Abstract
Mechanistic target of rapamycin (mTOR) is a serine-threonine kinase that coordinates nutrient and growth factor availability with cellular growth, division, and differentiation. Studies examining the roles of mTOR signaling in immune function revealed critical roles for mTOR in regulating T cell differentiation and function. However, few studies have investigated the roles of mTOR in early B cell development. In this study, we found that mTOR is highly activated during the pro- and pre-B stages of mouse B cell development. Conditional disruption of the mTOR coactivating protein Raptor in developing mouse B cells resulted in a developmental block at the pre-B cell stage, with a corresponding lack of peripheral B cells and loss of Ag-specific Ab production. Pre-B cell survival and proliferation were significantly reduced in Raptor-deficient mice. Forced expression of a transgenic BCR or a BclxL transgene on Raptor-deficient B cells failed to rescue B cell development, suggesting that pre-BCR signaling and B cell survival are impaired in a BclxL-independent manner. Raptor-deficient pre-B cells exhibited significant decreases in oxidative phosphorylation and glycolysis, indicating that loss of mTOR signaling in B cells significantly impairs cellular metabolic capacity. Treatment of mice with rapamycin, an allosteric inhibitor of mTOR, recapitulated the early B cell developmental block. Collectively, our data reveal a previously uncharacterized role for mTOR signaling in early B cell development, survival, and metabolism.
Introduction
Mechanistic target of rapamycin (mTOR) is an evolutionarily conserved serine/threonine kinase that coordinates nutrient and energy availability with cell metabolism, growth, and proliferation. mTOR is located within two discrete protein complexes, mTOR complex (mTORC)1 and mTORC2, which are characterized by the association of mTOR with one of two coactivator proteins, Raptor and Rictor, respectively. mTORC1 is positively regulated by a number of upstream signals, including abundant amino acids, growth factors, and energy levels. When activated, mTORC1 phosphorylates downstream targets p70 ribosomal protein S6 kinase 1 and eukaryotic translation initiation factor 4E-binding protein 1 (4EBP1), thereby promoting protein synthesis and cell growth. mTORC1 also promotes lipid synthesis, oxidative metabolism, and glycolysis to help fuel cell growth and proliferation while also inhibiting autophagy (1, 2). In contrast to mTORC1, mTORC2 is activated by growth factors independently of nutrients and predominantly functions to regulate the actin cytoskeleton and cell survival (1).
Studies examining mTORC1 functions in the immune system revealed important roles for mTOR in regulating the differentiation and function of specific T cell subsets. For example, the immunosuppressant rapamycin, used clinically to prevent organ transplant rejection, acts, in part, by suppressing T cell proliferation (3). More recently, it was shown that mTORC1 activity also controls Th1, Th2, and Th17 differentiation, T regulatory cell differentiation, and CD8 effector and memory cell development (4, 5). mTOR also has distinct roles in the generation and function of dendritic cells, myeloid cells, and innate lymphocytes (4, 6–8). However, there have been limited studies on the cell-specific roles of mTOR signaling in early B cell development.
During B cell development, B cell progenitors must successfully pass several checkpoints to ensure that mature B cells have a diverse repertoire of functional IgRs capable of recognizing a wide array of foreign proteins while exhibiting limited self-reactivity. Early committed B cells called progenitor B (pro-B) cells have their Ig H chain (IgH) and Ig L chain (IgL) genes in the germline configuration and rely on stromal cell–derived IL-7 to survive. Following productive in-frame VH-(DH)JH rearrangement of the IgH genes, Ig μ-H chain (Igμ) proteins are expressed on the cell surface in conjunction with the surrogate L chains lambda5 and VpreB and the signal-transducing Igα and Igβ proteins as the pre-BCR complex (Fig. 1A). Signaling through the pre-BCR promotes IgH allelic exclusion, clonal expansion of pre-B cells, and activation of IgL gene rearrangements and transcription. Following in-frame VL-JL rearrangements and IgL expression, IgL and IgH pair to form surface IgM, at which point immature B cells are tested for reactivity against self-antigens (central tolerance). B cells that react to self-antigens with high avidity are deleted (negative selection) or undergo receptor editing with expression of alternative IgLs. B cells with low-avidity interactions or no reactivity to self-antigens become anergic or are positively selected and migrate out of the bone marrow (BM) to the spleen where development continues (9, 10).
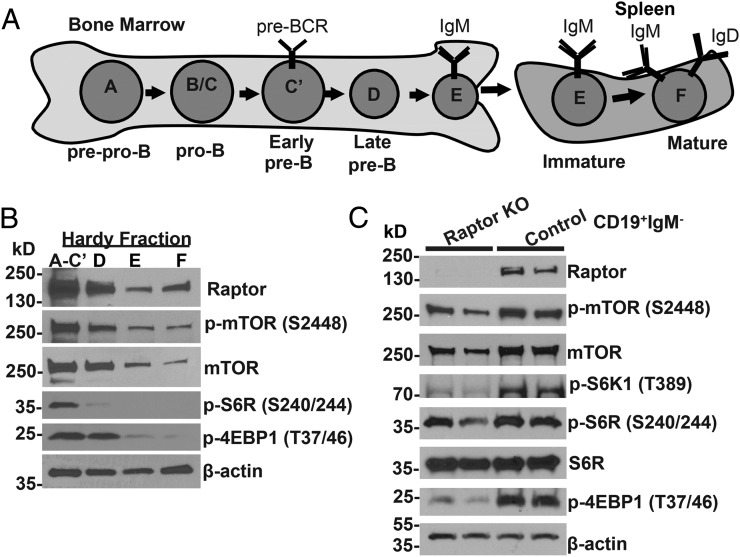
FIGURE 1.
mTOR signaling is normally activated in early B cell development and is decreased in Raptor-deficient mice. (A) Diagram of mouse B cell developmental stages with Hardy fraction notations (9). (B) BM B cells corresponding to Hardy fractions A–C′ (B220+CD43+), D (B220+CD43−IgM−), E (B220loCD43−IgM+), and F (B220hiCD43−IgM+) were purified from six WT mice and pooled. Immunoblotting was performed using Abs with specificity for the indicated proteins. β-actin was used as a loading control. (C) CD19+IgM− BM cells were isolated from two Raptor-null and two WT control mice. Immunoblotting was performed using Abs with specificity for the indicated proteins.
Pre-BCR and IL-7R activate PI3Ks, membrane-bound lipid kinases that can activate multiple signaling pathways (11, 12), including mTORC1 and mTORC2 (13). Gene-targeting studies in mice indicate that activation of PI3Ks is essential for B cell development beyond the pre-B cell stage (14–16). Deletion of Rictor revealed that mTORC2 is important for mature B cell development (17). However, B cell–specific roles for mTORC1 in early B cell development are unclear. In this study, we conditionally deleted the mTORC1 coactivator Raptor specifically in B cells during early B cell development in mouse BM using the Cre-LoxP system. Unlike deletion of mTOR (which targets mTORC1 and mTORC2), deletion of Raptor allowed us to selectively target mTORC1 during early B cell development. We found that mTORC1 signaling is essential for B cell development beyond the pre-B cell stage and plays a critical role in engendering IgH protein expression, pre-B cell survival, and optimal glycolytic and respiratory capacity required to fuel B cell division and Ab production.
Materials and Methods
Mice
Raptorfl/fl mice were described previously (18). Mice with Raptor deletion in the B cell lineage were generated by crossing Raptorfl/fl mice (obtained from The Jackson Laboratory) with transgenic Mb1-Cre mice (obtained from M. Reth, Max Planck Institute of Immunobiology and Epigenetics) (19). MD4(IgMHEL) mice, BclxL-transgenic mice, and Rag2−/−γc−/− mice were described previously (20–22). Mice were maintained in a specific pathogen–free facility at the University of Washington, and all procedures were reviewed and approved by the University of Washington Institutional Animal Care and Use Committee.
Flow cytometry
BM cells and splenocytes were stained with fluorescent-conjugated Abs with specificities for the following mouse Ags: B220 (RA3-6B2), IgM, CD43 (S7), CD22.2, CD25 (7D4), IAb (AF6-120.1), MHC class II, IgMa, IgMb, CD62L, Igμ, Ig κ-L chain (Igκ), heat stable Ag (HSA), or BP-1 (various fluorochromes). Mitochondrial staining was performed with MitoTracker Green FM and MitoSOX Red (Molecular Probes/Life Technologies). Flow cytometric data were acquired on a FACSCanto II or LSR II flow cytometer (BD Biosciences), and data were analyzed using FlowJo software.
Cell proliferation and cell viability
For in vivo BrdU-proliferation assays, mice were injected i.p. with 1 mg of BrdU (BD Pharmingen). BM cells were collected 16 h later, fixed, treated with DNase I, and stained with anti-BrdU Ab. For in vitro–proliferation assays, total BM cells were harvested, labeled with CFSE (Molecular Probes/Life Technologies), and cultured in the presence or absence of IL-7, stem cell factor (SCF), and Flt3 ligand (Flt3L) at 10 ng/ml for 3–4 d. Cells were then stained with anti-mouse B220 and IgM or CD43 fluorescent-conjugated Abs. For cell-viability assays, cells were stained with Annexin V (BD Biosciences) or CellEvent Caspase-3/7 (Molecular Probes/Life Technologies), and 7-aminoactinomycin D (7-AAD; BD Biosciences) or Ghost Dye Live/Dead Stain (Tonbo Biosciences).
Immunoblotting
Immunoblotting was performed, as previously described (23, 24), on FACS-sorted BM B cells corresponding to the pro/pre-B cell fraction B220loCD43+, Hardy fractions A–F, or MACS-sorted (Miltenyi Biotec) CD19+IgM− BM B cells. The following Abs were used: Raptor (Bethyl Laboratories), p-mTOR (S2448), mTOR, p-S6K1 (T389), phosphorylated ribosomal S6 protein (S6R; S240/244), p-4EBP1 (T37/46), p-AKT (T308), p-AKT (S473), AKT, p-Foxo1 (S256), Foxo1, p-ERK (T202/Y204), and ERK (Cell Signaling). β-actin (Sigma) was used as a loading control. Densitometry analysis was performed using ImageJ software.
Quantitative RT-PCR
B220+CD43+ pro/pre-B cells were FACS sorted using a BD FACSAria III. RNA was isolated using an Ambion RNAqueous-4PCR kit. cDNAs were made using Superscript III reverse transcriptase (Invitrogen). Quantitative RT-PCR was performed using SYBR Green Master Mix (ABI) on a Stratagene Mx3005 qPCR System. Primer sequences are listed in Supplemental Table I.
Immunization
Mice were immunized with keyhole limpet hemocyanin (KLH) protein (Calbiochem), and KLH-specific Ab production was measured 4 wk later using KLH-coated ELISA plates, as previously described (23).
Gene-rearrangement analysis
CD19+IgM− or B220+ BM cells were purified by MACS purification columns (Miltenyi Biotec), and DNA was extracted using a QIAGEN DNeasy kit. IgH and IgL PCR reactions were performed as previously described (25, 26). Amplification products were separated on a 1.5% agarose gel and transferred to a positively charged nylon membrane. The membrane was probed with biotin-labeled JH3 oligonucleotide and detected using the North2South Chemiluminescent Detection Kit (Thermo Scientific). Primers used are listed in Supplemental Table I.
Extracellular flux analysis
CD19+IgM− B cells were purified by MACS purification columns (Miltenyi Biotec) and cultured in complete medium for 16 h in the presence of IL-7 (10 ng/ml). Before measurements, medium was changed to unbuffered DMEM supplemented with 10 mM glucose, 2 mM glutamine, and 1 mM pyruvate. Cells were seeded at 1 × 106 cells/well in XF24 tissue culture plates precoated with Cell-Tak (BD Biosciences). Oligomycin, carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), rotenone, and antimycin A were added to the media sequentially. Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using a Seahorse XF24 analyzer (Seahorse Bioscience) and analyzed using WAVE software.
Rapamycin treatment
Rapamycin (LC Labs) was dissolved in ethanol, diluted in PEG400/Tween 80, and sterile-filtered though a 0.2-μM filter. C57BL/6 mice were treated i.p. daily for 3 wk with rapamycin (8 mg/kg) or vehicle alone. BM was harvested for flow cytometry at the end of 3 wk.
Statistical analysis
All statistical analyses were performed using GraphPad Prism software. Statistical differences were determined using the Student t test with Holm–Sidak correction for multiple comparisons or two-way ANOVA. All data are presented as mean ± SD.
Results
mTORC1 signaling is differentially activated during early B cell development
B lymphocyte development within the BM and fetal liver proceeds through a series of developmental stages that are defined by the differential expression of specific cell surface markers (27). B cells corresponding to Hardy fractions A–C′ consist of prepro B cells (fraction A; B220loCD43+HSAlo, BP1−, IgH−, IgL−), pro-B cells (fraction B; B220loCD43+HSAmid, BP1−, IgH−, IgL−), and large pre-B cells (fraction C/C′; B220loCD43+HSAmid/hi, BP1+, IgH+, IgL−) (Fig. 1A). B cells in fraction D correspond to late/small pre-B cells (B220loCD43−IgM−), whereas fractions E and F correspond to immature (B220loCD43−IgM+IgD−) and mature (B220hiCD43−IgM+IgD+) long-lived recirculating B cells, respectively. To determine how mTORC1 signaling is modulated during normal B cell development, we sorted mouse BM cells from wild-type (WT) mice and examined expression and activation of proteins involved in the mTORC1 signaling pathway. mTORC1 proteins mTOR and Raptor were expressed at higher levels in large pro-B, large pre-B, and small pre-B cells (fractions A–C′ and D) and were decreased in expression in small resting immature and mature B cells (fractions E and F) (Fig. 1B, Supplemental Fig. 1A). p-S6R and p-4EBP1, two downstream proteins that are phosphorylated by activated mTORC1, were also increased in fractions A–D. Thus, these results indicate that mTORC1 signaling is most active during the pro-B and pre-B stages of B cell development and declines as cells acquire expression of IgM and IgD during the immature and mature B cell stages.
Deletion of Raptor in the B cell lineage results in disruption of mTORC1 signaling and a block in B cell development
To determine the role of mTORC1 in B cell development, we deleted the mTORC1 activating protein Raptor specifically in B cells by breeding Raptorfl/fl mice (18) with Mb1-Cre mice, which express the Cre enzyme beginning at the pro-B cell stage under control of the B cell–specific Igα promoter (19). To confirm that deletion of Raptor disrupted mTORC1 signaling, we isolated CD19+IgM– pro-B and pre-B cells from WT and Raptorfl/flMb1-Cre BM and examined expression and activation of mTORC1 signaling proteins. As expected, we noted loss of Raptor protein expression in Raptorfl/flMb1-Cre cells (Fig. 1C, Supplemental Fig. 1A). In addition, we noted decreased total mTOR, p-S6K1, p-S6R, and p-4EBP1 in Raptor-null B cell progenitors compared with WT B cell progenitors. These results indicate that B cell–specific disruption of Raptor in Raptorfl/flMb1-Cre mice results in inhibition of mTORC1 signaling in B cell progenitors.
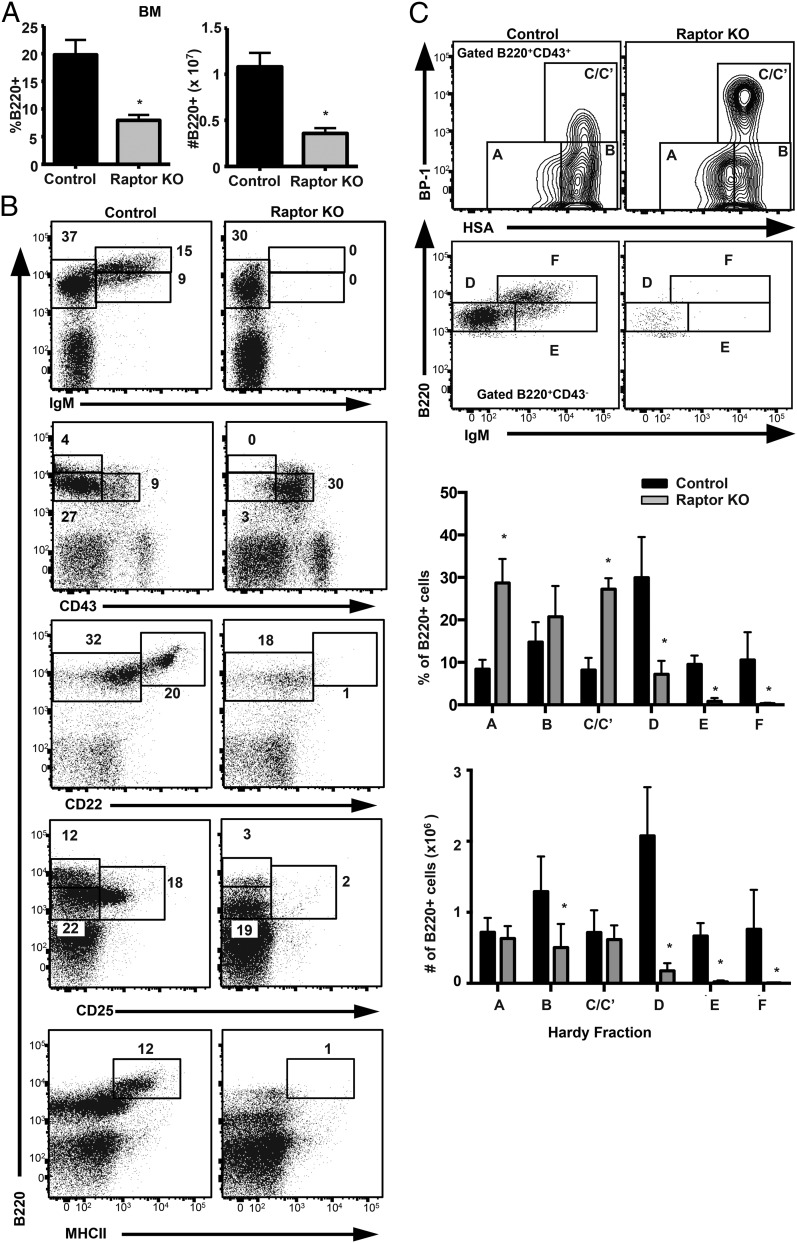
We then used flow cytometry to evaluate the consequences of Raptor disruption and inhibition of mTORC1 signaling on B cell development within the BM, spleen, and peritoneum of Raptorfl/flMb1-Cre mice relative to WT controls. Strikingly, Raptor deficiency resulted in reduced total BM B cells, which was characterized by a complete loss of immature (B220loIgM+) and mature (B220hiIgM+) B cell populations (Fig. 2A, 2B). Analysis of cell surface markers that are differentially expressed during B cell development revealed a block in development at the B220+CD43+CD22−CD25−MHCII− pre-B cell stage (Fig. 2B). Raptor-deficient BM B cells exhibited a decrease in the number and percentage of lymphocytes corresponding to Hardy fractions D–F (late pre-B to mature B cell), as well as increased percentages of prepro-B (fraction A) and early pre-B (fraction C/C′) cell populations (Fig. 2C). Our results indicate that loss of Raptor results in a B cell developmental block from the early/large (fraction C/C′) to late/small pre-B cell (fraction D) stage.
FIGURE 2.
mTOR signaling is required for pre-B cell development. (A) BM cells from Raptor-null and WT control mice (n ≥ 5 mice per genotype) were analyzed by flow cytometry. The percentage (left panel) and number (right panel) of B220+ lymphocytes were calculated for each genotype. Data are mean ± SD and were calculated from two independent experiments. (B) BM cells from Raptor-null and WT control mice were stained with fluorescent-conjugated Abs and analyzed by flow cytometry. Shown are representative plots (n ≥ 6 mice per genotype) from three independent experiments. (C) BM cells from Raptor-null and WT control mice were stained with fluorescent-conjugated Abs and analyzed by flow cytometry. Shown are representative plots (n ≥ 6 mice per genotype) from three independent experiments. The percentage and number of B220+ lymphocytes in each Hardy fraction were calculated for each genotype. Data are mean ± SD pooled from three independent experiments. *p < 0.05.
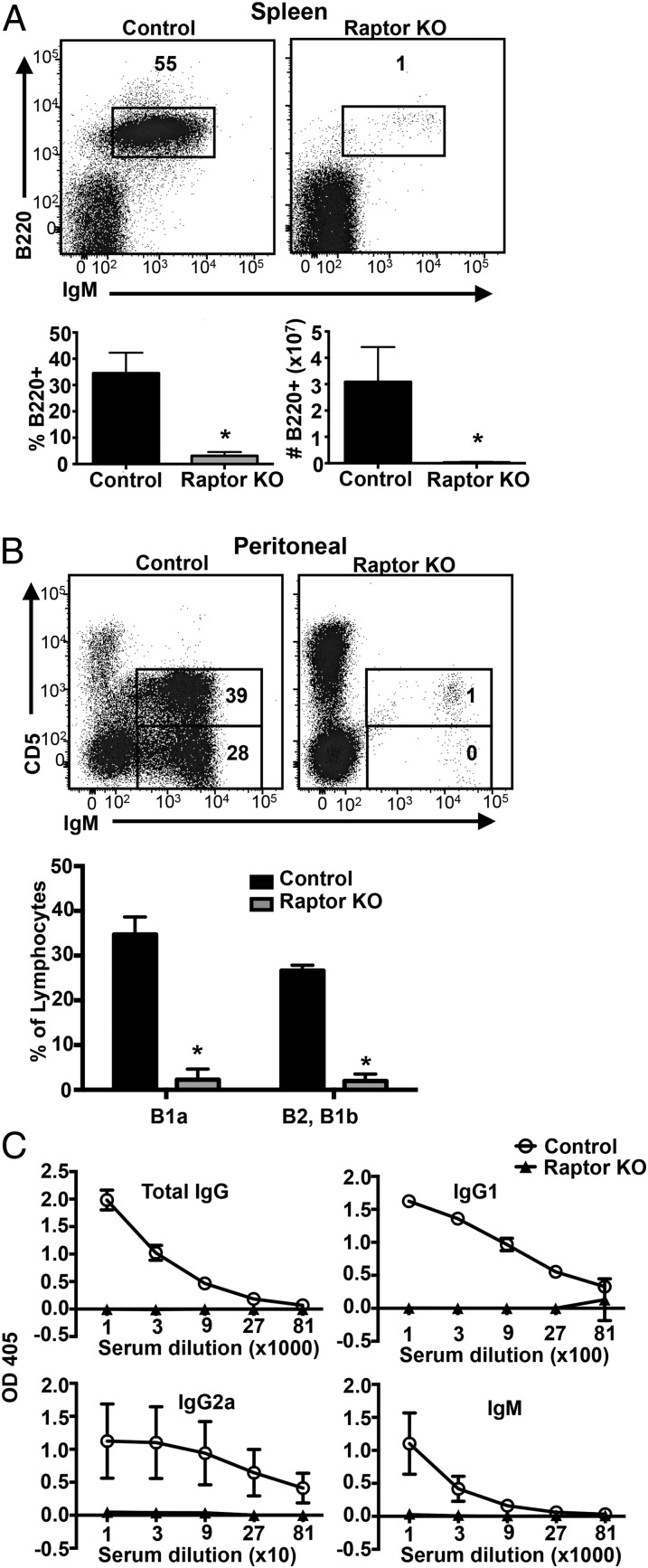
We further characterized the consequences of Raptor depletion on peripheral B cell development. Consistent with our findings in BM, flow cytometric examination of splenocytes revealed a marked decrease in the percentage and total number of B220+ splenic B cells in Raptorfl/flMb1-Cre mice versus WT control mice (Fig. 3A). Similarly, analysis of peritoneal lymphocytes revealed a marked decrease in conventional B2 and innate-like CD5+IgM+ B1a cells (Fig. 3B). Immunization of Raptorfl/flMb1-Cre mice and WT control mice with the T cell–dependent Ag KLH revealed the complete absence of KLH-specific total IgG, IgG1, IgG2a, and IgM in Raptor-deficient mice compared with control mice (Fig. 3C). Collectively, these results indicate that Raptor is required to promote B cell development from the large pre-B to small pre-B cell stage and for the generation of mature Ab-producing B cells and plasma cells.
FIGURE 3.
Raptor-deficient mice lack peripheral B cells and exhibit impaired Ab production. Splenocytes (A) and peritoneal cells (B) from Raptor-null and WT control mice were stained with fluorescent-conjugated Abs and analyzed by flow cytometry. Percentage and number of B220+ cells were calculated for each genotype. Data are mean ± SD (n ≥ 5 mice per genotype) pooled from three independent experiments. (C) Raptor-null (n = 5) and control (n = 5) mice were immunized with KLH Ag. Sera were collected 4 wk postimmunization and analyzed by ELISA for the indicated KLH-specific Igs. *p < 0.05.
We next focused on determining why Raptor is essential for B cell development beyond the pre-B cell stage. Previous studies showed that the PI3K–AKT–Foxo1 signaling pathway regulates expression of critical pre-B cell genes, including Rag1 and Rag2 (28, 29). Disruption of the PI3K–AKT–Foxo1 pathway results in impaired IgH gene rearrangement and expression (29), as well as a pre-B cell developmental block (14–16). We reasoned that, as a downstream effector of PI3K-AKT, mTORC1 may also play a role in regulating expression of pre-B cell genes required for normal development. We first examined whether mTORC1 signaling deficiency results in altered PI3K–AKT–Foxo1 activation. We found that Raptor-deficient B cells displayed similar levels of AKT activation, as shown by phosphorylation of AKT at T308 and S473 and phosphorylation of Foxo1 (Supplemental Fig. 1B). We next examined mRNA levels of factors known to be required for pre-BCR signaling in FACS-sorted B220+CD43+ cells. Quantitative RT-PCR was performed to determine transcript abundance of key factors required for IgH and IgL gene rearrangement and pre-B cell development (including Ebf1, Pax5, Foxp1, Rag1, Rag2, E2A, lambda5, Vpreb, CD79a, and CD79b). No significant deficiencies in transcript levels were found in Raptor-null versus WT pre-B cells (Supplemental Fig. 1C). Interestingly, expression of the pre-BCR components lambda5, Vpreb1, and Vpreb2 were increased in Raptor-null B cell progenitors compared with WT B cell progenitors (Supplemental Fig. 1C). This is consistent with increased percentages of pre-B cells in the B220+CD43+HSAhiBP-1hi fraction (Fig. 2C), which express the highest levels of these pre-BCR components (30).
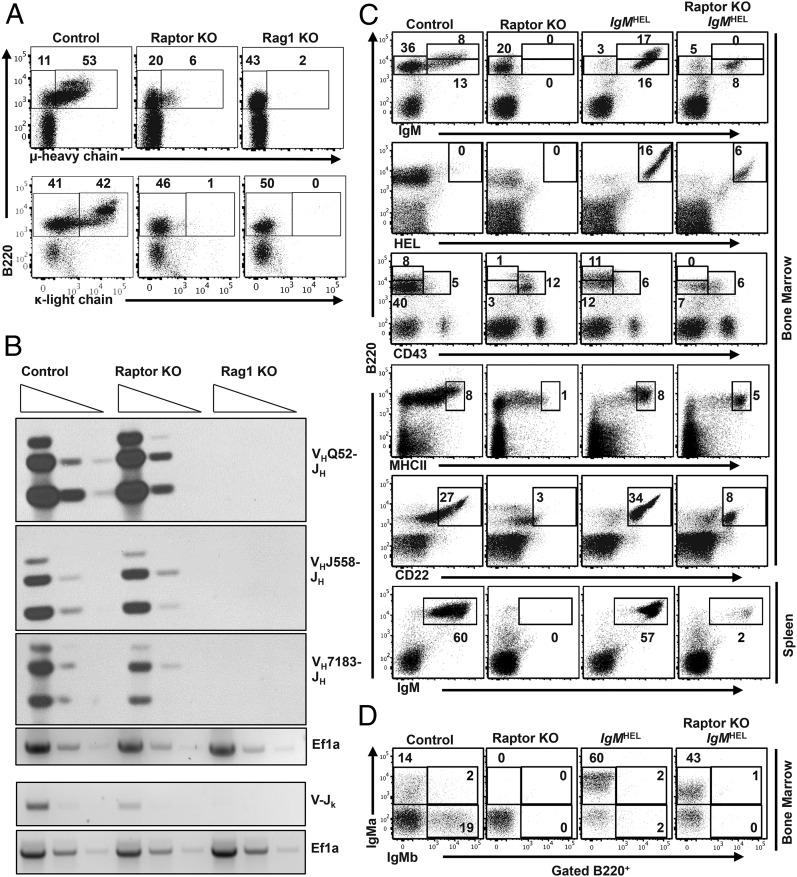
We then examined whether protein expression and/or rearrangement of the IgH or IgL gene was affected in Raptor-deficient B cells. We performed flow cytometry to examine the relative expression of intracellular (IC) Igμ and Igκ proteins. We found that expression of IC Igμ was markedly reduced, but still present, in Raptor-deficient B cells compared with WT B cells, whereas IC Igκ protein was completely absent (Fig. 4A). In contrast, surface lambda5 protein was present at elevated levels on Raptor-deficient B cell progenitors (Supplemental Fig. 1D), consistent with increased levels of lambda5 mRNA. We then tested whether IgH and IgL gene rearrangements are disrupted in Raptor-deficient B cells. Semiquantitative PCR analysis for VH-(DH)JH rearrangement revealed normal IgH gene rearrangement in Raptor-deficient B cells relative to WT positive controls and Rag1−/− negative control cells (Fig. 4B). Igκ gene rearrangement was reduced in Raptor-knockout (KO) cells relative to WT cells, consistent with a defect in pre-BCR signaling. These results suggest that Raptor-deficient B cells undergo normal VH-(DH)JH rearrangement but fail to properly express and/or respond to pre-BCR signals, resulting in a developmental block at the pre-B cell stage.
FIGURE 4.
Expression of Ig H and L chains are impaired in Raptor-deficient B cells, and enforced expression of IgMHEL transgene does not restore normal development. (A) BM cells from Raptor-null and WT control mice were stained with anti-B220 and anti-IC Igμ or Igκ Abs. Rag1-null mice were used as a control for loss of μ and κ expression. Data are representative of three independent experiments (n ≥ 5 mice per genotype). (B) DNA was isolated from CD19+IgM− pro- and pre-B cells from mice of the indicated genotypes. DNA was serially diluted, and PCR was used to amplify rearranged IgH and IgL genes. The EF1a gene was used as a loading control. Data shown are representative of two independent experiments. (C and D) BM and splenocytes from WT control, Raptorfl/flMb1-Cre, IgMHEL, and Raptorfl/flMb1-Cre IgMHEL mice were stained with the indicated Abs and analyzed by flow cytometry. Shown are representative plots (n = 4 mice per genotype) from two independent experiments.
To further test whether impaired pre-BCR signaling is responsible for the B cell developmental block, we crossed Raptorfl/flMb1-Cre mice to mice expressing transgenic IgM H and L chain proteins specific for the hen egg lysozyme (HEL) Ag (IgMHEL) (20). We then examined whether enforced expression of surface IgMHEL, which mimics pre-BCR formation, could drive B cell maturation in the absence of mTOR activation. Expression of IgMHEL in Raptor-deficient B cells partially stimulated maturation of a small population of Raptor-null cells, as determined by downregulation of CD43, as well as upregulation of CD22 and MHC class II expression (Fig. 4C). However, expression of IgMHEL on Raptorfl/fl Mb1-Cre pre-B cells almost completely failed to rescue immature B cell development in the spleen (Fig. 4C, Supplemental Fig. 2A), suggesting that signaling responses downstream of the pre-BCR are impaired.
To determine whether allelic exclusion occurs normally following pre-BCR expression in Raptorfl/fl Mb1-Cre mice, we determined whether expression of IgMHEL (IgMa isoform) could efficiently extinguish expression of endogenous IgM (IgMb isoform) in the presence or absence of Raptor. We found that allelic exclusion occurred normally in Raptorfl/flMb1-Cre IgMHEL mice, as seen by the exclusive expression of the “a” isotype of IgM expressed in IgMHEL+ animals, compared with the mixture of endogenous “a” and “b” isotypes expressed in the WT littermate controls (Fig. 4D). Expression of IgMHEL also extinguished endogenous VH-(DH)JH gene rearrangement in the presence and absence of Raptor, as determined by semiquantitative PCR in Raptorfl/flMb1-Cre IgMHEL mice compared with WT controls and Raptor-KO B cell progenitors (Supplemental Fig. 2B). These results indicate that the block in B cell development observed in Raptorfl/flMb1-Cre mice occurs from a combination of impaired IgH protein expression and impaired pre-BCR signaling, which, when overcome by enforced IgM transgene expression, allows for a small population of B cell progenitors to proceed to the immature B cell stage but no further.
To determine whether the defect in B cell development is cell autonomous, we performed BM transplantation in which WT or Raptorfl/flMb1-Cre BM cells were transplanted into Rag2−/−γc−/− mice, which lack B, T, and NK lineage cells (22). As expected, WT cells reconstituted IgM+ B lineage cells normally in BM and spleen, whereas Raptorfl/flMb1-Cre cells remained blocked at the B220+CD43+IgM− pre-B cell stage (Supplemental Fig. 2C). These results, combined with the relative selectivity of Mb1-Cre for deletion of floxed alleles in B lineage cells suggests that conditional disruption of Raptor results in a block in B cell development at the pre-B cell stage in a cell-autonomous manner.
Raptor-deficient B cell progenitors have reduced proliferative capacity and undergo increased cell death
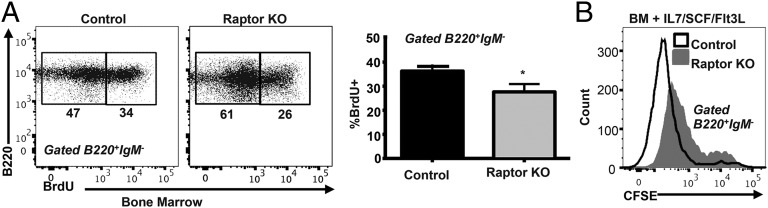
We next addressed whether Raptorfl/flMb1-Cre B cells exhibit defective cell-proliferative capacity and/or increased cell death, both of which could account for the failure to progress beyond the pre-B cell stage. We first evaluated cell proliferation in vivo using a BrdU-incorporation assay. Compared with WT control mice, Raptor-deficient pro-B/pre-B cells exhibited a small, but significant, decrease in BrdU incorporation, indicative of decreased cell proliferation (Fig. 5A). We then cultured CFSE-labeled BM cells from Raptorfl/flMb1-Cre mice and WT mice for 3–4 d in the presence of IL-7, SCF, and Flt3L. Both control and Raptor-deficient B cells proliferated in response to cytokine stimulation (Fig. 5B, Supplemental Fig. 3A); however, despite normal levels of IL-7R expression (Supplemental Fig. 3B), disruption of Raptor resulted in less robust proliferation, as reflected by reduced numbers of cell divisions.
FIGURE 5.
Raptor-deficient B cells exhibit decreased proliferation. (A) Raptor-null and WT control mice were injected with BrdU. BM was collected 16 h later, stained with fluorescent-conjugated cell surface markers against B220 and IgM, fixed, permeabilized, and stained with anti-BrdU Ab. Data are mean ± SD (n ≥ 4 mice per genotype) pooled from two independent experiments. (B) BM cells from Raptor-null and WT control mice were labeled with CFSE and cultured for 4 d in media containing IL-7/SCF/Flt3L cytokines. B220+IgM− cells were analyzed for CFSE dilution. Data shown (n = 3 mice per genotype) are representative of two independent experiments. *p < 0.05.
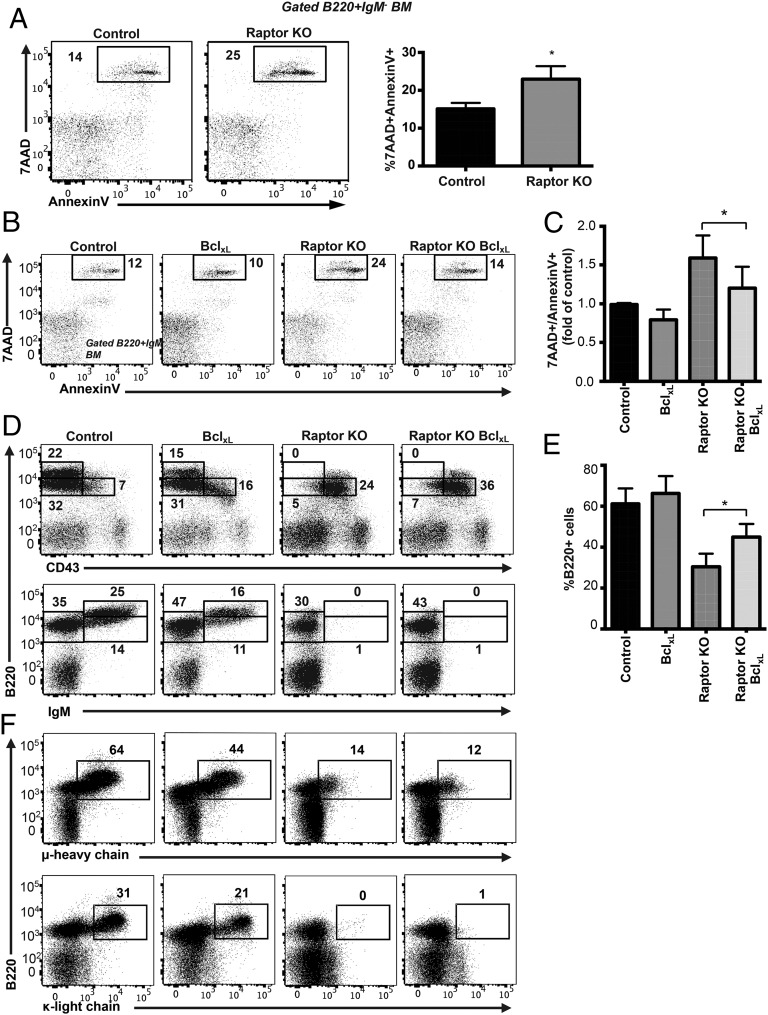
To examine the effects of Raptor deficiency on B cell survival, we stained BM cells with Annexin V and 7-AAD and analyzed cell survival of gated B220+IgM− pro-B/pre-B cells. Compared with control B cells, Raptor-deficient B cells had significantly elevated Annexin V and 7-AAD staining, indicating greater cell death (Fig. 6A). To determine whether increased cell death was responsible for the block in B cell development in Raptor-KO mice, we crossed Raptorfl/flMb1-Cre mice with mice expressing the anti-apoptotic BclxL transgene under the control of the IgH promoter (21). We found that expression of the BclxL transgene increased pre-B cell survival in Raptorfl/flMb1-Cre BclxL mice compared with Raptorfl/flMb1-Cre mice, as shown by a reduction in Annexin V/7-AAD+ staining (Fig. 6B, 6C). However, although the percentage and total number of B220+IgM− cells increased in Raptorfl/flMb1-Cre BclxL mice compared with Raptorfl/flMb1-Cre mice (Fig. 6D, 6E), we did not observe any restoration in B cell development pass the early pre-B cell stage in these mice (Fig. 6D, 6F). These results indicate that loss of mTOR signaling prevents further maturation of B cells beyond the pre-B cell stage in a BclxL-independent manner.
FIGURE 6.
Inhibiting apoptosis does not rescue B cell development of Raptor-deficient B cells. (A) BM cells and splenocytes from Raptor-null and WT control mice were stained with Annexin V and 7-AAD to assess cell death and analyzed by flow cytometry. Data are mean ± SD (n ≥ 4–6 mice per genotype) calculated from two independent experiments. (B and C) BM cells from mice of the indicated genotypes were stained with Annexin V and 7-AAD to assess cell death and analyzed by flow cytometry. Data are mean ± SD (n ≥ 5 mice per genotype) and are representative of at least four independent experiments. (D and E) BM cells from mice of the indicated genotypes were stained with fluorescent-conjugated Abs and analyzed by flow cytometry. Data are mean ± SD (n ≥ 5 mice per genotype) from at least two independent experiments. (F) BM cells from mice of the indicated genotypes were stained with anti-B220 and anti-IC Igμ or Igκ Abs. Data are representative of at least two independent experiments (n ≥ 5 mice per genotype). *p < 0.05.
Alterations in cellular bioenergetics in Raptor-deficient B cells
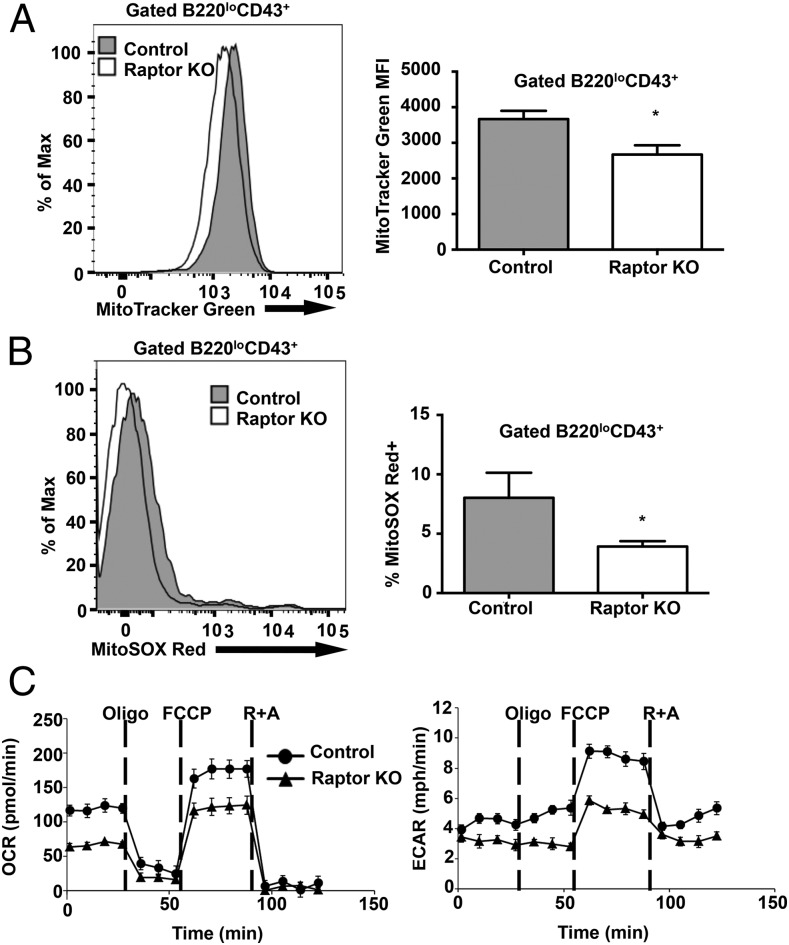
mTOR signaling plays a fundamental role in mitochondrial biogenesis, ATP production, and overall cellular metabolism, which can have profound effects on cell division and cell fate determination (31, 32). To determine whether loss of Raptor in pre-B cells affects mitochondrial biogenesis and function, we measured mitochondrial membrane potential in B cell progenitors from WT and Raptorfl/flMb1-Cre mice using the mitochondrial-selective probe MitoTracker Green FM. Pro-B and early pre-B cells from Raptor-deficient mice exhibited decreased MitoTracker Green staining compared with control mice, suggesting decreased mitochondrial mass (Fig. 7A). In addition, loss of Raptor resulted in decreased percentages of pro-B/pre-B cells containing high levels of mitochondrial superoxide, as measured by MitoSOX Red (Fig. 7B). To determine whether Raptor-deficient cells display reduced metabolic function, as suggested by these results, we measured oxidative phosphorylation and glycolytic potential in WT and Raptorfl/flMb1-Cre purified pre-B cells using the Seahorse XF24 Metabolic analyzer. We found that Raptor-deficient pre-B cells had reduced basal OCR and reduced respiratory capacity following treatment with the mitochondrial uncoupler FCCP, which is indicative of decreased oxidative phosphorylation. In addition, Raptor-deficient pre-B cells exhibited reductions in ECAR following treatment with oligomycin, which suggests decreased glycolytic potential. These results are consistent with reduced basal and maximal mitochondrial capacity following disruption of mTOR signaling in Raptor-null pre-B cells.
FIGURE 7.
Raptor-deficient B cells display altered metabolism. (A) BM cells from WT control and Raptor-null mice were stained with MitoTracker Green FM to analyze mitochondrial membrane potential. Data are mean ± SD (n ≥ 3 mice per enotype) and are representative of two independent experiments. (B) BM cells from WT control and Raptor-null mice were stained with MitoSOX Red to analyze mitochondrial reactive oxygen species. Data are mean ± SD (n ≥ 5 mice per genotype) pooled from two independent experiments. (C) CD19+IgM− B cells were isolated from BM of Raptor and WT control mice. OCR and ECAR were analyzed with a Seahorse Bioscience extracellular flux analyzer. Cells were cultured in the presence of IL-7 (10 ng/ml) for 24 h prior to analysis. Oligomycin (Oligo), FCCP, and rotenone and antimycin A (R+A) were added to the culture at the indicated time points. Data are mean ± SD (n = 3 mice per genotype) and are representative of two independent experiments. *p < 0.05.
Rapamycin inhibits B cell development in vivo
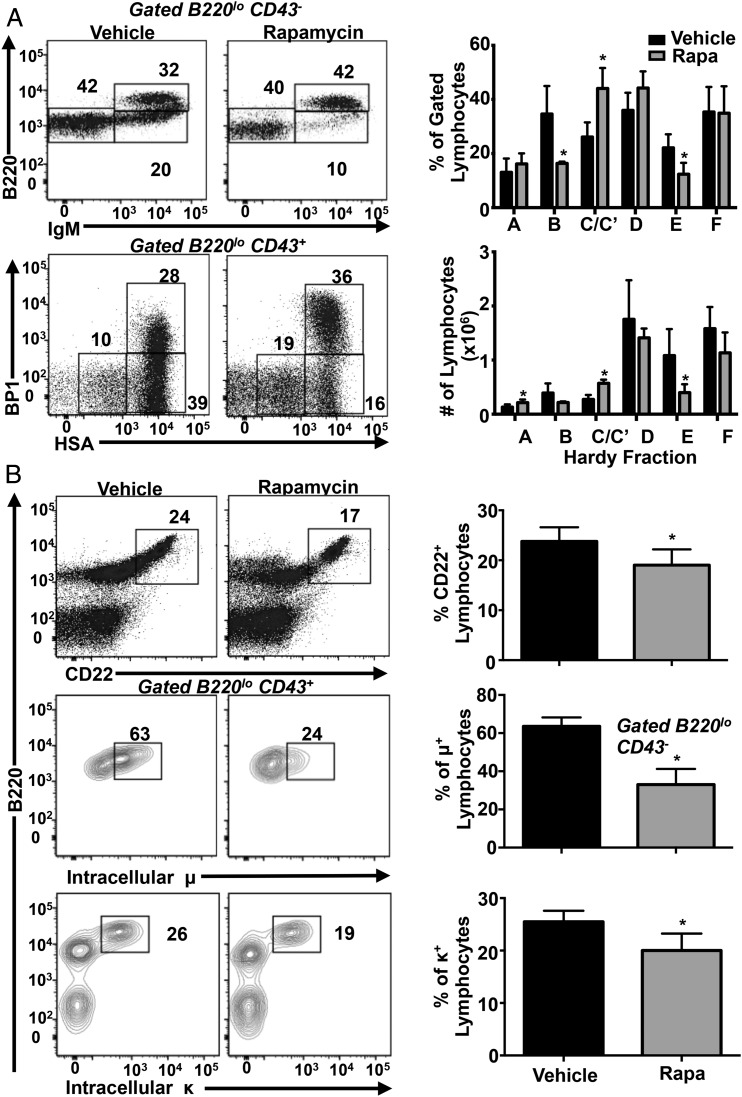
The classical mTOR inhibitor rapamycin acts, in part, by binding FKBP12 and allosterically inhibiting mTORC1 (2, 3). To determine whether rapamycin inhibits early B cell development, we treated WT mice with rapamycin or vehicle daily for 3 wk and analyzed B cell development in the BM. Rapamycin treatment resulted in a significant decrease in the percentage and total number of BM immature B (fraction E) cell populations (Fig. 8A). Similar to Raptorfl/flMb1-Cre mice, rapamycin-treated animals accumulated cells in the early pre-B cell stage (fraction C/C′). Furthermore, the percentages of surface CD22+ and IC Igμ+ and Igκ+ cells also were reduced in rapamycin-treated mice (Fig. 8B). These results, as well as similar previous findings (33), indicate that rapamycin inhibits early B cell development in mice.
FIGURE 8.
Rapamycin inhibits B cell development in vivo. (A) and (B) WT C57BL/6 mice were injected i.p. daily for 3 wk with vehicle or 8 mg/kg rapamycin (n = 5 mice per treatment). BM cells were stained with the indicated Abs and analyzed by flow cytometry. Representative dot plots are shown. Bar graphs show mean ± SD. *p < 0.05.
Discussion
In this study, we examined the role of mTORC1 signaling in early B cell development using a murine model of B cell–specific Raptor deficiency. Previous studies examined mTOR functions in B cells using mice with constitutive hypomorphic alleles of mTOR in all tissues (34), conditional deletion of Raptor in all tissues (35), or conditional deletion of mTOR or Raptor in later stages of B cell maturation (36, 37). These previous studies identified partial inhibition of B cell development in the BM, as well as deficits in marginal zone B cell and humoral immune responses following disruption of mTOR signaling (34–36). In contrast, our study reveals a complete block in development at the pre-B cell stage in mice with early B cell–specific disruption of Raptor. Importantly, deletion of Raptor using Mb1-Cre selectively targeted mTORC1, whereas previous studies using hypomorphic or floxed alleles of mTOR targeted mTORC1 and mTORC2. Our results identify a critical cell-autonomous role for mTORC1 signaling in promoting pre-B cell development, survival, proliferation, and metabolism and support the existence of a metabolic checkpoint controlling the development of B cell progenitors.
Our analysis of endogenous mTORC1 signaling in various stages of B cell development revealed stage-specific expression and activation of mTORC1 during the pro-B and pre-B cell stages, consistent with a critical role for mTOR signaling in early pre-B cell development. The pre-B cell stage marks an important transition point during which IL-7R signaling and pre-BCR signaling synergize to ensure that proliferation and Ig gene rearrangements are mutually exclusive (12). The IL-7R signaling pathway is required for proliferation and survival of pro-B and early pre-B cells, as well as for IgH gene rearrangement and expression (38). IL-7R signaling strongly activates the PI3K/AKT pathway, a major upstream regulator of mTORC1 signaling (1). In this study, we find that IgH gene rearrangements and expression of IL-7–dependent genes, including Ebf1, Rag1, Rag2, Pax5, Bcl2, BclxL, lambda5, and VpreB, occurred normally in mTORC1-deficient B cells. However, IL-7–induced cell proliferation and expression of IgH protein were impaired in pro/pre-B cells from mTORC1-deficient mice. These results suggest that mTORC1-dependent signaling may be required for IL-7–dependent cell growth and IgH protein expression or stabilization.
In addition to IL-7R signaling, pre-BCR signals are required for passage through the pre-B cell checkpoint. Formation of the pre-BCR signals successful in-frame rearrangement of the IgH gene segments leading to termination of further IgH gene rearrangement (allelic exclusion), a burst of cell proliferation, and IgL gene transcription and rearrangement (10). Aggregation of the pre-BCR results in activation of the Src-family protein tyrosine kinase Lyn and the Syk family protein tyrosine kinase Syk, which then phosphorylate ITAM motifs on target proteins, including the signal transducing receptors Igα and Igβ. This leads to recruitment of the adaptor protein BLNK, which further couples to downstream signaling pathways, including the Ras/Raf/Erk pathway and the PI3K/AKT/mTOR pathway. Targeted disruption of the genes encoding the PI3K catalytic subunits p110α and p110δ (16) or the regulatory subunit p85α (14, 15) results in blocks in B cell development at the pre-B cell stage. Similarly, targeted disruption of Foxo1 (28), a key transcription factor that is negatively regulated by AKT phosphorylation, also results in an early B cell developmental block due to the requirement of Foxo1 to activate transcription of critical pre-B cell genes, including Rag1 and Rag2 (39). Recently, disruption of PDK1, which phosphorylates and activates AKT downstream of PI3K, was also shown to result in reduced mTORC1 signaling, reduced IgH protein expression, and a block in B cell development at the pre-B cell stage (29, 40). In this article, we show that deletion of Raptor recapitulates the block in B cell development seen with disruption of the PI3K/AKT pathway, suggesting that mTORC1 may act downstream of PI3K/AKT signaling to promote IgH expression and pre-B cell development. However, although PI3K signaling–deficient pre-B cells fail to downregulate Rag1 and Rag2 expression, resulting in impaired allelic exclusion (16), we find that disruption of mTORC1 does not result in increased expression of Rag1 and Rag2, does not alter allelic exclusion, nor expression of other Foxo1 target genes including FasL, Il7r, and Bcl2l11. These results suggest that mTORC1 and Foxo1 may act independently to stimulate pre-B cell development downstream of PI3K.
Despite reduced levels of IgH protein and absent IgL protein in mTORC1-deficient mice, transgenic expression of surface IgM (which mimics pre-BCR formation) only minimally rescues B cell development; mTOR-deficient B cells are able to express IgMHEL transgene (albeit at reduced percentages) but fail to migrate out of the BM and complete B cell maturation in the spleen. These results contrast strikingly with expression of a transgenic BCR in Rag1−/− or Rag2−/− pro-B cells, which restores normal splenic B cell numbers relative to BCR-transgenic mice alone (24, 41, 42). mTORC1-deficient pre-B cells also exhibit reduced survival that we were able to rescue, in part, by expression of a BclXL transgene. Inhibiting cell death by expression of prosurvival genes, such as BclXL and Bcl-2, was shown to rescue B cell developmental blocks occurring at the pro-B (43) and immature (44) stages of development. However, despite an ∼40% increase in pre-B cell survival, the BclxL transgene failed to rescue development of IgM+ Raptor-null B cells. These results indicate that mTORC1 is required for optimal expression of IgH protein and cell survival, as well as for normal pre-B cell differentiation downstream of the pre-BCR.
Treatment of mice with the allosteric mTORC1 inhibitor rapamycin inhibited early B cell development, as shown by an accumulation of cells corresponding to Hardy fractions C/C′ and depletion of fractions B and E. Notably, rapamycin treatment recapitulated the characteristic buildup of B220+CD43+HSAhiBP1hi cells that we observed in Raptorfl/fl Mb1-Cre mice. Rapamycin, as well as newer ATP-competitive mTOR catalytic site inhibitors, are used or being investigated for use as chemotherapeutic and immunosuppressive agents (45, 46). Our data suggest that chronic use of rapamycin may result in inhibition of normal B cell development in humans. Catalytic site inhibitors of mTOR, which target mTORC1 and mTORC1, are thought to have lesser effects on normal lymphocyte proliferation relative to rapamycin (47). Future studies examining dose-dependent in vivo effects of mTOR inhibitors on B cell development and function will be of interest to determine whether clinically relevant doses of these drugs recapitulate our findings in Raptor-deficient mice.
mTOR signaling is known to control T cell metabolism and differentiation. mTOR signaling promotes the shift from primarily oxidative metabolism to glycolytic metabolism during effector T cell differentiation (48). High mTORC1 activity stimulates glycolysis, which favors the generation of T effector cells, whereas low mTORC1 activity shifts metabolism primarily to oxidative metabolism and enhances T regulatory and T memory differentiation (5). Our data show that loss of mTORC1 signaling results in global metabolic changes leading to reductions in oxidative phosphorylation and glycolysis during pre-B cell generation. These results suggest that the pre-BCR checkpoint may require one or both metabolic pathways during development and that these metabolic activities are dependent upon intact mTORC1 signaling. Consistent with this finding, previous studies highlighted the importance of glycolysis in promoting B cell development (49). Specifically, early pre-B cells were shown to be particularly sensitive to inhibition of glycolysis with 2-deoxyglucose, resulting in fewer late pre-B and immature B cells (49). Given the importance of cell growth and the metabolic requirements for clonal expansion and the generation of bioprecursors required for Ab production, it may not be surprising that disruption of a key driver of protein, lipid, nucleotide, and mitochondrial biogenesis would impair B cell development. However, because the block in B cell development was nearly complete in the absence of Raptor, our studies support the existence of a metabolic checkpoint whereby developing pre-B cells are tested for sufficient activation of anabolic pathways before cell division and differentiation are permitted. Of note, the oncoprotein c-Myc, a potent driver of glycolysis and protein synthesis in B cells, is also activated by IL-7 and the pre-BCR and is required for normal pre-B cell development (23, 50–52). Hence, the developmental potential of B cells may be determined, in part, by the abilities of mTORC1 and c-Myc to stimulate glycolysis and synthesize sufficient IgH protein and other unidentified proteins required for pre-BCR signaling. Defining the differences in the proteome between pre-B cells from Raptor-null and WT mice may provide clues as to how B cell progenitors are tested for metabolic fitness.
Supplementary Material
Acknowledgments
We thank Jacky Chan, Lim Kang, Janella Kang, and Davina Kang for managing the mouse colony and genotyping mice.
This work was supported by National Institutes of Health Grants 1R25 OD010450 (to T.N.I. and B.M.I.); R21AI109020-01, RO1AI092093-01, and 1R56AI092093 (to B.M.I.); and 5K01OD010554-05 (to J.A.R.).
The online version of this article contains supplemental material.
- BM
- bone marrow
- 4EBP1
- eukaryotic translation initiation factor 4E-binding protein 1
- ECAR
- extracellular acidification rate
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- Flt3L
- Flt3 ligand
- HEL
- hen egg lysozyme
- HSA
- heat stable Ag
- IC
- intracellular
- IgH
- Ig H chain
- IgL
- Ig L chain
- Igκ
- Ig κ-L chain
- Igμ
- Ig μ-H chain
- KLH
- keyhole limpet hemocyanin
- mTOR
- mechanistic target of rapamycin
- mTORC
- mTOR complex
- OCR
- oxygen consumption rate
- pro-B
- progenitor B
- SCF
- stem cell factor.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Laplante M., Sabatini D. M. 2012. mTOR signaling in growth control and disease. Cell 149: 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimobayashi M., Hall M. N. 2014. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell Biol. 15: 155–162. [DOI] [PubMed] [Google Scholar]
- 3.Abraham R. T., Wiederrecht G. J. 1996. Immunopharmacology of rapamycin. Annu. Rev. Immunol. 14: 483–510. [DOI] [PubMed] [Google Scholar]
- 4.Powell J. D., Pollizzi K. N., Heikamp E. B., Horton M. R. 2012. Regulation of immune responses by mTOR. Annu. Rev. Immunol. 30: 39–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu C., Chapman N. M., Karmaus P. W. F., Zeng H., Chi H. 2015. mTOR and metabolic regulation of conventional and regulatory T cells. J. Leukoc. Biol. 97: 837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donnelly R. P., Loftus R. M., Keating S. E., Liou K. T., Biron C. A., Gardiner C. M., Finlay D. K. 2014. mTORC1-dependent metabolic reprogramming is a prerequisite for NK cell effector function. J. Immunol. 193: 4477–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei J., Yang K., Chi H. 2014. Cutting edge: discrete functions of mTOR signaling in invariant NKT cell development and NKT17 fate decision. J. Immunol. 193: 4297–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marçais A., Cherfils-Vicini J., Viant C., Degouve S., Viel S., Fenis A., Rabilloud J., Mayol K., Tavares A., Bienvenu J., et al. 2014. The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat. Immunol. 15: 749–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardy R. R., Hayakawa K. 2001. B cell development pathways. Annu. Rev. Immunol. 19: 595–621. [DOI] [PubMed] [Google Scholar]
- 10.Herzog S., Reth M., Jumaa H. 2009. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat. Rev. Immunol. 9: 195–205. [DOI] [PubMed] [Google Scholar]
- 11.Reth M., Nielsen P. 2014. Signaling circuits in early B-cell development. Adv. Immunol. 122: 129–175. [DOI] [PubMed] [Google Scholar]
- 12.Clark M. R., Mandal M., Ochiai K., Singh H. 2014. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat. Rev. Immunol. 14: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Limon J. J., Fruman D. A. 2012. Akt and mTOR in B cell activation and differentiation. Front. Immunol. 3: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki H., Terauchi Y., Fujiwara M., Aizawa S., Yazaki Y., Kadowaki T., Koyasu S. 1999. Xid-like immunodeficiency in mice with disruption of the p85alpha subunit of phosphoinositide 3-kinase. Science 283: 390–392. [DOI] [PubMed] [Google Scholar]
- 15.Fruman D. A., Snapper S. B., Yballe C. M., Davidson L., Yu J. Y., Alt F. W., Cantley L. C. 1999. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85alpha. Science 283: 393–397. [DOI] [PubMed] [Google Scholar]
- 16.Ramadani F., Bolland D. J., Garcon F., Emery J. L., Vanhaesebroeck B., Corcoran A. E., Okkenhaug K. 2010. The PI3K isoforms p110alpha and p110delta are essential for pre-B cell receptor signaling and B cell development. Sci. Signal. 3: ra60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee K., Heffington L., Jellusova J., Nam K. T., Raybuck A., Cho S. H., Thomas J. W., Rickert R. C., Boothby M. 2013. Requirement for Rictor in homeostasis and function of mature B lymphoid cells. Blood 122: 2369–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polak P., Cybulski N., Feige J. N., Auwerx J., Rüegg M. A., Hall M. N. 2008. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 8: 399–410. [DOI] [PubMed] [Google Scholar]
- 19.Hobeika E., Thiemann S., Storch B., Jumaa H., Nielsen P. J., Pelanda R., Reth M. 2006. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. USA 103: 13789–13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartley S. B., Crosbie J., Brink R., Kantor A. B., Basten A., Goodnow C. C. 1991. Elimination from peripheral lymphoid tissues of self-reactive B lymphocytes recognizing membrane-bound antigens. Nature 353: 765–769. [DOI] [PubMed] [Google Scholar]
- 21.Fang W., Mueller D. L., Pennell C. A., Rivard J. J., Li Y. S., Hardy R. R., Schlissel M. S., Behrens T. W. 1996. Frequent aberrant immunoglobulin gene rearrangements in pro-B cells revealed by a bcl-xL transgene. Immunity 4: 291–299. [DOI] [PubMed] [Google Scholar]
- 22.Garcia S., DiSanto J., Stockinger B. 1999. Following the development of a CD4 T cell response in vivo: from activation to memory formation. Immunity 11: 163–171. [DOI] [PubMed] [Google Scholar]
- 23.Habib T., Park H., Tsang M., de Alborán I. M., Nicks A., Wilson L., Knoepfler P. S., Andrews S., Rawlings D. J., Eisenman R. N., Iritani B. M. 2007. Myc stimulates B lymphocyte differentiation and amplifies calcium signaling. J. Cell Biol. 179: 717–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park H., Staehling K., Tsang M., Appleby M. W., Brunkow M. E., Margineantu D., Hockenbery D. M., Habib T., Liggitt H. D., Carlson G., Iritani B. M. 2012. Disruption of Fnip1 reveals a metabolic checkpoint controlling B lymphocyte development. Immunity 36: 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlissel M. S., Baltimore D. 1989. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell 58: 1001–1007. [DOI] [PubMed] [Google Scholar]
- 26.Schlissel M. S., Corcoran L. M., Baltimore D. 1991. Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J. Exp. Med. 173: 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardy R. R., Carmack C. E., Shinton S. A., Kemp J. D., Hayakawa K. 1991. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 173: 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dengler H. S., Baracho G. V., Omori S. A., Bruckner S., Arden K. C., Castrillon D. H., DePinho R. A., Rickert R. C. 2008. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat. Immunol. 9: 1388–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venigalla R. K., McGuire V. A., Clarke R., Patterson-Kane J. C., Najafov A., Toth R., McCarthy P. C., Simeons F., Stojanovski L., Arthur J. S. 2013. PDK1 regulates VDJ recombination, cell-cycle exit and survival during B-cell development. EMBO J. 32: 1008–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dul J. L., Argon Y., Winkler T., ten Boekel E., Melchers F., Mårtensson I. L. 1996. The murine VpreB1 and VpreB2 genes both encode a protein of the surrogate light chain and are co-expressed during B cell development. Eur. J. Immunol. 26: 906–913. [DOI] [PubMed] [Google Scholar]
- 31.Schieke S. M., Phillips D., McCoy J. P., Jr., Aponte A. M., Shen R.-F., Balaban R. S., Finkel T. 2006. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J. Biol. Chem. 281: 27643–27652. [DOI] [PubMed] [Google Scholar]
- 32.Morita M., Gravel S.-P., Chénard V., Sikström K., Zheng L., Alain T., Gandin V., Avizonis D., Arguello M., Zakaria C., et al. 2013. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 18: 698–711. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y., Hu T., Hua C., Gu J., Zhang L., Hao S., Liang H., Wang X., Wang W., Xu J., et al. 2014. Rictor is required for early B cell development in bone marrow. PLoS One 9: e103970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang S., Readinger J. A., DuBois W., Janka-Junttila M., Robinson R., Pruitt M., Bliskovsky V., Wu J. Z., Sakakibara K., Patel J., et al. 2011. Constitutive reductions in mTOR alter cell size, immune cell development, and antibody production. Blood 117: 1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoshii T., Tadokoro Y., Naka K., Ooshio T., Muraguchi T., Sugiyama N., Soga T., Araki K., Yamamura K., Hirao A. 2012. mTORC1 is essential for leukemia propagation but not stem cell self-renewal. J. Clin. Invest. 122: 2114–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S., Pruitt M., Tran D., Du Bois W., Zhang K., Patel R., Hoover S., Simpson R. M., Simmons J., Gary J., et al. 2013. B cell-specific deficiencies in mTOR limit humoral immune responses. J. Immunol. 191: 1692–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Limon J. J., So L., Jellbauer S., Chiu H., Corado J., Sykes S. M., Raffatellu M., Fruman D. A. 2014. mTOR kinase inhibitors promote antibody class switching via mTORC2 inhibition. Proc. Natl. Acad. Sci. USA 111: E5076–E5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corfe S. A., Paige C. J. 2012. The many roles of IL-7 in B cell development; mediator of survival, proliferation and differentiation. Semin. Immunol. 24: 198–208. [DOI] [PubMed] [Google Scholar]
- 39.Amin R. H., Schlissel M. S. 2008. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat. Immunol. 9: 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baracho G. V., Cato M. H., Zhu Z., Jaren O. R., Hobeika E., Reth M., Rickert R. C. 2014. PDK1 regulates B cell differentiation and homeostasis. Proc. Natl. Acad. Sci. USA 111: 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young F., Ardman B., Shinkai Y., Lansford R., Blackwell T. K., Mendelsohn M., Rolink A., Melchers F., Alt F. W. 1994. Influence of immunoglobulin heavy- and light-chain expression on B-cell differentiation. Genes Dev. 8: 1043–1057. [DOI] [PubMed] [Google Scholar]
- 42.Spanopoulou E., Roman C. A., Corcoran L. M., Schlissel M. S., Silver D. P., Nemazee D., Nussenzweig M. C., Shinton S. A., Hardy R. R., Baltimore D. 1994. Functional immunoglobulin transgenes guide ordered B-cell differentiation in Rag-1-deficient mice. Genes Dev. 8: 1030–1042. [DOI] [PubMed] [Google Scholar]
- 43.Strasser A., Harris A. W., Corcoran L. M., Cory S. 1994. Bcl-2 expression promotes B- but not T-lymphoid development in scid mice. Nature 368: 457–460. [DOI] [PubMed] [Google Scholar]
- 44.Amanna I. J., Dingwall J. P., Hayes C. E. 2003. Enforced bcl-xL gene expression restored splenic B lymphocyte development in BAFF-R mutant mice. J. Immunol. 170: 4593–4600. [DOI] [PubMed] [Google Scholar]
- 45.Schenone S., Brullo C., Musumeci F., Radi M., Botta M. 2011. ATP-competitive inhibitors of mTOR: an update. Curr. Med. Chem. 18: 2995–3014. [DOI] [PubMed] [Google Scholar]
- 46.Wander S. A., Hennessy B. T., Slingerland J. M. 2011. Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. J. Clin. Invest. 121: 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Janes M. R., Limon J. J., So L., Chen J., Lim R. J., Chavez M. A., Vu C., Lilly M. B., Mallya S., Ong S. T., et al. 2010. Effective and selective targeting of leukemia cells using a TORC1/2 kinase inhibitor. Nat. Med. 16: 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang K., Shrestha S., Zeng H., Karmaus P. W. F., Neale G., Vogel P., Guertin D. A., Lamb R. F., Chi H. 2013. T cell exit from quiescence and differentiation into Th2 cells depend on Raptor-mTORC1-mediated metabolic reprogramming. Immunity 39: 1043–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kojima H., Kobayashi A., Sakurai D., Kanno Y., Hase H., Takahashi R., Totsuka Y., Semenza G. L., Sitkovsky M. V., Kobata T. 2010. Differentiation stage-specific requirement in hypoxia-inducible factor-1alpha-regulated glycolytic pathway during murine B cell development in bone marrow. J. Immunol. 184: 154–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iritani B. M., Eisenman R. N. 1999. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc. Natl. Acad. Sci. USA 96: 13180–13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schuhmacher M., Staege M. S., Pajic A., Polack A., Weidle U. H., Bornkamm G. W., Eick D., Kohlhuber F. 1999. Control of cell growth by c-Myc in the absence of cell division. Curr. Biol. 9: 1255–1258. [DOI] [PubMed] [Google Scholar]
- 52.Le A., Lane A. N., Hamaker M., Bose S., Gouw A., Barbi J., Tsukamoto T., Rojas C. J., Slusher B. S., Zhang H., et al. 2012. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 15: 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.