Abstract
The Center for Expanded Data Annotation and Retrieval is studying the creation of comprehensive and expressive metadata for biomedical datasets to facilitate data discovery, data interpretation, and data reuse. We take advantage of emerging community-based standard templates for describing different kinds of biomedical datasets, and we investigate the use of computational techniques to help investigators to assemble templates and to fill in their values. We are creating a repository of metadata from which we plan to identify metadata patterns that will drive predictive data entry when filling in metadata templates. The metadata repository not only will capture annotations specified when experimental datasets are initially created, but also will incorporate links to the published literature, including secondary analyses and possible refinements or retractions of experimental interpretations. By working initially with the Human Immunology Project Consortium and the developers of the ImmPort data repository, we are developing and evaluating an end-to-end solution to the problems of metadata authoring and management that will generalize to other data-management environments.
Keywords: datasets as topic, data curation, data collection, standards, biological ontologies
INTRODUCTION
The scientific method requires nothing less than that experiments be reproducible and that the data be available for other scientists to examine and reinterpret. In an era when data are generated at rates and in quantities never before imaginable, there is an urgent need to understand the structure of datasets, the experimental conditions under which they were produced, and the information that other investigators may need to make sense of the data.1 The ultimate Big Data challenge lies not in the data, but in the metadata—the machine-readable descriptions that provide data about the data. It is not enough to simply put data online; data are not usable until they can be “explained” in a manner that both humans and computers can process.
There has been a groundswell of effort to develop and promote metadata standards that scientists can use to annotate their results. Biomedical organizations such as the Global Alliance for Genomics and Health2 and FORCE11,3 and more general associations such as the Research Data Alliance,4 work to evangelize the essential role that metadata plays in data sharing. Activities to collect and define community-driven standards,5 such as BioSharing,6 offer important resources not only to biomedical researchers, but also to journal editors and to biomedical curators who seek guidance regarding which standards to use.7 Despite a growing set of guidelines and templates for defining metadata and numerous ontologies from which metadata authors can select standard terms for describing their experiments, the barriers to authoring the metadata needed for sharing, analyzing, and interpreting big datasets are tremendously high.8 It takes time and effort to create well-specified metadata, and investigators view the task of metadata authoring (or data annotation) to be a burden that may benefit other scientists, but not the team that did the work in the first place.
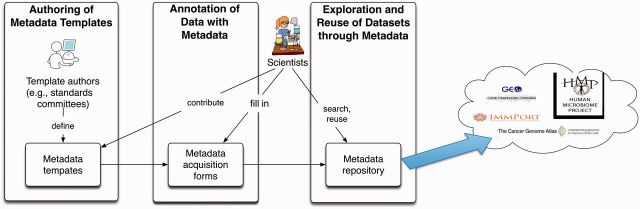
The Center for Expanded Data Annotation and Retrieval (CEDAR) was established in the autumn of 2014 to develop computer-assisted approaches to overcome the impediments to creating high-quality biomedical metadata.9 Our overarching plan is to create a computational ecosystem for development, evaluation, use, and refinement of biomedical metadata. Our approach centers on the use of metadata templates, which define sets of data elements needed to describe particular types of biomedical experiments (or assays). The templates include value sets (controlled terms and synonyms) for specific data elements. They also may indicate constraints on the use of the value sets when filling in data elements of the template. CEDAR will use a library of such templates to help scientists—the original researchers or data curators—to author new metadata for the submission of annotated datasets to appropriate online data repositories. CEDAR is developing methods to support and accelerate an end-to-end process whereby community-based organizations collaborate to create metadata templates, investigators or curators use the templates to define the metadata for individual experiments, and other scientists search the metadata to access and analyze the corresponding online datasets (Figure 1).
Figure 1:
The CEDAR ecosystem for metadata management. Communities of biomedical scientists author metadata templates (and template components), which are stored in an online template repository (left panel). Investigators annotate their experimental data by assembling composite templates and by filling in the templates using metadata-acquisition forms to create collections of experimental metadata (center panel). The metadata are both stored in a CEDAR metadata repository (right panel) and exported along with the primary data to archives such as ImmPort, GEO, and the Stanford Digital Repository. Analysis of the CEDAR metadata repository (right panel) will reveal patterns in the metadata that will enable the tools for metadata acquisition (center panel) to use predictive data entry to ease the task of filling out the templates.
Our methods support the notion of metadata as evolving descriptions of biomedical experiments. As a result, metadata need to change as datasets are revised and re-explored, as experimental results are re-interpreted in light of other findings, and as new publications appear in the scientific literature. The metadata thus will expand as investigators re-examine the primary data and as new papers and datasets appear online. Our emerging Web-based tools will enable investigators to learn both from our growing collection of metadata and from the primary datasets that the metadata describe.
THE METADATA PROBLEM
CEDAR does not represent the first attempt to address the problem of metadata quality or to make experimental data more discoverable. Since the turn of the current century, the scientific community has been the subject of legislative mandates and executive orders attempting to make experimental data created at public expense openly available and interpretable by other investigators.10 Despite explicit federal directives, the actual amount of data sharing has been relatively modest. The biomedical community is consequently limited in its ability to confirm past conclusions and to mine the data to generate new inferences.11 While popular magazines such as The Economist run cover stories on “How Science Goes Wrong”12—specifically citing the inability of biomedical researchers to examine one another’s data and to replicate one another’s work—investigators worry that the Big Data revolution will fizzle if it continues to be difficult or impossible for scientists to locate their colleagues’ experimental datasets online, to glean how the experiments actually were performed, and to understand how the data should be interpreted.
Workers in biomedicine understand the importance of making data available publicly to confirm scientific conclusions and to perform new analyses.13 The explosion of interest in dry-bench biomedical research and in the exciting discoveries that can emerge from the examination of large, online datasets is palpable.14 If there is one obstacle to the sharing of Big Data in biomedicine and to the breakthroughs that will result from large-scale exploration of online datasets, it is very simple to understand: people hate to author metadata.
The problems are both technical and cultural.8 Technically, there is a need to ease the hassles of creating high-quality metadata to annotate experimental results. Culturally, there is a need to educate scientists about the benefits of publishing their data—and the metadata needed to make their data useful to others. CEDAR is developing methods, tools, and training experiences that will simplify the process by which biomedical investigators annotate their experimental data with high-quality metadata, making possible the indexing, retrieval, integration, and analysis of Big Data repositories in ways that to date have been impossible. Although we are keeping an open eye to other kinds of biomedical metadata (such as those used to structure electronic health records or to serve as common data elements for clinical trials), our goal is to advance biomedical science by enhancing the authoring and downstream use of high-quality metadata that describe laboratory experiments—particularly experiments that will lead to the generation of the large datasets at the core of the NIH Big Data to Knowledge initiative.15
CEDAR has been founded with the conviction that the combined use of services from the National Center for Biomedical Ontology (NCBO)16 and machine learning from a large repository of biomedical metadata that we ourselves will create in the course of our work will offer an opportunity to reduce the frustration that investigators feel when they define metadata. Although the technology that CEDAR is developing may not make researchers enjoy authoring metadata, we anticipate that the overall ecosystem that we are creating will demonstrate that the authoring of high-quality metadata need not be onerous and that science has much to gain if experimental datasets are accompanied by first-rate annotations that can actually explain the associated data.
AN ECOSYSTEM FOR METADATA AUTHORING AND MANAGEMENT
The approach that we are taking in CEDAR recognizes the growing trend among investigators in many disciplines to define templates that structure metadata.17 The hundreds of minimal information guidelines and formats served by BioSharing6 are testament to the value that many biomedical scientists see in this approach and to the need to offer investigators help in navigating and selecting from the wealth of existing resources.13 We know, however, that templates are not enough. The Gene Expression Omnibus,18 for example, which requests that investigators provide metadata conformant with the Minimal Information About a Microarray Experiment standard,19 struggles to get data submitters to fill out even a fraction of the “minimally required” fields. CEDAR collaborators in the Human Immunology Project Consortium (HIPC),20 who are busily creating metadata templates for their own data-sharing purposes, argue that most of the “minimal information sets” are not sufficiently minimal for most investigators to use. Experience within the ISA Commons7 suggests that metadata templates should be pieced together from smaller components, taking into consideration the type of the experiment, the technology and assay employed, the experimental condition, the organism or tissue studied, and so on. Currently, developers must configure templates manually using a dedicated component of the ISA software suite,21 drawing on their knowledge of which minimal information sets and terminologies should be pieced together. In the CEDAR project, we are creating a comprehensive repository of template components that investigators will be able to assemble as needed using special-purpose tools to create frameworks for the metadata specifications for new experiments (see Figure 1). The template components will be stored in an extended version of the NCBO BioPortal repository.16 Whenever possible, linkages between the data elements of the template components and the archive of biomedical ontologies maintained in BioPortal will suggest to the authors of new metadata how the data elements in the templates should be filled in with terms from the designated ontologies.
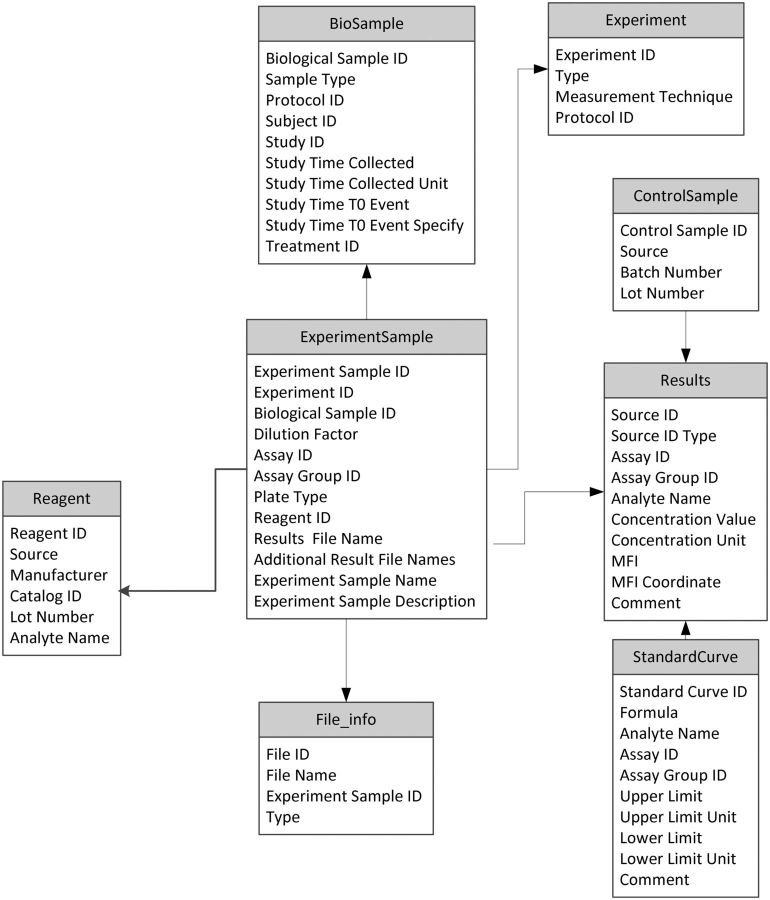
CEDAR’s collaborators from HIPC are developing templates for structuring metadata regarding experiments in human immunology.22 HIPC takes seriously the goals of putting all its data online, and of annotating the data with comprehensive metadata (Figure 2). We intend to archive these templates in our repository and to develop mechanisms to ease the manner in which HIPC investigators will fill out the templates to define specific instances of experimental metadata (Figure 3).
Figure 2:
HIPC metadata template. The Human Immunology Project Consortium creates templates such as this one (for annotating the results of multiplex bead array assays) to standardize all its experimental metadata. HIPC templates are providing the initial test of the CEDAR template-management technology.
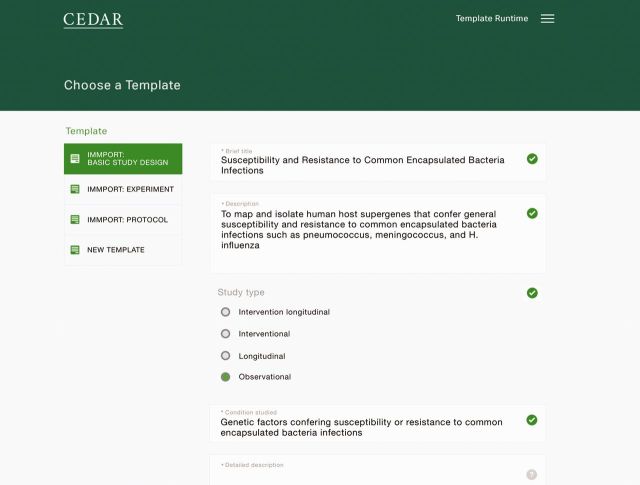
Figure 3:
Prototype user interface for template selection and instantiation. Here, the end user has selected the “ImmPort Basic Study Design” template, and she has filled in values for the template’s slots for brief title, description, study type, and condition studied. The enumerated value sets for slots such as “study type” are taken from ontologies stored in the NCBO BioPortal repository.
CEDAR will benefit from HIPC’s long-term commitment to developing templates for a wide range of experimental metadata. All the HIPC datasets are themselves archived in ImmPort,23 the designated repository for all experimental data generated by grantees of the Division of Allergy, Immunity, and Transplantation of the National Institute of Allergy and Infectious Diseases. HIPC and ImmPort are providing CEDAR with a laboratory in which to study the metadata problem from end to end—from the authoring of metadata templates, to computer-assisted assembly and instantiation of those templates to create new metadata specifications, to the archiving of datasets and their associated metadata in an online repository.
Ultimately, CEDAR will experiment with other end-to-end platforms for archiving data and their associated metadata, including the Stanford Digital Repository,24 a growing collection of digital records, both from the collection of Stanford University Libraries and from the laboratories of Stanford faculty members. Through our partnership with the ISA community and the BioSharing initiative, we will have ready access to machine-readable community guidelines and formats that we can put to use in the authoring of metadata for a great variety of biomedical datasets.
Simply amassing a large library of metadata templates in electronic form and linking the template slots to elements of biomedical ontologies will not be sufficient for solving the “metadata problem,” however. Investigators are going to want active assistance in filling out such templates. Accordingly, CEDAR is studying a variety of techniques to ease the work of entering metadata into the template fields. For example, we will use NCBO technology to facilitate the selection of ontology terms from pick lists.16 Similarly, we are encouraged by the potential of natural-language processing to assist in the completion of CEDAR templates for describing experimental conditions. There has been considerable work to extract certain kinds of metadata automatically from the text of Web pages.25 Analogous processing of the “Methods” section of online publications could inform the specification or enhancement of some of the metadata elements needed to annotate the datasets described in the corresponding articles.26 We will use the NCBO Annotator27 and other natural-language techniques to drive the recommendation of ontology terms from such narrative text. As we amass our archive of filled-out metadata templates (see Figure 1), we will use machine-learning techniques to identify patterns in the metadata that can facilitate both predictive entry of new metadata and metadata quality assurance. Through a multipronged approach, we hope to make it simple, and maybe even fun, for scientists to annotate their experimental data in ways that will ensure their value to the scientific community.
As Edwards and colleagues28 emphasize, the collection of data about experimental data does not end with the initial publication of a dataset and its associated annotations. Scientists may discuss the dataset in follow-on publications, in letters to the editor, in annotations in PubMed Commons, in interchanges at conferences, and even in e-mail to one another. All of these additional forms of expression are themselves metadata, and need to be captured in order to provide a complete picture for interpreting the primary dataset. CEDAR plans to do just that, making public our repository of all the metadata created and collected using our tools, and growing that repository to include whatever additional annotations regarding the initial dataset can be gleaned from online sources. Although we may or may not be able to update the version of the metadata archived in whatever online repositories are being used to store the primary dataset, the mirrored metadata that we will maintain in our own metadata repository will expand over time. The CEDAR metadata repository will provide not only an enriched source of information about the experiments that users have described using our tools, but also a collection of scientific information that, in its own right, can be explored by users, mined for new associations, and put to use to simplify the work of the authors of new metadata as they fill in metadata templates.
COMPETING INTERESTS
None.
FUNDING
CEDAR is supported by grant U54 AI117925 awarded by the National Institute of Allergy and Infectious Diseases through funds provided by the trans-NIH Big Data to Knowledge (BD2K) initiative (www.bd2k.nih.gov).
CONTRIBUTORS
MAM, CAB, KHC, MMD, KAD, OG, AG-B, PK, SHK, MJO’C, YP, PR-S, S-AS, and JAW are responsible for the conception and design of this work. MAM is responsible for drafting of the initial manuscript. KHC and YP are each responsible for the preparation of a figure for manuscript. MAM, CAB, KHC, MJO’C, and S-AS provided critical revision of the manuscript. MAM, CAB, KHC, MMD, KAD, OG, AG-B, PK, SHK, MJO’C, YP, PR-S, S-AS, and JAW gave final approval of the manuscript. All authors agree to be accountable for all aspects of the work.
REFERENCES
- 1.Borgman CL. The conundrum of sharing research data. J Am Soc Inform Sci Technol. 2012;63(6):1059–1078. [Google Scholar]
- 2.Global Alliance for Genomics & Health. http://genomicsandhealth.org. Accessed March 23, 2015.
- 3.FORCE11. The future of research communications and e-scholarship. https://www.force11.org. Accessed March 23, 2015.
- 4.Research Data Alliance: research data sharing without barriers. https://rd-alliance.org. Accessed March 23, 2015.
- 5.Yarmey L, Baker KS. Towards standardization: a participatory framework for scientific standard-making. Int J Digit Curation. 2013;8(1):157–172. [Google Scholar]
- 6.Biosharing. http://www.biosharing.org. Accessed March 23, 2015.
- 7.Sansone S-A, Rocca-Serra P, Field D, et al. Toward interoperable bioscience data. Nat Genet. 2012;44(2):121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tenopir C, Allard S, Douglass K, et al. Data sharing by scientists: practices and perceptions. PLoS ONE. 2011;6(6):e21101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Center for Expanded Data Annotation and Retrival. http://metadatacenter.org. Accessed March 22, 2015. [DOI] [PMC free article] [PubMed]
- 10.Fischer EA. Public access to data from federally funded research: provisions in OMB Circular A-110. Report for Congress R42983. Congressional Research Service. March 1, 2013. [Google Scholar]
- 11.Vasilevsky NA, Brush MH, Paddock H, et al. On the reproducibility of science: unique identification of research resources in the biomedical literature. Peer J. 2013;1:e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.How science goes wrong. The Economist, October 19, 2013. http://www.economist.com/news/leaders/21588069-scientific-research-has-changed-world-now-it-needs-change-itself-how-science-goes-wrong
- 13.Tenenbaum JD, Sansone S-A, Haendel MA. A sea of standards for omics data: sink or swim? JAMIA. 2014;21(2):200–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Service RF. Biology’s dry future. Science. 2013;342(6155):186–189. [DOI] [PubMed] [Google Scholar]
- 15.Margolis R, Derr L, Dunn M, et al. The National Institutes of Health’s Big Data to Knowledge (BD2K) initiative: capitalizing on biomedical big data. JAMIA. 2014;21(6):957–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musen MA, Noy NF, Shah NH, et al. The National Center for Biomedical Ontology. JAMIA. 2012;19(2):190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenberg J. Understanding metadata and metadata schemes. Catalog Classification Quart. 2005;40(3/4):17–36. [Google Scholar]
- 18.Gene Expression Omnibus. http://www.ncbi.nlm.nih.gov/geo/. Accessed March 23, 2015.
- 19.Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat Genet. 2001;29(4):365–371. [DOI] [PubMed] [Google Scholar]
- 20.Human Immunology Project Consortium. http://www.immuneprofiling.org/hipc/page/show. Accessed March 23, 2015.
- 21.Rocca-Serra P, Brandizi M, Maquire E, et al. ISA software suite: supporting standards-compliant experimental annotation and enabling curation at the community level. Bioinformatics. 2010;26(18):2354–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brusic V, Gottardo R, Kleinstein SH, et al. Computational resources for high-dimensional immune analysis from the Human Immunology Project Consortium. Nat Biotechnol. 2014;32(2):146–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattacharya S, Andorf S, Dunn P, et al. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. 2014;58(2-3):234–239. [DOI] [PubMed] [Google Scholar]
- 24.Cramer T, Kott K. Designing and implementing second generation digital preservation services: a scalable model for the Stanford Digital Repository. D-Lib Magazine. 2010;16(9–10). doi:10.1045/september2010-cramer. [Google Scholar]
- 25.Greenberg J. Metadata extraction and harvesting: a comparison of two automatic metadata generation applications. J Internet Comput. 2004;6(4):59–82. [Google Scholar]
- 26.Chao TC. Mapping methods metadata for research data. Int J Digit Curation. 2015;10(1):82–94. [Google Scholar]
- 27.Whetzel T, Noy NF, Shah NH, et al. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acid Res., 2011;39(Web server issue):W541–W545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edwards PN, Mayernik MS, Batcheller AL, et al. Science friction: data, metadata, and collaboration. Soc Stud Sci. 2011;41(5):667–690. [DOI] [PubMed] [Google Scholar]