Abstract
MicroRNAs (miRs) have emerged as key biological effectors in human health and disease. These small noncoding RNAs are incorporated into Argonaute (Ago) proteins, where they direct post-transcriptional gene silencing via base-pairing with target transcripts. Although miRs have become intriguing biological entities and attractive therapeutic targets, the translational impacts of miR research remain limited by a paucity of empirical miR targeting data, particularly in human primary tissues. Here, to improve our understanding of the diverse roles miRs play in cardiovascular function and disease, we applied high-throughput methods to globally profile miR:target interactions in human heart tissues. We deciphered Ago2:RNA interactions using crosslinking immunoprecipitation coupled with high-throughput sequencing (HITS-CLIP) to generate the first transcriptome-wide map of miR targeting events in human myocardium, detecting 4000 cardiac Ago2 binding sites across >2200 target transcripts. Our initial exploration of this interactome revealed an abundance of miR target sites in gene coding regions, including several sites pointing to new miR-29 functions in regulating cardiomyocyte calcium, growth and metabolism. Also, we uncovered several clinically-relevant interactions involving common genetic variants that alter miR targeting events in cardiomyopathy-associated genes. Overall, these data provide a critical resource for bolstering translational miR research in heart, and likely beyond.
INTRODUCTION
MicroRNAs (miRs; ∼19–22 nucleotides) broadly regulate cellular gene expression at the post-transcriptional level, playing roles in nearly all biological processes. The human genome encodes ∼2000 miRs, many of which are highly conserved across species (1). miRs are excised from stem–loop transcripts and subsequently incorporated into Ago proteins, producing sequence-guided effector complexes capable of base-pairing with and repressing target transcripts via translational inhibition and mRNA destabilization (2). Canonically, miRs bind to target mRNA 3′-untranslated regions (3′-UTRs) containing short sequence elements complementary to their ‘seed’ region (5′-miR nucleotides 2–8). As with transcription factors, this minimal degree of sequence recognition allows miRs to regulate an array of transcripts, offering powerful means to coordinate cellular responses by targeting multiple genes among complex biological pathways. However, with this comes a significant challenge in deciphering meaningful miR:target interactions among the plethora of possible base-pairing events across vast transcriptomes.
Currently, researchers rely primarily on computational prediction of miR binding sites as a first step towards identifying biologically-relevant targets. Although bioinformatic approaches have proven valuable, they remain limited by their inherent tendency to overpredict sites, inability to accurately model target site accessibility (RNA structure or competitive protein binding), and failure to reliably predict non-canonical (e.g. partial seed or 3′-guided) binding events. These limitations, among others, support the need for generating complementary wet-lab data to facilitate the discovery of bona-fide interactions. For this, studies profiling gene expression after miR overexpression or inhibition have pointed to functional relationships between miRs and mRNA transcripts, but this fails to discern direct and indirect interactions. To address this, recent work has focused on using high-throughput methods [e.g. HITS-CLIP, PAR-CLIP, iCLIP and RISC-seq (3–8)] to biochemically isolate miR:target interactions on a transcriptome-wide scale. For example, HITS-CLIP, the RNA equivalent of CHIP-seq, can be used to capture Ago:RNA complexes, allowing researchers to characterize the Ago-bound miRs and the mRNA fragments to which they are engaged. This allows for fine-resolution mapping of miR:target interactions across the transcriptome in virtually any given cell or tissue.
In mice, HITS-CLIP has been applied to query miR targeting in several tissues (3,9); however, human data have predominantly derived from cell culture systems (3,6), and there remains a significant need to produce high-throughput miR targeting data spanning the breadth of human tissues. Although primary human tissues are not well-suited for methods that necessitate treatment with photo-activated nucleotide analogs or miR mimics/inhibitors (e.g. PAR-CLIP and RISC-seq), their miR targeting profiles can be assessed using HITS-CLIP and iCLIP procedures. Recently, our group employed Ago2 HITS-CLIP in human brain samples, generating the first transcriptome-wide miR binding map in a primary human tissue (5). We have since expanded our efforts to the heart, where there is a broad interest in the biological and therapeutic relevance of cardiac miRs. Indeed, many studies have demonstrated that miRs are critical regulators of cardiogenesis, contractile function and pathological and protective responses to cardiovascular disease (10–12). However, progress in the field has slowed due to the lack of a comprehensive, ‘omics’-level view of miR targeting events in cardiac tissues. To begin globally defining the regulatory targets of important heart-related miRs, we applied HITS-CLIP methodology to decipher the first transcriptome-wide map of miR binding sites in human myocardial tissue. We then highlight example utilities of the resulting data by evaluating underappreciated miR binding sties in coding regions and by exploring the intriguing interface among human genetic variations, miR biology, and cardiovascular disease.
MATERIALS AND METHODS
Samples
Left ventricular cardiac tissues were obtained from the Iowa Biobank for Cardiovascular Research (Supplementary Table S1). Advanced cardiomyopathy subjects donating tissue were consented under an approved IRB protocol. At the time of left ventricular assist device implantation or heart transplantation, tissues were procured, flash frozen and stored at −80°C until use.
Ago2 HITS-CLIP and data processing
HITS-CLIP was performed as previously described, with some modifications (5). In brief, fresh-frozen human heart tissues were pulverized and subjected to UV-irradiation to crosslink protein:nucleic acid interactions. Ago2-associated RNAs were immunoprecipitated and subjected to adapter ligation and RT-PCR to generate libraries for high-throughput sequencing, done on an Illumina Hi-Seq. Sequencing reads were pre-processed and mapped as described previously (5), with only minor changes. Adapter sequences were removed, and trimmed reads were mapped to the human genome (hg19). Identical alignments were collapsed in each sample to remove PCR replicates. Strand-specific read coverage was then calculated using the alignments from each of the six samples.
miR expression levels
Adapter-trimmed Ago2 reads of 15–27 nts were mapped to miR precursors (as annotated by miRBase (1)), and all valid alignments with up to one mismatch were reported. Mature miR expression levels were calculated as the sum of read counts having the same 5′ start position, normalized to the total number of miR-mapped reads in each sample. Seed family abundance was calculated as the total expression from all mature miR family members sharing the same seed sequence (5′-nts 2–8), based on TargetScan annotations (13).
Identification of statistically significant Ago2 clusters
We used zero-truncated negative binomial model (ZTNB) to calculate the significance of read coverage at each mapped genomic position (5). We assume that read heights for each gene are sampled from an underlying ZTNB distribution. For each sample, parameters for gene-specific ZTNB probability density functions were estimated using read heights measured at all positions within an annotated gene region. P-values were calculated as the probability of observing a read height as large as the height in question, and assigned to each position. Fisher's method was applied to summarize p-values at each genomic position across the six samples. Positions with an FDR <5% were deemed significant. Significant positions within 60 nts of one another were merged into a single contiguous interval, and resulting regions of <50 nts were symmetrically extended to 50 nts; this was done to account for the Ago binding footprint (3). Intragenic cluster positions were then annotated according to their overlapping gene structures (Ensembl annotations).
Correlation of Ago2-associated RNAs across samples
Correlation coefficients (Pearson r) were calculated for read heights in non-miR loci using the wigCorrelate application (14). Pairwise comparisons were made between the six samples, using a minimum read height of 10. For miRs, pairwise correlation coefficients were calculated using normalized expression levels for canonical mature miRs detected in any of the six samples.
PhastCons score
Sequence conservation in human cardiac Ago2 clusters was evaluated using the Conservation Plot tool (Cistrome Analysis Pipeline Module (15)).
Motif analyses
HOMER software was used to discover motifs enriched in Ago2 HITS-CLIP cluster sequences (16). Additionally, motif enrichment was performed using a sliding window approach, as done previously (5). Briefly, a single base offset sliding window of 7 nts was used to determine heptamer frequencies in cluster and background sequences. Enrichment data were calculated for all possible heptamers. Enrichment data for known miR seed families was calculated by combining enrichment of 7M8 and 7A1 sequences. Enrichment significance was calculated by Fisher's exact test, and p-values were transformed to Benjamini-Hochberg FDRs.
Functional validation of miR:target gene pairs
Microarray data for miR overexpression and inhibition studies were retrieved from the Gene Expression Omnibus database. For miR overexpression studies, the cumulative fractions of Ago2 HITS-CLIP target genes (with 3′UTR- or CDS-located clusters having the relevant miR seed sites) and background genes (i.e. no site) were generated for each microarray dataset for miR overexpression studies. Data were then grouped by discrete intervals (0.01 log2 fold-change increments) and summarized across all ten experiments by averaging the cumulative fraction values for targets and background sets at each interval. The miR inhibition study involved the pooled depletion of 25 highly expressed miRs in HEK293 cells (6). Among these, twelve corresponded to abundant cardiac miRs present in our data. Genes harboring 3′-UTR-located Ago2 HITS-CLIP clusters having sequences matching these miR seeds were used for the analysis, as were experimentally validated target genes for the same miRs (annotated by miRTarBase (17)).
Luciferase-based miR targeting assays were performed as previously described (5). Regions spanning Ago2 HITS-CLIP sites for the indicated target genes were PCR amplified and cloned into a dual luciferase plasmid (psiCheck2; Promega). Site-directed mutagenesis was used to introduce the relevant SNP alleles. HEK293 cells in 24-well plates were co-transfected in triplicate with 20 ng of luciferase reporter plasmids along with 25 nmol/l of synthetic pre-miRs (Ambion) or 20 ng of miR expression plasmids. Firefly and Renilla luciferase activities were measured 48 h later using the Dual-Glo Luciferase Assay System (Promega), and Renilla/Firefly activity ratios were normalized to pre-miR-Neg1 control for each 3′-UTR tested.
For testing CDS-targeted miR-29 binding sites, HT1080 or N2a cells cultured in 24-well plates were co-transfected in triplicate with 25 nmol/l of synthetic pre-miRs (Ambion) and with 100–300 ng of plasmids expressing the full-length coding regions of various target genes. After 48 h, protein lysates were harvested and evaluated by western blot to determine target gene expression levels by densitometry. Plasmids are described in the Supplementary Material (Detailed Methods).
RESULTS
Ago2 HITS-CLIP identifies miRs and their transcriptomic targets in human cardiac tissues
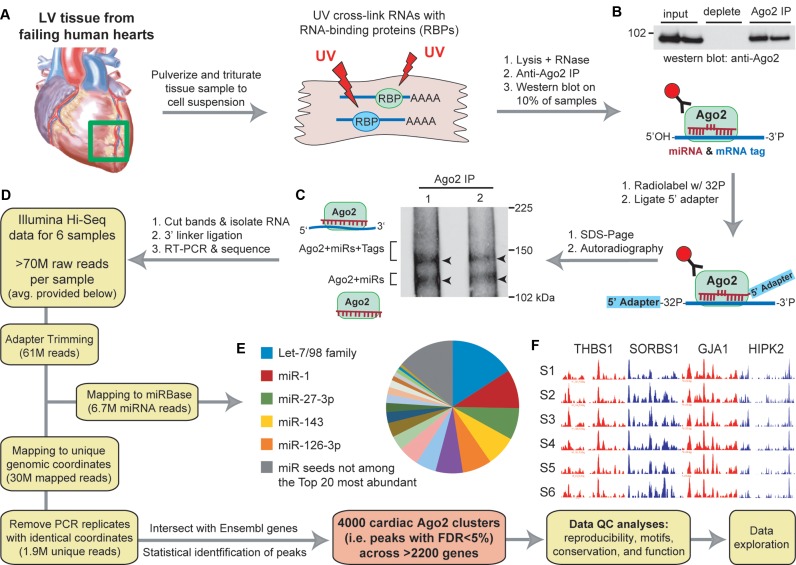
We adapted and applied Ago2 HITS-CLIP to six human left ventricular cardiac tissue samples obtained from cardiomyopathy patients (25–61 year-old males; Figure 1A and Supplementary Table S1). Biochemical assays revealed highly efficient and specific immunoprecipitation of Ago2 associating with radiolabeled cellular RNAs (Figure 1B and C). Evaluation of purified Ago2:RNA complexes by gel electrophoresis followed by autoradiography reproduced the expected doublet (based on prior observations (3,5)), signifying Ago2 bound to miRs (lower band) and Ago2:miR complexes engaged with target RNAs (upper band). Cardiac Ago2-associated RNA libraries were generated by adapter ligation and RT-PCR; subsequent high-throughput sequencing yielded ∼70 million reads for each sample, of which ∼7 million reads mapped to known human miRs (precursors annotated by miRBase (1)) and ∼30 million mapped to unique positions within the human genome (Figure 1D and Supplementary Table S2). After removing identical reads to eliminate PCR bias, each sample yielded ∼2 million uniquely-mapped unique reads, offering substantial coverage for downstream computational analyses.
Figure 1.
Workflow for performing Ago2 HITS-CLIP in human heart (A–F). (A) Illustration depicting the processing of left ventricular cardiac tissues (n = 6 separate individuals). (B) Western blot showing immunoprecipitation of human Ago2 from heart tissues. (C) Autoradiography of Ago2 CLIP isolated 32P-labeled RNA:protein complexes resolved by SDS-PAGE showing the expected doublet corresponding to the designated complexes. (D) Diagram indicating the workflow for analyzing high-throughput sequencing data to obtain miR read information and cardiac Ago2 clusters (i.e. binding sites). The average number of reads per sample at each filtering step is provided. (E) Pie chart showing the relative abundance of miRs (grouped into families based on shared seed sequence) across all samples. (F) UCSC Genome Browser images of positional read coverage for each sample (S1–S6) illustrate the cross-sample data reproducibility. Images span the 3′-UTRs of the indicated genes, covering ∼7400 nts in total. Red and blue indicate positive and negative strand orientation, respectively. Figure scheme adapted from previously published work (5).
Reads mapping to annotated mature miRs were highly reproducible across samples (median R2 > 0.9; Supplementary Figure S1A), especially among the most highly expressed miRs (Supplementary Figure S1B). Altogether, we detected ∼800 known, annotated miRs (Supplementary Table S3), with several of the most abundant being established cardiac-enriched miRs. These highly expressed miRs appear to span the various cell populations in the heart; for example, miR-1, miR-126 and miR-29 are abundant in cardiomyocytes, vasculature, and cardiac fibroblasts, respectively (11). Upon grouping miR read counts into their respective seed families, the top five most abundant Ago2-associated seed sequences accounted for nearly half of the total Ago2 ‘seed load’ across the samples (Figure 1E and Supplementary Figure S1B), and the top 20 seeds constituted almost 90%. These observations corroborate previous reports supporting that highly abundant miRs often comprise the vast majority of miRs in a given cell, and are thus the key drivers of miR-mediated gene regulation within (3,5,6,18).
Non-miR, uniquely-mapped unique reads were evaluated to identify transcriptomic miR binding sites among the samples. Pairwise correlations of the positional read counts (i.e. number of overlapping reads at each genomic position) for each sample supported high reproducibility of read coverage across individuals (median R2>0.95; Supplementary Figure S2A). Furthermore, the inter-sample data uniformity was visually evident across genes in coding and non-coding regions (Figures 1F and Supplementary Figure S2B). A statistically stringent peak-calling method was employed to identify Ago2 binding sites (i.e. peaks or clusters) within Ensembl-annotated genes, resulting in the identification of 4000 human cardiac Ago2 clusters (FDR < 5%) associating with >2200 genes (Supplementary Table S4). Remarkably, >95% of the clusters are ≤60-nts in length, meaning that most represent distinct non-overlapping miR binding events, while the others may harbor multiple sites not resolved as individual peaks.
Characterization of human cardiac Ago2 binding sites
The dependable use of HITS-CLIP and the resulting data requires several downstream measures of quality control to evaluate the reliability of the ascertained binding sites. For miR targeting, this includes checking the data, as a whole, for expected consistencies between miR levels and their cognate seed complements found within the Ago2 clusters. In addition, assessing overall cross-species conservation rates across the cluster regions can provide a surrogate measure of their biological significance. Beyond this, publically-available mRNA profiling datasets from miR manipulation studies (e.g. overexpression or inhibition) may be used in high-throughput fashion to detect functionally-responsive miR:target gene pairs among the data. Here, we assess these important QC measures prior to exploring the Ago2 HITS-CLIP interactome for biologically meaningful miR:target interactions.
Cluster sequences
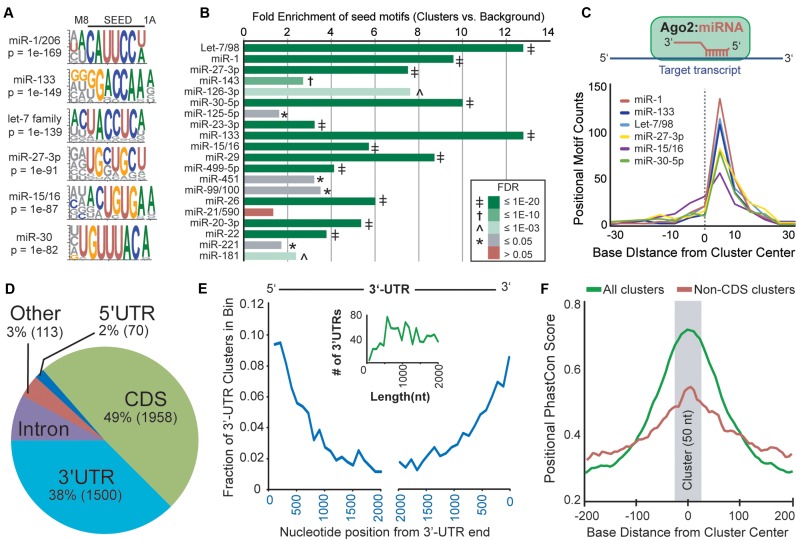
We analyzed cardiac Ago2 cluster sequence composition, compared to nearby flanking sequences, using unbiased motif enrichment analysis to determine the relationship between the human cardiac Ago2 clusters and miRs detected in the samples. Among the most significantly enriched motifs were sequences complementary to the most abundant miR seeds detected within the Ago2-miR-mRNA complexes (Figure 2A and Supplementary Table S5), including 7A1 (reverse complement of miR positions 2–7 followed by adenine base), 7M8 (reverse complement of miR positions 2–8) and some non-canonical seeds with G:U wobble base-pairs or bulged nucleotides. A heptamer-biased approach was also used to determine the specific enrichment of 7A1 and 7M8 sites for each known miR seed sequence, and sites corresponding to 19 of the top 20 most abundant miR seeds (based on miR reads) were significantly enriched (FDR < 5%; Figure 2B and Supplementary Table S6). It is important to note that the majority of the clusters primarily correspond to these most highly expressed miRs, as cluster seed counts were clearly diminished for miRs beyond the top 20 (Supplementary Figure S3A). That said, a strong correlation between miR abundance and significance of seed site enrichment in cluster sequences was observed for the top 50 most highly expressed miR seeds (Supplementary Figure S3B). Overall, the miR and motif enrichment data validate one another, meaning that the significant enrichments of target seed sites corroborates the observed abundance of seeds among the miR reads, and vice versa. The integrity of the data is further supported by the clear positional bias of the enriched seed motifs to be slightly right of center within the clusters (Figure 2C); this is the anticipated position of these sites when engaged by reverse complementary miR sequences within Ago2. Together, these findings mimic our previous data from brain (5), supporting the reproducible adaptation of our HITS-CLIP protocol across human tissues.
Figure 2.
Characterization of human cardiac Ago2 clusters. (A) Analysis of cluster sequences using the HOMER algorithm identified significantly enriched motifs corresponding to abundant cardiac miRs (sequence logos shown). (B) Relative combined enrichments of 7A1 and 7M8 sites corresponding to the top 20 most abundant miR seeds is shown. The significance of enrichment [Ago2 clusters vs. background (flanking sequences); FDR] is indicated by color key and symbol. (C) Plot summarizing the positional enrichment of motifs within clusters. The enriched motifs corresponding to abundant cardiac miRs are located to the right of center, consistent with miR:target recognition via reverse complementary Watson–Crick base-pairing, illustrated above. (D) The intragenic distribution of the 4000 human cardiac Ago2 clusters within mRNAs based on Ensembl gene annotations. CDS = coding regions. (E) The positional distribution of clusters within binned 3′-UTR locations is shown. The inset graph (green line) depicts the number of 3′-UTRs by length for genes containing 3′-UTR Ago2 clusters, supporting that the observed decrease in clusters distant from 3′-UTR ends is not length dependent. (F) The positional mean PhastCons scores within and near cardiac Ago2 clusters (∼50 nts, centered on zero) are shown for all clusters or non-CDS clusters only. For the latter, clusters overlapping coding regions were removed to avoid a potential bias for coding sequences showing increased conservation, particularly smaller exons flanked by less conserved intronic sequences.
We next annotated the clusters for the presence of crosslinking-induced mutations sites (CIMS (19)) and miR seeds to facilitate identifying miR:target gene pairs among the data. CIMS analysis identified precise protein-RNA crosslink sites in roughly 50% of the clusters (Supplementary Table S7). Computational miR seed site prediction using TargetScan (13) and PITA (20) was also performed on cluster sequences, and the top hits (up to 10 total) corresponding to highly-expressed miRs (i.e. top 100 seeds) are provided for each cluster, ranked by the abundance of the targeting miR (Supplementary Table S3 and S8). Enriched non-canonical sites for the most highly-expressed miRs (e.g. miR-1 and miR-133 bulged seeds) are also included. Overall, miR seed sites were identified in ∼95% of the clusters. To assess the translational utility of this resource, we determined the cross-species conservation of the identified miR sites between humans and mice (Supplementary Table S8), finding that ∼47% of the target seed sequences are conserved.
Positional distribution of clusters
The intragenic distribution of cardiac Ago2 clusters was determined based on Ensembl gene annotations, revealing that 97% reside within mRNA transcripts (>2200 genes). Surprisingly, the bulk of the Ago2 binding sites (49%) were found to overlap coding regions (CDS; Figure 2D), with some larger genes harboring many sites (e.g. 106 CDS sites in TTN alone). Numerous cardiac Ago2 binding sites (38%) also localized within 3′-UTRs, their canonical targeting domain. As observed in previous Ago HITS-CLIP studies (3,5), human cardiac Ago2 binding sites were preferentially located at the 5′ and 3′ ends of 3′-UTRs (Figure 2E), a hallmark feature associated with gene silencing (13). Further evidence supporting site functionality is gleaned from an increase in cross-species conservation spanning cardiac Ago2 clusters, relative to flanking genomic sequences (Figure 2F). Beyond mature mRNA regions, we also noted the presence of Ago2 binding events within introns (e.g. MYZAP and TTN), as well as long non-coding RNAs (Supplementary Figure S2B), offering a unique resource to stimulate the investigation of the functional relevance of these site-types, which remain largely understudied.
To better understand the biological relevance of the distribution of cardiac Ago2 binding across genes, we performed gene ontology enrichment analysis using the >2200 target genes among our data (Supplementary Figure S4A and Table S9). Interestingly, these genes were significantly enriched for RNA-binding proteins (n = 285), supporting the existence of a complex interplay between miRs and other post-transcriptional gene regulators. Other noteworthy enrichments included genes related to muscle development (n = 170), structure and function (n = 110), as well as TCA cycle (n = 89) and adrenergic signaling (n = 54). Gene set enrichment analysis was also performed using the specific miR:target gene pairs (for each miR) to evaluate whether our dataset points to novel or known functions for certain miRs (Supplementary Figure S4B and Table S10). The results reaffirm the well-established role for miR-29 in controlling extracellular matrix proteins (21) and hint at added miR-29 functionality in coordinating energy production, along with miR-103/107 and miR-15/16. The resulting associations also highlight miR-24 and miR-22 as regulators of muscle structure and contraction, and point to an intriguing interface between miR-133 and calpain signaling, both known mediators of cardiac hypertrophy (22–24).
Functionality of human cardiac Ago2 binding sites in 3′-UTRs and coding regions
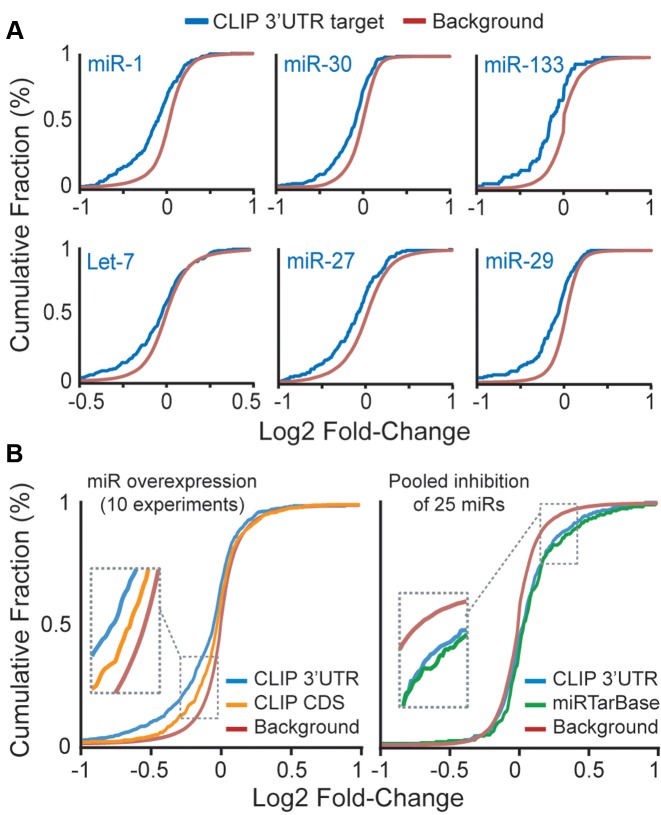
We next evaluated the functionality of cardiac miR:target gene pairs identified by Ago2 HITS-CLIP. First, we exploited publically-available microarray data from studies testing the effects of individual miR overexpression or pooled inhibition of abundant miRs in human cell cultures (6). We initially focused on 3′-UTR target sites, as recent data support that miR-mediated gene suppression via CDS occurs predominantly through translational repression (25). In each independent dataset, miR overexpression broadly down-regulated many of the respective targets identified by Ago2 HITS-CLIP (Figure 3A). Overall, 3′-UTR sites showed greater responsiveness compared to CDS-targeted sites (Figure 3B, left). By contrast, in miR inhibition studies, targets identified by Ago2 HITS-CLIP were generally up-regulated (Figure 3B, right), with the magnitude being equivalent to that observed for miR:target interactions previously validated in wet-lab experiments (e.g. RT-PCR and western blot), as catalogued by miRTarBase (17). These data reiterate our previously described observations in brain (5), lending further support to the cross-tissue reproducibility of this method.
Figure 3.
Functionality of miR:target gene pairs identified by cardiac Ago2 HITS-CLIP. Functionality of miR:target gene pairs was determined using gene expression microarray data from miR overexpression or miR inhibition studies in human cell lines. Cumulative fraction curves for miR:target gene pairs (e.g. genes having 3′-UTR or CDS Ago2 HITS-CLIP clusters or experimentally-validated miR:target genes recorded in miRTarBase; each corresponding to the overexpressed or inhibited miRs in the microarray experiments) are plotted. Curve shifts to the left or right, relative to background genes (i.e. no site), indicates a general down- or up-regulation of the target genes, respectively. (A) Plots showing the down-regulation of target genes in several individual miR overexpression datasets. (B) Plots summarizing data from ten miR overexpression studies (left) and a single dataset for pooled miR inhibition (right).
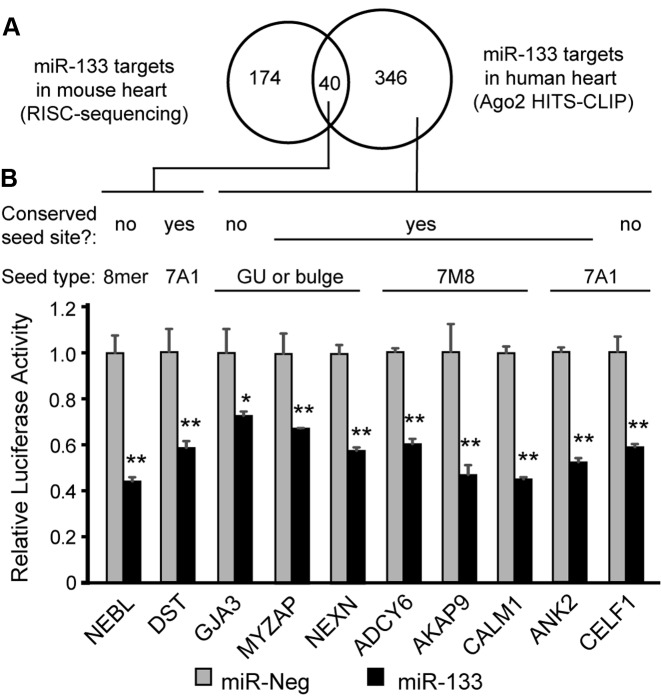
We also performed wet-lab experiments to assess the function of several miR:target interactions identified by our cardiac Ago2 HITS-CLIP. We first focused on targets of miR-133, a prominent cardiac-enriched miR that changes in many pathological heart conditions (11). This allowed for comparison to sites previously identified in miR-133 transgenic mice by cardiac RISC-seq, which identifies miR target transcripts by virtue of their enrichment in Ago-associated mRNA pools following miR overexpression (8). Interestingly, there was less than 20% overlap in miR-133 target genes between the two datasets (Figure 4A), with the human cardiac Ago2 HITS-CLIP data pointing to nearly 350 miR-133 targets not previously identified in the mouse RISC-seq studies; this is likely, in part, due to inter-species sequence differences (i.e. ∼40% of these miR-133 target seed sequences are not conserved in mice, Supplementary Table S8). Several of these new miR-133 targets were among a select group of 10 interactions that we experimentally tested for functionality using luciferase-based reporters. For this, 3′-UTRs of cardiac disease-related genes harboring miR-133 sites (deciphered by Ago2 HITS-CLIP) were cloned downstream of a luciferase gene and subsequently evaluated in cell culture studies. The tested genes included some for which mutations are known causes of human cardiomyopathies and arrhythmias (NEBL, NEXN, AKAP9, CALM1 and ANK2), as well as others associated with structural and functional cardiac abnormalities in mice (DST, GJA3, MYZAP and CELF1) (26–34). Notably, each of 10 3′-UTR reporters were repressed following miR-133 treatment, relative to negative control miR (∼25–55% silencing; Figure 4B).
Figure 4.
Cardiac miR-133 targets in human Ago2 HITS-CLIP and mouse RISC-seq data. (A) Venn diagram showing the intersection of mouse heart miR-133 target genes identified by RISC-seq (8) with miR-133 seed-containing human cardiac Ago2 clusters. (B) Ten miR-133 target genes were tested for functional responsiveness using luciferase-plasmids co-expressing Firefly luciferase (normalizer) and Renilla luciferase reporters containing 3′-UTRs corresponding to the indicated genes. Reporter plasmids were co-transfected with synthetic pre-miRs (negative control or miR-133) into HEK293 cells, and luciferase assays were performed 48 h later. Renilla/Firefly ratios were normalized to the pre-miR-Neg treatment for each 3′-UTR construct. Site information for the miR-133 seed is provided, including whether the seed sequence is conserved between human and mouse. Error bars depict standard deviation from the mean (n = 3 biological replicates). Two-tailed pairwise t-tests were used to compare miR-133 to miR-Neg for each construct, and P-values are provided; *P < 0.05, **P < 0.01.
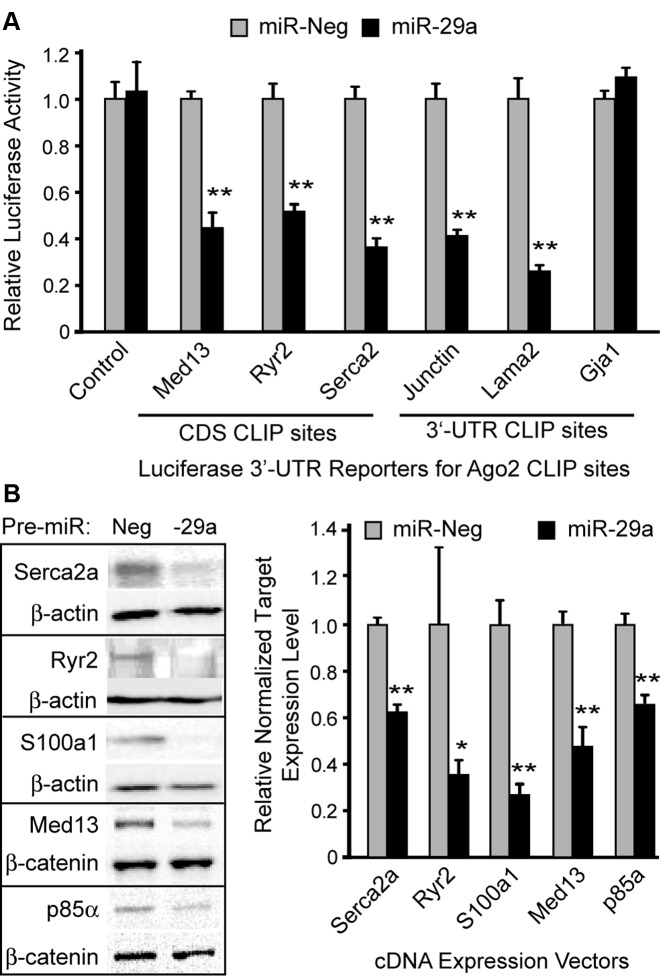
We next tested the functional responsiveness of several Ago2 HITS-CLIP targets corresponding to miR-29. These included both CDS- and 3′-UTR-targeted sites identified in genes related to cardiomyopathy, calcium regulation, hypertrophic growth and metabolic signaling in cardiomyocytes. To date, the major focus of miR-29 biology in the heart has been on its role in cardiac fibroblasts (21), however, our HITS-CLIP data point to new miR-29 functionality in cardiomyoctyes. In co-transfection studies, we found that miR-29 suppressed luciferase based reporters harboring miR-29 CLIP sites found in Ryr2, Serca2, and Junctin, key regulators of sarcoplasmic reticulum Ca2+ (Figure 5A); miR-29 sites from MED13 and LAMA2 genes were also found to be functionally responsive in this assay, but one non-canoncal site (G:U wobble) in GJA1 was non-functional. To account for the artificial nature of moving CDS sites to the 3′-UTR of luciferase reporter constructs, we performed additional experiments to evaluate suppression of protein synthesis derived from full-length cDNA expression plasmids, testing several target genes having a miR-29 site within their terminal coding exon, upstream of the stop codon. Co-transfection of these expression plasmids with miR-29 resulted in ∼40–70% reduction in target protein levels within 48 h (Figure 5B), further supporting functional interactions between miR-29 and regulators of cardiac calcium (e.g. Serca2a, Ryr2 and S100a1), and cardiomyocyte growth and metabolic signaling [PIK3R1 (p85-alpha) and Med13]. Together with the aforementioned microarray analyses, the wet-lab data for miR-133 and miR-29 establish that Ago2 HITS-CLIP is capable of effectively capturing functional cardiac miR:target interactions, including many unforeseen sites found in coding regions.
Figure 5.
Functional assessment of miR-29 sites in cardiomyocyte-related genes. (A) Six miR-29 target genes were tested for functional responsiveness using luciferase-plasmids co-expressing Firefly luciferase (normalizer) and Renilla luciferase reporters harboring cardiac Ago2 CLIP sites cloned into the 3′-UTR. Reporter plasmids were co-transfected with synthetic pre-miRs (negative control or miR-29a) into HEK293 cells, and luciferase assays were performed 48 h later. Renilla/Firefly ratios were normalized to the pre-miR-Neg treatment for each construct. (B) Silencing of genes harboring miR-29 CDS-targeted binding sites was assessed by co-transfecting cultured cells with synthetic pre-miRs and plasmids expressing the full-length coding regions of the various target genes. At 48 h post-transfection, western blot analysis was performed to measure target gene expression levels, using β-actin or β-catenin as loading controls (left). Band intensities were quantified by densitometry, and normalized data are plotted relative to miR-Neg1 for each target (right). Error bars depict standard deviation from the mean (n = 3 biological replicates). Two-tailed pairwise t-tests were used to compare miR-29a to miR-Neg for each construct; ** and * P-value <0.01 and <0.05, respectively.
Identification of clinically-relevant human cardiac Ago2 binding sites
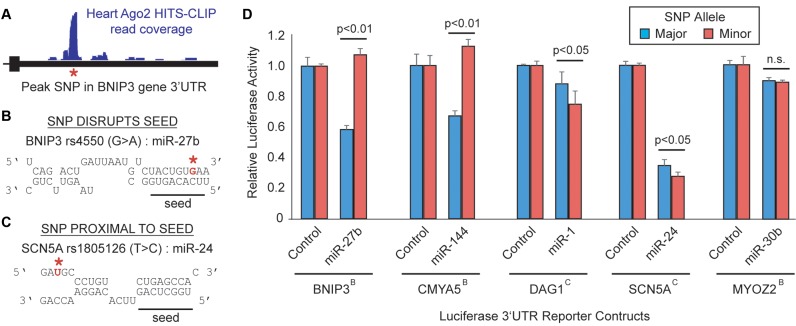
In our prior work in brain, we demonstrated that Ago2 HITS-CLIP is capable of fine-resolution mapping of miR binding sites, allowing one to decipher the potential interface of miR biology with human genetics and disease (5). To assess this interface in heart, we searched the cardiac Ago2 clusters for disease-relevant miR:target interactions overlapping genetic variants (Figure 6A), finding >500 common single-nucleotide polymorphism (SNPs) among the data (Supplementary Table S11). Further evaluation of these interactions revealed possible clinically-relevant miR target sites within genes linked to various heart conditions (Table 1). We focused this analysis on miR:target:SNP interfaces for which (i) the SNPs are positioned within or immediately proximal to seed sites matching abundant cardiac miRs (Figure 6B and C and Supplementary Figure S5), and (ii) existing evidence supports a relationship for the miRs and/or target genes with disease. For example, in the 3′-UTR of the BNIP3 gene, we identified a miR-27 binding site overlapping a common polymorphism which may have interesting relevance to ischemic heart disease (Figure 6A and B). BNIP3 encodes an autophagy-related protein that promotes cardiomyocyte death in response to ischemia (35), and elevated expression resulting from disrupted miR activity might worsen clinical outcomes resulting from atherosclerosis and myocardial infarction.
Figure 6.
Biological influence of SNPs within or near miR binding sites. (A) Schematic highlighting an example SNP (red asterisk) underlying a cardiac Ago2 HITS-CLIP peak; shown is SNP rs4550, located in the BNIP3 3′-UTR. Evaluation of miR:target hybridization reveals that SNPs (denoted in red) may reside within (B) or proximal to (C) miR seeds present in Ago2 clusters. The impact of SNPs on miR-mediated gene regulation was evaluated using 3′-UTR luciferase reporters, testing several of the disease-relevant miR:target:SNP interactions listed in Table 1 (D). Dual-luciferase plasmids containing the indicated 3′-UTR, harboring either the major or minor SNP allele, were co-transfected into HEK293 cells along with synthetic pre-miRs or miR expression plasmids (miR-1). Luciferase assays were performed at 48 h post-transfection and Renilla/Firefly ratios were normalized to the control miR for each 3′-UTR construct. The position of the SNP, either within (B) or proximal to the seed (C), is indicated in superscript for each gene. Error bars depict standard deviation from the mean (n = 3 biological replicates). Two-tailed pairwise t-tests for the indicated comparisons were performed and p-values provided; n.s. = not significant.
Table 1. Summary of disease-relevant SNPs in human cardiac Ago2 clusters.
| Gene | Disease relevance | SNP ID | Alleles (MAF*) | Minor allele effect | Comments |
|---|---|---|---|---|---|
| BNIP3 | MI and HF | rs4550 | G/A (32%) | Disrupts miR-27 seed | BNIP3 is elevated in ischemic heart failure and promotes cell death |
| CMYA5 | Cardiomyopathy | rs11323646 | T/- (28%) | Disrupts miR-144 seed | Gene encodes myospryn (interacts with z-discs and calcineurin) |
| DAG1 | DCM | rs1801143 | C/T (22%) | Enhances miR-1 3′ pairing | Mutations in this dystroglycan gene cause dilated cardiomyopathy |
| GJA1 | DCM & Arrhythmia | rs111878880 | T/C (6%) | Adjacent to miR-30 seed Disrupts miR-29 seed | Encodes Cx43, a gap junction protein that is critical for cardiac conductance |
| LAMA2 | DCM | rs1049476 | T/C (21%) | Disrupts miR-126 seed Adjacent to miR-29 seed | Mutations in this extracellular laminin protein cause dilated cardiomyopathy |
| LPL | Atherosclerosis | rs1059611 | T/C (13%) | Disrupts miR-136 seed | 3′UTR SNP associated with serum lipid levels in several GWAS studies |
| MYOZ2 | HCM | rs9995277 | G/A (29%) | Disrupts miR-30 seed | Mutations in this z-disc and calcineurin-tethering protein cause HCM |
| NEXN | DCM and HCM | rs145017889 | AGTGTTG/- (7%) | Disrupts miR-181 seed | Mutations in this F-actin binding protein destabilize cardiac z-discs |
| SCN5A | Brugada syndrome, QT, QRS interval | rs1805126 | T/C (49%) | Adjacent to miR-24 site | Synonymous CDS SNP associated with heart rhythm changes in GWAS studies |
| TTN | DCM and HCM | rs16866531 rs2303832 | A/G (3.9%) A/T (3.0%) | Disrupts miR-1 seed Disrupts miR-133 seed | These SNPs may serve a modifiers of the many pathogenic mutations in titin |
*MAF = minor allele frequency, MI = myocardial infarction, HF = heart failure, DCM = dilated cardiomyopathy, HCM = hypertrophic cardiomyopathy.
Several other interesting SNPs were located within structural heart genes encoding proteins associating with sarcomeric z-discs (e.g. myospryn, myozenin 2, nexilin and titin). Rare mutations in some of these genes are associated with dilated and hypertrophic cardiomyopathies (36), and these common genetic variants may modify disease progression and outcomes. Also, altered regulation of sarcomeric genes could influence the onset and clinical course of heart failure in patients (37).
Another notable SNP emerging from these analyses has been associated with changes in electrocardiographic measures in independent genome-wide association studies (GWAS) (38,39). This SNP, located in the terminal coding region of the SCN5A gene, is a synonymous variant and has been dismissed as potentially causal. However, our data point to the possibility that this polymorphism may modulate a nearby miR-24 site, altering cardiac sodium channel expression. Moreover, this common genetic variant could be a genetic modifier of the many SCN5A mutations known to cause Brugada syndrome (40), an inherited disease with incomplete penetrance associated with sudden cardiac death.
To assess the functional impact of these polymorphisms on miR-mediated gene regulation, we performed 3′-UTR reporter luciferase assays for several of the disease-relevant miR:target:SNP interactions listed in Table 1. In four of five cases tested, the SNP significantly altered miR-induced gene suppression (Figure 6D). Notably, SNPs located within miR seed sites completely abrogated silencing (e.g. BNIP3 and CMYA5), whereas those proximal to seeds modestly enhanced silencing, perhaps by imparting RNA structural changes improving seed site accessibility (20). These results reiterate the utility of coupling SNP analysis with Ago2 HITS-CLIP to identify translationally-relevant interactions that interface with human genetics, fostering the development of novel hypotheses concerning the effects of genetic variants on disease. As such, we evaluated this interface on a broader-level, identifying 572 common SNPs present within the human cardiac Ago2 clusters (Supplementary Table S11). We evaluated whether these SNPs are known to associate with host-gene expression levels in human tissues, and found 77 SNPs that are annotated cis-eQTLs [Genotype-Tissue Expression (GTEx) project (41)]. We next used computational means to determine if and how these SNPs may alter miR seeds or regional mRNA structures (based on PITA predictions). Notably, the resulting data support our findings in Figure 6D, as the minor alleles for both the SCN5A and DAG1 SNPs are predicted to enhance miR target site accessibility, encouraging increased silencing. To promote further use of this resource, we have tabulated this information in Supplementary Table S11, supporting future interrogations of polymorphisms that influence human phenotypes and disease outcomes through altered miR function and gene dosage mechanisms.
DISCUSSION
A vast literature supports that miRs play protective and pathogenic roles in heart, and thus, delineating their targets in affected cardiac tissues is critical for translating miR research to the clinic. However, few efforts have been made to generate comprehensive in vivo miR targeting data by high-throughput biochemical means (5,9,42). With respect to the heart, some progress to systematically define Ago-associated cellular transcripts has been made using RISC-seq technology (8), which is useful in models amenable to miR manipulation (e.g. cell culture and animals) to evaluate dynamic changes in Ago:mRNA interactions in response to altered miR activities. Although RISC-seq has proven valuable for identifying enriched Ago-associated mRNAs in transgenic mouse hearts overexpressing specific cardiac miRs, this approach is not well-suited for application in primary human tissues.
Ago HITS-CLIP is an established state-of-the-art technique for comprehensively identifying transcriptome- and miRnome-wide miR:target interactions at high resolution. We therefore adapted and applied this methodology to generate a transcriptome-wide map of miR binding sites in human heart tissue. Our efforts yielded 4000 Ago2 binding sites, many harboring sequences complementary to highly expressed cardiac miRs. As with other immunoprecipitation-based techniques, HITS-CLIP is biased towards capturing abundant miRs complexed with the highly expressed target transcripts, and this was indeed observed in our current dataset (e.g. genes expressed in heart versus genes captured by cardiac Ago2 HITS-CLIP have median RNA-seq FPKMs of 9.63 and 39, respectively; Supplementary Figure S6). For miRs, this can be considered acceptable, as miR functions depend on their stoichiometric capacity to bind the many potential target sites across the transcriptome. Indeed, it is broadly accepted that the most abundant miRs within cells are the most important biological players, as they are able to outcompete ‘dilution effects’ (18). However, this differs for target transcripts, as some lowly abundant transcripts are known to encode proteins having critical cellular functions, even at low dosages. To generate adequate coverage on these meekly expressed transcripts, future HITS-CLIP efforts may require increased sequencing depth or the incorporation of targeted sequence capture technologies. Another notable limitation with applying Ago CLIP approaches to primary human tissues is the inability to discern which cell types the miR-target interactions occur within. As a first step towards addressing this caveat, one can use pre-existing data from rodent studies that have defined miR and mRNA populations within the two predominant cell types of the heart, cardiomyocytes and fibroblasts [GSE58453, (43)]. To assist end-users of this resource we have included columns in Supplementary Tables S3 and S4 to annotate the relative abundances of the miR and target mRNAs in these two cell types. While this is the best-available approach, there are caveats pertaining to potential cross-species differences and the possibility that certain miRs and mRNAs may change during cell isolation procedures. Nevertheless, these data mostly align with our findings, as additional quality control analyses yielded little-to-no evidence for unexplainable interactions (e.g. cardiomyocyte-specific miRs binding with fibroblast-specific mRNAs) among our data, lending further support to the integrity of this resource.
Interestingly, our comparative analyses revealed a minimal overlap between miR-133 targets identified by Ago2 HITS-CLIP (human) and those found by RISC-seq (mouse). Although these two approaches are methodologically distinct, this miR targeting divergence is likely species-related, as was previously observed in human and mouse brains (5). While there is certainly some level of cross-species conservation among miR binding sites, these data highlight the importance of complementing rodent studies with the query of miR targeting in human tissues, particularly when considering translational aspects of miR biology. For example, among the miR-133 targets found in human but not mouse heart, several have clinical relevance to heart disease (e.g. genes for which known mutations cause cardiomyopathies). Despite these differences, continued efforts to thoroughly define the overlap in cardiac miR targeting events in both rodent models and human tissues are warranted to improve our interpretation of how rodent studies may inform our understanding of miR functions in human heart health and disease.
Overall, the human cardiac miR:target interactome represents a valuable resource for accelerating our understanding of miR functions in the cardiovascular system, and likely beyond. Although we subjected failing human heart samples to this analysis, we anticipate that these data will broadly inform the study of miRs in cardiac biology, in both normal and diseased hearts. Indeed, we predict that a case-control comparative study would reveal that the majority of the Ago2 binding sites are shared across healthy and diseased samples, but that the extent to which they are bound may vary with disease. In other words, peak heights will change but not their identities. This expected large degree of case-control overlap is supported by others’ observations that only ∼5% of genes are differentially expressed in normal and failing human cardiac tissues (GEO datasets: GSE1145, GSE5406 and GSE57345).
Even beyond heart, general interrogation of miR biology using this resource is highly encouraged, as many of the detected miRs and target genes are broadly expressed across tissues. In exploring the data ourselves, we identified several clinically-relevant interactions that interface with human genetic variations. Further studies are needed to more thoroughly characterize the impacts of these SNPs on miR-mediated regulation of the host gene and how this may influence its overall expression, with consideration for other regulatory controls and biological contexts. Our data also point to new and interesting roles for miR-29 in cardiomyocyte growth and calcium handling, which may have significant clinical relevance to cardiac hypertrophy and contractile dysfunction in cardiomyopathy patients. Interestingly, prior studies have reported alterations in miR-29 levels in hypertrophic cardiomyopathy and atrial fibrillation (44,45). Together, these findings support the need to expand the study of miR-29 in heart, particularly beyond cardiac fibroblasts. Additional avenues for future investigation could include querying the cardiac Ago2 HITS-CLIP data for other biologically-relevant miR binding sites within coding regions, particularly those which may elicit splice-isoform specific gene suppression. This data resource may also guide future interrogations of intriguing Ago2 bindings sites within intronic regions, long non-coding RNAs (lncRNAs) and perhaps even circular RNAs (circRNAs) (46), as the functional relevance of these sites remains largely unknown. Although our luciferase data mostly point to gene suppression mechanisms for the tested miR binding sites, these experiments need to be followed-up in more biologically-relevant contexts, keeping in mind that some interactions may function as miR ‘sponges’ in competing endogenous RNA (ceRNA) expression networks (47). Indeed, the bias of our data towards highly-expressed transcripts may be advantageous for evaluating these alternative mechanisms. Together, our work—both in brain and heart—and the utility of the resulting data provides a foundation for applying Ago2 HITS-CLIP to comprehensively characterize miR:target interactomes across the broad spectrum of human tissues, with future considerations for sub-anatomical structures and head-to-head comparisons of normal and diseased samples.
ACCESSION NUMBERS
Data have been deposited to the NCBI Gene Expression Omnibus (GSE83410).
Supplementary Material
Acknowledgments
We thank Dr Jay Nelson (OHSU) for providing the Ago2 antibody, as well as Dr Jonathan Lytton (Calgary), Dr Christopher George (Cardiff), and Dr Chad Grueter (Iowa) for providing Serca2, Ryr2 and Med13-GFP expression plasmids. We also thank Gabrielle Abouassaly for assistance with molecular cloning, Dr Kevin Knudtson and his staff of the University of Iowa DNA Facility for sequencing-related services and Dr Barry London's (Iowa) lab for feedback on the data and manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
American Heart Association [14SDG18590008 to R.L.B.]; Roy J. Carver Trust [University of Iowa to B.L.D.]; McLaughlin Trust [University of Iowa to F.L.J.]; National Institutes of Health [R01 NS076631 to B.L.D., T32 HL007638 to R.M.S., T32 HL007121 to J.M.M]. Funding for open access charge: Start-up funds.
Conflict of interest statement. None declared.
REFERENCES
- 1.Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eichhorn S.W., Guo H., McGeary S.E., Rodriguez-Mias R.A., Shin C., Baek D., Hsu S.H., Ghoshal K., Villen J., Bartel D.P. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell. 2014;56:104–115. doi: 10.1016/j.molcel.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chi S.W., Zang J.B., Mele A., Darnell R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zisoulis D.G., Lovci M.T., Wilbert M.L., Hutt K.R., Liang T.Y., Pasquinelli A.E., Yeo G.W. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boudreau R.L., Jiang P., Gilmore B.L., Spengler R.M., Tirabassi R., Nelson J.A., Ross C.A., Xing Y., Davidson B.L. Transcriptome-wide discovery of microRNA binding sites in human brain. Neuron. 2014;81:294–305. doi: 10.1016/j.neuron.2013.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jr, Jungkamp A.C., Munschauer M., et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konig J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B., Turner D.J., Luscombe N.M., Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matkovich S.J., Van Booven D.J., Eschenbacher W.H., Dorn G.W. 2nd. RISC RNA sequencing for context-specific identification of in vivo microRNA targets. Circ. Res. 2011;108:18–26. doi: 10.1161/CIRCRESAHA.110.233528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schug J., McKenna L.B., Walton G., Hand N., Mukherjee S., Essuman K., Shi Z., Gao Y., Markley K., Nakagawa M., et al. Dynamic recruitment of microRNAs to their mRNA targets in the regenerating liver. BMC Genomics. 2013;14:264. doi: 10.1186/1471-2164-14-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boettger T., Braun T. A new level of complexity: the role of microRNAs in cardiovascular development. Circ. Res. 2012;110:1000–1013. doi: 10.1161/CIRCRESAHA.111.247742. [DOI] [PubMed] [Google Scholar]
- 11.Small E.M., Frost R.J.A., Olson E.N. MicroRNAs Add a New Dimension to Cardiovascular Disease. Circulation. 2010;121:1022–U1066. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olson E.N. MicroRNAs as therapeutic targets and biomarkers of cardiovascular disease. Sci. Transl. Med. 2014;6:239ps233. doi: 10.1126/scitranslmed.3009008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhead B., Karolchik D., Kuhn R.M., Hinrichs A.S., Zweig A.S., Fujita P.A., Diekhans M., Smith K.E., Rosenbloom K.R., Raney B.J., et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu T., Ortiz J.A., Taing L., Meyer C.A., Lee B., Zhang Y., Shin H., Wong S.S., Ma J., Lei Y., et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu S.D., Lin F.M., Wu W.Y., Liang C., Huang W.C., Chan W.L., Tsai W.T., Chen G.Z., Lee C.J., Chiu C.M., et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39:D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullokandov G., Baccarini A., Ruzo A., Jayaprakash A.D., Tung N., Israelow B., Evans M.J., Sachidanandam R., Brown B.D. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods. 2012;9:840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang C., Darnell R.B. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat. Tiotechnol. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 21.van Rooij E., Sutherland L.B., Thatcher J.E., DiMaio J.M., Naseem R.H., Marshall W.S., Hill J.A., Olson E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. U.S.A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Care A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P., Bang M.L., Segnalini P., Gu Y., Dalton N.D., et al. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 23.Matkovich S.J., Wang W., Tu Y., Eschenbacher W.H., Dorn L.E., Condorelli G., Diwan A., Nerbonne J.M., Dorn G.W. 2nd. MicroRNA-133a protects against myocardial fibrosis and modulates electrical repolarization without affecting hypertrophy in pressure-overloaded adult hearts. Circ. Res. 2010;106:166–175. doi: 10.1161/CIRCRESAHA.109.202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heidrich F.M., Ehrlich B.E. Calcium, calpains, and cardiac hypertrophy: a new link. Circ. Res. 2009;104:e19–e20. doi: 10.1161/CIRCRESAHA.108.191072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hausser J., Syed A.P., Bilen B., Zavolan M. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 2013;23:604–615. doi: 10.1101/gr.139758.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purevjav E., Varela J., Morgado M., Kearney D.L., Li H., Taylor M.D., Arimura T., Moncman C.L., McKenna W., Murphy R.T., et al. Nebulette mutations are associated with dilated cardiomyopathy and endocardial fibroelastosis. J. Am. Coll. Cardiol. 2010;56:1493–1502. doi: 10.1016/j.jacc.2010.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassel D., Dahme T., Erdmann J., Meder B., Huge A., Stoll M., Just S., Hess A., Ehlermann P., Weichenhan D., et al. Nexilin mutations destabilize cardiac Z-disks and lead to dilated cardiomyopathy. Nat. Med. 2009;15:1281–1288. doi: 10.1038/nm.2037. [DOI] [PubMed] [Google Scholar]
- 28.Chen L., Marquardt M.L., Tester D.J., Sampson K.J., Ackerman M.J., Kass R.S. Mutation of an A-kinase-anchoring protein causes long-QT syndrome. Proc. Natl. Acad. Sci. U.S.A. 2007;104:20990–20995. doi: 10.1073/pnas.0710527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nyegaard M., Overgaard M.T., Sondergaard M.T., Vranas M., Behr E.R., Hildebrandt L.L., Lund J., Hedley P.L., Camm A.J., Wettrell G., et al. Mutations in calmodulin cause ventricular tachycardia and sudden cardiac death. Am. J. Hum. Genet. 2012;91:703–712. doi: 10.1016/j.ajhg.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohler P.J., Schott J.J., Gramolini A.O., Dilly K.W., Guatimosim S., duBell W.H., Song L.S., Haurogne K., Kyndt F., Ali M.E., et al. Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature. 2003;421:634–639. doi: 10.1038/nature01335. [DOI] [PubMed] [Google Scholar]
- 31.Boyer J.G., Bhanot K., Kothary R., Boudreau-Lariviere C. Hearts of dystonia musculorum mice display normal morphological and histological features but show signs of cardiac stress. PLoS One. 2010;5:e9465. doi: 10.1371/journal.pone.0009465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chi N.C., Bussen M., Brand-Arzamendi K., Ding C., Olgin J.E., Shaw R.M., Martin G.R., Stainier D.Y. Cardiac conduction is required to preserve cardiac chamber morphology. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14662–14667. doi: 10.1073/pnas.0909432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seeger T.S., Frank D., Rohr C., Will R., Just S., Grund C., Lyon R., Luedde M., Koegl M., Sheikh F., et al. Myozap, a novel intercalated disc protein, activates serum response factor-dependent signaling and is required to maintain cardiac function in vivo. Circu. Res. 2010;106:880–890. doi: 10.1161/CIRCRESAHA.109.213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang G.S., Kearney D.L., De Biasi M., Taffet G., Cooper T.A. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J. Clin. Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diwan A., Krenz M., Syed F.M., Wansapura J., Ren X., Koesters A.G., Li H., Kirshenbaum L.A., Hahn H.S., Robbins J., et al. Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J. Clin. Invest. 2007;117:2825–2833. doi: 10.1172/JCI32490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seidman C.E., Seidman J.G. Identifying sarcomere gene mutations in hypertrophic cardiomyopathy: a personal history. Circ. Res. 2011;108:743–750. doi: 10.1161/CIRCRESAHA.110.223834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong S.W., Hu Y.W., Ho J.W., Ikeda S., Polster S., John R., Hall J.L., Bisping E., Pieske B., dos Remedios C.G., et al. Heart failure-associated changes in RNA splicing of sarcomere genes. Circ. Cardiovasc. Genet. 2010;3:138–146. doi: 10.1161/CIRCGENETICS.109.904698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfeufer A., Sanna S., Arking D.E., Muller M., Gateva V., Fuchsberger C., Ehret G.B., Orru M., Pattaro C., Kottgen A., et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat. Genet. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holm H., Gudbjartsson D.F., Arnar D.O., Thorleifsson G., Thorgeirsson G., Stefansdottir H., Gudjonsson S.A., Jonasdottir A., Mathiesen E.B., Njolstad I., et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat. Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 40.Wilde A.A., Brugada R. Phenotypical manifestations of mutations in the genes encoding subunits of the cardiac sodium channel. Circ. Res. 2011;108:884–897. doi: 10.1161/CIRCRESAHA.110.238469. [DOI] [PubMed] [Google Scholar]
- 41.Consortium G.T. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chi S.W., Hannon G.J., Darnell R.B. An alternative mode of microRNA target recognition. Nat. Struct. Mol. Biol. 2012;19:321–327. doi: 10.1038/nsmb.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matkovich S.J., Edwards J.R., Grossenheider T.C., de Guzman Strong C., Dorn G.W. 2nd. Epigenetic coordination of embryonic heart transcription by dynamically regulated long noncoding RNAs. Proc. Natl. Acad. Sci. U.S.A. 2014;111:12264–12269. doi: 10.1073/pnas.1410622111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dawson K., Wakili R., Ordog B., Clauss S., Chen Y., Iwasaki Y., Voigt N., Qi X.Y., Sinner M.F., Dobrev D., et al. MicroRNA29: a mechanistic contributor and potential biomarker in atrial fibrillation. Circulation. 2013;127:1466–1475. doi: 10.1161/CIRCULATIONAHA.112.001207. [DOI] [PubMed] [Google Scholar]
- 45.Roncarati R., Viviani Anselmi C., Losi M.A., Papa L., Cavarretta E., Da Costa Martins P., Contaldi C., Saccani Jotti G., Franzone A., Galastri L., et al. Circulating miR-29a, among other up-regulated microRNAs, is the only biomarker for both hypertrophy and fibrosis in patients with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2014;63:920–927. doi: 10.1016/j.jacc.2013.09.041. [DOI] [PubMed] [Google Scholar]
- 46.Boeckel J.N., Jae N., Heumuller A.W., Chen W., Boon R.A., Stellos K., Zeiher A.M., John D., Uchida S., Dimmeler S. Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA. Circ. Res. 2015;117:884–890. doi: 10.1161/CIRCRESAHA.115.306319. [DOI] [PubMed] [Google Scholar]
- 47.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.