Abstract
Modern biomedical data collection is generating exponentially more data in a multitude of formats. This flood of complex data poses significant opportunities to discover and understand the critical interplay among such diverse domains as genomics, proteomics, metabolomics, and phenomics, including imaging, biometrics, and clinical data. The Big Data for Discovery Science Center is taking an “-ome to home” approach to discover linkages between these disparate data sources by mining existing databases of proteomic and genomic data, brain images, and clinical assessments. In support of this work, the authors developed new technological capabilities that make it easy for researchers to manage, aggregate, manipulate, integrate, and model large amounts of distributed data. Guided by biological domain expertise, the Center’s computational resources and software will reveal relationships and patterns, aiding researchers in identifying biomarkers for the most confounding conditions and diseases, such as Parkinson’s and Alzheimer’s.
Keywords: big; biomedical; data; resource; discovery; science, neuroscience (ja); big data; analytics; BD2K; discovery science; Parkinson's disease; Alzheimer's disease (ID)
INTRODUCTION
The overarching goal of the Big Data for Discovery Science (BDDS) BD2K Center (www.bd2k.ini.usc.edu) is to enrich biomedical data by linking or integrating domains—and, in so doing, better understand how these data interact with each other. This integration of proteomic, genomic, phenomic, and clinical data will empower scientists to unearth new hypotheses and discover new insights. To achieve this goal, the BDDS Center is developing tools to accelerate the interactive integration and exploration of multi-omic and clinical “big data”, thus enabling scientists to extract and exploit knowledge that is currently trapped under layers of complexity. These tools are organized around 3 primary thrusts, focused on the management, manipulation and analyses, and modeling of big data, and will be delivered to the community via a new BDDS Portal.
While our Center will focus on areas of neuroscience, we expect this “-ome to home” approach will be applicable to other biomedical investigations outside the neurosciences. Ultimately, the BDDS system will connect diverse types of biomedical data—from molecular to behavioral—enabling the systematic analysis and discovery of new patterns and correlations that lead to actionable possibilities that can be used to improve health.
MANAGING BIG DATA
There is a wide disparity between the enormous potential of big data and their realization as practically usable resources for everyday use.1 Scientists currently spend much of their research time managing and aggregating data rather than doing science, with self-reported values of 90% being common.2 Furthermore, Nobel Laureate Oliver Smithies noted that experimental information “isn’t science until you make it available to others so that they can build on it.”3
The difficulties associated with integrating diverse data are exacerbated by the distributed nature of big data and the frequent use of ad hoc and even simplistic data organization and management methods. For example, data are often stored in file systems with metadata coded in file names or within file-specific formats.4,5 Thus, unified searching or organization around metadata values is difficult, requiring complex bespoke scripts and detailed knowledge of data layout on multiple storage systems.
We will address these obstacles to discovery by creating unified big-data digital asset management software6 that will systematically enable researchers to discover organize, search, integrate, and link data from diverse sources and types and from diverse locations, including the cloud, data repositories,7 and other big data resources (e.g., Storage Resource Manager (SRM)8, Intregrated rule oriented data system (iRODS),9 Modern Era Retrospective-Analysis for Research and Applications (MERRA)10).
What BDDS has done
We have validated one of our overall data management approaches by deploying a global effort in a BDDS-supported project. This powerful example, called Global Alzheimer's Association Interactive Network (GAAIN), provides access to a vast collection of Alzheimer's disease research data, sophisticated analytical tools, and computational resources. GAAIN separates the management of the metadata from the data itself, and represents an example of an asset-based approach to complex data management. The main outcome is a successful federated data repository that extends search and analytic capabilities so that researchers worldwide can find an ever-expanding collection of data coupled to a library of sophisticated tools to analyze and relate images, genetic information, and clinical and biological data. Figure 1 shows the cohort discovery interface of GAAIN called the GAAIN Scoreboard (www.gaain.org/explore).
Figure 1:
The Scoreboard provides a summary view of the data matching search criteria in the Global Alzheimer's Association Interactive Network (GAAIN) federated system. The far-left column shows each of GAAIN’s Data Partners. The top row shows the data attributes in GAAIN. A user can select any number of these data attributes to determine a collective total number of subject data available from GAAIN Data Partners.
What BDDS plans to do
Building on experiences integrating data from diverse sources, such as the Image Data Archive (IDA),11 the Global Alzheimer's Association Interactive Network (GAAIN)12 13 (see Figure 1), and Facebase,14 we will develop a system to deliver sophisticated and easy-to-use data management applications to anyone. This system will enable representation of configurable “entities” that store related metadata. It will allow files, directories, and other data storage mechanisms to be remotely indexed alongside arbitrary metadata extracted and/or supplied by users. Interactive tools will guide users through the process of incorporating new data, characterizing it with appropriate metadata; automatically extracting descriptive parameters, such as image quality or anatomical feature size; and establishing linkages with related data. Sophisticated search and navigation tools will allow users to identify and aggregate data into hypothesis-specific datasets by filtering data based on metadata, statistical characterizations, or other relevant properties. We will integrate diverse existing neuroimaging and genetics datasets into the platform to enable use of these tools for an initial investigation into creating new risk models for Parkinson’s disease.
We are developing application programming interfaces, a graphical user-focused knowledge discovery system, and interactive tools for data fusion, model-based metadata data integration, and searching across distributed data collections.15 Such techniques will enable dynamic search and navigation, curation of meta-data and promote semantic data understanding.
MANIPULATING AND ANALYZING BIG DATA
The transformation of big data into biomedical knowledge frequently requires big data transfers and large-scale computations, in which multi-component computational pipelines execute on many terabytes of distributed data and many hundreds or thousands of processors. More importantly, individual types of data need to be standardized and viewed in the context of biological domain expertise. Integrating 2 or more data types provides even richer possibilities for understanding biology and disease leading to the identification of dynamical biological networks. Understanding dynamical disease-perturbed networks can also provide fundamental insights into disease mechanisms, early diagnostic biomarkers and early therapeutic targets. Discovery science requires this type of useful data integration and it remains one of the grand challenges of systems biology.
What BDDS has done
BDDS investigators have applied Globus services16 to a range of biomedical domains, deploying and operating services that have been used to move and share imaging, genomic, and proteomic data. We have prototyped integration with analysis services and biomedical digital asset management system17 services to explore the ability to provide higher-level abstractions via a distributed data cloud. We have prototyped metadata extraction capabilities for common genomic and imaging formats such as Variant Call Format (VCF), Binary Version of Sequence Alignment File (BAM), Neuroimaging Technology Initiative (NIfTI), and Digital Imaging the Communications in Medicine (DICOM).18,19 These prototypes will form the basis of the distributed biomedical data cloud development undertaken in this thrust.
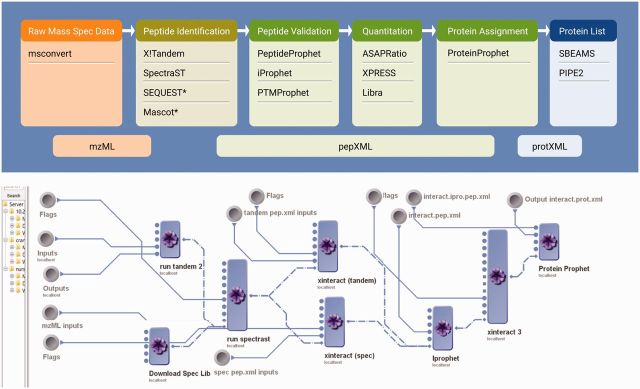
We are also working on data-fusion and resource interoperability methods that will enable the push-pull interactions required to drive and draw knowledge, expertise, and resources between omics data generated by genomics and proteomics researchers, tools developed by biomedical engineers, and cloud services supported by multiple organizations. Figure 2 illustrates an example of the Trans-Proteomic Pipeline20–22 implemented as platform-agnostic local, distributed, and cloud-based infrastructures. This pipeline highlights the ability to process distributed data via an optimized cloud-based analysis platform.
Figure 2:
Workflow representations of complex Trans-Proteomic Pipeline computational protocols implemented as platform-agnostic local, distributed, and cloud-based infrastructures.
What BDDS plans to do
BDDS will address additional big data manipulation issues by creating an adaptive and extensible distributed data access system to accommodate the fact that biomedical data is frequently large, heterogeneous, and distributed. We have and are developing platforms that allow us to standardize both genomic and proteomic data. These systems will support remote access to data located in a wide variety of locations and storage systems—thus creating a ‘distributed biomedical data cloud’ through which users will be able to seamlessly access their data and raise the level of abstraction employed when interacting with big data. Our extensible data access system will support metadata access as well as subsetting, feature extraction, and other operations on remote data. This system will also provide for rapid and reliable transfer of data to other locations when required—for example, for data integration or because a storage system does not support local analysis. Our system builds upon the successful Globus research data management services, which researchers worldwide have used to transfer over 77PB of data in more than 3 billion files.
To meet user computational needs, BDDS will create an adaptive and extensible workflow system to accommodate diverse data types, different data formats, varied sources of processing, and simplicity in operation. We have created a system that records each and every operation and step to provide comprehensive provenance needed for replication. The distinct processing steps needed to make data usable in the same model necessitate a flexible and open architecture. Our system builds upon the successful workflow architecture called the Laboratory of Neuro Imaging (LONI) Pipeline.23 Its intuitive interface and Extensible Markup Language (XML) descriptive language are well suited to such applications. Adapting this system to accommodate big data and the breadth of data domains is one focus of this thrust.
The BDDS Center will create and integrate biomedical data cloud services, including analytical tools, and provide transparent distributed data access. In subsequent years, the Center’s milestones will include developing remote subsetting and visualization capabilities, integration of HTTP access to distributed data, and remotely invoked BDDS metadata extraction methods. We will also develop approaches to synchronize metadata in the BDDS portal and enhance our data transformation capabilities.
MODELING BIG DATA
The first 2 BDDS thrusts focus on mechanisms that streamline and improve the effectiveness of the biomedical researcher. The third thrust is directed toward the creation of new modeling and analysis methods for knowledge extraction, capable of linking across diverse biomedical data types.
Analytic methods for big data form a clear bottleneck in many applications, both due to lack of scalability in the underlying algorithms and the complexity of the data to be analyzed. We will work to accelerate and optimize the entire end-to-end analysis process, from the development phase through to large-scale application. One explicit challenge is the realization that biological networks operate at all levels—genetic, molecular, cellular, organ, and even social networks. How do we capture these networks and then integrate them into a seamless whole that explains complex biological processes?
Big data modeling challenges already arise in the first stages of the analytic process, when a researcher is developing a new analysis. It is considered good practice to use “black box” testing that confirms expected transformations of exemplary inputs (“unit tests”). This practice can become impractical when dealing with big data, particularly as code units are executed in large numbers, on different systems, and requiring growing resource levels (disk space, memory, network connectivity). Thus, new methods are required to develop and evaluate modeling and analytical pipelines when applied to big data.
What BDDS has done
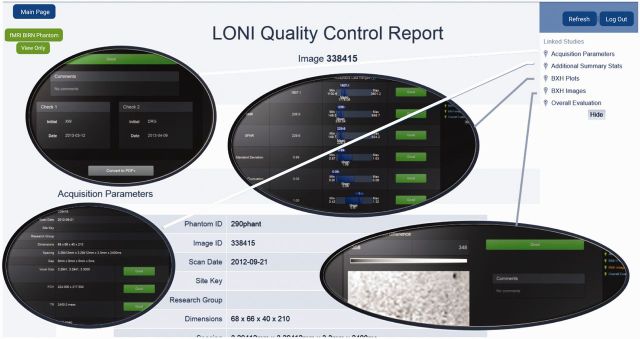
A tool to improve the quality control (QC) of big data is under development. Data analysis pipelines, for instance, often undergo development concurrently with the research that depends on them, and validating these pipelines is essential to ensure the integrity of research results. The Pipeline QC web-service (http://QC.loni.usc.edu) provides an illustrative example of modeling by providing a semi-automated QC system for multidimensional neuroimaging data. This service enables collaborators to initiate the automated QC processing pipeline protocols on the web, review the results (quantitative, qualitative, and graphical outputs), and assess the caliber of the structural, functional and diffusion volumes. As large-scale brain imaging studies involve multiple sites collecting, processing, and interpreting heterogeneous data, our QC system allows different (authorized) users to upload imaging data, run a standardized QC pipeline workflows, inspect the reported data characteristics, and annotate the properties of the metadata and imaging data (see Figure 3).
Figure 3:
LONI Quality Control (QC) system for high-throughput semi-supervised curation of multidimensional neuroimaging data.
What BDDS plans to do
To enable the formation of new, data-driven linkages between and among genomics, proteomics, phenomics, and clinical data and across spatial, temporal, and other scales, BDDS plans on coupling workflow technologies to modern computation resources. BDDS will develop methods for the joint processing of multi-modal data. We will adapt the LONI Pipeline to perform the proteomics, genomics, and image processing, as we describe in the following, along with experimental design logic.
The processing of proteomics data will be based on the widely used Trans-Proteomics Pipeline set of tools20–22 which include the PeptideProphet24 and ProteinProphet25 algorithms. The LONI Pipeline implementation will enable users to build complex workflows that perform analysis from raw instrument files through search engine processing, post-search validation, protein inference, and abundance quantification. The results may then be integrated with genomics and imaging data processed in the same LONI Pipeline environment.
The genomic data analysis will be based on our current analysis pipeline, incorporating further modules as they are developed. This pipeline includes components for detecting deletions and copy number variants by comparison to pre-computed Reference Coverage Profiles26; characterization of structural variation junctions; confidence filtering of observed variants based on error profiles pre-computed from thousands of whole genomes; annotation of observed variants for functional impact and for frequency in the populations27; computation of polygenic risk scores and comparison to the corresponding multigenome distributions. We have extensive experience developing systems for interactive, visual analysis of large-scale genomic sequences (the GESTALT Workbench,28 http://db.systemsbiology.net/gestalt/), integrative genomic resources (Kaviar,27), multi-genome analysis algorithms that improve the quality of interpretation of individual genomes26 and algorithms for interpretation of genomes in the context of family pedigrees and rare variant association29.
We will create a user-focused, knowledge discovery, graphical system for presenting data search results and interpreting data. One inherent challenge with big data is enabling researchers with deep expertise in one domain to grasp quickly the nuances important in another domain in which they are not expert. For instance, a geneticist reviewing proteomic data must be able to understand salient attributes revealed by an analysis of that data. BDDS will aid this crucial step by developing novel methods for extracting, understanding, and implementing actionable knowledge. In particular, BDDS will develop novel methods for data visualization and other tools to meet the data exploration, linkage, and modeling challenges faced by researchers.
BDDS is developing a post-hoc testing framework that can be applied to the entire analytical pipeline or to any subcomponent thereof. The framework automatically 1) learns the structure of the analysis outputs, 2) models distributions, and 3) identifies outliers. These outliers are then evaluated as either possible analytical failures, or as possible novel findings of interest.
BDDS will create new “pipelets” for each domain and connect to new data sources. In subsequent years, the Center will create full provenance capabilities to accommodate new data, develop modeling software to graphically change parameters and variables and test and validate capabilities on new disease models.
The ultimate objective for BDDS is to be able to convert data into knowledge. This requires the data management, integration, analysis, and modeling discussed above, posing fascinating challenges for the future.
FUNDING
This work was supported by National Institutes of Health grant 1U54EB020406-01.
COMPETING INTERESTS
The authors have no competing interests to declare.
CONTRIBUTORS
Each of the listed authors in this submission – Arthur W. Toga, Ian Foster, Carl Kesselman, Ravi Madduri, Kyle Chard, Eric W. Deutsch, Nathan D. Price, Gustavo Glusman, Benjamin D. Heavner, Ivo D. Dinov, Joseph Ames, John Van Horn, Roger Kramer and Leroy Hood – meet the authorship criteria identified by International Committee of Medical Journal Editors (ICMJE).
Substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data for the work; AND
Drafting the work or revising it critically for important intellectual content; AND
Final approval of the version to be published; AND
Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
REFERENCES
- 1.Van Horn JD, Toga AW. Human neuroimaging as a “Big Data” science. Brain Imaging Behav. 2014;8(2):323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howe B, Cole G, Souroush E, et al. Database-as-a-service for long-tail science. Proceedings of the 23rd International Conference on Scientific and Statistical Database Management. Portland, OR: Springer-Verlag; 2011:480–489. [Google Scholar]
- 3.Smithies O. Science brick by brick. Nature. 2010;467(7317): S6–S6. [DOI] [PubMed] [Google Scholar]
- 4.Foster I, Voeckler J, Wilde M, Zhao Y. Chimera: a virtual data system for representing, querying, and automating data derivation. 14th International Conference on Scientific and Statistical Database Management. Edinburgh, Scotland; 2002. [Google Scholar]
- 5.Stef-Praun T, Clifford B, Foster I, Hasson U, Hategan M, Small SL, Wilde M, Zhao Y. Accelerating medical research using the swift workflow system. Stud Health Technol Inform. 2007;126:207–216. [PMC free article] [PubMed] [Google Scholar]
- 6.Schuler RE, Kesselman C, Czajkowski K. Digital asset management for heterogeneous biomedical data in an era of data-intensive science. Bioinformatics and Biomedicine (BIBM), 2014 IEEE International Conference on, IEEE, 2 Nov–5 Nov 2014, Belfast, United Kingdom. 2014. [Google Scholar]
- 7.Crawford KL, Toga AW. The Alzheimer's Disease Neuroimaging Initiative Informatics Core: A Decade in Review. Alzheimer's & Dementia. 2015. (In Press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shoshani A, Sim A, Gu J. Storage resource managers: Middleware components for grid storage. NASA Conference Publication. 2002;209–224. [Google Scholar]
- 9.Rajasekar A, Moore R, Hou C-Y, et al. iRODS Primer: integrated rule-oriented data system. Synthesis Lectures on Information Concepts, Retrieval, and Services. 2010;2(1):1–143. [Google Scholar]
- 10.Schnase JL, Duffy DQ, Tamkin GS, et al. MERRA analytic services: meeting the big data challenges of climate science through cloud-enabled climate analytics-as-a-service. Comput, Environ Urban Sys. 2014. (In Press). [Google Scholar]
- 11.Neu SC, Crawford KL, Toga AW. Practical management of heterogeneous neuroimaging metadata by global neuroimaging data repositories. Front Neuroinform. 2012;6(8):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toga AW, Bhatt P, Ashish N. Data sharing in Alzheimer's disease research. Alzheimer's Disease and Associated Disorders. 2015. (In press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toga AW, Neu SC, Bhatt P, Crawford KL, Ashish N. The Global Alzheimer’s Association Interactive Network. Alzheimer's & Dementia. 2015. (In Press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marazita ML, Hochheiser H, Murray JC. The FaceBase Hub: a resource for translational craniofacial genetics. Am J Med Genet Part A. Hoboken, NJ: Wiley-Blackwell; 2014. [Google Scholar]
- 15.Van Horn JD, Toga AW. Multisite neuroimaging trials. Curr Opin Neurol. 2009;22(4):370–378, pmc2777976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster I. Globus online: accelerating and democratizing science through cloud-based services. IEEE Internet Computing 2011;15(3):70–73. [Google Scholar]
- 17.Schuler RE, Kesselman C, Czajkowski K. An asset management approach to continuous integration of heterogeneous biomedical data. Data Integration in the Life Sciences. Switzerland, Springer; 2014. [Google Scholar]
- 18.Whitcher B, Schmid VJ, Thornton A. Working with the DICOM and NIfTI Data Standards in R. J Stat Softw. 2011;44(6):1–28. [Google Scholar]
- 19.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28(4):464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deutsch EW, Mendoza L, Shteynberg D, et al. A guided tour of the trans-proteomic pipeline. Proteomics. 2010;10(6):1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005: 1:2005.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slagel J, Mendoza L, Shteynberg D, Deutsch EW, Moritz RL. Processing shotgun proteomics data on the Amazon Cloud with the Trans-Proteomic Pipeline. Mol Cell Proteomics. 2014;14(2):399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinov ID, Lozev KM, Petrosyan P, et al. Neuroimaging study designs, computational analyses and data provenance using the LONI Pipeline. PLoS ONE. 2010;5(9):e13070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. [DOI] [PubMed] [Google Scholar]
- 25.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical Chem. 2003;75(17):4646–4658. [DOI] [PubMed] [Google Scholar]
- 26.Glusman G, Severson A, Dhankani V, et al. Identification of copy number variants in whole-genome data using Reference Coverage Profiles. FrontGenet. 2015;6:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glusman G, Caballero J, Mauldin DE, Hood L, Roach JC. “Kaviar: an accessible system for testing SNV novelty. Bioinformatics. 2011;27(22):3216–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glusman G, Lancet D. GESTALT: a workbench for automatic integration and visualization of large-scale genomic sequence analyses. Bioinformatics. 2000;16(5):482–483. [DOI] [PubMed] [Google Scholar]
- 29.Roach JC, Glusman G, Smit AFA, et al. Analysis of Genetic Inheritance in a Family Quartet by Whole Genome Sequencing. Science. 2010;328(5978):636–639. [DOI] [PMC free article] [PubMed] [Google Scholar]