Abstract
We studied the transcriptional response to an increase in DNA supercoiling in Streptococcus pneumoniae by using seconeolitsine, a new topoisomerase I inhibitor. A homeostatic response allowing recovery of supercoiling was observed in cells treated with subinhibitory seconeolitsine concentrations. Supercoiling increases of 40.7% (6 μM) and 72.9% (8 μM) were lowered to 8.5% and 44.1%, respectively. Likewise, drug removal facilitated the recovery of cell viability and DNA-supercoiling. Transcription of topoisomerase I depended on the supercoiling level. Also specific binding of topoisomerase I to the gyrase A gene promoter was detected by chromatin-immunoprecipitation. The transcriptomic response to 8 μM seconeolitsine had two stages. An early stage, associated to an increase in supercoiling, affected 10% of the genome. A late stage, manifested by supercoiling recovery, affected 2% of the genome. Nearly 25% of the early responsive genes formed 12 clusters with a coordinated transcription. Clusters were 6.7–31.4 kb in length and included 9–22 responsive genes. These clusters partially overlapped with those observed under DNA relaxation, suggesting that bacteria manage supercoiling stress using pathways with common components. This is the first report of a coordinated global transcriptomic response that is triggered by an increase in DNA supercoiling in bacteria.
INTRODUCTION
DNA is dynamically compacted (up to 1000-fold in the bacterial chromosome) in order to optimize its essential functions of DNA replication, chromosome segregation and gene expression (reviewed in (1)). The shape and extensiveness of the bacterial chromosome, or nucleoid, is determined by the supercoiling of the DNA strands, and by the binding of nucleoid-associated proteins or NAPs (2). These NAPs form a functional network that maintains the topology of the DNA by bending, wrapping or bridging (1). In addition, NAPs regulate transcription by constraining supercoils. Several NAPs have been characterized in the Gram-negative bacterium Escherichia coli. However, very few have been discovered in Gram-positive bacteria (3). In prokaryotes, transcription is regulated by a combination of cis factors, such as chromosomal topology and promoter DNA sequences, and trans factors, such as structural and regulatory proteins. These factors function either by facilitating or inhibiting the interaction of the RNA polymerase with specific promoter regions (4), or by targeting several genes, such as the NAPs (3).
In bacteria, the level of DNA supercoiling is maintained by the opposite activities of DNA topoisomerases that relax DNA, such as topoisomerase IV (Topo IV) and topoisomerase I (Topo I), and by gyrase, which introduces negative supercoils (5). Streptococcus pneumoniae has three topoisomerases: gyrase, Topo IV and Topo I. The first two are the targets of the clinically widely used fluoroquinolones. However, drugs targeting Topo I are scarce. Cheng et al. (6) have identified a phenantrene alkaloid that inactivates E. coli Topo I but did not significantly inhibit cell growth. We have discovered an alkaloid compound, seconeolitsine (SCN), which inhibits the relaxation activity of the S. pneumoniae Topo I enzyme at concentrations equivalent to those inhibiting cell growth. Modelling of pneumococcal Topo I based on the crystal structure of E. coli Topo I, together with docking calculations, indicated strong interactions of SCN with the nucleotide-binding site of the protein, which correlated with their inhibitory effect (7).
Homeostasis of DNA supercoiling occurs mainly via the regulation of transcription of topoisomerase genes. This type of control was first described in E. coli. In this bacterium, transcription of the Topo I gene (topA) decreases in response to DNA relaxation (8), while transcription of gyrase genes (gyrA, and gyrB) increases (9–11). Likewise, an increase of gyrase expression in response to relaxation has also been observed in other bacteria (12,13). In E. coli, the expression of gyrase and Topo I genes is also mediated by NAPs, which affect DNA supercoiling (14,15). Nevertheless, these regulatory mechanisms do not seem to apply to S. pneumoniae, which lacks most of the NAPs found in E. coli. Given the smaller size of the pneumococcal chromosome (∼2 Mb for S. pneumoniae versus ∼4.6 Mb for E. coli), a low number of NAPs is likely to suffices for cellular viability. In S. pneumoniae, the transcription of gyrB and topA genes is regulated by their chromosomal location in specific topological domains. Meanwhile, gyrA and parEC (encoding Topo IV) carry their own regulatory signals in their promoter regions. We have analyzed the transcription of fusions between the promoter regions of topoisomerase genes and a reporter gene cloned in plasmids in S. pneumoniae. Under DNA relaxation, expression from the chromosomal promoters was either up-regulated (PgyrB) or down-regulated (PtopA), however, expression from these promoters was down-regulated in the plasmid. Nevertheless, the topology-dependent expression of both PparE and PgyrA was equivalent in the plasmid or in the chromosome. In the case of gyrA, its promoter has a DNA bend that is sensitive to the level of supercoiling (16). This indicates that the topological organization and positioning of key genes is optimized to apply a stratified and systemic control of chromosome supercoiling.
In many bacterial species, DNA supercoiling varies in response to diverse physiological situations (17–19). A reduction in DNA supercoiling has been shown to have an effect on global transcription of the genome in E. coli (20,21), Haemophilus influenzae (22) and S. pneumoniae (23). In the latter, chromosome relaxation induced by the GyrB inhibitor novobiocin (NOV) alters transcription of about 14% of the genome (23). A majority of these differentially expressed genes (DEGs) are close to each other, and form topologically sensitive gene clusters that regulate expression at a level above the operon (23). We propose that the S. pneumoniae genome is organized according to four types of domains characterized by their transcriptional response to DNA relaxation: up-regulation (U domains), down-regulation (D domains), non-regulation (N domains), and flanking (F domains). F domains have a remarkably high AT content (>65%) and may play a structural role in the maintenance of DNA topology. Interestingly, a large fraction of the genes of U and D domains showed conservation. The gene-lack indexes (the number of genomes in which a gene is present divided by the total number of genomes analyzed) were lower on average in U (1.51) and D (1.65) domains than in the non-regulated genome (1.91). However, F domains showed very high gene-lack indexes (4.66). Altogether, DNA topology is a general transcriptional regulator that can be superimposed upon other types of more specific regulatory mechanisms. This is further supported by the fact that changes in the expression of a heterologous gene cassette, in response to relaxation, depend on the position of the cassette in the chromosome (16).
A general transcriptomic response to an increase in negative supercoiling has not yet been described. Hypernegative supercoiling has only been reported in E. coli topA mutants. However, all these mutants, with the exception of topA10, have acquired compensatory mutations in gyrase genes (24). In the topA10 mutant, a 22% increase in chromosomal negative supercoiling was detected (25). In this mutant, previous studies have detected an alteration in the relative abundance of 88 proteins (26), which represents 2% of the genome (27). On the other hand, Topo I plays a major role in transcription, because of its physical interaction with RNA polymerase (28). Hypernegative supercoiling stabilizes the RNA-DNA hybrid (R-loop) (29). This is supported by the observation that the growth defect of topA mutant can be partially compensated by RNase HI overproduction (30). Topo I prevents R-loop formation by relaxing the negative supercoiling induced by transcription (31–33). Therefore, the effects of hypernegative supercoiling in transcription would directly depend on the activity of Topo I.
In this report, we have studied the global transcriptomic response to an increase in DNA supercoiling in S. pneumoniae, as a model system. For this aim, we took advantage of the availability of SCN, a novel and unique inhibitor of its Topo I enzyme. We estimated the changes in supercoiling levels and established a relationship with cell viability. The transcriptomic response involved clusters of genes that were sensitive to changes in DNA topology.
MATERIALS AND METHODS
Analysis of the topology of covalently closed circles
S. pneumoniae was cultured at 37ºC in a casein hydrolysate-based liquid medium (AGCH) containing 0.2% yeast extract and 0.3% sucrose (34). Strain R6 containing plasmid pLS1, with a size of 4408 bp, was grown in the presence of 1 μg/ ml of tetracycline. In order to test viability of S. pneumoniae cells under various conditions, such as the addition of SCN, aliquots of the cultures were plated on Mueller-Hinton drug-free agar plates containing 5% sheep's blood. The plates were incubated at 37°C in an atmosphere of 5% CO2. Plasmid DNA was extracted as described (35). Agarose gels of plasmid DNA were run in neutral/neutral two-dimensional agarose gels. The first dimension was run in the presence of 5 μg/ ml and the second dimension was run in the presence of 15 μg/ ml of chloroquine. Circular DNA molecules were analyzed by Southern hybridization with a 240-bp specific pLS1 probe (23). The DNA linking number (Lk) was calculated by quantifying the amount of every topoisomer. The DNA supercoiling density (σ) was calculated using the equation σ = ΔLk/ Lk0. Changes in the linking number (ΔLk) were determined using the equation Lk = Lk − Lk0, in which Lk0 = N/10.5, N is the length of the DNA strand in bp, and 10.5 the number of bp per one complete turn in B-DNA.
RNA extraction and quantitative real time PCR (qRT-PCR) experiments
RNA was extracted from samples containing 2 to 4 × 108 cells with the RNeasy mini kit (Quiagen). cDNAs were synthesized using 5 μg of total RNA as the template (36). The resulting cDNAs were in turn used as the template in qRT-PCR amplifications (Chromo 4, Bio-Rad). The total volume in each reaction was 20 μl and contained 2 μl of cDNA, 0.4 μM of each primer, and 10 μl of Sso Advanced Universal SYBER Green Supermix (BioRad). The program had 42 cycles of three steps: denaturation (30 s at 94°C), annealing (30 s at 45–56°C), and elongation (30 s at 68°C). The following oligonucleotide pairs used were: GYRARTF/ GYRARTR (gyrA); GYRBRTF/ GYRBRTR (gyrB); TOPARTF/ TOPARTR (topA); PARE214/ PARE274R (parE) (16). Assays were performed in triplicate and normalized against the signal from a 16S rDNA internal control (23).
Western blot analysis
GyrA and Topo I proteins were purified as previously described (7,37). Polyclonal rabbit antibodies against GyrA were obtained after three subcutaneous injections at 3-week intervals of polyacrylamide portions containing a 50 kDa Nterminal-GyrA fragment. Blood was recovered and the serum stored at −80°C. Polyclonal antibodies against Topo I were obtained from Davids Biotechnologie and polyclonal antibodies against LytA were kindly provided by Ernesto García (CIB, CSIC, Madrid, Spain). Whole cell lysates were separated on Any KD™ Criterion TGX gels (Bio-Rad) and transferred to PVDF membranes with Trans-Blot Turbo (Bio-Rad) at 25 V, 1 A for 30 min. The protein-transferred membranes were blocked with Super Block T20 (TBS) blocking buffer (Thermo Scientific) for 1 h and probed with antibodies anti-GyrA (diluted 1:2000), anti-Topo I (diluted 1:2000) and anti-LytA (diluted 1:20000) in Super Block for 2 h. The anti-GyrA and anti-Topo I antibodies were previously purified by affinity using purified proteins bound to a PVDF membrane. Anti-rabbit IgG-Peroxidase antibody (Sigma-Aldrich) served as the secondary antibody. Super Signal West Pico chemiluminescent substrate (Thermo Scientific) was utilized to develop the signal and monitored with a ChemiDoc™ MP system (Bio-Rad). Image analysis was performed with Image Lab™ software (Bio-Rad).
Chromatin immunoprecipitation (ChIP)
ChIP with cultures grew as indicated in the legend of Figure 1C was carried out as described previously (38). Briefly, 70 ml of culture (5 × 109 cells) was mixed by inverting with 7 ml of fixing solution (50 mM Tris [pH 8.0], 100 mM NaCl, 0.5 mM EGTA, 1 mM EDTA, 11% [vol/vol] formaldehyde) and incubated at room temperature for 30 min. Cells were treated with 7 ml of quenching solution (1.25 M glycine, 50 mM Tris [pH 8.0], 100 mM NaCl, 0.5 mM EGTA, 1 mM EDTA) for 30 min at 4°C to stop crosslinking. Cells were then washed and sonicated 7 × 20 s with a Vibra Cell 75043 (Bioblock Scientific) with a microtip at 20% amplitude. A fraction containing 5 × 108 cells was kept as the whole cell extract (WCE) control. Fragmented DNA from 3 × 109 cells was immunoprecipitated with 4–5 μg of affinity purified and concentrated anti-GyrA or anti-Topo I pre-coupled to 100 μl protein G Dynabeads (Invitrogen). The resulting ChIP-DNA and the WCE were subjected to qRT-PCR analysis with the oligonucleotides pairs annealing at upstream regions of topA and gyrA (Figure 5A). Amplification of internal fragments of topA and gyrA was performed as a control using oligonucleotides pairs previously described (23).
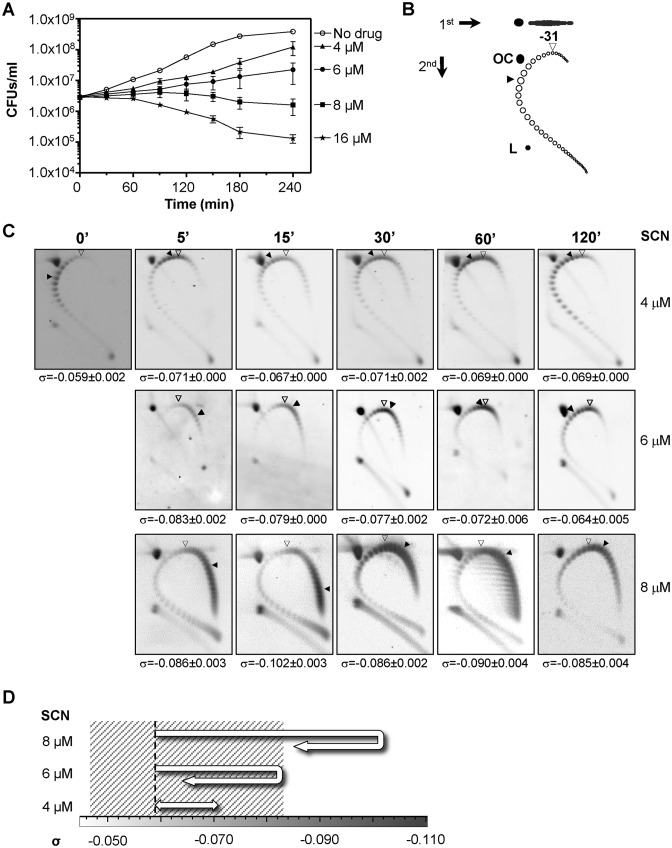
Figure 1.
SCN affects cell viability and induces supercoiling. (A) Cultures of R6 (pLS1) were grown at an OD620nm of 0.4 and diluted 100-fold in medium containing SCN from 4 to 16 μM. Samples were taken before the addition of the drug (zero time) and at the indicated times were plated in drug-free agar medium. Results are the average of three independent replicates ± SEM. (B) Cartoon illustrating topoisomer distribution of plasmid pLS1 subjected to 2D-agarose gel electrophoresis run in the presence of 5 and 15 μg/ ml chloroquine in the first and second dimensions, respectively. OC, open circle; L, linear form. It was previously determined that 15 μg/ ml of chloroquine introduces 31 positive supercoils. An open arrowhead indicates the topoisomer that migrated with ΔLk = 0 in the second dimension and that had a ΔWr of −31. A blackened arrowhead points to the more abundant topoisomer. (C) Cultures at an OD620nm of 0.4 were treated with 4–8 μM of SCN. Distribution of pLS1 topoisomers after treatment at the indicated SCN concentrations. Plasmid DNA was isolated and subjected to 2D-agarose gel electrophoresis as described under Materials and Methods. Indicated supercoiling density (σ) values are averages ± SD from three independent replicates. (D) Diagram showing the schematic trajectory of supercoiling density at the indicated SCN concentrations. A dashed line indicates supercoiling density of DNA coming from no treated cells. Shadowed area represents the supercoiling density interval where cells can survive.
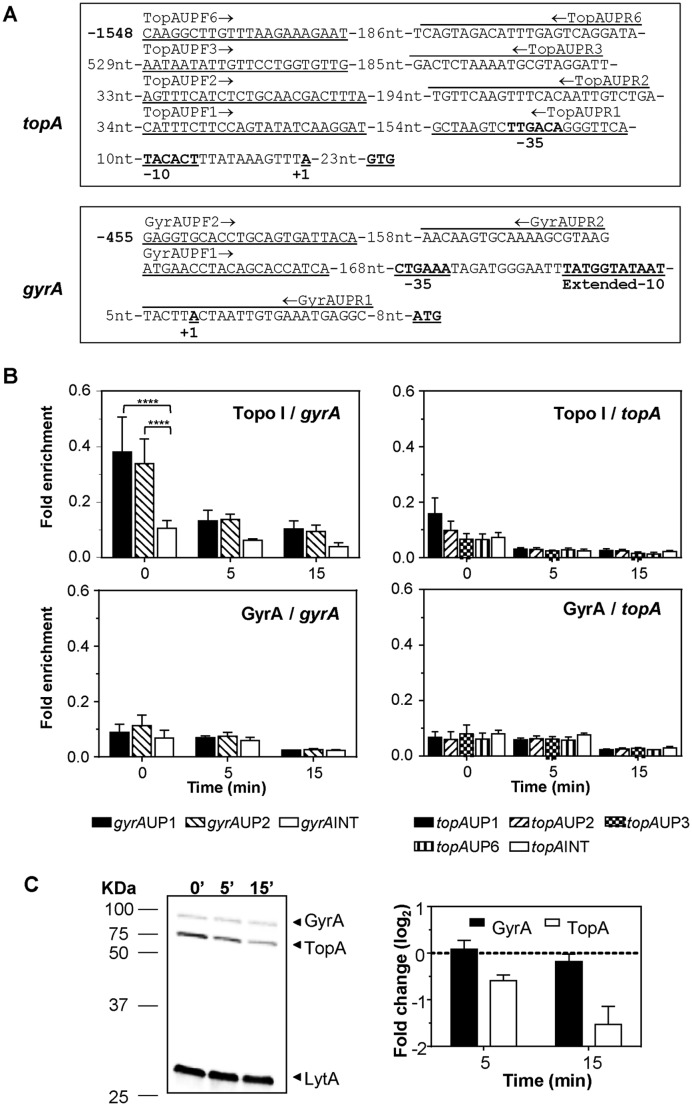
Figure 5.
Recruitment of Topo I and GyrA to topA and gyrA upstream sequences. (A) Sequence of the topA and gyrA upstream regions and localization of the oligonucleotides used in the qRT-PCR experiments. The −35 and −10 boxes, and the nucleotide in which transcription is initiated (+1) are indicated. (B) Determination of Topo I or gyrase binding on upstream topA or gyrA regions by ChIP-qRT-PCR. Exponentially growing cells were subjected to chromatin immunoprecipitation (ChIP) using anti-Topo I or anti-GyrA antibodies, and the pulled-down DNA was subsequently analyzed by qPCR. The graphs show pulldown efficiency (ChIP-DNA/input DNA) for each primer pair. Values are the average ±SD of three independent replicates. ****P < 0.0001. (C) Western blot analysis of GyrA and Topo I performed as described in the legend of Figure 4.
Microarray data normalization and analysis
The high density Agilent expression arrays of S. pneumoniae were processed at the Functional Genomics Core Facility, Institut de Recerca Biomèdica, Barcelona, Spain. The arrays included one or two sets of 17 probes (average size of 60 nucleotides) for each 2037 protein coding genes from S. pneumoniae R6. Using 25 ng of total RNA as template, cDNAs were obtained from using the Trans-Plex WTA2 kit (Sigma). These cDNAs were labelled with Cy3 using the Agilent Genomic DNA ULS™ (Universal Linkage System) kit. A total of 250 ng of labelled cDNA were used for hybridization (40 h at 65°C) and scanning was performed with a Roche MS200 scanner. The raw data were extracted and normalized using the Agilent Feature extraction software v11.5.1.1. The median expression of every probeset was calculated and the result was scaled by log2 and normalized according to quantile using the Bioconductor software (www.bioconductor.org/). A principal component analysis was then carried out using the Partek Genomics Suite 6.4. The significance of differential gene expression was determined using the ANOVA test. Each microarray experiment was carried out in duplicate using cDNA prepared from two independent cultures.
RNA library preparation for RNA-Seq
Total RNA (5 μg) obtained as described above was depleted of ribosomal RNA using the Ribo-Zero (Bacteria) Magnetic Kit (Illumina). Libraries for RNA sequencing (RNA-Seq) were prepared using the ScriptSeq v2 RNA-Seq system (Illumina). Briefly, 1 μg RNA samples were chemically fragmented. Using randomly primed cDNA synthesis, cDNAs that were tagged at the 5′ end (equivalent to the 3′ end of the original RNA) were synthesized. The cDNAs were then tagged at the 3′ end using Terminal-Tagging Oligos (TTO). These oligonucleotides randomly annealed to the cDNA, and were extended by DNA polymerase. The resulting di-tagged cDNAs were purified with the Ampure bead XP system. The enrichment and the barcoding of the purified di-tagged cDNAs were done with 15 cycles of PCR. The library size was determined with a 2100 Bioanalyzer Instrument (Agilent Technologies). qPCR quantifications were done with a Kapa Library quantification Kit (Kapa Biosystems). The sequencing was accomplished with a NextSeq 500 platform using a 1 × 75 base run. A total of 29.8–35.47 million passing filters reads were obtained per sample.
Accession number
All data are available at the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo) with via accession numbers GSE77748 (RNA-Seq) and GSE77587 (array). These data have been grouped under SuperSeries GSE77749.
Genome level alignments and annotation and measurement of expression level
For each pair of replicates in the microarray analysis, the median background was subtracted from the median intensity. The adjusted median intensity of the two replicates was then averaged. The differences in the intensity of the replicates were normalized using the fit function implemented in the limma package (39) of Bioconductor. The statistical significance was assessed using moderated t-statistics calculated by the empirical Bayes method (40), which was also implemented in the limma package. In order to analyze the results of the RNA-Seq, around 30M reads per replicate were mapped onto the genome of the R6 strain of S. pneumoniae using bowtie (41). Reads counts per gene were obtained using a perl script written in-house. The statistical significance of the counts was calculated using edgeR (42).
Detection of topological clusters
A two-step strategy was used. In the first step, a sliding window of 11 genes was applied to the whole genome. The percentage of DEGs in the window was assigned to the central gene that resulted from counting all the genes with the majority response, either up-regulated or down-regulated, and subtracting the contrary ones if present. Those clusters that contained more than 12 genes and ≥40% DEGs with the same response and were also assigned a P-value <0.05 using Fisher's exact test were selected for a second step of border refinement. In this step, closed cores at a distance of ≤5 genes were fused into one. Non-DEGs located at the borders of clusters were discarded. Conversely, those DEGs that were immediately adjacent and which had the same response as the defined cluster were included. Fisher's P-value was recalculated after domain extreme refinement. In addition, clusters that exclusively contained genes belonging to a unique co-transcriptional unit were considered functional rather than structural domains and were removed. Operons were predicted using the algorithm published (43). The P-value was calculated considering the observed number of concordant DEGs within a domain in comparison to the expected number according to the fraction of DEGs, either up-regulated or down-regulated, in the genome. P-value indicates the probability that the response observed does not match the null-hypothesis, i.e. that the transcriptional response is evenly distributed along the chromosome.
RESULTS
S. pneumoniae is able to survive increases of about 40% in the amount of supercoiling
The effects of SCN in the wild type R6 strain carrying plasmid pLS1 were assessed. Cell viability was affected in a concentration-dependent manner (Figure 1A). Although a reduction in the growth rate was observed at the subinhibitory concentrations of 4 μM (0.25× MIC), and 6 μM (0.375× MIC), no decrease in viable cells was observed. However, a reduction in cell viability of 28% and 42% was detected at 180 min and 240 min of treatment with 8 μM SCN (0.5× MIC), respectively. Likewise, a reduction in viability of more than 92% was observed at 180 and 240 min when increasing the concentration of SCN to 16 μM (1× MIC).
The alterations in supercoiling resulting from treatment with 4–8 μM of SCN were analyzed by quantifying the distribution of topoisomers of plasmid pLS1 by two-dimensional agarose gel electrophoresis. This plasmid is appropriate to study supercoiling, given that it replicates by a rolling circle mechanism (44) and all their genes are transcribed in the same direction, avoiding problems of transcription interference during replication (45). Those topoisomers with Lk values less than −31 were negatively supercoiled and migrated to the right of the bubble-shaped arc, and those with Lk values higher than −31 (positively supercoiled) migrate to the left (Figure 1B). The increase in negative supercoiling correlated with the increase in the concentration of SCN (Figure 1C). The increase in supercoiling also correlated with a decrease in cell viability. Modest increases in supercoiling densities (σ) with respect to the untreated culture between 15% and 18.3% were observed under treatment with 4 μM SCN. This result coincided with a weak effect on cell viability. With 6 μM SCN, the σ increased 40.7% at 5 min, 33.9% at 15 min, and 30.5% at 30 min. Nevertheless, the σ value recovered to 22% higher than the untreated culture at 60 min and to 8.5% higher than the untreated culture at 120 min. Treatment with 8 μM SCN resulted in higher and longer lasting increases in the σ value: 45.8% and 72.9% at 5 min and 15 min, respectively. A partial recovery was observed at 30 min, 45.8% with respect to time 0 min. However, σ values were maintained at latter times: 52% at 60 min and 44.1% at 120 min (Figure 1C and D). The σ values provided a good estimation of chromosomal supercoiling (25) and indicate that the inhibition of Topo I by SCN resulted in an increase of supercoiling in vivo. These results also showed that treatment with subinhibitory SCN concentrations (4–8 μM) permitted the recovery of supercoiling levels to a certain degree without affecting cell viability. In addition, we were able to demonstrate that cells could withstand an increase of up to 40.7% in supercoiling. Importantly, a complete recovery of both viability and supercoiling levels occurred in cells treated during 120 min with either 6 or 8 μM SCN upon removal of the inhibitor (Figure 2). We tested whether these effects were due to inactivation of SCN. Supernatants of cultures grew as in Figure 1, with either no drug or with 8 μM of SCN were taken after 30, 60, 120 or 240 min. These were inoculated with strain R6 (4 × 105 CFUs) and incubated at 37ºC with a 5% CO2 atmosphere during 18 h. While normal growth was observed (OD620nm in the range of 0.8–1.2) in supernatants obtained from cultures grown in the absence of SCN, no growth was observed in the supernatants obtained from SCN-treated cultures. These results indicated that SCN preserved its inhibitory effect for at least 240 min of incubation.
Figure 2.
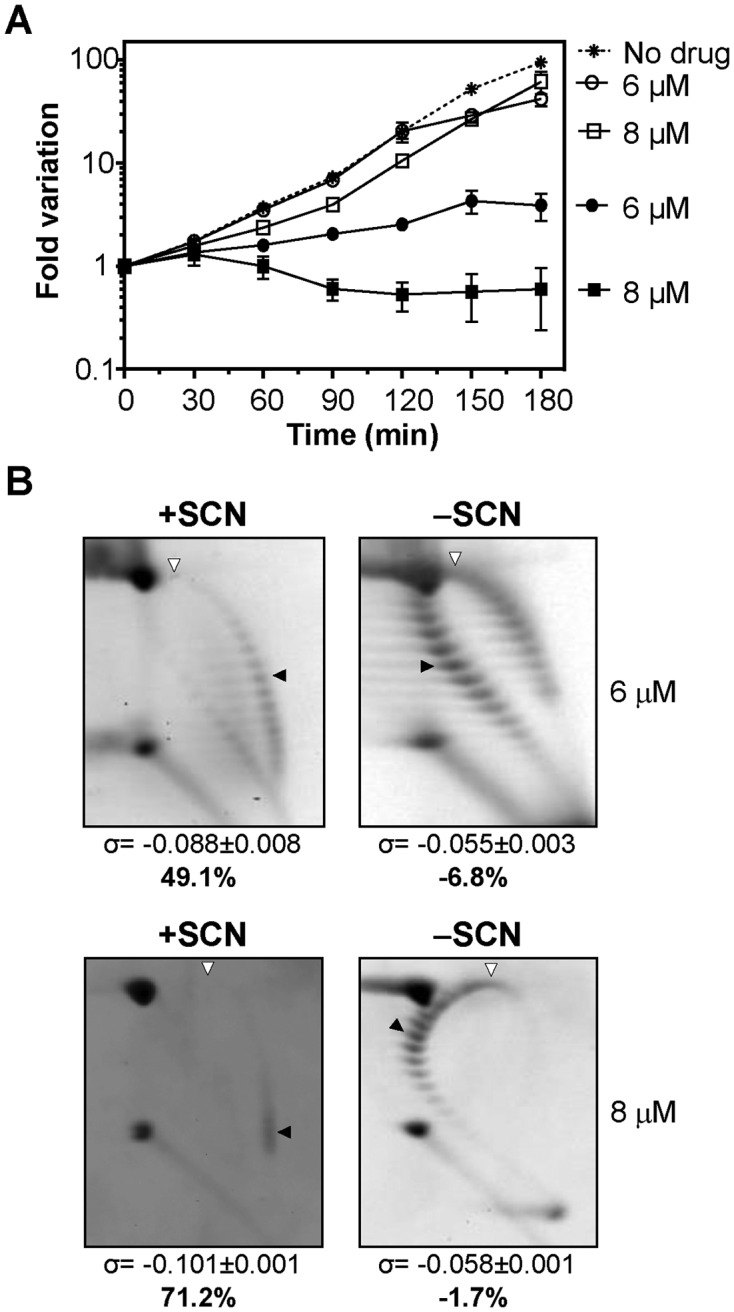
The removal of SCN led to a recovery in the cell viability and in the supercoiling density. (A) Viability of R6 (pLS1) cultures. Cultures were grown at an OD620nm of 0.4 and diluted 100-fold in medium containing either 6 or 8 μM of SCN. After 120 min growth in the presence of the indicated amounts of SCN, cells were centrifuged, washed, and suspended in either drug-free medium (empty symbols) or in medium containing the same SCN concentrations (6 or 8 μM SCN, full symbols) than in the previous treatment. As control, a culture was grown at OD620nm of 0.4, diluted 100-fold in drug-free medium and grown during 180 min (no drug). Samples were taken at the indicated times and plated in drug-free agar medium. Results are the average of three independent replicates ± SEM. (B) Distribution of pLS1 topoisomers in 2D-agarose gel electrophoresis. Electrophoresis conditions and symbols are as indicated in the legend of Figure 1. Samples were taken after 120 min treatment with SCN (+SCN) and 120 min after removal of the drug (−SCN). Results are the average of at least three independent replicates ± SD.
The variation in the level of supercoiling correlated with the level of topA transcription
We analyzed the levels in the transcription of the DNA topoisomerase genes that occurred during the reversal of the increase in the supercoiling. Two situations were studied. Firstly, cultures were treated with either 6 or 8 μM SCN (Figure 1). Secondly, cultures were treated with either 6 or 8 μM SCN followed by removal of the drug (Figure 2). The expression of the DNA topoisomerase genes was measured by qRT-PCR. Only the results from early time points (5, 15 and 30 min) were compared with the results of a previous study in which the transcriptomic response to DNA relaxation had been analyzed (23). At these time points, the most significant change observed was a reduction in the expression of topA. With 6 μM SCN, the most significant decrease in topA expression (1.8-fold) was observed at 5 min (Figure 3A), in tandem with the greatest increase in supercoiling (40.7%). More profound variations in the expression of topA were observed with 8 μM SCN: a 3.9-fold down-regulation at 5 min, a 2.5-fold at 15 min, and a 1.9-fold decrease and 30 min. In addition, there was a >2-fold increase in gyrB at 5 and 15 min and an unexpected increase in parEC at 15 min. No changes in gyrA transcription were detected. The results from the microarrays (see below) and the qRT-PCR with respect the DNA topoisomerase genes correlated well (R2 = 0.94, P < 0.0001, data not shown).
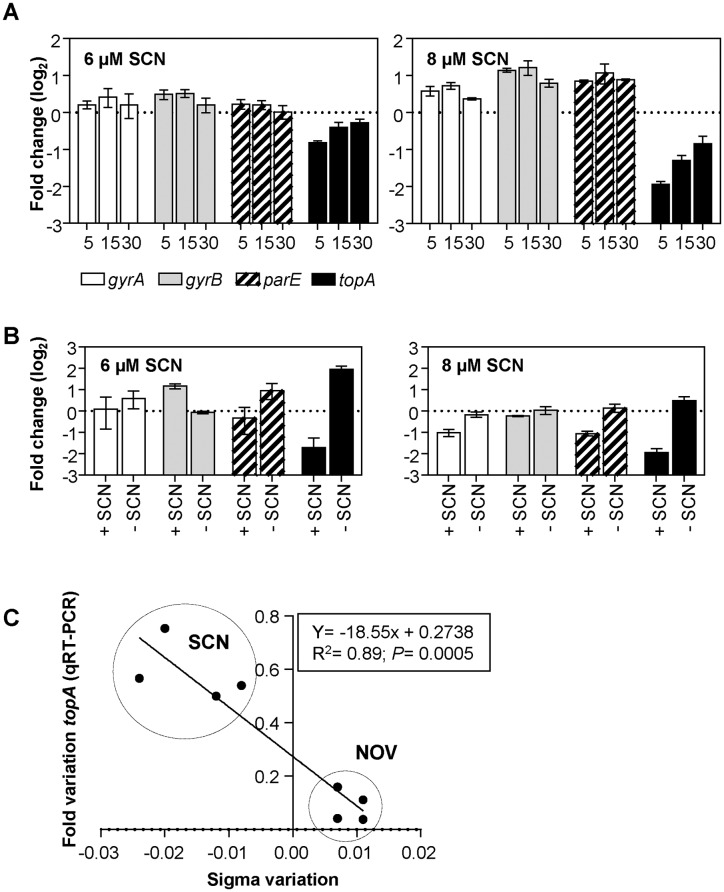
Figure 3.
Changes in supercoiling levels affect the transcription of topA as measured by qRT-PCR. (A) Cultures of the R6 strain at an OD620nm of 0.4 were treated with 6 or 8 μM of SCN. (B) Cultures were grown at an OD620nm of 0.4 and diluted 100-fold in medium containing either 6 μM or 8 μM of SCN. After 120 min of growth, cells were centrifuged, washed, and suspended either in medium alone (− SCN) or medium containing 6 or 8 μM of SCN (+ SCN). (C) Correlation between fold changes in supercoiling levels and changes in the transcription of topA. The data corresponds to samples treated with either SCN or NOV at concentrations that allowed cellular growth and recovery of DNA supercoiling: 4 and 6 μM (0.25× MIC and 0.375× MIC) for SCN; 0.5 and 1 μg/ml (0.5× MIC, and 1× MIC) for NOV. In both cases, time points of 15 and 30 minutes were used. The conditions of the qRT−PCR experiments and the normalization of the data are described in the Materials and Methods. In (A) and (B), values from three independent replicates (mean ± SEM, n = 3) were normalized against values obtained at time 0 min.
In experiments where treatment with SCN was followed by removal of the drug, the main affected gene was also topA (Figure 3B). The expression of topA decreased about 4-fold after 120 min in the presence of 6 and 8 μM SCN. This effect was reverted when the drug was removed thanks to a subsequent increase in topA transcription of 12.7-fold for 6 μM and 5.4-fold for 8 μM SCN.
We analyzed the relationship between topA transcription and supercoiling levels. Changes in topA transcription, as measured by qRT-PCR, that had been induced with either SCN or NOV (23) were plotted against the induced σ variation. A good correlation (R2 = 0.89, P = 0.0005) was observed (Figure 3C). Altogether, these data strongly suggest that Topo I is highly sensitive to changes in DNA supercoiling in S. pneumoniae.
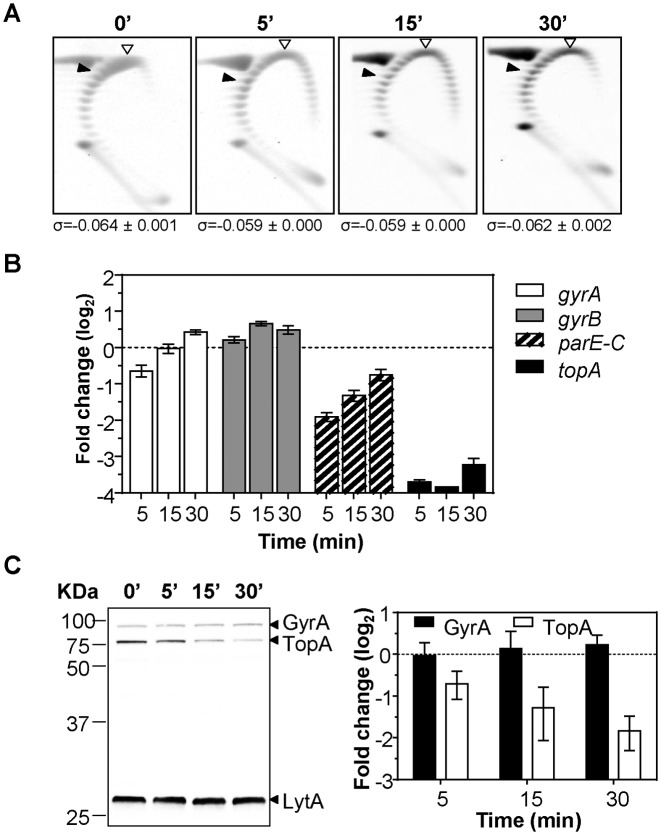
To gain further clues about topA regulation, we performed an experiment in which cultures were co-treated with subinhibitory concentrations of both NOV (0.5× MIC) and SCN (6 μM, 0.375× MIC). As expected, the level of supercoiling of the replicating plasmid did not change (Figure 4A). However, the transcription of parE and topA did show a change (Figure 4B) and these variations were additive to that observed with NOV alone (23) or with SCN alone (Figure 3A). The most significant decreases in expression were observed for topA: 13.0-fold, 14.3-fold and 9.3-fold at 5, 15 and 30 min, respectively. The parE gene also showed significant decreases of 3.8-fold at 5 min and 2.5-fold at 15 min. These results suggested, that, under conditions in which no detectable changes in global supercoiling were observed, local supercoiling changes could still affect the transcription of topA and parE. The decrease in topA expression correlated with a decrease in Topo I at the protein level, as showed by Western-blot analysis (Figure 4C).
Figure 4.
SCN and NOV together affect the transcription and translation of topA without changing the supercoiling density. A culture of R6 (pLS1) at an OD620 of 0.4 was co-treated with 6 μM of SCN and 0.8 μM of NOV. Samples were taken before the addition of drugs (zero time) and at the indicated times (A) plasmid DNA was isolated and subjected to 2D agarose gel electrophoresis as described under Materials and Methods. Indicated supercoiling density (σ) values are averages ± SD from two independent replicates. (B) RNA was isolated, cDNA synthesized and qRT-PCR performed for topoisomerase genes using the appropriated oligonucleotide pairs indicated in Materials and Methods. Values (mean ± SEM, n = 3) were normalized against those obtained at time 0 min. (C) Western blot analysis of GyrA and Topo I. Crude cell extracts were prepared from cells grown as in (A). Samples containing 20 μg of protein were separated by SDS-PAGE and blotted. The membrane was incubated with polyclonal anti-GyrA, anti-Topo I and anti-LytA antibodies. The results (mean relative to LytA and to time zero min ± SD) of three independent experiments are represented.
Topo I binds to gyrA upstream region
To ascertain if the promoter regions of topA and gyrA recruited Topo I or gyrase proteins, experiments of ChIP were performed using antibodies directed against the pneumococcal GyrA subunit and the Topo I enzyme. Cultures grew as described in Figure 1C were treated with 8 μM SCN and subjected to ChIP. To determine the specificity of binding to the upstream topA and gyrA regions, qRT-PCR reactions with oligonucleotides pairs covering gyrA and topA upstream regions were used (Figure 5A). These pairs covered region −1548 to −51 of topA (taking the first Topo I nucleotide as nt = 1) and −455 to −8 of gyrA. Two internal fragments of gyrA (positions 1431 to 1589) and topA (positions 371–513) were also used as controls. The initiation site of transcription of topA was inferred from the data of RNASeq (Supplementary Figure S1). No significant increase in the recruitment of gyrase was observed at gyrA or topA upstream regions in relation to the internal gene fragments. However, for Topo I, significant differences of 3.8-fold were observed in the binding to the region covered by oligonucleotide pair GyrAUP1 and 3.4-fold for GyrAUP2 in relation to the internal gyrA fragment used (Figure 5B). Treatment with SCN diminished Topo I recruitment, accordingly with the decrease in the amount of this protein (Figure 5C). These results suggest a specific binding of Topo I to the gyrA promoter region.
The global transcriptomic response to treatment with 8 μM SCN has two stages: an early response followed by recovery
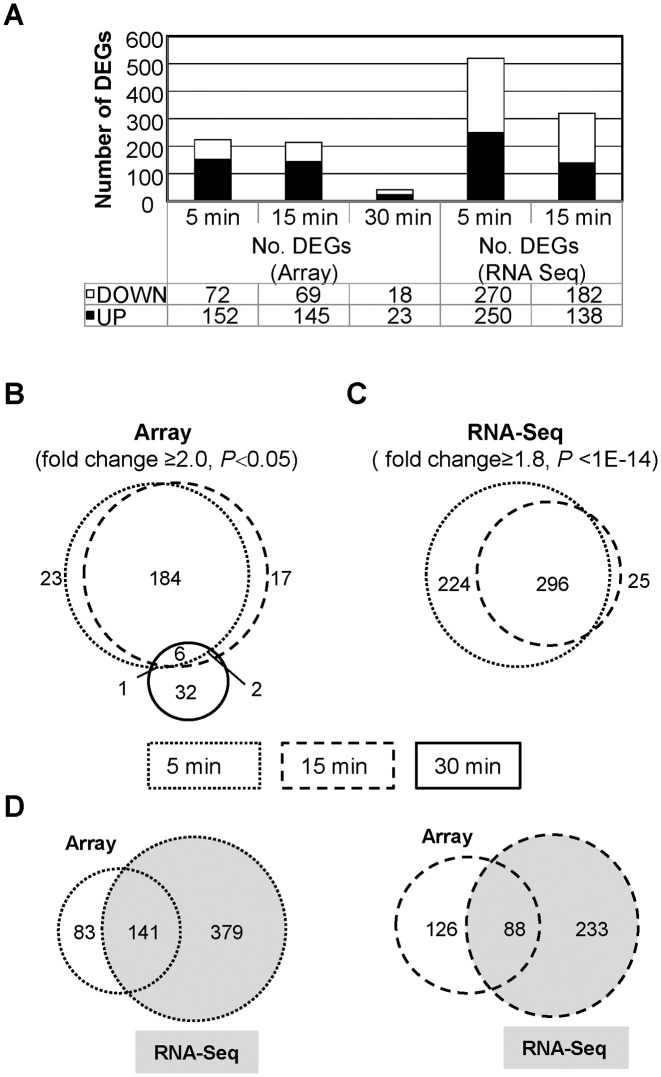
The expression of 224 genes was modified in R6 strain after treatment with 8 μM SCN for 5 min. This represented 10.9% of the genome. Meanwhile, 214 genes, or 10.5% of the genome, were modulated at 15 min, and only 41 genes, or 2.0% of the genome, were affected at 30 min (Figure 6A). An analysis of the DEGs that were detected in the microarrays (Figure 6B) suggested that there were two stages in the transcriptomic response to SCN. An early stage, including 5 and 15 min, with 184 common DEGs, that would represent an active response against increased supercoiling. A late stage, at 30 min, would represent a supercoiling recovery phase. All microarray data are shown in Supplementary Table S1.
Figure 6.
Gene expression analysis of R6 treated with SCN was performed using microarrays and the RNA-Seq method. (A) Categorization of DEGs in all conditions and times studied. (B) DEGs that were detected in microarrays in the 8 μM SCN condition represented in Venn diagrams. Each circle represents a single time interval. (C) DEGs detected using RNA-Seq in the 8 μM SCN. (D) DEGs detected by the two techniques.
In the early phase, transcriptional variations occurs in clusters
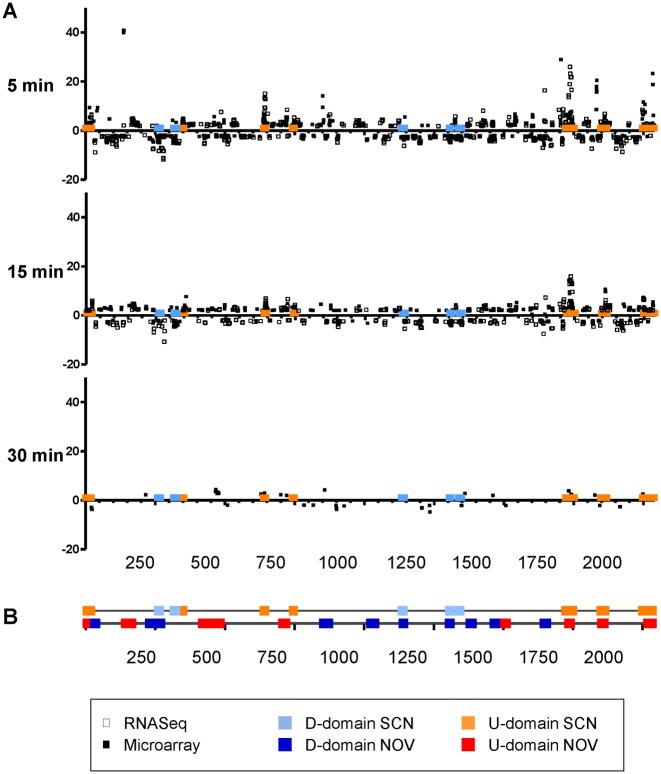
The transcriptomic response to 8 μM of SCN and the changes in supercoiling observed suggested that there was a homeostatic transcriptomic response that is mediated by the level of supercoiling. The localization of the DEGs suggested that the response depended on topologically sensitive gene clusters. A similar effect was observed during DNA relaxation induced by the inhibition of gyrase with NOV (23). In order to test this model, RNA sequencing (RNA-Seq) was performed. RNA-Seq quantifies the level of RNA transcription more accurately than microarrays (46). A common response was observed (Figure 6B, C), with 296 common DEGs between 5 min (520 DEGs) and 15 min (214 DEGs). The number of DEGs detected by RNA-Seq was 2-fold higher than the number detected by microarrays at 5 min and 1.5-fold higher at 15 min. More than half of the DEGs detected by microarrays were also detected by RNA-Seq at 5 min and almost half of them at 15 min (Figure 6D). Using statistical methods, DEGs detected either by microarrays or RNA-Seq at the 5 and 15 min time points of treatment with 8 μM SCN were grouped into topologically sensitive gene clusters. For purposes of comparison, a re-analysis was done of the DEGs detected by microarray at the time points of 5 min and 15 min after exposure to 0.5× MIC of NOV. Time points 5, 15 and 30 min after treatment with 40× MIC NOV were also re-analyzed. A two-step strategy was used to detect topological clusters: core detection and border refinement (see Materials and Methods). A total of 16 clusters were detected in the treatment with NOV. Seven of them were up-regulated while nine were down-regulated. A total of 12 clusters were revealed in the treatment with SCN. Seven of them were up-regulated while five were down-regulated. The characteristics of the topologically sensitive gene clusters are shown in Table 1. The clusters affected by treatment with NOV were larger than those affected by treatment with SCN. The former were dubbed NOV-clusters and the latter were dubbed SCN-clusters. NOV-clusters varied in size from 11.15 to 71.88 kb (average ± SD: 23.66 ± 15.82) and contained 8–44 responsive genes (average ± SD: 18.31 ± 10.92). SCN-clusters varied from 6.68 to 31.88 kb in size (average ± SD: 14.02 ± 7.58) and contained 9–22 responsive genes (average ± SD: 12.33 ± 3.80). More than 50% of the DEGs induced by NOV and more than 25% of those induced by SCN grouped in topological clusters. The location of NOV-clusters and SCN-clusters are shown in Figure 7.
Table 1. Characteristics of the topologically sensitive gene clusters.
| Size | |||||||
|---|---|---|---|---|---|---|---|
| Druga | Responseb | Cluster | Gene range (spr codes) | DEGsc | (No of genes) | (kb) | P value |
| NOV | U | 1 | 2022−0017 | 34 | 42 | 21.11 | 8.4E−10 |
| U | 2 | 0129−0161 | 30 | 33 | 31.78 | 4.1E−10 | |
| U | 3 | 0416−0484 | 44 | 69 | 71.88 | 1.2E−8 | |
| U | 4 | 0703−0721 | 12 | 19 | 19.12 | 6.9E−3 | |
| U | 5 | 1515−1536 | 17 | 22 | 24.69 | 4.8E−5 | |
| U | 6 | 1762−1775 | 11 | 14 | 13.61 | 1.8E−3 | |
| U | 7 | 1877−1888 | 9 | 12 | 16.51 | 1.2E−2 | |
| D | 8 | 0027−0042 | 12 | 16 | 12.62 | 1.0E−3 | |
| D | 9 | 0232−0277 | 33 | 45 | 48.11 | 4.7E−8 | |
| D | 10 | 0856−0884 | 21 | 29 | 23.54 | 1.2E−5 | |
| D | 11 | 1036−1048 | 8 | 13 | 23.91 | 4.1E−2 | |
| D | 12 | 1131−1145 | 12 | 15 | 11.15 | 7.0E−4 | |
| D | 13 | 1310−1325 | 14 | 16 | 11.77 | 4.9E−5 | |
| D | 14 | 1397−1408 | 9 | 12 | 15.81 | 1.2E−2 | |
| D | 15 | 1485−1498 | 12 | 14 | 15.99 | 4.0E−4 | |
| D | 16 | 1669−1687 | 15 | 19 | 16.99 | 2.0E−4 | |
| SCN | U | 1 | 2040−0019 | 18 | 26 | 21.65 | 2.0E−4 |
| U | 2 | 0342−0355 | 12 | 14 | 6.87 | 4.0E−4 | |
| U | 3 | 0628−0639 | 11 | 12 | 8.59 | 6.0E−4 | |
| U | 4 | 0744−0754 | 11 | 11 | 6.68 | 2.0E−4 | |
| U | 5 | 1750−1777 | 22 | 28 | 31.38 | 2.4E−6 | |
| U | 6 | 1876−1888 | 10 | 13 | 22.00 | 4.8E−3 | |
| U | 7 | 2002−2013 | 11 | 11 | 11.25 | 2.0E−4 | |
| D | 8 | 0254−0266 | 9 | 13 | 12.51 | 1.5E−2 | |
| D | 9 | 0314−0327 | 12 | 14 | 17.11 | 4.0E−4 | |
| D | 10 | 1129−1142 | 10 | 14 | 11.05 | 6.3E−3 | |
| D | 11 | 1314−1323 | 10 | 10 | 7.78 | 7.0E−4 | |
| D | 12 | 1353−1366 | 12 | 14 | 11.42 | 4.0E−4 | |
aNOV, novobiocin. SCN, seconeolitsine.
bU, genes were up-regulated. D, genes were down-regulated.
cDEGs included were those showing significant fold variations. These variations were detected either in microarrays (≥2 and P <0.05) or by RNA-Seq methods (≥1.8 and P <1E-14).
Figure 7.
Localization of DEGs in the S. pneumoniae chromosome. (A) Cultures of the R6 strain grown to an OD620nm of 0.4 were treated with 8 μM of SCN. Samples were taken before the addition of the drug (time 0 min), and at indicated times, total RNA was isolated and differential expression was analyzed either by microarrays or by the RNA-Seq method. The relative fold variation of each gene is represented alongside the 3′ location of each open reading frame in the S. pneumoniae R6 chromosome (ORFs 1 to 2043; bases 1 to 2 038 615). Boxes indicate the predicted topological clusters. (B) Localization of topological clusters detected as a result of treatment with either SCN or NOV.
DISCUSSION
Increase in DNA supercoiling induced by SCN, a specific inhibitor of Topo I (7), revealed a homeostatic response directed to the restoration of DNA supercoiling. A subinhibitory SCN concentration caused an increase in the σ at 5 and 15 min. At these times, an early transcriptomic response affecting ∼10% of the genome was observed. Meanwhile, recovery in supercoiling levels at 30 min was associated with the attenuation of the transcriptional response. With respect to the DNA topoisomerases, the only gene that was significantly affected was topA (Topo I), which was down-regulated in the early stage, while the level of supercoiling peaked.
In the late stage, the levels of supercoiling were partial recovered and the global transcriptomic response affected only 2% of the genome. Consequently, among the 41 genes that were responsive in the late phase, only eight were common to the early phase. This type of homeostatic response was also observed when DNA relaxation was induced with subinhibitory NOV concentrations. We applied the same statistical methods in order to identify the topological clusters of the early phase of both NOV and SCN. We found that >25% of DEGS induced by SCN and >50% induced by NOV could be grouped into topological clusters. The size of NOV-clusters is ±SD: 23.7 ± 15.8 and the average size of SCN-clusters is ±SD: 14.0± 7.6. These clusters are smaller than the 100-kb domains estimated in E. coli by means of chromosome relaxation in vivo (47,48). However, they are similar in size to the Salmonella enterica domains. These domains were estimated to be 25-kb by taking into account the site-specific recombination events that occurred between chromosomal sites that were distant to each other (49).The NOV- and SCN- clusters were also similar in size to the 10 kb E. coli domains predicted using transcriptional data (50). Noticeably, transcription of the genes coding for the primary metabolic machinery, those that tend to be highly expressed, are not affected by SCN (nor NOV). These highly expressed genes follow stochastic transcriptional bursts (51), given that they are necessary for physiological adaptation. We predict that our domains represent the cores of longer self-interacting domains similar to the ones observed in Caulobacter crescentus (52). Although the sizes of these self-interacting domains (30–400 kb) are larger than the ones observed in our study, relaxation induced by NOV reduces their size and sharpness. Although the NOV-clusters and SCN-clusters were not identical, their position in the chromosome essentially overlapped. This was unexpected, given the opposite nature of the topological challenges. These results support the idea that the chromosome is divided into topological domains that have a fixed location. Cell responds to an increase or a decrease in the level of supercoiling by modulating global transcription.
No evident DNA damage pathway was activated under SCN treatment. We found that only 3 out of 35 S. pneumoniae R6 genes encoding DNA repair proteins (Supplementary Table S2), dinF, hexA and recA were up-regulated in the presence of SCN. Moreover, these genes were also included within SCN domains U5 and U6, suggesting that their location in the genome has a tropism for regions where transcription is favoured under conditions of stress. S. pneumoniae lacks an SOS response. Competence, a transient period by which pneumococci concertedly uptake exogenous DNA and integrate it into the chromosome, has been suggested as a general stress response (53). Genes encoding relevant competence genes were up-regulated and located within SCN domain U1: comCDE (encoding the competence stimulating factor and the sensing machinery triggering the early response) and comX, activated by ComE, which triggers the late response. These genes were also up-regulated under DNA relaxation and are located in the NOV domain U1. Altogether, this indicates a very close relationship between topology, DNA stress and competence, which is likely to implicate a mixture of direct and indirect effects.
There are some differences between the responses during DNA relaxation and during an increase in supercoiling. Our previous studies showed that DNA relaxation led to the full recovery of the optimal supercoiling level (23). This recovery affected all the DNA topoisomerase genes: gyrA and gyrB were up-regulated and topA and parEC were down-regulated. When supercoiling is increasing, the changes of topA expression correlated with the level of supercoiling. However, at 15 min, gyrB was only a modestly up-regulated (2-fold) as was parE. These changes did not correlated with the level of supercoiling. The down-regulation of topA appears to be the only event necessary to allow recovery of supercoiling levels from an increased supercoiling of ∼73% to ∼43%. The regulation of topA depends on its location in a chromosomal D-domain, as defined both by NOV (23) or SCN treatments. This means that topA is down-regulated as a result of topological stress, originated either by a decrease or an increase in negative supercoiling. There was a good correlation between the amount of topA mRNA and the changes in σ values during the cellular homeostatic responses to relaxation or to increased supercoiling (R2 = 0.89, P = 0.0005). Although the roles of the other topoisomerases could not be excluded, this result indicates that the recovery of supercoiling levels in the cell mainly depends on the levels of topA expression. Inhibition of the opposing activities of gyrase and Topo I by treatment with subinhibitory concentrations of both NOV and SCN prevents the change in general supercoiling, as detected in the pLS1 plasmid. However, in this case, the transcription of both topA and parEC were also diminished. Although we do not have a clear explanation for this fact, local changes in supercoiling in chromosomal regions (that included these promoters) could not be ruled out. Likewise, studies on the regulation of transcription of topA of Mycobacterium showed that its optimal expression also requires the maintenance of an appropriate level of supercoiling in its promoter (54). The results of ChIP showed a preferential binding of Topo I to the gyrA promoter region. This region has an intrinsic DNA bend that is responsible for the supercoiling transcriptional response of gyrA (16). Our results support that Topo I plays an essential role in gyrA expression and that it is the main topoisomerase involved in the regulation of supercoiling in S. pneumoniae.
The recovery in supercoiling levels was incomplete in the case of SCN. This result suggests that S. pneumoniae is able to survive increases in supercoiling levels of up to ∼41%. Lower increases in supercoiling have been detected in nucleoids from an E. coli topA10 mutant strain carrying plasmid pBR322. The increase in chromosomal supercoiling in the mutant was 22±7% (average ± SD) and this corresponded to an increase of 14.2 ± 1.7% in pBR322 (25). The usage of antimicrobial drugs complements the utilization of mutants. SCN additions at short intervals close to the MIC enable a precise regulation of the effect. The survival of S. pneumoniae observed in response to increases in supercoiling levels (∼41%) is higher than the 25% observed for DNA relaxation (23). This indicates that S. pneumoniae, and very likely genetically-related bacteria, are more tolerant to hypernegative supercoiling than to hyper-relaxation. Likewise, the results from the experimental evolution in E. coli have revealed an increase in supercoiling that is associated with mutations in topA. This resulted in an increased bacterial fitness (55). A similar homeostatic mechanism associated to increased negative supercoiling might also be operative in bacteria that have a reverse gyrase enzyme. These bacteria keep DNA in a slightly overwound state to protect their genome against thermal melting (56).
In conclusion, we have detected topological domains responding similarly to alterations, both increase and decrease, in supercoiling. This suggests that there is a selective pressure for the location of key genes in the genome.
Supplementary Material
Acknowledgments
We thank Jorge B. Schvartzman (Centro de Investigaciones Biológicas, CSIC, Madrid, Spain) and Pedro A. Lazo-Zbikowski (Instituto de Biología Molecular y Celular del Cáncer, CSIC, Salamanca, Spain) for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Economía y Competitividad [BIO2014-55462-R to A.G.C.]. AJMG is the beholder of a Miguel Servet contract from the Spanish Ministry of Health. Funding for open access charge: Ministerio de Economía y Competitividad [BIO2014-55462-R to A.G.C.]. AJMG is the beholder of a Miguel Servet contract from the Spanish Ministry of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dorman C.J. Genome architecture and global gene regulation in bacteria: making progress towards a unified model? Nat. Rev. Microbiol. 2013;11:349–355. doi: 10.1038/nrmicro3007. [DOI] [PubMed] [Google Scholar]
- 2.Wang X., Montero Llopis P., Rudner D.Z. Organization and segregation of bacterial chromosomes. Nat. Rev. Genet. 2013;14:191–203. doi: 10.1038/nrg3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dillon S.C., Dorman C.J. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 4.Browning D.F., Busby S.J. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 5.Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 6.Cheng B., Liu I.F., Tse-Dinh Y.C. Compounds with antibacterial activity that enhance DNA cleavage by bacterial DNA topoisomerase I. J. Antimicrob. Chemother. 2007;59:640–645. doi: 10.1093/jac/dkl556. [DOI] [PubMed] [Google Scholar]
- 7.García M.T., Blázquez M.A., Ferrándiz M.J., Sanz M.J., Silva-Martín N., Hermoso J.A., de la Campa A.G. New alkaloid antibiotics that target the DNA topoisomerase I of Streptococcus pneumoniae. J. Biol. Chem. 2011;286:6402–6413. doi: 10.1074/jbc.M110.148148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tse-Dinh Y.-C. Regulation of the Escherichia coli DNA topoisomerase I gene by DNA supercoiling. Nucleic Acids Res. 1985;13:4751–4763. doi: 10.1093/nar/13.13.4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menzel R., Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983;34:105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- 10.Menzel R., Gellert M. Modulation of transcription by DNA supercoiling: a deletion analysis of the Escherichia coli gyrA and gyrB promoters. Proc. Natl. Acad. Sci. U.S.A. 1987;84:4185–4189. doi: 10.1073/pnas.84.12.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menzel R., Gellert M. Fusions of the Escherichia coli gyrA and gyrB control regions to the galactokinase gene are inducible by coumermycin treatment. J. Bacteriol. 1987;169:1272–1278. doi: 10.1128/jb.169.3.1272-1278.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiara A.S., Cundliffe E. Interplay of novobiocinresistant and –sensitive DNA gyrase activities in selfprotection of the novobiocin producer Streptomyces sphaeroides. Gene. 1989;81:65–72. doi: 10.1016/0378-1119(89)90337-5. [DOI] [PubMed] [Google Scholar]
- 13.Unniraman S., Chatterji M., Nagaraja V. DNA gyrase genes in Mycobacterium tuberculosis: a single operon driven by multiple promoters. J. Bacteriol. 2002;184:5449–5456. doi: 10.1128/JB.184.19.5449-5456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vora T., Hottes A.K., Tavazoie S. Protein occupancy landscape of a bacterial genome. Mol. Cell. 2009;35:247–253. doi: 10.1016/j.molcel.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Travers A., Muskhelishvili G. DNA supercoiling - a global transcriptional regulator for enterobacterial growth? Nat. Rev. Microbiol. 2005;3:157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- 16.Ferrándiz M.J., Arnanz C., Martín-Galiano A.J., Rodríguez-Martín C., de la Campa A.G. Role of global and local topology in the regulation of gene expression in Streptococcus pneumoniae. PloS One. 2014;9:e101574. doi: 10.1371/journal.pone.0101574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorman C.J. DNA supercoiling and environmental regulation of gene expression in pathogenic bacteria. Infect. Immun. 1991;59:745–749. doi: 10.1128/iai.59.3.745-749.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorman C.J., Corcoran C.P. Bacterial DNA topology and infectious disease. Nucleic Acids Res. 2009;37:672–678. doi: 10.1093/nar/gkn996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouafa Z.A., Reverchon S., Lautier T., Muskhelishvili G., Nasser W. The nucleoid-associated proteins H-NS and FIS modulate the DNA supercoiling response of the pel genes, the major virulence factors in the plant pathogen bacterium Dickeya dadantii. Nucleic Acids Res. 2012;40:4306–4319. doi: 10.1093/nar/gks014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peter B.J., Arsuaga J., Breier A.M., Khodursky A.B., Brown P.O., Cozzarelli N.R. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004;5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeong K.S., Xie Y., Hiasa H., Khodursky A.B. Analysis of pleiotropic transcriptional profiles: a case study of DNA gyrase inhibition. PLoS Genet. 2006;2:e152. doi: 10.1371/journal.pgen.0020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gmüender H., Kuratli K., Di Padova K., Gray C.P., Keck W., Evers S. Gene expression changes triggered by exposure of Haemophilus influenza to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 2001;11:28–42. doi: 10.1101/gr.157701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrándiz M.J., Martín-Galiano A.J., Schvartzman J.B., de la Campa A.G. The genome of Streptococcus pneumoniae is organized in topology-reacting gene clusters. Nucleic Acids Res. 2010;38:3570–3581. doi: 10.1093/nar/gkq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DiNardo S., Voelkel K.A., Sternglanz R., Reynolds A.E., Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982;31:43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- 25.Pruss G.J., Manes S.H., Drlica K. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell. 1982;31:35–42. doi: 10.1016/0092-8674(82)90402-0. [DOI] [PubMed] [Google Scholar]
- 26.Steck T.R., Franco R.J., Wang J.Y., Drlica K. Topoisomerase mutations affect the relative abundance of many Escherichia coli proteins. Mol. Microbiol. 1993;10:473–481. doi: 10.1111/j.1365-2958.1993.tb00920.x. [DOI] [PubMed] [Google Scholar]
- 27.Blattner F.R., Plunkett G., 3rd, Bloch C.A., Perna N.T., Burland V., Riley M., Glasner J.D., Rode C.K., Mayhew G.F., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 28.Cheng B., Zhu C.X., Ji C., Ahumada A., Tse-Dinh Y.C. Direct interaction between Escherichia coli RNA polymerase and the zinc ribbon domains of DNA topoisomerase I. J. Biol. Chem. 2003;278:30705–30710. doi: 10.1074/jbc.M303403200. [DOI] [PubMed] [Google Scholar]
- 29.Drolet M. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol. Microbiol. 2006;59:723–730. doi: 10.1111/j.1365-2958.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- 30.Drolet M., Phoenix P., Menzel R., Masse E., Liu L.F., Crouch R.J. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc. Natl. Acad. Sci. U.S.A. 1995;92:3526–3530. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drolet M., Bi X., Liu L.F. Hypernegative supercoiling of the DNA template during transcription elongation in vitro. J. Biol. Chem. 1994;269:2068–2074. [PubMed] [Google Scholar]
- 32.Phoenix P., Raymond M.A., Masse E., Drolet M. Roles of DNA topoisomerases in the regulation of R-loop formation in vitro. J. Biol. Chem. 1997;272:1473–1479. doi: 10.1074/jbc.272.3.1473. [DOI] [PubMed] [Google Scholar]
- 33.Masse E., Drolet M. Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J. Biol. Chem. 1999;274:16659–16664. doi: 10.1074/jbc.274.23.16659. [DOI] [PubMed] [Google Scholar]
- 34.Lacks S.A., López P., Greenberg B., Espinosa M. Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pLS1. J. Mol. Biol. 1986;192:753–765. doi: 10.1016/0022-2836(86)90026-4. [DOI] [PubMed] [Google Scholar]
- 35.Martín-Parras L., Lucas I., Martínez-Robles M.L., Hernández P., Krimer D.B., Hyrien O., Schvartzman J.B. Topological complexity of different populations of pBR322 as visualized by two-dimensional agarose gel electrophoresis. Nucleic Acids Res. 1998;26:3424–3432. doi: 10.1093/nar/26.14.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balsalobre L., de la Campa A.G. Fitness of Streptococcus pneumoniae fluoroquinolone-resistant strains with topoisomerase IV recombinant genes. Antimicrob. Agents Chemother. 2008;52:822–830. doi: 10.1128/AAC.00731-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernández-Moreira E., Balas D., González I., de la Campa A.G. Fluoroquinolones inhibit preferentially Streptococcus pneumoniae DNA topoisomerase IV than DNA gyrase native proteins. Microb. Drug Resist. 2000;6:259–267. doi: 10.1089/mdr.2000.6.259. [DOI] [PubMed] [Google Scholar]
- 38.Attaiech L., Minnen A., Kjos M., Gruber S., Veening J.W. The ParB-ParS chromosome segregation system modulates competence development in Streptococcus pneumoniae. mBio. 2015;6:e00662. doi: 10.1128/mBio.00662-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Applic. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 41.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price M.N., Huang K.H., Alm E.J., Arkin A.P. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 2005;33:880–892. doi: 10.1093/nar/gki232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de la Campa A.G., del Solar G.H., Espinosa M. Initiation of replication of plasmid pLS1. The initiator protein RepB acts on two distant DNA regions. J. Mol. Biol. 1990;213:247–262. doi: 10.1016/S0022-2836(05)80188-3. [DOI] [PubMed] [Google Scholar]
- 45.Olavarrieta L., Hernández P., Krimer D.B., Schvartzman J.B. DNA knotting caused by head-on collision of transcription and replication. J. Mol. Biol. 2002;322:1–6. doi: 10.1016/s0022-2836(02)00740-4. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Worcel A., Burgi E. On the structure of the folded chromosome of Escherichia coli. J. Mol. Biol. 1972;71:127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]
- 48.Sinden R.R., Pettijohn D.E. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc. Natl. Acad. Sci. U.S.A. 1981;78:224–228. doi: 10.1073/pnas.78.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higgins N.P., Yang X., Fu Q., Roth J.R. Surveying a supercoil domain by using the gamma delta resolution system in Salmonella typhimurium. J. Bacteriol. 1996;178:2825–2835. doi: 10.1128/jb.178.10.2825-2835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Postow L., Hardy C.D., Arsuaga J., Cozzarelli N.R. Topological domain structure of the Escherichia coli chromosome. Genes Deve. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chong S., Chen C., Ge H., Xie X.S. Mechanism of transcriptional bursting in bacteria. Cell. 2014;158:314–326. doi: 10.1016/j.cell.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Le T.B., Imakaev M.V., Mirny L.A., Laub M.T. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013;342:731–734. doi: 10.1126/science.1242059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prudhomme M., Attaiech L., Sanchez G., Martin B., Claverys J.P. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science. 2006;313:89–92. doi: 10.1126/science.1127912. [DOI] [PubMed] [Google Scholar]
- 54.Ahmed W., Menon S., PV D.N.B.K., Nagaraja V. Autoregulation of topoisomerase I expression by supercoiling sensitive transcription. Nucleic Acids Res. 2016;44:1541–1552. doi: 10.1093/nar/gkv1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crozat E., Philippe N., Lenski R.E., Geiselmann J., Schneider D. Long-term experimental evolution in Escherichia coli. XII. DNA topology as a key target of selection. Genetics. 2005;169:523–532. doi: 10.1534/genetics.104.035717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogawa T., Yogo K., Furuike S., Sutoh K., Kikuchi A., Kinosita K., Jr Direct observation of DNA overwinding by reverse gyrase. Proc. Natl. Acad. Sci. U.S.A. 2015;112:7495–7500. doi: 10.1073/pnas.1422203112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.