Abstract
The Big Data to Knowledge (BD2K) Center for Causal Discovery is developing and disseminating an integrated set of open source tools that support causal modeling and discovery of biomedical knowledge from large and complex biomedical datasets. The Center integrates teams of biomedical and data scientists focused on the refinement of existing and the development of new constraint-based and Bayesian algorithms based on causal Bayesian networks, the optimization of software for efficient operation in a supercomputing environment, and the testing of algorithms and software developed using real data from 3 representative driving biomedical projects: cancer driver mutations, lung disease, and the functional connectome of the human brain. Associated training activities provide both biomedical and data scientists with the knowledge and skills needed to apply and extend these tools. Collaborative activities with the BD2K Consortium further advance causal discovery tools and integrate tools and resources developed by other centers.
Keywords: Big Data to knowledge (BD2K), center of excellence, causal discovery, biomedical knowledge, biomedical science
INTRODUCTION
Much of science consists of discovering and modeling causal relationships in nature. With rapid advancements in technology and networking, biomedical scientists increasingly generate multiple complex data types for a large number of samples, each of which has an enormous number of measurements recorded. Although statistical and machine learning methods can predict the value of a variable X from observed predictors, the best predictors of X are often poor models of the causes of X (hence the slogan “correlation is not causation”), which motivated the development of algorithms specifically devoted to the discovery of valid causal models.
Indeed, tremendous progress has been made in developing computational methods for representing and discovering causal knowledge from data.1–6 These causal discovery methods have found applications in a wide range of fields, including econometrics, education, epidemiology, climate research, medicine, and biology.2,7 Current capabilities include 1) the representation of existing causal knowledge as a graphical network model with precisely defined semantics, 2) the discovery of causal networks of relationships from a combination of prior knowledge and experimental and observational data, and 3) the use of causal networks to suggest how changing one variable (e.g., a drug binding to a signaling protein and blocking a pathway) is likely to influence the state of another variable (e.g., cell apoptosis). While much progress has been made in the development of these computational methods and their application in biomedical science8–35 and other fields, they are not sufficiently efficient to analyze big datasets nor easy for biomedical scientists to access or apply to their data.
To fill this gap, the Center for Causal Discovery (CCD) is building on the extensive code base of causal modeling and discovery (CMD) algorithms that we have developed and implemented over the past 25 years1,2,4,36–38 and integrating new or improved algorithms as they are reported in the literature. Software products from the Center will allow biomedical and data scientists to select and apply one or more data-appropriate causal discovery algorithms to their biomedical datasets and compare the causal relationships that each algorithm predicts.
ORGANIZATION
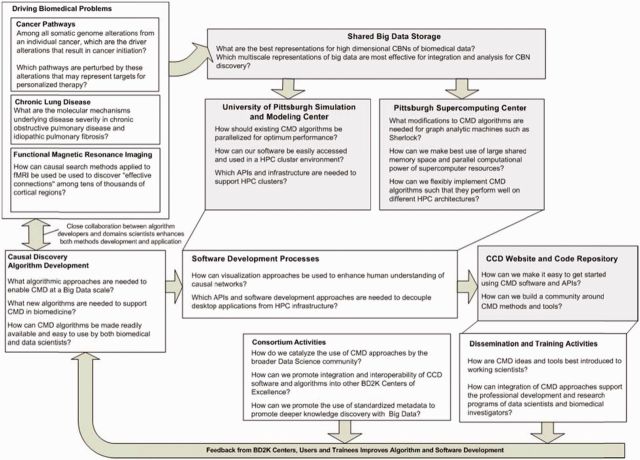
The CCD integrates the efforts of 5 main teams of experts: Algorithm Development, Software and Systems Architecture, Driving Biomedical Projects Training and Dissemination, and Consortium Activities. Figure 1 summarizes the overall impact of the CCD in relation to the types of problems we are solving through each component of the center. As noted above, our ultimate goal is to provide CMD tools with which end users can efficiently search for and characterize causal relationships responsible for an observed phenotype or phenomenon using large and often merged omics, imaging, and clinical datasets—and to tailor training to each end-user constituency.
Figure 1:
Center for Causal Discovery (CCD) organization and workflow optimized for the development of causal modeling and discovery (CMD) algorithms and tools designed to help address causal discovery in biomedicine from big data.
The CCD is a joint effort of approximately 40 investigators at the University of Pittsburgh (Pitt), Carnegie Mellon University, the Pittsburgh Supercomputing Center, and Yale University. We also have consulting investigators at the California Institute of Technology, New York University, Rutgers University, Stanford University, the University of Crete, and the University of North Carolina. Drs Gregory Cooper, Ivet Bahar, and Jeremy Berg serve as the Center Principal Investigators who direct Center activities with guidance from the Executive Committee and Internal and External Advisory Boards.
ALGORITHM DEVELOPMENT
Algorithm development and optimization is at the core of CCD efforts, and we are focusing on the discovery of structural causal relationships that can be represented by causal Bayesian networks (Table 1). We are using 2 main classes of algorithms that model hidden variables and sample selection and have the ability to discover them based on observational data, data from experimental interventions, or both: constraint-based algorithms, which use tests of conditional independence, and Bayesian algorithms, which allow the specification of structure and parameter prior probabilities.
Table 1:
Several key causal relationships in a causal Bayesian network.
| Graphical representation | Causal relationship |
|---|---|
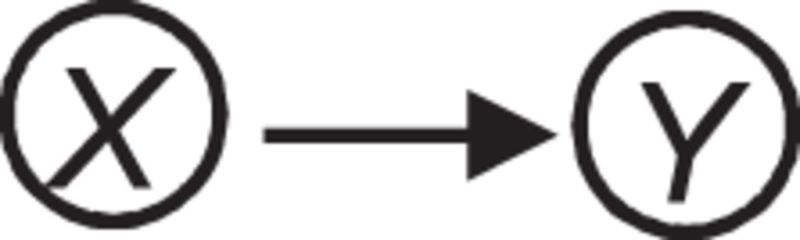 |
Direct cause |
 |
An endogenous latent variable (H) |
 |
A latent confounding variable (H) |
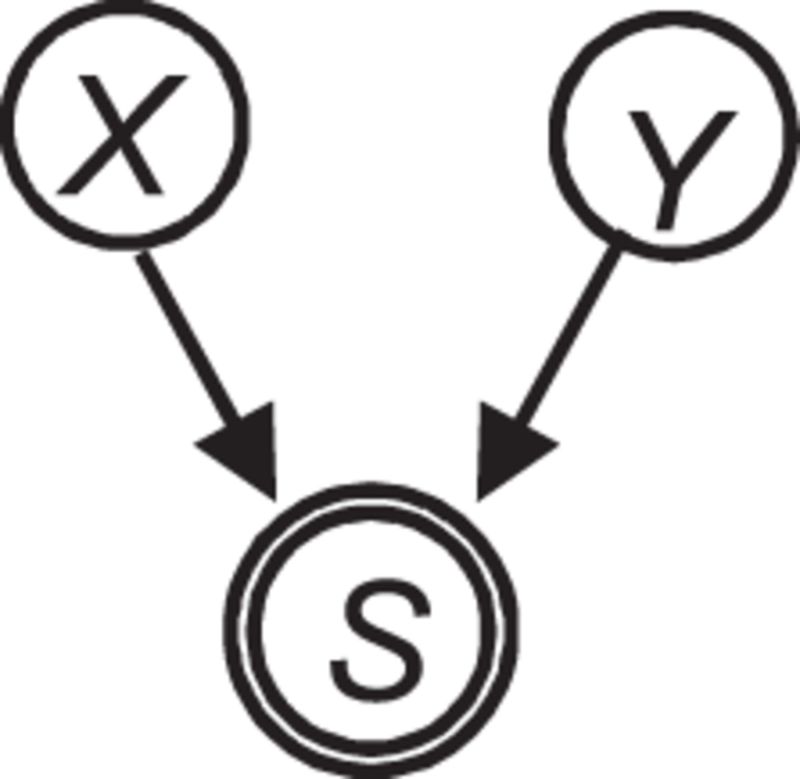 |
Selection |
X, Y, and Z denote measured variables. H denotes a hidden (latent) variable. The variable S surrounded by double circles denotes selection in which the values of X and Y influence whether a sample appears in the dataset.
Some additional key characteristics of causal discovery problems are shown in Table 2. No current causal discovery algorithms can optimally address all these issues, though algorithms exist for addressing important subsets of the issues,6 and additional algorithms are being developed in the CCD and elsewhere to more fully address them. We are in the process of implementing and making available the best CMD algorithms as well as developing new algorithms to address pressing needs of causal discovery in biomedicine. Our Algorithm Development team is working closely with the Software and Systems Architecture groups to generate algorithms that are highly efficient and parallelized so that they can analyze very large datasets in a practical amount of computing time.
Table 2:
Some key characteristics of causal discovery problems.
| Topic | Characteristics |
|---|---|
| Prior knowledge | none; deterministic; probabilistic |
| Variable types | discrete; continuous; both |
| Temporal dynamics | none |
| stationary time series; non-stationary time series | |
| discrete time; continuous time | |
| Distributions | parametric; non-parametric |
| linear; non-linear | |
| additive noise; non-additive noise | |
| Feedback cycles | absent; present |
| Latent confounders | absent; present |
| Selection bias | absent; present |
| Datasets | single; multiple datasets on same variables; multiple datasets on overlapping variables |
SOFTWARE AND SYSTEMS DEVELOPMENT
Our goal is to make CMD algorithms accessible and useful to a wide variety of biomedical researchers who might not otherwise take advantage of them. These algorithms will be freely available as open-source application programming interfaces, which will facilitate their use by other biomedical and data scientists. We aim to provide “one-stop shopping” for scientists who wish to incorporate causal discovery methods into their research. To do so, we are, in parallel with algorithm development, creating a computational platform that supports the continual accumulation, refinement, integration, documentation, and dissemination of causal discovery algorithms.
We are also developing an interactive computer system that facilitates the application of the CMD algorithms to biomedical data (Figure 2). Such a system will have a graphical user interface that can run on a desktop computer. Backend processing of causal analyses can take place on the desktop machine for tasks that are not too computationally demanding, while more demanding tasks are automatically relayed to and performed on a high-performance computer cluster. We are fortunate to engage data scientists at the Pittsburgh Supercomputing Center for this work, and resources available through this and other high-performance computer centers are available to investigators across the country seeking to apply CMD tools to their big data through the Extreme Science and Engineering Discovery Environment (XSEDE), https://www.xsede.org.
Figure 2:
Basic workflow of the causal modeling and discovery (CMD) system under development in the Center for Causal Discovery (CCD). End users will interact with wizards that help them select the appropriate methods at each step in the workflow.
DRIVING BIOMEDICAL PROJECTS
To ensure our methods are broadly applicable, we selected 3 very different driving biomedical projects (DBPs) to drive the development of our CMD tools and algorithms. Teams of bench scientists, who generate biomedical big data through their ongoing research, and data scientists involved in algorithm and software development, meet biweekly to ensure close collaboration on the iterative development and improvement of our CMD methods and tools.
The Cancer Signaling Pathways DBP seeks to discover the genomic drivers of tumors and the altered cell signaling pathways that result in cancer.39,40 The ability to discover and model these causal relationships accurately is key to more fully realizing precision cancer diagnosis, prognosis, and therapy. We are analyzing public data sources, including The Cancer Genome Atlas (TCGA) data41, which is mirrored locally in real time, and internal research and electronic health record data on a variety of cancer types, with an initial focus on breast cancer. The data include measurements of somatic mutations, copy number alterations, mRNA expression, and protein activation, as well as phenotype and clinical outcome. Computational predictions will be tested and validated in cell line and xenograft models as part of a broader program aimed at discovering new therapeutic targets.
The Chronic Lung Disease DBP aims to discover the cellular factors that lead to susceptibility and progression of chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis.42 We are analyzing data from the Lung Genomics Research Consortium and the Lung Tissue Research Consortium to discover and model causal relationships between molecular variables, clinical variables (∼80 per patient), and image features to characterize disease mechanisms and predict disease severity. The data include high-resolution images of lung tissue for which single nucleotide polymorphisms (SNPs), DNA methylation, mRNA expression, and microRNA expression are concurrently measured.
The Brain Functional Connectivity DBP seeks to discover the causal influences among small spatial regions of the human brain using fMRI data representing the activity of ∼2 mm3 regions (voxels) about every 2 s. These regions define thousands of variables that we analyze to generate a causal network of functional influence.43,44 Currently, we are performing this analysis on functional magnetic resonance imagining (fMRI) data from individuals with autism spectrum disorder and neurotypical individuals. We seek to characterize causal-network differences between these groups as well as differences among individuals with autism spectrum disorder (ASD). Our goal is to improve sub-classification of ASD subjects using the causal patterns evidenced in response to a variety of stimuli. Individuals within a sub-classification may or may not be homogenous with regard to ultimate causes of their condition, but we hope to reduce variance. We plan a similar investigation in individuals with schizophrenia. These efforts, like those for the molecular mechanisms for cancer and lung disease, typically involve problems in which the number of variables and possible relationships among them is much higher than the sample size. Such analyses are possible because our search algorithms allow us to identify causal structure when the number of variables is orders of magnitude larger than the number of samples.2,45
While we anticipate that new biomedical discoveries will be made in each of these problem areas using the methods developed by the CCD, the broader impact will be the development of the methods and tools themselves, which will be applicable to a wide spectrum of biomedical research.
TRAINING AND DESSEMINATION
The Training component of the CCD is dedicated to training researchers in both biomedical science and data science. For biomedical scientists, we teach the conceptual underpinnings of CMD methods, the application of those methods to biomedical problems (including an understanding of what kinds of problems the methods should or should not be applied to), and the use of software developed by the Center. We are teaching data scientists how to understand and incorporate CMD methods into computational workflows and how to develop new algorithms, software, and systems for CMD.
We will provide training resources in the form of downloadable materials that can be used for asynchronous learning or as part of established courses; online courses (both credit and noncredit), workshop videos, and discussion groups; and in-person workshops, short courses, graduate courses, internships, and hackathons. Our CMD training will rely heavily on the TETRAD program (http://www.phil.cmu.edu/tetrad/) developed by CCD investigators from Carnegie Mellon University. These activities are intended for undergraduate and graduate students, postdoctoral fellows, young investigators, and established investigators from academia and industry, both within and beyond the Big Data to Knowledge (BD2K) Centers of Excellence.
We will also maintain at our website (http://ccd.pitt.edu) online interfaces and tutorials for CCD software as well as libraries of algorithms, software tools, and datasets to allow one-stop shopping for any scientist interested in causal discovery.
CONSORTIUM COLLABORATION
Our Consortium component has two main goals: to encourage and facilitate the use of CCD tools by other scientists, both inside and outside the BD2K Consortium, and to design and implement intra-Consortium projects with other BD2K Centers.
Our Technical Catalyst will make brief site visits on a rotating basis to other BD2K Centers to learn how our Center can better serve their needs and how their products can be integrated into our workflow; we will prepare Technical Reports summarizing each site visit and opportunities for synergy. Our Scientific Catalyst program engages leading biomedical and data scientists who have agreed to promote the use of CCD tools in their respective scientific communities and to solicit feedback on how the CCD can better meet their needs.
In addition, we currently are pursuing two intra-Consortium projects. The first is in partnership with the Patient-centered Information Commons: Standardized Unification of Research Elements (PIC-SURE) at Harvard to access and analyze datasets containing genetic, environmental, imaging, behavioral, and clinical data on a large number of individual patients. Together, we will apply CMD methods to explore new hypotheses about the relationships between risk factors, diseases, and outcomes. In a second project, we are working with the Stanford Center for Expanded Data Annotation and Retrieval (CEDAR) to use their metadata methods to support our CMD analyses and to use our knowledge about CMD in developing metadata descriptions in (CEDAR).
SUMMARY
The CCD seeks to discover, optimize, apply, and disseminate methods and tools for CMD with large and complex data to generate new biomedical knowledge and to encourage and train both data and biomedical scientists in their use. We will serve as a central resource for anyone seeking causal discovery algorithms, software tools, training, and collaboration.
Key Personnel
University of Pittsburgh: Joseph Ayoob, Ivet Bahar, Michael Barmada, Michael Becich, Panayiotis Benos, Jeremy Berg, Chakra Chennubhotla, Maria Chikina, Gregory Cooper, Panos Chrysanthis, Nathan Clark, Michael Davis, Roger Day, Jeremy Espino, Harry Hochheiser, Rebecca Crowley Jacobson, Xia Jiang, Michelle Kienholz, Alexandros Labrinidis, Adrian Lee, Xinghua Lu, Frank Schneider, William Shirey, and Shyam Visweswaran. Carnegie Mellon University: David Danks, Clark Glymour, Joseph Ramsey, Richard Scheines, and Peter Spirtes. Pittsburgh Supercomputing Center: Nicholas Nystrom. Yale University: Naftali Kaminski.
FUNDING
Research reported in this publication was supported by grant U54HG008540 awarded by the National Human Genome Research Institute through funds provided by the trans-NIH Big Data to Knowledge initiative (www.bd2k.nih.gov). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
COMPETING INTERESTS
None.
CONTRIBUTIONS
G.F.C. wrote the manuscript. C.G., I.B., M.J.B., M.L.K., P.B., and R.C.J. reviewed and revised the manuscript. All authors read and approved the final manuscript.
REFERENCES
- 1.Glymour C, Cooper GF, eds. Computation, Causation, and Discovery. Cambridge, MA: MIT Press; 1999. [Google Scholar]
- 2.Spirtes P, Glymour C, Scheines R. Causation, Prediction, and Search. Cambridge, MA: MIT Press; 2000. [Google Scholar]
- 3.Pearl J. Causality: Models, Reasoning, and Inference. Cambridge, U.K: Cambridge University Press; 2009. [Google Scholar]
- 4.Spirtes P. Introduction to causal inference. J Mach Learn Res. 2010;11:1643-1662. [Google Scholar]
- 5.Illari PM, Russo F, Williamson J, eds. Causality in the Sciences. Oxford, UK: Oxford University Press; 2011. [Google Scholar]
- 6.Kalish M, Buhlmann P. Causal structure learning and inference: a selective review. Qual Technol Quant Manag. 2014;11:3–21. [Google Scholar]
- 7.Shipley B. Cause and Correlation. Cambridge, UK: Cambridge University Press; 2000. [Google Scholar]
- 8.Stekhoven DJ, Moraes I, Sveinbjoernsson G, Hennig L, Maathuis MH, Buehlmann P. Causal stability ranking. Bioinformatics. 2012;28(21):2819–2823. [DOI] [PubMed] [Google Scholar]
- 9.Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. [DOI] [PubMed] [Google Scholar]
- 10.Chen LS, Emmert-Streib F, Storey JD. Harnessing naturally randomized transcription to infer regulatory relationships among genes. Genome Biol. 2007;8(10):R219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramsey JD, Hanson SJ, Glymour C. Multi-subject search correctly identifies causal connections and most causal directions in the DCM models of the Smith et al. simulation study. Neuroimage. 2011;58(3):838–848. [DOI] [PubMed] [Google Scholar]
- 12.Zhang K, Hyvärinen A. Distinguising causes from effects using nonlinear acyclic causal models. JMLR Workshop Conf Proc. 2008;6:157–164. [Google Scholar]
- 13.Schadt EE, Lamb J, Yang X, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37(7):710–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hageman RS, Leduc MS, Korstanje R, Paigen B, Churchill GA. A Bayesian framework for inference of the genotype-phenotype map for segregating populations. Genetics. 2011;187(4):1163–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aliferis CF, Statnikov A, Tsamardinos I, Mani S, Koutsoukos XD. Local causal and Markov blanket induction for causal discovery and feature selection for classification – part II: analysis and extensions. J Mach Learn Res. 2010;11:235–284. [Google Scholar]
- 16.Aliferis CF, Statnikov A, Tsamardinos I, Mani S, Koutsoukos XD. Local causal and Markov blanket induction for causal discovery and feature selection for classification – part I: algorithms and empirical evaluation. J Machine Learning Res. 2010;11:171–234. [Google Scholar]
- 17.Rockman MV. Reverse engineering the genotype-phenotype map with natural genetic variation. Nature. 2008;456(7223):738–744. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Zhu J, Lum PY, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452(7186):429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkinson DJ. Bayesian methods in bioinformatics and computational systems biology. Brief Bioinform. 2007;8(2):109–116. [DOI] [PubMed] [Google Scholar]
- 20.Needham CJ, Bradford JR, Bulpitt AJ, Westhead DR. Inference in Bayesian networks. Nat Biotechnol. 2006;24(1):51–53. [DOI] [PubMed] [Google Scholar]
- 21.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37(4):382–390. [DOI] [PubMed] [Google Scholar]
- 22.Pournara I, Wernisch L. Reconstruction of gene networks using Bayesian learning and manipulation experiments. Bioinformatics. 2004;20(17):2934–2942. [DOI] [PubMed] [Google Scholar]
- 23.Friedman N. Inferring cellular networks using probabilistic graphical models. Science. 2004;303(5659):799–805. [DOI] [PubMed] [Google Scholar]
- 24.Gagneur J, Stegle O, Zhu C, et al. Genotype-environment interactions reveal causal pathways that mediate genetic effects on phenotype. PLoS Genet. 2013;9(9):e1003803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pe'er D, Hacohen N. Principles and strategies for developing network models in cancer. Cell. 2011;144(6):864–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akavia UD, Litvin O, Kim J, et al. An integrated approach to uncover drivers of cancer. Cell. 2010;143(6):1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berndt SI, Gustafsson S, Magi R, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45(5):501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz SM, Schwartz HT, Horvath S, Schadt E, Lee SI. A systematic approach to multifactorial cardiovascular disease: causal analysis. Arterioscler Thromb Vasc Biol. 2012;32(12):2821–2835. [DOI] [PubMed] [Google Scholar]
- 29.Schadt EE, Bjorkegren JL. NEW: network-enabled wisdom in biology, medicine, and health care. Sci Transl Med. 2012;4(115):115rv1. [DOI] [PubMed] [Google Scholar]
- 30.Schadt EE. Molecular networks as sensors and drivers of common human diseases. Nature. 2009;461(7261):218–223. [DOI] [PubMed] [Google Scholar]
- 31.Le TD, Liu L, Liu B, et al. Inferring microRNA and transcription factor regulatory networks in heterogeneous data. BMC Bioinformatics. 2013;14:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang K, Saito M, Bisikirska BC, et al. Genome-wide identification of post-translational modulators of transcription factor activity in human B cells. Nat Biotechnol. 2009;27(9):829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dojer N, Gambin A, Mizera A, Wilczynski B, Tiuryn J. Applying dynamic Bayesian networks to perturbed gene expression data. BMC Bioinformatics. 2006;7:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stingo FC, Chen YA, Vannucci M, Barrier M, Mirkes PE. A Bayesian graphical modeling approach to microRNA regulatory network inference. Ann Appl Stat. 2010;4(4):2024–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran LM, Zhang B, Zhang Z, et al. Inferring causal genomic alterations in breast cancer using gene expression data. BMC Syst Biol. 2011;5:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooper GF, Herskovits EH. A Bayesian method for the induction of probabilistic networks from data. Machine Learning. 1992;9:309–347. [Google Scholar]
- 37.Spirtes P, Cooper GF. An experiment in causal discovery using a pneumonia database. Proceedings of the International Workshop on Artificial Intelligence and Statistics, 1999, Fort Lauderdale, Florida. [Google Scholar]
- 38.Yoo C, Cooper GF. Discovery of causal relationships in a gene-regulation pathway from a mixture of experimental and observational DNA microarray data. Proceedings of the Pacific Symposium on Biocomputing, 2002, Kauai, Hawaii. [DOI] [PubMed] [Google Scholar]
- 39.Lu S, Jin B, Cowart LA, Lu X. From data towards knowledge: Revealing the architecture of signaling systems by unifying knowledge mining and data mining of systematic perturbation data. PLoS One. 2013;8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu S, Lu X. Integrating genome and functional genomics data to reveal perturbed signaling pathways in ovarian cancers. AMIA Summits Transl Sci Proc. 2012;2012:72–78. [PMC free article] [PubMed] [Google Scholar]
- 41.The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Noth I, Garcia JG, Kaminski N. A variant in the promoter of MUC5B and idiopathic pulmonary fibrosis. N Engl J Med. 2011;364(16):1576–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramsey JD, Hanson SJ, Hanson C, Halchenko YO, Poldrack RA, Glymour C. Six problems for causal inference from fMRI. Neuroimage. 2010;49(2):1545–1558. [DOI] [PubMed] [Google Scholar]
- 44.Mumford J, Ramsey JD. Bayesian networks for fMRI: a primer. Neuroimage. 2014;86:573–582. [DOI] [PubMed] [Google Scholar]
- 45.Bühlmann P, van de Geer S. Statistics for High-Dimensional Data: Methods, Theory and Applications. Springer; 2011. [Google Scholar]