Abstract
We describe here the vision, motivations, and research plans of the National Institutes of Health Center for Excellence in Big Data Computing at the University of Illinois, Urbana-Champaign. The Center is organized around the construction of “Knowledge Engine for Genomics” (KnowEnG), an E-science framework for genomics where biomedical scientists will have access to powerful methods of data mining, network mining, and machine learning to extract knowledge out of genomics data. The scientist will come to KnowEnG with their own data sets in the form of spreadsheets and ask KnowEnG to analyze those data sets in the light of a massive knowledge base of community data sets called the “Knowledge Network” that will be at the heart of the system. The Center is undertaking discovery projects aimed at testing the utility of KnowEnG for transforming big data to knowledge. These projects span a broad range of biological enquiry, from pharmacogenomics (in collaboration with Mayo Clinic) to transcriptomics of human behavior.
Keywords: BD2K, Network analysis, genomics, big data, scalable
INTRODUCTION
Biology in the 21st century has emerged as a “big data” science on par with physics or astronomy. Widespread adoption of high throughput “omics” technologies has created massive amounts of data, yet there is a consensus that the floodgates have only barely opened.1 The explosive growth of data volume has fostered intense research in the development of informatics tools to store, manage, and analyze such data.2 However, the scale and efficiency of analysis are lagging behind the generation of data, a fact recognized by the major national funding agencies, which results in the true potential of the data to accelerate biological discovery not being realized.
Analysis of biological data today is hampered by 2 major bottlenecks:
(1) Integration. Different biotechnological tools record different kinds of cellular activities that provide complementary views of the same underlying biological phenomena. However, it has proved extremely difficult to integrate those partial descriptions into a well-organized whole, even though the advantages of such an integrative analysis of diverse data types are well recognized.3
(2) Scalability. The challenge of integrative data analysis is generally met with the most heavy-duty machine learning techniques of the day, which typically do not scale well with data size. Biology needs analysis tools that can handle the data deluge of its modern omics era.
The recently established National Institutes of Health (NIH) Center for Excellence in Big Data Computing at the University of Illinois at Urbana-Champaign (UIUC) is addressing the issues of integrative analysis and scalability associated with big data analysis in biology. This Center named “KnowEnG, a Scalable Knowledge Engine for Large-Scale Genomic Data” (http://knoweng.org), will build a new E-science infrastructure from the ground up, laying its algorithmic foundations, engineering the scalable systems that form its skeleton frame, and creating the human-computer interface that makes it hospitable.
KNOWLEDGE-GUIDED ANALYSIS OF USER DATA
A common paradigm of genomic data analysis today is to perform statistical analysis of an experimental data set and then to interpret the results in the light of prior knowledge from the literature or from publicly available genomic data sets. Using BLAST (Basic Local Alignment Search Tool)4 to identify related genes and then importing annotations of those genes is perhaps the most common example of this paradigm. Gene set enrichment analysis (exemplified by the GSEA tool5) is another popular example. Services such as DAVID (Database for Annotation, Visualization and Integrated Discovery)6 have gained widespread acceptance to facilitate this paradigm of knowledge-guided analysis; a survey in 2008 identified at least 68 such services.7 A large number of genomics studies perform “Gene Ontology”8 enrichments as a first look at the associated data. A web resource called Pathguide9 lists 547 online resources, cataloging biological pathways of different types or from different contexts. There has clearly been an outstanding worldwide push towards creating these knowledge bases. Yet, the ability of a biomedical scientist or clinician to use this incredible wealth of community knowledge in analyzing their data sets is limited mainly to the above-mentioned strategy of gene set enrichment tests or to manual navigation of the community knowledge with their gene of interest as the starting point. The general paradigm of knowledge-guided analysis is far from reaching its full potential. A few tools such as GENEMANIA10 have made valuable contributions in taking the paradigm towards more advanced analysis.
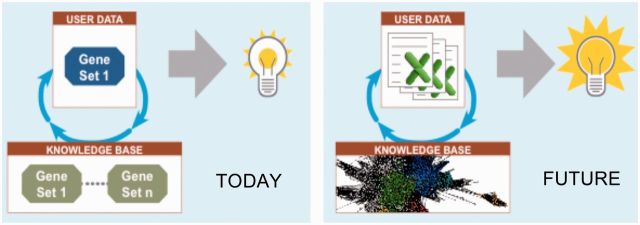
The Center’s goal is to build an E-science system called “Knowledge Engine for Genomics” (KnowEnG, pronounced “knowing”), founded upon this paradigm of combining the statistical analysis of experimental data with the context of existing community knowledge to extract the most useful insights from the experimental data. A key feature of the KnowEnG system will be the integration of prior knowledge during analysis of experimental data rather than the 2-step approach in vogue today where analysis is restricted to the user’s data and the results are then tested for associations with prior knowledge. In the KnowEnG framework, every user will be able to analyze their own experimental data sets (stored as spreadsheets) in light of the vast knowledge base of public-domain omics data sets available today (figure 1).
Figure 1:
The today figure illustrates a user’s gene set being tested for overlap with community gene sets. The future figure, KnowEnG, illustrates a user’s spreadsheets being analyzed in the light of a heterogeneous knowledge network, representing community data sets, leading to deeper insights.
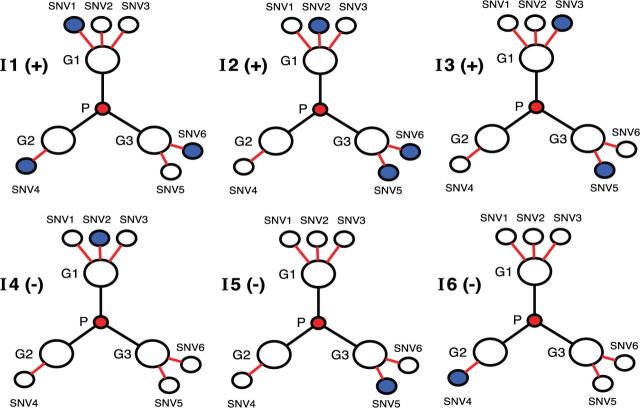
An oversimplified example of the rationale for the proposed approach is shown in figure 2 , where the user seeks to discriminate between 3 patients who respond to a drug (individuals I1, I2, I3) from 3 nonresponding patients (individuals I4, I5, I6), using information on variants they carry at 3 gene loci (G1, G2, G3). No individual gene locus cleanly separates the 2 groups of patients in terms of the presence of variants. However, the user observes that the 3 genes are members of the same pathway P; and when considering these 3 related genes, a simple discriminative rule appears—the number of variants at gene loci associated with pathway P is ≥ 2 in responding patients and ≤ 1 in nonresponding patients. This implicates pathway P as a potential determinant of drug response. In short, systematic inclusion of prior knowledge (in this case, pathway information) leads to an improved analysis of the user’s data set. This illustrates the basic premise of the KnowEnG system.
Figure 2:
Example of a knowledge-guided analysis. Prior knowledge that genes G1, G2, and G3 belong to the same pathway P allows discrimination of individuals I1, I2, and I3 from I4, I5, and I6 based on variants they carry (blue circles).
INNOVATIONS AT THE KNOWENG CENTER
User-data analysis in the context of prior knowledge, represented as a massive heterogeneous “Knowledge Network” (KN), will require efficient algorithms and implementations that scale well with the already voluminous and rapidly growing knowledge base of public data sets. The KnowEnG framework will bring the latest algorithmic technologies in large-scale network mining11 and machine learning,12 as well as the trending solutions to scalable computing13 and high performance computing, to bear on the most pressing bioinformatics needs of today. A central thesis of the Center’s research activities is that by abstracting a diverse set of bioinformatics tasks into a common framework and common informatics challenges, we will be able to streamline the development of core technology for biological big data analysis. This strategy will allow us and other data mining and information science experts to channel their expertise and innovations to the burning questions in biology today without being embroiled in the minutia of different data sets.
Our focus in building KnowEnG is not on the federation of different data sets and providing a visual interface to them; existing efforts have been quite successful in this regard.7,9,14,15 Instead, we will focus on the analysis of user-specific, modestly sized data sets in the context of massive, integrative collections of data in the public domain. We aim to go beyond access to support analysis. Just as the widely popular Galaxy Suite16–18 democratized scripting for genomics, KnowEnG aims to democratize analysis. Taking inspiration from the manner in which the Galaxy Suite has developed, the KnowEnG framework will be made available through a web portal linked to a commercial cloud and also will be installable on local cloud infrastructures of the user’s organization.
To address the scalability issue related to using big data, we are exploring ways to compute a projection of the KN into a problem-specific, low-dimensional space that captures the information most relevant to the user data set. Relevance is key to enable scalability because even though the data may grow quickly, the part of the big data relevant to a specific analysis tends to remain tractable. Examples of ideas to be pursued here include the following: (1) extracting the neighborhood of KN nodes representing the genes most informative for the user’s analysis; (2) precomputing indexes of such neighborhood information; (3) techniques for dimensionality reduction on the KN such as Diffusion Component Analysis;19 and (4) pathway-informed methods such as SPIA20 and Paradigm.21 The KnowEnG analysis framework is being designed from the ground up as a system for scalable computation with massive community knowledge bases, with a targeted deployment on cloud and cluster infrastructures.
User interfaces play a crucial role in the adoption of any research tool. Accordingly, the new framework will allow the biologist to perform different types of knowledge-assisted analyses of their data through a single, familiar interface rather than learn to use a different environment for each type of analysis. The focus is on (1) ease of use by using HUBzero,22 a heavily tested platform for web portal design; (2) innovative visualizations to be created by experts in user interface design; and (3) novel text-mining components that include automated literature summarization and “wikification” to support follow-up investigation of analysis results. Not only will users have access to different operations to analyze their data, there will be various means to help them decide which operations to use in a particular situation. The HUBzero platform will be utilized to provide robust and secure user access to the system, and to support analysis, visualization, collaboration, and training in a common setting.
The KnowEnG Center is also engaged in extensive and multiple evaluations of the new E-science framework in order to ensure usability and wide appeal. Evaluations are in the form of 3 major ongoing research projects, each with their own significant data sets, related to topics of great biomedical significance—cancer pharmacogenomics (in collaboration with Mayo Clinic), transcriptomics of social behavior, and discovery of novel antibiotics. For instance, the cancer pharmacogenomics application will allow biomedical scientists and clinicians to understand the molecular basis of drug response in different individuals, identify subtypes of a disease, and predict whether a particular drug is likely to be effective for a specific patient. By heavily engaging these active research projects for evaluation rather than relying solely on existing benchmarks, we hope to achieve an evaluation strategy that goes beyond assessing computational accuracy to judging biological relevance and suggesting modifications of the very goals of the analysis system.
SPECIFIC AIMS OF THE KNOWENG SYSTEM
The development and assessment of the KnowEnG system is being conducted through 7 major projects described below.
Collating and organizing data sets for building the KN. The KnowEnG framework will represent community data sets as a massive heterogeneous network, called the KN, composed primarily of genes and their annotations as well as mutual relationships. The community data sets will be obtained from popular resources such as STRING (Search Tool for the Retrieval of Interacting Genes/Proteins),14 GO (Gene Ontology),8 MSigDB (Molecular Signatures Database),23 Reactome,24 IntAct,25 InterPro,26 TCGA (The Cancer Genome Atlas),27 GDSC (Genomics of Drug Sensitivity in Cancer),28 etc. This project aims primarily to identify all relevant data sources, to download current versions, to setup a regular update schedule, and to construct the large graph representing the KN.
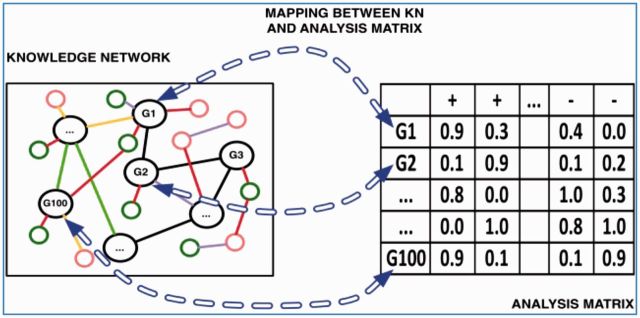
Analytics Suite: Core operations for knowledge-guided analysis of user data. The Analytics Suite is the collection of core operations for analyzing the user’s experimental data sets in the light of publicly available knowledge bases. The user data, provided as a spreadsheet, will be called the “Analysis Matrix.” Columns of the analysis matrix will correspond to macroscopic entities such as patients, species, or tissue types while rows will correspond to molecular entities, typically genes. Data in the analysis matrix will represent measurements of the microscopic entities (eg, genes) in each macroscopic entity (patients, species, tissue type, etc), eg, expression measurements on genes in each patient or presence/absence of genes in each species. Molecular entities such as genes, represented by rows of the analysis matrix, also feature as nodes in the KN, thus establishing a connection between the user data and prior knowledge (figure 3). The user will have the option of selecting which parts of the KN are most relevant to their analysis, and precomputed estimates of the relative value of different data sources for a given class of analysis will help the user in utilizing heterogeneous types of public data in their analysis. We will also address the challenge of potential discrepancies among different sources through suitable techniques of “truth finding”29–31 and data cleaning. The core operations constituting the Analytics Suite will act on the analysis matrix and the KN in an integrated manner and will be chosen so that a wide range of bioinformatics tasks are special cases of these operations.
Cloud-based, scalable implementation of the Analytics Suite. Robust software engineering practices are being employed to implement the Analytics Suite algorithms developed by the Center. The focus is on efficient implementations that scale well with the large and rapidly growing KN. The final deployment will be based on a commercial cloud platform accessible to users worldwide.
User interfaces and visualization of analysis results. KnowEnG will provide both text-based and graphical interfaces to the Analytics Suite. This includes a querying system and a Web-based front end tailored to access scalable computational resources and heterogeneous software components. Visualization tools will be incorporated with an emphasis on intuitive interactions with complex analytics. The user interface is being developed and tested in close collaboration with clinicians (from Mayo Clinic) and life science researchers (from Illinois).
Cancer pharmacogenomics. A major part of the evaluation of KnowEnG framework is through the formulation of new hypotheses based on the data emerging from 2 well-established translational studies in cancer pharmacogenomics: the Mayo Clinic BEAUTY breast cancer clinical trial (led by Dr Judy Boughey and Dr Matthew Goetz), and a model system consisting of 300 lymphoblastoid cell lines (led by Dr Liewei Wang). Hypotheses generated using the KnowEnG framework will be experimentally tested by researchers at Mayo Clinic.
Comparative transcriptomics. A second major evaluation project involves exploration and discovery of novel patterns in transcriptomic data, with a special emphasis on comparative transcriptomics. Transcriptomic data sets generated at the Institute of Genomic Biology at UIUC that explore the relationships between gene expression and behavioral states in humans, mice, stickleback fish, and honeybees will be subjected to analysis through KnowEnG.
Prediction of organismal phenotypes from annotated genomes. The KnowEnG system will be used also for prediction of phenotypic traits from fully or partially assembled and annotated genomes and their evolutionary relationships. Bacteria of the genus Streptomyces are capable of producing tens of thousands of chemicals secreted into their environment, many of which have medically useful properties such as antibiotic activity. The presence of the enzymes and metabolic pathways required for the synthesis of these compounds can be deduced from the presence of signature operons in the bacterial genomes. Such inference will be mapped into the core operations of the Analysis Suite, and KnowEnG will be used for a systematic genomics-based approach to antibiotic screening.
Figure 3:
Rows in Analysis Matrix (user’s spreadsheet) map to nodes in Knowledge Network, making joint analysis possible.
CONCLUSION
The KnowEnG framework will provide a common portal for genomics data analysis where the scientist will come with their own data sets in the form of spreadsheets and ask KnowEnG to analyze those data sets in the light of a massive knowledge base of community data sets. The Center is engaged in demonstrating that this paradigm of community knowledge-guided data analysis is applicable to a very wide range of biological and biomedical studies. The KnowEnG framework will empower biomedical scientists and clinicians with state-of-the-art algorithms and scalable systems that computer scientists can offer today. Its utility will be demonstrated on problems of immediate as well as long-term health relevance.
FUNDING
This work was supported by the NIGMS, grant 1U54GM114838, through funds provided by the trans-NIH Big Data to Knowledge (BD2K) initiative (www.bd2k.nih.gov).
COMPETING INTERESTS
None.
CONTRIBUTORS
The text of this manuscript has been borrowed from the proposal for grant 1U54GM114838. All authors contributed to writing the grant proposal and this manuscript.
REFERENCES
- 1.Pennisi E. Human genome 10th anniversary. Will computers crash genomics [published online ahead of print February 12, 2011]? Sci. 2011;331(6018):666–668. doi: 10.1126/science.331.6018.666. [DOI] [PubMed] [Google Scholar]
- 2.Ouzounis CA. Rise and demise of bioinformatics? Promise and progress. PLoS Computational Biol. 2012;8(4):e1002487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goble C, Stevens R. State of the nation in data integration for bioinformatics. J Biomed Inform. 2008;41(5):687–693. [DOI] [PubMed] [Google Scholar]
- 4.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. [DOI] [PubMed] [Google Scholar]
- 5.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery [published online ahead of print May 8, 2003]. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 7.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium [published online ahead of print May 10, 2000]. Nat Genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list [published online ahead of print December 31, 2005]. Nucleic Acids Res. 2006;34(Database issue):D504–D506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4 doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Y, Han J. Mining Heterogeneous Information Networks: Principles and Methodologies. Morgan & Claypool Publishers; 2012. [Google Scholar]
- 12.Witten IH, Frank E, Hall MA. Data Mining: Practical Machine Learning Tools and Techniques. Morgan Kaufmann; 2011. [Google Scholar]
- 13.White T. Hadoop: The Definitive Guide. O'Reilly Media; 2012. [Google Scholar]
- 14.Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database issue):D561–D568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Z, Hung JH, Wang Y, et al. VisANT 3.5: multi-scale network visualization, analysis and inference based on the gene ontology [published online ahead of print May 26, 2009]. Nucleic Acids Res. 2009;37(Web Server issue):W115–W121. doi: 10.1093/nar/gkp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giardine B, Riemer C, Hardison RC, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15(10):1451–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences [published online ahead of print August 27, 2010]. Genome Biol. 2010;11(8):R86 doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blankenberg D, Von Kuster G, Coraor N, et al. Galaxy: a web-based genome analysis tool for experimentalists [published online ahead of print January 14, 2010]. Curr Protocol Mol Biol. 2010; Chapter 19:Unit 19.10.1-21. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho H, Berger B, Peng J. Diffusion component analysis: unraveling functional topology in biological networks. In: Przytycka TM, ed. Research in Computational Molecular Biology. Springer International Publishing; 2015: 62–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tarca AL, Draghici S, Khatri P, et al. A novel signaling pathway impact analysis. Bioinform. 2009;25(1):75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaske CJ, Benz SC, Sanborn JZ, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinform. 2010;26(12):i237–i245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLennan M. Managing data within the HUBzero platform. OMICS. 2011;15(4):247–249. [DOI] [PubMed] [Google Scholar]
- 23.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0 [published online ahead of print May 7, 2011]. Bioinform. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croft D, Mundo AF, Haw R, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42(Database issue):D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hermjakob H, Montecchi-Palazzi L, Lewington C, et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32(Database issue):D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apweiler R, Attwood TK, Bairoch A, et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29(1):37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research N, Weinstein JN, Collisson EA, Mills GB, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genetics. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41(Database issue):D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin X, Han J, Yu PS. Truth discovery with multiple conflicting information providers on the web. IEEE Trans Knowledge Data Eng. 2008;20(6):796–808. [Google Scholar]
- 30.Bleiholder J, Naumann F. Data fusion. ACM Computing Surveys. 2009;41(1):1–41. doi: 10.1145/1456650.1456651. [Google Scholar]
- 31.Zhao B, Rubinstein BIP, Gemmell J, Han J. A Bayesian approach to discovering truth from conflicting sources for data integration. Proc VLDB Endowment. 2012;5(6):550–561. [Google Scholar]