Abstract
The past decade has appreciated rapid advance in identifying the once elusive intestinal stem cell (ISC) populations that fuel the continual renewal of the epithelial layer. This advance was largely driven by identification of novel stem cell marker genes, revealing the existence of quiescent, slowly‐ and active‐cycling ISC populations. However, a critical barrier for translating this knowledge to human health and disease remains elucidating the functional interplay between diverse stem cell populations. Currently, the precise hierarchical and regulatory relationships between these ISC populations are under intense scrutiny. The classical theory of a linear hierarchy, where quiescent and slowly‐cycling stem cells self‐renew but replenish an active‐cycling population, is well established in other rapidly renewing tissues such as the haematopoietic system. Efforts to definitively establish a similar stem cell hierarchy within the intestinal epithelium have yielded conflicting results, been difficult to interpret, and suggest non‐conventional alternatives to a linear hierarchy. While these new and potentially paradigm‐shifting discoveries are intriguing, the field will require development of a number of critical tools, including highly specific stem cell marker genes along with more rigorous experimental methodologies, to delineate the complex cellular relationships within this dynamic organ system.
Abbreviations
- BrdU
5‐bromo‐2′‐deoxyuridine
- CBC
crypt base columnar
- FACS
fluorescence‐activated cell sorting
- GFP
green fluorescent protein
- ISC
intestinal stem cell
- LRC
label‐retaining cell
- mTert
mouse telomerase reverse transcriptase
- YFP
yellow fluorescent protein
Introduction
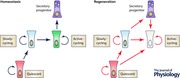
The intestinal epithelium serves critical functions for sustaining life: it provides an expansive surface area for nutrient uptake, mediates immune homeostasis, and maintains a contiguous barrier to the external environment (Peterson & Artis, 2014). The epithelial layer must be continuously renewed to safeguard against accumulation of physical and mutational injury (Stappenbeck et al. 1998; Wong et al. 1999). This renewal is fueled by a proliferative stem cell compartment tightly regulated to maintain discrete stem and progenitor cell pools. The proliferative zone of the small intestine's epithelial compartment is housed in protective invaginations – crypts of Lieberkühn – which line the floor of the organ and surround the base of the villus protrusions lined with differentiated epithelium (Wong et al. 1999, Henning & von Furstenberg, 2016). Within the protective crypt niche, the stem cell populations reside among regulatory crypt epithelium and surrounding stromal cells (Booth & Potten, 2000; Brittan & Wright, 2002). At least three types of stem cells have been identified in the intestine: quiescent ISCs (stem cells that do not divide at homeostasis; Montgomery et al. 2011), slowly‐cycling ISCs (stem cells that rarely divide during homeostasis; Sangiorgi & Capecchi, 2008; Takeda et al. 2011) and active‐cycling ISCs (stem cells that rapidly proliferate during homeostasis; Barker et al. 2007). These populations are hypothesized to be coordinately regulated, to exist in an ordered hierarchy, and to ultimately give rise to progenitor populations (immature cells with lineage commitment), transit‐amplifying cells (rapidly proliferating cells that increase epithelial numbers), and differentiated epithelial lineages (enterocytes, goblet, enteroendocrine, tuft and Paneth cells) (Cheng & Leblond, 1974 c; Karam, 1999; Barker et al. 2007; Sangiorgi & Capecchi, 2008; Gerbe et al. 2012) (Fig. 1). However, the exact relationship between identified ISC populations is not completely clear (Abstract figure). There are data supporting a structural hierarchy among stem and progenitor cells, but contradictory evidence also exists indicating that ISCs may reversibly transit between states of variable competency. Further, hierarchical relationships that exist during homeostasis may change in response to stimulation by a regenerative microenvironment. These intriguing scientific challenges represent the frontier of ISC biology: to understand the complex cellular interplay within the ISC niche.
Figure 1. Intestinal stem cell hierarchy .
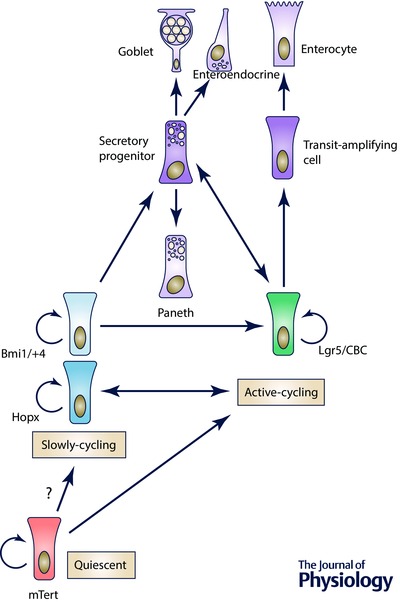
Evidence for multiple functional populations of stem cells support the existence of quiescent, slowly‐cycling, and active‐cycling stem cell classifications. The dynamic relationships between intestinal stem cells, progenitors, and differentiated lineages has evolved to reflect potential plasticity of the Lgr5+ stem cell and secretory progenitors, and the Hopx stem cell population.
Historical ISC identification: label retention and morphology
The intestinal epithelium undergoes continual renewal (Leblond & Stevens, 1948), with the continuum of proliferation to differentiation represented along the crypt–villus axis. Early studies used radioisotopes to label proliferating cells within the crypt and track their fates (Leblond & Stevens, 1948; Leblond & Messier, 1958; Walker & Leblond, 1958). Such labelling techniques were designed to gain insight into the unknown cell population underlying epithelial turnover. The existence of a cell population capable of self‐renewal (i.e. a stem cell pool) with multiple lineage potential residing among the undifferentiated cells within the crypt base was elegantly demonstrated in the mouse colon using radioautographic tracing (Chang & Leblond, 1971) and in the small intestine with 3H‐thymidine and ‘phagosome’ tracing (Cheng, 1974 a,b; Cheng & Leblond, 1974 a,b,c). The meticulous and detailed cataloguing of proliferative crypt base columnar cells (CBCs, slender cells localized between Paneth cells in the crypt base) and their direct progeny revealed that these CBCs were multipotent progenitors or stem cells (Cheng & Leblond, 1974 a,c). Notably, these first experiments in the small intestine indicated that the immature proliferative cells occupy the nine lowest cell positions in the crypt. Not surprisingly, because of the short window of isotope labelling used in these studies, only active‐cycling cells were analysed (therefore thought to be progenitors) and not long‐lived stem cell populations in the crypt. Due to the general acceptance that a true stem cell must rarely divide, this active‐cycling CBC population was largely dismissed as a candidate stem population for over 30 years.
In later studies by Christopher Potten, a label‐retaining stem cell population was identified using a long‐term labelling strategy with 3H‐thymidine and later with 5‐bromo‐2′‐deoxyuridine (BrdU) (Potten et al. 1974), localized to the +4 cell position of the intestinal crypt, and hypothesized to be a facultative stem cell. In agreement with the classical stem cell characteristics, these cells rarely divided, were resistant to cytotoxic stress, and were capable of regenerating injured epithelium (Rizvi & Wong, 2005). Thus, for three decades, the ISC field was solely focused on the +4 stem cell population.
The era of marker‐based identification of ISCs
The lack of functional stem cell markers and assays for demonstrating stemness stagnated the ISC field for years. A major breakthrough occurred when Hans Clevers’ group, using an elegant microarray study to compare normal and tumour intestinal epithelium, identified the Wnt target gene Lgr5 (Van der Flier et al. 2007) as a specific marker for the active‐cycling intestinal CBC population (Barker et al. 2007). Lgr5, a G‐protein‐coupled receptor, serves a critical function in ISC regulation by binding the Wnt agonist, R‐spondin, to amplify the local Wnt signal (Carmon et al. 2011). A knock‐in mouse expressing green fluorescent protein (GFP) and inducible Cre recombinase from the endogenous Lgr5 locus was generated and demonstrated strong GFP expression in the proliferative CBCs (Barker et al. 2007). Further, in vivo lineage tracing in the Lgr5 reporter mouse intestine demonstrated functional stemness of this cell population down the length of the intestine, as evidenced by stripes of LacZ‐expressing epithelial cells resident on crypt–villus units which encompassed all of the differentiated epithelial cell lineages (Barker et al. 2007). With the development of in vitro growth conditions supporting proliferation and differentiation of single Lgr5‐GFP‐expressing intestinal epithelial cells, Clevers’ group provided an ex vivo assay as a second approach to demonstrate a cell's stem potential (Sato et al. 2009). These critical advances energized the field, which resulted in the identification and validation of numerous additional markers of active‐cycling ISCs (Table 1).
Table 1.
Newly discovered stem cell populations
| Overlapping expression | ||
|---|---|---|
| Active‐cycling | with other cell populations | |
| Lgr5 | (Barker et al. 2007) | Y |
| Ascl2 | (Van der Flier et al. 2009) | NR |
| Olfm4 | (Van der Flier et al. 2009) | NR |
| Lrig1 | (Wong et al. 2012) | Y |
| Sox9lo | (Formeister et al. 2009) | NR |
| CD24lo | (von Furstenburg et al. 2011) | NR |
| Upper SP | (von Furstenburg et al. 2014) | NR |
| CD44+CD24loCD166+GRP78lo | (Wang et al. 2013) | NR |
| Smoc2 | (Munoz et al. 2012) | NR |
| Troy | (Fafilek et al. 2013) | NR |
| Overlapping expression | ||
| Slowly‐cycling/quiescent | with other cell populations | |
| Bmi1 | (Sangiorgi & Capecchi, 2008) | Y |
| mTert | (Montgomery et al. 2011) | NR |
| Hopx | (Takeda et al. 2011) | Y |
| Lrig1 | (Powellet al. 2012) | NR |
| Dclk1 | (May et al. 2008) | Y |
| Sox9hi | (Roche et al. 2015) | NR |
| Lower SP | (von Furstenburg et al. 2014) | NR |
| LRC | (Buczacki et al. 2013) | NR |
| Wip1 | (Demidov et al. 2007) | NR |
| Krt19 | (Asfaha et al. 2015) | NR |
NR, not reported; Y, yes.
Establishing specific strategies to identify and isolate rare slowly‐cycling ISCs, which were historically described to be located at the +4 cell position in the crypt, was more challenging. Characterization of a Bmi1‐Cre;R26R‐Yellow fluorescent protein (YFP) lineage mouse by Mario Capecchi's group revealed an expression pattern consistent with Potten's label‐retaining, slowly‐cycling ISC population (Sangiorgi & Capecchi, 2008). As expected, crypt‐based Bmi1‐expressing cells displayed slow cycling dynamics at homeostasis, but expanded and demonstrated enhanced lineage tracing capacity in response to regenerative injury (Yan et al. 2012). Fluorescence‐activated cell sorting (FACS)‐isolated Bmi1‐expressing cells initiate growth of intestinal enteroids under conventional Sato culture conditions (Sato et al. 2009), further validating their role in stem cell homeostasis (Yan et al. 2012). As with the active‐cycling stem cell population, this discovery set the stage for identification of numerous slowly‐cycling ISC populations, including those expressing Hopx (Takeda et al. 2011), as well as one population that appears to be more quiescent than slowly‐cycling, expressing mouse telomerase reverse transcriptase (mTert) (Montgomery et al. 2011) (Table 1). However, it remains unclear whether each of these stem cell populations, defined by marker gene expression, represents distinct or overlapping populations. Further, it remains controversial whether these populations exist in hierarchical relationships during homeostatic conditions and if these relationships are maintained during tissue regeneration.
Hierarchical relationships between ISC populations
To begin to explore hierarchical relationships between the numerous newly discovered stem cell populations within the intestinal epithelium, we first turn to other organ systems with similar dynamic renewal properties, such as the skin (Hsu et al. 2014), germ cells (Nakagawa et al. 2010), and the haematopoietic system (Kondo et al. 2003). In these systems, a classical stem cell hierarchy is well established. Rare slowly‐cycling stem cells are positioned ‘upstream’ of both the active‐cycling stem cell populations and committed lineage progenitors. This general strategy, where a subset of stem cells are slowly‐cycling but capable of rejuvenating an active‐cycling population, protects against accumulated mutations and transformation of the active‐cycling stem cells, which are more prone to genotoxic and cytotoxic stresses (Li & Clevers, 2010). Because the intestinal epithelium has active‐ (CBC, Lgr5), slowly‐cycling (Bmi1, Hopx), and quiescent (mTert) stem cell populations, it is likely that such a hierarchy exists, but this theory has proven difficult to conclusively demonstrate.
A recent landmark study provides compelling evidence that a stem cell hierarchy exists between two of the identified ISC populations, Bmi1+ and Lgr5+. This study employed an elegant diphtheria toxin approach – harnessing expression of the extracellular protein of Corynebacterium dipththeriae that inhibits protein synthesis and kills susceptible cells – to specifically ablate the Lgr5+ ISC population. The approach revealed that this active‐cycling pool is dispensable for maintenance of normal epithelial architecture and homeostasis. Remarkably, ablation of the Lgr5+ ISC population resulted in both expansion and enhanced lineage tracing capacity from the Bmi1+ ISC population (Tian et al. 2011). The authors went on to demonstrate direct and definitive lineage tracing of the Bmi1+ ISC to an Lgr5+ ISC under homeostatic conditions, using a β‐gal indicator for Bmi1+ ISC lineage tracing and GFP to mark the Lgr5+ population. These findings hierarchically position the Bmi1+ ISC upstream of the Lgr5+ ISC under homeostatic conditions (Fig. 1). Interestingly, earlier studies exploiting diphtheria toxin to ablate the Bmi1+ ISC resulted in collapse of crypt architecture in a subset of crypts (Sangiorgi & Capecchi, 2008). In retrospect, these findings may indicate that the Bmi1+ population provides important renewal of the active‐cycling ISC population.
In addition to direct hierarchical relationships, studies in regenerating crypts also revealed that apparent lineage‐committed progenitor cells possess plasticity. In response to injury, these cells display the ability to dedifferentiate toward a stem cell fate. The idea that committed lineage progenitors can be coaxed back into a stem cell fate is not entirely novel, as it has been described and studied in other systems such as the haematopoietic system (Graf, 2002). Indeed, within the intestine, it appears that chromatin marks between active‐cycling Lgr5+ ISCs and progenitor cell types are nearly identical, supporting the notion that various intestinal epithelial cells, regardless of their differentiation status, have the capacity to rapidly change their cellular expression programme in response to environmental stimuli (Kim et al. 2014). In one such example, Doug Winton's group demonstrated that intestinal H2B‐YFP label‐retaining cells (LRCs), thought to represent quiescent secretory lineage precursors, are committed to mature into differentiated secretory cells (Paneth and enteroendocrine) rather than an active‐cycling ISC (Buczacki et al. 2013). However, using a novel genetic mouse model, the authors demonstrate that these LRCs can be coaxed into re‐acquiring stem cell function after injury. Supporting this concept, an independent study from the Clevers’ group found that secretory progenitors marked by the Notch ligand Dll1 have a similar stem cell capacity after tissue damage (van Es et al. 2012). These studies highlight that hierarchical relationships between different ISC pools may not be simplistically linear and therefore is not easily defined. Furthermore, an exciting study by Jonathan Epstein's laboratory provides the first evidence that hierarchical relationships between ISC populations can be bidirectional and not merely linear. Using a complex tri‐transgenic mouse cross to lineage trace from the Hopx loci, while maintaining the ability to detect Lgr5+ populations, the authors identified double‐marked cells within the crypt epithelium indicating that Hopx+ ISCs could give rise to Lgr5+ ISCs and vice versa (Takeda et al. 2011; Li et al. 2014). This intriguing concept suggests that the intestinal epithelium has built in contingency plans for robust maintenance of tissue homeostasis in the event of ablation of discrete ISC populations.
Challenges with marker‐based definitions for stem cell populations
The majority of currently identified stem cell populations within the intestinal epithelium are based solely upon gene or protein expression (Table 1). Multiple challenges exist with this approach in clarifying relationships between different populations. Unlike the haematopoietic field where the development and organization of multiplexed antibodies to cell surface antigens facilitated the identification, characterization and manipulation of discrete populations and related subpopulations (Kondo et al. 2003), these types of tools have drastically lagged behind in the ISC field. To compensate, the field has relied upon the generation of stem cell‐reporter mouse lines, a cumbersome and expensive approach to exploring relationships between ISC populations. The fact that the majority of ISC‐reporter mice were generated with GFP prevents combinatorial analyses of multiple stem cell populations. Further, unlike in the haematopoietic system, intestinal lineage progenitors have been primarily identified by histological hallmarks (Cheng, 1974 b). The inability to dissect populations down a lineage differentiation pathway and identify intermediate populations prevents the resolution of underlying mechanisms that drive these processes. Furthermore, understanding whether or not interconvertible relationships between progenitors, stem cells and differentiated lineages underlies homeostasis or regenerative mechanisms cannot be adequately and clearly addressed.
Reliance on gene or protein expression complicates and limits useful tracking of stem cell populations. Gene expression within discrete stem cell populations may reflect a physiological response to their environment. For example, while Lgr5 expression on the active‐cycling CBC functions to mediate a proliferative Wnt signal to this cell population, homozygous knockout of this protein in embryonic intestines that were transplanted and matured in kidney capsule xenografts retained normal tissue architecture, despite complete loss of Lgr5 expression (Tian et al. 2011). Interestingly the void of cells was filled with unknown, non‐Lgr5‐expressing CBC populations (Tian et al. 2011). This finding strongly suggests that either a subpopulation non‐Lgr5‐expressing CBCs can compensate, or that CBCs can harness a bypass mechanism to gain independence from Lgr5‐mediated Wnt signalling. This bypass mechanism could involve either upregulation of different Wnt‐regulatory machinery or transition into an entirely different cell state that can proliferate independently of Wnt. For example, Lrig1 is reported to be expressed on a variety of cells within the crypt, with overlapping expression in Lgr5+ (Wong et al. 2012) and +4‐positioned (Powell et al. 2012) ISCs. Because of its broader expression pattern, as reported by Wong et al., and its biological function as a negative regulator of the ErbB receptor family, it is likely that Lrig1 expression reflects a transient cell signalling state rather than marking a discrete homogeneous stem cell population.
Further complicating marker‐based identification of cells, studies interchangeably define populations based on their RNA expression or protein expression (Munoz et al. 2012). While Bmi1 RNA may be expressed at very low levels in most cells within the crypt base (Munoz et al. 2012), the Bmi1 protein is expressed in +4‐positioned cells, as determined within the Bmi1‐reporter mouse intestine (Tian et al. 2011). A cell's identity might be based both on its function from its protein expression, and its potential as reflected by its RNA status, but this notion has not been formally addressed.
Finally, many of the protein markers have broad expression patterns that are not restricted to a single ‘population’ of cells within the intestinal epithelium. Prime examples of this are that the stem cell marker Dclk1, which is also expressed on the villus and in cells co‐expressing differentiated enteroendocrine or tuft markers (Levin et al. 2010; Gerbe et al. 2012). Additionally, Bmi1 is not only expressed within the crypt compartment, but also by a subset of differentiated villus cells (Takeda et al. 2011; Munoz et al. 2012; Li et al. 2014). This heterogeneous expression found from many stem cell markers complicates the ability to fine tune characterization of discrete functional ISC populations. For instance, Lgr5 is expressed on active‐cycling stem cell populations, but was recently described to be expressed on secretory progenitor cells (Buczacki et al. 2013; Grun et al. 2015). Hopx, which is expressed in a subset of active‐cycling ISCs, the slowly‐cycling ISC population, secretory progenitors and mature Paneth cells, serves as an additional example of heterogeneous expression (Li et al. 2014). Current experimental methodologies cannot easily differentiate two populations in the in vivo studies involving lineage tracing. While marker‐based identification of ISCs has moved the field forward, the next step for ISC manipulation should be towards a unified, cell surface antigen‐based approach for isolation of ISC populations. This approach has served the haematopoietic stem cell field well in defining discrete populations, their relationships and their overlapping function within the blood.
Caveats with current assays to determine stemness
In addition to challenges identifying and manipulating ISC populations, undoubtedly the biggest hurdle is the lack of definitive assays to determine functional stemness of discrete, putative stem cell populations. Four primary ISC assays are routinely employed by the field: (a) reporter mice, (b) lineage tracing, (c) gene expression profiling, and (d) in vitro enteroid cultures. The location of the proliferative stem cell niche in relation to the differentiated cells on the adjacent villi provide a convenient secondary architecture to readily appreciate the continuum of cell proliferation to differentiation (Stappenbeck et al. 1998), and to discover new progenitor and ISC populations based on protein expression. These studies are typically initiated with observations from RNA or protein expression patterns discovered in immunohistochemical analyses, but then followed with the generation of a reporter mouse line for confirmation. While these studies function to identify putative populations, they do not definitively confirm stem cell behaviour.
The ability to lineage trace from a discrete cell population, that is to identify all progeny derived from a single cell, was originally established as a developmental biology tool but has more recently been harnessed in stem cell biology to assay the stem cell potential of various populations within a tissue (Kretzschmar & Watt, 2012). To investigate the lineage‐propagating potential of a putative ISC, a mouse line that expresses inducible Cre recombinase from the putative ISC promoter is generated and then crossed to a Cre‐reporter mouse line (for lineage tracing) or an inducible diphtheria toxin mouse line (for ablation of the population). While the ability to inducibly activate Cre provides an important scientific tool, most of these reporters rely on tamoxifen for activation, which is known to differentially affect the viability of various ISC pools (Zhu et al. 2013). Therefore, results using such approaches must be interpreted with caution. Further, most of these mice require complicated breeding schemes to generate bi‐ or tri‐transgenic/knock‐in mouse lines. Therefore, these studies are inefficient, costly, time consuming, and complex to analyse. One primary caveat with analyses of the in vivo lineage studies is that many of the ISC markers harbour heterogeneous expression patterns (e.g. expressed in differentiated, progenitor, and ISCs). Specifically, if the marker is expressed in slowly‐cycling and differentiated populations, the interpretation of the results may be different from that if the marker were represented in a single population. These studies complicate interpretation, as it is difficult to determine if lineage tracing originates from a differentiated population (i.e. suggesting that differentiated cells have the plasticity to convert to a stem cell state) or if it originates from a rare undifferentiated subpopulation with stem cell capacity. Often the extent of marker heterogeneity is not appreciated in publications describing the initial ‘validation’ of a stem cell marker.
Bioinformatics of gene expression data sets have been heavily leveraged to gain insight into the relationships between stem cell populations (Munoz et al. 2012; Li et al. 2014; Grun et al. 2015). While these types of studies have yielded amazing breakthroughs in the ISC field (Barker et al. 2007; Van der Flier et al. 2007), most of the analyses are based upon RNA expression and this raises the concern about the validity of RNA vs. protein expression patterns. Further, interpretation of the clustering and modelling is always superimposed upon what appears logical within the current state of stem cell interactions and could carry some bias.
Due to the lack of available experimental tools, very few studies demonstrate direct lineage relationships through temporal analysis. Instead, they rely on circumstantial evidence to make inferences regarding hierarchy. For example, thorough gene and protein expression analyses of five FACS‐isolated Lgr5‐GFP populations based on GFP signal intensity compared levels of quiescent (mTert) and slowly‐cycling ISC marker gene expression (including Lrig1, Bmi1 and Hopx). Relative gene expression levels (but not protein levels) between these arbitrary populations led to the conclusion that all Lgr5+ ISCs express Bmi1 (van der Flier et al. 2009; Munoz et al. 2012).
Finally, while the field is fortunate to have an in vitro assay system to grow single FACS‐isolated cells into 3‐dimensional enteroid structures that recapitulate stem and differentiated domains (Sato et al. 2009), it must be acknowledged that the growth conditions mimic an activated, regenerative or cancer‐like microenvironment. Therefore, this assay system cannot be used to address questions involving normal homeostasis (due to lack of signalling gradients and domains) or to resolve regulation of signalling pathways to discrete populations within the enteroids due to excess growth factors that are primarily geared towards supporting the Wnt‐dependent active‐cycling Lgr5+ ISC pool (Sato et al. 2009). Further, we lack the necessary culture conditions to understand the regulatory microenvironment for quiescent and slowly‐cycling ISCs. Importantly, taken together, the field lacks an in vivo reconstruction assay, which was instrumental in elevating the level of discovery for the haematopoietic stem cell system (Jacobson et al. 1951; Becker et al. 1963; Weissman & Shizuru, 2008).
Future directions for the ISC field
Over the past decade, breakthrough discoveries have elevated the ISC field, facilitating rapid advancement in identifying markers of the once elusive ISC populations – quiescent, slowly‐ and active‐cycling – and have laid the foundation for unravelling their complex inter‐relationships during homeostasis, in response to regenerative cues, and in disease. Understanding the dynamic interplay between ISC populations and how they are related and inter‐regulated represents a critical threshold for translating our stem cell‐based knowledge to patient therapeutics. Understandably, evolution of this nascent field brings with it rekindled controversy as we seek clarity into ISC relationships and hierarchy, highlighting that there are still many unanswered questions.
Emerging issues with ISC population heterogeneity and overlap of ISC markers require elucidation. While it is clear that ISC populations distinct from the active‐cycling Lgr5+ pool exist, their exact identities and functions during tissue homeostasis and repair remain unclear. Multiple markers of putative quiescent and slowly‐cycling ISCs have been identified (Table 1), but it is increasingly clear that these markers represent heterogeneous populations. Currently, it appears that none of the identified +4 ISC markers successfully distinguish a homogeneous population, as marker‐positive cells are located on the differentiated villi and do not lineage trace. This indicates that a rare subset of marker‐positive, crypt‐based cells may harbour stem cell characteristics. Therefore, development of novel single cell technologies to dissect this heterogeneity will address the degree of overlap between all ISC markers. These approaches will uncover new ISC markers, that when implemented in a combinatorial fashion with current markers, will support identification schemes for increased specificity for homogeneous ISC populations. Advances have been made in these technologies in recent years, including single cell gene expression analysis of FACS‐isolated ISC and crypt cell populations (Dalerba et al. 2011; Rothenberg et al. 2012; Grun et al. 2015; Roche et al. 2015), as well as developing platforms to assay stemness of single ISC populations (Gracz et al. 2015). Unfortunately, the majority of ISC markers are represented by intracellular proteins or proteins where functional antibodies have not been successfully derived. Therefore, the field requires a focused development of ISC population‐specific cell surface antibodies to translate discoveries in mice to humans. Overall, these approaches will lead to greater population specificity and facilitate future studies to address the functional relevance of marker expression to ultimately illuminate the hierarchical relationships between distinct ISCs.
Complicating the traditional views of a stem cell hierarchy is the recent evidence that lineage‐committed progenitors possess a level of plasticity allowing them to revert to a stem cell state. In the intestine, this plasticity has been demonstrated by lineage‐tracing of ‘committed cells’ (van Es et al. 2012; Buczacki et al. 2013) and highlights the existence of alternative cellular mechanisms to regenerate the stem cell niche after injury. It is important to note that these exciting findings do not disprove the existence of a more traditional stem cell hierarchy, as both mechanisms may co‐exist. Instead, these discoveries may allude to the amazing redundancy orchestrated within the ISC niche to ensure maintenance of a functional epithelium. We propose that these different views of ‘hierarchy’ likely depend upon microenvironmental context (i.e. homeostasis vs. injury or disease).
With current experimental tools and methodologies, it is impossible to distinguish the relative contribution of direct hierarchy vs. progenitor plasticity in restoration of tissue homeostasis after injury. In order to elucidate the functional importance of these diverse cellular mechanisms, a greater understanding of the cell–cell signalling interactions within the niche responsible for regulating the maintenance or stimulation of the quiescent and slowly‐cycling ISC populations is paramount. This elucidation will require continued development of novel technologies for querying multiple cell signalling readouts derived from discrete cell populations (Simmons et al. 2015), with further advancement of real‐time in vivo intravital imaging (Ritsma et al. 2014), in vitro culture methods that support quiescent and slowly‐cycling ISC propagation, and identification of specific culture conditions that stimulate interconversion between ISC populations.
With the exciting advancements in ISC marker identification and initial studies into the hierarchical relationships between distinct ISC populations, a complex picture is emerging. It appears that the intestinal epithelium does not follow a simplistic linear stem cell hierarchy model. Instead, a model of hierarchy with built‐in redundancy likely exists (Abstract figure) whereby different types of cells with stem potential are called upon depending on the physiological context. For example, during homeostasis, the slowly‐cycling ISC population provides moderate renewal of the active‐cycling niche. However, in response to injury, this activity is enhanced and plasticity of lineage‐committed progenitors is appreciated (Abstract figure). These diverse mechanisms illustrate an exquisite evolutionary safety net built into a dynamic epithelium to ensure robust tissue renewal and rapid restoration of homeostasis after insult, properties that are absolutely essential for survival.
Additional information
Competing interests
None declared.
Author contributions
All authors contributed to the writing and editing of this review.
Funding
This work was supported by NIH/NIDDK (U01DK085525).
Biographies
Nicholas R. Smith is a postdoctoral fellow at Oregon Health & Science University. His work on differential regulation of quiescent and active‐cycling stem cells in in vitro and in vivo systems provides new mechanisms to maintain cellular homeostasis.
Alexandra C. Gallagher is a research intern at Oregon Health & Science University with expertise on the Lgr5‐expressing stem cell population.
Melissa H. Wong is an associate professor at Oregon Health & Science University. She has contributed to the intestinal stem cell field with work on Wnt signalling and the stem cell hierarchy.
This review was presented at the “Gastrointestinal Tract XVI: GI homeostasis, the microbiome and the barrier, development and disease” FASEB Summer Research, Steamboat Springs, Colorado, USA between 2–7 August 2015.
References
- Asfaha S, Hayakawa Y, Muley A, Stokes S, Graham TA, Ericksen RE, Westphalen CB, von Burstin J, Mastracci TL, Worthley DL, Guha C, Quante M, Rustgi AK & Wang TC (2015). Krt19+/Lgr5− cells are radioresistant cancer‐initiating stem cells in the colon and intestine. Cell Stem Cell 16, 627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ & Clevers H (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5 . Nature 449, 1003–1007. [DOI] [PubMed] [Google Scholar]
- Becker AJ, McCulloch EA & Till JE (1963). Cytological demonstration of the clonal nature of spleen colonies derived from transplanted mouse marrow cells. Nature 197, 452–454. [DOI] [PubMed] [Google Scholar]
- Booth C & Potten CS (2000). Gut instincts: thoughts on intestinal epithelial stem cells. J Clin Invest 105, 1493–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittan M & Wright NA (2002). Gastrointestinal stem cells. J Pathol 197, 492–509. [DOI] [PubMed] [Google Scholar]
- Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R & Winton DJ (2013). Intestinal label‐retaining cells are secretory precursors expressing Lgr5. Nature 495, 65–69. [DOI] [PubMed] [Google Scholar]
- Carmon KS, Gong X, Lin Q, Thomas A & Liu Q (2011). R‐spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/β‐catenin signaling. Proc Natl Acad Sci USA 108, 11452–11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WW & Leblond CP (1971). Renewal of the epithelium in the descending colon of the mouse. I. Presence of three cell populations: vacuolated‐columnar, mucous and argentaffin. Am J Anat 131, 73–99. [DOI] [PubMed] [Google Scholar]
- Cheng H (1974. a). Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. II. Mucous cells. Am J Anat 141, 481–501. [DOI] [PubMed] [Google Scholar]
- Cheng H (1974. b). Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. IV. Paneth cells. Am J Anat 141, 521–535. [DOI] [PubMed] [Google Scholar]
- Cheng H & Leblond CP (1974. a). Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. I. Columnar cell. Am J Anat 141, 461–479. [DOI] [PubMed] [Google Scholar]
- Cheng H & Leblond CP (1974. b). Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. III. Entero‐endocrine cells. Am J Anat 141, 503–519. [DOI] [PubMed] [Google Scholar]
- Cheng H & Leblond CP (1974. c). Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. Am J Anat 141, 537–561. [DOI] [PubMed] [Google Scholar]
- Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA, Sim S, Okamoto J, Johnston DM, Qian D, Zabala M, Bueno J, Neff NF, Wang J, Shelton AA, Visser B, Hisamori S, Shimono Y, van de Wetering M, Clevers H, Clarke MF & Quake SR (2011). Single‐cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol 29, 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidov ON, Timofeev O, Lwin HN, Kek C, Appella E & Bulavin DV (2007). Wip1 phosphatase regulates p53‐dependent apoptosis of stem cells and tumorigenesis in the mouse intestine. Cell Stem Cell 1, 180–190. [DOI] [PubMed] [Google Scholar]
- Fafilek B, Krausova M, Vojtechova M, Pospichalova V, Tumova L, Sloncova E, Huranova M, Stancikova J, Hlavata A, Svec J, Sedlacek R, Luksan O, Oliverius M, Voska L, Jirsa M, Paces J, Kolar M, Krivjanska M, Klimesova K, Tlaskalova‐Hogenova H & Korinek V (2013). Troy, a tumor necrosis factor receptor family member, interacts with Lgr5 to inhibit Wnt signaling in intestinal stem cells. Gastroenterology 144, 381–391. [DOI] [PubMed] [Google Scholar]
- Formeister EJ, Sionas AL, Lorance DK, Barkley CL, Lee GH & Magness ST (2009). Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol 296, G1108–G1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F, Legraverend C & Jay P (2012). The intestinal epithelium tuft cells: specification and function. Cell Mol Life Sci 69, 2907–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracz AD, Williamson IA, Roche KC, Johnston MJ, Wang F, Wang Y, Attayek PJ, Balowski J, Liu XF, Laurenza RJ, Gaynor LT, Sims CE, Galanko JA, Li L, Allbritton NL & Magness ST (2015). A high‐throughput platform for stem cell niche co‐cultures and downstream gene expression analysis. Nat Cell Biol 17, 340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T (2002). Differentiation plasticity of hematopoietic cells. Blood 99, 3089–3101. [DOI] [PubMed] [Google Scholar]
- Grun D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, Clevers H & van Oudenaarden A (2015). Single‐cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525, 251–255. [DOI] [PubMed] [Google Scholar]
- Henning SJ & von Furstenberg RJ (2016). GI stem cells – new insights into roles in physiology and pathophysiology. J Physiol 594, 4769–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Li L & Fuchs E (2014). Emerging interactions between skin stem cells and their niches. Nat Med 20, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson LO, Simmons EL, Marks EK & Eldredge JH (1951). Recovery from radiation injury. Science 113, 510–511. [DOI] [PubMed] [Google Scholar]
- Karam SM (1999). Lineage commitment and maturation of epithelial cells in the gut. Front Biosci 4, D286–298. [DOI] [PubMed] [Google Scholar]
- Kim TH, Li F, Ferreiro‐Neira I, Ho LL, Luyten A, Nalapareddy K, Long H, Verzi M & Shivdasani RA (2014). Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature 506, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, Beilhack GF, Shizuru JA & Weissman IL (2003). Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol 21, 759–806. [DOI] [PubMed] [Google Scholar]
- Kretzschmar K & Watt FM (2012). Lineage tracing. Cell 148, 33–45. [DOI] [PubMed] [Google Scholar]
- Leblond CP & Messier B (1958). Renewal of chief cells and goblet cells in the small intestine as shown by radioautography after injection of thymidine‐H3 into mice. Anat Rec 132, 247–259. [DOI] [PubMed] [Google Scholar]
- Leblond CP & Stevens CE (1948). The constant renewal of the intestinal epithelium in the albino rat. Anat Rec 100, 357–377. [DOI] [PubMed] [Google Scholar]
- Levin TG, Powell AE, Davies PS, Silk AD, Dismuke AD, Anderson EC, Swain JR & Wong MH (2010). Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology 139, 2072–2082.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L & Clevers H (2010). Coexistence of quiescent and active adult stem cells in mammals. Science 327, 542–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Yousefi M, Nakauka‐Ddamba A, Jain R, Tobias J, Epstein JA, Jensen ST & Lengner CJ (2014). Single‐cell analysis of proxy reporter allele‐marked epithelial cells establishes intestinal stem cell hierarchy. Stem Cell Reports 3, 876–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May R, Riehl TE, Hunt C, Sureban SM, Anant S & Houchen CW (2008). Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase‐like‐1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells 26, 630–637. [DOI] [PubMed] [Google Scholar]
- Montgomery RK, Carlone DL, Richmond CA, Farilla L, Kranendonk ME, Henderson DE, Baffour‐Awuah NY, Ambruzs DM, Fogli LK, Algra S & Breault DT (2011). Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci USA 108, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S, Volckmann R, Kung KS, Koster J, Radulescu S, Myant K, Versteeg R, Sansom OJ, van Es JH, Barker N, van Oudenaarden A, Mohammed S, Heck AJ & Clevers H (2012). The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J 31, 3079–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Sharma M, Nabeshima Y, Braun RE & Yoshida S (2010). Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science 328, 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson LW & Artis D (2014). Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol 14, 141–153. [DOI] [PubMed] [Google Scholar]
- Potten CS, Kovacs L & Hamilton E (1974). Continuous labelling studies on mouse skin and intestine. Cell Tissue Kinet 7, 271–283. [DOI] [PubMed] [Google Scholar]
- Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE, Guo Y, Shyr Y, Aronow BJ, Haigis KM, Franklin JL & Coffey RJ (2012). The pan‐ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 149, 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritsma L, Ellenbroek SI, Zomer A, Snippert HJ, de Sauvage FJ, Simons BD, Clevers H & van Rheenen J (2014). Intestinal crypt homeostasis revealed at single‐stem‐cell level by in vivo live imaging. Nature 507, 362–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi AZ & Wong MH (2005). Epithelial stem cells and their niche: there's no place like home. Stem Cells 23, 150–165. [DOI] [PubMed] [Google Scholar]
- Roche KC, Gracz AD, Liu XF, Newton V, Akiyama H & Magness ST (2015). SOX9 maintains reserve stem cells and preserves radioresistance in mouse small intestine. Gastroenterology 149, 1553–1563.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg ME, Nusse Y, Kalisky T, Lee JJ, Dalerba P, Scheeren F, Lobo N, Kulkarni S, Sim S, Qian D, Beachy PA, Pasricha PJ, Quake SR & Clarke MF (2012). Identification of a cKit+ colonic crypt base secretory cell that supports Lgr5+ stem cells in mice. Gastroenterology 142, 1195–1205.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangiorgi E & Capecchi MR (2008). Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 40, 915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ & Clevers H (2009). Single Lgr5 stem cells build crypt‐villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. [DOI] [PubMed] [Google Scholar]
- Simmons AJ, Banerjee A, McKinley ET, Scurrah CR, Herring CA, Gewin LS, Masuzaki R, Karp SJ, Franklin JL, Gerdes MJ, Irish JM, Coffey RJ & Lau KS (2015). Cytometry‐based single‐cell analysis of intact epithelial signaling reveals MAPK activation divergent from TNF‐α‐induced apoptosis in vivo . Mol Syst Biol 11, 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck TS, Wong MH, Saam JR, Mysorekar IU & Gordon JI (1998). Notes from some crypt watchers: regulation of renewal in the mouse intestinal epithelium. Curr Opin Cell Biol 10, 702–709. [DOI] [PubMed] [Google Scholar]
- Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM & Epstein JA (2011). Interconversion between intestinal stem cell populations in distinct niches. Science 334, 1420–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD & de Sauvage FJ (2011). A reserve stem cell population in small intestine renders Lgr5‐positive cells dispensable. Nature 478, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Flier LG, Sabates‐Bellver J, Oving I, Haegebarth A, De Palo M, Anti M, Van Gijn ME, Suijkerbuijk S, Van de Wetering M, Marra G & Clevers H (2007). The intestinal Wnt/TCF signature. Gastroenterology 132, 628–632. [DOI] [PubMed] [Google Scholar]
- van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, van Es JH, Barker N, Peters PJ, van de Wetering M & Clevers H (2009). Transcription factor achaete scute‐like 2 controls intestinal stem cell fate. Cell 136, 903–912. [DOI] [PubMed] [Google Scholar]
- van Es JH, Sato T, van de Wetering M, Lyubimova A, Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J, Martens AC, Barker N, van Oudenaarden A & Clevers H (2012). Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol 14, 1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Furstenberg RJ, Buczacki SJ, Smith BJ, Seiler KM, Winton DJ & Henning SJ (2014). Side population sorting separates subfractions of cycling and non‐cycling intestinal stem cells. Stem Cell Res 12, 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Furstenberg RJ, Gulati AS, Baxi A, Doherty JM, Stappenbeck TS, Gracz AD, Magness ST & Henning SJ (2011). Sorting mouse jejunal epithelial cells with CD24 yields a population with characteristics of intestinal stem cells. Am J Physiol Gastrointest Liver Physiol 300, G409–G417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BE & Leblond CP (1958). Sites of nucleic acid synthesis in the mouse visualized by radioautography after administration of C14‐labelled adenine and thymidine. Exp Cell Res 14, 510–531. [DOI] [PubMed] [Google Scholar]
- Wang F, Scoville D, He XC, Mahe MM, Box A, Perry JM, Smith NR, Lei NY, Davies PS, Fuller MK, Haug JS, McClain M, Gracz AD, Ding S, Stelzner M, Dunn JC, Magness ST, Wong MH, Martin MG, Helmrath M & Li L (2013). Isolation and characterization of intestinal stem cells based on surface marker combinations and colony‐formation assay. Gastroenterology 145, 383–395.e1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman IL & Shizuru JA (2008). The origins of the identification and isolation of hematopoietic stem cells, and their capability to induce donor‐specific transplantation tolerance and treat autoimmune diseases. Blood 112, 3543–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong MH, Stappenbeck TS & Gordon JI (1999). Living and commuting in intestinal crypts. Gastroenterology 116, 208–210. [DOI] [PubMed] [Google Scholar]
- Wong VW, Stange DE, Page ME, Buczacki S, Wabik A, Itami S, van de Wetering M, Poulsom R, Wright NA, Trotter MW, Watt FM, Winton DJ, Clevers H & Jensen KB (2012). Lrig1 controls intestinal stem‐cell homeostasis by negative regulation of ErbB signalling. Nat Cell Biol 14, 401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, Sangiorgi E, Capecchi MR & Kuo CJ (2012). The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci USA 109, 466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Huang YF, Kek C & Bulavin DV (2013). Apoptosis differently affects lineage tracing of Lgr5 and Bmi1 intestinal stem cell populations. Cell Stem Cell 12, 298–303. [DOI] [PubMed] [Google Scholar]


