Abstract
The essential mycobacterial transcriptional regulators RbpA and CarD act to modulate transcription by associating to the initiation complex and increasing the flux of transcript production. Each of these factors interacts directly with the promoter DNA template and with RNA polymerase (RNAP) holoenzyme. We recently reported on the energetics of CarD-mediated open complex stabilization on the Mycobacterium tuberculosis rrnAP3 ribosomal promoter using a stopped-flow fluorescence assay. Here, we apply this approach to RbpA and show that RbpA stabilizes RNAP-promoter open complexes (RPo) via a distinct mechanism from that of CarD. Furthermore, concentration-dependent stopped-flow experiments with both factors reveal positive linkage (cooperativity) between RbpA and CarD with regard to their ability to stabilize RPo. The observation of positive linkage between RbpA and CarD demonstrates that the two factors can act on the same transcription initiation complex simultaneously. Lastly, with both factors present, the kinetics of open complex formation is significantly faster than in the presence of either factor alone and approaches that of E. coli RNAP on the same promoter. This work provides a quantitative framework for the molecular mechanisms of these two essential transcription factors and the critical roles they play in the biology and pathology of mycobacteria.
INTRODUCTION
The regulation of gene expression serves as a gateway between genotype and phenotype. By modulating the output of specific genes, cells tune their molecular makeup to best suit environmental conditions. Much of this regulation is enacted by modulating the rates of transcription initiation to control the flux of RNA production. In bacteria, the basal transcriptional machinery is composed of RNA polymerase holoenzyme which consists of a catalytic core enzyme (ββ’α2ω) and a dissociable sigma factor (σ) that directs promoter recognition. Transcription initiation proceeds via the binding of RNAP holoenzyme to promoter DNA to form closed complex (RPc) followed by the spontaneous unwinding of approximately a turn of DNA to form open complex (RPo). In RPo, the single-stranded DNA template is correctly positioned in the polymerase active site, incoming ribonucleotides may bind and RNA polymerization may ensue. The polymerization of the initial ribonucleotides leads to promoter escape, the formation of a stable elongation complex and the production of transcript. In Escherichia coli, transcription initiation mechanisms have been well-studied and multiple kinetic intermediates have been identified between the initial DNA-holoenzyme closed complex and open complex (1). These details are of crucial importance in understanding the structural transitions that the complexes go through during the isomerization to open complex. However, a minimal kinetic scheme that describes promoter binding and opening in two reversible steps (R+P ↔ RPc ↔ RPo) has proven to be a useful and practical starting point when investigating mechanisms of open-complex formation and regulation (2).
The regulation of transcription initiation involves factor-dependent tuning of the stabilities of transcription intermediates and the rates of interconversion between these states. Examples include modulation of polymerase-promoter affinity, changing the equilibrium between RPc and RPo and influencing the rate of NTP-dependent promoter escape (3–5). It has recently become apparent that initiation in Mycobacterium tuberculosis (Mtb) is controlled via mechanisms that are distinct from those found in E. coli. For example, mycobacteria lack Fis (6), DksA (7) and AT-rich upstream activating elements (8,9). Furthermore, mycobacteria possess unique transcription factors not found in E. coli including CarD (10–12) and RbpA (13–15).
CarD is an RNAP- and DNA-binding protein that is essential for growth in M. tuberculosis, other mycobacteria (10–12). Furthermore, CarD is required for the response of Mtb to oxidative stress, some antibiotics and infection of mice (10). In vitro, CarD binds to initiation complexes and stimulates the production of RNA by stabilizing the relatively unstable RPo generated by mycobacterial RNAP holoenzyme on ribosomal promoters (11,12,16). In initiation complexes, it makes direct molecular interactions with the β1 lobe of the RNAP β subunit through its N-terminal RNAP-interaction domain (RID) (10,17,18) (Figure 1A and B). CarD also binds DNA non-specifically through its C-terminal domain (CTD) (19, Srivastava: 2013ga; 20), which contains a tryptophan residue that interacts with the upstream edge of the transcription bubble to stabilize RPo (18). Mutations in the RID, CTD or tryptophan lead to distinct in vivo and in vitro effects suggesting that full CarD activity requires each of these three functional modules (12,17,20). Based on these studies and previous analyses by our group, our working model for how CarD stabilizes open complex and stimulates transcription consists of a two-tiered, concentration-dependent mechanism. The model predicts that at low concentrations (i.e. <100 nM), CarD binds RPo and slows the rate of bubble collapse by conformational selection, while at higher concentrations, CarD binds RPc and accelerates the rate of DNA unwinding by induced fit (12). We expect both of these kinetic effects to play roles in vivo where the concentration of CarD is well in excess of 100 nM (12).
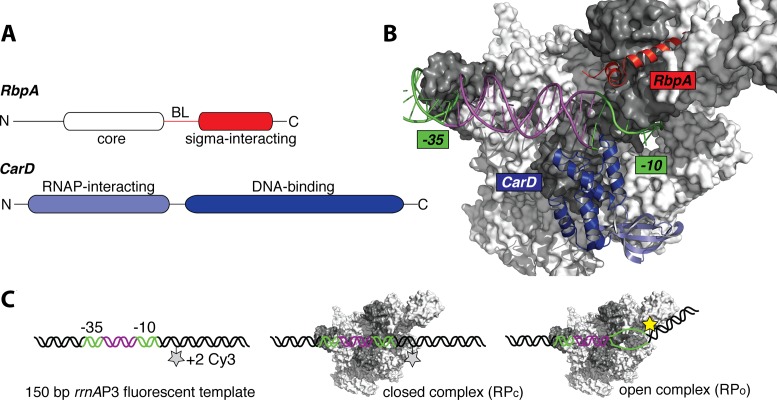
Figure 1.
Mycobacterial transcription factors and a fluorescence assay for following open complex formation. (A) CarD is made up of an N-terminal RNAP-interaction domain (RID, light blue) and a C-terminal DNA-binding domain (dark blue). RbpA consists of a core domain (white), a basic linker domain (BL) and a sigma-interacting domain (SID, red). (B) Ribbon representations of both factors are shown in the context of an open complex structure. Both domains of CarD and the SID of RbpA are modeled together based on the structure of CarD bound to open complex (4XLS (18)) and the structure of the RbpA SID domain bound to domain 2 of σA (4X8K (15)). (C) Fluorescence assay for following the formation of open complexes. A Cy3 fluorophore attached to the +2 non-template dT undergoes fluorescence enhancement upon open-complex formation.
RbpA is also an essential RNAP- and DNA-binding protein found in M. tuberculosis and other Actinobacteria, but not in E. coli (13,15). RbpA consists of an unstructured N-terminal tail, a central core β-barrel domain, a 15 amino acid basic linker (BL) and a C-terminal sigma-interacting domain (SID) (15,21) (Figure 1A). The SID binds to the second domain of sigma (σ2, domains 1.2, 2.3 and the non-conserved region) in both the presence and absence of the core RNAP enzyme and has specificity to the housekeeping sigma factor σA and the stress-response sigma factor σB (15,21,22). A crystal structure of a BL/SID construct bound to σA2 has been solved (15) and can be used to position RbpA in the initiation complex (Figure 1B). RbpA's sigma specificity has led to proposals that RbpA plays a role in the competition of sigma factors for RNAP core (14,23). RbpA potentially stabilizes RNAP holoenzyme by binding both sigma and either β2 or another region of the core subunits (14,15). Furthermore, RbpA can increase the affinity of holoenzyme to the promoter, presumably via the interaction of the BL with DNA (15,24). An arginine residue (R79) in the BL is thought to play a role in RbpA's ability to bind DNA, although it is unknown whether this interaction may contribute to RbpA's promoter specificity (15). RbpA stimulates transcription in vitro and the BL/SID region of the protein are sufficient for partial stimulation (15). However, the mechanism of transcriptional stimulation by RbpA remain unclear.
Here, we perform a mechanistic analysis of the function of Mtb RbpA during open complex formation on the Mtb rrnAP3 promoter using a real-time fluorescence assay (25) and show that RbpA stabilizes open complex. Furthermore, RbpA accelerates the approach to equilibrium providing insight into the kinetic mechanism at play. Interestingly, this trend is qualitatively distinct from that of CarD, suggesting that the two factors function in fundamentally different ways.
Furthermore, stopped-flow fluorescence experiments performed in the presence of both factors reveal positive linkage between the effect of CarD and RbpA on RPo stability at the Mtb rrnAP3 promoter. The observation that the two factors bind cooperatively demonstrates for the first time that RbpA and CarD can act on the same initiation complex simultaneously. This is consistent with the prediction that CarD and RbpA have distinct binding sites on initiation complexes (15,18). We present a thermodynamic analysis to quantitatively describe the positive linkage between the two factors. Lastly, we observe a dramatic acceleration in observed rates when both factors are present, and the approach to equilibrium under these conditions resembles that of E.coli RNAP acting on the same promoter.
The data and analysis presented here reveal important details regarding the mechanistic differences between CarD and RbpA and provide a kinetic framework for the function of these two essential mycobacterial transcription factors both independently and cooperatively. This work brings us a step closer to understanding the functional logic of transcription regulation in a pathogenic bacteria that represents a significant burden to human health worldwide.
MATERIALS AND METHODS
Protein purification
Mbo RNAP, Mbo σA and Mtb CarD were prepared as previously described (12). Mtb RbpA was cloned into a pET-SUMO-His6 vector and introduced into E. coli BL21(DE3) cells. After growth at 37°C to an OD600 of 0.8, protein over-expression was induced by the addition of 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). The tagged protein was purified by nickel affinity chromatography (HP HiTrap, GE Healthcare) and the SUMO tag was cleaved overnight with His-tagged Ulp1 protease. Pure untagged RbpA was collected in the flow through of a second nickel affinity column. The protein was stored in 20 mM Tris (pH 8.0), 150 mM NaCl and 1 mM beta-mercaptoethanol at −80°C.
Promoter DNA
A total of 150 base-pair Mtb rrnAP3 promoter fragments with a Cy3 label on the +2 non-template dT were prepared as previously described (12), with one notable exception: residues flanking the promoter sequence were replaced with those native to the Mtb genome (H37Rv coordinates 1 471 577–1 471 727, Supplemental Sequence) (26,27). Control experiments indicated that this change had no effect on previously published results describing the effect of CarD on open-complex stabilization on the same promoter with random flanking sequence (12).
Stopped-flow fluorescence assay
The stopped-flow assay was performed as previously described (12). In brief, Mbo RNAP core was mixed with 3- to 6-fold molar excess Mbo σA at 25°C to form holoenzyme. Holoenzyme was mixed with transcription factor(s) or an equivalent volume of transcription factor storage buffer such that the proteins were initially at twice the desired final reaction concentrations. Promoter DNA was also prepared at twice the desired final reaction concentration. Equal volume mixing was performed in a stopped-flow apparatus (Applied Photophysics SX-20, total shot volume 150 μl, dead time < 2 ms), so the initial protein and DNA solutions were each diluted by half in order to reach their final reaction concentrations. All experiments were performed using a final MboRNAP holoenzyme concentration of 225 nM and a DNA concentration of 10 nM unless otherwise noted. Excitation light was provided by a 510 nm LED light source, as opposed to a 515 nm light source from an arc lamp passed through a monochromator. Emission was collected at 570+ nm using a long-pass filter. Control experiments indicated that this subtle change in excitation wavelength had no effect on previously published results describing the effect of CarD on mycobacterial open-complex stabilization. All experiments were performed at 25°C in the following final solution conditions: 14 mM Tris pH 8.0, 120 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA and 10% glycerol by volume.
Two to three traces were collected per condition. Traces for each condition were averaged and plotted as fold-change over DNA by first subtracting the buffer signal from all traces and then plotting the data as (F−F0)/F0, where F0 is the signal for DNA alone and F is the signal for DNA mixed with protein. The fold-change traces were fit to a triple exponential from 0.1–1200 s using the ProData Viewer software from Applied Photophysics:
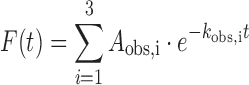 |
(1) |
where Aobs,i and kobs,i are the amplitude and observed rate of the ith kinetic phase. To facilitate consistency in assignment of fast, intermediate and slow phases, traces were anchored to the intermediate and slow phases using either a single or double exponential before fitting the fast phase of the trace. Fractional amplitudes were calculated according to:
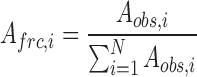 |
(2) |
In order to estimate an overall rate for the approach to equilibrium, an amplitude-averaged rate was calculated using the intermediate and slow phases according to:
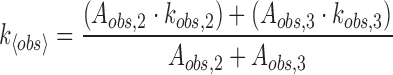 |
(3) |
Conditions that were repeated multiple times on different days were used to estimate standard error of the mean (SEM). An average SEM was used to estimate uncertainty for specific conditions that were only repeated multiple times on the same day to better estimate the actual error for these points.
In vitro aborted transcription assay
CarD and RbpA used in this assay were diluted into 1× dialysis buffer (20 mM Tris pH 8.0, 150 mM NaCl and 1 mM BME). Mbo σA was mixed in 8-fold molar excess with core RNAP to reconstitute the RNAP holoenzyme. A total of 85 bp overlapping primers (IDT) were annealed and extended to prepare a linear fragment of dsDNA Mtb Erdman strain genomic DNA containing nucleotides 1 470 151 to 1 470 300 which includes the Mtb rrnAP3 promoter. The final reaction conditions were: 225 nM Mbo σA-holo RNAP, 1–2 μM CarD or equivalent volume of buffer, 1–2 μM RbpA or equivalent volume of buffer, 10 nM linear DNA template, 210 μM GpU dinucleotide, 21 μM UTP, 0.1 μL [α - 32 P]-UTP, 14 mM Tris pH 8.0, 100 mM NaCl, 10.2 mM MgCl2, 5% (vol/vol) glycerol, 1 mM DTT and 0.1 mg/ml BSA (NEB) in a total volume of 20 μl. Mbo core and σA were incubated for 10 min at room temperature. CarD, RbpA and/or dialysis buffer were added to the polymerase and the proteins were incubated for 10 more minutes at room temperature. The DNA template was added and the reactions were diluted to 17.5 μl, followed by an additional 10 min at room temperature. Reactions were initiated with addition of a 2.5 μl mixture containing GpU, UTP and the radiolabeled UTP. After 20 min incubation at room temperature, the reactions were stopped with 2x formamide buffer [98% (vol/vol) formamide, 5 mM EDTA] and run on a 22% urea PAGE gel.
RESULTS
RbpA stabilizes rrnAP3 open complexes
To study the effect of RbpA on transcription open complexes (RPo) we used a fluorescence assay that reports on RPo formation in real-time as previously reported (12,25). For these experiments, we use M. bovis (Mbo) RNAP which is virtually identical to MtbRNAP differing only in the 69th amino acid of the β’ subunit which is a proline in MboRNAP and an arginine in MtbRNAP. A stopped-flow apparatus was used to mix Mbo σA-saturated MboRNAP with Cy3-labeled Mtb rrnAP3 promoter, and fluorescence intensity was monitored over time. The dye is conjugated to the +2 non-template thymine and exhibits enhanced fluorescence intensity upon RPo formation, providing a way to measure the kinetics of open complex formation as well as the amount of open complex at equilibrium (Figure 1C). Several control experiments support our interpretation of Cy3 fluorescence enhancement as a reporter of promoter-opening. First, promoter-less templates do not show fluorescence enhancement, demonstrating sequence dependence (12). Second, experiments performed at 10°C, 25°C and 37°C showed increasing fluorescence enhancement by RNAP with increasing temperature, consistent with the known temperature dependence of promoter-melting (12). Third, at high concentrations of RNAP, where promoters are saturated with holoenzyme, CarD leads to a large fluorescence enhancement showing that CarD specifically affects a step after holoenzyme binds the promoter (i.e. promoter opening). Lastly, the CarD-dependent fluorescence enhancement follows the same trend of open-complex stabilization observed in potassium permanganate experiments (12,16).
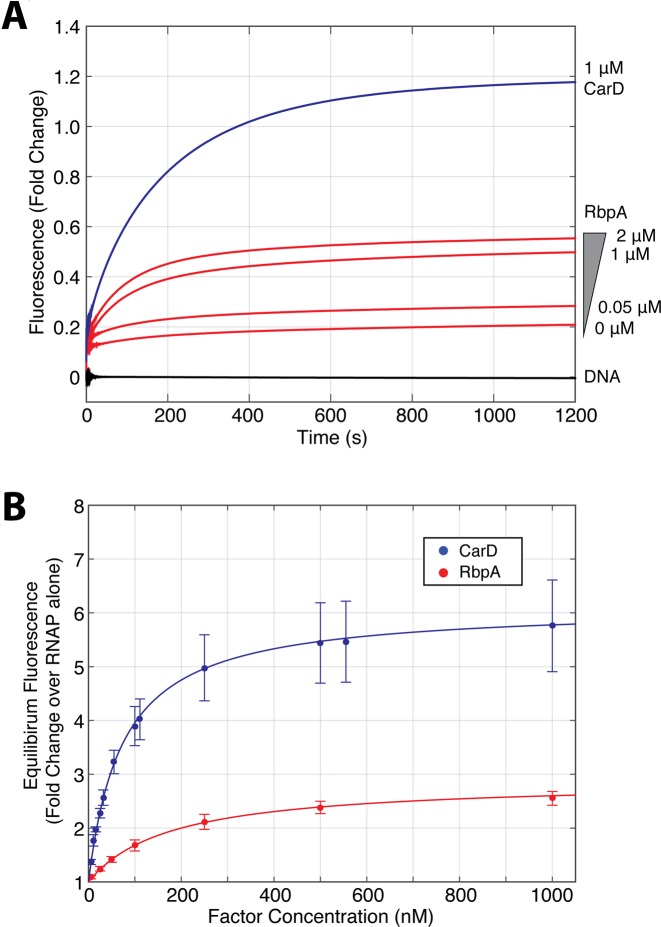
Incubating MboRNAP with increasing concentrations of wild-type RbpA results in increasing equilibrium fluorescence, demonstrating that RbpA stabilizes RPo at the rrnAP3 promoter (Figure 2A). The R79A mutant of RbpA, which is known to play an important role in transcriptional activation (15), results in approximately half the increase in equilibrium fluorescence when added at a concentration at which WT RbpA saturates (2 μM, Supplementary Figure S1). A fit of the equilibrium fluorescence fold-change generated by WT RbpA normalized to RNAP alone to the binding isotherm 1 + A*[RbpA]/([RbpA]+Keff) gives a value of the concentration of half-maximal effect, Keff = 177 ± 23 nM. This Keff is approximately 2-fold weaker than that of CarD (73 ± 4 nM). At saturation, RbpA produces ∼3 times (A = 2.9) more fluorescence enhancement than MboRNAP alone, compared to CarD which saturates at ∼6-fold (A = 6.1) over RNAP (Figure 2B). Assuming equilibrium fluorescence enhancement correlates with the extent of RPo stabilization, the lower equilibrium fluorescence generated by RbpA at saturation suggests that RbpA stabilizes RPo to a lesser extent than CarD. However, given that Cy3 fluorescence enhancement is sensitive to local changes in the dye's microenvironment, we do not exclude the possibility that the assay reports on multiple open complexes, each with subtle conformational differences leading to different fluorescence enhancements. With this in mind, it is possible that the RbpA-stabilized RPo produces lower fluorescence enhancement than the RPo stabilized by CarD. If this were the case, the extent of fluorescence enhancement would not correspond to the extent of open complex stabilization, but instead would report on the average fluorescence enhancement of an ensemble of open complexes. Clearly, the difference in fluorescence enhancements contains information regarding the mechanisms of open complex stabilization by the two factors. Here, we can only speculate on the molecular details that lead to the observed enhancements, however, these details do not impact the conclusions drawn from the analyses that follow.
Figure 2.
RbpA and CarD stabilize open complex. (A) Fluorescent fold-change is plotted over time from mixing MboRNAP and Mtb factors with a +2 T Cy3-labeled Mtb rrnAP3 promoter template. DNA alone mixed with buffer (black), a titration of RbpA from 0–2 μM (red) and 1 μM CarD (blue) are shown. (B) The equilibrium fluorescence fold change over 225 nM RNAP alone is plotted versus the concentration of either RbpA (red) or CarD (blue). The data are fit with the binding model A*[factor]/([factor]+Keff) to extract amplitudes (ACarD = 6.1 ± 0.1 and ARbpA = 2.9 ± 0.1) and effective binding constants (Keff,RbpA = 177 ± 23 nM and Keff,CarD = 73 ± 4 nM). Two to three shots were collected for each condition and error bars represent standard error of the mean.
RbpA stabilizes open complexes through a different kinetic mechanism than CarD
Although RbpA and CarD are unique proteins that bind the transcription initiation complex at distinct sites, the two factors share several important structure-function properties. Specifically, both factors bind RNAP-holoenzyme and DNA, and both factors stabilize RPo. For these reasons, we considered the possibility that RbpA stabilizes open-complexes using a similar kinetic mechanism as CarD (12). Curves in the presence of either RpbA or CarD were well-fit by a triple exponential (see Materials and Methods) and the phases were separable by approximately an order of magnitude. The low fractional amplitude of kobs,1 made it challenging to measure, so we cannot exclude the possibility that RbpA or CarD influence the kinetics of the fastest observed phase.
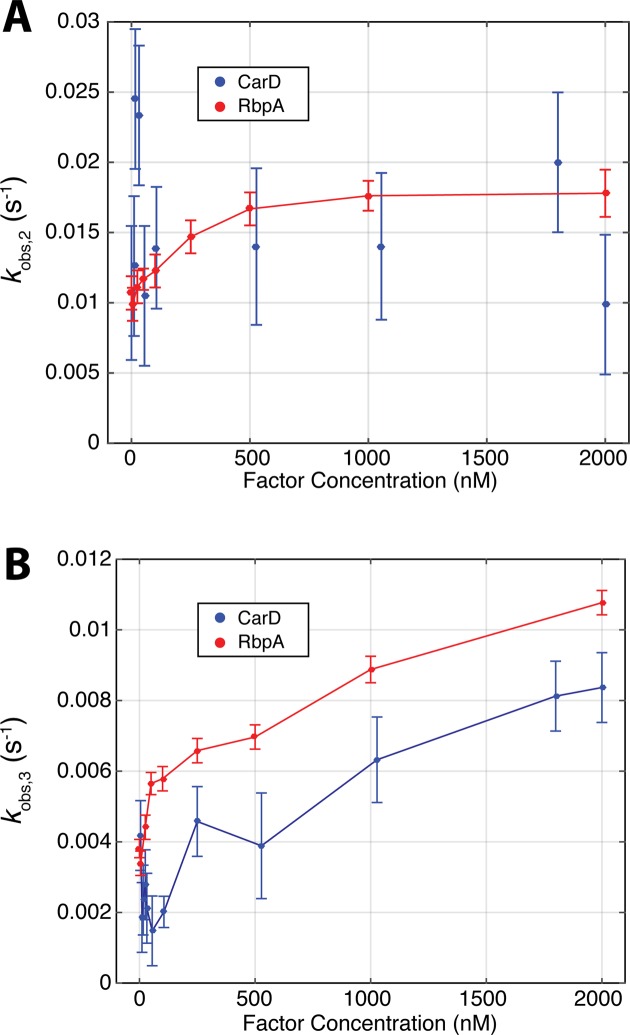
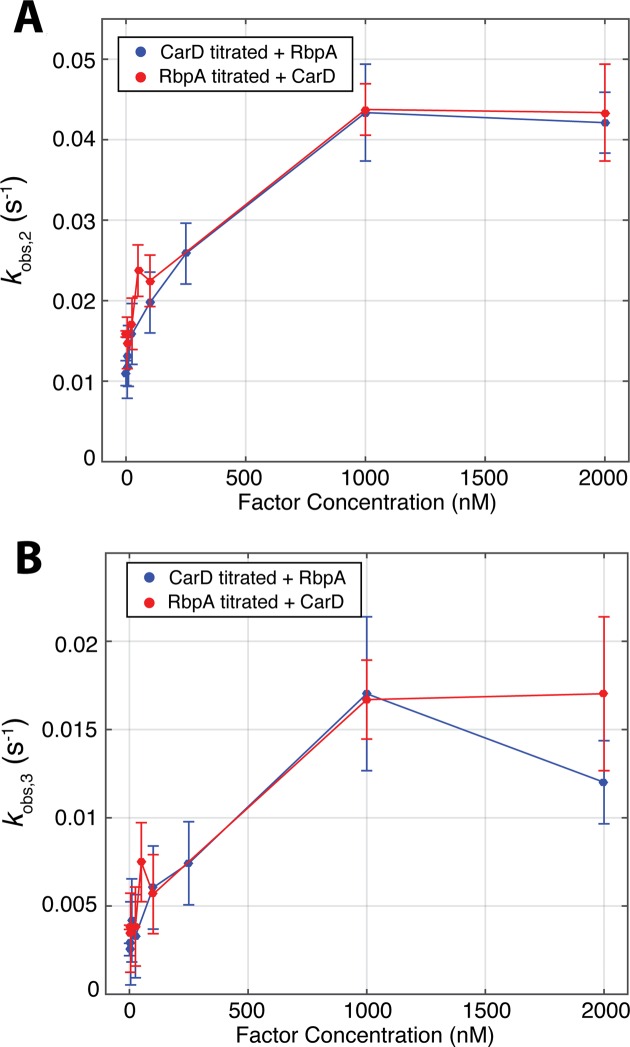
We observed dramatic differences between RbpA and CarD in the intermediate and slow phases. CarD traces are dominated by the slowest phase (kobs,3,), whereas RbpA traces contain significant contributions from both the intermediate and slowest phases (kobs,2 and kobs,3, respectively) (Supplementary Figure S2). Analysis of the trends in the observed rates themselves also indicated differences between RbpA and CarD (Figure 3B and C). We observe monotonic and saturable increases in kobs,2 and kobs,3 with increasing RbpA concentration (Figure 3B). Conversely, we were unable to discern a systematic trend in kobs,2 as a function of CarD concentration (Figure 3B). Furthermore, kobs,3 displays a non-monotonic trend with increasing CarD concentration (Figure 3C and Supplementary Figure S3). Thus, RbpA accelerates the approach to equilibrium at all concentrations tested while CarD decelerates equilibration at low concentrations and accelerates equilibration at high concentrations.
Figure 3.
Concentration dependence of observed rates. (A) The intermediate phase of stopped-flow time courses is different between RbpA and CarD. For RbpA (red), kobs,2 increases in a monotonic and saturable manner with increasing concentration. For CarD, kobs,2, does not show any systematic concentration dependence. (B) RbpA and CarD exhibit different concentration-dependent trends in kobs,3. The slowest phase of time courses increases monotonically with increasing RbpA concentration (red), unlike CarD (blue), which decelerates at low concentrations and accelerates at higher concentrations. Two to three shots were collected for each condition and error bars represent standard error of the mean.
We considered the mechanistic implications of the distinct kinetic trends observed for RbpA and CarD. CarD's non-monotonic trend in kobs,3 can be explained in the context of a two-step reversible model coupled to factor binding (12) (Supplementary Figure S4A). However, the observation of multiple saturable observed rates in the presence of RbpA demonstrates that the simple two-step reversible mechanism of open complex formation cannot account for the observed kinetics. Therefore, our data suggest that the mechanism of RpbA open complex stabilization must involve more states. Since RbpA is known to bind sigA, RNAP core and holoenzyme, one possibility is that RbpA's protein-binding interactions contribute to the observed kinetics (Supplementary Figure S4B). Another possibility is that RbpA interacts with additional intermediates on pathway to RPo (Supplementary Figure S4B). These possibilities are not mutually exclusive, and a detailed analysis of the kinetic mechanism of RbpA's stabilization of RPo remains an active area of investigation.
RbpA and CarD cooperatively stabilize open complex
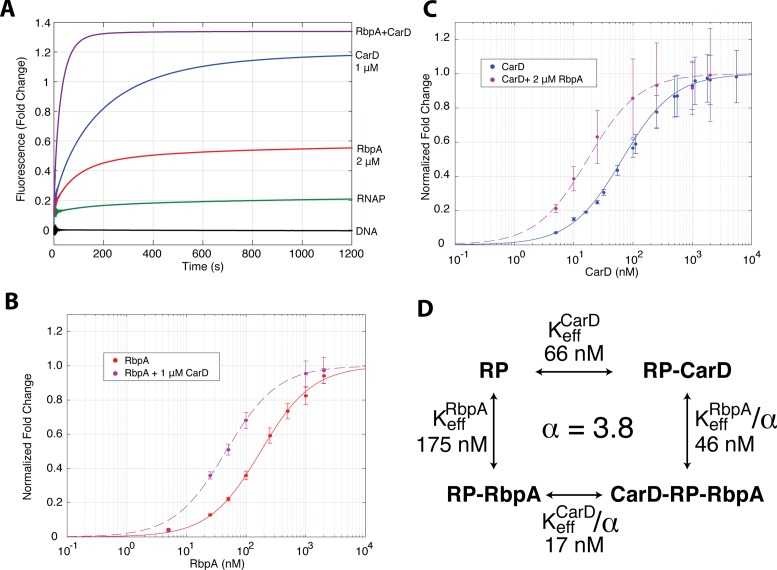
As each factor stabilizes RPo and structural modeling suggests that they could both bind RP complexes concurrently (15), we tested the effect of RbpA and CarD together on open-complex stability. Since RbpA generates a lower fluorescence enhancement compared to CarD, we reasoned that if RbpA competed with CarD for binding to the initiation complex, the addition of both factors would lead to an intermediate enhancement. However, in the presence of both factors at saturating concentrations, the equilibrium fluorescence fold change is actually greater than that for saturating CarD alone (Figure 4A).
Figure 4.
RbpA and CarD bind to initiation complexes cooperatively. (A) Fluorescent fold change as a function of time for DNA alone (black), MboRNAP alone (green), 2 μM RbpA (red), 1 μM CarD and 2 μM RbpA + 1 μM CarD (magenta). (B) Normalized fluorescence fold-change as a function of RbpA concentration in the absence (red) and presence (magenta) of 1 μM CarD. (C) Normalized fluorescence fold-change as a function of CarD concentration in the absence (red) and presence (magenta) of 2 μM RbpA. (D) A thermodynamic cycle between apo-RP, CarD-bound RP, RbpA-bound RP and the doubly-bound RP. Here, RP represents the total population of DNA-bound polymerase and includes RPc and RPo. The four titrations shown in (B) and (C) were globally fit with three parameters: Keff,CarD, Keff,RbpA and a cooperativity factor α which relates the ratio of Keff of a factor in the presence and absence of the other. The fits are shown in (B) and (C) along with the values for Keff. The fit cooperativity factor, α = 3.8 ± 0.4, corresponds to a ΔΔG = –RTln(α) = −3.3 kJ/mol (−0.8 kcal/mol). Two to three shots were collected for each condition and error bars represent standard error of the mean.
If RbpA and CarD bind concurrently to the rrnAP3 promoter-RNAP complex, one expects to measure positive linkage or cooperativity between the two factors as both stabilize the same conformation, namely open complex. To test this model, we performed titrations of each factor in the presence of the other factor at saturation and asked whether we observed a shift in Keff relative to each factor alone. Indeed, titrating RbpA in the presence of 1 μM CarD results in a Keff (48 ± 10 nM) that is lower than that obtained from a titration of RbpA alone (177 ± 23 nM), suggesting that the presence of CarD increases the binding affinity of RbpA to transcription initiation complexes (Figure 4B). Likewise, a CarD titration in the presence of 2 μM RbpA results in a Keff (16 ± 2 nM) that is lower than a titration of CarD alone (73 ± 4 nM), indicating that the presence of RbpA allows CarD to interact with the complex at lower concentrations (Figure 4C). This observation of heterotropic, positive linkage between RbpA and CarD demonstrates that they act cooperatively on transcription initiation complexes at the Mtb rrnAP3 promoter.
To provide an overall quantification of the observed linkage, we consider the thermodynamic cycle involving four states: (i) RP, (ii) RP-CarD, (iii) RP-RbpA and (iv) RP-RbpA-CarD, where RP represents promoter-bound RNAP and includes an ensemble of states (i.e. RPc, RPo and all intermediates) (Figure 4D). We globally fit the four experimental binding curves (CarD titrations ± 2 μM RbpA and RbpA titrations ± 1 μM CarD) to a model where each factor has an effective affinity (Keff) and linkage is captured by the cooperativity factor α (Supplementary Figure S5). In the context of this model, the effective affinity of a factor in the presence on the other is given by Keff/α. A fit of the data with this three-parameter model results in values of Keff,CarD = 66 ± 7 nM, Keff,RbpA = 175 ± 20 nM and α = 3.8 ± 0.4. All four Keff values are within error of the values obtained from fits of the individual titrations alone. Thus, a simple model of cooperativity captures the positive linkage between RbpA and CarD at initiation complexes.
Given that RbpA and CarD cooperatively stabilize open complex, we hypothesized that they would also stimulate transcription above the level of either factor acting alone. To test this hypothesis, we performed an aborted transcription assay in which we measured the production of a three nucleotide transcript from the rrnAP3 using a dinucleotide primer (GpU) and a radio-labeled UTP. RbpA and CarD acting individually at saturating concentrations stimulate transcription of the 3-nt product over RNAP alone (Supplementary Figure S6). The presence of both factors results in a further increase in 3-nt production over either factor acting alone. These results are consistent with a model in which CarD and RbpA cooperatively stabilize a transcription-competent open complex.
RbpA and CarD act together to accelerate the approach to RPo equilibrium
To identify any cooperative kinetic effects stemming from the presence of both RbpA and CarD on open complex equilibration, we used a triple exponential to fit the raw data traces and examined the manner in which the observed rates (Figure 5B and C) and the fractional amplitudes (Supplementary Figure S7) depended on transcription factor concentration with both factors present.
Figure 5.
Kinetic effects of cooperative stabilization. (A) The intermediate phase (kobs,2) increases monotonically with concentration for either RbpA titrated in the presence of CarD (red) or CarD titrated in the presence of RbpA (blue). (B) The slowest phase (kobs,3) increases monotonically with concentration for either RbpA titrated in the presence of CarD (red) or CarD titrated in the presence of RbpA (blue). Two to three shots were collected for each condition and error bars represent standard error of the mean.
Compared to RbpA and CarD individually, we found that kobs,2 and kobs,3 were faster when both factors were present. While the presence of CarD did not affect the dependence of the observed rates on RbpA concentration, the presence of RbpA did affect the dependence of the observed rates on CarD concentration. Titrations of CarD performed in the presence of RbpA at saturation indicated that both kobs,2 (Figure 5B) and kobs,3 (Figure 5C) now increase in a saturable and monotonic manner. This trend is quite different than what is observed for CarD titrations performed in the absence of RbpA, in which kobs,2 does not exhibit a discernible trend, and kobs,3 exhibits non-monotonic behavior.
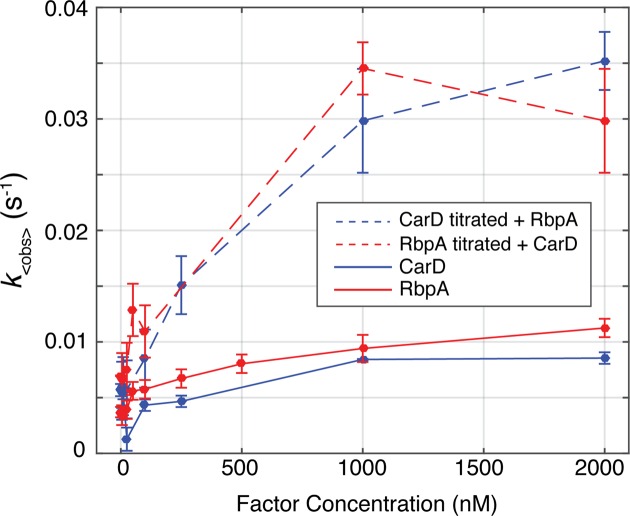
To facilitate a general kinetic comparison of RbpA and CarD acting individually and together, we calculated an amplitude-averaged rate as a means to quantify an overall, apparent rate of open-complex equilibration (Materials and Methods). This analysis supports the conclusion that open-complex equilibration is faster in the presence of both factors (Figure 6). Specifically, the averaged rate (k<obs>) observed with saturating concentrations of both factors is at least 3 times faster than in the presence of either factor alone. Interestingly, the presence of both factors leads to traces with similar kinetics and equilibrium fluorescence to those obtained using the RNAP holoenzyme from E. coli (Figure 7).
Figure 6.
The kinetics of equilibration are accelerated by the presence of both factors. An amplitude-averaged rate is plotted as a function of factor concentration for RbpA titrations (red) and CarD titrations (blue) and in the absence (solid lines) and presence (dashed lines) of the other factor at saturation. Two to three shots were collected for each condition and error bars represent standard error of the mean.
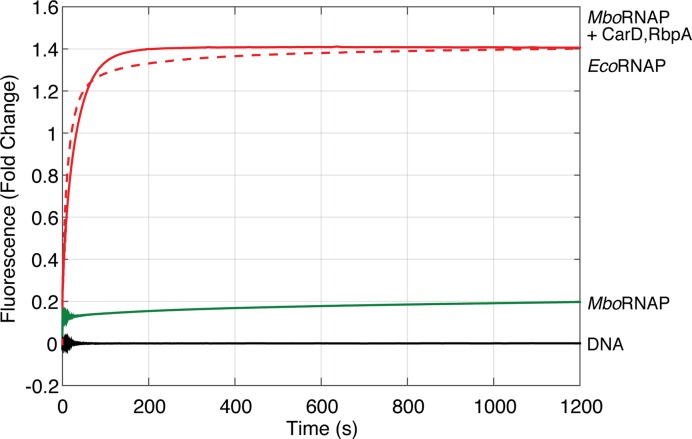
Figure 7.
The kinetics of open complex formation for MboRNAP in the presence of RbpA and CarD resemble those for EcoRNAP on the rrnAP3 promoter. Fluorescence time courses collected with RbpA and CarD (solid red) dramatically increase the fluorescence signal at equilibrium and accelerate the kinetics compared to RNAP alone (green). The kinetics of MboRNAP with RbpA and CarD approaches that of 85 nM EcoRNAP (dashed red), which equilibrates to the same fluorescence fold-change (1.4).
DISCUSSION
We have presented a study of RbpA and CarD acting individually and together to stabilize mycobacterial transcription open complexes. Like CarD, we found that RbpA stabilizes RPo at the Mtb rrnAP3 promoter, albeit via a different kinetic mechanism. Using a stopped-flow fluorescence assay that reports on open-complex formation in real time, we observed that, compared to CarD, RbpA generates a lower equilibrium fluorescence at saturating concentrations and has a weaker apparent affinity for initiation complexes. In addition, RpbA exhibits two observed rates (kobs,2 and kobs,3) with appreciable fractional amplitudes in contrast to the one dominant observed rate (kobs,3) detected in the presence of CarD. The magnitudes of both of these rates show monotonic and saturable acceleration with increasing RbpA concentration. These results all suggest that the kinetic mechanism by which RbpA stabilizes mycobacterial open complexes is distinct from that of CarD.
A two-step reversible model (R+P ↔ RPc ↔ RPo) can capture the kinetics of open complex formation under certain conditions. However, this model is almost certainly an over-simplification of the real mechanism. For example, we have shown that kinetic traces of open complex formation by MboRNAP alone exhibit three observed rates which cannot be accounted for in the context of the two-step reversible model. Furthermore, the kinetic mechanism of open-complex formation by E. coli RNAP includes multiple closed and open intermediates (1,28–30) and transcription with mycobacterial RNAP has previously been analyzed with E. coli derived models (31). Therefore, the kinetic mechanism of mycobacterial open complex formation likely involves intermediates between the initial closed complex and the final transcription-competent open complex. We hypothesize that the distinct kinetic signatures of RbpA and CarD are due, in part, to their differential affinities with these intermediates. Furthermore, intermediate open complexes may have different fluorescence properties leading to the differential enhancements observed between RbpA and CarD. Another possible way to expand the two-step reversible model to account for the complexity of the observed kinetics is to add the interaction of these factors with free polymerase in the absence of DNA. RbpA is known to bind sigma factor, core RNAP and holoenzyme (14,15,22) and CarD binds to RNAP (10,18,19). In light of these possibilities, analysis using kinetic models that include multiple intermediates and DNA-independent assembly states remains an ongoing research direction (Supplementary Figure S4B).
In addition to studying each factor individually, we performed experiments using both RbpA and CarD together. Stopped-flow titrations indicated that the presence of one factor increases the apparent affinity of the other factor to the RNAP-promoter complex, demonstrating positive linkage between the two factors. The observation of positive linkage demonstrates that not only can CarD and RbpA bind the initiation complex simultaneously, but also that they do so cooperatively. Cooperativity between transcription factors has been observed for transcription initiation (32–34), elongation (35) and termination (36). This cooperativity can lead to activation or repression and affords complexity and tunability to gene regulation (37). Furthermore, cooperativity amongst transcription factors can occur between the same factors (homotropic) or different factors (heterotropic) (32–34). In Mtb specifically, cooperative binding of DevR factors to their DNA sites plays a role in the activation of a regulon required for the induction of dormancy in response to hypoxic conditions (38,39). However, to our knowledge the cooperative association of RbpA and CarD is the first example of heterotropic cooperativity between transcription factors that directly bind initiation complexes to stabilize open complex.
When speculating about the mechanism of cooperativity between RbpA and CarD, we consider two general possibilities that are not mutually exclusive: (i) a ‘direct’ mechanism in which RbpA and CarD physically interact, so that the presence of one factor provides an additional binding surface for the other; and (ii) an ‘allosteric’ mechanism where the binding of one factor results in conformational changes in the initiation complex that lead to higher affinity binding of the second factor (i.e. conformational selection). Although a ‘direct’ interaction between RbpA and CarD is not predicted by structural modeling, we do not exclude the possibility that direct interactions between the two factors contribute to the observed positive linkage. In particular, the location of RbpA's core domain in the initiation complex is unknown and predicted to be in close proximity to the C-terminal domain of CarD. The allosteric mechanism, however, must contribute to the observed positive linkage through conformational selection of RPo. RbpA and CarD each must have a higher affinity to RPo than to RPc as they both stabilize RPo. Thus, the binding of one factor shifts the population of initiation complexes to the higher affinity open-complex conformation(s), resulting in a lower Keff for the other factor. Whether the allosteric mechanism of conformational selection accounts for all or just some of the positive linkage remains an active area of investigation.
When studying the effect of both factors acting together on the kinetics of open-complex equilibration, we noticed a substantial acceleration in kobs,2 and kobs,3, demonstrating that the overall approach to equilibrium is faster compared to either factor acting alone (Figures 5 and 6). Based on this observation, we hypothesize that the interactions of both factors with the initiation complex accelerate distinct forward rates on the pathway to open-complex formation. The pathway to open complex determined with the E. coli system involves many structural intermediates including DNA bending and wrapping, DNA unwinding and loading of the template strand, assembly of the clamp/jaw domains and closing of the clamp/jaw domains around the downstream DNA (1,30). More specifically, the bending of downstream duplex into the polymerase cleft has been linked with the nucleation of DNA melting (30). The basic linker of RbpA interacts with the extended −10 motif (15) and tryptophan 85 in CarD's DNA-binding domain interacts with the upstream edge of the transcription bubble at the junction between the double-stranded −12 base-pair and the single-stranded −11 position (11,18), precisely where nucleation of promoter melting occurs (Figure 1B). Taken together, these structural considerations suggest the possibility that RbpA and CarD cooperate to facilitate promoter bending and nucleation of promoter-melting. This combined mechanism of RbpA and CarD may be similar to the induced-fit mechanism of bending and opening used to describe the effect of the transcription factor Mtf1 on mitochondrial open-complex kinetics (40). Importantly, this model is also compatible with the proposed conformational-selection mechanism in which CarD acts to prevent bubble collapse after the promoter DNA has been opened (11,12,18).
We also show that RbpA and CarD jointly lead to transcriptional dynamics for MboRNAP similar to E. coli RNAP (Figure 7). This result leads us to speculate that RbpA and CarD play the role of general transcription factors for Mtb, at least at housekeeping σA-dependent promoters. In light of the cooperativity between the two factors, the preference of RbpA (14,22) for σA and σB holoenzymes and the presumed sigma-independence of CarD raise interesting questions regarding the regulatory logic behind each of these essential factors. For example, the cooperativity may provide a mechanism for the preferential recruitment of CarD to RbpA-dependent promoters. Alternatively, the association of CarD with other sigma factors may lead to RbpA binding at non-σA or σB promoters. In addition, our data predict that the overall effect on open complex equilibrium at initiation complexes containing both factors will be greater than those containing either factor alone. This effect could lead to the different regulatory outcomes depending on which factors are present at specific promoters.
In summary, we describe the kinetics and concentration dependencies of RbpA and CarD acting individually and cooperatively on open-complex formation at the Mtb rrnAP3 promoter. We expect the work presented here to provide a quantitative framework that can be used to develop mechanistic models of RbpA and CarD. We hypothesize that the concurrent binding and positive linkage between these essential transcription factors play important roles in mycobacterial gene regulation in that they result in (i) the more efficient recruitment of transcription factors to initiation complexes and (ii) the rapid formation of a more stable open complex. Further studies are needed to describe the detailed kinetics of both factors on all phases of transcription initiation to understand their overall effect on transcriptional flux at promoters throughout the mycobacterial genome.
Supplementary Material
Acknowledgments
The authors would like to thank Dr Tim Lohman for the use of his stopped-flow spectrophotometer and Dr Tom Ellenberger for the use of the HPLC and FPLC in his laboratory. We also thank Dr Roberto Galletto, Dr Lohman, Dr Ellenberger and Dr Tomasz Heyduk for scientific input at various stages of this work.
Author contributions: J.R. performed the fluorescence experiments. E.A.G. and C.L.S. conceived the research. J.R. and E.A.G. planned the experiments, analyzed the data and interpreted the results. J.R. and A.R.M. purified all reagents. J.P. and A.L.G. cloned MtbRbpA and performed the transcription assays. J.R., C.L.S. and E.A.G. wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01GM107544 to E.A.G. and C.L.S.]; Sigma Aldrich Predoctoral Fellowship [to J.R.]; NIGMS Cell and Molecular Biology Training Grant [GM007067 to J.P. and A.L.G.]; Stephen I. Morse Graduate Fellowship [to A.L.G.]. Funding for open access charge: NIH [R01GM107544].
Conflict of interest statement. None declared.
REFERENCES
- 1.Saecker R.M., Record M.T., Jr, deHaseth P.L. Mechanism of bacterial transcription initiation: Promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 2011;412:754–771. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McClure W.R. Rate-limiting steps in RNA chain initiation. Proc. Natl. Acad. Sci. U.S.A. 1980;77:5634–5638. doi: 10.1073/pnas.77.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee D.J., Minchin S.D., Busby S.J.W. Activating transcription in bacteria. Annu. Rev. Microbiol. 2012;66:125–152. doi: 10.1146/annurev-micro-092611-150012. [DOI] [PubMed] [Google Scholar]
- 4.Rutherford S.T., Villers C.L., Lee J.-H., Ross W., Gourse R.L. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:236–248. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schröder O., Wagner R. The bacterial DNA-binding protein H-NS represses ribosomal RNA transcription by trapping RNA polymerase in the initiation complex. J. Mol. Biol. 2000;298:737–748. doi: 10.1006/jmbi.2000.3708. [DOI] [PubMed] [Google Scholar]
- 6.Kahmann R., Rudt F., Koch C., Mertens G. G inversion in bacteriophage Mu DNA is stimulated by a site within the invertase gene and a host factor. Cell. 1985;41:771–780. doi: 10.1016/s0092-8674(85)80058-1. [DOI] [PubMed] [Google Scholar]
- 7.Kang P.J., Craig E.A. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J. Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao L., Ross W., Appleman J.A., Gaal T., Leirmo S., Schlax P.J., Record M.T., Gourse R.L. Factor independent activation of rrnB P1. An ‘extended’ promoter with an upstream element that dramatically increases promoter strength. J. Mol. Biol. 1994;235:1421–1435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- 9.Newlands J.T., Josaitis C.A., Ross W., Gourse R.L. Both fis-dependent and factor-independent upstream activation of the rrnB P1 promoter are face of the helix dependent. Nucleic Acids Res. 1992;20:719–726. doi: 10.1093/nar/20.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stallings C.L., Stephanou N.C., Chu L., Hochschild A., Nickels B.E., Glickman M.S. CarD is an essential regulator of rRNA transcription required for mycobacterium tuberculosis persistence. Cell. 2009;138:146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srivastava D.B., Leon K., Osmundson J., Garner A.L., Weiss L.A., Westblade L.F., Glickman M.S., Landick R., Darst S.A., Stallings C.L., et al. Structure and function of CarD, an essential mycobacterial transcription factor. Proc. Natl. Acad. Sci. U.S.A. 2013;110:12619–12624. doi: 10.1073/pnas.1308270110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rammohan J., Ruiz-Manzano A., Garner A.L., Stallings C.L., Galburt E.A. CarD stabilizes mycobacterial open complexes via a two-tiered kinetic mechanism. Nucleic Acids Res. 2015;43:3272–3285. doi: 10.1093/nar/gkv078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dey A., Verma A.K., Chatterji D. Role of an RNA polymerase interacting protein, MsRbpA, from Mycobacterium smegmatis in phenotypic tolerance to rifampicin. Microbiology. 2010;156:873–883. doi: 10.1099/mic.0.033670-0. [DOI] [PubMed] [Google Scholar]
- 14.Hu Y., Morichaud Z., Chen S., Leonetti J.P., Brodolin K. Mycobacterium tuberculosis RbpA protein is a new type of transcriptional activator that stabilizes the A-containing RNA polymerase holoenzyme. Nucleic Acids Res. 2012;40:6547–6557. doi: 10.1093/nar/gks346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubin E.A., Tabib-Salazar A., Humphrey L.J., Flack J.E., Olinares P.D.B., Darst S.A., Campbell E.A., Paget M.S. Structural, functional, and genetic analyses of the actinobacterial transcription factor RbpA. Proc. Natl. Acad. Sci. U.S.A. 2015;112:7171–7176. doi: 10.1073/pnas.1504942112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis E., Chen J., Leon K., Darst S.A., Campbell E.A. Mycobacterial RNA polymerase forms unstable open promoter complexes that are stabilized by CarD. Nucleic Acids Res. 2015;43:433–445. doi: 10.1093/nar/gku1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiss L.A., Harrison P.G., Nickels B.E., Glickman M.S., Campbell E.A., Darst S.A., Stallings C.L. Interaction of CarD with RNA Polymerase Mediates Mycobacterium tuberculosis Viability, Rifampin Resistance, and Pathogenesis. J. Bacteriol. 2012;194:5621–5631. doi: 10.1128/JB.00879-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bae B., Chen J., Davis E., Leon K., Darst S.A., Campbell E.A. CarD uses a minor groove wedge mechanism to stabilize the RNA polymerase open promoter complex. Elife. 2015;4 doi: 10.7554/eLife.08505. doi:10.7554/eLife.08505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulten G., Sacchettini J.C. Structure of the Mtb CarD/RNAP β-lobes complex reveals the molecular basis of interaction and presents a distinct DNA-binding domain for Mtb CarD. Structure. 2013;21:1859–1869. doi: 10.1016/j.str.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garner A.L., Weiss L.A., Manzano A.R., Galburt E.A., Stallings C.L. CarD integrates three functional modules to promote efficient transcription, antibiotic tolerance and pathogenesis in mycobacteria. Mol. Microbiol. 2014;93:682–697. doi: 10.1111/mmi.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bortoluzzi A., Muskett F.W., Waters L.C., Addis P.W., Rieck B., Munder T., Schleier S., Forti F., Ghisotti D., Carr M.D., et al. Mycobacterium tuberculosis RNA polymerase-binding protein A (RbpA) and its interactions with sigma factors. J. Biol. Chem. 2013;288:14438–14450. doi: 10.1074/jbc.M113.459883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y., Morichaud Z., Sudalaiyadum Perumal A., Roquet-Baneres F., Brodolin K. Mycobacterium RbpA cooperates with the stress-response σB subunit of RNA polymerase in promoter DNA unwinding. Nucleic Acids Res. 2014;40:6547–6557. doi: 10.1093/nar/gku742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manganelli R. Sigma factors: Key molecules in mycobacterium tuberculosis physiology and virulence. Microbiol. Spectr. 2014;2:1–23. doi: 10.1128/microbiolspec.MGM2-0007-2013. [DOI] [PubMed] [Google Scholar]
- 24.Verma A.K., Chatterji D. Dual role of MsRbpA: transcription activation and rescue of transcription from the inhibitory effect of rifampicin. Microbiology. 2014;160:2018–2029. doi: 10.1099/mic.0.079186-0. [DOI] [PubMed] [Google Scholar]
- 25.Ko J., Heyduk T. Kinetics of promoter escape by bacterial RNA polymerase: effects of promoter contacts and transcription bubble collapse. Biochem. J. 2014;463:135–144. doi: 10.1042/BJ20140179. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez-y-Merchand J.A., Colston M.J., Cox R.A. The rRNA operons of Mycobacterium smegmatis and Mycobacterium tuberculosis: comparison of promoter elements and of neighbouring upstream genes. Microbiology. 1996;142:667–674. doi: 10.1099/13500872-142-3-667. [DOI] [PubMed] [Google Scholar]
- 27.Lew J.M., Kapopoulou A., Jones L.M., Cole S.T. Tuberculosis. Tuberculosis. 2011;91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Buc H., McClure W.R. Kinetics of open complex formation between Escherichia coli RNA polymerase and the lac UV5 promoter. Evidence for a sequential mechanism involving three steps. Biochemistry. 1985;24:2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- 29.Roe J.H., Burgess R.R., Record M.T. Kinetics and mechanism of the interaction of Escherichia coli RNA polymerase with the lambda PR promoter. J. Mol. Biol. 1984;176:495–522. doi: 10.1016/0022-2836(84)90174-8. [DOI] [PubMed] [Google Scholar]
- 30.Ruff E., Record M., Jr, Artsimovitch I. Initial events in bacterial transcription initiation. Biomolecules. 2015;5:1035–1062. doi: 10.3390/biom5021035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.China A., Tare P., Nagaraja V. Comparison of promoter-specific events during transcription initiation in mycobacteria. Microbiology. 2010;156:1942–1952. doi: 10.1099/mic.0.038620-0. [DOI] [PubMed] [Google Scholar]
- 32.Ackers G.K., Johnson A.D., Shea M.A. Quantitative model for gene regulation by lambda phage repressor. Proc. Natl. Acad. Sci. U.S.A. 1982;79:1129–1133. doi: 10.1073/pnas.79.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vossen K.M., Stickle D.F., Fried M.G. The mechanism of CAP-lac repressor binding cooperativity at the E. coli lactose promoter. J. Mol. Biol. 1996;255:44–54. doi: 10.1006/jmbi.1996.0005. [DOI] [PubMed] [Google Scholar]
- 34.Lemon B., Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- 35.Mogridge J., Mah T.F., Greenblatt J. A protein-RNA interaction network facilitates the template-independent cooperative assembly on RNA polymerase of a stable antitermination complex containing the lambda N protein. Genes Dev. 1995;9:2831–2845. doi: 10.1101/gad.9.22.2831. [DOI] [PubMed] [Google Scholar]
- 36.Nehrke K.W., Platt T. A quaternary transcription termination complex. Reciprocal stabilization by Rho factor and NusG protein. J. Mol. Biol. 1994;243:830–839. doi: 10.1006/jmbi.1994.1685. [DOI] [PubMed] [Google Scholar]
- 37.Michel D. Progress in biophysics and molecular biology. Prog. Biophys. Mol. Biol. 2010;102:16–37. doi: 10.1016/j.pbiomolbio.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 38.Chauhan S., Tyagi J.S. Cooperative binding of phosphorylated DevR to upstream sites is necessary and sufficient for activation of the Rv3134c-devRS operon in Mycobacterium tuberculosis: implication in the induction of DevR target genes. J. Bacteriol. 2008;190:4301–4312. doi: 10.1128/JB.01308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gautam U.S., Chauhan S., Tyagi J.S. Determinants outside the DevR C-terminal domain are essential for cooperativity and robust activation of dormancy genes in Mycobacterium tuberculosis. PLoS One. 2011;6:e16500. doi: 10.1371/journal.pone.0016500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang G.-Q., Deshpande A.P., Patel S.S. Transcription factor-dependent DNA bending governs promoter recognition by the mitochondrial RNA polymerase. J. Biol. Chem. 2011;286:38805–38813. doi: 10.1074/jbc.M111.261966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.