SUMMARY
The Zinc finger protein of the cerebellum 2 (Zic2) is one of the vertebrate homologs of the Drosophila pair-rule gene odd-paired (opa). Our molecular and biochemical studies demonstrate that Zic2 preferentially binds to transcriptional enhancers and is required for the regulation of gene expression in embryonic stem cells. Detailed genome-wide and molecular studies reveal that Zic2 can function with Mbd3/NuRD in regulating the chromatin state and transcriptional output of genes linked to differentiation. Zic2 is required for proper differentiation of ES cells, similar to what has been previously reported for Mbd3/NuRD. Our study identifies Zic2 as a key factor in the execution of transcriptional fine-tuning with Mbd3/NuRD in ES cells through interactions with enhancers. Our study also points to the role of the Zic family of proteins as enhancer-specific binding factors functioning in development.
INTRODUCTION
Pluripotent cells possess nearly unlimited capacity for replicating themselves, while maintaining the potential to give rise to a broad spectrum of differentiated progenies (Gurdon and Melton, 2008; Hanna et al., 2010; Jaenisch and Young, 2008). The diverse interplay of transcription factors and enhancers in the permissive chromatin of embryonic stem (ES) cells is the primary determinant of pluripotency, leading to spatiotemporal specificity in gene expression upon differentiation (Bulger and Groudine, 2011; Lin et al., 2013). Enhancers are a specific group of cis-acting regulatory DNA elements that orchestrate the expression of their associated genes independently of distance and orientation (de Laat and Duboule, 2013; Levine, 2010; Smith and Shilatifard, 2014; Spitz and Furlong, 2012; Visel et al., 2009).
Recent genome-wide studies in pluripotent cells have revealed that active enhancers are marked with the histone modifications H3 lysine 4 monomethylation (H3K4me1) and histone H3 lysine 27 acetylation (H3K27ac), while poised enhancers are associated with H3K4me1 alone or with H3K27 trimethylation (H3K27me3) (Creyghton et al., 2010; Heintzman et al., 2009; Rada-Iglesias et al., 2011). The different signatures of enhancers are set up through crosstalk among chromatin modifying enzymes. The COMPASS family members Mll3 and Mll4 are required for the maintenance of H3K4me1 at enhancers and enhancer promoter communication during development (Ardehali et al., 2011; Herz et al., 2014; Herz et al., 2012; Hu et al., 2013; Lee et al., 2013). The H3K4 demethylase LSD1 functions together with the nucleosome remodeling and histone deacetylation (NuRD) complex in deactivating enhancers during cell stage transitions (Whyte et al., 2012). NuRD is a unique chromatin-remodeling complex possessing both chromatin opening and closing activities implemented by Chd4 and Hdac1/2 activities, respectively (Allen et al., 2013; Wade et al., 1998; Xue et al., 1998). Mbd3, a core component of NuRD, is essential for the pluripotency of mouse ES cells (Kaji et al., 2006; Kaji et al., 2007).
Prior to the function of the histone modifiers, DNA binding transcription factors typically occupy enhancers at different stages, building the landscape of the cis-regulatory elements, entailing the recruitment of chromatin remodelers and transcriptional modulators (Deng and Blobel, 2010; Levine, 2010; Ostuni et al., 2013; Pennacchio et al., 2013; Zaret and Carroll, 2011). For instance, the pioneer transcription factor FoxA1 was found to direct estrogen receptor alpha (ERα) to genomic regions in mammary epithelial cells (Hurtado et al., 2011). The Brg1 chromatin-remodeling complex is recruited to oligodendrocyte-specific enhancers predefined by Olig2 to activate the expression of myelination-associated genes (Yu et al., 2013). The activation of latent enhancers in terminally differentiated macrophages also requires stimulus-activated transcription factors (Ostuni et al., 2013).
Zinc finger protein of the cerebellum (Zic) family proteins have previously been implicated as major regulators of neuroectodermal development from Drosophila to humans (Houtmeyers et al., 2013). In this study, we demonstrate that Zic2 is enriched at the enhancers of both active and poised genes in ES cells. We also biochemically identify Zic2 as a cofactor of the Mbd3-NuRD complex, co-occupying enhancer regions genome-wide and functioning together in regulating the ES cell chromatin state and gene expression. While Mbd3/NuRD is essential in the maintenance of ES cell pluripotency, depletion of Zic2 in ES cells leads to impaired lineage commitment. The identification of the DNA binding factor Zic2 as a novel interaction partner of NuRD in ES cells reveals Zic2 as a critical factor for NuRD’s role in chromatin modification in ES cells and in the regulation of cellular differentiation, while also providing a novel direction in exploring the etiology human diseases such as holoprosencephaly resulting from mutation of Zic2 (Brown et al., 1998).
RESULTS
Zic2 occupies enhancers in mouse ES cells
The Zic family, comprised of Zic1-5 in mammals, is a group of C2H2-type zinc finger proteins that play versatile roles during development and disease pathogenesis (Houtmeyers et al., 2013). Zic2 is a master regulator of neurogenesis, however, the expression of Zic2 can be easily detected in the early embryo stage of fertilized zygotes, largely overlapping with the expression pattern of the pluripotent marker Oct3/4, well before the induction of the neuronal system (Brown and Brown, 2009). To investigate the function of Zic2 in early development, we carried out Zic2 ChIP-seq in mouse ES cells. Our analysis identified 10,273 significant binding sites, 83% of which are present in TSS distal regions (Figure 1A). Zic2 is found with the active enhancer signature of p300, H3K4me1 and H3K27ac at enhancers of the active Tbx3 gene, or with H3K4me1 alone at enhancers of the poised gene Gpr83 in ES cells (Figure 1B). The depletion of Zic2 in ES cells leads to the derepression of both of these genes (Figure S1B). Zic2 also occupies the regulatory elements near the key stem cell genes Pou5f1, Nanog, Sox2 and Klf4 (Figure S1A), although their expression is largely unaffected by Zic2 knockdown (Figure S1B). As a result, the ability of ES cells to form alkaline phosphatase positive clones is not affected by Zic2 knockdown (Figure S1C). Using the H3K27ac mark to distinguish active and poised enhancers, we find that Zic2 occupies both classes, with 44% of non-TSS Zic2 peaks having an active enhancer signature (Figure 1C). Gene ontology analysis demonstrates that genes nearest to non-TSS Zic2 peaks are involved in various developmental processes including embryonic morphogenesis and neuron differentiation (Figure 1D).
Figure 1. Zic2 occupies enhancers in mouse ES cells.
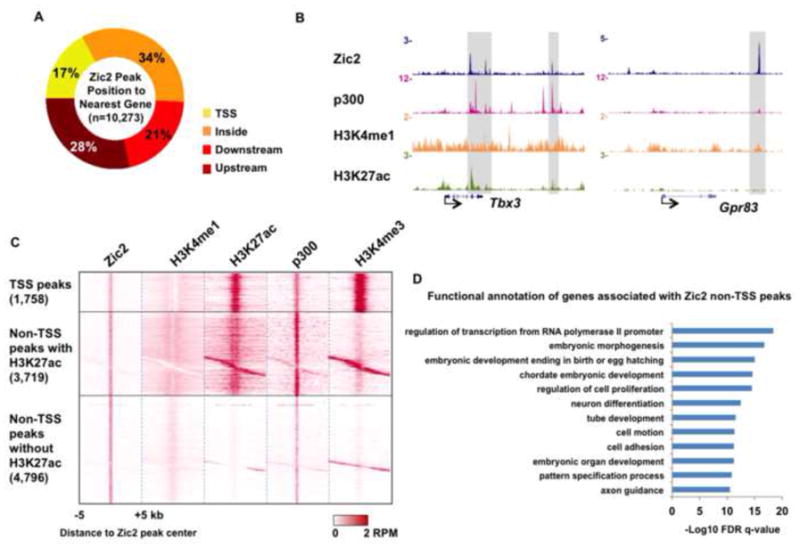
(A) Pie chart demonstrating the percentage of Zic2 peaks overlapping with a transcription start site (TSS), residing within a gene (inside), or upstream or downstream of the nearest gene. (B) Genome browser track examples of the colocalization of Zic2 with the enhancer marks H3K4me1, H3K27ac and/or p300 at active Tbx3 and inactive Gpr83 genes. The co-bound regions are highlighted with gray bars. (C) Heat map of binding profiles in mouse embryonic stem cell for Zic2, H3K4me1, H3K27ac, p300 (Creyghton et al., 2010) and H3K4me3 (Mikkelsen et al., 2007) are shown within 5 kb of the center of Zic2 peaks, which were partitioned into three groups, TSS, non-TSS with the presence of H3K27ac, and non-TSS without H3K27ac. (D) GO term analysis of genes bound by Zic2 non-TSS peaks were performed with DAVID (Dennis et al., 2003). Benjamini-Hochberg FDR was used to determine significance of enrichment of terms. The peaks were assigned to the nearest genes regardless of the actual distance. Zic2 ChIP-seq was done in duplicates. The common peaks detected in the duplicate studies were used for genome-wide studies. See also Figure S1.
Biochemical interactions of Zic2 with the Mbd3-containing NuRD complex in ES cells
To investigate the potential mechanism by which Zic2 functions in regulating transcription, we sought to identify Zic2 interacting proteins in mouse ES cells. We generated a stable ES cell line expressing Zic2 protein with a Flag tag under a tetracycline-inducible promoter. The Benzonase nuclease was used during all purifications to avoid DNA and RNA-dependent interactions. Flag-Zic2 and control purifications were analyzed by silver staining (Figure 2A). Analysis of five independent purifications using Multidimensional Protein Identification Technology (MudPIT) resulted in the identification of all of the core components of the Mbd3-containing NuRD complex (Le Guezennec et al., 2006), including Chd4, Mta1/2 and Hdac1/2, at similar levels (Figure 2B and S2A). The co-purification of Chd4, Sall4, Mta2 and Mbd3 with Zic2 in ES cells was also validated by Western blotting (Figure S2B). Furthermore, reciprocal co-immunoprecipitation experiments confirmed the endogenous interaction between Zic2 and NuRD subunits (Figure 2C–D). Since Mbd3-NuRD is an abundant complex, and since relatively low levels of NuRD are purified with Zic2, we performed reciprocal co-immunoprecipitation experiments and confirmed the endogenous, but substochiometric interaction between Zic2 and NuRD subunits (Figure 2C–D). We also examined the co-fractionation of Zic2 with components of NuRD including Mta2, Mbd3 and Mbd2. The elution pattern of Mta2 and Mbd3 appeared to be similar, but only overlapped that of Mbd2 to a limited extent (Figure 2E). This is consistent with previously published studies indicating that Mbd2 and Mbd3 are mutually exclusive in NuRD (Gunther et al., 2013; Le Guezennec et al., 2006) and that Mta2 can preferentially associate with the Mbd3 form of NuRD (Zhang et al., 1999). The co-fractionation of Zic2 with Mbd3 is much more prominent than with Mbd2 (Figure 2E), which is in line with the MudPIT data showing that all of the six Mbd3 peptides identified in the Zic2 purification are unique to Mbd3 (Figure S2A), while no Mbd2 peptides are detected in Zic2 purifications. This is further supported by the detection of Zic2 in Mbd3, but not Mbd2, immunoprecipitates (Figure 2C). Unlike Mbd3, which is required for the assembly of the NuRD complex and for the stability or expression of some of its subunits (Kaji et al., 2006), Zic2 knockdown has no apparent effect on the stability of NuRD subunits (Figure S2C–D).
Figure 2. Zic2 interacts with the Mbd3-containing NuRD complex in embryonic stem cells.
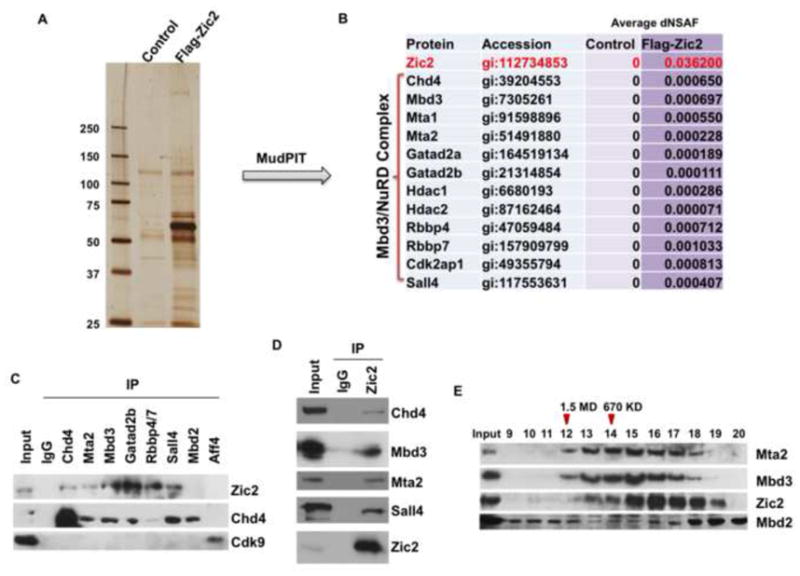
(A) A clonal ES cell line expressing Flag-tagged Zic2 was generated, and the resulting protein complexes were affinity purified and analyzed by SDS-PAGE, silver staining and mass spectrometry. (B) MudPIT analysis of the relative abundance of Zic2 and Mbd3-NuRD subunits are shown. dNSAF values represent the distributed normalized spectral abundance factor. All the core components of Mbd3-containing NuRD (Chd4, Mbd3, Mta1/2, Gatad2a/b, Hdac1/2, Rbbp4/7 (Allen et al., 2013) as well as known NuRD-associated proteins Cdk2ap1 and Sall4 (Kloet et al., 2014; Lu et al., 2009; Spruijt et al., 2010) were reproducibly identified in the Flag-Zic2 purifications. (C–D) Endogenous immuneprecipitations (IP) followed by Western blotting confirm the interaction of Zic2 with subunits of the Mbd3-NuRD complex. (C) NuRD subunit IP followed by Zic2 Western blotting. Aff4, which interacts with Cdk9 within the Super Elongation Complex, served as a negative control for the IP experiments. (D) Zic2 IP followed by Mbd3-NuRD subunit Western blotting. (E) Size exclusion chromatography of mouse ES cell nuclear extracts demonstrated that the elution pattern of Zic2 overlaps to a larger extent with Mbd3 and Mta2 than with Mbd2. Elution profile corresponding to 1.5 MDa and 670 KDa are indicated with arrowheads. See also Figure S2.
Zic2 and NuRD-dependent transcriptional activity in mouse ES cells
To further gain insight into the function of Zic2 and Mbd3-NuRD, we mapped the genomic distribution of the core component Mbd3 and the remodeling factor Chd4 in ES cells. Both Chd4 and Mbd3 are predominantly found at non-TSS regions (Figure S3A–D). Through the ChIP-seq analyses, we found that 11,058 and 7,421 genomic sites are bound by Chd4 and Mbd3 in ES cells, respectively (Figure S3A–B). 3,773 peaks are co-occupied by both Chd4 and Mbd3 (Figure S3E). However, about half of the Mbd3 peaks and two thirds of the Chd4 peaks are mutually exclusive of each other (Figure S3E). This could result from the fact that multiple versions of NuRD could exist, potentially consisting of different combinations of Chd3 or Chd4; Mbd2 or Mbd3; Mta1, Mta2, or Mta3. Therefore, it is conceivable that Chd4 and Mbd3 are independent of each other at some genomic loci. Additionally, chromatin regulators such as NuRD can be difficult to ChIP due to their indirect interaction with DNA and transient nature of their interaction with chromatin during remodeling of nucleosomes (Hu and Wade, 2012; Ram et al., 2011). Therefore low signal-to-noise ratios obtained by Chd4 and Mbd3 ChIP-seq might lead to an underestimation of the convergence between these two factors (Figure S3C–D). Nonetheless, of the 3,773 Chd4-Mbd3 co-bound sites, 2,047 of these are also bound by Zic2 (Figure S3E). Zic2-Mbd3-Chd4 co-bound sites were found at both active and poised/inactive genes with H3K27me3 (Figure 3A). Sites with Zic2, but without Mbd3-Chd4, are more enriched for H3K27me3 (Figure 3A).
Figure 3. Zic2 colocalizes with Mbd3-NuRD Complex in ES Cells and functions in regulating gene expression.
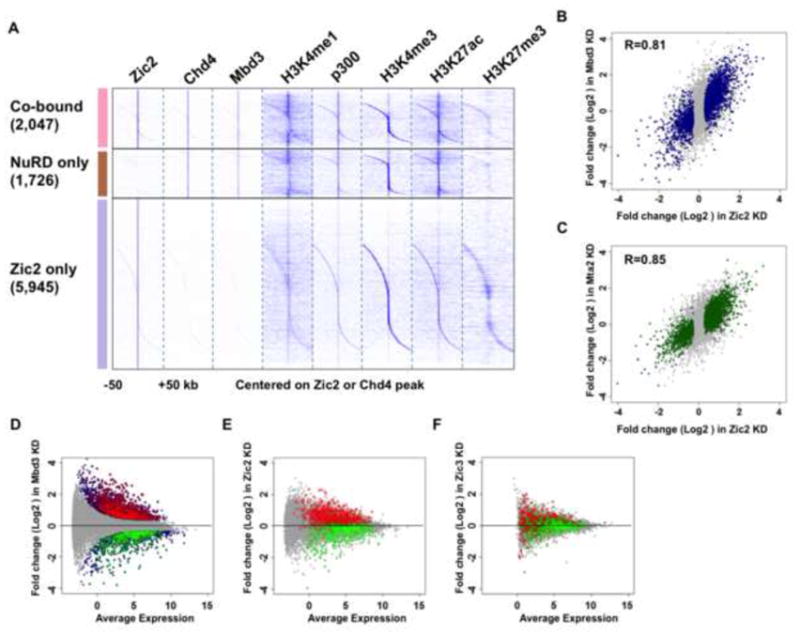
(A) Binding profiles in mouse ES cells demonstrating the presence of the indicated histone modifications at Zic2, Chd4 and Mbd3 co-bound regions (2,047 regions, highlighted in pink), Chd4-Mbd3 only regions (1,726 regions, highlighted in brown) and Zic2 only regions (5,945 regions, highlighted in purple) within 50 kb around the Zic2 or Chd4 peak. H3K4me1 and p300 data are from (Creyghton et al., 2010), and H3K4me3 and H3K27me3 data are from (Mikkelsen et al., 2007). (B–C) Scatter plots demonstrating differentially expressed genes, as assessed by RNA-seq in Zic2 knockdown versus Mbd3 (B) and Mta2 (C) knockdowns. Genes significantly misregulated are shown in blue and green, respectively. Correlation coefficients are shown. RNA-seq after Zic2 knockdown was performed and analyzed in four replicates. RNA-seq after Mbd3 or Mta2 knockdown was performed and analyzed in duplicates. (D–F) MA plots demonstrating differentially expressed genes after (D) Mbd3, (E) Zic2 and (F) Zic3 knockdown in ES cells. Significantly changed genes after Mbd3 knockdown are shown in blue. Genes up-regulated in both Mbd3 knockdown and knockout datasets are highlighted with red circles, and genes down-regulated in both Mbd3 knockdown and knockout datasets are highlighted with green circles. Genes regulated by Mbd3 were changed in the same direction after Zic2, but not Zic3, knockdown. See also Figure S3 and Table S1.
To further investigate the functional relationship between Zic2 and the Mbd3-NuRD complex, we measured differential gene expression using RNA-seq upon Mbd3 or Mta2 knockdown. Known Mbd3-repressed genes, including Tbx3, Htra1 and Ppp2r2c, are also up-regulated after Mta2 and Zic2 depletion (Reynolds et al., 2012b) (Figure S1B and S3F–G). The correlation in gene expression between the Zic2 knockdown and Mbd3 or Mta2 ranged from 0.81–0.85 (Figure 3B–C). In order to validate our differential gene expression analyses, we analyzed published mRNA-Seq data from Mbd3 wild-type and knockout ES cells (Reynolds et al., 2012a). The 891 genes up-regulated in both Mbd3 knockout and knockdown datasets show a generally increasing trend in expression after Zic2 RNAi, with 512 out of the 891 genes (~58%) being significantly up-regulated after Zic2 RNAi. Similarly, a large portion of genes (43%) down-regulated in both datasets show significantly reduced expression after Zic2 RNAi (Figure S3H). The genes regulated by Mbd3 were changed in the same direction upon knockdown of Zic2, but not upon knockdown of Zic3, another Zic family member that is highly expressed in ES cells (Figures 3D–F). Furthermore, the fold changes in gene expression in Zic2 knockdown and Mbd3 knockout cells also show a high concordance (Figure S3I).
The majority of Zic2-Chd4-Mbd3 co-bound genes that were significantly up or down regulated upon Zic2 loss were changed in the same direction after either Mbd3 or Mta2 knockdown (Figure S3J). 44% of the Zic2 and Mbd3 co-regulated genes are bound by Zic2 and/or Mbd3 only at their nearest enhancer regions, or they are found at both the enhancer and TSS regions. However, less than 10% of co-regulated genes are occupied by either of these two factors only at the TSS region (Table S1). This suggests a dominant role for Zic2/NuRD’s occupancy at enhancers for the regulation of gene expression.
Consistent with a previous study showing that H3K27me3 levels at bivalent genes are reduced in Mbd3 null ES (Reynolds et al., 2012b), knockdown of Zic2 also leads to reduced H3K27me3 (Figure 4A). Importantly, H3K27me3 changes resulting from depletion of these two factors in ES cells are highly correlated (Figure 4B). Prominent examples of loci marked with H3K27me3 in ES cells are the Hox clusters. We observed that upon knockdown of Zic2, H3K7me3 is broadly reduced in the Hox C cluster (Figure 4C). In contrast, H3K27ac is increased at the region between Hoxc12 and Hoxc13 that could be regulated through putative enhancers in the Hox C cluster bound by Zic2/NuRD (Figure 4C). However, H3K4me1, which is also enriched across the Hox C cluster in ES cells, is unchanged upon Zic2 knockdown (Figure 4C). Furthermore, bulk levels of H3K4me1, H3K27me3 and H3K27ac remained unchanged in Zic2 knockdown ES cells (Figure S4). Together these data indicate that Zic2 is a major co-repressor through its biochemical association with NuRD-bound enhancers.
Figure 4. Similar to Mbd3, Zic2 is required for the maintenance of H3K27me3 chromatin state and transcriptional repression of the homeotic cluster in ES cells.
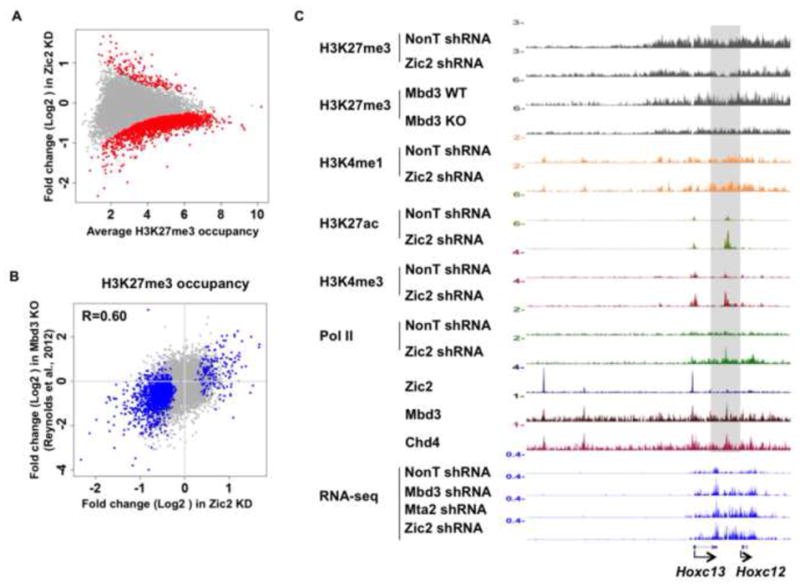
(A) MA plot demonstrating differential enrichment of H3K27me3 after Zic2 knockdown in ES cells. Significantly changed H3K27me3 bound regions are shown in red. (B) Scatter plot demonstrating differential occupancy of H3K27me3 in Zic2 knockdown versus Mbd3 knockout (Reynolds et al., 2012b). The H3K27me3 bound regions significantly misregulated are shown in blue. Correlation coefficients are shown. (C) Histone H3K27me3 is broadly reduced across the Hox clusters. The Hox C cluster is shown as an example. ChIP-seq track files of H3K27me3 occupancy in control (NonT) and Zic2 knockdown cells, and Mbd3 WT and KO cells in the posteriors Hox C region. Other histone marks and Pol II occupancy in control and Zic2 knockdown cells are also shown. RNA-seq track files of a transcript located within the posterior Hox C region with upregulated expression in Mbd3, Mta2 and Zic2 RNAi-treated cells are also shown. H3K27me3 ChIP-seq after Zic2 knockdown was performed and analyzed in duplicates. H3K27me3 ChIP-seq in Mbd3 WT and KO ES cells (Reynolds et al., 2012b) were analyzed in duplicates. See also Figure S4.
Zic2 is essential for the pluripotency of ES cells
We identified the previously known nervous system target of Zic2, EphA4, (Escalante et al., 2013), as well as neuronal lineage genes Fgf5 and Otx2, as being co-occupied by Zic2 and NuRD in ES cells (Figure 5A). In order to investigate whether Zic2 phenocopies the function of Mbd3-NuRD in lineage specification, the control, Zic2, or Mbd3-depleted ES cells were differentiated into embryoid bodies (EBs) for 2, 4 and 8 day periods, and expression levels were measured by RT-qPCR. We find that the differentiation-induced expression of these genes is significantly impaired by the depletion of either Zic2 or Mbd3, suggesting a shared function in pluripotency (Figure 5B). Furthermore, our genome wide analyses indicate that more than 33% of the 2,862 genes induced in day 5 EBs (Lin et al., 2013) are bound by Zic2 at the undifferentiated states, suggesting the possibility that Zic2 has a broad role in marking the enhancers of genes in the ES state which are to be activated later in the development. To specifically evaluate the requirement of Zic2 in neural differentiation, control and Zic2 knockdown ES cells were cultured in N2B27 media, which can induce the differentiation of ES cells towards neuronal lineage (Ying et al., 2003). After 5 days of culture in N2B27 media, the expression of the pluripotency gene Pou5f1 is largely reduced, while markers of the neuronal lineage such as Tubb3 and Nestin are activated (Figure S5A). However, knockdown of Zic2 leads to cell death during the N2B27 differentiation process, indicating a critical role of Zic2 in neural differentiation (Figure S5B).
Figure 5. Zic2 and ES cells pluripotency.
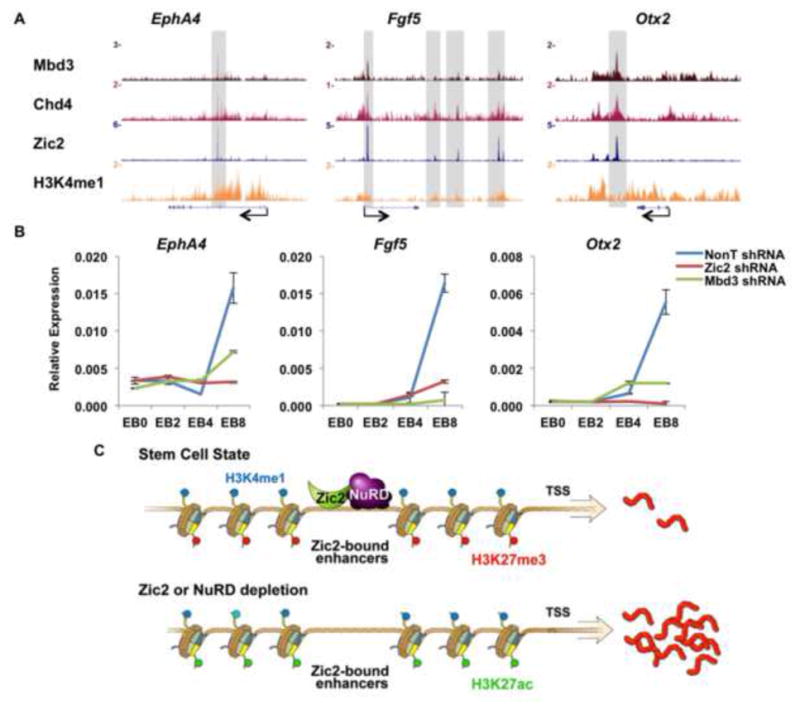
(A) ChIP-seq track examples of Zic2, Chd4, Mbd3-occupied lineage-specific genes in the ES cell state. Zic2 is found with NuRD at putative enhancers for EphA4, Fgf5 and Otx2. (B) qRT-PCR analyses of the activation kinetics of lineage-specific genes in control (NonT), Zic2, or Mbd3 knockdown embryoid bodies (EBs). Control, Zic2, or Mbd3 knockdown ES cells were induced to form EB for the indicated time points. The expression levels were normalized to Actb. Error bars represent the s.d. of independent measurements of a representative experiment. (C) Model: In the stem cell state (top panel), the zinc finger-containing protein Zic2 functions together with the NuRD repressor complex, setting a poised, yet to be activated, state of developmental genes. Knockdown of Zic2 (bottom panel) and/or NuRD subunits leads to derepression of the same group of developmental genes. See also Figure S5.
DISCUSSION
Here, we report our identification of Zic2 as an enhancer-bound factor that biochemically interacts with the Mbd3-NuRD complex in ES cells, and regulates ES cell chromatin and gene expression (Figure 5C). Specifically, we demonstrated that (1) over 80% of Zic2-bound sites are non-TSS sites exhibiting a chromatin signature of enhancers; (2) purification of Zic2 and subsequent proteomic analyses identified core components of Mbd3-NuRD, as well as NuRD-associated proteins Cdk2ap1 and Sall4 as interactors; (3) knockdown of Zic2 affects the expression of known ES cell targets of Mbd3-NuRD; (4) changes in H3K27me3 in Zic2 RNAi ES cells are correlated with changes in H3K27me3 observed in Mbd3 knockout ES cells; and (5) Zic2 and Mbd3 are required for proper expression of lineage specific genes.
The five members of the Zic family in mammals are characterized by possessing a conserved zinc finger domain encompassing five zinc fingers which mediate interactions with DNA and proteins and have partially overlapping expression patterns in tissues of developing embryos and adults, particularly in the nervous system (Houtmeyers et al., 2013). In mouse ES cells both Zic2 and Zic3 are highly expressed. However, our genome wide binding and expression analyses demonstrate that these two members function non-redundantly despite the high level of homology in the zinc finger regions. Therefore, the Zic family of proteins may have functional diversity even in cells/tissues where their expression patterns overlap. In both human and mouse models, heterozygous mutations in Zic2 lead to holoprosencephaly, indicating an indispensable function of Zic2 in the development of the brain (Houtmeyers et al., 2013). There is only one Zic family member in Drosophila, encoded by the pair-rule gene odd-paired (opa) (Houtmeyers et al., 2013). Unlike other pair rule genes, which are expressed in alternating parasegments, opa is equally expressed throughout the segmented region of the embryo. This suggests that Opa coordinates with more spatially restricted transcription factors to positively and negatively regulate gene expression (Benedyk et al., 1994).
Our biochemical and genome-wide data reveal a role for Zic2 in functioning with Mbd3-Chd4-NuRD in ES cells. However, multiple versions of NuRD can exist, for example, Mbd2 vs. Mbd3-containing NuRD or Chd3 vs. Chd4-containing NuRD. In mouse ES cells the expression level of Mbd2 and Chd3 is lower than Mbd3 and Chd4. Therefore, the Mbd3-Chd4 version of NuRD could be a major form of NuRD in ES cells. Although we are not able to detect the interaction of Zic2 with Mbd2 or Chd3 in ES cells, we cannot rule out the possibility that Zic2 may partner with Mbd2 or Chd3 in tissues in which these proteins are highly expressed or modified in a way that can interact with Zic2.
Both our biochemical data and genome-wide occupancy studies indicate that Zic2 and NuRD are found together at a subset of genomic loci (Figure S3E). Only a low percentage of NuRD is pulled down with Zic2 (Figure 2A–B) and that Zic2 co-elutes with a fraction of Mbd3 and Mta2 by size exclusion chromatography (Figure 2D). The existence of NuRD only peaks is consistent with NuRD’s interacting with numerous other DNA-binding transcription factors. Zic2 “only” peaks tend to be found at genomic regions with low levels of H3K4me1, p300 and H3K27ac (Figure 3A), and these might be cis-regulatory elements that might function at later developmental stages. In contrast, the Zic2 “together with NuRD” elements are most likely to be responsible for the transcriptional and chromatin changes observed upon depletion of either Zic2 or Mbd3-NuRD subunits.
Previously it was proposed that Mbd3-NuRD facilitates the recruitment of PRC2 to deposit and maintain H3K27me3 in ES chromatin. However, a biochemical/physical interaction between PRC2 and NuRD was not detected in a previous study (Reynolds et al., 2012b). Likewise, although our genome-wide H3K27me3 ChIP-seq analysis shows remarkable correspondence between Zic2 and NuRD depletion, we did not find components of PRC2 in our Zic2 purifications. An interaction with PRC2 could be dynamic or dependent on co-occupancy on chromatin without direct protein-protein interactions between Zic2 and PRC2 components. These potential modes of interaction would be lost due to our high salt buffer and benzonase treatments used throughout the nuclear protein extraction and biochemical purifications.
Zic2 and NuRD also co-localize at active enhancers marked with H3K27ac. NuRD has previously been implicated in the decommissioning of enhancers during ES cell differentiation (Whyte et al., 2012). It is possible that the observed co-occupancy of Zic2 with the Mbd3-NuRD at these active enhancers facilitates the decommissioning of these enhancers during lineage specification. Furthermore, Zic2 was previously found to function as a transcriptional repressor of dopamine receptor D1 in the cerebellum (Yang et al., 2000) and the downstream targets of the β-catenin-TCF4 complex (Pourebrahim et al., 2011). Our identification of a functional linkage between Zic2 and NuRD provides a possible mechanism for Zic2’s regulation of the expression pattern of neuronal-linage genes in adults, and potentially the etiology of holoprosencephaly caused by mutation of Zic2.
A recent study suggests that the transcription factor Otx2 is essential in directing Oct4 to the Fgf5 enhancers to upregulate its transcription during differentiation of mouse ES cells into epiblast-like cells. At the ES cell stage, when the epiblast-like cell-specific enhancers of Fgf5 are unmarked by Otx2 (Buecker et al., 2014), Zic2 is already found occupying these regions (Figure 5A). Furthermore, these enhancers are enriched for the DNA binding motif for the Zic family members (Buecker et al., 2014). This, together with the data showing that Zic2 in ES cells already occupies these and other non-TSS loci lacking H3K4me1 and p300 (Figure 1C), suggests an early bookmarking function of Zic2 in addition to its roles later in development. Our study also suggests that transcriptional fine-tuning in ES cells, as seen for the role of Zic2/NuRD in restricting transcription between the Hoxc12 and Hoxc13 loci (Figure 4C), is a process that is regulated at the enhancer level. Further studies of these factors should shed further light on their role in regulating this intricate molecular process through their activities at cis-regulatory elements/enhancers.
EXPERIMENTAL PROCEDURES
ES Cell Culture and Differentiation
Mouse ES cells V6.5 were cultured on irradiated mouse embryonic fibroblast (MEF) feeder layers in 0.1% gelatin-coated tissue culture plate. Cells were grown in DMEM (D6546, Sigma) supplemented with 15% ES-certified fetal bovine serum (Hyclone), 2 mM L-glutamine, 0.1 mM nonessential amino acids, 0.1 mM β-mercaptoethanol, and recombinant LIF (Millipore). Zic2 cDNA was amplified and cloned into pBS31 with N terminal Flag tag. The expression vector was transfected in KH2 cells and followed by selection with hygromycin. The expression of Flag-Zic2 protein was induced with 1 μg/ml tetracycline for 24 to 36 h. Mouse Zic2 (SHCLNG-NM_009574), Zic3 (SHCLNG-NM_003413), Mbd3 (SHCLNG-NM_013595) and Mta2 (SHCLNG-NM_011842) shRNA constructs were purchased from Sigma. The Non-targeting (NonT) shRNA construct (SHC002) was purchased from Sigma. For ChIP and RNA analysis, cells were grown for one passage off feeders on tissue culture plates for 30 minutes. Lentivirual infection, embryoid bodies (EBs) formation and N2B27 monolayer neuronal differentiation were performed as previously described (Lin et al., 2013; Ying et al., 2003).
Antibodies
The antibodies to: Zic2 (ab150404), Zic3 (ab80966), H3K27ac (ab4729), Chd4 (ab70469), Sall4 (ab29112), Mbd2 (ab38646) were purchased from Abcam. The antibodies to: Mbd3 (A302-528A), Gatad2b (A301-283A), Chd4 (A301-081A), and Mta2 (A300-395A) were purchased from Bethyl. The antibodies to Rbbp4/7 (sc-33170) and RNA Pol II (sc-899) were purchased from Santa Cruz. Antibodies against H3K4me1, H3K4me3, Aff4 and Cdk9 were generated in our laboratory and were described previously (Hu et al., 2013; Lin et al., 2010; Luo et al., 2012). H3 (antigen full length yeast H3) and H3K27me3 (antigen: LASKAARK(me3)SAPST) were also generated in our laboratory.
Immunoprecipitation (IP)
Cells were lysed in a high-salt lysis buffer containing 420 mM NaCl and proteinase inhibitors (Roche) for 30 min at 4°C. After centrifugation, the balance buffer (20 mM HEPES [pH 7.4], 1 mM MgCl2, 10 mM KCl) was added to the supernatant to make the final NaCl concentration 300 mM. The lysate was then incubated with antibodies and protein A beads for 4 h at 4°C. The beads were spun down and washed three times with wash buffer before boiling in SDS loading buffer.
Flag Purification, MudPIT Analysis
Anti-Flag M2 agarose was purchased from Sigma. Nuclear extract preparation and Flag affinity purifications were performed in the presence of Benzonase (Sigma). Trichloroacetic acid-precipitated protein mixtures from the Flag purifications were digested with endoproteinase Lys-C and trypsin (Roche) and analyzed by MudPIT (Lin et al., 2010).
Size Exclusion Chromatography
Nuclear extract from ES cells was subjected to Superose 6 size exclusion chromatography (GE Healthcare) with size exclusion buffer (40 mM HEPES [PH 7.5], 350 mM NaCl, 10% glycerol and 0.1% Tween-20). Fractions were analyzed by Western blotting.
Quantitative RT-PCR
Total RNA samples were isolated with RNeasy (Qiagen), treated with RNase free DNase I (NEB), and re-purified with the RNeasy column. cDNAs were synthesized with the High Capacity RNA-to-cDNA Kit (Applied Biosystems). The expression levels were measured on MyIQ (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad). Relative expression to Actb was calculated. Primers used in the qRT-PCR assays: Fgf5-forward: ACCCGGATGGCAAAGTCAA; Fgf5-reverse: CAATCCCCTGAGACACAGCAA; EphA4-forward: TGGAATTTGCGACGCTGTCA; EphA4-reverse: CACTTCCTCCCACCCTCCTT; Otx2-forward: TGGGCTGAGTCTGACCACTT; Otx2-reverse: GCCCTAGTAAATGTCGTCCTCTC; Pou5f1-forward: AGCCGACAACAATGAGAACC; Pou5f1-reverse: TGGTCTCCAGACTCCACCTC; Nestin-forward: CTGCAGGCCACTGAAAAGTT; Nestin-reverse: GACCCTGCTTCTCCTGCTC; Tubb3 forward: AGGTGCGTGAGGAGTACCC; Tubb3-reverse: AGGGCTTCATTGTCGATGCAG.
ChIP-Seq
5×107 cells were used per ChIP assay according to a previously described protocol (Lee et al., 2006). Briefly, cells were cross-linked with 1% paraformaldehyde for 10 min at room temperature, quenched with glycine and fixed chromatin was sonicated and immunoprecipitated with specific antibodies. Libraries were prepared with Illumina’s ChIP-Seq sample prep kit for next-generation sequencing.
Accession to NGS data
ChIP-seq and expression data have been deposited at GEO under the accession number GSE61188.
Total RNA-Seq
ES cells (V6.5) were infected with lentivirus carrying Non-targeting (NonT) shRNA, Zic2 shRNA, Mbd3 shRNA, or Mta2 shRNA. 24 hours later, ES cells were selected with puromycin (2 ug/ml) for 48 hours. Total RNA was extracted with the RNeasy kit (Qiagen), treated with DNase I (NEB), and re-purified with RNeasy. For RNA-seq, total RNA was depleted of rRNA using Ribozero (Illumina) before library preparation performing Tru-seq sample prep (Illumina).
ChIP-Seq Analysis
Sequencing data was acquired through the default Illumina pipeline using Casava v1.8. Reads were aligned to the mouse genome (UCSC mm9) using the Bowtie aligner v0.12.9 allowing uniquely mapping reads only and allowing up to two mismatches (Langmead et al., 2009). Reads were extended to 150 bases toward the interior of the sequenced fragment and normalized to total reads aligned (reads per million; RPM). External sequencing data was acquired from GEO and ArrayExpress as raw reads and aligned in the same way as internally sequenced samples.
Peak detection was performed with MACS v1.4.2 (Zhang et al., 2008). Associated control samples were used to determine statistical enrichment at a p < 1e-8 and FDR < 0.05. The enrichments in external data were determined at p < 1e-5 and FDR < 0.05. The enrichment for H3K27me3 was called using the broad domain peak detector SICER v1.1 (Zang et al., 2009) at the FDR < 1e-8, window size of 200, and gap size of 600.
The overlapping high-confidence peak regions of two biological replicates were used for further analysis. The overlapping peaks were merged into one region as the union of all. Co-bound peaks were called if the peak regions of two samples are overlapping each other. Occupancies were calculated as the mean coverage under each peak region. R package edgeR 3.0.8 was used to perform differential occupancy analysis at p-value < 1e-5.
For ChIP-Seq enrichment profiles, regions of interest are shown for each factor as a binary value of enriched/not enriched and rows were sorted by the shortest distance of peak center to an annotated TSS. Regions shown are oriented 5′ to 3′ corresponding to the orientation of the nearest gene. Regions spanning 50 kb on either side of the indicated feature were binned into 200 bp windows. Each line represents a peak (peak centered). For ChIP-Seq heatmaps, regions of interest were centered at the center of each peak and sorted by the shortest distance of peak center to an annotated TSS. Regions shown are oriented 5′ to 3′ corresponding to the orientation of the nearest gene. Regions spanning 5 kb on either side of the indicated feature were binned into 25 bp windows.
Gene annotations and transcript start site information were from Ensembl 67 utilizing only Refseq mRNA entries. Non-TSS peaks are the peaks not overlapping with any TSS of the gene annotations. GO analysis was performed with the gene list using DAVID (Huang da et al., 2009a, b).
Total RNA-Seq Analysis
Sequencing data was acquired through the default Illumina pipeline using Casava v1.8. Reads were aligned to the mouse genome UCSC mm9 and to gene annotations from Ensembl 67 using TopHat v2.0.9 (Trapnell et al., 2009), using option –g 1 and allowing up to two mismatches. R package edgeR 3.0.8 was used to perform differential expression analysis at p-value < 1e-5 (Robinson et al., 2010) and p-value < 0.05 for external RNA-seq data.
Data generated for this study includes ChIP-seq data for Zic2, Zic3, Chd4, Mbd3, H3K27me3, H3K4me1, H3K4me3, H3K27ac and Pol II, and the RNA-seq data. Other data sets come from previously published studies: p300, H3K4me1 and H3K27ac ChIP-seq data are from GEO accession number GSE24164 (Creyghton et al., 2010); H3K4me3 and H3K27me3 ChIP-seq data are from GEO accession number GSE12241 (Mikkelsen et al., 2007); H3K27me3 ChIP-seq in Mbd3 WT and KO ES cells are from the ArrayExpress repository under accession E-MTAB-888 (Reynolds et al., 2012b); RNA-seq in Mbd3 WT and KO ES cells are from the ArrayExpress repository under accession E-MTAB-997 (Reynolds et al., 2012a); Chd4 ChIP-seq is from GEO accession number GSE27841(Whyte et al., 2012); and Mbd3 ChIP-seq is from GEO accession number GSM1246867 (Hnisz et al., 2013).
The parameters for next generation sequencing are reported in Table S1.
Supplementary Material
Acknowledgments
We thank the Molecular Biology core facility at the Stowers Institute for creating and sequencing libraries for next-generation sequencing and the Stowers Tissue Culture facility for large-scale cell culture and establishment of stable cell lines. We thank Alex Garruss for the initial analysis of the ChIP-seq and RNA-seq data. We thank Laura Shilatifard and Lisa Kennedy for editorial assistance. Z.L. is a Fellow of the Leukemia and Lymphoma Society. These studies were supported in part by grants R01GM069905 and R01CA89455 from National Cancer Institute to A.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen HF, Wade PA, Kutateladze TG. The NuRD architecture. Cell Mol Life Sci. 2013;70:3513–3524. doi: 10.1007/s00018-012-1256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 2011;30:2817–2828. doi: 10.1038/emboj.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedyk MJ, Mullen JR, DiNardo S. odd-paired: a zinc finger pair-rule protein required for the timely activation of engrailed and wingless in Drosophila embryos. Genes & development. 1994;8:105–117. doi: 10.1101/gad.8.1.105. [DOI] [PubMed] [Google Scholar]
- Brown L, Brown S. Zic2 is expressed in pluripotent cells in the blastocyst and adult brain expression overlaps with makers of neurogenesis. Gene expression patterns: GEP. 2009;9:43–49. doi: 10.1016/j.gep.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Brown SA, Warburton D, Brown LY, Yu CY, Roeder ER, Stengel-Rutkowski S, Hennekam RC, Muenke M. Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nature genetics. 1998;20:180–183. doi: 10.1038/2484. [DOI] [PubMed] [Google Scholar]
- Buecker C, Srinivasan R, Wu Z, Calo E, Acampora D, Faial T, Simeone A, Tan M, Swigut T, Wysocka J. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- Deng W, Blobel GA. Do chromatin loops provide epigenetic gene expression states? Curr Opin Genet Dev. 2010;20:548–554. doi: 10.1016/j.gde.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Escalante A, Murillo B, Morenilla-Palao C, Klar A, Herrera E. Zic2-dependent axon midline avoidance controls the formation of major ipsilateral tracts in the CNS. Neuron. 2013;80:1392–1406. doi: 10.1016/j.neuron.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Gunther K, Rust M, Leers J, Boettger T, Scharfe M, Jarek M, Bartkuhn M, Renkawitz R. Differential roles for MBD2 and MBD3 at methylated CpG islands, active promoters and binding to exon sequences. Nucleic Acids Res. 2013;41:3010–3021. doi: 10.1093/nar/gkt035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB, Melton DA. Nuclear reprogramming in cells. Science. 2008;322:1811–1815. doi: 10.1126/science.1160810. [DOI] [PubMed] [Google Scholar]
- Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Hu D, Shilatifard A. Enhancer Malfunction in Cancer. Mol Cell. 2014;53:859–866. doi: 10.1016/j.molcel.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes & development. 2012;26:2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtmeyers R, Souopgui J, Tejpar S, Arkell R. The ZIC gene family encodes multi-functional proteins essential for patterning and morphogenesis. Cell Mol Life Sci. 2013;70:3791–3811. doi: 10.1007/s00018-013-1285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Gao X, Morgan MA, Herz HM, Smith ER, Shilatifard A. The MLL3/MLL4 Branches of the COMPASS Family Function as Major Histone H3K4 Monomethylases at Enhancers. Molecular and cellular biology. 2013;33:4745–4754. doi: 10.1128/MCB.01181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Wade PA. NuRD and pluripotency: a complex balancing act. Cell Stem Cell. 2012;10:497–503. doi: 10.1016/j.stem.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nature genetics. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Kaji K, Nichols J, Hendrich B. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007;134:1123–1132. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- Kloet SL, Baymaz HI, Makowski M, Groenewold V, Jansen PW, Berendsen M, Niazi H, Kops GJ, Vermeulen M. Towards elucidating the stability, dynamics and architecture of the nucleosome remodeling and deacetylase complex by using quantitative interaction proteomics. The FEBS journal. 2014 doi: 10.1111/febs.12972. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, Lasonder E, Stunnenberg HG. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Molecular and cellular biology. 2006;26:843–851. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Wang C, Xu S, Cho YW, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife. 2013;2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. Transcriptional enhancers in animal development and evolution. Current biology: CB. 2010;20:R754–763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Garruss AS, Luo Z, Guo F, Shilatifard A. The RNA Pol II elongation factor Ell3 marks enhancers in ES cells and primes future gene activation. Cell. 2013;152:144–156. doi: 10.1016/j.cell.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Jeong HW, Kong N, Yang Y, Carroll J, Luo HR, Silberstein LE, Yupoma, Chai L. Stem cell factor SALL4 represses the transcriptions of PTEN and SALL1 through an epigenetic repressor complex. PLoS One. 2009;4:e5577. doi: 10.1371/journal.pone.0005577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Lin C, Guest E, Garrett AS, Mohaghegh N, Swanson S, Marshall S, Florens L, Washburn MP, Shilatifard A. The SEC family of RNA Polymerase II elongation factors: gene target specificity and transcriptional output. Molecular and cellular biology. 2012 doi: 10.1128/MCB.00182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. Enhancers: five essential questions. Nature reviews Genetics. 2013;14:288–295. doi: 10.1038/nrg3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourebrahim R, Houtmeyers R, Ghogomu S, Janssens S, Thelie A, Tran HT, Langenberg T, Vleminckx K, Bellefroid E, Cassiman JJ, et al. Transcription factor Zic2 inhibits Wnt/beta-catenin protein signaling. J Biol Chem. 2011;286:37732–37740. doi: 10.1074/jbc.M111.242826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Latos P, Hynes-Allen A, Loos R, Leaford D, O’Shaughnessy A, Mosaku O, Signolet J, Brennecke P, Kalkan T, et al. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012a;10:583–594. doi: 10.1016/j.stem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Salmon-Divon M, Dvinge H, Hynes-Allen A, Balasooriya G, Leaford D, Behrens A, Bertone P, Hendrich B. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J. 2012b;31:593–605. doi: 10.1038/emboj.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. Enhancer biology and enhanceropathies. Nature structural & molecular biology. 2014;21:210–219. doi: 10.1038/nsmb.2784. [DOI] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nature reviews Genetics. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Spruijt CG, Bartels SJ, Brinkman AB, Tjeertes JV, Poser I, Stunnenberg HG, Vermeulen M. CDK2AP1/DOC-1 is a bona fide subunit of the Mi-2/NuRD complex. Mol Biosyst. 2010;6:1700–1706. doi: 10.1039/c004108d. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Current biology: CB. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Yang Y, Hwang CK, Junn E, Lee G, Mouradian MM. ZIC2 and Sp3 repress Sp1-induced activation of the human D1A dopamine receptor gene. J Biol Chem. 2000;275:38863–38869. doi: 10.1074/jbc.M007906200. [DOI] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LM, Mao M, et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248–261. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes & development. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes & development. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


