Abstract
A new technology, genetic alphabet expansion using artificial bases (unnatural bases), has created high-affinity DNA ligands (aptamers) that specifically bind to target proteins by ExSELEX (genetic alphabet Expansion for Systematic Evolution of Ligands by EXponential enrichment). We recently found that the unnatural-base DNA aptamers can be stabilized against nucleases, by introducing an extraordinarily stable, unique hairpin DNA (mini-hairpin DNA) and by reinforcing the stem region with G–C pairs. Here, to establish this aptamer generation method, we examined the stabilization of a high-affinity anti-VEGF165 unnatural-base DNA aptamer. The stabilized aptamers displayed significantly increased thermal and nuclease stabilities, and furthermore, exhibited higher affinity to the target. As compared to the well-known anti-VEGF165 RNA aptamer, pegaptanib (Macugen), our aptamers did not require calcium ions for binding to VEGF165. Biological experiments using cultured cells revealed that our stabilized aptamers efficiently inhibited the interaction between VEGF165 and its receptor, with the same or slightly higher efficiency than that of the pegaptanib RNA aptamer. The development of cost-effective and calcium ion-independent high-affinity anti-VEGF165 DNA aptamers encourages further progress in diagnostic and therapeutic applications. In addition, the stabilization process provided additional information about the key elements required for aptamer binding to VEGF165.
INTRODUCTION
DNA and RNA aptamers that specifically bind to target molecules are expected to become an alternative to protein-based antibodies for pharmaceutical applications (1–9). They are initially generated by an evolutionary engineering method in a test tube (Systematic Evolution of Ligands by EXponential enrichment (SELEX)) (10,11), and then chemically synthesized for subsequent large-scale preparation. DNA aptamers are considered to be more advantageous in terms of cost, as compared to RNA aptamers and antibodies. However, some issues with DNA aptamers still remain, such as their relatively low affinity to targets and poor stability against nuclease digestion.
Although several post-SELEX modification methods to stabilize aptamers have been reported (12–17), there are fewer opportunities for modifying DNA aptamers to confer increased resistance against nucleases without a loss of target affinity and an increase in cost. The most established method is the modification of the 2′-position of the ribose moieties in aptamers with fluoro and methoxy groups (18–20). Since these 2′-modified nucleotides can be introduced into RNA by transcription (21–23), 2′-modified RNA aptamers can be directly generated by SELEX (18,24,25). However, applying these 2′-modifications to DNA aptamers is often restricted, because of the different sugar conformation of the 2′-deoxyribose moieties in DNA from those of the 2′-modified ribose moieties, as well as the bulkiness of 2′-mothxy modifications (26). In addition, the present post-stabilization methods are laborious, because many aptamer candidates with numerous combinations of modification types and positions have to be screened extensively. At present, only an anti-VEGF165 RNA aptamer, pegaptanib (Macugen), modified with 2′-fluoro and methoxy groups, has been approved for the treatment of neovascular age-related macular degeneration (19,27,28). Although several improvements have been reported (29–31), no DNA aptamers have been approved as drugs yet.
Genetic alphabet expansion using unnatural base pairs (32,33) provides a new SELEX method (genetic alphabet Expansion for Systematic Evolution of Ligands by EXponential enrichment (ExSELEX)) for generating nucleic acid aptamers containing unnatural bases (34–37). We created an unnatural base pair between hydrophobic Ds and Px bases that functions as a third base pair in replication (38–40), and applied the Ds–Px pair to SELEX using Ds-containing DNA libraries, by which we generated high-affinity Ds-containing DNA aptamers (34). The presence of only a few Ds bases in the generated unnatural-base DNA aptamers imparts a substantial improvement in their affinities to target proteins.
In addition to ExSELEX, we recently found that a Ds-containing DNA aptamer can be stabilized, by introducing an extraordinarily stable mini-hairpin DNA sequence and by placing reinforcing G–C pairs in the stem regions of their secondary structures (41). DNA fragments with GCGNAGC, CCGNAGG, GCGNNAGC and CCGNNAGG sequences (N = A, G, C or T) form compact hairpin-like structures (named mini-hairpins) containing two G–C and GNNA or GNA loops, with a sheared G–A pair (42–45). The melting temperatures of the GCGAAGC and GCGAAAGC fragments are as high as 76°C in 0.1 M NaCl, and these fragments are also quite resistant to nuclease digestion. We demonstrated the stabilization of an anti-interferon-γ DNA aptamer containing Ds bases by introducing the mini-hairpin sequences (41). Thus, our present objective is to explore the versatility of this stabilization method and to establish a new aptamer generation method toward pharmaceutical applications, by applying it to other aptamers.
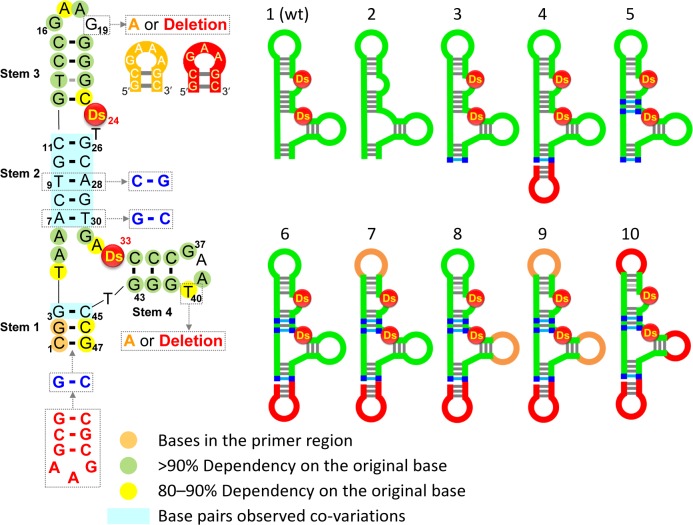
Here, we report the stabilization of a Ds-containing DNA aptamer that was previously generated by ExSELEX targeting VEGF165 (34). The sequence and the secondary structure of the original aptamer (Aptamer 1) are shown in Figure 1. The secondary structure was presumed by the sequence analysis obtained from a doped re-selection (34). The initial aptamer, Aptamer 1 (47-mer), contains two Ds bases, four stems (Stems 1–4) and GAAG and GAAT loops, and exhibits a low picomolar KD value (KD = ∼1 pM). The replacement of the Ds bases with A substantially reduced the binding ability of the DNA aptamer (Aptamer 2) (KD = 347 pM). Our stabilization method significantly improved the thermal stability and nuclease resistance of the Ds-containing aptamers, without loss of affinity. We also confirmed the biological activity of the improved aptamers, by comparison with that of pegaptanib.
Figure 1.
Secondary structures of the original anti-VEGF165 unnatural-base DNA aptamer (1) and its variants (2–10). The sequence of Aptamer 1 is shown on the left, and the base positions with more than 90 or 80–90% base-dependency, calculated by using the data obtained from the previous doped re-selection (Supplementary Figure S1), are indicated with green or yellow colored circles, respectively. Each variant is schematically represented on the right.
MATERIALS AND METHODS
Oligonucleotides
DNA fragments were purchased from Gene Design or chemically synthesized with an Oligonucleotide Synthesizer nS-8 (Gene Design), using CE phosphoramidites of the natural, modified and Ds bases (Glen Research). The chemically synthesized DNA fragments were purified by gel electrophoresis.
Competition assays
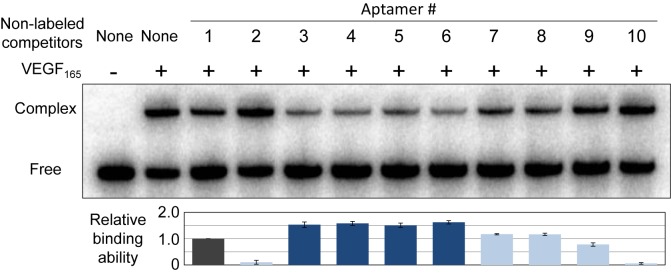
To compare the binding abilities of the aptamer variants to human VEGF165, competitive binding assays were performed by native-gel electrophoresis. The 32P-labeled Aptamer 1 (final concentration: 100 nM) was mixed with an equal amount of each non-labeled aptamer variant (100 nM), and then mixed with human VEGF165 (Peprotech, 100 nM) in 1× PBS (1 mM KH2PO4, 3 mM Na2HPO4 and 155 mM NaCl, pH 7.4, Gibco) supplemented with 0.005% (vol/vol) Nonidet P-40. After an incubation (20 μl solution) at 37°C for 30 min, 5 μl of 25% glycerol was added and the reactions were immediately subjected to 10% non-denaturing polyacrylamide gel electrophoresis, to isolate and detect the complex of the labeled Aptamer 1 with VEGF165. The gel was dried and visualized with a Bio-imaging analyzer, BAS2500 (Fuji Film). The band intensities were quantified using the Multi Gauge software, and the gel shift rate was determined. By normalizing the competitive inhibition rate of each non-labeled aptamer variant in comparison with non-labeled Aptamer 1, the relative binding abilities were calculated. The experiments were repeated three times, and the averaged data are plotted on the graph in Figure 2.
Figure 2.
Binding analysis of each anti-VEGF165 aptamer variant by a competition assay. 32P-labeled Aptamer 1 (100 nM) was incubated with VEGF165 (100 nM), in the presence of each non-labeled variant as a competitor (100 nM), at 37°C for 30 min. The aptamer-VEGF165 complexes were separated from the free aptamers on a native 10% polyacrylamide gel, and the relative binding abilities were calculated from their band intensities, normalized by that of Aptamer 1.
SPR measurements
The KD values of Aptamer 1, Aptamer 4 and Pegaptanib 34 were determined with a BIAcore T200 (GE Healthcare) at 25 or 37°C, in running buffer (20 mM Tris-HCl, 150 mM NaCl, 1 mM MgCl2, 0.05% (vol/vol) Nonidet P-40, pH 7.7) in the presence or absence of 1 mM CaCl2. Each biotinylated aptamer was immobilized on a Sensor chip SA (GE Healthcare), and the interaction of the aptamer with human VEGF165 was detected by monitoring injections of 0.156 to 5 nM VEGF165 (diluted with running buffer) in the Kinetic Injection mode. Measurement conditions were 100 μl/min flow rate, 150 s for protein injection time and 450 s for dissociation monitoring time. After each injection, the sensor surface was regenerated with a 5 μl injection of 50 mM NaOH/1 M NaCl, and the aptamer refolding was subsequently accomplished by equilibration with the running buffer for 10 min. To determine the KD values, sensorgrams of both a reference cell (no aptamer immobilization) and a measurement with the running buffer injection were subtracted from each sensorgram of the aptamers. The data were fitted with a two-state reaction binding model (Pegaptanib 34, without calcium ions) or a 1 : 1 binding model, using the BIA evaluation T200 software, version 1.0 (GE Healthcare).
Inhibitory experiments
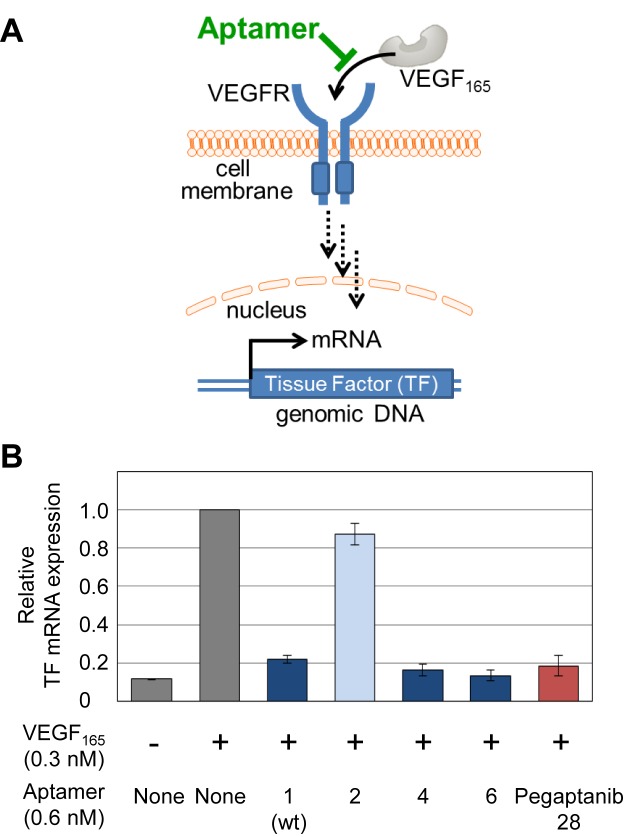
Each aptamer's inhibitory effect on VEGF165 was evaluated by a VEGF-induced tissue factor (TF) expression assay, using human umbilical vein endothelial cells (HUVECs, LONZA). HUVECs were cultured in Endothelial Cell Growth Medium (LONZA) in 24-well plates at 37°C, under a 5% CO2 atmosphere. The cells were washed with OPTI-MEM (Life Technologies) twice, and pre-incubated in OPTI-MEM (0.5 ml/well) for 2 h. The medium was then aspirated, and 0.5 ml of OPTI-MEM containing 0.3 nM VEGF165 and 0.6 nM of each aptamer was freshly added to each well. After a 1 h incubation, total RNA was extracted by using an RNeasy Mini Kit (QIAGEN). The cDNAs were prepared by reverse transcription, using an Oligo dT primer (15-mer) and M-MLV Reverse Transcriptase RNase H Minus, Point Mutant (Promega), and were used as the templates for real time PCR analysis of TF mRNA expression, using a KAPA SYBR fast qPCR kit (Kapa Bio Systems). The expression of beta actin was used as the reference, and the TF mRNA expression level was normalized to the beta actin expression. In each plate, the experiment under the same conditions was duplicated, and the relative TF mRNA expression was calculated by comparison with that in the presence of VEGF165 and without any aptamers. The final data were obtained as the average of three data sets, and are shown in Figure 6.
Figure 6.
Inhibition of the interaction between VEGF165 and its receptor by anti-VEGF165 aptamers. (A) Schematic illustration of the inhibition of the VEGF165-induced cellular signaling pathway. (B) Tissue Factor (TF) mRNA expression inhibition by the aptamers. Relative TF mRNA expression levels in HUVECs treated with VEGF165 in the presence of each aptamer are shown.
RESULTS
To apply the stabilization method to the anti-VEGF165 Ds-containing DNA aptamer (Aptamer 1), we estimated the modifiable base positions and regions from the base-dependency of each base in the predicted secondary structure of Aptamer 1 for the target binding. The base-dependency (%) was determined from the base conservation data previously obtained by re-selection of the initial anti-VEGF165 DNA aptamer, using a partially randomized (doped) library containing 55% of the initial base and 15% of each of the other three natural bases at each position, except for the unnatural base positions (Supplementary Figure S1) (34). The secondary structure was also estimated from the data of the co-variation among the natural standard base pairs by the doped re-selection (Figure 1).
Based on the secondary structure, including the base-dependency and co-variation data of Aptamer 1, we designed several modified variants through our stabilization process (Figure 1 and Supplementary Table S1). Aptamer 3 has an extra G–C pair in Stem 1, and Aptamer 4 has the mini-hairpin sequence at the 3′-terminus of Aptamer 3. The 3′-mini-hairpin DNA protects the aptamer from 3′-exonucleases, and the base stacking between the 3′-terminal G in the mini-hairpin and the 5′-G1 of the aptamer also protects it from 5′-exonucleases. In Aptamer 5, the A–T pairs in Stem 2 of Aptamer 3 were replaced with G–C pairs. Aptamer 6 has the mini-hairpin DNA at the 3′-terminus of Aptamer 5. The two loops, 16GAAG19 and 37GAAT40, in Aptamer 1 are both very similar to the mini-hairpin loop sequences, and the base-dependencies of the fourth bases, G19 and T40 (40.7 and 80.2%, respectively), in the loops are relatively low as compared to those (90%) of most bases in the single-stranded regions. Thus, to introduce the mini-hairpin sequence into Aptamer 6, G19 and T40 were each or both replaced with A to make the mini-hairpin GAAA loop (Aptamers 7, 8 and 9) or deleted to make the mini-hairpin GAA loop (Aptamer 10).
We first surveyed the binding abilities of these aptamer variants to VEGF165, by a gel-shift competition assay (Figure 2). The 32P-labeled Aptamer 1 was mixed with the equivalent amount of each variant, and the labeled Aptamer 1 that competitively bound to VEGF165 was quantified on a native polyacrylamide gel. Surprisingly, as compared to Aptamer 1, the binding abilities of Aptamers 3–6 were significantly improved. In contrast, the GAAA-loop mini-hairpin replacement of either one of the internal stem-loop structures (Aptamers 7 and 8) did not clearly improve their affinities, but these aptamers still exhibited high affinities similar to that of Aptamer 1. However, the full replacement with the GAAA- (Aptamer 9) or the GAA-loop mini-hairpin (Aptamer 10) severely reduced the binding ability. In the mini-hairpin structures, the conformations of the other bases, especially the highly base-dependent G16, A18, G37 and A39 bases, in the loops might be changed by the sheared G–A pair formation. The adverse effect of the GAAA-loop mini-hairpin introduced into one of the internal hairpins in Aptamers 7 and 8 might be compensated by the stabilization effect of Stems 1 and 2. Thus, introducing the 3′-mini-hairpin structure and increasing the G–C pairs in Stems 1 and 2 were most effective in increasing both the stability and affinity.
The KD values of Aptamers 1 and 4 were measured by surface plasmon resonance (SPR) (Figure 3). We also measured the modified 34-mer RNA aptamer derivative of pegaptanib (Pegaptanib 34) (Supplementary Figure S2). Although Pegaptanib 34 tightly binds to VEGF165 in the presence of calcium ions, removing the divalent ions significantly reduced the binding ability. In contrast, Aptamers 1 and 4 tightly bound to VEGF165 without requiring calcium ions. In the presence of 1 mM CaCl2 at 37°C, the binding ability of Aptamer 4 (KD = 1.2 pM) was superior to those of Pegaptanib 34 (KD = 11 pM) and Aptamer 1 (KD = 7.5 pM).
Figure 3.
Binding analysis of anti-VEGF165 aptamers by a BIAcore T200 at 25°C or 37°C, in the presence and absence of calcium ions in the running buffer (20 mM Tris-HCl, 150 mM NaCl, 1 mM MgCl2, 0.05% Nonidet P40, pH 7.7), against VEGF165 (0.156 to 5 nM).
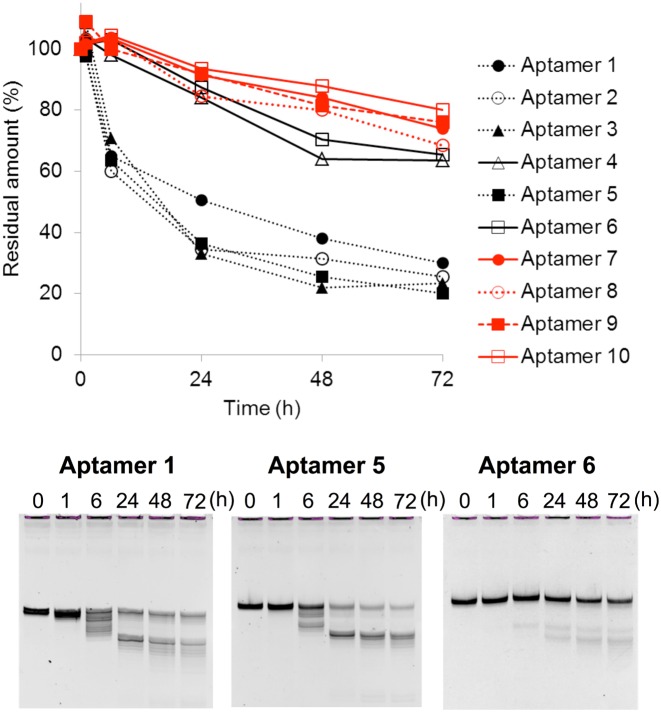
Next, we examined the stability of the variants against nuclease digestion, using human serum at 37°C (Figure 4 and Supplementary Figure S3). Attaching the 3′-mini-hairpin structure to block the exonuclease activities in the serum greatly enhanced the nuclease resistance. More than 65% of the aptamers (Aptamers 4 and 6–10) survived after 72 h in serum. By introducing the mini-hairpin DNA at the 3′-terminus, the amounts of the specific shorter bands on the gel were significantly reduced, and thus these shorter bands are likely to result from exonuclease digestion of the aptamers.
Figure 4.
Nuclease resistance of anti-VEGF165 aptamers in 96% human serum at 37°C for up to 72 h. Aptamers indicated with red lines contain the internal mini-hairpin DNA.
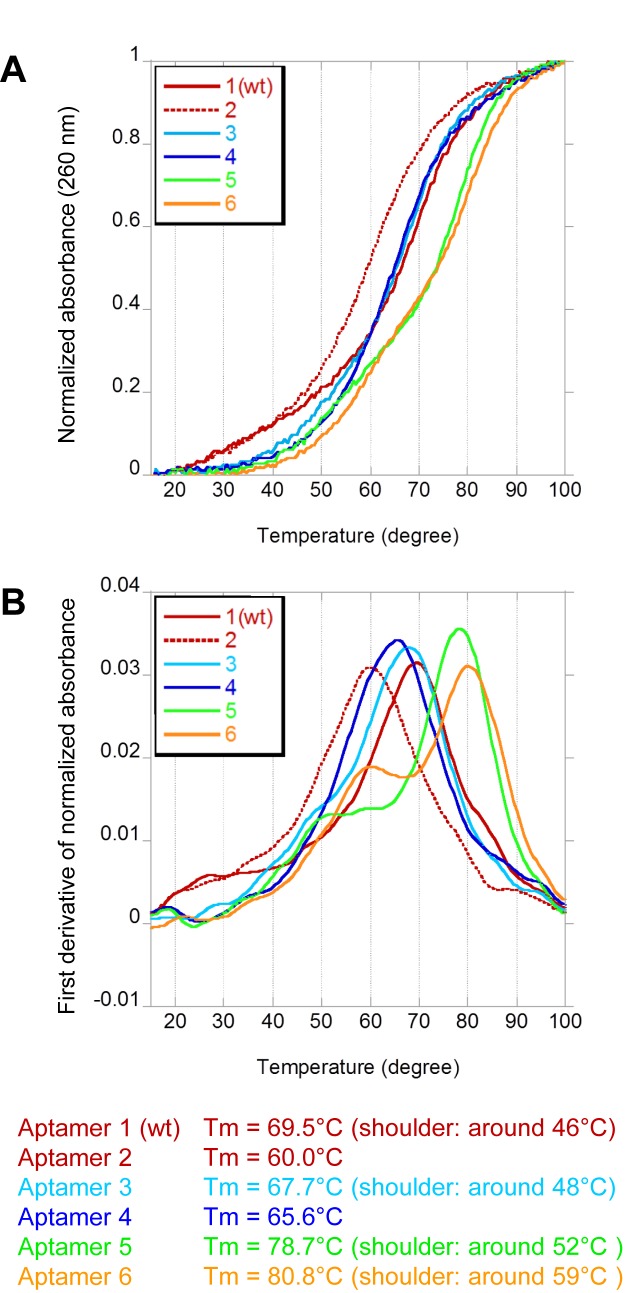
We also measured the thermal stabilities of Aptamers 1–6 (Figure 5). Even though the original aptamer (Aptamer 1) exhibited high stability (Tm = 69.5°C), the stabilization further increased the Tm value of Aptamer 6 (Tm = 80.8°C) by ∼10°C. We also observed another peak or shoulders at lower temperatures on the first derivatives of the Tm profiles. These shoulders might result from the denaturation of the Stem 1 and/or 2 regions, and the 3′-mini-hairpin structure might stabilize Stem 1 by the stacking interaction. More importantly, the Tm curve indicated that Aptamer 6 maintained its structure at around 37°C.
Figure 5.
Thermal stabilities of DNA aptamer variants. The UV melting profiles of aptamer variants were monitored, using a SHIMADZU UV-2450 spectrometer equipped with a temperature controller (TSMPC-8). The absorbance of each sample (2 μM in 1× PBS) was monitored at 260 nm from 15°C to 100°C, at a heating rate of 0.5°C/min. Each melting temperature was calculated by the first derivative of the melting curve, using the IGOR Pro software (WaveMetrics, Inc.). The melting profiles, (A) normalized by the absorbance at 15°C and 100°C, and (B) the first derivatives of the normalized absorbance are shown.
Finally, the biological activities of Aptamers 1, 2, 4 and 6 were compared to that of a 28-mer pegaptanib derivative with the conventional 3′-3′ inverted deoxythymidine (Pegaptanib 28) to protect the 3′-terminus (Supplementary Figure S2). The expression of the VEGF-induced (TF) mRNA was assayed using human umbilical vein endothelial cells (HUVECs), in the absence or presence of each aptamer and VEGF165 (Figure 6). Aptamers 4 and 6 efficiently inhibited the interaction between VEGF165 and its receptor, and their activities were the same or slightly higher than that of Pegaptanib 28. The activity of Aptamer 1 was also high, but the Ds-to-A variant (Aptamer 2) exhibited much lower activity. Since the degradation profiles of Aptamers 1 and 2 were very similar (Supplementary Figure S3), the difference in these activities was attributed to their binding abilities to VEGF165.
DISCUSSION
The strengthened aptamer (Aptamer 6), with greatly increased stability, was generated by adding the mini-hairpin DNA fragment at the 3′-terminus and reinforcing Stems 1 and 2 by the G–C pair introduction. We found that the stabilization method also enhanced the binding ability of the aptamer to the target. Although the sequences of the two loop structures of GAAG and GAAT in Aptamer 1 are quite similar to that of the GAA or GAAA loop of the mini-hairpin DNA, our data revealed that these two loops are not compatible with the mini-hairpin loop for the binding. Without the replacements of these loop regions with the mini-hairpin DNA sequences, Aptamer 6 was sufficiently stabilized in the serum. In the cell culture assay, Aptamer 6 efficiently inhibited the VEGF activity as well as or better than the pegaptanib derivative. Since the stabilized aptamer is composed of natural base nucleotides with no chemical modifications, except for only two Ds bases, it is advantageous in terms of cost and safety.
This simple stabilization method consists of three steps: (i) addition of the mini-hairpin DNA to the 3′-terminus, (ii) replacement of the A–T pairs in the terminal and internal stem regions with the G–C pairs, and (iii) mini-hairpin DNA replacement of the internal stem-loop structures, if each base in the loop has the low base-dependency. Thus, this stabilization method requires information about the replaceable bases and the secondary structures of Ds-containing DNA aptamers, predicted by using the data obtained from doped re-selection (34,46–48).
Our results also provided valuable information about the binding sites of the aptamer to the target. The two loop regions might directly interact with the target protein. The doped selection data revealed the low base-dependency of G19 and T40 in Aptamer 1 (34). However, the double mutation of G19A and T40A (Aptamer 9) reduced the binding ability. This is due to the conformational changes of the highly conserved G16, A18, G37 and A39 base moieties by the GAAA-loop mini-hairpin formation in Aptamer 9. In the mini-hairpin structures, these bases cannot interact with the target protein, since they participate in the sheared base pairs of G16–A19 and G37–A40 and in the stacking formation of A18•A19 and A39•A40. In fact, in the doped re-selection data, the two GNNA loop sequences of the double mutant of G19A and T40A appeared only 18 times among 43 719 sequences (34). In addition, the variant with two GAA loops (Aptamer 10), in which all of the bases tightly stack upon each other in the loops, lost a significant amount of the binding ability. Thus, the flexibility of the G16, A18, G37 and A39 base moieties is important for the interaction with VEGF165. However, in the competition experiments, the affinities of Aptamers 7 and 8 were still as high as that of Aptamer 1, and the nuclease resistances of these aptamers were very high, as compared to that of Aptamer 6. Thus, Aptamers 7 and 8 could also be used under conditions with high nuclease concentrations.
Stems 1–4 of the aptamer might be structurally important as a scaffold, to form the entire tertiary structure for effective binding. In particular, the stabilization of Stem 1, by adding the G–C pair and the mini-hairpin DNA, increased the binding ability and stability. The replacement of the A–T pairs in Stem 2 with G–C pairs increased the nuclease resistance, without any loss of the binding ability. Furthermore, this stabilization method also improved the aptamer affinity to the target, and thus the rigidity of the tertiary structure of the Ds-containing DNA aptamer might be important for target binding.
We still do not know the exact roles of the two Ds bases. One possibility is that the hydrophobic Ds bases directly interact with VEGF165, and another is that they contribute to form a unique conformation of the tertiary structure by stacking with neighboring bases. Either way, the structural similarity between the two moieties, Stem 3 + GAAG loop + Ds24 and Ds33 + Stem 4 + GAAT loop, suggests that each moiety may bind to each monomer of the dimerized VEGF165.
This stabilization method could be extensively applied to Ds-containing DNA aptamers. So far, we have generated nine different Ds-containing DNA aptamers targeting proteins and cancer cells by ExSELEX (data not shown). All of them have a stem structure between the 5′- and 3′-terminal regions. We confirmed that the A–T pairs in the terminal stem structure of each aptamer can be replaced with the G–C pairs, and attaching the mini-hairpin DNA to the 3′-terminal can retain the high affinity of the aptamers to their targets. Since Ds-containing DNA aptamers seem to form relatively rigid tertiary structures, a stabilization method that increases the rigidity might be very effective for tight binding to targets. Other types of conventional nucleic acid aptamers with defined structures could also be stabilized by this method.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology [KAKENHI 26248043 to I.H.];grants for projects focused on developing key technologies for discovering and manufacturing drugs for next-generation treatment and diagnosis from the Ministry of Economy, Trade and Industry [to I.H.]; Japan Science and Technology Agency (JST) Precursory Research for Embryonic Science and Technology (PRESTO) [to M.K.]. Funding for open access charge: Institute of Bioengineering and Nanotechnology (IBN).
Conflict of interest statement. M.K. and I.H. own stock in TagCyx Biotechnologies.
REFERENCES
- 1.Zhou J., Rossi J.J. The therapeutic potential of cell-internalizing aptamers. Curr. Top. Med. Chem. 2009;9:1144–1157. doi: 10.2174/156802609789630893. [DOI] [PubMed] [Google Scholar]
- 2.Keefe A.D., Pai S., Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor A.I., Arangundy-Franklin S., Holliger P. Towards applications of synthetic genetic polymers in diagnosis and therapy. Curr. Opin. Chem. Biol. 2014;22:79–84. doi: 10.1016/j.cbpa.2014.09.022. [DOI] [PubMed] [Google Scholar]
- 4.Wu X., Chen J., Wu M., Zhao J.X. Aptamers: active targeting ligands for cancer diagnosis and therapy. Theranostics. 2015;5:322–344. doi: 10.7150/thno.10257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma H., Liu J., Ali M.M., Mahmood M.A., Labanieh L., Lu M., Iqbal S.M., Zhang Q., Zhao W., Wan Y. Nucleic acid aptamers in cancer research, diagnosis and therapy. Chem. Soc. Rev. 2015;44:1240–1256. doi: 10.1039/c4cs00357h. [DOI] [PubMed] [Google Scholar]
- 6.Bruno J.G. Predicting the uncertain future of aptamer-based diagnostics and therapeutics. Molecules. 2015;20:6866–6887. doi: 10.3390/molecules20046866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen A., Yang S. Replacing antibodies with aptamers in lateral flow immunoassay. Biosens. Bioelectron. 2015;71:230–242. doi: 10.1016/j.bios.2015.04.041. [DOI] [PubMed] [Google Scholar]
- 8.Toh S.Y., Citartan M., Gopinath S.C., Tang T.H. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens. Bioelectron. 2015;64:392–403. doi: 10.1016/j.bios.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 9.Lao Y.H., Phua K.K., Leong K.W. Aptamer nanomedicine for cancer therapeutics: barriers and potential for translation. ACS Nano. 2015;9:2235–2254. doi: 10.1021/nn507494p. [DOI] [PubMed] [Google Scholar]
- 10.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 11.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 12.Lapa S.A., Chudinov A.V., Timofeev E.N. The toolbox for modified aptamers. Mol. Biotechnol. 2016;58:79–92. doi: 10.1007/s12033-015-9907-9. [DOI] [PubMed] [Google Scholar]
- 13.Meek K.N., Rangel A.E., Heemstra J.M. Enhancing aptamer function and stability via in vitro selection using modified nucleic acids. Methods. 2016 Mar 21; doi: 10.1016/j.ymeth.2016.03.008. doi:10.1016/j.ymeth.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Gao S., Zheng X., Jiao B., Wang L. Post-SELEX optimization of aptamers. Anal Bioanal Chem. 2016;408:4567–4573. doi: 10.1007/s00216-016-9556-2. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y., Ren X., Schluesener H.J., Zhang Z. Aptamers: selection, modification and application to nervous system diseases. Curr. Med. Chem. 2011;18:4159–4168. doi: 10.2174/092986711797189646. [DOI] [PubMed] [Google Scholar]
- 16.Keefe A.D., Cload S.T. SELEX with modified nucleotides. Curr. Opin. Chem. Biol. 2008;12:448–456. doi: 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 17.Wang R.E., Wu H., Niu Y., Cai J. Improving the stability of aptamers by chemical modification. Curr. Med. Chem. 2011;18:4126–4138. doi: 10.2174/092986711797189565. [DOI] [PubMed] [Google Scholar]
- 18.Pagratis N.C., Bell C., Chang Y.F., Jennings S., Fitzwater T., Jellinek D., Dang C. Potent 2′-amino-, and 2′-fluoro-2′-deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. Nat. Biotechnol. 1997;15:68–73. doi: 10.1038/nbt0197-68. [DOI] [PubMed] [Google Scholar]
- 19.Ruckman J., Green L.S., Beeson J., Waugh S., Gillette W.L., Henninger D.D., Claesson-Welsh L., Janjic N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998;273:20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 20.Waters E.K., Genga R.M., Thomson H.A., Kurz J.C., Schaub R.G., Scheiflinger F., McGinness K.E. Aptamer BAX 499 mediates inhibition of tissue factor pathway inhibitor via interaction with multiple domains of the protein. J. Thromb. Haemost. 2013;11:1137–1145. doi: 10.1111/jth.12201. [DOI] [PubMed] [Google Scholar]
- 21.Chelliserrykattil J., Ellington A.D. Evolution of a T7 RNA polymerase variant that transcribes 2′-O-methyl RNA. Nat. Biotechnol. 2004;22:1155–1160. doi: 10.1038/nbt1001. [DOI] [PubMed] [Google Scholar]
- 22.Meyer A.J., Garry D.J., Hall B., Byrom M.M., McDonald H.G., Yang X., Yin Y.W., Ellington A.D. Transcription yield of fully 2′-modified RNA can be increased by the addition of thermostabilizing mutations to T7 RNA polymerase mutants. Nucleic Acids Res. 2015;43:7480–7188. doi: 10.1093/nar/gkv734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ibach J., Dietrich L., Koopmans K.R., Nobel N., Skoupi M., Brakmann S. Identification of a T7 RNA polymerase variant that permits the enzymatic synthesis of fully 2′-O-methyl-modified RNA. J. Biotechnol. 2013;167:287–295. doi: 10.1016/j.jbiotec.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Burmeister P.E., Lewis S.D., Silva R.F., Preiss J.R., Horwitz L.R., Pendergrast P.S., McCauley T.G., Kurz J.C., Epstein D.M., Wilson C., et al. Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem. Biol. 2005;12:25–33. doi: 10.1016/j.chembiol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 25.Waters E.K., Genga R.M., Schwartz M.C., Nelson J.A., Schaub R.G., Olson K.A., Kurz J.C., McGinness K.E. Aptamer ARC19499 mediates a procoagulant hemostatic effect by inhibiting tissue factor pathway inhibitor. Blood. 2011;117:5514–5522. doi: 10.1182/blood-2010-10-311936. [DOI] [PubMed] [Google Scholar]
- 26.Diener J.L., Daniel Lagasse H.A., Duerschmied D., Merhi Y., Tanguay J.F., Hutabarat R., Gilbert J., Wagner D.D., Schaub R. Inhibition of von Willebrand factor-mediated platelet activation and thrombosis by the anti-von Willebrand factor A1-domain aptamer ARC1779. J. Thromb. Haemost. 2009;7:1155–1162. doi: 10.1111/j.1538-7836.2009.03459.x. [DOI] [PubMed] [Google Scholar]
- 27.Ng E.W.M., Shima D.T., Calias P., Cunningham E.T., Guyer D.R., Adamis A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 28.Drolet D.W., Green L.S., Gold L., Janjic N. Fit for the eye: Aptamers in ocular disorders. Nucleic Acid Ther. 2016;26:127–146. doi: 10.1089/nat.2015.0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He W., Elizondo-Riojas M.A., Li X., Lokesh G.L., Somasunderam A., Thiviyanathan V., Volk D.E., Durland R.H., Englehardt J., Cavasotto C.N., et al. X-aptamers: a bead-based selection method for random incorporation of druglike moieties onto next-generation aptamers for enhanced binding. Biochemistry. 2012;51:8321–8323. doi: 10.1021/bi300471d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gold L., Ayers D., Bertino J., Bock C., Bock A., Brody E.N., Carter J., Dalby A.B., Eaton B.E., Fitzwater T., et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yatime L., Maasch C., Hoehlig K., Klussmann S., Andersen G.R., Vater A. Structural basis for the targeting of complement anaphylatoxin C5a using a mixed L-RNA/L-DNA aptamer. Nat. Commun. 2015;6:6481. doi: 10.1038/ncomms7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malyshev D.A., Romesberg F.E. The expanded genetic alphabet. Angew. Chem. Int. Ed. Engl. 2015;54:11930–11944. doi: 10.1002/anie.201502890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirao I., Kimoto M. Unnatural base pair systems toward the expansion of the genetic alphabet in the central dogma. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2012;88:345–367. doi: 10.2183/pjab.88.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimoto M., Yamashige R., Matsunaga K., Yokoyama S., Hirao I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol. 2013;31:453–457. doi: 10.1038/nbt.2556. [DOI] [PubMed] [Google Scholar]
- 35.Sefah K., Yang Z., Bradley K.M., Hoshika S., Jimenez E., Zhang L., Zhu G., Shanker S., Yu F., Turek D., et al. In vitro selection with artificial expanded genetic information systems. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1449–1454. doi: 10.1073/pnas.1311778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L., Yang Z., Sefah K., Bradley K.M., Hoshika S., Kim M.J., Kim H.J., Zhu G., Jimenez E., Cansiz S., et al. Evolution of functional six-nucleotide DNA. J. Am. Chem. Soc. 2015;137:6734–6737. doi: 10.1021/jacs.5b02251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimoto M., Matsunaga K., Hirao I. DNA aptamer generation by genetic alphabet expansion SELEX (ExSELEX) using an unnatural base pair system. Methods Mol. Biol. 2016;1380:47–60. doi: 10.1007/978-1-4939-3197-2_4. [DOI] [PubMed] [Google Scholar]
- 38.Kimoto M., Kawai R., Mitsui T., Yokoyama S., Hirao I. An unnatural base pair system for efficient PCR amplification and functionalization of DNA molecules. Nucleic Acids Res. 2009;37:e14. doi: 10.1093/nar/gkn956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamashige R., Kimoto M., Takezawa Y., Sato A., Mitsui T., Yokoyama S., Hirao I. Highly specific unnatural base pair systems as a third base pair for PCR amplification. Nucleic Acids Res. 2012;40:2793–2806. doi: 10.1093/nar/gkr1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okamoto I., Miyatake Y., Kimoto M., Hirao I. High fidelity, efficiency and functionalization of Ds-Px unnatural base pairs in PCR amplification for a genetic alphabet expansion system. ACS Synth. Biol. 2016 Feb 05; doi: 10.1021/acssynbio.5b00253. doi:10.1021/acssynbio.5b00253. [DOI] [PubMed] [Google Scholar]
- 41.Matsunaga K., Kimoto M., Hanson C., Sanford M., Young H.A., Hirao I. Architecture of high-affinity unnatural-base DNA aptamers toward pharmaceutical applications. Sci. Rep. 2015;5:18478. doi: 10.1038/srep18478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirao I., Nishimura Y., Naraoka T., Watanabe K., Arata Y., Miura K. Extraordinary stable structure of short single-stranded DNA fragments containing a specific base sequence: d(GCGAAAGC) Nucleic Acids Res. 1989;17:2223–2231. doi: 10.1093/nar/17.6.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirao I., Kawai G., Yoshizawa S., Nishimura Y., Ishido Y., Watanabe K., Miura K. Most compact hairpin-turn structure exerted by a short DNA fragment, d(GCGAAGC) in solution: an extraordinarily stable structure resistant to nucleases and heat. Nucleic Acids Res. 1994;22:576–582. doi: 10.1093/nar/22.4.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshizawa S., Ueda T., Ishido Y., Miura K., Watanabe K., Hirao I. Nuclease resistance of an extraordinarily thermostable mini-hairpin DNA fragment, d(GCGAAGC) and its application to in vitro protein synthesis. Nucleic Acids Res. 1994;22:2217–2221. doi: 10.1093/nar/22.12.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshizawa S., Kawai G., Watanabe K., Miura K., Hirao I. GNA trinucleotide loop sequences producing extraordinarily stable DNA minihairpins. Biochemistry. 1997;36:4761–4767. doi: 10.1021/bi961738p. [DOI] [PubMed] [Google Scholar]
- 46.Hirao I., Harada Y., Nojima T., Osawa Y., Masaki H., Yokoyama S. In vitro selection of RNA aptamers that bind to colicin E3 and structurally resemble the decoding site of 16S ribosomal RNA. Biochemistry. 2004;43:3214–3221. doi: 10.1021/bi0356146. [DOI] [PubMed] [Google Scholar]
- 47.Knight R., Yarus M. Analyzing partially randomized nucleic acid pools: straight dope on doping. Nucleic Acids Res. 2003;31:e30. doi: 10.1093/nar/gng030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duclair S., Gautam A., Ellington A., Prasad V.R. High-affinity RNA aptamers against the HIV-1 protease inhibit both in vitro protease activity and late events of viral replication. Mol. Ther. Nucleic Acids. 2015;4:e228. doi: 10.1038/mtna.2015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.