Abstract
Objective To review and evaluate available software tools for electronic health record–driven phenotype authoring in order to identify gaps and needs for future development.
Materials and Methods Candidate phenotype authoring tools were identified through (1) literature search in four publication databases (PubMed, Embase, Web of Science, and Scopus) and (2) a web search. A collection of tools was compiled and reviewed after the searches. A survey was designed and distributed to the developers of the reviewed tools to discover their functionalities and features.
Results Twenty-four different phenotype authoring tools were identified and reviewed. Developers of 16 of these identified tools completed the evaluation survey (67% response rate). The surveyed tools showed commonalities but also varied in their capabilities in algorithm representation, logic functions, data support and software extensibility, search functions, user interface, and data outputs.
Discussion Positive trends identified in the evaluation included: algorithms can be represented in both computable and human readable formats; and most tools offer a web interface for easy access. However, issues were also identified: many tools were lacking advanced logic functions for authoring complex algorithms; the ability to construct queries that leveraged un-structured data was not widely implemented; and many tools had limited support for plug-ins or external analytic software.
Conclusions Existing phenotype authoring tools could enable clinical researchers to work with electronic health record data more efficiently, but gaps still exist in terms of the functionalities of such tools. The present work can serve as a reference point for the future development of similar tools.
Keywords: phenotyping, electronic health records, phenotype algorithm authoring tool, clinical research, review
INTRODUCTION
The widespread adoption of electronic health record (EHR) systems offers considerable potential for secondary use of clinical data,1–3 especially for clinical research.4,5 For example, as individual genetic variants often have weak correlations to complex diseases,6 large sample sizes are needed for genome-wide association studies in order to obtain significant results.7–9 The cost of assessing and identifying patients with a given disease or characteristic (a process referred to here as “phenotyping”) in a large number of patients is very high.4,10 However, the use of the data in EHR systems for such research could be a cost-effective solution.4,11 As the EHR captures data in the delivery of care, researchers can use it to identify patient cohorts with conditions or events that are relevant to the study.12 This can be achieved by defining study specific inclusion and exclusion criteria based on the EHR-based data fields—referred to as phenotype algorithms—and subsequently executing those algorithms on top of EHR systems.13
However, several challenges exist when attempting to use EHRs for scalable phenotyping. First, the use of EHR data requires complex processing because EHR is a result of clinical practices and operations, so that it is in general multi-dimensional and temporal,14,15 and contains different data types.16 The different types of data in EHRs, such as diagnostic codes, laboratory results, and clinical notes, have varied availability; they may come in the form of structured data, semi-structured data, or un-structured data, and the same data may be collected in different formats across organizations, or even between different clinical specialties in the same organization.16 Second, EHRs are typically optimized for data on single patients but not for the aggregation across cohorts of patients, thus the specification of queries can be challenging.11,17 Finally, EHRs usually contain a high volume of data points, increasing their complexity.15,18 As a result of these challenges, it is necessary to have knowledge of how the data are structured and represented in order to accurately formulate queries that define accurate phenotypes.19,20 Furthermore, additional experience is needed to create a phenotype definition that is portable across multiple institutions with different EHR systems.
Clinical researchers often rely on expert database analysts to perform queries in order to identify patient cohorts according to their needs. This can be a time-consuming, error-prone, and inflexible process.21,22 As discussed by Zhang et al.,23 the traditional model of phenotype extraction involves a data analyst who mediates between the clinical researcher and the clinical database. The clinical researcher has to communicate the phenotype algorithm—typically in human readable pseudo-code—to the data analyst who then translates it into a computable form. The clinical researcher and the data analyst may need to go through multiple cycles of communication in order for the request to be correctly translated. Miscommunication can lead to mismatch errors between the data analyst’s computable algorithm and the researcher’s desired algorithm. In addition, scarcity of data analyst resources may result in research bottlenecks as requests from clinical researchers grow in complexity and volume.24 One of the solutions to this problem is to design an intuitive phenotype algorithm authoring tool so that clinical researchers can directly define the algorithm criteria unambiguously, preferably using the same data elements that are typically available within the EHR systems. Such an approach has the potential to significantly reduce the level of iterations and repeated interactions between researchers and data analysts.
The purpose of this study was to review available software tools for authoring EHR-driven phenotype algorithms and evaluate their functionalities using the current literature and feedback from the developers of these tools. By evaluating state-of-the-art tools, this study aimed to identify the gaps in phenotyping workflow support and provide insights to improve the throughput of this process. Identifying and rectifying these gaps will ultimately be necessary to facilitate wide-scale adoption of phenotyping authoring and execution tools, thus enabling clinical researchers to work more productively with data analysts, and also directly with EHR data.
METHOD
Phenotyping tools identification
The overall strategy involved two stages: (1) literature review to identify the existing tools and their features and (2) survey developers of existing tools to confirm our assessment of tool capabilities.
Literature search strategy
Online database searches were performed between April and May 2014, for relevant articles. In order to discover as many relevant tools as possible, a set of broad search terms was used. These search terms included: “electronic medical records,” “electronic health records,” “EHR,” “Medical Records Systems, Computerized,” “clinical research,” “translational research,” “graphical,” “visual,” “interface,” “query,” “platform,” and combinations of these terms. The term “phenotyping” was not used because its vagueness may have limited the comprehensiveness of the search.25 Initial searches were conducted using databases including PubMed, Embase, Web of Science, and Scopus. A follow-up search was conducted using Google Scholar to identify relevant papers in the reference lists. Google was also searched using the same terms, limiting to the website domains of.edu or.org.
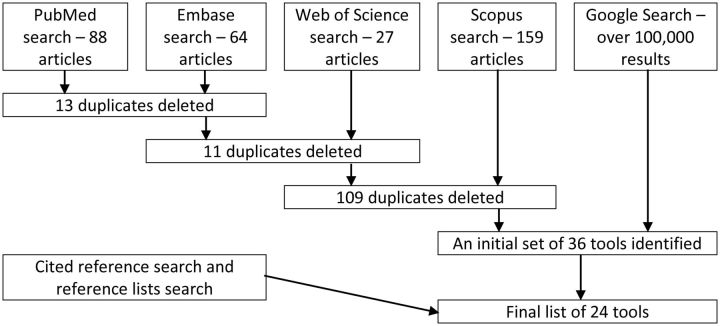
Results of each academic literature database search were recorded and saved to an EndNote library. Search results were reviewed by one of the authors (PS) and the citation information of those results meeting inclusion criteria was entered into a spreadsheet. For every result entered into the spreadsheet, an additional Google search was conducted for the specific tool in attempt to discover if a user interface or project description of the tool were available. Figure 1 shows the literature search workflow.
Figure 1:
Flow diagram of the literature search.
Inclusion and exclusion criteria
The inclusion criteria included: (1) the tool provides a query function for users to identify patient cohorts; (2) the tool does not require users to use a programming language or a database-specific query language (e.g., SQL) to author a query; (3) the tool works with EHRs or a database that is fully or partially derived from EHRs; (4) the tool is an academic application rather than a commercially available software; and (5) the publication or documentation associated with the tool was written in English. The exclusion criteria included: (1) local extensions of an application (such as SHRINE,26 Galaxy,27 and FURTHeR,28 since the difference among the infrastructures of these extensions and the Informatics for Integrating Biology and the Bedside platform is minimal) and (2) generic tools that provide query building capabilities beyond the healthcare domain, such as SAS29 and KNIME.30
Tool evaluation survey
Survey items
The research team opted to survey the developers of the tools rather than evaluate each tool individually because (1) it was not possible for the research team to obtain access to all the identified tools and conduct thorough evaluations and (2) an evaluation based on publications or documentations may not reflect the current state of the tools.
The survey was designed based on a previously conducted phenotyping tool review and evaluation studies.31,32 The survey had 30 questions in total, which were grouped into nine sections for information regarding (1) algorithm representation, (2) Boolean operator functions, (3) temporal operator functions, (4) other operation functions, (5) data support and software extensibility, (6) search functions, (7) user interface, (8) data output features, and (9) other features. Most of the questions (28 out of 30) were “yes/no” questions with an open-ended comment field. The other two questions were a multiple-choice question and an open-ended question. A description or example was provided for each question, if applicable, to clarify the meaning of the question. Table 1 shows the sections, questions, and the corresponding description/examples provided in the survey.
Table 1:
Phenotyping tools capabilities survey questions
| Section | Question | Clarification/examples |
|---|---|---|
| Algorithm representation | The algorithm can be represented in non-programming language, such as natural languages, charts, or diagrams. (Non-programming language) | The graphical editor can generate a visual flowchart, or natural language representation of the algorithm. |
| The algorithm can be represented as computable language. (Computable language) | The underlying definition is something that a computer can understand and execute, to return results. | |
| The translation between the non-computable and computable language can be done automatically by the platform. (Automatic representation translation) | ||
| The system represents algorithms as relational queries. (Relational queries) | ||
| Boolean operators | Algorithms can be written to exclude entities (patients, events, etc.) that have or do not have certain properties. (Boolean operations) | “Exclude patients with an ICD9 code of 250.01.” |
| The system can perform Boolean operations, including negation, on properties and combinations of other logic. (Nested Boolean logic) | “Find all patients that are not deceased, and were seen in the past two years.” | |
| The system supports unlimited complexity of nested Boolean logic. (Exclusion) | Nesting of Boolean operators can go down an infinite number of levels. | |
| Temporal operators | Allow you to specify the reference date to use for temporal operations. (Temporal operations) | Can relate something to the “first occurrence,” or the “date documented,” etc. |
| Allow you to relate to a specific date. (Relate to a date) | “Number of A&E admission due to fever on 25 December 2013.” | |
| Allow you to relate to arbitrary time interval. (Relate to a time interval) | “Number of A&E admission due to fever in the past 6 months.” | |
| Allow you to relate items occurring at the same time. (Relate co-occur items) | “Number of patients who had a diagnosis of diabetes and were on insulin at the time of diagnosis.” | |
| Allow you to relate items occurring before/after each other. (Relate sequential items) | “Patients who had a FNA biopsy and later underwent surgery” | |
| Temporal relationships/operators can be nested at any level of the definition. (Nested temporal operators) | “Diagnosis X at least 6 months before (Procedure Y OR Procedure Z).” | |
| Other operation functions | Algorithms may include arithmetic operations, which may be nested at any level of the definition. (Arithmetic operations) | “Has at least 6 fasting glucose lab results OR at least 10 random glucose lab results.” |
| Can specify what level of information/entity to relate against (events, patients, etc.) (Specify entity to relate to) | “Find all patients with age >30, with at least one event that occurred in the past month”—relates age to patient, and occurrence date to event. | |
| Data support and software extensibility | Supports any type of structured data element, in any terminology (even ad hoc). (Structured data element) | Lists of diagnoses can be in ICD-9 or ICD-10; age may be represented as ad-hoc categories of age (i.e., 0-9, 10-19); biobank status may be an institutional value based on type of sample(s) available (i.e., blood, saliva, tissue). |
| Allows defining criteria for text/unstructured data sources as part of the algorithm definition. (Unstructured data sources) | Apply regular expressions. Find a list of CUIs within a particular section of a clinical note. | |
| Supports data from any EHR system. (Any EHR) | Some platforms are designed to work with specific EHR systems, while others can be used with different EHR systems. | |
| Support plug-ins or external software algorithms, such as machine learning, statistical computations, or natural language processing. (Extensibility) | Allows you to specify that the algorithm should call out to an external system to perform some additional analysis. | |
| Search functions | Support searching by codes. (Codes) | “Number of patients who are diagnosed Cerebral Hemorrhage coded 430, 442.81, 421.” |
| Support searching by keywords. (Keywords) | “Number of patients who are diagnosed with ‘Cerebral Hemorrhage’.” | |
| Support advanced search. (Advanced search) | “Code ranges/find all codes 442.*-443.0, wildcards/find terms like ‘cereb* hemm*’.” | |
| User interface | How is the system accessed and used? (Web-based/ desktop-based/ native mobile application) | |
| Supports drag-and-drop to build the algorithm. (Drag-and-drop operation) | ||
| Includes documentation. (Documentation) | Help guides, tutorials, etc. | |
| Data output features | Can export an algorithm definition in a human-readable format. (Human-readable format) | Create/save a PDF or HTML document containing the criteria for the algorithm. |
| Can export an algorithm definition in a computable format. (Computable format) | Allow exporting the definition of an algorithm so that it can be imported into another system (or another instance of the same system). | |
| What is returned from the search/query? (Return from the query) | For example, patient counts, lists of patients, any clinical data (events, labs). | |
| Other features | Is your system open source? (Open source) | |
| Please list any other features that your platform offers that have not already been discussed. |
Survey administration
The protocol of this survey study was reviewed and approved by Northwestern University’s Institutional Review Board. Potential developers of the phenotyping tools were identified through publications or official websites associated with the tools. Emails were sent to these individuals to ask if they were the developers of the tools and if they were willing to participate in the study, or if they could recommend someone else to fill out the survey if they were not the developer or not available. The survey data were collected and managed using REDCap electronic data capture tools.33
Survey result verification
After receiving the survey results from the developers of the phenotyping tools, the research team conducted additional verifications of the survey answers using the following information associated with each of the corresponding tool: journal publications, conference proceedings, help documentations, written or video tutorials, and software trials or demos.
RESULTS
List of phenotype algorithm authoring tools
A total number of 24 phenotype algorithm authoring tools were included in this review and evaluation. Please refer to Table 2 for the list of the tools and the brief descriptions for each tool.
Table 2:
The list of the phenotyping tools identified from the literature and web search and their brief descriptions
| Phenotyping tool | Description |
|---|---|
| Advanced Screening for Active Protocols (ASAP)34 | ASAP is a tool designed for identifying and pre-screening patients for clinical trial eligibility based on the Ohio State University Medical Center's Information Warehouse (IW) that contains data from electronic health record (EHR) systems and billing and administrative systems. |
| Biomedical Translational Research Information System and its de-identified query tool35–37 | Biomedical Translational Research Information System is a clinical research data repository developed at the US National Institutes of Health to consolidate data from multiple databases, including EHR systems, and provide query functions for data retrieval in the dataset. |
| DANBIO38 | DANBIO is Denmark's nation-wide research database and EHR system for rheumatoid arthritis, ankylosing spondylitis, and psoriatic arthritis patients. The user can perform queries in the system to derive subset of data for research. |
| DBMap39 | DBMap is a data visualization and knowledge discovery framework implemented in the University of California, San Francisco's Brain Research Registry. Its user interface allows the users to query the database and returns the results visualized as a color-coded map. |
| Duke Enterprise Data Unified Content Explorer (DEDUCE)21,40 | DEDUCE is a query platform developed to support data exploration, cohort identification, and data extraction from Duke University's enterprise data warehouse, which stores clinical data from a number of hospitals and clinics of the university’s health care system. |
| Electronic Health Records for Clinical Research (EHR4CR)41,42 | EHR4CR is a European platform aims to improve clinical research with EHRs by supporting clinical protocol feasibility, patient identification and recruitment, clinical trial execution, and adverse event reporting. For patient cohort identification, a formal query language is developed to enable queries to be executed to heterogeneous EHRs. |
| electronic Primary Care Research Network (ePCRN) Research Workbench43–45 | ePCRN is an electronic infrastructure that offers a database of clinical information and a research portal to support the conduction of randomized control trials. Its Research Workbench enables users to create queries on the EHR data to identify eligible patient cohorts for research. |
| Electronic Medical Records and Genomics (eMERGE) Network and its Record Counter (eRC)46 | eRC is a research tool designed for research planning purpose and feasibility assessment for the genotyped patients in the eMERGE subject pool. This tool supports functions for users to construct queries base on diagnosis codes. |
| Eureka! Clinical Analytics47 | Eureka! Clinical Analytics is part of the Analytic IW software system developed at Emory University that enables users to upload a data source, specify patient cohort definitions as temporal patterns, and derive the cohort matches into an instance of i2b2. |
| Feasibility Assessment and Recruitment System for Improving Trial Efficiency (FARSITE)48 | FARSITE aims to support clinical trial feasibility assessment and recruitment in the UK. Its query interface provides assessments of the size of patient cohorts returned from the user specified search criteria to assist the evaluation of clinical trial feasibility. |
| Harvest15 | Harvest is a software toolkit designed for building web-based application to perform custom query of a dataset for data discovery and reporting purpose. This toolkit is developed by the Children’s Hospital of Philadelphia Research Institute and optimized for biomedical research use. |
| Informatics for Integrating Biology and the Bedside (i2b2)11,26 | The i2b2 platform is based on Research Patient Data Registry, which is developed in Partners HealthCare. The software allows users to perform queries on an EHR system and identify patient cohorts that fit the research criteria. A project data mart can be created for the selected patient cohorts for further processing and analysis. |
| Integrative Platform for Translational Research (IPTrans) and its query tool49 | IPTrans is the user interface level of Chado, which is a modular ontology-oriented database model and it supports the management of clinical and socio-demographic data, project management, and microarray assays and biomaterials management. It enables users to author a set of clinical or socio-demographic characteristics criteria to identify patient cohorts. |
| Measure Authoring Tool (MAT)50 | The MAT is designed to author electronic Clinical Quality Measures using the Quality Data Model (QDM), which is aligned with Meaningful Use standards. It is possible to author phenotype algorithms that are compliant with QDM. |
| Multi-Modality, Multi-Resource Information Integration environment (Physio-MIMI) and Visual Aggregator and Explorer (VISAGE)23 | VISAGE is a query interface and a component of Physio-MIMI. It provides query building, managing and exploring functionalities to assist users with hypothesis generation and patient cohort identification activities. |
| Phenotype Builder51 | Phenotype Builder is prototype software tool designed for the users to author phenotype algorithms by manipulating data elements in a graphical user interface. |
| RetroGuide/HealthFlow19,52,53 | RetroGuide is an EHR query authoring system that utilizes a flowchart analytical paradigm for clinical researchers to perform queries. HealthFlow is a package that integrates RetroGuide with FlowGuide, which is a prospective version that communicates with EHR in real time. |
| SemanticDB and Semantic Research Assistant (SRA)54 | SemanticDB is a clinical research platform developed in Cleveland Clinic and it consists of three main components: a clinical data repository, a query interface, and a data analysis and reporting interface. The SRA is the query interface that allows the users to use natural language to construct queries for patient cohort identification. |
| Stanford Translational Research Integrated Database Environment (STRIDE) and Anonymous Patient Cohort Discovery Tool55 | STRIDE is an informatics platform that consists of a clinical data warehouse, a data management application development framework, and a biospecimen data management system, developed in Stanford University. The users can use the query tool called the Anonymous Patient Cohort Discovery Tool to identify potential research patient cohorts. |
| Synthetic Derivative (SD) and its query tool9 | SD is a de-identified clinical information database derived from Vanderbilt University Medical Center’s EHR system and has a link to the corresponding DNA biobank (BioVU). Its query interface allows users to create queries to identify patient cohorts. |
| Translational Research Platform for colorectal cancer (crcTRP)56 | crcTRP is a software platform designed to support colorectal cancer research. It provides a solution to collect data in multiple sources, including EHRs and integrate clinical and omics data, and a web portal for data query and data visualization. |
| TrialViz57 | TrialViz is a query system that works with Clinical Practice Research Datalink database in the UK. This tool enables the users to author queries for selecting patient cohorts, examine the quality of the extracted data, and visualize the results of the queries. |
| University of Virginia's (UVa) Clinical Data Repository (CDR)58,59 | CDR is a data warehouse that contains data derived from multiple UVa clinical and administrative patient information systems and Virginia Department of Health. It provides a web interface for the users to conduct queries for patient cohort identification. |
| Utah Population Database Limited (UPDBL) and its query tool60 | UPDBL is a research platform at the University of Utah that includes data from multiple sources such as EHRs, vital records, driver license records, voter registration, etc. Its query tool allows users to build and run queries and view aggregated results. |
Tool evaluation survey results
In total, we received the responses from the developers of 16 out of 24 tools, with one survey response collected for each tool. The response rate accounted for 67%. The 16 tools included in the evaluation included: Advanced Screening for Active Protocols, Biomedical Translational Research Information System and its de-identified query tool, DANBIO, Duke Enterprise Data Unified Content Explorer, Electronic Medical Records and Genomics Network and its Record Counter, Harvest, Informatics for Integrating Biology and the Bedside, Integrative Platform for Translational Research and its query tool, Measure Authoring Tool, Phenotype Builder, Multi-Modality Multi-Resource Information Integration environment and Visual Aggregator and Explorer, RetroGuide/HealthFlow, Synthetic Derivative and its query tool, Stanford Translational Research Integrated Database Environment and Anonymous Patient Cohort Discovery Tool, TrialViz, and Utah Population Database Limited and its query tool. There was no missing data presented in the “yes/no” questions. Please refer to the online supplementary document (Appendix Table A1 and Table A2) for the details of the evaluation for each tool against the survey items. Please refer to Figure 2–5 for the results from the “yes/no” questions. According to the responses, in the 16 tools that were evaluated, seven (44%) only support defining criteria from the EHR systems that the tool was designed to use with (“specific EHR” group in the figures); the rest of them (56%) were designed to support any EHR systems (“any EHR” group in the figures).
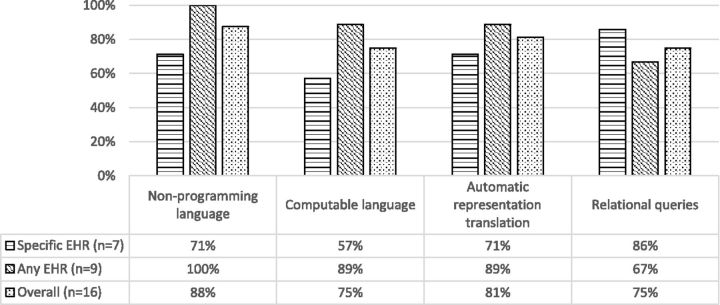
Figure 2:
Percentages of phenotype authoring tools that support the corresponding algorithm representation features.
For the algorithm representation features, 11 out of 16 tools (69%) were reported to be able to represent the algorithm in both noncomputable (defined as a format that is optimized for review by a human, that a computer is not able to also understand) and computable formats (those that can be interpreted and executed by a computer). The rest of the tools were reported to be only able to represent the algorithm to be either noncomputable or computable.
For the data support features, five of out the 16 tools (31%) were reported to support both structured data and un-structured data.
In terms of the results returned from the execution of the algorithm, 11 tools (69%) were able to report patient counts and seven tools (44%) were able to report patient/encounter list; 9 tools (56%) were able to report some sort of summary statistics of the patient cohort, and 4 tools (25%) were able to generate detailed report according to user specification.
DISCUSSION
Algorithm representation
It was found that a high percentage (88% overall) of the surveyed tools are able to represent the algorithm in a noncomputable format. The survey results also indicated that for the tools that are able to represent algorithms in both formats, all of them can translate the algorithm between these two formats automatically. Representing algorithms in a human-readable format is very useful in phenotyping. Creating phenotype algorithm involves knowledge level authoring (e.g., inclusion/exclusion criteria) and data level authoring (e.g., specific value ranges of a data field).61 For clinical researchers, usually the knowledge level is the first step of the authoring process, and it can be better represented in noncomputable formats such as flow charts or natural language. In addition, the noncomputable representation is also more likely to be used in communication between clinical researchers themselves, between clinical researchers and data analysts, or even between institutions where computable algorithms may be represented differently due to the difference in software platforms. As such, the use of noncomputable, human-readable algorithm representation can potentially increase the portability of the algorithm among these entities. However, the lack of a standardized representation of the algorithms still remains a challenge. Although quality standards such as the National Quality Forum Quality Data Model62 and HL7 Health Quality Measures Format63 can be used, they are not comprehensive for complex phenotype algorithms.64 There is a need for developing a standard mechanism for phenotype algorithm representation.
Boolean operators, temporal operators, and other operation features
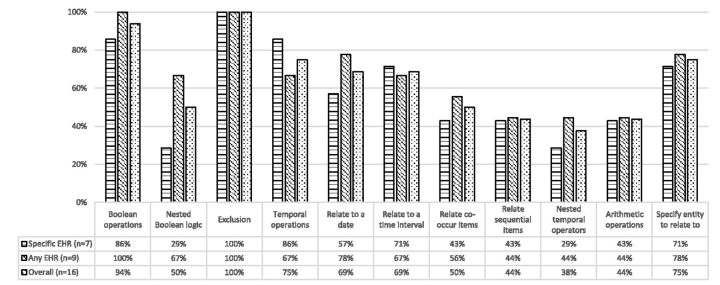
Intuitive query authoring tools with graphical interface can make clinical researchers who are not expert data analysts able to perform data queries themselves; however, those tools may not provide sufficient capability in authoring complex queries that completely satisfies the requirements of clinical research.19 In the evaluation, it was found that some operational features had low implementation rate among the surveyed tools: nested Boolean logic (50% overall, and 29% for specific EHR group), relate co-occur items (50% overall), nested temporal operators (38% overall), and arithmetic operations (44% overall).
It may still be possible to author simple algorithms based on basic Boolean operators and temporal operators. However, many algorithms require complex operations. An analysis of the phenotype algorithms on the electronic Medical Records and Genomics Network65 indicated that most of the algorithms require complex Boolean and temporal logic.66 For example, the algorithms for diabetic retinopathy, hypothyroidism, resistant hypertension, and type 2 diabetes all require nested Boolean logic and complex temporal logic.66
The implementations of various temporal operation are not very high (see Figure 3) among the surveyed tools. However, as noted by Albers et al.,67 human phenotypes are inherently time-dependent and dynamic (e.g., the probability of acquiring a disease and physical characteristic can change over time); but this is somewhat neglected in many phenotype algorithms. This situation may be caused by the inherent difficulty in authoring dynamic phenotype algorithms with temporal components or lack of required tools to finish the task.
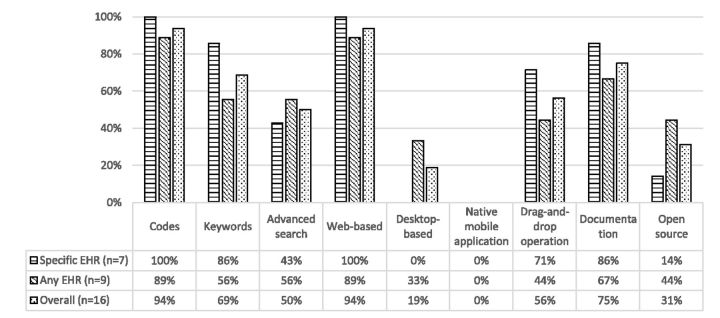
Figure 4:
Percentages of phenotype authoring tools that support the corresponding search functions, user interface, and other features.
Figure 5:
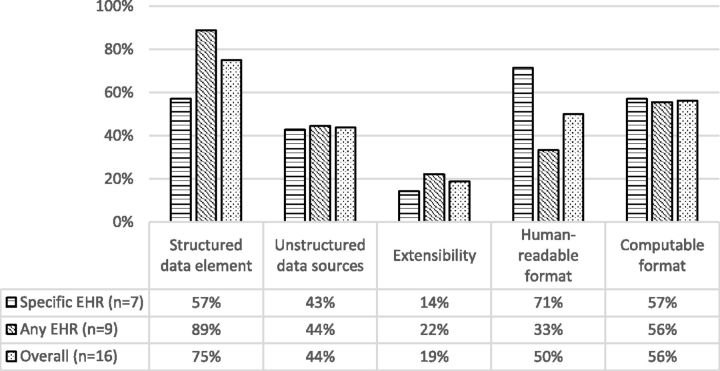
Percentages of phenotype authoring tools that support the corresponding data support and software extensibility and data output features.
Figure 3:
Percentages of phenotype authoring tools that support the corresponding Boolean operators, temporal operators, and other operation features.
Data support and software extensibility
Many of the surveyed tools (75% overall) support defining queries that utilize structured data while less than half of them (44% overall) provide such support for un-structured data. Structured data may be an accurate way of storing and extracting data;68 for example, the positive predictive value of using billing codes to identify acute myocardial infarction was reported to be higher than 90%.69 However, using only structured data may negatively influence the accuracy of cohort definition in some cases.34,70 For example, a study comparing using structured International Classification Of Diseases - 9 (ICD-9) code and natural language processing (NLP) processed un-structured data for clinical trials pre-screening concluded that using a combination of both types of data would yield the best results.71 In an another study, the researchers found that NLP-based techniques showed higher sensitivity and positive predictive value than ICD9 code-based techniques in identifying individuals in need of testing for celiac disease.72
A solution to the lack of the processing capability for un-structured data is to allow defining a phenotype algorithm that (when executed) may invoke external software, such as a NLP plug-in, to be able to be used in the system. Unfortunately, most of the tools that did not support un-structured data also did not support the use of plug-ins. Another problem associated with not supporting plug-ins is that novel approaches in phenotyping may not be used without modifying the core system. While a review article indicated that there were machine learning and statistical analysis approaches being using as phenotype algorithms in research,25 these functions are rarely supported by existing phenotyping tools.
It was found that only about half of the evaluated tools could be used with different EHR systems to facilitate portability of phenotyping algorithms between institutions. This is a significant design challenge as not all of the EHR databases provide mappings to standardized terminology systems, such as ICD-9/10, RxNorm, or LOINC, as recommended.73 In designing a phenotyping tool that supports portability, the algorithm should be able to employ standardized terminologies and at the same time accommodate non-standardized when possible.
User interface and other features
Almost all the evaluated tools (94% overall) provide a web interface for convenient access. Also most tools offer basic search functions for codes in medical terminologies. However, only about half of the tools (50%) provided advanced search functions (e.g., wildcard matching), and this may reduce the usability for certain search scenarios.
In terms of data return from queries, there were variations among the tools. Some tools only provide a simple patient count, while others provide a detailed patient list, an encounter list, and detailed reports depending on the specifications of the user. This may relate to the different scenarios for which the tools were designed. For example, a query tool may be designed for assessing the size of the patient cohort given the algorithm before starting a clinical trial, and due to patient privacy issues, no further information will be shown other than a simple patient count; another system may be designed for a de-identified database and able to provide all the data fields for the user to conduct complex data analysis. For these different cases, the designers may need to study the specific user needs in order to design the best form of data output.
Limitations
The main limitation of this study was that the evaluation survey was only filled out by developers of 67% of the tools. It is therefore possible that the results are biased towards the tools for which we were able to obtain developers’ feedback.
CONCLUSION
This review and evaluation of existing EHR-driven phenotype algorithm authoring tools provided an overview of the current state of the available tools. Overall, these phenotyping tools can provide interfaces that are relatively accessible for the clinical researchers who may not have high expertise in database and query coding. Most of the evaluated tools can enable users to author simple algorithms. However, important gaps also exist: many of the evaluated tools do not support the complex logic specifications, un-structured data processing, and external analytic software. These problems are obstacles for the users to author more complex algorithms. Future development of phenotyping tools should focus on improving capabilities in these areas.
FUNDING
This work was funded primarily by R01-GM105688 from the National Institute of General Medical Sciences. Additional contribution came from the eMERGE Network sites funded by the National Human Genome Research Institute through the following grants: U01-HG004610 and U01-HG006375 (Group Health Cooperative/University of Washington); U01-HG004608 (Marshfield Clinic); U01-HG04599 and U01-HG06379 (Mayo Clinic); U01-HG004609 and U01-HG006388 (Northwestern University); U01-HG006389 (Essentia Institute of Rural Health); U01-HG04603 and U01-HG006378 (Vanderbilt University); and U01-HG006385 (Vanderbilt University serving as the Coordinating Center). LVR received additional support from NCATS grant UL1TR000150.
COMPETING INTERESTS
The authors have no competing interests to declare.
CONTRIBUTORS
JCD, EM, JP, and WKT provided leadership for the project; P.S. conducted systematic literature review for the candidate tools; LVR, JX, RCK, JP, and WKT refined and reviewed the list of tools; LVR and JX designed and administered the survey; LVR, JX, JCD, and JP recruited participants for the survey; JX analyzed the survey data; LVR, JX, and HM performed verification for the survey responses. JX and LVR drafted the manuscript; all authors contributed expertise and edits.
SUPPLEMENTARY MATERIAL
Supplementary material is available online at http://jamia.oxfordjournals.org/.
REFERENCES
- 1.Safran C, Bloomrosen M, Hammond WE, et al. Toward a national framework for the secondary use of health data: an American Medical Informatics Association White Paper. JAMIA. 2007;14(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Embi PJ, Payne PR. Clinical research informatics: challenges, opportunities and definition for an emerging domain. JAMIA. 2009;16(3):316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Clercq E, Van Casteren V, Jonckheer P, et al. Research networks: can we use data from GPs' electronic health records? Stud Health Technol Inform. 2006;124:181–186. [PubMed] [Google Scholar]
- 4.Murphy S, Churchill S, Bry L, et al. Instrumenting the health care enterprise for discovery research in the genomic era. Genome Res. 2009;19(9):1675–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. JAMIA. 2012;20(1):117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manolio TA, Bailey-Wilson JE, Collins FS. Genes, environment and the value of prospective cohort studies. Nat Rev Genet. 2006;7(10):812–820. [DOI] [PubMed] [Google Scholar]
- 7.Burton PR, Clayton DG, Cardon LR, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–885. [DOI] [PubMed] [Google Scholar]
- 9.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Therap. 2008;84(3):362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spivey A. Gene–environment studies: who, how, when, and where? Environ Health Persp. 2006;114(8):A466–A467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). JAMIA. 2010;17(2):124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richesson RL, Hammond WE, Nahm M, et al. Electronic health records based phenotyping in next-generation clinical trials: a perspective from the NIH Health Care Systems Collaboratory. JAMIA. 2013;20(e2):e226–e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li D, Endle CM, Murthy S, et al. Modeling and executing electronic health records driven phenotyping algorithms using the NQF Quality Data Model and JBoss® Drools engine. AMIA Ann Symp Proc. 2012;2012:532-541. [PMC free article] [PubMed] [Google Scholar]
- 14.D'Avolio LW, Farwell WR, Fiore LD. Comparative effectiveness research and medical informatics. Am J Med. 2010;123(12):e32–e37. [DOI] [PubMed] [Google Scholar]
- 15.Pennington JW, Ruth B, Italia MJ, et al. Harvest: an open platform for developing web-based biomedical data discovery and reporting applications. JAMIA. 2013;21(2):379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denny JC. Mining electronic health records in the genomics era. PLoS Comput Biol. 2012;8(12):e1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy S. Data warehousing for clinical research. In: Liu LÖ, Tamer M, eds. Encyclopedia of database systems. New York, NY: Springer; 2009:679–684. [Google Scholar]
- 18.Hey AJ, Trefethen AE. The data deluge: An e-science perspective. In: Berman F, Fox G, Hey T, eds. Grid Computing: Making the Global Infrastructure a Reality. New York, NY: John Wiley & Sons, Inc; 2003. [Google Scholar]
- 19.Huser V, Narus SP, Rocha RA. Evaluation of a flowchart-based EHR query system: A case study of RetroGuide. J Biomed Inform. 2010;43(1):41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadkarni PM. Data extraction and ad hoc query of an entity—Attribute—Value database. JAMIA. 1998;5(6):511–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horvath MM, Rusincovitch SA, Brinson S, Shang HC, Evans S, Ferranti JM. Modular design, application architecture, and usage of a self-service model for enterprise data delivery: The Duke Enterprise Data Unified Content Explorer (DEDUCE). J Biomed Inform. 2014;52:231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy SN, Gainer V, Chueh HC. A visual interface designed for novice users to find research patient cohorts in a large biomedical database. AMIA Ann Symp Proc. 2003;2003:489-493. [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang G-Q, Siegler T, Saxman P, et al. VISAGE: a query interface for clinical research. AMIA Summits Transl Sci Proc. 2010;2010:76. [PMC free article] [PubMed] [Google Scholar]
- 24.Hruby GW, Boland MR, Cimino JJ, et al. Characterization of the biomedical query mediation process. AMIA Summits Transl Sci Proc. 2013;2013:89–93. [PMC free article] [PubMed] [Google Scholar]
- 25.Shivade C, Raghavan P, Fosler-Lussier E, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. JAMIA. 2013;21(2):221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber GM, Murphy SN, McMurry AJ, et al. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. JAMIA. 2009;16(5):624–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Center for Research Informatics at the University of Chicago. Self Service Data Analysis Environment. http://cri.uchicago.edu/?page_id=1202. Accessed Janurary 5, 2015.
- 28.Narus SP, Schultz ND, Livne OE, Bradshaw RL, Mitchell JA. Going FURTHeR with i2b2. http://wiki.siframework.org/file/view/Going%20FURTHeR%20with%20i2b2.pdf/290575597/Going%20FURTHeR%20with%20i2b2.pdf. Accessed Janurary 5, 2015. [Google Scholar]
- 29.Institute S. SAS/STAT 12. 1 User's Guide: Survey Data Analysis. Cary, NC: SAS Institute; 2012.
- 30.Berthold MR, Cebron N, Dill F, et al. KNIME-the Konstanz information miner: version 2.0 and beyond. ACM SIGKDD Explorations Newsletter. 2009;11(1):26–31. [Google Scholar]
- 31.Rasmussen LV, Xu J, Liu R, et al. Evaluation of existing phenotype authoring tools for clinical research. Paper presented at: AMIA 2014 Annual Symposium 2014; Washington D.C. [Google Scholar]
- 32.Mo H, Thompson W, Rasmussen LV, et al. Towards A Desiderata for Phenotyping Algorithm Authoring Languages. Paper presented at: AMIA 2014 Annual Symposium 2014; Washington D.C. [Google Scholar]
- 33.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pressler TR, Yen P-Y, Ding J, Liu J, Embi PJ, Payne PR. Computational challenges and human factors influencing the design and use of clinical research participant eligibility pre-screening tools. BMC Med Informat Decis Mak. 2012;12(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cimino JJ, Ayres EJ, Remennik L, et al. The National Institutes of Health’s Biomedical Translational Research Information System (BTRIS): Design, contents, functionality and experience to date. J Biomed Inform. 2014;52:11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cimino JJ, Ayres EJ. The clinical research data repository of the US National Institutes of Health. Stud Health Technol Inform. 2010;160(Pt 2):1299. [PMC free article] [PubMed] [Google Scholar]
- 37.Cimino JJ, Ayres EJ, Beri A, Freedman R, Oberholtzer E, Rath S. Developing a self-service query interface for re-using de-identified electronic health record data. Stud Health Technol Inform. 2013;192:632–636. [PMC free article] [PubMed] [Google Scholar]
- 38.Hetland ML. DANBIO—powerful research database and electronic patient record. Rheumatology. 2011;50(1):69–77. [DOI] [PubMed] [Google Scholar]
- 39.Zhang M, Zhang H, Tjandra D, Wong ST. DBMap: a space-conscious data visualization and knowledge discovery framework for biomedical data warehouse. Inform Technol Biomed, IEEE Transactions on. 2004;8(3):343–353. [DOI] [PubMed] [Google Scholar]
- 40.Horvath MM, Winfield S, Evans S, Slopek S, Shang H, Ferranti J. The DEDUCE Guided Query tool: providing simplified access to clinical data for research and quality improvement. J Biomed Inform. 2011;44(2):266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ouagne D, Hussain S, Sadou E, Jaulent M-C, Daniel C. The Electronic Healthcare Record for Clinical Research (EHR4CR) information model and terminology. Stud Health Technol Inform. 2012;180:534–538. [PubMed] [Google Scholar]
- 42.Doods J, Bache R, McGilchrist M, Daniel C, Dugas M, Fritz F. Piloting the EHR4CR feasibility platform across Europe. Methods Inform Med. 2014;53(4):264–268. [DOI] [PubMed] [Google Scholar]
- 43.Peterson KA, Fontaine P, Speedie S. The Electronic Primary Care Research Network (ePCRN): a new era in practice-based research. J Am Board Fam Med. 2006;19(1):93–97. [DOI] [PubMed] [Google Scholar]
- 44.Feyisetan S, Tyson G, Taweel A, Vargas-Vera M, Van Staa T, Delaney B. ePCRN-IDEA2: An Agent-Based System for Large-Scale Clinical Trial Recruitment. Proceedings of the 11th International Conference on Autonomous Agents and Multiagent Systems. Richland, SC: International Foundation for Autonomous Agents and Multiagent Systems; 2012:19-28. [Google Scholar]
- 45.Delaney BC, Peterson KA, Speedie S, Taweel A, Arvanitis TN, Hobbs FR. Envisioning a learning health care system: the electronic primary care research network, a case study. Ann Fam Med. 2012;10(1):54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Danciu I, Cowan JD, Basford M, et al. Secondary use of clinical data: the Vanderbilt approach. J Biomed Inform. 2014;52:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Post AR, Krc T, Rathod H, et al. Semantic ETL into i2b2 with Eureka! AMIA Summits Transl Sci Proc. 2013;2013:203-207. [PMC free article] [PubMed] [Google Scholar]
- 48.Köpcke F, Prokosch H-U. Employing computers for the recruitment into clinical trials: a comprehensive systematic review. J Med Internet Res. 2014;16(7):e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyoshi NSB, Pinheiro DG, Silva WA, Felipe JC. Computational framework to support integration of biomolecular and clinical data within a translational approach. BMC Bioinformatics. 2013;14(1):180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Centers for Medicare & Medicaid Services. Measure Authoring Tool. https://www.emeasuretool.cms.gov/web/guest/mat-home. Accessed December 1, 2014.
- 51.Peissig P, Miller A, Yoder N, et al. Simplifying High Throughput Electronic Phenotyping. Poster presented at AMIA Annual Symposium 2011; Washington D.C. [Google Scholar]
- 52.Huser V, Rocha R. Analyzing medical data from multi-hospital healthcare information system using graphical flowchart models. Paper presented at: Biomedical Informatics and Cybernetics Symposium 2007; Orlando, FL. [Google Scholar]
- 53.Huser V, Rasmussen LV, Oberg R, Starren JB. Implementation of workflow engine technology to deliver basic clinical decision support functionality. BMC Med Res Methodol. 2011;11(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pierce CD, Booth D, Ogbuji C, Deaton C, Blackstone E, Lenat D. SemanticDB: a semantic Web infrastructure for clinical research and quality reporting. Curr Bioinformatics. 2012;7(3):267–277. [Google Scholar]
- 55.Lowe HJ, Ferris TA, Hernandez PM, Weber SC. STRIDE–An integrated standards-based translational research informatics platform. AMIA Annual Symposium Proceedings. 2009;2009:391-395. [PMC free article] [PubMed] [Google Scholar]
- 56.Deng N, Zheng L, Liu F, Wang L, Duan H. CrcTRP: a translational research platform for colorectal cancer. Comput Math Methods Med. 2013;2013:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tate AR, Beloff N, Al-Radwan B, et al. Exploiting the potential of large databases of electronic health records for research using rapid search algorithms and an intuitive query interface. JAMIA. 2014;21(2):292–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mullins IM, Siadaty MS, Lyman J, et al. Data mining and clinical data repositories: Insights from a 667,000 patient data set. Comput Biol Med. 2006;36(12):1351–1377. [DOI] [PubMed] [Google Scholar]
- 59.Scully KW, Einbinder JS, Pates RD, et al. Web-accessible patient data warehouse at the University of Virginia. Proc AMIA Symp. 1999;1999:1216. [Google Scholar]
- 60.Hurdle JF, Haroldsen SC, Hammer A, et al. Identifying clinical/translational research cohorts: ascertainment via querying an integrated multi-source database. JAMIA. 2012;20(1):164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernández-Breis JT, Maldonado JA, Marcos M, et al. Leveraging electronic healthcare record standards and semantic web technologies for the identification of patient cohorts. JAMIA. 2013;20(e2):e288–e296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.National Quality Forum. Quality Data Model. http://www.qualityforum.org/QualityDataModel.aspx. Accessed Janurary 27, 2015.
- 63.Health Level Seven. The Health Quality Measures Format. http://www.hl7.org/implement/standards/product_brief.cfm?product_id=97. Accessed Janurary 27, 2015.
- 64.Rasmussen LV, Kiefer RC, Mo H, et al. A modular architecture for electronic health record-driven phenotyping. The AMIA 2015 Joint Summits on Translational Science; 2015; San Francisco, CA. [PMC free article] [PubMed] [Google Scholar]
- 65.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Conway M, Berg RL, Carrell D, et al. Analyzing the heterogeneity and complexity of Electronic Health Record oriented phenotyping algorithms. AMIA Annu Symp Proc. 2011;2011:274–283. [PMC free article] [PubMed] [Google Scholar]
- 67.Albers D, Elhadad N, Tabak E, Perotte A, Hripcsak G. Dynamical phenotyping: using temporal analysis of clinically collected physiologic data to stratify populations. PloS One. 2014;9(6):e96443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davis MF, Sriram S, Bush WS, Denny JC, Haines JL. Automated extraction of clinical traits of multiple sclerosis in electronic medical records. JAMIA. 2013;20(e2):e334–e340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fan J, Arruda-Olson AM, Leibson CL, et al. Billing code algorithms to identify cases of peripheral artery disease from administrative data. JAMIA. 2013;20(e2):e349–e354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bazarian JJ, Veazie P, Mookerjee S, Lerner EB. Accuracy of Mild Traumatic Brain Injury Case Ascertainment Using ICD - 9 Codes. Acad Emerg Med. 2006;13(1):31–38. [DOI] [PubMed] [Google Scholar]
- 71.Li L, Chase HS, Patel CO, Friedman C, Weng C. Comparing ICD9-encoded diagnoses and NLP-processed discharge summaries for clinical trials pre-screening: a case study. AMIA Ann Symp Proc. 2008;2008:404-408. [PMC free article] [PubMed] [Google Scholar]
- 72.Ludvigsson JF, Pathak J, Murphy S, et al. Use of computerized algorithm to identify individuals in need of testing for celiac disease. JAMIA. 2013;20(e2):e306–e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hellenman J, Goossen WTF. Modeling nursing care in health level 7 reference information model. Comput Inform Nurs. 2003;21(1):37–45. [DOI] [PubMed] [Google Scholar]