Abstract
Mobile sensor data-to-knowledge (MD2K) was chosen as one of 11 Big Data Centers of Excellence by the National Institutes of Health, as part of its Big Data-to-Knowledge initiative. MD2K is developing innovative tools to streamline the collection, integration, management, visualization, analysis, and interpretation of health data generated by mobile and wearable sensors. The goal of the big data solutions being developed by MD2K is to reliably quantify physical, biological, behavioral, social, and environmental factors that contribute to health and disease risk. The research conducted by MD2K is targeted at improving health through early detection of adverse health events and by facilitating prevention. MD2K will make its tools, software, and training materials widely available and will also organize workshops and seminars to encourage their use by researchers and clinicians.
Keywords: mobile health (mHealth), wearable sensors, big data, data science research, knowledge discovery
INTRODUCTION
Complex and common disorders such as cancer, cardiovascular diseases, obesity, diabetes, depression, asthma, and addiction represent the major burden of disease in the United States and globally.1 These disorders are caused by the interactions of multiple risk factors, rather than any single genetic, behavioral, social, or environmental source.2,3 To reach the next level of biomedical understanding, it is critical for researchers to be able to monitor the health states of patients in their natural environments and to quantify the complex temporal dynamics of key physical, biological, behavioral, psychological, social, and environmental factors that contribute to health and disease risks. Such activities will substantially improve physicians’ ability to predict person-specific disease risk and treatment response and will enable researchers to develop more efficacious prevention and treatment strategies, thus moving the field closer to a fully realized vision of the Precision Medicine initiative.3,4
Rapid advances in technology are leading to mobile sensing devices that now make collecting “natural environment” data feasible.5 While ongoing efforts focused on the analysis of “big data” in the areas of genomics, imaging, and electronic health records are making significant strides, data analytics tools specific to the unique features of mobile sensor data need to be developed and disseminated, so that this wealth of mobile sensor data (characterized by high volume, velocity, variety, variability, versatility, and semantic gap) can be converted into information, knowledge, and, ultimately, action. Investing in a strong, open, scientific, and computational infrastructure for mobile sensor big data at this early stage promises outsized returns that will advance science and improve health.
Given the diversity of the challenges of addressing mobile sensor big data, developing a comprehensive solution requires a truly transdisciplinary approach in which end-to-end solutions are developed jointly by experts in sensor design, mobile systems, machine learning, pattern mining, big data computing, health informatics, experiment design, clinical research, and health research. Because mobile health (mHealth) is a young discipline, the necessary expertise is not readily available at a single institution. Therefore, the Center of Excellence for Mobile Sensor Data-to-Knowledge (MD2K) brings together experts in computer science, engineering, medicine, behavioral science, and statistics from 11 universities (Cornell Tech; Georgia Tech; Northwestern University; Ohio State University; Rice University; University of California, Los Angeles; University of California, San Diego; University of California, San Francisco; University of Massachusetts Amherst; University of Memphis; University of Michigan) and Open mHealth (a nonprofit organization). The MD2K investigators not only cover all the necessary areas of expertise, but they are also national leaders in their fields, with proven track records in mHealth research. We describe herein the driving biomedical applications of MD2K, the mobile sensors being used, the data science research and training activities being pursued, and MD2K’s anticipated scientific and societal impacts.
DRIVING BIOMEDICAL APPLICATIONS OF MD2K
To guide the development and evaluation of our data analytics methods and tools and to demonstrate the broad utility of MD2K, we selected two driving biomedical applications: 1) improving smoking cessation (including reducing lapses in smoking cessation) among smokers and 2) reducing hospital readmissions among congestive heart failure (CHF) patients.
Cigarette Smoking
Cigarette smoking is the leading preventable cause of death in the United States, responsible for 1 in 5 deaths annually.6 Recent research regarding mobile sensing technology that can infer whether or not an individual is smoking by monitoring their respiration7 and arm movements8 demonstrates the feasibility of automatically and unobtrusively detecting patients’ lapses in smoking cessation. Combined with other research on detecting high-risk triggers for smoking (eg, stress9), these works open up the possibility of using mobile devices for sensor-triggered just-in-time adaptive interventions.10 For example, upon detecting a rapid rise in an individual’s stress level, via data collected from wearable sensors, an intervention can be triggered to provide an abstinent smoker with mobile-based social support on their smart phone, to prevent a lapse in their smoking cessation. This approach can potentially address other behavioral risk factors, such as impulsive eating, alcoholism, and illicit drug use.
Congestive Heart Failure
CHF affects nearly 6 million people in the United States, with 670 000 new cases diagnosed annually. It is a leading cause of preventable hospitalization and, thus, unnecessary healthcare expenditure. Current technological approaches have failed to reduce the rate of readmission after hospitalization for CHF,11–13 in part due to their low sensitivity or poor positive predictive value.14 Implanted device-based diagnostics15 and an implanted hemodynamic monitor16,17 are promising solutions, but their applicability may be limited to all but the most advanced CHF patients. Our recent work on the EasySense sensor,18 which uses a wideband radio frequency to measure the accumulation of lung fluid, provides the first-ever opportunity to monitor the worsening of lung congestion in a non-invasive manner. In addition, sensor-based detection and prediction of potentially risky behaviors that could lead to an episode of heart failure, such as eating sodium-rich meals (eg, fast food) or medication nonadherence, creates opportunities for mobile technologies to identify when the patient is at risk and deliver just-in-time adaptive interventions to clinicians and patients to avert an exacerbation of CHF. This approach can potentially address other chronic conditions, such as hypertension, diabetes, chronic obstructive pulmonary disease, and asthma.
MOBILE SENSORS IN MD2K
The data science research being conducted by MD2K is intended to be generalizable to a variety of sensors and for a wide range of biomedical applications. To demonstrate the feasibility of these approaches, MD2K is using five sources of mobile sensor data that are directly applicable to the two biomedical applications described in the preceding. Data from each sensor suite are collected by the smart phone in real-time (via wireless communication if the sensor is not embedded in the smart phone) (see Figure 1):
The AutoSense chestband,19 which collects electrocardiogram (ECG), respiration, and accelerometry data, can be used to infer stress9 (from the ECG and/or respiration data), whether or not a patient is smoking7 (from the respiration data), and patient drug use20 (from the ECG data).
A smart watch with inertial sensors (3-axis accelerometers and 3-axis gyroscopes) that can infer when a patient is smoking and eating8 (by tracking arm movements).
A radio-frequency-based micro-radar sensor (called EasySense18) that can non-invasively measure heart activity and lung fluid volume in CHF patients (by analyzing the echo and absorption from ultra-wideband radio frequency probes).
Smart eyeglasses,21 for capturing video in the direction of the wearer’s gaze and inferring, from that data, exposure to smoking cues, such as seeing a cigarette advertisement, while simultaneously assessing the state of the eye (eg, fatigue) by monitoring the eye itself.
Global Positioning System data from the smart phone that can be used to infer geoexposure, for factors such as proximity to a point-of-sale for tobacco or to fast food (ie, sodium-rich) restaurants.
Figure 1:
Five sources of mobile sensor data in mobile sensor data-to-knowledge applications. Each sensor (not embedded in the phone) transmits the data it collects over a wireless channel to the smart phone.
DATA SCIENCE RESEARCH IN MD2K
While mobile sensors offer tremendous opportunities for accelerating biomedical discoveries and optimizing care delivery, they also present substantial transdisciplinary data analytics challenges. Although several ongoing initiatives focus on extracting actionable biomedical knowledge from very large amounts of data in a variety of applications, mobile sensor data presents a different and higher-level challenge, due to its unique qualities, ie, its high volume, velocity, variety, variation, versatility, and semantic gap (see Table 1). Consequently, the MD2K Center brings together world-class mHealth experts from the data science research and biomedical research fields to address these major barriers to processing complex mobile sensor data.
Table 1:
Major data science research challenges in converting mobile sensor data into health information, knowledge, and action
| Issue | Challenge |
|---|---|
| Volume | 14.5 GB of data per individual daily, for 10 h of wearing MD2K sensors, presents big data computational challenges for population-scale processing. |
| Velocity | 30 kB/s of data, generated by the wearable sensors (eg, EasySense), present significant computational and battery life challenges for real-time processing on the mobile device (eg, for just-in-time intervention). |
| Variety | Data from a wide variety of sensors must be combined (eg, EasySense, accelerometers, eyeglasses, and global positioning system-derived measures for congestive heart failure monitoring). |
| Variability | Sensor data quality varies dynamically due to attachment degradation, changes in sensor placement, wireless losses, and battery depletion. |
| Semantic Gap | Sensors produce generic data (eg, 0s and 1s) that require sophisticated processing to obtain interpretable health-related measures. For example, arm movements produced by the action of smoking should be distinguished from those produced by the action of eating or talking. Likewise, change in lung fluid due to a change in posture should not raise alarm. |
| Versatility | Sensor data can reveal private social behaviors. For example, electrocardiogram data can be used to monitor and manage stress, but can also reveal that a patient is using cocaine. |
The data science research of MD2K is organized in four thrusts. Thrust 1 (Mobile Sensor Data-to-Information or MD2I) is developing general principles and computational methods for inferring markers (ie, measures) of patient health as well as markers of behavioral, physical, social, and environmental risk factors that are robust to wide variability in subjects’ behaviors, an array of known and unknown confounders, errors in self-report data, and the variable quality and availability of sensor data. MD2I is developing a mobile sensor data processing toolkit as open-source software that implements the data analytic steps required for computing robust markers. Data science researchers can use this toolkit to develop new markers. For the two driving biomedical applications, MD2I is producing a variety of markers that can be used directly by biomedical researchers. They include detecting a lapse in smoking cessation, detecting the onset of congestion in CHF patients (eg, lung water volume), risk factors for lapses in smoking cessation (eg, stress), and risk factors for the development of CHF (eg, eating fast food), all from sensor data. Applying the computational methods developed by Thrust 1 to the data collected by mobile sensors converts this time series of sensor data to a time series of robust markers.
Thrust 2 (Mobile Sensor Information-to-Knowledge or MI2K) is developing discriminative latent variable models to discover patterns in multivariate time series of markers to detect intermediate health outcomes (eg, a lapse in smoking cessation) and generate alerts for patients and care providers about the surrounding context in ways that can inform care decisions. MI2K is developing frequent pattern mining and Granger causality models, to discover predictors of adverse health events from the time series of markers, as well as a discovery dashboard, to engage biomedical researchers in the discovery process. Thrust 2 is also developing learning algorithms for the online adaptation of rules for deciding the content and timing of sensor-triggered, just-in-time adaptive interventions. The computational methods for robust marker development (by MD2I) and the computational methods for time series pattern mining (by MI2K) are being incorporated in a new big data computing platform so that these methods can be applied to large, population-scale mobile sensor data.
The big data computing platform created by Thrust 3 (MD2K-Computation) is supporting data science and biomedical researchers to efficiently process vast amounts of dense mobile sensor data for data science research and biomedical discovery. It also enables the application of these models to individual-scale data for just-in-time intervention delivery on mobile devices. To provide a responsive user experience to researchers, care providers, and individuals, Thrust 3 builds on our ongoing research on big data computational methods, such as Iterative Map-Reduce.22 We are also developing computational mechanisms and software for managing participants’ privacy and identity by leveraging our ongoing work on sensor data vaults23 and recent developments in fully homomorphic encryption.
To make MD2K extensible to new mobile data sources and applicable to other biomedical applications, Thrust 3 is building on the Open mHealth initiative to standardize all the data types, analytic modules, visualization modules, and storage modules, developed in MD2K, so that they are interoperable, extensible, and generalizable.24–26 Development of Open mHealth-compliant modules will facilitate both integration of new device data streams into MD2K systems and the adoption of MD2K-developed techniques by the broader research and clinical community. Many mHealth research tools do not get reused and repurposed, because the data they produce have inconsistent formats, making it very hard to effectively use them beyond initial experimentation. Such challenges are even greater in the clinical realm, in which understanding the true meaning of data can be critical. Open mHealth is collaborating with clinical experts, to define and publish a standard language for mHealth data, and with technical experts and consumer-app designers, to design and build out an open developer platform. Open mHealth has developed a set of open data schemas that provide guidelines for optimally structuring different types of digital health data for clinical use. Using a common set of data schemas is critical for the seamless exchange of different types of data among different platforms and systems, and will ultimately enable personally generated data to be integrated and used alongside clinically generated data from electronic health records and other health information technology.
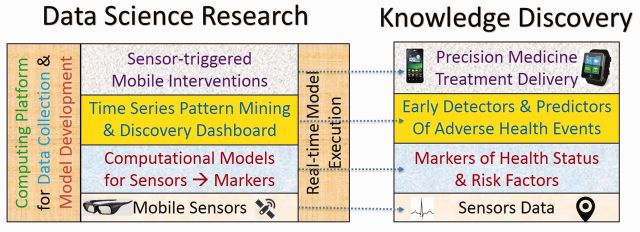
Each year (for 3 years), Thrust 4 (MD2K-Application) will be conducting user studies that support our use cases, involving, respectively, a new pool of 75 smokers before and immediately after an attempt to quit and 75 CHF patients in the hospital and 30 days post-discharge. These studies will evaluate the accuracy of markers and the feasibility of sensor-triggered, just-in-time interventions. Newly discovered markers and intervention triggers will be incorporated into the studies in succeeding years. Our goal for the MD2K Center is to help realize the Precision Medicine initiative by creating a bidirectional rapid feedback loop among Thrusts 1-4 of data science research, so that biomedical applications will inform the development of the technology, and vice versa, by using design thinking expertise27 and participatory design. Figure 2 summarizes the key data science research and knowledge discovery outcomes targeted by MD2K.
Figure 2:
Major outcomes of mobile sensor data-to-knowledge include both data science research and knowledge discovery, which are tightly coupled. Application of data science research, on the left, to the data collected by mobile sensors results in knowledge discovery, on the right.
TRAINING ACTIVITIES IN MD2K
MD2K also supports multidisciplinary training activities to enable the broader biomedical and data science research communities to use MD2K data analytic tools for biomedical discovery in biomedical applications. Its aim is also to stimulate the data science research community to build upon and advance the science of MD2K by equipping data science researchers with datasets; open-source software; documentation; as well as online forums, online tutorials, training videos, virtual seminars, and a comprehensive web-based resource library called mHealthHUB (see http://mhealth.md2k.org/). Finally, a yearly, week-long boot camp will be organized in order to train new and young investigators for transdisciplinary collaborations (see https://md2k.org/events/traininginstitute/).
SCIENTIFIC AND SOCIETAL IMPACT OF MD2K
The ubiquity of mobile phones (90%28 in the United States in 2014) and the emergence of mass-market mobile devices with embedded sensors (eg, smart watches) offer great opportunities to both assess and improve health. Adoption of mobile sensor technology in the next generation of scientific studies, such as the United States’s Precision Medicine initiative (which will recruit over a million participants to a cohort for long-term medical monitoring3), will enable the continuous collection of individual-level behavioral, social, and environmental data. With rich monitoring and user engagement capabilities (eg, via display screens on smart phones and smart watches), mobile technology can assess and improve adherence to and outcomes of personalized treatment plans in the delivery of precision medicine. Big data analytic tools developed by MD2K will be an essential component of precision medicine, which will enable the collection, integration, management, visualization, analysis, and interpretation of health data generated by mobile sensors. In conclusion, MD2K will advance the science of mHealth and make significant contributions to the societal goals of reducing healthcare costs and improving individual and population health outcomes.
CONTRIBUTORS
All 24 authors of this manuscript have made substantial contributions to the conception and design of the work being presented, have directly contributed or critically revised the manuscript, have approved the final version, and are accountable for the work. The order of the authors is alphabetical following the lead author. The specific contributions of each author are described as follows: Santosh Kumar led all aspects of the work, including conception, design, writing, revision, and submission; Gregory Abowd contributed to Thrust 1; William Abraham contributed to the congestive heart failure section and Thrust 4; Mustafa al’Absi contributed to the smoking cessation section and Thrust 4; J. Gayle Beck contributed to the training activities section; Duen Horng Chau contributed to Thrust 2; Tyson Condie contributed to Thrust 3; David Conroy contributed to smoking cessation in Thrust 4; Emre Ertin contributed to AutoSense, EasySense, and smart watch sensors, and Thrust 1; Deborah Estrin contributed to Open mHealth in Thrust 3; Deepak Ganesan contributed to the smart eyeglasses sensor and Thrust 1; Cho Lam contributed to the smoking cessation section and Thrust 4; Benjamin Marlin contributed to Thrust 1; Clay Marsh contributed to the Introduction and Thrust 4; Susan Murphy contributed to Thrusts 2 and 4; Inbal Nahum-Shani contributed to Thrust 4; Kevin Patrick contributed to the Introduction and Thrust 4; James Rehg contributed to all aspects of the data science research section, as the lead of data science research activities; Moushumi Sharmin contributed to Thrust 2; Vivek Shetty contributed to the training activities section; Ida Sim contributed to the Introduction and Open mHealth in Thrust 3; Bonnie Spring contributed to the smoking cessation section and Thrust 4; Mani Srivastava developed Table 1 and lead the description of Thrust 3; David Wetter contributed to the smoking cessation section and Thrust 4.
FUNDING
This research was supported by grant U54EB020404 awarded by the National Institute of Biomedical Imaging and Bioengineering through funds provided by the trans-National Institutes of Health Big Data to Knowledge initiative (www.bd2k.nih.gov).
COMPETING INTERESTS
None.
REFERENCES
- 1.World Health Organization. World Health Statistics 2011. http://www.who.int/whosis/whostat/2011. Accessed February 27, 2015.
- 2.Collins F. The case for a US prospective cohort study of genes and environment. Nature. 2004;429(6990):475–477. [DOI] [PubMed] [Google Scholar]
- 3.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015. http://www.nejm.org/doi/full/10.1056/NEJMp1500523#t=article. Accessed February 27, 2015. [Google Scholar]
- 4.Mirnezami R, Nicholson J, Darzi A. Preparing for precision medicine. N Engl J Med. 2012;366(6):489–491. [DOI] [PubMed] [Google Scholar]
- 5.Kumar S, Nilsen W, Pavel M, Srivastava M. Mobile health – Revolutionizing health via transdisciplinary research. IEEE Computer Magazine, Cover Feature. 2013;46(1):28–35. [Google Scholar]
- 6.Center for Disease Control and Prevention, Office on Smoking and Health, National Center for Chronic Disease Prevention and Health Promotion. Smoking and Tobacco Fast Facts. Atlanta, Georgia. 2011. http://www.cdc.gov/tobacco/data_statistics/fact_sheets/fast_facts/. Accessed February 27, 2015.
- 7.Ali AA, Hossain SM, Hovsepian K, Rahman MM, Kumar S. SmokeTrack: Automated detection of cigarette smoking in the mobile environment from respiration measurements. Proceedings of ACM Conference on Information Processing in Sensor Networks (IPSN); 2012 Beijing, China, pp 269–280. [Google Scholar]
- 8.Parate A, Chiu M, Chadowitz C, Ganesan D, Kalogerakis E., RisQ: recognizing smoking gestures with inertial sensors on a Wristband. Proceedings of the12th International Conference on Mobile Systems, Applications and Services (MobiSys 2014), pp. 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plarre K, Raij AB, Hossain M, et al. Continuous inference of psychological stress from sensory measurements collected in the natural environment. Proceedings of 2011 ACM Information Processing in Sensor Networks (IPSN); 2011; Chicago, Illinois, pp. 97–108. [Google Scholar]
- 10.Patrick K, Intille S, Zabinski MF. An ecological framework for cancer communication: implications for research. J Med Internet Res. 2005;7(3):e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaudhry SI, Mattera JA, Curtis JP, et al. Telemonitoring in patients with heart failure. N Engl J Med. 2010;36:2301–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koehler F, Winkler S, Schieber M, et al. Impact of remote telemedical management on mortality and hospitalizations in ambulatory patients with chronic heart failure: the telemedical interventional monitoring in heart failure study. Circulation. 2011;123:1873–1880. [DOI] [PubMed] [Google Scholar]
- 13.van Veldhuisen DJ, Braunschweig F, Conraads V, et al. Intrathoracic impedance monitoring, audible patient alerts, and outcome in patients with heart failure. Circulation. 2011;124:1719–1726. [DOI] [PubMed] [Google Scholar]
- 14.Abraham WT, Compton S, Haas G, et al. On behalf of the FAST Study Investigators. Intrathoracic impedance vs daily weight monitoring for predicting worsening heart failure events: results of the Fluid Accumulation Status Trial (FAST). Congest Heart Fail. 2011;17:51–55. [DOI] [PubMed] [Google Scholar]
- 15.Whellan DJ, Sarkar S, Koehler J, et al. Development of a method to risk stratify patients with heart failure for 30-day readmission using implantable device diagnostics. Am J Cardiol. 2013;111:79–84. [DOI] [PubMed] [Google Scholar]
- 16.Abraham WT, Adamson PB, Hasan A, et al. Safety and accuracy of a wireless pulmonary artery pressure monitoring system in patients with heart failure. Am Heart J. 2011;161:558–566. [DOI] [PubMed] [Google Scholar]
- 17.Abraham WT, Adamson PB, Bourge RC, et al. Wireless pulmonary artery haemodynamic monitoring in chronic heart failure: a randomised controlled trial. Lancet. 2011;377:658–666. [DOI] [PubMed] [Google Scholar]
- 18.Gao J, Ertin E, Kumar S, al'Absi M. Contactless sensing of physiological signals using wideband RF probes. Proceedings from 2013 Asilomar Conference on Signals, Systems, and Computers; 2013; Pacific Grove, California, pp. 86–90. [Google Scholar]
- 19.Ertin E, Stohs N, Kumar S, et al. AutoSense: unobtrusively wearable sensor suite for inferencing of onset, causality, and consequences of stress in the field. Proceedings from 2011 ACM Conference on Embedded Networked Sensing Systems; 2011; Seattle, Washington, pp. 274–287. [Google Scholar]
- 20.Hossain SM, Ali AA, Ertin E, et al. Identifying drug (cocaine) intake events from acute physiological response in the presence of free-living physical activity. Under review by 2014 ACM Information Processing in Sensor Networks (IPSN); 2014; Berlin, Germany, pp. 71–82. [PMC free article] [PubMed] [Google Scholar]
- 21.Mayberry A, Hu P, Marlin B, Salthouse C, Ganesan D. iShadow: design of a wearable, real-time mobile gaze tracker. Proceedings of the 12th International Conference on Mobile Systems, Applications and Services (MobiSys 2014); 2014, pp. 82–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Condie T, Mineiro P, Polyzotis N, Weimer M. Machine learning on big data. Proceedings of ACM SIGMOD Conference; 2013; New York, NY, pp. 939–942. [Google Scholar]
- 23.Chakraborty S, Shen C, Raghavan KR, Shoukry Y, Miller M, Srivastava MB. ipShield: a framework for enforcing context-aware privacy. Proceedings of USENIX Symposium on Networked Systems: Design and Implementation (NSDI); 2014; Seattle, WA, pp. 143–156. [Google Scholar]
- 24.Estrin DE, Sim I. Open mHealth architecture: an engine for healthcare innovation. Science. 2010;330:759–760. [DOI] [PubMed] [Google Scholar]
- 25.Chen C, Haddad D, Selsky J, et al. Making sense of mobile health data: An open architecture to improve individual and population level health. J Med Internet Res. 2012;14(4):e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Open mHealth [website]. http://openmhealth.org/. Accessed February 27, 2015.
- 27.Cross N. Design Thinking: Understanding How Designers Think and Work. London, United Kingdom: Bloomsbury Academic; 2011. [Google Scholar]
- 28.Pew data from 2014 indicate that 90% of US adults have a cell phone (http://www.pewinternet.org/three-technology-revolutions/). The latest (2015) estimate is that 64% have a smartphone (http://www.pewinternet.org/2015/04/01/us-smartphone-use-in-2015/).