Abstract
Objective Clinical documents made available for secondary use play an increasingly important role in discovery of clinical knowledge, development of research methods, and education. An important step in facilitating secondary use of clinical document collections is easy access to descriptions and samples that represent the content of the collections. This paper presents an approach to developing a collection of radiology examinations, including both the images and radiologist narrative reports, and making them publicly available in a searchable database.
Materials and Methods The authors collected 3996 radiology reports from the Indiana Network for Patient Care and 8121 associated images from the hospitals’ picture archiving systems. The images and reports were de-identified automatically and then the automatic de-identification was manually verified. The authors coded the key findings of the reports and empirically assessed the benefits of manual coding on retrieval.
Results The automatic de-identification of the narrative was aggressive and achieved 100% precision at the cost of rendering a few findings uninterpretable. Automatic de-identification of images was not quite as perfect. Images for two of 3996 patients (0.05%) showed protected health information. Manual encoding of findings improved retrieval precision.
Conclusion Stringent de-identification methods can remove all identifiers from text radiology reports. DICOM de-identification of images does not remove all identifying information and needs special attention to images scanned from film. Adding manual coding to the radiologist narrative reports significantly improved relevancy of the retrieved clinical documents. The de-identified Indiana chest X-ray collection is available for searching and downloading from the National Library of Medicine (http://openi.nlm.nih.gov/).
Keywords: information storage and retrieval, abstracting and indexing, radiography, medical records, biometric identification
BACKGROUND AND SIGNIFICANCE
Because of the difficulties and efforts needed to de-identify and distribute collections of clinical notes and images, only a few such collections have been made publicly available. The available collections either contain text reports—for example, the i2b2 collections, which include over 1500 provider notes and discharge summaries1,2—or radiology images,3 but there are no downloadable collections of images paired with their associated diagnostic reports. Our goal was to fill this gap by enhancing a set of chest X-ray studies that had been collected and de-identified for another purpose4 and making them publically available through the National Library of Medicine (NLM) image retrieval services (Open-i).5 Our enhancements included manual review of the narrative text to assess the adequacy of the de-identification and correct any failures, and the manual encoding of all positive findings reported in the Findings or Impression sections of the radiology reports, both clearly identified by named section headers within the diagnostic reports. Manual coding could add special value to the retrieval of radiology reports, because they are so rife with hedging and negation, i.e., assertions about findings/diseases that are absent.6 Researchers searching for images with a particular finding, e.g., nodules, will not want to pull studies whose report asserts “no nodules or masses.” Furthermore, in other contexts, e.g., MEDLINE® manual Medical Subject Heading® (MeSH®) indexing, manual coding is known to improve retrieval results.7
To test the hypothesis that coding the facts asserted in the reports improves retrieval results, we manually coded all salient findings and diagnoses that were reported as present and none of those reported to be absent. We then indexed the annotated reports by section with a publicly available search engine Lucene,8 and conducted retrieval experiments using real-life image search queries asked by clinicians and collected in the ImageCLEF medical image retrieval 2008–2013 evaluations.9 The goal of the experiments was twofold: to test if manual coding of the reports improved retrieval and to find the best combinations of report sections to be indexed for retrieval.
This paper presents all steps we took to acquire the collection and to make it searchable and accessible.
MATERIALS AND METHODS
Collecting Images and Reports
With IRB approval (OHSRP# 5357), the Indiana University (IU) investigators pulled narrative chest x-ray reports for posterior–anterior (PA) chest x-ray examinations from 2 large hospital systems within the Indiana Network for Patient Care10 database. We limited the retrieval to 4000 such reports each from a different patient and took 2000 from each institution. We limited the total number to 4000 because that was the most we could manually review. In no case did we take more than one study per patient. We targeted outpatient studies because one use of the collection was to serve as a training set for automatic identification of abnormal chest images in an outpatient setting in Africa.4 We used the accession numbers carried in the narrative reports to pull the corresponding chest x-ray images in Digital Imaging and Communications in Medicine (DICOM) format (International Organization for Standardization (ISO) 12052)11 from the hospitals’ picture archiving system (PACS), then linked the images to the reports with dummy identifiers and automatically removed all Health Insurance Portability and Accountability Act (HIPAA) patient identifiers (including the accession numbers) from both the images and their associated reports. We used the Regenstrief Scrubber12 to de-identify the text reports; and the Radiologic Society of North America’s Clinical Trials Processor13 and the DICOM supplement 142 Clinical Trials De-identification methodology14 to de-identify all of the DICOM files (headers and images). The IU investigators then passed the collection of images and associated reports to the National Institutes of Health (NIH) team.
From the starting set of 4000 studies, the NIH team only included those whose diagnostic reports had labeled sections for: a) the reason for the study, b) findings, and c) the impression, or the final diagnosis, and whose images included at least a PA and a lateral view of the chest. All but 4 of the studies met these criteria, leaving us with a study set of 3996 narrative reports and 8121 images
Verifying that the De-identification Was Complete
The HIPAA of 1996 requires removal of information that can identify an individual and his or her relatives, household members, and employers. De-identification is deemed adequate only if any remaining information cannot be used to identify the individual.15 Note, however, that even the most accurate automatic de-identification techniques fail on occasion to remove all possible instances of personally identifiable information (PII).16 Because we planned to make these studies publicly available, we wanted to verify that the automatic de-identification processes removed all identifiers, so we manually inspected every narrative report and every DICOM image for identifying information.
We inspected the DICOM content, including both headers and images, using freely available DICOM viewers, Osirix for Mac OS X17 and MicroDICOM for Windows,18 and engaged 8 reviewers to examine this DICOM content. Each DICOM header and each of the 8121 images were independently inspected by 2 of the 8 reviewers who looked for: 1) definite HIPAA identifiers (e.g., patient names, hospital numbers, complete dates); 2) implanted medical devices that revealed a unique identification number and potential identifiers including: a) jaw outline/partial skull, b) teeth, and c) jewelry. The reviewers also tallied all of the HIPAA and potential identifiers by category. Though the potential identifiers are not HIPAA identifiers, we planned to go the extra mile and exclude studies that revealed what we classed as potential identifiers from our public release.
We also inspected the text of all of the de-identified diagnostic reports for identifiers that might have escaped notice during the automatic scrubbing effort. Two independent reviewers looked for scrubbing failures in each diagnostic report during the same pass in which they coded the positive findings in these reports (see below).
If the 2 reviewers disagreed on the classification of any definite or theoretically possible patient identifier present in the DICOM header, image, or narrative report, they met to try to reconcile their differences. If after this reconciliation either reviewer still thought the image or the report contained PII, the image or the report and corresponding images were removed from the collection.
Annotation of Radiology Reports
We encoded the findings and diagnoses recorded in the radiology reports with MeSH19,20 codes supplemented by Radiology Lexicon® (RadLex)®21,22 codes, as needed to cover imaging terminology that was outside of MESH’s purview.
Two coders trained in medical informatics (S.E.S., a medical librarian and L.R., an MD) independently coded the positive findings in each report without any automated support. When the 2 could not agree on the coding of a given finding, the decision was adjudicated in a meeting that included the two plus a third annotator, D.D.F. (MD trained in medical informatics).
We produced the annotations in 2 passes. At the first pass, the annotators simply classified the reports into: normal and not normal. Acute or chronic disease findings, implanted medical devices, or surgical instruments were all considered not normal. The reports for the normal chest x-rays were labeled and subsequently indexed “normal,” so that people looking for normal x-rays could easily find these studies.
In the second pass, we coded the type of abnormality for each study classified as “not normal.” We coded the concepts in the not normal radiology reports according to the principles outlined in NLM Indexing Manual and Technical Memoranda.23 As in MEDLINE indexing, we use descriptors (headings) and subheadings—standard qualifiers added to descriptors to narrow down the topic. For example, we represented the RadLex term “upper lobe of left lung” combining MeSH term “Lung” with qualifiers left and upper lobe. An Impression that contains this term, for example, “Impression: left upper lobe infiltrate,” was coded as infiltrate/lung/upper lobe/left. The annotation guidelines are provided in Supplementary Appendix A.
We first coded the concepts (excluding negatives) that radiologists recorded in the Impression section. We used the Findings section of the report to: a) clarify an ambiguity in the impression section; b) discover synonymy; and c) identify minor, historic or incidental conditions that alter the appearance of the x-rays. Any such finding in c), even if minor, would throw the report into the non-normal category. We handled uncertainty as follows: when the hedging term indicated the pathology was present and the uncertainty was in the specifics, we ignored the hedging terms and coded the salient finding. We coded old findings if they were discussed in the report. For example, we coded “Calcified hilar lymph XXXX” as calcinosis/lung/hilum/lymph nodes. Figure 1 presents an example of an annotated report.
Figure 1:
A sample radiology report with manual and MTI annotations. Terms removed by the automatic text scrubber are replaced with XXXX. “COPD” in the impression section is annotated with the MeSH term “Pulmonary Disease, Chronic Obstructive.” “Scarring” is translated to MeSH term “Cicatrix.”
In addition to manual encoding described above, we also coded the same sections with an automatic encoding system to obtain a baseline against which to assess the incremental advantage that manual encoding provides to retrieval systems.
The automatic encoding was produced by Medical Text Indexer (MTI), a system currently used by NLM to index some PubMed/MEDLINE citations.24 MTI does not discriminate between positive and negative assertions about findings. So we ran the terms extracted by MTI through MetaMap25 and discarded the MTI-suggested terms that were characterized as negation by the Neg-Ex6 algorithm implemented in MetaMap.
Retrieval Experiments
We embedded the results of the manual and automatic coding of findings into two new “sections” of the report which we labeled “manual” and “MTI,” respectively, as shown in Figure 1. So for analysis purposes, the reports contained five distinct sections: Indications, Findings, Impression, Manual encoding, and MTI encoding. We indexed the resulting documents using Lucene-4.6.08 customized to include the Unified Medical Language System® (UMLS®) synonymy for query expansion. The creation of these sections as different “fields” in Lucene allowed us to limit searching to one or more sections and to determine the sections’ incremental contribution to searching success—for example, how much manual coding added to free text searching.
In addition to a document collection, retrieval experiments require a set of test queries, which we obtained by selecting all requests potentially relevant to chest x-rays from the questions asked by doctors and collected in the ImageCLEF medical image retrieval evaluations. Thirty distinct ImageCLEF queries shown in Supplementary Appendix B met our criteria. We used the queries and Lucene syntax to evaluate the contribution of the manually coded findings to text searching; searching the manual and the automatic coding alone and in combinations of the sections listed in Table 2.
Table 2:
Search results averaged over the 30 ImageCLEF medical image retrieval queries.
| Citation areas searched | Precision at 10 | Inferred Average Precision |
|---|---|---|
| Text content of the Impressions and Findings sections | 0.370 | 0.393 |
| MTI (automated) codes | 0.240 | 0.208 |
| MTI automated codes + Text | 0.410 | 0.417 |
| Only manual codes | 0.393 | 0.315 |
| Manual codes + Text | 0.470* | 0.536* |
| Manual codes + MTI automated codes + Text | 0.473* | 0.524* |
Asterisks indicate significant improvements over searching the impression and finding sections of narrative reports. Word “text” in the first column implies Impression + Finding sections.
Relevance of the retrieved reports to each query was evaluated independently by two judges (L.R. and S.E.S.) using the post hoc pooling method developed for the Text Retrieval Conference evaluations 26: the top 10 distinct reports as ranked by Lucene’s default scoring model found by each of the searches for each topic were combined automatically and sent to the judges who were blinded to the method used to find a given report. We evaluated relevance on a three-point scale: relevant; maybe relevant (e.g., if it is not clear if the patient has atelectasis: patchy left lower lobe airspace disease, possibly atelectasis or pneumonia); and not relevant (e.g., if the impression stated “no pneumonia”). Using the trec_eval 9.0 evaluation software,27 we evaluated the number of relevant reports in the top 10 reports delivered by each search (precision at 10, P@10) and inferred average precision that takes into account recall (the number of relevant documents in the collection that were found by the search), and distinguishes between irrelevant and unjudged documents in the search results. We combined the judgments and used them to evaluate the results as follows: for the report to be relevant, it needed one relevant judgment and another maybe relevant judgment. We measured statistical significance in the differences between the searches using the Wilcoxon signed-rank test.28
RESULTS
Quality of Automatic Text De-Identification
Manual review of the text in 3996 radiology reports de-identified at IU revealed no scrubbing failures. This was not surprising because the Regenstrief Scrubber errs very strongly on the side of sensitivity (recall). The mean number of words per report was 77.1; of these, 9.5 (12%) were removed by the Regenstrief Scrubber. Non-normal reports carried a mean of 84.5 words, of which the scrubber removed 9.9 (11.5%) per report. However, most of the removed words, 8 per report, were dates or words contained within the report footers (see Figure 1), and irrelevant to the clinical content of the report. The mean number of words in the Findings and Impression sections was 39, of which the scrubber removed only one per report (2.5%). The Indications section carried a mean of 6 words per report and the scrubber removed 0.9 (15%) of these.
Quality of Automatic De-Identification of the DICOM Header and Images
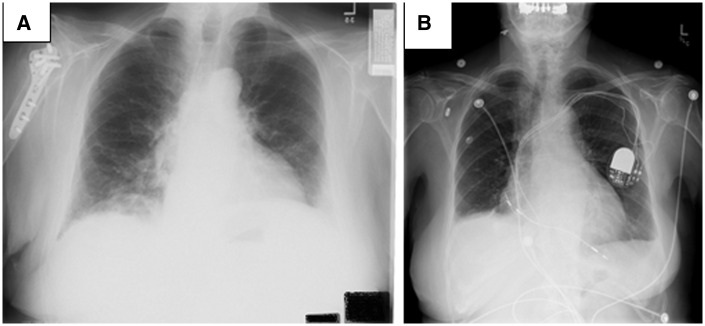
Review of the DICOM headers and images took about 1–2 min per each image and header, with most of the time spent reading DICOM headers. We found no PII in the automatically de-identified DICOM headers. The DICOM de-identifier appears not to address the content of the scanned images. We found four images from 2 patients (0.05% of 3996) with PII. The images for one patient showed full patient addressograph information, and the other, a full date (see Figure 2A; the actual tag data in the figure has been obscured for privacy reasons). Both were originally film-based images that were scanned into a DICOM PACS.
Figure 2:
Images with (A) hospital tag in the lower right-hand corner (the actual tag data has been obscured for privacy reasons) and (B) partially visible Medtronic device-specific radiopaque alphanumeric code and jaw outline and teeth.
Initially we had been concerned that devices implanted in the chest might carry unique device identifiers that could be traced back to the patient. Images for 107 (2.6%) patients showed pacemakers/defibrillators, and 72 (67%) of these did reveal short alphanumeric identifiers, but these were not identifiers that could be directly traced to the patient (see Figure 2B). They were product identifiers through which providers could get more information about the product and the patient from the manufacturers.29 A total of 432 patients had at least one image that revealed teeth or jaw outline (375 patients), face and/or skull (18 patients), or jewelry (39 patients). These are not formal HIPAA identifiers, but we removed the images and their corresponding reports from our public collection to avoid any possible risk of re-identification based on this content.
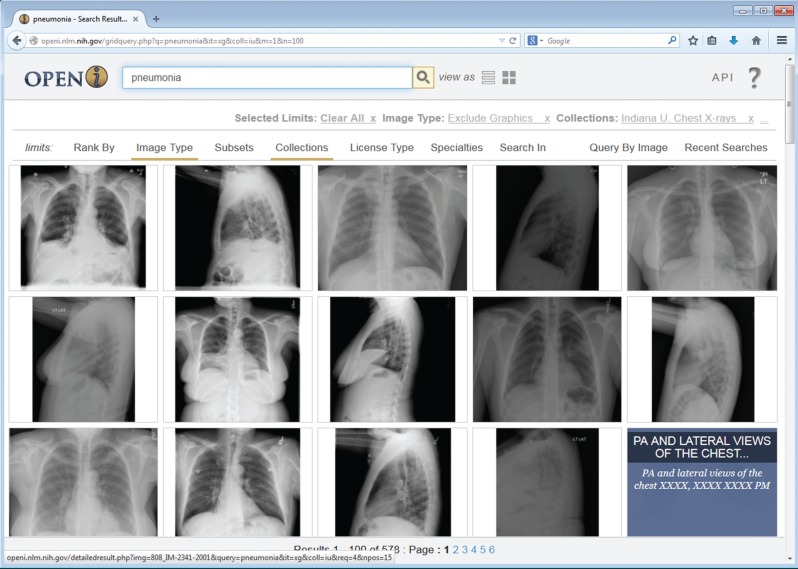
For public access we removed the 2 studies that revealed PII, and all images that showed teeth, partial jaw, jewelry, or partial skull. Because most studies included at least one image that did not reveal such findings, we only had to remove 41 (1%) of the studies entirely, leaving a subset of 7470 DICOM images and 3955 associated reports for our public subset. The public subset is now available for browsing or downloading via Open-i,30 an NLM service (see Figure 3) which is accessed by more than 20 000 distinct users daily, about 20% of whom are returning users. We believe these images and reports will be useful to educators, and imaging researchers as exemplified by Jaeger’s work4 and others.
Figure 3:
Grid view of search results for pneumonia in the radiology collection indexed in Open-i.
Outcome of the Encoding
Of the full set of 8121 images and 3996 reports, 3087 images and 1526 (38 %) reports were normal as judged by 2 coders. We used a total of 101 MeSH codes and 76 RadLex codes to represent the content of the Impressions and Findings sections in the 2470 not normal reports. The 50 most frequently occurring codes (taking MeSH and RadLex codes together) covered 4708 (68.1%) of the 6907 coded findings in all non-normal chest X-rays. The 10 most frequently assigned codes and the number of non-normal records to which the term was assigned is shown in Table 1. Note that though the preferred term from MeSH does not always reflect the phrasing radiologists would use, the radiologist phrasings are almost always included among the alternative terms in MeSH (e.g., cicatrix includes alternatives of: Cicatrization, Scar, Scars, and Scarring). All assigned terms and counts are provided in Supplementary Appendix C.
Table 1:
The 10 most frequently assigned codes, not counting the 1526 “normal” labels.
| Code | Source(s) | # Coded reports (% of 2470 non-normal reports) |
|---|---|---|
| Cardiomegaly | MeSH | 375 (15.1) |
| Pulmonary atelectasis | MeSH | 347 (14.0) |
| Calcified granuloma | MeSH/RadLex | 284 (11.5) |
| Aorta/ tortuous | MeSH/RadLex | 253 (10.2) |
| Lung/hypoinflated | MeSH/RadLex | 245 (9.9) |
| Opacity/lung base | RadLex | 203 (8.2) |
| Pleural effusion | MeSH | 172 (6.9) |
| Lung/ hyperinflation | MeSH/RadLex | 164 (6.6) |
| Cicatrix/lung | MeSH | 148 (5.9) |
| Calcinosis/lung | MeSH | 141 (5.7) |
The first column represents preferred terms from MESH and RadLex. The source of each of post-coordinated terms is given in the second column.
Retrieval Results
Overall, 30 queries (See Supplementary Appendix B for the list of all queries) retrieved 841 distinct records. The number of records returned per query by the Lucene search engine ranged from 2 to 73. Two authors (L.R. and S.E.S.) independently judged whether each record was relevant to the query. The inter-annotator agreement between the 2 judges regarding the relevance of the report to the query on a three-point scale (relevant, partially relevant and not relevant) was very good (Cohen’s κ = 0.85). For the sharper distinction about whether the report was relevant or definitely not relevant, the agreement was even better (κ = 0.9). Table 2 shows the results of searching the Impression and Findings sections of narrative reports using only a text search or text search augmented by the codes produced by automatic and manual annotations.
Searches that accessed both manual coding and narrative text were significantly more successful than searches that used the text alone according to both metrics. Searches based on manual coding alone were not better than those based only on text. Searching MTI alone was the least successful and using MTI codes in addition to narrative text did not provide an advantage over text searches alone. Some queries had only a few relevant reports in the collection, which kept the overall results from being even better.
DISCUSSION
Preparing collections of clinical documents for public use is a valuable but labor-intensive process. In addition to using the studies provided by IU in our own line of research,4 we prepared a subset for public distribution by manually verifying the results of automatic de-identification of the DICOM headers and images and the associated text reports. We also manually coded the impressions and findings in the diagnostic reports. Although these were the most time-consuming steps of the process, our study shows that checking the images was necessary and coding diagnostic findings/diagnoses was desirable.
Our verification of automatic de-identification found a very small (4) number of images carrying HIPAA identifiers. These were all scanned images with identifiers embedded in the picture—as is common for film-based x-rays. DICOM systems can distinguish scanned from digitally acquired images, and the DICOM de-identification system should either exclude scanned images or require manual review and manual processing of scanned films potentially showing PII. With the latter approach, scanned images can be identified and manually reviewed before they are included in a de-identified DICOM image set. Given the ubiquity of character recognition it might also be possible to find and remove characters embedded in the image,31 but to retain the single letters (R, L) used to signify laterality.
We defined a category of possible PII that went beyond strict HIPAA requirements. These included images that showed jaw with teeth because recent research reports 94.3% accuracy in human identification based on 2–3 adjacent molars.32 To be on the safe side, we also excluded studies that showed partial skull images and jewelry, though we do not know the true risk of re-identification they pose.
Augmenting text searches with manual annotation of the reports with the most salient concepts (manual + Text in Table 2) significantly improved retrieval results compared to searching text alone. P@10 rose from 37% to 47% (P ≤ .05) and inferred average precision from 39.3% to 53.6% (P ≤ .01). Text searches for pneumonia, however, were slightly better without manual annotation. Both judges judged the following impression relevant to pneumonia: “Focal airspace disease in the right middle lobe. This is most concerning for pneumonia,” but the manual annotation had only one code: Airspace Disease/lung/middle lobe/right/focal. Pneumonia is mentioned only once in this report and in the field that has less weight than the manual codes; therefore the report was ranked too low to contribute to the results of text searches with manual annotation. MTI indexing of the impressions and findings that excluded negated codes did not significantly improve the text search results. Manual coding showed better results than automatic MTI coding for several reasons: 1) MTI has access only to MeSH, which lacks codes for many radiology findings, whereas the coders used a coding system that included both RadLex codes and MeSH codes as needed; 2) The automatic method did misidentify some negated concepts and hedging as true findings; and 3) Coders do not need an exact string match to identify a code. For example, the coders assigned “Thoracic Vertebrae/degenerative” to “There are severe degenerative changes of the thoracic spine,” but MTI missed this term because “thoracic vertebrae” does not have “thoracic spine” as a synonym in MeSH. The explicit coding of normal chest studies as such by the manual coders facilitated searches intended to include or exclude normal studies. Searching the text “normal” would not provide the same results because many reports contain both normal and not normal findings.
We have created the first publicly available collection of chest x-ray studies (images and report), and made it available to a wide range of researchers through Open-i. Other publicly available collections of clinical documents contain either text or image data and are mostly small and their existence is often known only to special interest groups such as participants in i2b2 challenges.1 Some collections that were public in the past are no longer available—for example, the Bioscope corpus33 that contains medical text is cited by only 20 PubMed articles and is no longer available. Many have complained that available public collections are not accessible for easy browsing as reflected in one researcher’s comment: “it would be good to [see] … at least a sample of the documents in the collection to decide if I want to request it.”34 A necessary first step in providing access to collections of clinical and biomedical data is to catalog them (e.g., in the table of NIH-supported data repositories).35 Better would be a single method for browsing image collections. We have loaded the IU collection for chest x-ray studies in Open-i (http://openi.nlm.nih.gov/) where the images and reports can easily be browsed, searched, and/or downloaded along with images in Open-i from other sources.
CONCLUSIONS
The IU collection of clinical radiology images and text reports was prepared for development of clinical decision support algorithms. Manual examination of automatically de-identified reports and images showed that aggressive de-identification of radiology reports reliably removed all personally identifiable information. Automatically de-identified images failed to find two of the nearly 4000 patients whose images revealed PII. Overall, we removed 651 images (8% of the original set), most of which did not have personally identifiable information according to the current definition, but contained some information that might be highly specific, such as teeth or custom jewelry.
Manual annotations have significantly improved search results over searching only the text of the reports. The public, and doubly de-identified, collection is searchable and downloadable from the NLM image retrieval service (Open-i) that also provides access to more than 2.6 million images and enriched MEDLINE citations from over 700 000 PubMedCentral articles.
Since its public release, our collection has attracted two research groups that have obtained the data using the Open-i API at http://openi.nlm.nih.gov/services.php. The original DICOM images are available at http://openi.nlm.nih.gov/contactus.php
FUNDING
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors. This work was supported by the Intramural Research Program of the National Library of Medicine, National Institutes of Health.
COMPETING INTERESTS
The authors have no competing interests to declare.
CONTRIBUTORS
All authors contributed to the conception of the work, interpretation of data and revising of the manuscript. M.K. and M.R. acquired the data. S.E.S., L.R., and D.D.F. adapted the controlled vocabularies and annotated the reports. G.T. and S.A. supervised manual de-identification validation of DICOM images. D.D.F., M.R., and C.M. drafted the manuscript. All authors have approved the final version of the manuscript. All authors are accountable for the parts of the work they have done and are able to identify which co-authors are responsible for specific other parts of the work. All authors have confidence in the integrity of the contributions of their co-authors.
Acknowledgments
Many thanks to our collaborators at the Indiana Network for Patient Care and IU Radiology who assisted with collecting the data. This work was supported by the Intramural Research Program of the National Library of Medicine, National Institutes of Health.
SUPPLEMENTARY MATERIAL
Supplementary material is available online at http://jamia.oxfordjournals.org/.
REFERENCES
- 1.i2b2 NLP Research Datasets. https://www.i2b2.org/NLP/DataSets/Main.php. Accessed February 11, 2015.
- 2.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. JAMIA. 2013;20(5):806–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Imaging Archive. http://www.cancerimagingarchive.net/. Accessed February 11, 2015.
- 4.Jaeger S, Karargyris A, Candemir S, et al. Automatic tuberculosis screening using chest radiographs. IEEE Trans Med Imaging. 2014;33(2):233–245. [DOI] [PubMed] [Google Scholar]
- 5.Demner-Fushman D, Antani S, Simpson MS, Thoma GR. Design and development of a multimodal biomedical information retrieval system. JCSE. 2012;6(2):68–177. [Google Scholar]
- 6.Chapman WW, Bridewell W, Hanbury P, et al. Evaluation of negation phrases in narrative clinical reports. Proc AMIA Symp. 2001:105–109. [PMC free article] [PubMed] [Google Scholar]
- 7.Hersh W. Information Retrieval: A Health and Biomedical Perspective. Heidelberg: Springer; 2008. [Google Scholar]
- 8.Apache Lucene.™ http://lucene.apache.org/. Accessed February 11, 2015.
- 9.Kalpathy-Cramer J, de Herrera AGS, Demner-Fushman D, et al. Evaluating performance of biomedical image retrieval systems—An overview of the medical image retrieval task at ImageCLEF 2004–2013 . Comput Med Imaging Graph. 2015;39:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Indiana Network for Patient Care. http://www.ihie.org/indiana-network-for-patient-care Accessed February 11, 2015.
- 11.The DICOM Standard. http://medical.nema.org/standard.html. Accessed February 11, 2015.
- 12.Friedlin FJ, McDonald CJ. A software tool for removing patient identifying information from clinical documents. JAMIA. 2008;15:601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.RSNA Clinical Trials Processor. https://www.rsna.org/ctp.aspx. Accessed February 11, 2015.
- 14.DICOM Clinical Trial De-identification Profiles. ftp://medical.nema.org/medical/dicom/final/sup142_ft.pdf. Accessed February 11, 2015.
- 15.Department of Health and Human Services. Modifications to the HIPAA Privacy, Security, Enforcement, and Breach Notification Rules Under the Health Information Technology for Economic and Clinical Health Act and the Genetic Information Nondiscrimination Act; Other Modifications to the HIPAA Rules; Final Rule. Federal Register Part II, Vol. 78, No. 17, January 25, 2013. http://www.gpo.gov/fdsys/pkg/FR-2013-01-25/pdf/2013-01073.pdf. Accessed February 11, 2015. [PubMed]
- 16.El Emam K, Jonker E, Arbuckle L, et al. A systematic review of re-identification attacks on health data. PLoS One. 2011;6(12):e28071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.OsiriX Imaging Software. http://www.osirix-viewer.com. Accessed February 11, 2015.
- 18.MicroDicom - free DICOM viewer for Windows. http://www.microdicom.com. Accessed February 11, 2015.
- 19.Rogers FB. Medical subject headings. Bull Med Libr Assoc. 1963;51:114–116. [PMC free article] [PubMed] [Google Scholar]
- 20.Medical Subject Headings. http://www.nlm.nih.gov/mesh/meshhome.html. Accessed February 11, 2015.
- 21.Langlotz CP. RadLex: a new method for indexing online educational materials. Radiographics. 2006;26(6):1595–1597. [DOI] [PubMed] [Google Scholar]
- 22.RadLex RSNA Informatics. http://www.radlex.org/. Accessed February 11, 2015.
- 23.MEDLINE Indexing Online Training. http://www.nlm.nih.gov/bsd/indexing/training/INT_030.html. Accessed February 11, 2015.
- 24.Mork JG, Jimeno-Yepes AJ, Aronson AR. The NLM Medical Text Indexer System for Indexing Biomedical Literature. BioASQ Workshop, Valencia, Spain, September 27, 2013. [Google Scholar]
- 25.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. JAMIA. 2010;17(3):229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voorhees EM, Harman DK, eds. TREC: Experiment and Evaluation in Information Retrieval. Vol. 1 Cambridge: MIT Press; 2005. [Google Scholar]
- 27.trec_eval, NIST. http://trec.nist.gov/trec_eval/. Accessed February 11, 2015.
- 28.Siegel S, Castellan NJ, Nonparametric Statistics for the Behavioral Sciences, 2nd ed New York: McGraw-Hill; 1988. [Google Scholar]
- 29.Jacob S, Shahzad MA, Maheshwari R, et al. Cardiac rhythm device identification algorithm using X-Rays: CaRDIA-X. Heart Rhythm. 2011;8(6):915–922. [DOI] [PubMed] [Google Scholar]
- 30.Open-I API services. http://openi.nlm.nih.gov/contactus.php. Accessed February 11, 2015.
- 31.Vcelak P, Kleckova J. Automatic real-patient medical data de-identification for research purposes. Int J Sci, Eng Technol. 2011;52:506–510. [Google Scholar]
- 32.Lin PL, Lai YH, Huang PW. Dental biometrics: Human identification based on teeth and dental works in bitewing radiographs. Pattern Recogn. 2012;45(3):934–946. [Google Scholar]
- 33.Vincze V, Szarvas G, Farkas R, et al. The BioScope corpus: biomedical texts annotated for uncertainty, negation and their scopes. BMC Bioinformatics. 2008;9(Suppl 11):S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruch P. TREC Medical 2013. Mailing List. TREC-MED. NIST. September 16, 2013. [Google Scholar]
- 35.NIH Data Sharing Repositories. http://www.nlm.nih.gov/NIHbmic/nih_data_sharing_repositories.html. Accessed February 11, 2015.