and centrifuged for 16 h at 33 000 rpm in a SW60 Ti rotor (Beckman). Fractions of 200 μl were collected manually.
Five microliters of the GraFix purified UTPB sample (0.1–0.3 mg/ml) was applied to freshly glow-discharged, carbon-coated grids and negatively stained with 2% uranyl acetate. EM images were acquired on a Tecai T12 transmission EM (FEI) operated at 120 kV with a nominal magnification of 42,000 and a pixel size of 2.49 Å. Micrographs were recorded on a CCD camera with defocus values of 0.8–2 μm. Then, 5812 particles were picked from 108 micrographs with e2boxer.py and corrected for contrast transfer function in EMAN2 (33). Reference-free 2D classification was conducted with RELION (34).
RESULTS
Domain–domain interactions of UTPB proteins
A two-hybrid interaction network has been derived for six yeast UTPB proteins (15). However, two-hybrid interactions can be artificial and which domain of each protein mediates the interaction is still mostly unknown. To determine the major interactions that assemble six UTPB proteins into a complex, we assessed two-hybrid interactions for individual domains of yeast UTPB proteins in a non-systemic manner (Figure 1B). The two-hybrid strain was grown at three different stringency conditions to assess the strength of the interaction. We also examined many of the detected two-hybrid interactions with in vitro pull-down assays using recombinant proteins from S. cerevisiae (sc) or C. thermophilum (ct) (Figure 1C, D and data not shown). C. thermophilum is a thermophilic yeast that can grow at up to 60°C and its proteins often show favorable properties for biochemical and structural studies (35). The interaction data from the two organisms, if acquired, are fully consistent with each other.
The two-hybrid interactions that were detected only under the lowest stringency conditions, if analyzed, were not confirmed with pull-down assay, suggesting that they are truly weak or non-specific. In contrast, the two-hybrid interactions detected at medium or strong stringency conditions were largely confirmed. As an exception, we were unable to detect the interaction between Utp21 and the Utp12 WD domain at the protein level. The N-terminal region of Utp6 (residues 1–91 in scUtp6) has been shown to bind Utp18 (15), but it is unknown which part of Utp18 is involved. We showed that residues 1–112 of ctUtp18 have a strong interaction with residues 1–139 of ctUtp6 (Figure 1C).
In summary, our data showed that Utp1, Utp21, Utp12 and Utp13 interact with each other via their CTDs, and that Utp6 and Utp18 associate through their N-terminal regions. In a previous study, we have determined that a region N-terminal to the Utp18 WD domain (residues 100–190 in scUtp18) interacts with the WD domain of Utp21. Thus, we identified the primary interactions at the domain level that link all six UTPB proteins together. Based on the interaction results, we further carried out crystallographic analysis of individual domains and subcomplexes of UTPB using S. cerevisiae and C. thermophilum proteins.
Structure of the CTD complex
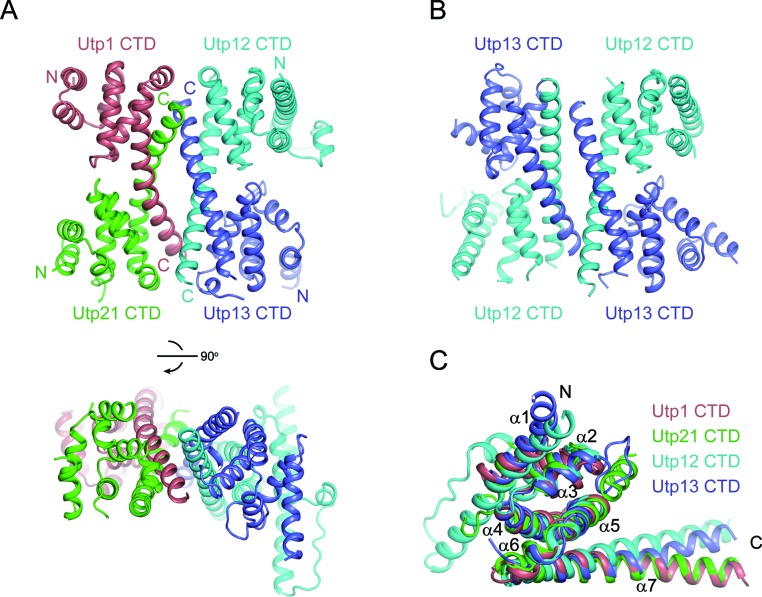
We found that the CTDs of Utp12 and Utp13 formed a complex when they were co-expressed in E. coli (Supplementary Figure S1A and C), thus confirming their two-hybrid interaction. We determined the crystal structure of the ctUtp12 and ctUtp13 CTD complex with Se-SAD phasing and refined the structure to 3.7 Å resolution and an Rwork/Rfree of 0.275/0.299 (Figure 2B and Supplementary Table S1). We further found that the CTDs of Utp12, Utp13, Utp1 and Utp21 formed a homogenous complex and co-eluted during gel filtration (Supplementary Figure S1B and D). The size of the complex is about 100 kDa as compared to protein standards, suggesting that the four CTDs form a tetramer. The crystal structure of the four CTDs of C. thermophilum proteins was subsequently determined by a combination of Se-SAD phasing and molecular replacement using the dimeric structure of ctUtp12 and ctUtp13 CTDs as a search model. The structure revealed a CTD heterotetramer and was refined to 3.5 Å resolution and an Rwork/Rfree of 0.256/0.279 (Figure 2A). The dimer structure of the ctUtp12 and ctUtp13 CTDs is highly similar to the corresponding part in the heterotetramer structure (rmsd = 1.19 Å for 231 Cα pairs). The N-terminal α1 helix of Utp13 displays different conformations in the two structures, likely due to different crystal packing environments. Interestingly, in crystals of the ctUtp12 and ctUtp13 CTDs, the asymmetric unit contains two dimers that pack against each other as in the heterotetramer structure (Figure 2B). The self-association of the Utp12 and Utp13 CTD dimer is likely physiologically irrelevant, because a mixture of the four CTDs of Utp12, Utp13, Utp1 and Utp12 led to a homogenous heterotetramer (Supplementary Figure S1B). Our discussion is focused on the heterotetramer structure.
Figure 2.
Structure of the ctUtp12, ctUtp13, ctUtp21 and ctUtp1 CTDs. (A) Ribbon representation of the CTD tetramer structure shown in two orthogonal orientations. Utp21, Utp1, Utp12 and Utp13 are colored green, salmon, cyan and blue, respectively. The N- and C-termini of each domain are labeled. (B) Structure of the ctUtp12 and ctUtp13 CTD dimer. Two copies of the dimer in the ASU pack into a tetramer. The dimer on the right has the same orientation as its counterpart in the tetramer structure shown in A. (C) Structural alignment of the four CTDs by MatchMaker in Chimera.
Remarkably, the four CTDs adopt a similar all-α fold composed of seven α-helices (α1–α7). The α5 and α7 helices are connected by a short α6 helix. These α-helices are arranged into a right-handed superhelix or solenoid of 1.75 turns. As a result, the N-terminal α1 helix is nearly perpendicular to the C-terminal α7 helix. The longest α7 helix at the C-terminus projects outward from the structural core and mediates major intermolecular interactions. The CTD structure resembles helical repeat structures adopted by Armadillo and HEAT motifs, but is shorter and less regular in terms of size and orientation of helices.
The four CTD structures superimposed with a root-mean-square deviation (rmsd) of 1–1.3 Å for 50–70 Cα atom pairs (Figure 2C). The size of each α-helix is generally conserved among different CTDs, but the α1 helix of Utp12 is much longer than its counterpart in other proteins.
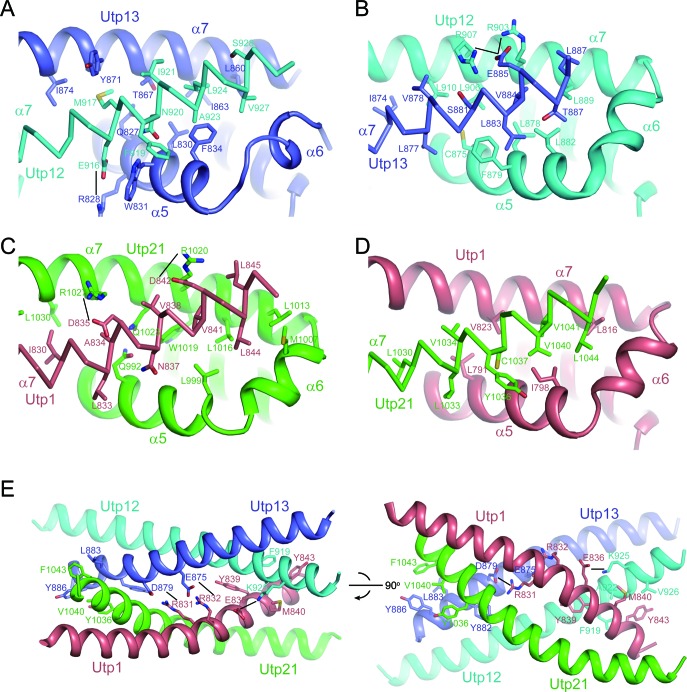
The tetramer can be described as a dimer of dimers. Utp12 and Utp13 associate into one dimer, and Utp1 and Utp21 combine into the other dimer. Due to the pseudo dyad symmetry in each dimer, the complex contains four similar dimer interfaces (Figure 3A–D). In each interface, the C-terminal half of the α7 helix of one subunit packs against a groove formed by the α5 and α7 helices of the other subunit in the dimer. These interfaces are stabilized by many hydrophobic, electrostatic and van der Waals interactions. The four interfaces are organized in a similar way, but differ in specific interactions. This would ensure the association specificity between the four homologous domains.
Figure 3.
Intermolecular interactions in the CTD tetramer. (A–D) Four dimer interfaces. The C-terminal region of the α7 helices are shown as Cα trace. The interacting residues are shown as sticks and labeled. Oxygen is colored red, nitrogen is colored blue and carbon is colored by proteins. Potential salt bridge pairs are connected by lines. (E) The tetramer interface shown in two orthogonal views.
The α7 helices of two subunits in each dimer form an anti-parallel coiled-coil. The coiled-coils of two dimers further pack into a four-helix bundle to mediate the tetramer formation (Figure 3E). At the tetramer interface, Utp21 primarily contacts Utp13 and Utp1 primarily contacts Utp12. The tetramer interface is stabilized mainly by hydrophobic and van der Waals interactions that involve many aromatic residues. In addition, two salt bridges connect the α7 helices of Utp1 and Utp13. The residues that mediate intermolecular interactions are mostly conserved (Supplementary Figure S2).
Structure of Utp6
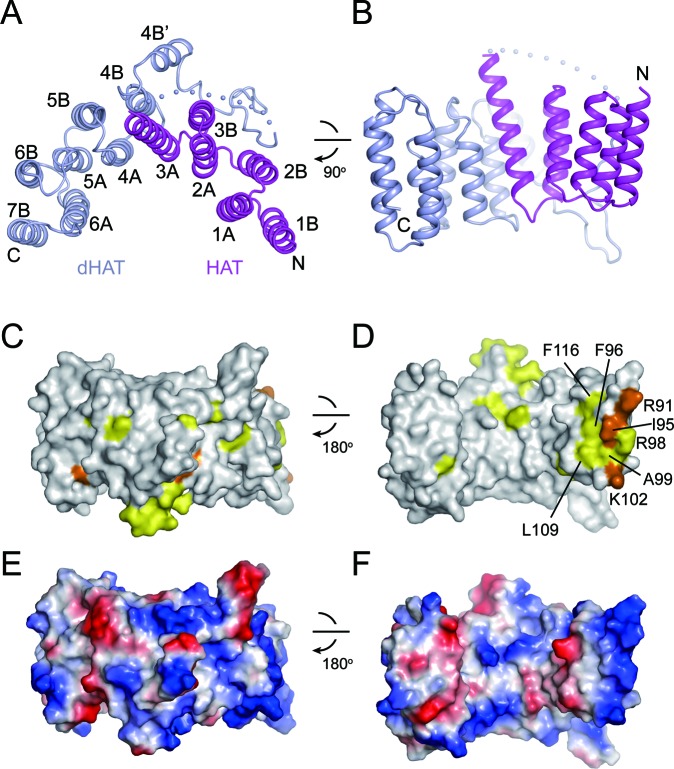
Utp6 contains three HAT repeats (residues 86–196 in ctUtp6) at the central region. We crystallized a fragment of ctUtp6 (residues 81–414) including the HAT repeats and the residues to the C-terminus. The structure was determined with Se-SAD phasing and refined to 3.3 Å resolution and an Rwork/Rfree of 0.205/0.255 (Figure 4A and B; Supplementary Table S1). A HAT repeat folds into two antiparallel helices A and B, which are characterized by specific signature residues (18,36). The three HAT repeats of Utp6 start with a B helix and fold into 6 α-helices (1B to 3A). These helices pack into a superhelix or solenoid structure that closely resembles other HAT domain structures (17,18).
Figure 4.
Structure of ctUtp6. (A and B) Ribbon representation of ctUtp6 structure shown in two orthogonal orientations. The HAT domain is colored magenta and the C-terminal deviant HAT domain is colored lightblue. The helices are labeled and the N- and C-termini are indicated. The dots stand for disordered residues. (C and D) Conservation surface of the Utp6 structure shown in two opposite views. Residues that are conserved in 100 and 80% of the sequences aligned in Supplementary Figure S3 are colored orange and yellow, respectively. The structures shown in D and B have the same orientation. (E and F) Charge surface in two opposite views. The surface is colored blue to red for positively to negatively charged region.
Interestingly, despite lacking the conserved HAT sequence motif, the sequences C-terminal of the HAT repeats also fold into a HAT-like helical structure, called the deviant HAT domain (dHAT). The HAT and dHAT domains each contribute six and seven helices to form an integral superhelix that spans approximately a half circle. The two domains are interrupted by a long loop and a 4B' helix that bind at the convex surface of the superhelix. The sequences of the dHAT domain are poorly conserved (Figure 4C and D; Supplementary Figure S3) and also functionally dispensable (15), suggesting that the dHAT domain plays a minor role in Utp6 structure and function.
The G99E mutation of scUtp6 has been shown to inhibit yeast growth and disrupt the interaction with Utp21 (15). The equivalent A99 residue in ctUtp6 is exposed and situated in a highly conserved region around helix 1B (Figure 4D). The conserved region likely constitutes the binding site for Utp21 or RNA as it is highly positively charged (Figure 4E and F).
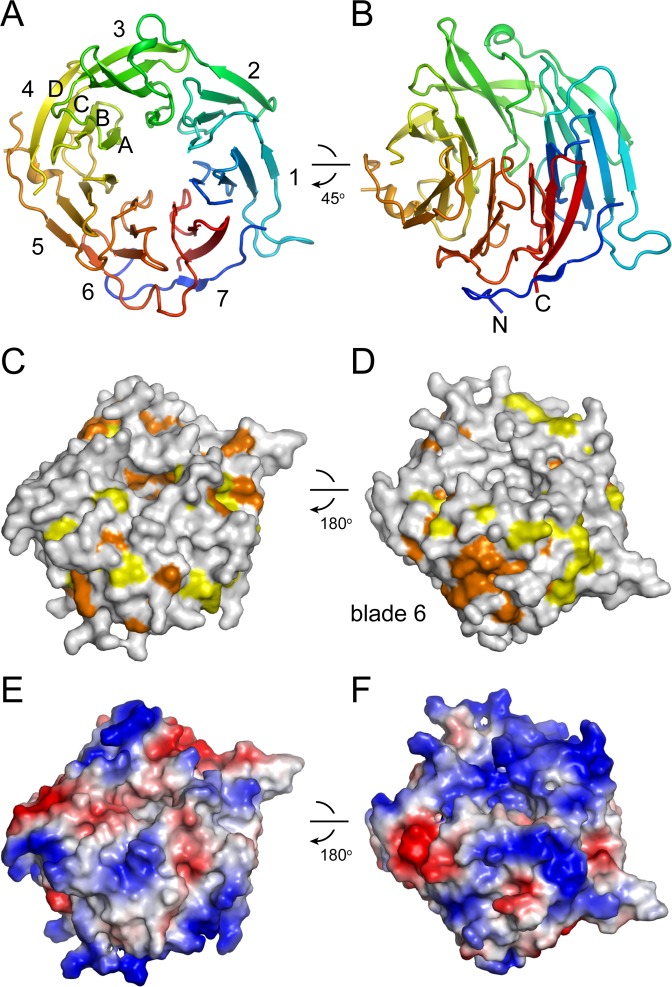
Structure of the ctUtp18 WD domain
The crystal structure of the WD domain of ctUtp18 (residues 254–617) was determined with Se-SAD phasing and refined to an Rwork/Rfree of 0.195/0.244 at 2.33 Å resolution (Figure 5A and B; Supplementary Table S1). The WD domain adopts a typical seven-bladed β-propeller structure with each propeller composed of four antiparallel β strands. The side surface at blade 6 is highly conserved and probably functions as a binding site for other molecules (Figure 5C and D; Supplementary Figure S4). The top surface is positively charged and likely binds to RNA (Figure 5E and F).
Figure 5.
Structure of the ctUtp18 WD domain. (A and B) Ribbon representation of the ctUtp18 WD domain structure. B is a 45° rotation of A along the horizontal axis. Seven blades and the four stands of one blade are labeled and the N- and C-termini are indicated. (C and D) Conservation surface in two opposite views. Residues that are conserved in 100 and 80% of six sequences aligned in Supplementary Figure S4 are colored orange and yellow, respectively. The structures shown in D and B have the same orientation. (E and F) Charge surface in two opposite views. The surface is colored blue to red for positively to negatively charged region.
Docking of WD domains with crosslinking distance restraints
The UTPB complex has been previously analyzed with CXMS and ∼50 intramolecular and 21 intermolecular crosslinks were identified (24) (Figure 6A). The intermolecular crosslinks provide valuable spatial restraints on the structural organization of UTPB. The crosslinks among the four CTDs are fully consistent with the CTD tetramer structure (Figure 6B); namely, the inter-Cα distances between the equivalent ct residues of the crosslinked lysine pairs are all <20 Å (mostly 9–13 Å). This underscores the reliability of the crosslinking data. The other intermolecular crosslinks mainly occur within two groups of WD domains: the WD domains of Utp1, Utp21 and Utp18 and the WD domains of Utp12 and Utp13. We attempted to assemble each of the WD domain groups by rigid body docking of individual domains in the presence of distance restraints for crosslinked residues (see ‘Materials and Methods’ section). Except for the WD domain of scUtp21, whose crystal structure has been determined (22), other structures of S. cerevisiae WD domains were built with homology modeling. The calculated lowest-energy structure ensembles are shown in Figure 6C and D and a representative structure of each complex is shown in Figure 7D.
Figure 6.
Assembly of UTPB WD domains with crosslinking distance restrains. (A) Intermolecular crosslinks detected by (24) are shown as connections between the primary structures of UTPB proteins. No intermolecular crosslinks were detected for Utp6. The diagrams of Utp12 and Utp13 are flipped. The detected strong two-hybrid interactions between individual domain are also indicated. (B) Crosslinks are mapped to the CTD tetramer structure and shown as Cα–Cα distances. (C and D) Ensembles of the Utp1, Utp21 and Utp18 WD domain complex (C) and the Utp12 and Utp13 WD domain complex (D). Thirty structures are shown as Cα traces for each complex and aligned to Utp21 or Utp12. The residue pairs with distance restraints are connected with lines.
Figure 7.
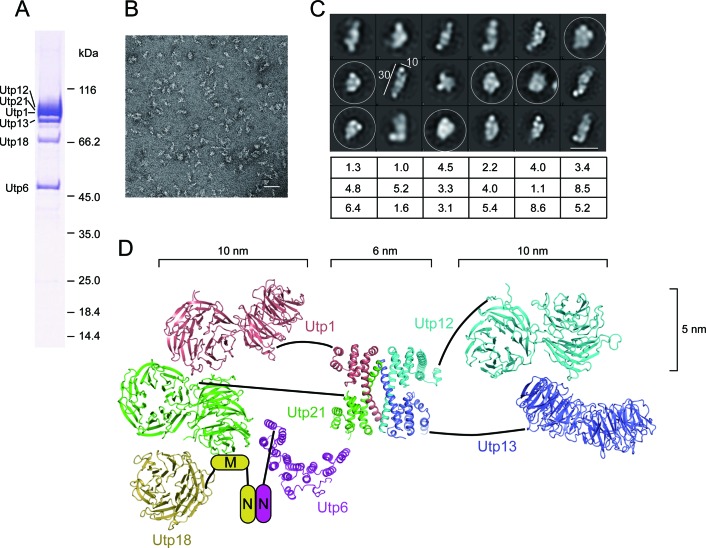
Overall structure of UTPB studied by single-particle EM. (A) An SDS-PAGE gel image of purified UTPB complex. The migrating positions of protein markers are shown on the right. (B) Electron micrograph of negatively stained UTPB. Scale bar, 50 nm. (C) Representative class averages of UTPB. The views of compacted conformations are circled. Scale bar at the bottom-right corner, 25 nm. The dimensions of one elongated class average are labeled with 10 and 30 nm bars. The percentages of total particles used for calculating each class are listed at the bottom. (D) Structural model of UTPB. The N-terminal regions of Utp6 and Utp18 with unknown structure are shown as cartoons. The major dimension of individual domains are indicated.
The results showed that the relative positions of the domains were not well defined by the distance restraints, particularly for the Utp12 and Utp13 WD domain complex. One reason is that the number of available distance restraints was small. Nevertheless, the rough positions of each domain can be determined from such calculation. The WD domains of Utp18 and Utp1 bind to the approximately opposite sides of the WD domain of Utp21. The C-terminal WD domain of Utp12 docks at the junction between the two WD domains of Utp13.
Overall structure of UTPB
We next examined the overall structure of UTPB with single particle negative stain EM (Figure 7A–C). The class averages showed that the UTPB complex predominately adopts elongated conformations, with the longest dimension of ∼30 nm and a width of ∼10 nm (Figure 7C). As a comparison, a tandem WD domain is about 10 nm long, 5 nm wide and 3 nm thick and the CTD tetramer has dimensions of about 6 × 6 × 3 nm. The extended conformations most probably correspond to the two groups of WD domains placed at the opposite sides of the CTD tetramer (Figure 7D). In addition, there are a few compacted views that probably resulted from different conformations of the complex in which the two WD domain groups were folded. The different conformations of UTPB suggest a flexible linker between the tandem WD domain and the CTD in Utp1, Utp21, Utp12 and Utp13.
DISCUSSION
We analyzed the structure of UTPB, a large component of the 90S pre-ribosome, by using hybrid approaches. By integrating all available structural data we depicted a structural model for UTPB (Figure 7D). At the heart of the UTPB structure is the tetrameric complex of the Utp1, Utp21, Utp12 and Utp13 CTDs. Projecting out from the two sides of the CTD tetramer are two arms: one composed of the WD domains of Utp12 and Utp13 and the other composed of the WD domains of Utp1 and Utp21. The latter arm is further enlarged by Utp18, which binds Utp21, and by Utp6, which associates with Utp18 and Utp21. Utp12 and Utp13 have been shown to form a dimer that is dissociable from the remaining part of the UTPB complex (23), which is consistent with the dimer-of-dimers organization of Utp1, Utp21, Utp12 and Utp13.
The two arms appear to preferentially point to opposite directions, giving rise to predominantly the extended conformations of UTPB seen in EM. Several lines of evidence also suggest that the tandem WD domain is flexibly connected to the CTD in Utp1, Utp21, Utp12 and Utp13. The two kinds of domains are independently folded and have no detectable interaction and few crosslinks between them. In addition, flexible linkers are consistent with the multiple conformations of the complex seen by EM. We were unable to reconstruct a reliable 3D model from EM data probably due to the flexibility of the complex. The structural flexibility may be important to allow UTPB to mediate the dynamic assembly process of 90S.
The CXMS and molecular docking analysis showed that the WD domains of Utp1, Utp12 and Utp18 are close in space and as are the WD domains of Utp12 and Utp13. However, interactions between these isolated WD domains could not be detected with two-hybrid and in vitro binding assays (data not shown), suggesting that their association is weak and dependent on the primary interactions mediated by the nearby CTDs of Utp1, Utp21, Utp12 and Utp13 and the M domain of Utp18.
We have previously shown that Utp18 primarily uses the M domain (residues 100–190 in scUtp18) to associate with the tandem WD domain of Utp21 (22). Here, we show that Utp6 and Utp18 associate with their N-terminal regions. The structure of these interacting parts, which account for ∼7% of UTPB mass, remains to be determined. The HAT domain of Utp6 has been shown to bind a peptide from the Utp21 WD domain (Kd = 10 μM) (15). This interaction was weak in our two-hybrid assay and could not be confirmed by pull-down assay, thus suggesting that it is not the major interaction that recruits Utp6 to UTPB.
Our results reveal that Utp1, Utp21, Utp12 and Utp13 are homologous proteins. They share not only an N-terminal tandem WD domain, but also a structurally similar CTD. The homology of their CTDs is not evident at the sequence level. The genes of the four proteins apparently evolved from duplication of an ancestor gene. All four proteins are essential in yeast, indicating their functions have been specified during evolution. We also showed that the CTD functions as a protein tetramerization domain.
Several lines of evidence support that the CTD tetramer structure is not resulted from crystallographic packing artifacts. First, the crosslinking data of reconstituted UTPB complex provide strong support for the tetrameric organization (Figure 6A). Five identified crosslinks between the CTDs of Utp12 and Utp13, one between the CTDs of Utp1 and Utp12 and one between the CTDs of Utp12 and Utp21 are fully consistent with the structure. Second, the gel filtration data show that the CTDs of Utp1, Utp21, Utp12 and Utp13 form a homogenous heterotetrameric complex in solution (Supplementary Figure S1B). The complex has the same degree of oligomerization in solution and crystal. There is little chance that the four proteins would associate differently in solution and crystal. Third, the dimerization mode between Utp12 and Utp13 is conserved in the two different crystals of the Utp12 and Utp13 CTD complex and the four protein CTD complex (Figure 2A and B). Finally, this kind of coiled-coil interaction is commonly seen in mediating protein–protein interaction.
Interestingly, despite its large size, the UTPB complex is built up by rather simple all-β or all-α folds. The complex contains a total of nine WD domains that are exclusively composed of β strands. Except for the N-terminal sequences of Utp18 and Utp6, for which we have no structural information, the remaining sequences, including the CTDs of Utp1, Utp21, Utp12 and Utp13 and the HAT and dHAT domains of Utp6, all adopt helical solenoid structures.
The abundant WD domains of UTPB are expected to mediate interactions with proteins within the UTPB complex and in the 90S. Our data showed that, except for the tandem WD domain of Utp21 that is responsible for recruiting Utp18, most of the WD domains play only a minor role in assembling UTPB itself. Alternatively, most of the WD domains likely function to bind external proteins. Unfortunately, few interactions with other proteins have been reported for UTPB proteins in the literature. Recently, a short motif at the N-terminal region of Utp18 has been found to bind Mtr4 that recruits the exosome for degrading the cleaved 5′ ETS product (37). Although the WD domain is considered as a classical protein-protein interaction domain, it has also been shown to bind RNA. For example, the WD domain of Rrp9/U3-55K recognizes the B/C motif of U3 snoRNA (38–40) and the tandem WD domain of Gemin5 directly interacts with snRNA (41). It cannot be ruled out that some WD domains of UTPB actually bind the pre-rRNA and U3 snoRNA. Identification of the protein and RNA binding targets of UTPB would be important for understanding its function in 90S assembly and pre-rRNA processing.
ACCESSION NUMBERS
The coordinates and structural factors have been deposited into the Protein Data Bank with accession codes 5IC7 (Utp18), 5IC8 (Utp6), 5IC9 (Utp12/Utp13) and 5ICA (Utp12/Utp13/Utp21/Utp1).
Supplementary Material
Acknowledgments
We are grateful to the staff in the Beamline BL17U at SSRF for assistance in data collection and Wanzhong He for great help in EM data collection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Strategic Priority Research Program of the Chinese Academy of Sciences [XDB08010203]; National Natural Science Foundation of China [31430024, 31325007, 91540201]; Beijing Municipal Government. Funding for open access charge: Chinese Academy of Sciences [XDB08010203].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ebersberger I., Simm S., Leisegang M.S., Schmitzberger P., Mirus O., von Haeseler A., Bohnsack M.T., Schleiff E. The evolution of the ribosome biogenesis pathway from a yeast perspective. Nucleic Acids Res. 2014;42:1509–1523. doi: 10.1093/nar/gkt1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woolford J.L., Jr, Baserga S.J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henras A.K., Soudet J., Gerus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turowski T.W., Tollervey D. Cotranscriptional events in eukaryotic ribosome synthesis. Wiley Interdiscip. Rev. RNA. 2014;6:129–139. doi: 10.1002/wrna.1263. [DOI] [PubMed] [Google Scholar]
- 5.Kressler D., Hurt E., Bergler H., Bassler J. The power of AAA-ATPases on the road of pre-60S ribosome maturation - Molecular machines that strip pre-ribosomal particles. Biochim. Biophys. Acta. 2011;1823:92–100. doi: 10.1016/j.bbamcr.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grandi P., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schafer T., Kuster B., Tschochner H., Tollervey D., et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 7.Dragon F., Gallagher J.E., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y., et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osheim Y.N., French S.L., Keck K.M., Champion E.A., Spasov K., Dragon F., Baserga S.J., Beyer A.L. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 9.Schafer T., Strauss D., Petfalski E., Tollervey D., Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2003;22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phipps K.R., Charette J.M., Baserga S.J. The SSU processome in ribosome biogenesis—progress and prospects. Wiley Interdiscip. Rev. RNA. 2011;2:1–21. doi: 10.1002/wrna.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krogan N.J., Peng W.T., Cagney G., Robinson M.D., Haw R., Zhong G., Guo X., Zhang X., Canadien V., Richards D.P., et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 12.Watkins N.J., Segault V., Charpentier B., Nottrott S., Fabrizio P., Bachi A., Wilm M., Rosbash M., Branlant C., Luhrmann R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 13.Perez-Fernandez J., Martin-Marcos P., Dosil M. Elucidation of the assembly events required for the recruitment of Utp20, Imp4 and Bms1 onto nascent pre-ribosomes. Nucleic Acids Res. 2011;39:8105–8121. doi: 10.1093/nar/gkr508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez-Fernandez J., Roman A., De Las Rivas J., Bustelo X.R., Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell. Biol. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Champion E.A., Lane B.H., Jackrel M.E., Regan L., Baserga S.J. A direct interaction between the Utp6 half-a-tetratricopeptide repeat domain and a specific peptide in Utp21 is essential for efficient pre-rRNA processing. Mol. Cell. Biol. 2008;28:6547–6556. doi: 10.1128/MCB.00906-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Champion E.A., Kundrat L., Regan L., Baserga S.J. A structural model for the HAT domain of Utp6 incorporating bioinformatics and genetics. Protein Eng. Des. Sel. 2009;22:431–439. doi: 10.1093/protein/gzp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Legrand P., Pinaud N., Minvielle-Sebastia L., Fribourg S. The structure of the CstF-77 homodimer provides insights into CstF assembly. Nucleic Acids Res. 2007;35:4515–4522. doi: 10.1093/nar/gkm458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai Y., Auperin T.C., Chou C.Y., Chang G.G., Manley J.L., Tong L. Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors. Mol. Cell. 2007;25:863–875. doi: 10.1016/j.molcel.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 19.Dosil M., Bustelo X.R. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J. Biol. Chem. 2004;279:37385–37397. doi: 10.1074/jbc.M404909200. [DOI] [PubMed] [Google Scholar]
- 20.Gallenberger M., Meinel D.M., Kroeber M., Wegner M., Milkereit P., Bosl M.R., Tamm E.R. Lack of WDR36 leads to preimplantation embryonic lethality in mice and delays the formation of small subunit ribosomal RNA in human cells in vitro. Hum. Mol. Genet. 2011;20:422–435. doi: 10.1093/hmg/ddq478. [DOI] [PubMed] [Google Scholar]
- 21.Bartelt-Kirbach B., Wuepping M., Dodrimont-Lattke M., Kaufmann D. Expression analysis of genes lying in the NF1 microdeletion interval points to four candidate modifiers for neurofibroma formation. Neurogenetics. 2009;10:79–85. doi: 10.1007/s10048-008-0154-0. [DOI] [PubMed] [Google Scholar]
- 22.Zhang C., Lin J., Liu W., Chen X., Chen R., Ye K. Structure of Utp21 tandem WD domain provides insight into the organization of the UTPB complex involved in ribosome synthesis. PLoS One. 2014;9:e86540. doi: 10.1371/journal.pone.0086540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poll G., Li S., Ohmayer U., Hierlmeier T., Milkereit P., Perez-Fernandez J. In vitro reconstitution of yeast tUTP/UTP A and UTP B subcomplexes provides new insights into their modular architecture. PLoS One. 2014;9:e114898. doi: 10.1371/journal.pone.0114898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang B., Wu Y.J., Zhu M., Fan S.B., Lin J., Zhang K., Li S., Chi H., Li Y.X., Chen H.F., et al. Identification of cross-linked peptides from complex samples. Nat. Methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 25.Tarassov K., Messier V., Landry C.R., Radinovic S., Serna Molina M.M., Shames I., Malitskaya Y., Vogel J., Bussey H., Michnick S.W. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Marti-Renom M.A., Stuart A.C., Fiser A., Sanchez R., Melo F., Sali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 30.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang C., Clore G.M. A simple and reliable approach to docking protein-protein complexes from very sparse NOE-derived intermolecular distance restraints. J. Biomol. NMR. 2006;36:37–44. doi: 10.1007/s10858-006-9065-2. [DOI] [PubMed] [Google Scholar]
- 32.Kastner B., Fischer N., Golas M.M., Sander B., Dube P., Boehringer D., Hartmuth K., Deckert J., Hauer F., Wolf E., et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods. 2008;5:53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 33.Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 34.Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amlacher S., Sarges P., Flemming D., van Noort V., Kunze R., Devos D.P., Arumugam M., Bork P., Hurt E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 36.Preker P.J., Keller W. The HAT helix, a repetitive motif implicated in RNA processing. Trends Biochem. Sci. 1998;23:15–16. doi: 10.1016/s0968-0004(97)01156-0. [DOI] [PubMed] [Google Scholar]
- 37.Thoms M., Thomson E., Bassler J., Gnadig M., Griesel S., Hurt E. The exosome is recruited to RNA substrates through specific adaptor proteins. Cell. 2015;162:1029–1038. doi: 10.1016/j.cell.2015.07.060. [DOI] [PubMed] [Google Scholar]
- 38.Zhang L., Lin J., Ye K. Structural and functional analysis of the U3 snoRNA binding protein Rrp9. RNA. 2013;19:701–711. doi: 10.1261/rna.037580.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clery A., Senty-Segault V., Leclerc F., Raue H.A., Branlant C. Analysis of sequence and structural features that identify the B/C motif of U3 small nucleolar RNA as the recognition site for the Snu13p-Rrp9p protein pair. Mol. Cell. Biol. 2007;27:1191–1206. doi: 10.1128/MCB.01287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Granneman S., Pruijn G.J., Horstman W., van Venrooij W.J., Luhrmann R., Watkins N.J. The hU3-55K protein requires 15.5K binding to the box B/C motif as well as flanking RNA elements for its association with the U3 small nucleolar RNA in Vitro. J. Biol. Chem. 2002;277:48490–48500. doi: 10.1074/jbc.M206631200. [DOI] [PubMed] [Google Scholar]
- 41.Lau C.K., Bachorik J.L., Dreyfuss G. Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat. Struct. Mol. Biol. 2009;16:486–491. doi: 10.1038/nsmb.1584. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.