Abstract
Objective To develop an open-source temporal relation discovery system for the clinical domain. The system is capable of automatically inferring temporal relations between events and time expressions using a multilayered modeling strategy. It can operate at different levels of granularity—from rough temporality expressed as event relations to the document creation time (DCT) to temporal containment to fine-grained classic Allen-style relations.
Materials and Methods We evaluated our systems on 2 clinical corpora. One is a subset of the Temporal Histories of Your Medical Events (THYME) corpus, which was used in SemEval 2015 Task 6: Clinical TempEval. The other is the 2012 Informatics for Integrating Biology and the Bedside (i2b2) challenge corpus. We designed multiple supervised machine learning models to compute the DCT relation and within-sentence temporal relations. For the i2b2 data, we also developed models and rule-based methods to recognize cross-sentence temporal relations. We used the official evaluation scripts of both challenges to make our results comparable with results of other participating systems. In addition, we conducted a feature ablation study to find out the contribution of various features to the system’s performance.
Results Our system achieved state-of-the-art performance on the Clinical TempEval corpus and was on par with the best systems on the i2b2 2012 corpus. Particularly, on the Clinical TempEval corpus, our system established a new F1 score benchmark, statistically significant as compared to the baseline and the best participating system.
Conclusion Presented here is the first open-source clinical temporal relation discovery system. It was built using a multilayered temporal modeling strategy and achieved top performance in 2 major shared tasks.
Keywords: natural language processing, electronic medical record, temporal relation discovery, document creation time, narrative container, Allen's temporal interval relations
BACKGROUND AND SIGNIFICANCE
Temporality is crucial for a deeper understanding of the course of clinical events in a patient’s electronic medical records.1,2 A large part of it is recorded in the electronic medical records free text. Automatic temporal relation discovery has the potential to dramatically increase the understanding of many medical phenomena such as disease progression, longitudinal effects of medications, and a patient’s clinical course. Extraction and interpretation of temporal relations has many clinical applications such as question answering,3,4 clinical outcomes prediction,5 and the recognition of temporal patterns and timelines.6
Despite the research progress and established temporal corpora and shared tasks in the general domain,7–11 active work in comprehensive temporal relation discovery from clinical free text has emerged only in the last 5 to 6 years. Given that a clinical note is written by physicians who have very limited time to express the details of the patient-physician encounter, nonstandard expressions, abbreviations, assumptions, and domain knowledge are used, which make the text hard to understand outside of the medical community, let alone by automated systems. The signals that are needed for extracting the temporal information embedded in the clinical free text are thus both domain-specific and complex.
After pilot clinical temporal annotations12,13 showed initial success, the 2012 Informatics for Integrating Biology and the Bedside (i2b2) Challenge established the first clinical corpus annotated with temporal information. Eighteen teams participated in the challenge through the development of their temporal relation discovery systems.14 However, the 2012 i2b2 challenge focused on select pairwise relations between clinical events and time expressions (described in the Methods section below). In other work, Pustejovsky and Stubbs15 proposed the concept of a narrative container that addresses temporal granularity and can significantly reduce the complexity of annotation and temporal reasoning. The Temporal Histories of Your Medical Events (THYME) corpus16,17 was then created and annotated with the incorporation of the narrative container concept and was adopted by SemEval 2015 Task 6: Clinical TempEval shared task as the official corpus.18
In this paper, we present our multilayered temporal modeling approach and methods for its automatic tagging. “Multilayered,” in this context, refers to the varied layers of granularity of temporal relations. We tested our approach against 2 publicly available corpora: the 2012 i2b2 challenge and the SemEval 2015 Task 6 shared task. Our work is a significant contribution over past reported results14,19 in that it presents a multilayered model of temporality (from rough to fine-grained temporal relations), thus capturing the nature of temporal granularity.
MATERIALS AND METHODS
Multilayered temporal model
Conventional temporal relation discovery includes the identification of events, time expressions, and temporal relations. In the clinical domain, events usually describe diseases/disorders, signs/symptoms, procedures, medications, and laboratory values, as well as general events such as a discussion of potential medication side effects, planning of a procedure, etc. Some examples of time expressions are tomorrow, postoperative, and Nov-11-2011. Typical temporal relations are those defined by the Allen set20—Before, Meets, Overlaps, Starts, During, Finishes, Is_Equal_To—and typical approaches attempt to assign temporal relations between all pairs of events and time expressions.
The verbose discovery of temporal relations over every possible pair is redundant, tedious, and inefficient because many temporal relations could be easily inferred. Take for example the following scenarios:
(Scenario 1) if concept A is before concept B and B is before concept C, then A is before C;
(Scenario 2) if a group of concepts, denoted by GA, is before another group of concepts, GB, then every concept temporally contained in GA is before every concept temporally contained in GB.
In order to efficiently identify the minimal linguistically derivable temporal relation set and maintain the maximal inferentially derivable relations, we devise a multilayered temporal model which is informed by the Allen set but is also a departure from it.
At the most coarse level, we link each clinical event to the document creation time (DCT). We call this relation Document Time Relation (DocTimeRel), with possible values such as Before, After, Overlap, and Before_Overlap. The difference between Before_overlap and Overlap is that the former emphasizes an event that started before DCT and is continuing through the present, often expressed in English in the present perfect tense. For example, in “He has had a [fever],” fever has a DocTimeRel of Before_overlap; while in “He has a [fever],” fever has a DocTimeRel of Overlap. By using DocTimeRel, events can be grouped into coarse temporal buckets.
At the intermediate level, we model a special type of concept named Narrative Container (NC).15 Narrative Containers are concepts that are central to the discourse and temporally contain 1 or more other concepts as, for example, within the following clinical text:
(Example 1) The patient had a fever during his recovery of his initial surgery on December 17th to remove the adenocarcinoma.
In this example, there are 2 NCs: (1) recovery, which contains fever; and (2) December 17th, which includes the events of surgery and remove. The advantage of introducing the NC concept is that NC matches the structural reality of narratives. Usually, clinicians cluster their discussions of clinical events around a given time. Therefore, it is often easy and natural to place events into containers and use a few relations to link the containers. Many detailed relations could be derived through posthoc inference, which simplifies both annotation and downstream learning tasks.15,21 Note that concepts serving as NCs are not explicitly annotated as containers per se—it is the event or time expression that temporally contains other events or times that functions as the conceptual NC. A NC is thus a central hub of multiple Contains relations.
At the most granular level of our multilayered temporal model, we represent a subset of the classic Allen relations—Before, Overlap, Begins-On, and Ends-On.
Corpora
In our current work, we used 2 publicly available data sets of clinical notes: (1) the THYME corpus (sets 1–200), which was used in SemEval 2015 Task 6: Clinical TempEval,16–18,22,23 and (2) the 2012 i2b2 challenge14 data set.
This subset of the THYME corpus contains 200 colon cancer patients with 3 notes per patient—1 oncology, 1 pathology, and 1 treatment note per patient. Of all 600 (200 × 3) notes, only 440 were annotated. These 440 notes were used by the Clinical TempEval and our work, with 293 for training and 147 for testing. The gold standard annotations contain DocTimeRel for events and temporal relations of type Before, Overlap, Begins-On, Ends-On, and Contains (representing the narrative container). In addition to clinical events and unlike the i2b2 corpus, the THYME corpus annotates general events as well (eg, discuss in We discussed alternative treatments).
The 2012 i2b2 challenge corpus14 consists of 310 discharge summaries—190 summaries for training and 120 for testing. Within each document, 2 types of temporal relations are annotated: (1) event-section time, which link every event from the patient history section to the admission date and every event from the hospital course section to the discharge date; and (2) the other relation links events/times either from the same sentence or from different sentences using Before, After, and Overlap relations. One peculiarity of the i2b2 corpus is that many cross-sentence Overlap relations are between events that are coreferential.24
Methods for coarse-level temporality (DocTimeRel)
In the general domain, the state-of-the-art results for DocTimeRel discovery are around 0.8 F1.9,10,25 In our current study, we investigate a DocTimeRel model for the clinical domain. The DocTimeRel labels in the THYME corpus are Before, After, Overlap, and Before_overlap. The DocTimeRel labels in the i2b2 data set are Before, After, Overlap.
We developed a supervised approach—multiclass support vector machine (SVM)26—for DocTimeRel automatic tagging. The instance for classification was every event, represented by a group of features (described in a later section). Support vector machine models were trained on the data with gold DocTimeRel labels. Model performance was evaluated on the testing data.
Each THYME document was stamped with its creation time that we used as the DCT. Each i2b2 document had 2 section times that could be viewed as variants of the DCT: one was the admission date and the other was the discharge date. The same DocTimeRel model—similar features (for the i2b2 data, the Section ID feature was replaced by event position feature, see online supplement) and the same algorithm—was used to train 2 separate classifiers for recognizing temporal relations between events and the admission time and relations between events and the discharge time.
Methods for both medium-grained level and fine-grained level temporality
Due to the different properties of event-time and event-event relations,27 we trained 2 multiclass SVM classifiers for recognizing temporal relations within the same sentence: (1) a classifier for relations between 2 events and (2) a classifier for relations between a time expression and a clinical event.
We generated all gold event-event and event-time pairs within the same sentence as candidates for event-event and event-time classifiers, respectively. As a result, a large number of negative training instances was generated that would cause class imbalance issues for classification. Some previous systems28 applied heuristics to focus on the classification on “likely” candidates. We instead applied cost-sensitive learning. In order to counterbalance the effect of the dominating classes, the weight for each class wi was adjusted inversely proportionally to class frequencies. As a result, the penalty factor C in SVM training26 was adjusted to C×wi for each class i.29 For example, if Overlap, Before, NULL categories had instances of 100, 10, 1000, respectively, the weights for these 3 classes would be 1, 10, 0.1. Thus, the majority class of NULL would not have a dominating effect and the minority classes would have balanced predictions. Even though there were not many training instances of minority classes, like “BEGINS-ON” or “ENDS-ON,” the model could still predict testing instances as minority classes.
A large number of implicit pairs were intentionally left unlabeled in the gold standard because their relations could be inferred. However, during training, the classifier cannot distinguish between implicit relations and nonrelations and this may harm learning. Therefore, we expanded the gold relations by calculating the closure sets of all possible relations in a clinical document. For instance, in the sentence—
(Example 2) In 2004 the patient was diagnosed with ascending colon cancer.
—2004, diagnosed, and cancer are the gold time expression and events. The gold standard will only mark Contains (2004, diagnosed) and Contains (diagnosed, cancer), while 2004 and cancer are left unlinked. Through closure, we get Contains (2004, cancer), which is added to the training set. For event-event relations, closure usually generates 55% more instances. For event-time relations, closure usually generates 82% more instances.
The i2b2 corpus and the THYME corpus take different approaches to finding the span of text for an event. The i2b2 corpus annotates the full noun phrase describing an event, while the THYME corpus annotates only the headword. So in Example 2, i2b2 annotators would annotate ascending colon cancer as the event, while THYME annotators would annotate only cancer as the event.
From a computational perspective, both approaches have their merits. Including full phrases means the model can learn patterns for very specific events, while including only headwords means the model can better generalize across similar events. To gain the advantages of both approaches, we propose a technique to expand gold events to enrich the training set with more temporal relations. We look for all possible Unified Medical Language System (UMLS30) entities whose spans overlap gold standard events. For example, in Example 2—“In 2004 the patient was diagnosed with ascending colon cancer”—there are 2 annotated events, “diagnosed” and “cancer.” Clinical Text Analysis and Knowledge Extraction System (cTAKES), through UMLS dictionary lookup, identifies 3 UMLS entities: “ascending colon cancer,” “colon cancer,” and “cancer.” Their spans all overlap with the gold event “cancer.” In the THYME data, we find UMLS entities covering gold headword events, while in the i2b2 data, we find UMLS entities covered by gold phrasal events. Gold events that do not have associated UMLS entities, such as “diagnosed” in example 2, are general domain events. Otherwise, gold events with associated UMLS entities, such as “cancer” in example 2, are clinical events.31 For each UMLS entity overlapping a clinical event, and for any temporal relation that the event participates in, we generate a new temporal relation that is identical to the original relation but with the event replaced by the UMLS entity. For Example 2, we get the following additional relations after event expansion:
Contains (2004, cancer)
Contains (2004, colon cancer)
Contains (2004, ascending colon cancer)
Contains (diagnosed, cancer)
Contains (diagnosed, colon cancer)
Contains (diagnosed, ascending colon cancer)
We only generated additional relations through the event-expansion technique on the training data, not on the test data. This event-expansion technique has been validated in the development sets, and we will describe its effect and usability in greater detail in a separate paper.
Classifier features and learning algorithm
Once the training pairs are generated, we represent each pair using a pool of features with reference to the top performing i2b2 temporal systems and general domain relation extraction literature.21,24,27,28,32 For different tasks (DocTimeRel, event-time, and event-event), we start with all top performing feature groups as reported in the literature and use ablation test to discard unnecessary groups. Based on ablation experiments, we describe the most discriminative features for each type of relation in Table 1. The description of the complete feature set used by each relation model is listed in our supplementary material; those additional features are motivated by the literature21,24,27,28,32 and are easy to extract as all the necessary preprocessing is conducted within cTAKES by the preceding modules. In order to maintain a clean open-source system, we removed perplexing features we have tried, such as constituency tree features and dependency tree features.
Table 1.
Core features per type of relation
| Feature | Description | DocTimeRel | Event-Event Relations | Event-Time Relations |
|---|---|---|---|---|
| Tokens | The first and the last word of each concept, all words covered by a concept as a bag, bag-of-words around each concept for a window of [−3, 3], bag-of-words between 2 concepts, and the number of words between 2 concepts (for the THYME corpus, the headword event is expanded to the immediately enclosing NP and the NP becomes the anchor for the token features) | ✓ | ✓ | ✓ |
| Part-of-speech tags | The Penn Treebank POS tags of each concept as a bag | ✓ | ||
| Event attributes | All event-related attributes such as polarity, modality, and type. Note that DocTimeRel is also an event attribute, and is used for reasoning on the within-sentence relations. | ✓ | ✓ | ✓ |
| UMLS feature | UMLS semantic types of each concept as features | ✓ | ✓ | |
| Dependency path | The dependency path between 2 concepts and the number of dependency nodes in-between | ✓ | ||
| Overlapped head | If 2 concepts share the same headword | ✓ | ||
| Temporal attributes | The class type of a time expression, eg, “Date,” “Time,” “Duration,” etc. | ✓ | ||
| Special words | Any special words from the time lexicon developed by the NRCC24 that the concepts or the context in-between contain | ✓ | ||
| Nearest flag | If the event-time pair in question is the closest among all pairs in the same sentence | ✓ | ||
| Conjunction feature | If there is any conjunction word between the arguments | ✓ | ||
| Nearby verb’s part-of-speech tag | The Penn Treebank POS tags of the verbs within the same sentence | ✓ | ||
| Section ID | The header of the section containing the target concept | ✓ | ||
| Closest verb | The tokens and Penn Treebank POS tags of the closest verb to the target concept within the same sentence | ✓ | ||
| TimeX feature | The tokens and attributes of the closest time expression in the same sentence | ✓ |
Abbreviations: DocTimeRel, document time relation; NP, Noun Phrase; THYME, temporal histories of your medical events; POS, part-of-speech; UMLS, unified medical language system; NRCC, National Research Council Canada; ID, Identifier.
All features are extracted using Apache cTAKES (The Apache Software Foundation, Forest Hill, Maryland).33,34
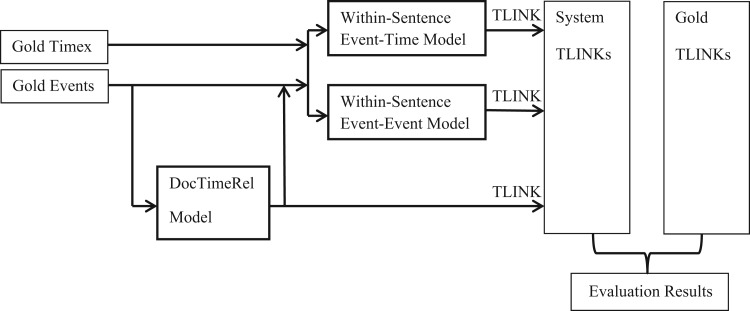
We use the L2-regularized L2-loss dual SVM as implemented by LIBLINEAR26 as the main learning algorithm. We use L2-based regularization because we want all features to play a role in decision making. L1 regularization and explicit feature selection mute features and would decrease the performance in our experiments. The whole processing flow (including DocTimeRel, event-time, and event-event models) is illustrated in Figure 1.
Figure 1:
The cTAKES-Temporal System architecture (predictions of the “coarse” level model, DocTimeRel, are used as features for the fine-grained models).
I2b2-specific learners
The 2012 i2b2 challenge data set annotates a relatively large amount of cross-sentence temporal relations. Therefore, we devised 3 additional cross-sentence learners. The first one is a cross-sentence event-event learner that pairs up main events in consecutive sentences. Main events are defined as the first and last event within a sentence.28 The learner uses the same feature set as the within-sentence event-event model and SVM as the algorithm. The second learner is a cross-sentence event-time classifier that pairs up every event and time expression in consecutive sentences. It uses the same feature set as the within-sentence event-time model and SVM as the algorithm. The third learner implements rules for directly linking coreferenced events as Overlap. Coreferenced pairs include the following: (1) 2 time expressions sharing the exact same tokens, eg, 2 mentions of “this morning” in adjacent sentence are considered coreferent; (2) event pairs that share the same headwords; and (3) we also created rules to link mentions of “admission” and “admission date,” mentions of “admission” and “admitted,” and mentions of “discharge” and “discharge date.”
Evaluation
The F1 score was used as the primary evaluation metric (the standard in the domain). Precision is calculated as the percentage of system links that can be verified in the transitive closure of the gold standard links (ie, closure is only computed on the gold standard links). Recall is calculated as the percentage of gold standard links that can be found in the transitive closure of the system links (ie, closure is only computed on the system links).35 The final F1 score was calculated on the transitive-closure-processed precision and recall as both i2b2 challenge 2012 and Clinical TempEval took such closure-enhanced setup.14 This modified F1 calculation is useful for evaluating temporal relations as they are difficult to exhaustively annotate, and this metric reduces the penalty for system annotation of annotator-missed relations. We used the official i2b2 2012 and Clinical TempEval evaluation scripts for evaluating our system so that our results would be directly comparable to the outcomes of the i2b2 2012 challenge and Clinical TempEval. Please note that the Clinical TempEval evaluation script takes cross-sentence relations into consideration, while our system annotates only within-sentence relations on THYME data. Clinical TempEval included only DocTimeRel and Contains.
RESULTS
Table 2 presents the performance of our temporal system on both the 2012 i2b2 challenge test set and Clinical TempEval test set. Please note that we did not participate in either of the shared tasks and thus enjoyed several advantages that the task participants did not have—we had access to the data and were not under significant time pressure to develop our system. For the i2b2 challenge (top 3 rows), our system was compared to the top 2 performing systems in that challenge, Vanderbilt University and the National Research Council Canada.24,28 Our overall system had the best recall, a better balance between recall and precision, and was on par with the best systems of the i2b2 challenge. For Clinical TempEval (bottom 3 rows), our system was compared to the best participating system (“BluLab”) and the baseline system that links a time expression to the closest event.18,23 Our system had the best F1 score. Using a similar document-by-document comparison,24 F1 scores of our system were significantly higher than both the baseline and “BluLab” (Wilcoxon signed rank test36; P < .05 ).
Table 2.
Comparison of system results for the temporal relation track of the 2012 i2b2 challenge and Clinical TempEval
| F1 Score | Precision | Recall | Method Summary | Challenges | ||
|---|---|---|---|---|---|---|
| Overall evaluation for all components | ||||||
| cTAKES-Temporal System | 0.695 | 0.697 | 0.693 | SVM + rules for coreferent pairs | 2012 i2b2 | |
| Vanderbilt University | 0.693 | 0.714 | 0.673 | Heuristic candidate generation + CRF + SVM | ||
| National Research Council Canada | 0.692 | 0.750 | 0.643 | MaxEnt + SVM + rule based | ||
| DocTimeRel F1 score | Overall scores for both event-time and event-event | |||||
| THYME System | 0.807 | 0.321 | 0.526 | 0.231 | SVM | 2015 Clinical TempEval (CONTAINS only) |
| Best participating System | 0.791 | 0.181 | 0.140 | 0.254 | Will be described in an upcoming publication by the BluTeam | |
| Baseline | NA | 0.260 | 0.554 | 0.170 | TIMEX to closest Event | |
Abbreviations: cTAKES, clinical text analysis and knowledge extraction system; SVM, support vector machine; CRF, Conditional Random Fields; MaxEnt, Maximum Entropy classifier; DocTimeRel, document time relation; THYME, temporal histories of your medical events; NA, not available. The best scores are marked bold.
We also wrote our own evaluation script that calculates the same closure-enhanced recall, precision, and F1 score but removes cross-sentence relations from the gold standard and splits apart the evaluation of event-event and event-time relations so that we may have a better sense of our system performance given only within-sentence relations. Table 3 shows our DocTimeRel, event-event, and event-time results on THYME test sets using our evaluation script.
Table 3.
Within-sentence event-time and event-event models results on SemEval 2015 THYME test set
| Precision | Recall | F1 Score | No. of Gold Instances | Relation Type | Model |
|---|---|---|---|---|---|
| 0.834 | 0.834 | 0.834 | 19234 | OVERALL | DocTimeRel (all features) |
| 0.816 | 0.773 | 0.794 | 1986 | AFTER | |
| 0.850 | 0.830 | 0.840 | 7231 | BEFORE | |
| 0.740 | 0.469 | 0.574 | 1007 | BEFORE/OVERLAP | |
| 0.832 | 0.891 | 0.861 | 9010 | OVERLAP | |
| 0.677 | 0.679 | 0.678 | 2213 | OVERALL | Event-Time (all features) |
| 0.39 | 0.299 | 0.338 | 67 | BEFORE | |
| 0.563 | 0.138 | 0.222 | 65 | BEGINS-ON | |
| 0.715 | 0.784 | 0.748 | 1803 | CONTAINS | |
| 0.625 | 0.278 | 0.385 | 54 | ENDS-ON | |
| 0.400 | 0.205 | 0.271 | 224 | OVERLAP | |
| 0.470 | 0.306 | 0.371 | 4632 | OVERALL | Event-Event (all features) |
| 0.375 | 0.241 | 0.293 | 758 | BEFORE | |
| 0.333 | 0.074 | 0.121 | 202 | BEGINS-ON | |
| 0.493 | 0.510 | 0.501 | 2378 | CONTAINS | |
| 0.300 | 0.092 | 0.141 | 65 | ENDS-ON | |
| 1 | 0.002 | 0.004 | 1229 | OVERLAP |
Abbreviation: DocTimeRel, document time relation.
We further split the training set of the THYME data (75/25), thereby creating a development set. We trained on the training split and tested on the development set for ablation tests. Each time a feature group was removed from the whole feature sets to test its contribution. Table 4 is the ablation study results on the development set (for a detailed feature description, see supplementary material). We did a feature-ablation study for only the CONTAINS even-event cases as the low system performance on other types could distort the results.
Table 4.
Ablation test for major features on SemEval 2015 THYME development seta
| Precision | Recall | F Score | ΔF Score | Model | |
|---|---|---|---|---|---|
| All | 0.633 | 0.657 | 0.645 | Event-Time | |
| Remove Tokens | 0.608 | 0.577 | 0.592 | −0.053 | |
| Remove Dependency Path | 0.615 | 0.627 | 0.621 | −0.024 | |
| Remove Temporal Attributes | 0.622 | 0.643 | 0.632 | −0.012 | |
| Remove Event Attributes | 0.658 | 0.612 | 0.634 | −0.011 | |
| Remove Nearest Flag | 0.621 | 0.654 | 0.637 | −0.008 | |
| Remove Conjunction feature | 0.632 | 0.649 | 0.640 | −0.004 | |
| Remove Special Words | 0.626 | 0.658 | 0.642 | −0.003 | |
| All | 0.830 | 0.830 | 0.830 | DocTimeRel | |
| Remove Tokens | 0.771 | 0.771 | 0.771 | −0.059 | |
| Remove Section ID | 0.802 | 0.802 | 0.802 | −0.028 | |
| Remove TimeX feature | 0.825 | 0.825 | 0.825 | −0.005 | |
| Remove Event Attributes | 0.825 | 0.825 | 0.825 | −0.005 | |
| Remove Closest Verb | 0.826 | 0.826 | 0.826 | −0.004 | |
| Remove Nearby Verbs' POS tags | 0.828 | 0.828 | 0.828 | −0.002 | |
| Remove UMLS features | 0.828 | 0.828 | 0.828 | −0.002 | |
| All | 0.658 | 0.576 | 0.614 | Event-Event CONTAINS | |
| Remove Tokens | 0.501 | 0.469 | 0.484 | −0.130 | |
| Remove Event Attributes | 0.647 | 0.562 | 0.602 | −0.013 | |
| Remove UMLS features | 0.65 | 0.572 | 0.609 | −0.006 | |
| Remove Part-of-speech tags | 0.668 | 0.561 | 0.610 | −0.004 | |
| Remove Dependency Path | 0.649 | 0.58 | 0.613 | −0.002 | |
| Remove Overlapped head | 0.662 | 0.572 | 0.614 | −0.001 |
Abbreviations: DocTimeRel, document time relation; ID, Identifier; UMLS, unified medical language system.
aEach result indicates the removal of a feature from the All Feature set. Features are sorted by the F score, with the most important feature at the top.
DISCUSSION
The temporal relation discovery system we present in this paper is part of the open-source Apache cTAKES33,34-temporal module, version 3.2.1 (the newest models are in the release of 3.2.2). The Apache cTAKES-temporal module is an end-to-end temporal solution that includes event and time expression detection, both with around 0.85 accuracy. We believe that our released system is the first open-source, end-to-end temporal system in the clinical domain with a state-of-the-art performance.
We designed a multilayered temporal relation discovery scheme from the most coarse level (DocTimeRel) to the intermediate level (narrative containers as marked by Contains relations) to the most granular relations. Table 3 shows that our models have good performance on both coarse and intermediate level relations (DocTimeRel F1 = 0.834; event-time Contains F1 = 0.748; event-event Contains F1 = 0.501), outperforming the state-of-the-art and establishing a new benchmark. These 2 coarse and intermediate models provide rough event temporality, thus achieving a “macro” success in temporal relation discovery. Even with the most granular microlevel relations, our system is still on par with the best i2b2 2012 challenge participants (Table 2). We believe the relative low relation discovery rate for micro relations is due to insufficient training instances (notice in Table 3 some categories such as Begins-On and Ends-On have only tens of instances). With more training examples becoming available in the future, we expect to see improved performance on these relations.
One may wonder if we can develop the models on a combined i2b2 and THYME corpora to improve the discovery rate for micro relations. We would like to point out that this is not possible because of the following: (1) i2b2 and the THYME corpus have different labels. The i2b2 labels are “BEFORE,” “AFTER,” and “OVERLAP,” while in THYME we have additional “BEGINS-ON,” “ENDS-ON,” and especially “CONTAINS” relations but no “AFTER” relations. Joint training would be hard due to the conflicts between the 2 labeling systems. (2) For the i2b2 data, an event is marked by a phrase; while for the THYME data, an event is marked by its headword. The different annotation strategies would affect how we extract contextual features as well as how we would apply event-expansion techniques.
Despite the heterogeneity of the 2 corpora, we did try to develop a generic approach that could work with both corpora without much customization. We applied the same classifiers for within sentence event-time and event-event classifications, similar classifiers for DocTimeRel and event-section time. The only difference was that for the i2b2 data, there were 2 additional classifiers for cross-sentence relations. The feature sets used for the 2 systems were almost the same with the exception of some changes to accommodate the differences between the 2 corpora.
In the meantime, we noted that coarse temporality could be sufficient to do meaningful inference, which could be then applied to biomedical use cases. In a methotrexate-induced liver toxicity study, we successfully identified rheumatoid arthritis patients who took methotrexate within the 3 months before their liver function abnormality37 with the help of DocTimeRel information. Through an extrinsic evaluation of clinical question-answering problems, it was also interesting to note that a large amount of time-sensitive clinical questions could be answered by coarse temporal relations like DocTimeRel (a separate in-progress publication). Conceptually, if we could put clinical concepts into coarse temporal bins correctly, the system-discovered temporality would not be too far from the reality even if the finer-grained local relations were incorrect. These inconsistencies could potentially be managed by an intelligent global inference scheme, taking into account classifier accuracy and confidence at different granularities to obtain the highest probability patient timeline.
The event-event relation discovery was more difficult than event-time relation discovery (0.371 vs 0.678 in F score, Table 3). Of an 81.6% chance, there was 1 time expression within a sentence. It was thus easier to reason many-to-one links between event-time than to reason the many-to-many links between event-event. We also experimented with limiting our event-event temporal relation discovery to medical events only, removing all general events from consideration. The result with that setup on the THYME corpus was a 0.531 F score, which was more comparable with the event-time result.
Table 4 shows the contribution of the most important features (Table 1, the core features) for each relation that can work well for both data sets. For each learner, the most discriminative features are usually 6 or 7 core features (Table 1). They are basic features describing the context of the target concepts (eg, token features), the attributes of the concepts, and syntactico-semantic relationships (such as dependencies) between a pair of concepts.
Table 4 shows that the most contributing feature for all 3 types of relation classifiers is the token feature. This is very different from the general domain, where more generalizable linguistic features such as the part-of-speech tags are more commonly beneficial.9–11 This phenomenon could be explained by the nature of clinical narratives which do not conform to the formal grammar and standard structures of the general-domain texts.38 Such characteristics require clinical natural language processing systems focused on that type of text.6,12 Therefore, token features are more useful in this sense, with the cost of less generalizability. In the future, we plan to explore methods for making the clinical-domain lexical features more generalizable. Using word embeddings39 trained on the clinical domain is one possible approach.
We also ran a feature ablation test on the event-time model of the i2b2 data. Results are shown in Supplementary Table 2. We observed a similar feature contribution pattern, except for the temporal attribute features. One possible reason is there is a new TIMEX3 type “PREPOSEXP" in the THYME data that covers terms like “preoperative," “postoperative," and “intraoperative.”31 This special type was not annotated in the i2b2 data and could be very informative in reasoning about event-time relationships.
Our error analysis shows that the major errors that are general to event-event, event-time, and DocTimeRel relations are the following: (1) Annotation errors or inconsistent annotations. Annotation errors would be when the same event or relation was labeled multiple times with different labels. Inconsistent annotations would be when the same or similar relation was labeled differently on different occasions. They would both end up with very similar feature vectors with different labels, which would confuse the models. (2) Conflicting, misleading, or missed cues. There could be competing/misleading evidence. For example, the preposition “in” usually suggests “OVERLAP,” while a nearby verb “will” is suggesting “AFTER.” In “History of severe COPD," “History” suggests “BEFORE,” while COPD is chronic and suggests “OVERLAP.” Important cues could also be out of scope or undiscovered. In “history of multiple cardiac [stents],” “history” would be a cue for “BEFORE;” however, it is out of the 3-window context of the event “stents.”
An error that is specific to the DocTimeRel model is that events appearing in the section headings oftentimes “OVERLAP” with the DCT. We do not have a feature to capture this header information, which leads to errors.
Errors that are specific to the event-time and event-event models are long-distance relations. The THYME annotation, especially the event-time relation annotation, focuses on short-distance relations. (On the development set, 81.4% of the gold event-time relations and 64.4% event-event relations have at most 6 words between their 2 arguments.) Without closure, the models would therefore be focused on short-distance relations (87.9% of system event-time predictions and 65.5% of event-event predictions on the development set are within 6 words between 2 arguments). For long-distance relations, the chance of picking up misleading or conflicting temporal evidence increases, which imposes challenges for building reliable models. Our hope for solving long-distance relations is through closure if both the short-distance event-time and event-event relations are correctly recognized. However, the performance on event-event relations, especially non-CONTAINS relations, is low. Some long-distance relations may not be established because of event-event errors. In the future, we will work on improving the accuracy of non-CONTAINS event-event relations, such as by incorporating more training instances in related categories.
A logical question is how these state-of-the-art results can be improved. In the future, we are planning to explore joint inference for all types of relations and compare it to our state-of-the-art pipeline solution. Even though we did make use of DocTimeRel info to help reasoning about event-time and event-event relations, and used event-time relations to help reasoning about event-event relations, we still classified each candidate pair separately. While computationally efficient, this approach can be globally optimized by taking all relational constraints (including coreference) into consideration.40–42 We have tested tree features and kernels43–47 in the past21,43 and will further investigate them. Our basic intention for exploring tree features is to identify useful patterns in syntactic structures that may be strong signals for temporality, especially for long-distance argument pairs. Long-distance candidates could have more misleading temporal surface evidence. Syntactic tree structures offer the potential to “filter” surface noise and help identify new patterns for inference.
Conclusion
In this study, we describe an open-source clinical temporal relation discovery system. Empowered by a multilayered modeling strategy, our system can take advantage of automatic inference, greatly reduce the complexity of temporal reasoning, and improve accuracy, especially at the macro level. Enhanced by class-wise weighting and event-expansion techniques, our system was on par with the best 2012 i2b2 challenge systems and achieved state-of-the-art performance on the 2015 Clinical TempEval corpus. Furthermore, some of its best performing components have proven useful in real biomedical informatics applications.
CONTRIBUTORS
CL and SB are co-first authors. All authors contributed to the design, experiments, analysis, and writing of the manuscript.
COMPETING INTERESTS
GKS is on the advisory board of Wired Informatics, LLC, which provides services and products for clinical NLP applications.
Acknowledgments
Thanks to Pei Chen and Sean Finan for technically supporting the experiments. The study was funded by R01 LM 10090 (THYME), U54LM008748 (i2b2), R01GM103859 (PGx), and U24CA184407 (DeepPhe). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
SUPPLEMENTARY MATERIAL
Supplementary material is available online at http://jamia.oxfordjournals.org/.
REFERENCES
- 1.Hripcsak G, Soulakis ND, Li L, et al. Syndromic surveillance using ambulatory electronic health records. J Am Med Inform Assoc 2009;16(3):354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Combi C, Shahar Y. Temporal reasoning and temporal data maintenance in medicine: issues and challenges. Comput Biol Med 1997:27(5):353–368. [DOI] [PubMed] [Google Scholar]
- 3.Das AK, Musen MA. A comparison of the temporal expressiveness of three database query methods. In Annual Symposium on Computer Applications in Medical Care 1995. IEEE Computer Society Press. [PMC free article] [PubMed] [Google Scholar]
- 4.Kahn MG, Fagan LM, Tu S. Extensions to the time-oriented database model to support temporal reasoning in medical expert systems. Methods Inform Med 1990;30(1):4–14. [PubMed] [Google Scholar]
- 5.Schmidt R, Ropele S, Enzinger C, et al. White matter lesion progression, brain atrophy, and cognitive decline: the Austrian stroke prevention study. Ann Neurol 2005;58(4):610–6. [DOI] [PubMed] [Google Scholar]
- 6.Zhou L, Hripcsak G. Temporal reasoning with medical data—a review with emphasis on medical natural language processing. J Biomed Inform 2007;40(2):183–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saurí R, Littman J, Gaizauskas R, et al. TimeML Annotation Guidelines, Version 1.2.1. Citeseer, 2006. [Google Scholar]
- 8.Pustejovsky J, Hanks P, Sauri R, et al. The timebank corpus. In: Corpus Linguistics. 2003. [Google Scholar]
- 9.Verhagen M, Gaizauskas R, Schilder F, et al. Semeval-2007 task 15: Tempeval temporal relation identification. In: Proceedings of the 4th International Workshop on Semantic Evaluations. Association for Computational Linguistics; 2007. [Google Scholar]
- 10.Verhagen M, Sauri R, Caselli T, et al. SemEval-2010 task 13: TempEval-2. In: Proceedings of the 5th International Workshop on Semantic Evaluation. Association for Computational Linguistics; 2010. [Google Scholar]
- 11.UzZaman N, Llorens H, Derczynski, L, et al. SemEval-2013 task 1: TempEval-3: evaluating time expressions, events, and temporal relations. In: Second Joint Conference on Lexical and Computational Semantics (*SEM), Volume 2: Proceedings of the Seventh International Workshop on Semantic Evaluation (SemEval 2013). Atlanta, GA: Association for Computational Linguistics; 2013. [Google Scholar]
- 12.Savova G, Bethard S, Styler W, et al. Towards temporal relation discovery from the clinical narrative. In: AMIA Annual Symposium Proceedings. San Francisco, USA: American Medical Informatics Association; 2009. [PMC free article] [PubMed] [Google Scholar]
- 13.Galescu L, Blaylock N. A corpus of clinical narratives annotated with temporal information. In: Proceedings of the 2nd ACM SIGHIT International Health Informatics Symposium. Miami, FL, USA: ACM; 2012. [Google Scholar]
- 14.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. J Am Med Inform Assoc 2013;20(5):806–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pustejovsky J, Stubbs A. Increasing informativeness in temporal annotation. In: Proceedings of the 5th Linguistic Annotation Workshop. Portland, Oregon, USA: Association for Computational Linguistics; 2011. [Google Scholar]
- 16.THYME, http://thyme.healthnlp.org. accessed 29 Jul 2015. [Google Scholar]
- 17.Styler W, Bethard S, Finan S, et al. Temporal annotations in the clinical domain. Trans Assoc Comput Linguist 2014;2:143–54. [PMC free article] [PubMed] [Google Scholar]
- 18.Bethard S, Derczynski L, Savova G, et al. Semeval-2015 task 6: Clinical tempeval. In: Proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015). Denver, Colorado, USA: Association for Computational Linguistics; 2015. [Google Scholar]
- 19.Raghavan P, Fosler-Lussier E, Elhadad N, et al. Cross-narrative temporal ordering of medical events. In: Proceedings of the 52nd Annual Meeting of the Association for Computational Linguistics (ACL). Baltimore, Maryland, USA: Association for Computational Linguistics; 2014. [Google Scholar]
- 20.Allen JF. An interval-based representation of temporal knowledge. In: IJCAI. 1981. San Francisco, USA: Vancouver, Canada: Morgan Kaufmann Publishers, pp. 221–226. [Google Scholar]
- 21.Miller TA, Bethard S, Dligach D, et al. Discovering narrative containers in clinical text. In ACL 2013. Sofia, Bulgaria; p. 18. [Google Scholar]
- 22.Bethard S, Derczynski L, Pustejovsky J, et al. Clinical TempEval. 2014;arXiv preprint arXiv:1403.4928. [Google Scholar]
- 23.TempEval C. http://alt.qcri.org/semeval2015/task6/. accessed 29 Jul 2015. [Google Scholar]
- 24.Cherry C, Zhu X, Martin J, et al. À la Recherche du Temps Perdu: extracting temporal relations from medical text in the 2012 i2b2 NLP challenge. J Am Med Inform Assoc 2013; p. amiajnl-2013-001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Llorens H, Saquete E, Navarro B. Tipsem (English and Spanish): evaluating CRFs and semantic roles in tempeval-2. In: Proceedings of the 5th International Workshop on Semantic Evaluation. Uppsala, Sweden: Association for Computational Linguistics; 2010. [Google Scholar]
- 26.Fan R, Chang K, Hsieh C, et al. LIBLINEAR: A Library for Large Linear Classication. J Mach Learn Res 2008;9:4. [Google Scholar]
- 27.Xu Y, Wang Y, Liu T, et al. An end-to-end system to identify temporal relation in discharge summaries: 2012 i2b2 challenge. J Am Med Inform Assoc 2013;20(5):849–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang B, Wu Y, Jiang M, et al. A hybrid system for temporal information extraction from clinical text. J Am Med Inform Assoc 2013; p. amiajnl-2013-001635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben-Hur A, Weston J. A user's guide to support vector machines. Methods Mol Biol 2010;609:223–39. [DOI] [PubMed] [Google Scholar]
- 30.Unified Medical Language System (UMLS). http://www.nlm.nih.gov/research/umls/. accessed 29 July 2015. [Google Scholar]
- 31.Styler WF, IV, Bethard S, Finan S, et al. Temporal annotation in the clinical domain. Trans Assoc Comput Linguist 2014;2:143–54. [PMC free article] [PubMed] [Google Scholar]
- 32.Dligach D, Bethard S, Becker L, et al. Discovering body site and severity modifiers in clinical texts. J Am Med Inform Assoc 2013: p. amiajnl-2013-001766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savova G, Masanz J, Ogren P, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17(5):507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Apache Clinical Text Analysis and Knowledge Extraction System (cTAKES). http://ctakes.apache.org, accessed 29 Jul 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.UzZaman N, Allen JF. Temporal evaluation. In: Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies: short papers-Volume 2. Stroudsburg, PA, USA: Association for Computational Linguistics; 2011. [Google Scholar]
- 36.Miller E. The signed-rank (Wilcoxon) test. Lancet 1969;1(7590):371. [DOI] [PubMed] [Google Scholar]
- 37.Lin C, Karlson EW, Dligach D, et al. Automatic identification of methotrexate-induced liver toxicity in patients with rheumatoid arthritis from the electronic medical record. J Am Med Inform Assoc 2014; p. amiajnl-2014-002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irvine AK, Haas SW, Sullivan T. TN-TIES: A system for extracting temporal information from emergency department triage notes. AMIA Annu Symp Proc 2008:328–32. [PMC free article] [PubMed] [Google Scholar]
- 39.Nikfarjam A, Sarker A, O'Connor K, et al. Pharmacovigilance from social media: mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. J Am Med Inform Assoc 2015; p. ocu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrett J, Diggle P, Henderson R, et al. Joint modelling of repeated measurements and time-to-event outcomes: flexible model specification and exact likelihood inference. J R Stat Soc Series B Stat Methodol 2015;77(1):131–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Q, Pan J, Belcher J. Bayesian inference for joint modelling of longitudinal continuous, binary and ordinal events. Stat Methods Med Res 2014: p. 0962280214526199. [DOI] [PubMed] [Google Scholar]
- 42.Wu L, Hu XJ, Wu H. Joint inference for nonlinear mixed-effects models and time to event at the presence of missing data. Biostatistics 2008;9(2):308–20. [DOI] [PubMed] [Google Scholar]
- 43.Lin C, Miller T, Kho A, et al. Descending-path convolution Kernel for syntactic structures. In Proceedings of the 52nd Annual Meeting of the Association for Computational Linguistics (ACL). 2014: Baltimore, Maryland, USA: pp. 81–86. [Google Scholar]
- 44.Miller T, Bethard S, Dligach D, et al. Discovering temporal narrative containers in clinical text. In: Proceedings of the 2013 Workshop on Biomedical Natural Language Processing. Sofia, Bulgaria: Association for Computational Linguistics; 2013. [Google Scholar]
- 45.Collins M, Duffy N. Convolution kernels for natural language. In: Advances in Neural Information Processing Systems. 2001. Vancouver, British Columbia, Canada. [Google Scholar]
- 46.Haussler D. Convolution Kernels on Discrete Structures. 1999. University of California at Santa Cruz: Technical report, Department of Computer Science; 1999. Technical Report UCSC-CRL-99-10. [Google Scholar]
- 47.Chowdhury FM, Lavelli A, Moschitti A. A study on dependency tree kernels for automatic extraction of protein-protein interaction. In: Proceedings of BioNLP 2011 Workshop. Portland, Oregon, USA: Association for Computational Linguistics; 2011. [Google Scholar]