Abstract
A majority of large-scale bacterial genome rearrangements involve mobile genetic elements such as insertion sequence (IS) elements. Here we report novel insertions and excisions of IS elements and recombination between homologous IS elements identified in a large collection of Escherichia coli mutation accumulation lines by analysis of whole genome shotgun sequencing data. Based on 857 identified events (758 IS insertions, 98 recombinations and 1 excision), we estimate that the rate of IS insertion is 3.5 × 10−4 insertions per genome per generation and the rate of IS homologous recombination is 4.5 × 10−5 recombinations per genome per generation. These events are mostly contributed by the IS elements IS1, IS2, IS5 and IS186. Spatial analysis of new insertions suggest that transposition is biased to proximal insertions, and the length spectrum of IS-caused deletions is largely explained by local hopping. For any of the ISs studied there is no region of the circular genome that is favored or disfavored for new insertions but there are notable hotspots for deletions. Some elements have preferences for non-coding sequence or for the beginning and end of coding regions, largely explained by target site motifs. Interestingly, transposition and deletion rates remain constant across the wild-type and 12 mutant E. coli lines, each deficient in a distinct DNA repair pathway. Finally, we characterized the target sites of four IS families, confirming previous results and characterizing a highly specific pattern at IS186 target-sites, 5′-GGGG(N6/N7)CCCC-3′. We also detected 48 long deletions not involving IS elements.
INTRODUCTION
With advances in massively parallel sequencing (next generation sequencing, NGS) technologies, whole genome shotgun sequencing (WGSS) has become an affordable approach to genome-wide characterization of genetic variation in bacterial and eukaryotic genomes (1–3). In addition to small-scale variations (e.g. single nucleotide substitutions and insertion/deletions of a few base pairs), large-scale variation due to genome rearrangements (e.g. inversions, transpositions and segmental duplications/deletions) in bacterial genomes are commonly observed in both natural isolates (4) and laboratory stocks (5,6). Such events have been shown to play important roles in genome evolution and environmental adaptation (7). For example, large-scale rearrangements arose in the Escherichia coli genome during a long-term evolution experiment in a recent study (8). Combining WGSS with optical mapping technology, they identified a total of 110 large-scale rearrangements (including 82 deletions, 19 inversions and 9 duplications) in which a majority of the events involved insertion sequence (IS) elements, a class of simple transposable elements in bacterial genomes (9). This result confirms the crucial roles of IS elements in mediating large-scale variation in bacterial genome evolution (10).
Similar to small-scale genetic mutations, large-scale genome rearrangements accumulate spontaneously in a selection-free environment. However, no estimate has been made for the rate of genome rearrangements in bacterial genomes, primarily because it is not easy to eliminate the effects of selection, even in a controlled environment. In recent years, a mutation accumulation (MA) strategy has been used to capture spontaneous mutations by allowing mutations to accumulate over many generations in the near-absence of selection (11). An MA experiment is achieved by initiating a number of lines (MA lines) from a single founder individual and repeatedly taking each of the lines through a population bottleneck of a single individual. This bottlenecking procedure results in an effective population size of Ne ≈ 1 for each line, allowing the power of random genetic drift to dominate that of selection and effectively removing the effects of selection on MA (12). When combined with high-throughput NGS sequencing of many replicate lines, the MA approach enables a direct measurement of the spontaneous genome-wide mutation rate as well as the mutation spectrum for single base-pair mutations and short indels (13–16). We have adopted this approach to investigate the mutation rates and spectra for wild-type (WT) E. coli and many derived DNA repair-deficient strains (16,17).
Our studies here focus on the large-scale variations involving IS elements occurring in MA lines, because these are primary remodelers of bacterial genomes (18) and account for the vast majority of the large-scale events we observed. ISs are the simplest type of bacterial selfish element; they are compact (0.7–2.5 kb in length), encoding only 1–2 genes (usually one) needed for transposition, and include the cis-acting sites upon which the transposase acts (19). Cis-sites usually, but not always, consist of inverted terminal repeats of a few dozen base pairs (19). Furthermore, most ISs duplicate a small number of base pairs at their insertion site such that an IS insertion is flanked by short direct repeats (19). While IS elements can transpose by a number of mechanisms, the elements in our study are of the most common copy-paste or cut-and-paste types; while the details differ, an important result is that transposition is or appears replicative, i.e. the resulting genome retains the donor insertion and a novel insertion (19). Thus IS lost is rare. IS insertions can have several implications for genome structure and function. In this study our focus is genome structural stability, but new IS insertions can play regulatory roles as well (20). IS-dependent events can have important implications for evolution (21), medicine (22,23) and industry (24,25). Important to this study, IS propagation generates large patches of homology scattered throughout the genome, which are substrates for homologous recombination.
Compared to small-scale events affecting only a single or a few bases, large-scale variation is more difficult to infer from WGSS data because current NGS techniques typically generate short reads (e.g. ∼100 bps for Illumina sequencers) (26) that rarely span the entire segment involved in a genome rearrangement. In this paper, we report a thorough analysis of large-scale variations in the genome of E. coli K-12 that occurred in previously published MA experiments (16,17). We implemented a novel software tool, GRASPER (Graph-based Rearrangement Analysis from Short and Paired-End Reads), that uses a graph-based algorithm (27) capable of detecting most large-scale variation involving repetitive regions, including novel IS insertions, in bacterial genomes. In order to obtain a comprehensive picture of the spontaneous rate and spectrum of genome rearrangement events involving IS elements (i.e. insertions, deletions and recombinations), we have extended GRASPER into a general purpose tool that reports most large-scale variations involving repetitive sequences. We analyzed 520 E. coli MA lines (for a total of ∼2.2 million generations) (16,17) using GRASPER, and identified 857 large-scale variations (shown on Figure 1; Supplementary Tables S7 and S8) involving IS elements, consisting of 758 insertions, 1 excision and 98 recombinations between two homologous IS elements. Further analyses of these events revealed several interesting and novel findings: (i) the rate of IS insertion in a selection-free environment is 3.5 × 10−4 insertions per genome per generation, while the rate of IS homologous recombination is 4.5 × 10−5 recombinations per genome per generation; (ii) the most active IS elements are from the four abundant E. coli IS families IS1, IS2, IS5 and IS186, which contributed ∼97.7% of all new IS insertions, whereas IS elements in the families IS3, IS4, IS30 and IS150 rarely transpose, and IS30, IS609, ISX, ISZ and IS600 never transpose; (iii) the rates of IS insertion and recombination are nearly constant across multiple E. coli lines, each deficient for a distinct DNA repair pathway, indicating that the transposition mechanisms of these elements are independent of these host functions; (iv) transpositions are biased to local insertions, in particular for IS1 and IS5 elements, but IS2 shows target-site immunity; (v) homologous recombination occurs predominantly between near-by IS elements (1–3 kb); and (vi) some IS elements have specific target sites in the E. coli genomes; in particular, IS186 has a specific sequence pattern flanking its target-site duplications (TSDs): 5′-GGGG(N6/N7)CCCC-3′.
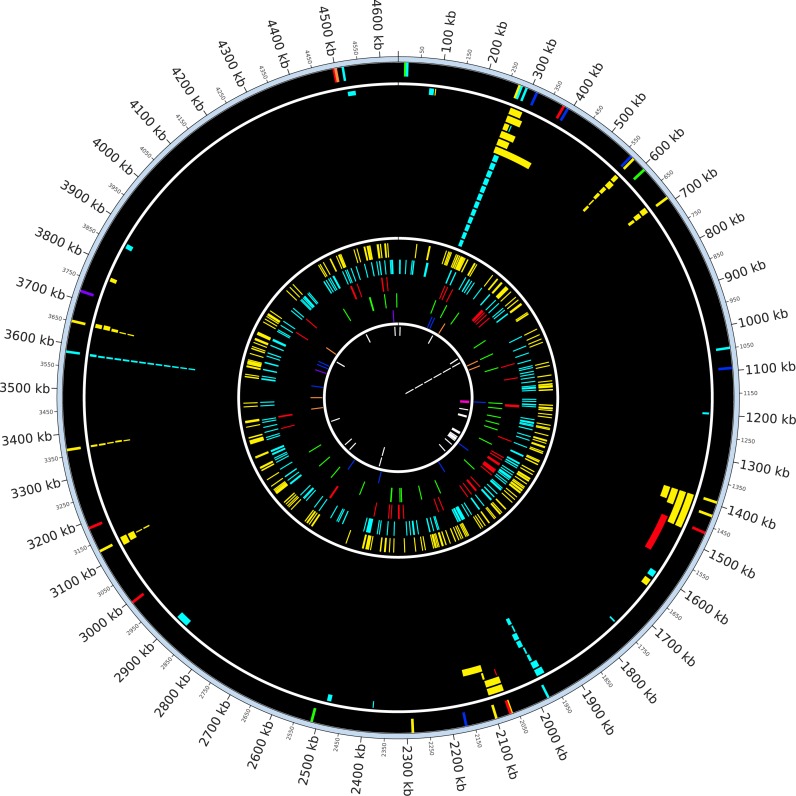
Figure 1.
Circos plot displaying large-scale rearrangements. Starting from the outer-most section to the inner-most section (each section separated by white ring), Circos plot displaying insertion sequence (IS) insertions in the founding strain, IS-associated deletions, novel IS insertions (each major IS family is drawn in an individual track) and other deletions (not associated with IS elements) recovered from mutation accumulation (MA) data. Colors indicate different IS families (IS5: yellow, IS1: cyan, IS2: red, IS186: green, IS3: blue, IS4: orange, IS150: purple, other deletions: white). A magenta band in the other deletion section indicates an e14 deletion that occurred 21 times and it is drawn as single band due to space limitation. Spatial clustering of IS-associated deletions anchored around preexisting IS insertions can be seen.
Together, these findings expand our knowledge of the dynamic role of IS elements in bacterial evolution.
MATERIALS AND METHODS
MA lines and whole genome shotgun sequencing
All methods and data are from (16) and (17). The raw WGSS data from these lines have been deposited into NCBI Short Read Archive (study accession SRP013707). Among the lines included in (17), there are lines that have shared mutations that were removed and counted only once. But for simplicity in this study we randomly chose only one line among the lines that have shared mutations. Unlike base-pair substitution (BPS),large-scale rearrangements are much less frequent, so that accidentally counting one event twice can cause greater errors in the estimation of the overall rate.
Pre-processing of sequencing data
Experiments have widely varying read lengths (2 × 90, 2 × 91, 2 × 99, 2 × 101 and 2 × 150 bp) as they were carried out at various times. To keep the read length consistent across different experiments, all the reads were trimmed to 90 bp. Sequencing reads were quality filtered by using Trimmomatic (28) (Trimmomatic options used: ILLUMINACLIP:<adaptersequence>:2:30:10 CROP:90 LEADING:3 SLIDINGWINDOW:4:15 MINLEN:70). After the reads are aligned to the reference E. coli K-12 MG1655 genome using BWA 0.7.9a (29), median and MAD(median absolute deviation) values for insert size distribution for each MA line were estimated based on uniquely mapped read pairs and supplied to GRASPER with a reference Al-Bruijn graph(l = 90, error rate of 5%).
GRASPER algorithm
GRASPER takes advantage of repeat information embedded in an Al-Bruijn graph representation of the reference genome to detect rearrangement events involving repeats, given paired-end WGSS data (27). It mainly uses paired-end signatures that are well established for classifying rearrangement events, looking at discordant signals, but it also incorporates read count information embedded in edges on Al-Bruijn graph to help resolve ambiguities in paired-end signatures. GRASPER adopts the coverage estimation approach used in (30) to embed the read count information on Al-Bruijn graphs, where 100 bp non-overlapping windows are used to keep track of the number of reads mapped to each window. GRASPER can accurately detect (i) transposition (insertion), (ii) deletion of non-repetitive region, (iii) deletion of repetitive region, (iv) deletion of non-repetitive regions bounded by repeats (via homologous recombination), (v) inversion (except those flanked by repeats longer than the library insert size) and (vi) tandem-duplication.
GRASPER first obtains a list of clusters of discordant read pairs, then tries to assign rearrangement events to the clusters. GRASPER also generates a list of all non-overlapping segments with depleted read counts, where a depletion signal is determined by setting a cutoff value for the number of reads obtained, using the Lander-Waterman model (31). When checking for depleted read count signals, any repetitive edge with a sufficient read count is ignored, because repetitive edges can still recruit reads unless all repeat copies are deleted in a sample genome.
For each breakpoint cluster with simple deletion signatures, GRASPER checks all possible paths, following the thread from the longest to the shortest, because there are multiple paths if any one end of the deletion boundary is repetitive. For each path, it checks if there is a depletion signal, and if there is, reports the path as a deletion event. This ensures that GRASPER always reports the longest deletion with support of a depletion signal. More complex rearrangements requiring two breakpoint clusters, such as transposition and inversion are called using the signatures shown in Supplementary Figure S5.
Among the remaining signals, deletion events of repetitive elements and tandem duplications are called. Any remaining clusters are reported as breakpoint signals without having events assigned to them. Also, the upstream and downstream regions of each remaining segment with a depletion signal are examined. If such a segment is surrounded by direct repeats, GRASPER calls the segment as deletion mediated by homologous recombination. This is especially easy with the repeat structure information embedded in Al-Bruijn graphs. Note that if the repeat length is longer than the insert size, no breakpoint signal is available.
GRASPER is implemented as a JAVA program and is available under the GPL v3 license as open-source software at https://github.com/COL-IU/GRASPER.
Permutation test for local hopping of IS elements
For the four most active IS elements (IS1A, IS2, IS5A and IS186) we tested whether their insertions are more likely to occur close (within a distance threshold) to a preexisting copy. All positions of pre-existing IS elements were randomly chosen and the distance to the closest preexisting copy was calculated for each observed insertion. This process was repeated 1000 times to obtain the expected number of insertions within d of the closest preexisting copies (d ∈ {1, 5, 10, 100, 200, 300 Kb}). For each d, a p-value was estimated by performing a binomial test (one-tailed t-test; see Table 2).
Table 2. Number of IS insertions landing within d of closest preexisting copy.
| IS1A | IS5A | IS2 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| d | Observed | Expected | P | Observed | Expected | P | Observed | Expected | P |
| 1000 | 5 | 0.5 | 1.8 × 10−4 | 19 | 1.5 | 4.2 × 10−15 | 1 | 0.2 | 0.19 |
| 5000 | 21 | 2.6 | 4.3 × 10−13 | 42 | 7.8 | 1.6 × 10−18 | 1 | 1.0 | 0.64 |
| 10 000 | 23 | 5.1 | 2.7 × 10−9 | 53 | 15.5 | 8.3 × 10−15 | 1 | 2.0 | 0.86 |
| 50 000 | 37 | 25.1 | 0.011 | 119 | 72.7 | 7.4 × 10−9 | 6 | 8.8 | 0.89 |
| 100 000 | 58 | 48.0 | 0.064 | 181 | 131.5 | 7.2 × 10−9 | 11 | 16.3 | 0.95 |
| 200 000 | 91 | 87.6 | 0.35 | 245 | 216.5 | 1.2 × 10−3 | 46 | 29.2 | 4.8 × 10−5 |
| 300 000 | 122 | 120.2 | 0.43 | 298 | 271.2 | 4.6 × 10−4 | 65 | 39.5 | 3.5 × 10−11 |
| Total | 233 | 361 | 71 | ||||||
Reconstruction of target-site duplications (TSDs)
All the putative insertions detected using GRASPER are further analyzed to obtain TSDs. Given an insertion, reads from both flanking regions are separately assembled to form contigs. The longest exact match between two contigs is found, to assign the TSD of the insertion.
RESULTS
IS elements in the E. coli founder genome
We first applied GRASPER to WGSS data from the E. coli MA lines to determine the set of genome rearrangement events fixed in the progenitor PFM2 strain that was used as the founder strain in all MA experiments considered here (16,17). As shown in Supplementary Table S1, PFM2 contains 41 copies of IS elements from 14 families.
There are only two IS differences between PFM2 and the reference E. coli K-12 strain MG1655: the PFM2 genome has one less copy of an IS1 element that is located at position 1 976 527, upstream of the promoter region of the flhCD operon (and is associated with hypermotility (32)). On the other hand, the PFM2 genome contains an extra IS186 insertion at ∼1.87 Mb, inside the yeaR gene, a regulator of nitrogen metabolism (33). We note that the IS families shown here were classified by GRASPER automatically, so that IS elements with almost identical sequences (below 5% divergence) are classified into the same family. But as a result, some conventional families (such as IS1 and IS5) were split into two, because elements in these families have high sequence divergence. GRASPER is designed to detect large-scale rearrangements (e.g. novel insertions involving repeats such as IS elements), but cannot distinguish which of the original copies of a repeat family is the parental donor of an event.
Detection of large-scale genome rearrangement events in E. coli MA lines
We then used GRASPER to detect all large-scale rearrangement events involving IS elements, including novel insertions, excisions and recombinations, using paired-end WGSS data from a total of 520 MA lines spanning ∼2.2 million generations (16,17) (see ‘Methods and Methods’ section for details). Each line has an average 70–80× sequencing coverage, which is sufficient to detect large-scale events with high confidence using GRASPER (27). In summary, we detected a total of 857 large-scale events involving IS elements: 758 novel insertions, 1 excision and 98 recombinations (Table 1). We selected an arbitrary subset of these detected events for polymerase chain reaction (PCR) verification. 100% (56/56 insertions and 21/21 recombinations) of the PCR results agreed with our predictions (See Supplementary Tables S5 and S6 for the list of PCR primers used), giving us high confidence that our results are accurate.
Table 1. Number of novel IS insertions detected in E. coli MA lines.
| IS name | IS family/group | No. of copies | No. of novel insertions | No. of recombinations |
|---|---|---|---|---|
| IS1A | IS1/- | 5 | 233 | 43 |
| IS1B | IS1/- | 1 | 3 | 1 |
| IS2 | IS3/IS2 | 6 | 71 | 2 |
| IS3 | IS3/IS3 | 5 | 10 | 0 |
| IS4 | IS4/IS4 | 1 | 8 | 0 |
| IS5A | IS5/IS5 | 10 | 361 | 52 |
| IS5B | IS5/IS5 | 1 | 2 | 0 |
| IS30 | IS30/- | 3 | 0 | 0 |
| IS150 | IS3/IS150 | 1 | 2 | 0 |
| IS186 | IS4/IS231 | 4 | 68 | 0 |
| IS609 | IS200/IS605 | 1 | 0 | 0 |
| ISX | IS3-like | 1 | 0 | 0 |
| ISZ | IS4-like | 1 | 0 | 0 |
| IS600 | IS3/IS3 | 1 | 0 | 0 |
| Total | 41 | 758 | 98 | |
Novel insertions of IS elements (i.e. transpositions) are the most frequent events, constituting 88.4% (758 of 857) of all detected events. Table 1 summarizes the number of novel insertions for each IS family present in PFM2. Among the 14 IS families, 5 families (IS30, IS609, ISX, ISZ and IS600) have no detected novel insertions in any MA line, suggesting that these families are completely inactive in the E. coli genome under our growth conditions (note that there is only one copy for each of these families in PFM2 and MG1655, except for IS30, which has three copies). Among the nine active IS families, IS5A and IS1A are the most active: their transpositions make up 78.4% (IS5A: 47.6% and IS1A: 30.7%) of all novel IS insertions. IS2 and IS186 also have significant activity, while only a small number of insertions were detected for the five families IS3, IS4, IS1B, IS5B and IS150.
GRASPER also detects homologous recombination events between two IS copies of the same family. There are two major types of homologous recombinations: those between direct copies, and those between inverted copies (See Supplementary Figure S1 illustration). Recombination between direct copies of an IS deletes the intervening genomic segment, leaving a single recombinant copy of the IS at the deletion junction; recombination between inverted copies of an IS inverts the intervening segment leaving a copy of the element at each end of the inversion. Because the insert size of the paired-end reads in WGSS data (typically 500 bp) is much shorter than the length of an IS element and no novel IS-flanking junctions are gained or lost, an inversion involving long inverted repeats (i.e. two copies of an IS element) does not create a diagnostic signal in paired-end reads and thus cannot be detected using WGSS data. Hence, the events reported in this paper are limited to deletions between direct copies of IS elements. While we cannot know how many recombination events between inverted IS copies occurred in our MA lines, one might expect a number roughly equal to the number between direct IS copies. Finally, tandem duplications via unequal crossing-over between proximal IS copies can be detected based on read coverage, but we did not observe such events in our data, and these structures are not expected to be stable even if formed (34).
A total of 98 deletion events between IS elements were detected in the 520 MA lines. All these events involved elements in the IS5(IS5A only), IS1(both IS1A and IS1B) and IS2 families (Table 1). Similar to novel insertions, IS5A and IS1A are the IS elements most involved in deletion formation, contributing to 96.9% (95 out of 98) of all deletion events. Finally, only one IS excision event was detected in the 520 MA lines, (a copy of IS3 element located at 566 015–567 258), indicating that IS excision is rare, even in a selection-free environment.
Deletion events are of three types: (i) between IS copies both of which exist in the founder strain (denoted as E + E); (ii) between an IS copy present in the founder strain and another, novel copy assumed to have arisen during the MA experiment (E + N); and (iii) between two novel copies (N + N). Among these three types, the E + N type dominates: 73.5% (72 out of 98) of observed deletion events are of this type whereas only 14 E + E and 12 N + N events were observed. Among the 14 E + E events, 12 are a recurrent deletion event between two copies of IS1A in a cryptic prophage (see below), and two are the same deletion event between two copies of IS5A (located at 2 064 183 and 2 099 773) that results in deletion of a segment containing 30 host genes and the entire CP4-44 prophage. Note that N + N events create a deletion with a novel IS element at the junction; thus we cannot eliminate the possibility that the deletion is caused by a single aberrant insertion event (35).
Transpositions of IS elements are biased to nearby—or distant—insertions
As shown in Figure 1, at the genomic scale we did not observe specific hotspots for IS element insertion. However, we found that novel insertion sites are often proximal to a preexisting copy of the same element. Although we do not have direct evidence that the proximal element is the parent element that induced the duplicative transposition, a high frequency of such cases would suggest that IS elements tend to transpose to a location close to the parent donor. To test if the nearby insertions are indeed significantly frequent, we counted the number of novel insertions whose distance to their closest preexisting element is less than certain threshold d (e.g. d = 1 Kb) for each of the four active IS families, IS1A, IS2, IS5A and IS186. Figure 2 shows the cumulative counts for the three families plotted against various values of d.
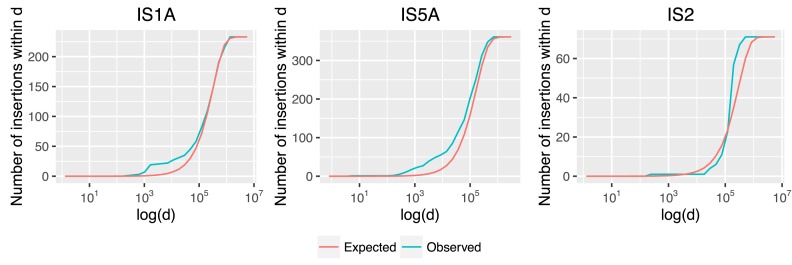
Figure 2.
Cumulative distribution of IS insertions within a distance threshold d. Insertions recovered in MA data are shown in blue (observed) and average counts using the 1000-permutation test are shown in red (expected). IS1A and IS5A exhibit bias for nearby insertions (small values of d in bp on x-axis; drawn in log scale) and IS2 shows bias for distant insertions.
Comparing the observed number to the expected number of corresponding insertion events (estimated using a permutation test; see ‘Materials and Methods’ section), IS1A and IS5A families are significantly biased to loci near to a preexisting element. For example, as shown in Table 2, we observed five novel insertions of an IS1A element within 1 Kb of a preexisting IS1A element, whereas the expected number is 0.5 (p < 0.0002). Similarly, we observed 19 novel insertions of a IS5A element within 1 Kb from a preexisting element, whereas the expected number is 1.5 (p < 10−14). These results indicate that IS1A and IS5A transpose to relatively near-by sites.
On the other hand, we did not observe significantly more-than-expected IS2 insertions within a 1 Kb from a preexisting element; but we did observe significantly more insertions within 200 Kb distance (p < 10−4; see Table 2). This result suggests that IS2 exhibits a level of target-site immunity (36), with insertions enriched in the regions right outside the perimeter of immunity (i.e. 200–300 Kb). Finally, we did not observe any bias for near-by transposition for the IS186 elements consistent with our observation that IS186 elements target specific genomic sites that contain a sequence pattern flanking their TSDs (see below).
Recombination occurs most frequently between two closely located copies of IS elements
We investigated the length distribution of the deleted intervals in the 98 cases of recombination between direct copies of an IS element. As shown in Figure 3, a substantial fraction (45 out of 98; 45.9%) of the deleted segments are ≤3 kb. There are two possible reasons for this high frequency of deletions between near-by IS elements: recombination between close IS elements occurs more frequently, or relatively short deletions are less likely to cause deleterious effects (lethality) in MA experiments. Given that the majority of deletions involve one or two novel insertions (see above and Figure 3), localized transposition must also play a role (see below).
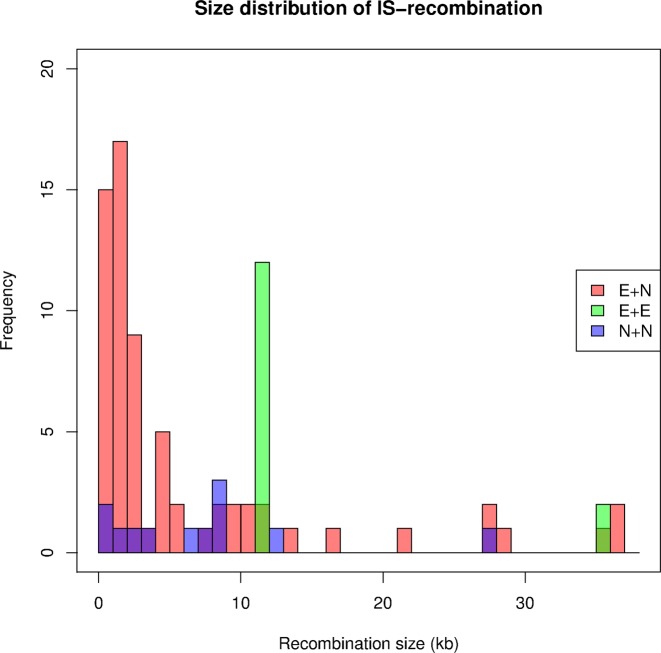
Figure 3.
Size distribution of IS-mediated recombination events. Distribution of all deletions (E + N, E + E and N + N types) mediated by recombination are plotted except for five recombination events of sizes larger than 37 kb (48.3, 81.3, 81.3, 91.9 and 106.1 kb). The peak in the 11–12 kb bin is caused by recurrent deletion of a part of the CP4-6 cryptic prophage. Note that darker colors (dark purple and dark green) indicates overlap of distributions. Dark purple indicates an overlap of E + N and N + N and dark green indicates an overlap of E + N and E + E.
The surprisingly large number (14 out of 98; 14.3%) of deletions that are 11–12 kb (Figure 3) are all due to the recurrent recombination event between two IS1A copies, both present in the reference genome (located around the 278.5–290.5 Kb region). This recombination event deletes ∼11.5 kb segment in the CP4-6 cryptic prophage (37), and thus is not expected to have a fitness effect. This specific deletion was observed in 12 MA lines across four different MA experiments (PFM2m, PFM40, PFM101 and PFM130) and had been reported as a fixed variation in E. coli strain NCM3722 (38).
Rates of insertion and recombination of IS elements are constant across different mutant backgrounds
We calculated the IS element insertion and homologous recombination rates for each of 15 MA experiments.
The genetic backgrounds used in these experiments include lines defective in major DNA repair pathways, including mismatch repair (mutL), nucleotide excision repair (uvrA), and base excision repair (several mutants) (Table 3).
Table 3. Insertions and recombinations involving IS elements detected in E. coli MA lines.
| Generations | Insertion | Recombination | ||||||
|---|---|---|---|---|---|---|---|---|
| Strain | Disrupted pathway* | Description | No. of Lines | per line | No. of Rate∧ (× 10−4) | No. of Rate∧ (× 10−5) | ||
| PFM2 | WT @3K gen | 38 | 3080 | 21 | 1.79 | 1 | 0.85 | |
| PFM2 | WT @6K gen | 21 | 6356 | 39 | 2.92 | 2 | 1.59 | |
| PFM2 | WT on min. medium | 44 | 6166 | 83 | 3.10 | 8 | 2.99 | |
| PFM5 | MMR | mutL | 34 | 375 | 10 | 7.84 | 1 | 7.84 |
| PFM101 | TLS | umuDC dinB | 39 | 6078 | 95 | 4.03 | 12 | 5.09 |
| PFM133 | TLS | umuDC dinB polB | 43 | 6204 | 108 | 4.07 | 12 | 4.52 |
| PFM35 | NER + TCR | uvrA | 23 | 6350 | 63 | 4.37 | 7 | 4.85 |
| PFM40 | ALK | alkA tagA | 37 | 6225 | 81 | 3.56 | 16 | 7.04 |
| PFM88 | ALK | ada ogt | 47 | 6269 | 113 | 3.88 | 16 | 5.50 |
| PFM180 | BER | xthA nfo | 40 | 3155 | 37 | 2.95 | 12 | 9.58 |
| PFM61 | OXDR | mutT | 25 | 599 | 6 | 4.62 | 0 | |
| PFM6 | OXDR | mutY | 25 | 1972 | 14 | 2.96 | 2 | 4.23 |
| PFM22 | BER | nth nei | 50 | 920 | 6 | 1.43 | 0 | |
| PFM91 | BER | nfi | 29 | 6308 | 65 | 3.60 | 5 | 2.77 |
| PFM94 | OXDR | mutY mutM | 25 | 1916 | 17 | 3.70 | 4 | 8.71 |
| Total | 520 | 4186 | 758 | 3.5 | 98 | 4.5 | ||
*MMR: mismatch repair, TLS: translesion synthesis, NER: nucleotide excision repair, TCR: transcription-coupled repair, ALK: alkylation damage repair, BER: base excision repair, OXDR: oxidative damage repair; ∧ per genome per generation.
We have previously reported the mutation rates and spectra of BPSs in these experiments (16,17). We estimate the overall insertion rate of IS elements to be ∼3.5 × 10−4 per genome per generation, about 1/3 the rate of BPS in WT E. coli (i.e. ∼1 × 10−3 per genome per generation). The recombination rate of IS elements is estimated to be ∼4.5 × 10−5 per genome per generation, a magnitude lower than the IS insertion rate.
The rates of insertions and recombinations of all IS elements in each MA experiment are summarized in the Table 3. Using each MA experiment as a data point, we plotted the number of insertion events versus the summed number of generations for each experiment. As shown on Figure 4A, the IS insertion rate is roughly constant (∼3.5 × 10−4 insertions per genome per generation, R2 = 0.93, p = 2.78 × 10−9) across all MA experiments in this study. Considering that BPS mutation rates differ by as much as 100-fold between these strains, depending on their genetic background (17), the IS insertion rate does not seem to be affected by deficiencies in multiple DNA repair pathways. The insertion rate remains constant across MA experiments for each of the four most active IS families (IS1, IS2, IS5 and IS186 (Supplementary Figure S2); insertion rates of these families are estimated to be 1.7 × 10−4, 1.2 × 10−4, 0.4 × 10−4 and 0.4 × 10−4 per genome per generation for IS5, IS1, IS186 and IS2, respectively.
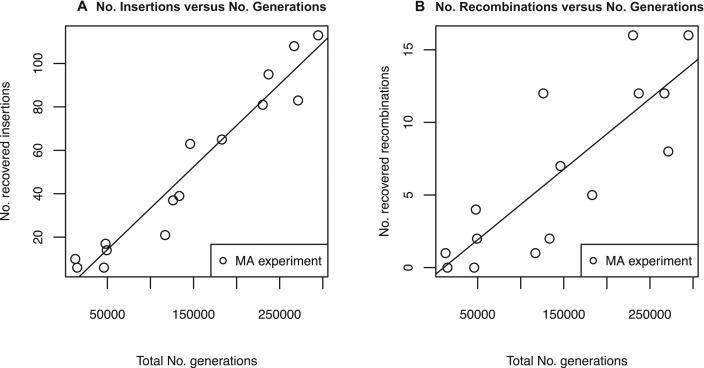
Figure 4.
IS insertion and recombination rates remain constant across MA experiment. Each data point represents the total number of novel IS insertions (A) and recombinations (B) detected in all MA lines in a single MA experiment (y-axis) versus the the total number of generations of these MA lines (Table 3). A linear regression line for insertion rate (A) is shown with a slope of ∼3.5 × 10−4 (R2 = 0.93, p = 2.78 × 10−9) and a linear regression line for recombination rate (B) is shown with a slope of ∼4.5 × 10−5 (R2 = 0.67, p = 1.59 × 10−4).
IS-associated deletions also exhibit a linear relationship with the number of generations across different MA experiments (see Figure 4B), although the data is noisier because fewer recombination events than insertions were observed in each MA experiment. Again, this implies that the IS-associated deletion rate remains constant across genetic background (∼4.5 × 10−5 recombinations per genome per generation, R2 = 0.67, p = 1.59 × 10−4).
Target-site duplication and insertion hotspots
For each detected insertion of an IS element of the four most active IS families (IS1A, IS2, IS5A and IS186) we reconstructed the TSD by performing a local assembly of reads that mapped to each end of the IS insertion and extracting the longest substring between the assembled contigs (see ‘Materials and Methods’ section). Out of the 733 detected insertions of IS1A, IS2, IS5A and IS186, we attempted to reconstruct the TSDs of 635 insertion sites where the inserted IS elements remain intact (i.e. not being deleted through recombination). Four hundred eighty-seven TSDs were successfully reconstructed, including 150 IS1A, 58 IS2, 220 IS5A and 59 IS186 transpositions. We then used the TSDs to produce a sequence logo for each IS family, using WebLogo 3.4 (39). No clear patterns were observed for IS2 and IS186. The TSDs of IS5 revealing a strong motif of 5′-(C/T)TA(A/G)-3′ (Supplementary Figure S4C), which is consistent with the 5′-YTAR-3′ pattern reported previously (9). IS1 has been characterized to have a TSD of 8 or 9 bps upon insertion, but no clear sequence motif has been previously reported, except a preference to insert at AT-rich regions (40); our analysis is consistent with these results (Supplementary Figure S4A and B).
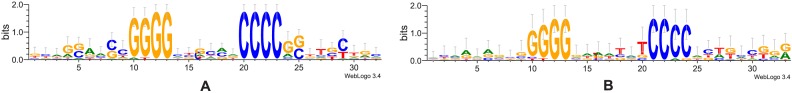
While IS186 does not have a conserved TSD sequence, there is a clear sequence motif flanking its insertion sites, 5′-GGGG(N6/N7)CCCC-3′, which consists of the palindromic 4mer (GGGG/CCCC) flanking the 6 or 7 core nucleotides, which are duplicated, (Figure 5). Among the 68 detected insertions of IS186 elements, we recovered the sequences of 59 insertion sites, in which 41 sites contain a 6-bp core, 16 have a 7-bp core, 1 have a 4-bp core and 1 have a 5-bp core; only 7 of them differ—by one or two bases—in the flanking palindrome (see Supplementary Table S4). Such a sequence pattern is rare in the E. coli genome: only 19 loci have 5′-GGGG(N6)CCCC-3′ and 35 loci have 5′-GGGG(N7)CCCC-3′. 37 IS186 insertion events occurred at 5′-GGGG(N6)CCCC-3′ sites and 13 insertion events occurred at 5′-GGGG(N7)CCCC-3′ sites. Not every putative insertion site (with the canonical motif) was hit by an insertion event while some sites were targeted multiple times. For instance, 16 independent insertion events occurred at a single 5′-GGGG(CCGCAA)CCCC-3′ site at 4 541 627 (see Supplementary Table S4 for details). Among the 19 5′-GGGG(N6)CCCC-3′ sites, 10 are found in non-coding regions and 9 in coding regions. Two insertion hotspots are in protein coding genes: 11 insertions in the menC gene and 16 insertions in the fimA gene. Because the flanking pattern (GGGG/CCCC) is palindromic, insertion events of IS186 occur on both strands (i.e. in either orientation) at a site (Supplementary Table S4). We conclude that unlike IS5 elements, the IS186 transposase recognizes the GC-rich flanking region, rather than the TSD sequence.
Figure 5.
Sequence logos of the reconstructed TSDs of IS186. Sequence logos with the 6 (A) and 7 (B) bps core are shown. The core sequences start at position 14 for both logos.
Insertion of IS elements is biased to non-coding regions
Among the 758 detected insertions, 470 occurred in protein coding sequences and 288 occurred in non-coding regions. Since only a small fraction (∼15%) of the E. coli genome is non-coding, this suggests IS elements insert more frequently in non-coding regions. To test the significance of this phenomenon, we used a binomial test to compare the actual results to the null hypothesis that IS insertion is equally likely in coding versus non-coding regions.
The results show that IS insertion is strongly biased to non-coding regions (p ∼0). Table 4 shows the insertion bias of the most active IS families, IS1A, IS2, and IS5A, are strongly biased to non-coding regions, and these families are most active in E. coli. In the case of IS5, the most active element, this bias is partially explained by the 2-fold enrichment of its insertion site motif, 5′-YTAR-3′, in coding regions (Table 5).
Table 4. IS insertions in coding versus non-coding regions.
| IS family | Total insertions | Coding | Non-coding | Biased to non-coding (P-value) |
|---|---|---|---|---|
| IS1A | 233 | 157 | 76 | 7.9 × 10−12 |
| IS1B | 3 | 1 | 2 | 0.06 |
| IS2 | 71 | 42 | 29 | 9.8 × 10−8 |
| IS3 | 10 | 6 | 4 | 0.05 |
| IS4 | 8 | 4 | 4 | 0.02 |
| IS5A | 361 | 206 | 155 | 1.1 × 10−37 |
| IS5B | 2 | 2 | 0 | 1 |
| IS150 | 2 | 1 | 1 | 0.27 |
| IS186 | 68 | 51 | 17 | 0.02 |
| Total | 758 | 470 | 288 | 5.9 × 10−55 |
Table 5. Occurrences of 5′-YTAR-3′ in coding versus non-coding regions.
| Total sites | CTAA | TTAG | CTAG | TTAA | Total | |
|---|---|---|---|---|---|---|
| Coding | 3 934 671 | 5617 | 5543 | 562 | 15 787 | 27 509 |
| Non-coding | 684 551 | 1938 | 1912 | 323 | 5389 | 9562 |
| Coding | 5′-YTAR-3′ per kb | 1.43 | 1.41 | 0.14 | 4.01 | 6.99 |
| Non-coding | 5′-YTAR-3′ per kb | 2.83 | 2.79 | 0.47 | 7.87 | 13.97 |
IS1 and IS2 elements do not have a specific insertion site motif.
Large-scale variations not involving IS elements
We also detected 48 long (>100 bps) deletions (2.2 × 10−5 per genome per generation) not involving IS elements in the MA experiments (Table 6; shown as white bands in Figure 1). Among these events, precise (3) deletion of the lambdoid cryptic prophage e14 occurred independently 21 times; the e14 prophage is known to excise and reintegrate via a 11-bp direct repeat cross-over site (41), which is what we observed. Among the 27 remaining deletions (1.2 × 10−5 per genome per generation), the sizes range from 126 to 11 337 bp, 20 were under 600 bp and most were under 300 bp. Fifteen out of 20 of these small deletions occurred in regions where tRNA genes or extragenic palindromic sequences (REP/RIP) elements are located.
Table 6. Deletions >100 bp detected in E. coli MA lines.
| Strain | Description | Deletions* (>100 bp) | e14 cryptic phage |
|---|---|---|---|
| PFM2 | WT @3K gen | 1 | 1 |
| PFM2 | WT @6k gen | 3 | 0 |
| PFM2 | WT on min medium | 9 | 5 |
| PFM5 | mutL | 0 | 0 |
| PFM101 | umuDC dinB | 2 | 2 |
| PFM133 | umuDC dinB polB | 3 | 2 |
| PFM35 | uvrA | 6 | 4 |
| PFM40 | alkA tagA | 4 | 0 |
| PFM88 | ada ogt | 6 | 0 |
| PFM180 | xthA nfo | 7 | 4 |
| PFM61 | mutT | 0 | 0 |
| PFM6 | mutY | 0 | 0 |
| PFM22 | nth nei | 1 | 1 |
| PFM91 | nfi | 4 | 2 |
| PFM94 | mutY mutM | 2 | 0 |
| Total | 48 | 21 |
*includes e14 cryptic phage deletions.
DISCUSSION
Through analysis of a large collection of WGSS data from MA lines we provide the first estimates of the genome-wide rates of three types of large-scale genome rearrangement in E. coli in a selection-free environment: 3.5 × 10−4 per genome per generation for novel IS insertions, 4.5 × 10−5 per genome per generation for deletions induced by recombination between homologous IS elements and 2.2 × 10−5 per genome per generation for deletions not involving IS elements. Novel IS insertions and IS-induced deletions estimates serve as baseline parameters for measuring the effects of selection in evolutionary studies. These are the two major types of genome rearrangements affecting genome size; other events are either very rare compared to these two events (e.g. only one excision event was detected throughout all MA lines over 3 million generations), or will not substantially change the genome size (e.g. BPS, short indels or genome inversions).
Although the IS insertions are almost 10-fold more frequent than IS-induced deletion, the deleted segments are typically longer than the average IS element. Our results are in contrast to the previously reported study of Touchon et al. (42), where they find gene gains are larger than gene loss, but losses occur more frequently. We suggest that their observations show the power of selection to bias fixation of the intrinsic mutation rate we have characterized. In fact, throughout all MA lines analyzed here, the total length of all novel IS insertions is ∼1.29 Mbps, while the total length of all deleted segments (induced by IS recombination) is ∼1.45 Mbps, 12.4% greater than the total insertion length. Thus, in the absence of selection, the E. coli genome would tend to shrink. Our results demonstrate cases of both local hopping and target-site immunity (reviewed in (36)). IS1A and IS5A transpose to relatively near-by sites, with new insertions showing significant clustering near pre-existing copies, frequently within 1 Kb of the assumed doner IS element. IS2 shows a different but particularly interesting pattern: insertions are rare near preexisting insertions, but are over-represented beyond a perimeter of 200 Kb. This pattern suggests a mechanism for long-range target-site immunity, similar to the target-site immunity of Tn7 which is at least 190 KB (43), but then a preference for insertion just beyond this perimeter, a pattern we are not aware of previously having been observed. Since the vast majority of IS-generated deletions are the product of one preexisting insertion and a new insertion (E + N), the distribution of these events to short intervals (Figure 3) is largely explained by local hopping, and does not require additional restraints.
We report that certain IS insertions preferentially target non-coding regions. This can be partially explained by the fact that IS target sites (e.g. IS186) are enriched in non-coding regions, which could be the consequence of an evolutionary interaction between IS elements and host (E. coli). Nevertheless, even after eliminating this bias, there is still an excess of IS insertions in non-coding regions, suggesting that selection against insertion (as well as IS-induced recombinations) occurred during the MA experiments. In the same MA experiments there was no bias between BPSs in coding versus non-coding regions, or between synonymous versus non-synonymous mutations (16). The greater selection against IS insertions and recombinations may be because these events inactivate the target genes completely, and are thus more likely to be deleterious. Alternatively, the preference for non-coding targets could also arise from some aspect of transcription or genomic structure. Thus our estimated rates of IS insertions/recombinations serves as lower bounds for the rates of these events in completely selection-free environments.
A striking observation in our study is that rates of IS insertions and recombinations are constant across the MA experiments with 13 E. coli strains, including the WT and 12 mutant strains each defective in a major DNA repair pathway (Table 3). Yet rates of BPSs of these strains differ by as much as 100-fold (16,17).
These results suggest that none of the DNA repair pathways has a substantial impact on the rates of IS insertion and recombination in E. coli. But these rates can be very different in different bacteria. We previously reported that in Deinococcus radiodurans the IS insertion rate is 2.7 × 10−3 per genome per generation (44), a magnitude higher than the rate in E. coli. The D. radiodurans genome contains ∼23 active IS elements, comparable to the number in E. coli. These species-specific differences in transposition remain to be explained; in E. coli we are determining rates for additional phylotypes, asking if there is within-species variation.
In this paper, we characterize the sequence pattern of the target sites of IS186 elements in E. coli. IS186 elements were previously known to insert into GC-rich regions (45), but the specificity of their insertion sites was unknown. The sequence pattern we characterized is surprisingly specific, and occurs only 54 times in the entire E. coli genome. Thus, it would seem relatively easy for the host genome to escape transposition of these elements by mutating these sites. In reality, however, the retained putative target sites the E. coli genome permit a relatively high activity of IS186 elements. It remains to be investigated if some of these putative target sites are selected for, and if IS186 insertion at these sites has a potential evolutionary advantage for E. coli.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US Army Research Office Multidisciplinary University Research Initiative Award [W911NF-09-1-0444 to P.L.F., in part]; National Science Foundation [DBI-1262588 to H.T.]. Funding for open access charge: US Army Research Office Multidisciplinary University Research Initiative Award [W911NF-09-1-0444 to P.L.F.].
Conflict of interest statement. None declared.
REFERENCES
- 1.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varshney R.K., Nayak S.N., May G.D., Jackson S.A. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol. 2009;27:522–530. doi: 10.1016/j.tibtech.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Brockhurst M.A., Colegrave N., Rozen D.E. Next-generation sequencing as a tool to study microbial evolution. Mol. Ecol. 2011;20:972–980. doi: 10.1111/j.1365-294X.2010.04835.x. [DOI] [PubMed] [Google Scholar]
- 4.Casjens S. The diverse and dynamic structure of bacterial genomes. Annu. Rev. Genet. 1998;32:339–377. doi: 10.1146/annurev.genet.32.1.339. [DOI] [PubMed] [Google Scholar]
- 5.Roth J.R., Benson N., Galitski T., Haack K., Lawrence J.G., Miesel L. Escherichia Coli and Salmonella: Cellular and Molecular Biology. Vol. 2. Washington D.C.: ASM Press; 1996. Rearrangements of the bacterial chromosome: formation and applications; pp. 2256–2276. [Google Scholar]
- 6.Naas T., Blot M., Fitch W.M., Arber W. Dynamics of IS-related genetic rearrangements in resting Escherichia coli K-12. Mol. Biol. Evol. 1995;12:198–207. doi: 10.1093/oxfordjournals.molbev.a040198. [DOI] [PubMed] [Google Scholar]
- 7.Romero D., Palacios R. Gene amplification and genomic plasticity in prokaryotes. Annu. Rev. Genet. 1997;31:91–111. doi: 10.1146/annurev.genet.31.1.91. [DOI] [PubMed] [Google Scholar]
- 8.Raeside C., Gaffé J., Deatherage D.E., Tenaillon O., Briska A.M., Ptashkin R.N., Cruveiller S., Médigue C., Lenski R.E., Barrick J.E., et al. Large chromosomal rearrangements during a long-term evolution experiment with Escherichia coli. Mbio. 2014;5:e01377–e01414. doi: 10.1128/mBio.01377-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahillon J., Chandler M. Insertion sequences. Microbiol. Mol. Biol. Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider D., Lenski R.E. Dynamics of insertion sequence elements during experimental evolution of bacteria. Res. Microbiol. 2004;155:319–327. doi: 10.1016/j.resmic.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Keightley P., Halligan D. Analysis and implications of mutational variation. Genetica. 2009;136:359–369. doi: 10.1007/s10709-008-9304-4. [DOI] [PubMed] [Google Scholar]
- 12.Phillips N., Salomon M., Custer A., Ostrow D., Baer C.F. Spontaneous mutational and standing genetic (co) variation at dinucleotide microsatellites in Caenorhabditis briggsae and Caenorhabditis elegans. Mol. Biol. Evol. 2009;26:659–669. doi: 10.1093/molbev/msn287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denver D.R., Dolan P.C., Wilhelm L.J., Sung W., Lucas-Lledó J.I., Howe D.K., Lewis S.C., Okamoto K., Thomas W.K., Lynch M., et al. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc. Natl. Acad. Sci. U.S.A. 2009;106:16310–16314. doi: 10.1073/pnas.0904895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ossowski S., Schneeberger K., Lucas-Lledó J.I., Warthmann N., Clark R.M., Shaw R.G., Weigel D., Lynch M. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sung W., Ackerman M.S., Miller S.F., Doak T.G., Lynch M. Drift-barrier hypothesis and mutation-rate evolution. Proc. Natl. Acad. Sci. U.S.A. 2012;109:18488–18492. doi: 10.1073/pnas.1216223109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee H., Popodi E., Tang H., Foster P.L. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E2774–E2783. doi: 10.1073/pnas.1210309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster P.L., Lee H., Popodi E., Townes J.P., Tang H. Determinants of spontaneous mutation in the bacterium Escherichia coli as revealed by whole-genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E5990–E5999. doi: 10.1073/pnas.1512136112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darmon E., Leach D.R. Bacterial genome instability. Microbiol. Mol. Biol. Rev. 2014;78:1–39. doi: 10.1128/MMBR.00035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hickman A.B., Dyda F. Mechanisms of DNA transposition. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0034-2014. doi:10.1128/microbiolspec.MDNA3-0034-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullany P., Allan E., Roberts A.P. Mobile genetic elements in Clostridium difficile and their role in genome function. Res. Microbiol. 2015;166:361–367. doi: 10.1016/j.resmic.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naito M., Pawlowska T.E. The role of mobile genetic elements in evolutionary longevity of heritable endobacteria. Mob. Genet. Elements. 2016;6:e1136375. doi: 10.1080/2159256X.2015.1136375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He S., Hickman A.B., Varani A.M., Siguier P., Chandler M., Dekker J.P., Dyda F. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. Mbio. 2015;6:e00762–e00815. doi: 10.1128/mBio.00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vincent A.T., Trudel M.V., Freschi L., Nagar V., Gagné-Thivierge C., Levesque R.C., Charette S.J. Increasing genomic diversity and evidence of constrained lifestyle evolution due to insertion sequences in Aeromonas salmonicida. BMC Genomics. 2016;17:1. doi: 10.1186/s12864-016-2381-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Renda B.A., Dasgupta A., Leon D., Barrick J.E. Genome instability mediates the loss of key traits by Acinetobacter baylyi ADP1 during laboratory evolution. J. Bacteriol. 2015;197:872–881. doi: 10.1128/JB.02263-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi J.W., Yim S.S., Kim M.J., Jeong K.J. Enhanced production of recombinant proteins with Corynebacterium glutamicum by deletion of insertion sequences (IS elements) Microb. Cell Fact. 2015;14:1. doi: 10.1186/s12934-015-0401-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker M. Structural variation: the genome's hidden architecture. Nat. Methods. 2012;9:133–137. doi: 10.1038/nmeth.1858. [DOI] [PubMed] [Google Scholar]
- 27.Lee H., Popodi E., Foster P.L., Tang H. Detection of structural variants involving repetitive regions in the reference genome. J. Comput. Biol. 2014;21:219–233. doi: 10.1089/cmb.2013.0129. [DOI] [PubMed] [Google Scholar]
- 28.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon S., Xuan Z., Makarov V., Ye K., Sebat J. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 2009;19:1586–1592. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lander E.S., Waterman M.S. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- 32.Barker C.S., Prüß B.M., Matsumura P. Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon. J. Bacteriol. 2004;186:7529–7537. doi: 10.1128/JB.186.22.7529-7537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Constantinidou C., Hobman J.L., Griffiths L., Patel M.D., Penn C.W., Cole J.A., Overton T.W. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J. Biol. Chem. 2006;281:4802–4815. doi: 10.1074/jbc.M512312200. [DOI] [PubMed] [Google Scholar]
- 34.Sonti R.V., Roth J.R. Role of gene duplications in the adaptation of Salmonella typhimurium to growth on limiting carbon sources. Genetics. 1989;123:19–28. doi: 10.1093/genetics/123.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Avila P., Grinsted J., De La Cruz F. Analysis of the variable endpoints generated by one-ended transposition of Tn21. J. Bacteriol. 1988;170:1350–1353. doi: 10.1128/jb.170.3.1350-1353.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Craig N.L. Target site selection in transposition. Annu. Rev. Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 37.Retallack D.M., Johnson L.L., Friedman D.I. Role for 10Sa RNA in the growth of lambda-P22 hybrid phage. J. Bacteriol. 1994;176:2082–2089. doi: 10.1128/jb.176.7.2082-2089.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyons E., Freeling M., Kustu S., Inwood W. Using genomic sequencing for classical genetics in E. coli K12. PLoS One. 2011;6:e16717. doi: 10.1371/journal.pone.0016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer J., Iida S., Arber W. Does the insertion element IS1 transpose preferentially into A+ T-rich DNA segments? Mol. Gen. Genet. 1980;178:471–473. doi: 10.1007/BF00270502. [DOI] [PubMed] [Google Scholar]
- 41.Greener A., Hill C. Identification of a novel genetic element in Escherichia coli K-12. J. Bacteriol. 1980;144:312–321. doi: 10.1128/jb.144.1.312-321.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Touchon M., Hoede C., Tenaillon O., Barbe V., Baeriswyl S., Bidet P., Bingen E., Bonacorsi S., Bouchier C., Bouvet O., et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeBoy R.T., Craig N.L. Tn7 transposition as a probe of cis interactions between widely separated (190 kilobases apart) DNA sites in the Escherichia coli chromosome. J. Bacteriol. 1996;178:6184–6191. doi: 10.1128/jb.178.21.6184-6191.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Long H., Kucukyildirim S., Sung W., Williams E., Lee H., Ackerman M., Doak T.G., Tang H., Lynch M. Background mutational features of the radiation-resistant bacterium Deinococcus radiodurans. Mol. Biol. Evol. 2015;32:2383–2392. doi: 10.1093/molbev/msv119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chong P., Hui I., Loo T., Gillam S. Structural analysis of a new GC-specific insertion element IS186. FEBS Lett. 1985;192:47–52. doi: 10.1016/0014-5793(85)80040-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.