Abstract
Life-history theory predicts a trade-off between reproductive investment and self-maintenance. The negative association between fertility and longevity found throughout multicellular organisms supports this prediction. As an important exception, the reproductives of many eusocial insects (ants, bees, and termites) are simultaneously very long-lived and highly fertile. Here, we examine the proximate basis for this exceptional relationship by comparing whole-body transcriptomes of differently aged queens of the ant Cardiocondyla obscurior. We show that the sets of genes differentially expressed with age significantly overlap with age-related expression changes previously found in female Drosophila melanogaster. We identified several developmental processes, such as the generation of neurons, as common signatures of aging. More generally, however, gene expression in ant queens and flies changes with age mainly in opposite directions. In contrast to flies, reproduction-associated genes were upregulated and genes associated with metabolic processes and muscle contraction were downregulated in old relative to young ant queens. Furthermore, we searched for putative C. obscurior longevity candidates associated with the previously reported lifespan-prolonging effect of mating by comparing the transcriptomes of queens that differed in mating and reproductive status. We found 21 genes, including the putative aging candidate NLaz (an insect homolog of APOD), which were consistently more highly expressed in short-lived, unmated queens than in long-lived, mated queens. Our study provides clear evidence that the alternative regulation of conserved molecular pathways that mediate the interplay among mating, egg laying, and aging underlies the lack of the fecundity/longevity trade-off in ant queens.
Keywords: fecundity/longevity trade-off, transcriptome, aging, mating, social insect, RNA-Seq
Introduction
Why organisms age and die and why they do so at different rates are among the most fundamental and least understood phenomena in biology. Of the various mechanistic and evolutionary explanations for aging and death (e.g., Rose 1990; Hughes and Reynolds 2005), those that involve a trade-off between fecundity and longevity have gained considerable empirical support. Throughout multicellular organisms, including volvocine algae, Drosophila, Caenorhabditis and human beings, increased investment in early and current reproduction negatively affects longevity (Stearns 1989; Westendorp and Kirkwood 1998; Michod et al. 2006; Flatt 2011; Tabatabaie et al. 2011). On the molecular level, there is evidence that this trade-off might be mediated by (IIS) and downstream endocrine signals, for example, juvenile hormone (JH) in insects (Flatt et al. 2005; Flatt and Kawecki 2007). Mutations in the IIS pathway were shown to have antagonistic pleiotropic effects on lifespan and reproduction in Drosophila melanogaster, Caenorhabditis elegans, and mice (Tatar et al. 2001; Partridge et al. 2005).
Perennial eusocial insects, such as termites, ants, and many bees, and eusocial Fukomys mole-rats, are a striking exception: Dependent on their environment, individuals may grow into long-lived reproductives or short-lived, nonreproductive workers (Keller and Genoud 1997; Keller 1998; Jemielity et al. 2005; Heinze and Schrempf 2008; Schmidt et al. 2013). This suggests that, on the population level, the trade-off between fecundity and longevity is reversed in these organisms. In addition, mating might not be that detrimental for the female reproductives (queens) of social insects as for solitary insects (Partridge et al. 1987; Trevitt and Partridge 1991). The short mating period early in life and the storage of sperm by queens result in a lifelong pair bond of males and females. This predicts that males benefit from increasing the lifespan of their female partners, as was already shown in the ant Cardiocondyla obscurior (Schrempf et al. 2005).
Understanding how reproductives of eusocial animals evade the fecundity/longevity trade-off not only serves to identify idiosyncratic pathways that link mating, fecundity, and lifespan but also might provide fundamental insight into the evolution of aging in general. Hence, considerable efforts have been made to reveal the physiological, endocrine, and transcriptomic correlates of the different life expectancies of reproductives and nonreproductives (Parker et al. 2004; Corona et al. 2005, 2007; Grozinger et al. 2007; Haddad et al. 2007; Jemielity et al. 2007; Schneider et al. 2011). For example, experiments addressing the oxidative stress theory of aging (which considers reactive oxygen species as a cause of aging; e.g., Finkel and Holbrook 2000) consistently showed that antioxidant enzyme gene expression and activity are lower in queens than in workers (Parker et al. 2004; Corona et al. 2005; Schneider et al. 2011). This might be explained either by a lower generation of reactive oxygen species in queens or by the mediation of oxidative stress resistance through other molecules, such as vitellogenin (Vg; Seehuus et al. 2006; Havukainen et al. 2013). Honeybee queens indeed have a higher titer of vitellogenin, associated with lower JH titers and lower expression of insulin-like peptide and receptor genes compared with workers (Corona et al. 2007). As this observation disagrees with the opposing effects of IIS and JH on lifespan and reproduction in Drosophila melanogaster (Flatt et al. 2005), it has been suggested that the traditional positive relationships between nutrition and IIS, and between JH and Vg, are reversed in honeybee queens (Corona et al. 2007; Remolina and Hughes 2008).
However, comparisons between queens and workers are often confounded because the two female castes typically differ not only in fecundity but also in developmental, morphological, physiological, and behavioral traits. All of these might affect the tempo of aging and senescence. To disentangle the effects of variation in phenotype, mating status, fecundity, and resource availability on lifespan require alternative approaches, for example, a comparison among reproductives of different fecundity and longevity.
Here, we used the ant C. obscurior as a social insect model to investigate the proximate mechanisms underlying variation in lifespan independent of variation in genotype, development, and morphology. Its colonies are typically inbred because young queens mate in the nest with wingless males and stay there to reproduce (e.g., Heinze and Hölldobler 1993). Queens are relatively short-lived (approximately 6 months), which allows monitoring their total lifespan and lifetime reproductive success (Schrempf et al. 2005; Heinze and Schrempf 2012). We used two approaches to investigate the effects of age and mating on gene expression.
First, we compared the transcriptomes of young mated (4-week-old) and old mated (18-week-old) C. obscurior queens to identify general signatures of aging. We then compared these data with transcriptomes of female D. melanogaster of different age (Pletcher et al. 2002; Doroszuk et al. 2012).
Second, we contrasted transcriptome data among three different types of 18-week-old queens, which were subjected to different mating regimes known to affect future life expectancy and fecundity: 1) Virgin queens (short average lifespan and low average fecundity, 18.2 weeks and 6.8 eggs per week); 2) mated queens (long lifespan and high fecundity, 26.0 weeks and 20.5 eggs per week), and 3) queens mated to sterilized males (sham-mated queens, long lifespan and low fecundity, 25.8 weeks and 6.1 eggs per week; Schrempf et al. 2005).
Finally, we investigated whether mating-induced gene expression changes in C. obscurior match those previously found in female D. melanogaster and honeybees.
Our results reveal for the first time a comprehensive picture of gene expression patterns associated with age, mating, and fecundity in a social insect and indicate that conserved pathways involved with senescence in solitary species may experience a reversal in gene expression patterns. The commonality of aging found between two species with opposite life histories indicates a persistent action of developmental genes later in life.
Results
Gene Expression Patterns of All Four Types of Queens
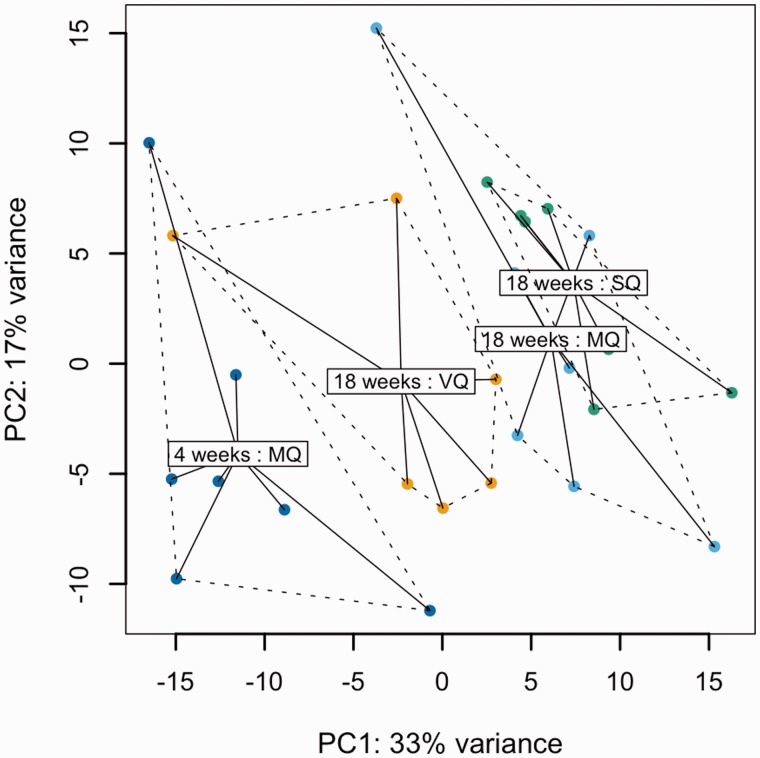
To reveal the effects of age and mating on gene expression, we analyzed transcriptomic data of four types of queens: MQ4 = 4-week-old mated queens, MQ18 = 18-week-old mated queens, SQ18 = 18-week-old sham-mated queens, VQ18 = 18-week-old virgin queens. We sequenced individual queens to account for biological variation across samples and achieved a final sample size of seven (MQ4, MQ18, SQ18) and six (VQ18) replicates. The principal component plot of all samples indicated a separation of MQ4 and VQ18 from all other groups and considerable overlap of SQ18 and MQ18 (fig. 1). To analyze age-specific expression, we first compared the transcriptomes of MQ4 and MQ18 and found 783 differentially expressed genes (DEGs) (adjusted P value < 0.05, supplementary file S1, Supplementary Material online).
Fig. 1.
Principal component analysis plot of the top 500 genes with highest variance across all samples illustrating variation within and between treatments. Variance stabilizing transformation of expression values was performed prior to analysis. The labels represent the center of mass of each group.
To disentangle the effects of mating and reproduction, we contrasted the gene expression profiles of MQ18, SQ18, and VQ18. VQ18 differed from MQ18 in 37 genes and from SQ18 in 350 genes, whereas SQ18 and MQ18 differed in five genes (adjusted P value < 0.05, supplementary file S1, Supplementary Material online).
Functional Annotation of Genes Differentially Expressed with Age and Reproduction (MQ4 vs. MQ18)
We used the corresponding D. melanogaster orthologs of all genes (determined by a reciprocal BLAST [Basic Local Alignment Search Tool]) in DAVID to test for a functional enrichment in all lists of DEGs. The 242 genes more highly expressed in MQ18 compared with MQ4 (160 orthologs) revealed an enrichment for Gene Ontology (GO) terms associated with reproduction, which reflects the higher rate of reproduction in older queens (supplementary file S2, Supplementary Material online). Clustering of these 102 GO annotations for biological processes (BP) resulted in five groups, which are best described by the terms cell cycle, cellular component assembly, female germ-line cyst encapsulation, germ cell development, and establishment or maintenance of cell polarity (table 1).
Table 1.
Enriched GO Terms for BP in Aged (MQ18) versus Young Mated Queens (MQ4).
| Enrichment Score | Representative GO Term | Count | % | P value | |
|---|---|---|---|---|---|
| Upregulated in MQ18 | |||||
| Annotation Cluster 1 | 2.3 | Cell cycle | 22 | 15 | 0.0001 |
| Annotation Cluster 2 | 2.0 | Cellular component assembly | 20 | 13 | 0.0013 |
| Annotation Cluster 3 | 2.0 | Female germ-line cyst encapsulation | 4 | 3 | 0.0005 |
| Annotation Cluster 4 | 1.8 | Germ cell development | 10 | 7 | 0.0042 |
| Annotation Cluster 5 | 1.7 | Establishment or maintenance of cell polarity | 7 | 5 | 0.0161 |
| Downregulated in MQ18 | |||||
| Annotation Cluster 1 | 3.3 | Cellular carbohydrate catabolic process | 9 | 3 | <0.0001 |
| Annotation Cluster 2 | 2.4 | Organic acid metabolic process | 19 | 6 | 0.0025 |
| Annotation Cluster 3 | 2.3 | Nucleoside monophosphate biosynthetic process | 7 | 2 | 0.0026 |
Note.—Annotation clusters are listed with their most significant GO term representing the biological meaning of the group.
KEGG pathway analysis of the 541 genes more highly expressed in MQ4 (336 orthologs) revealed a significant enrichment in fructose and mannose metabolism (P = 0.01), purine metabolism, pentose phosphate pathway, and metabolism of xenobiotics by cytochrome P450 (P < 0.05). The 37 significant GO terms also pointed to an association with mainly catabolic and biosynthetic processes. The most significant terms of the three generated clusters were cellular carbohydrate catabolic, organic acid metabolic, and nucleoside monophosphate biosynthetic process. Besides, genes involved in muscle contraction were overrepresented in the set of downregulated genes (P = 0.0009).
Functional Annotation of Genes Associated with Mating (VQ18 vs. MQ18 and SQ18, SQ18 vs. MQ18)
Neither the 33 genes with higher expression (19 orthologs) nor the 4 genes with lower expression (one ortholog) in VQ18 compared with MQ18 showed a significant functional enrichment. However, when we included eight genes for which homology was established by simple BLAST to D. melanogaster (e value < 10−05), GO analysis suggested an elevation in carbohydrate metabolic process in VQ18 (P = 0.008).
The 93 genes with lower expression in VQ18 compared with SQ18 (74 orthologs) were enriched for the GO terms protein localization in organelle (P = 0.004), organelle fission (P = 0.022), and 14 related BP. The 257 genes with lower expression in SQ18 (186 orthologs) were enriched for more diverse categories, which were summarized by the following terms: Neurological system process, muscle cell development, phototransduction, cyclic nucleotide metabolic process (P < 0.001), alcohol catabolic process, and homeostatic process (P < 0.01, a complete list of enriched terms is provided in supplementary file S2, Supplementary Material online).
An enrichment in nucleobase, nucleoside, nucleotide, and nucleic acid metabolism (P = 0.045) was found in the four genes more highly expressed in SQ18 compared with MQ18 (four orthologs).
Overlap of Age- and Mating-Associated Expression Patterns
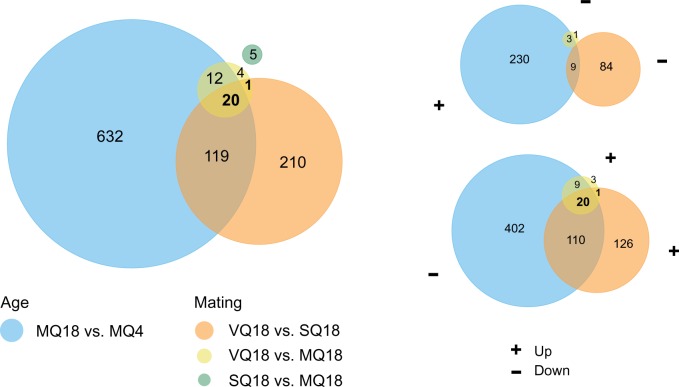
We determined the overlap of age- and mating-associated expression differences by comparing all four lists of DEGs (fig. 2). We found the highest overlap between genes with higher expression in MQ4 relative to MQ18 and genes with higher expression in VQ18 than in MQ18 or SQ18 (fig. 2, top right). Fewer genes showed the opposite pattern, that is, had lower expression in MQ4 and VQ18 than in MQ18 (bottom right).
Fig. 2.
Overlap of age- and mating-associated expression patterns. Venn diagrams showing the general overlap between all four comparisons (left) and specifically the overlap of genes upregulated with age in the mated queen type and downregulated in virgin compared with mated or sham-mated queens (top right) as well as vice versa (bottom right).
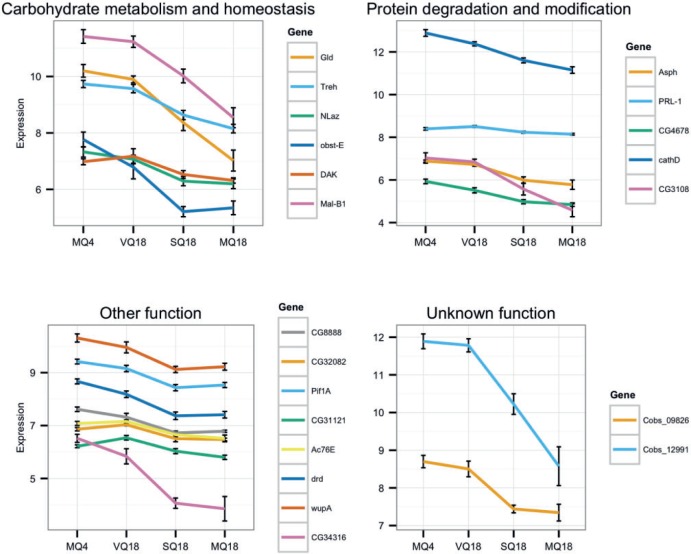
Twenty-one genes were differentially expressed in both the comparison of VQ18–MQ18 and VQ18–SQ18. They presumably reflect physiological changes as consequence of mating rather than of fertilization. All of them had higher transcript abundances in the shorter-lived phenotype, and all except PRL-1 were more highly expressed in MQ4 than in MQ18 (fig. 3). Thirteen genes have orthologs in D. melanogaster (table 2). Including five genes with putative homologs in D. melanogaster yielded a significant enrichment in carbohydrate metabolic process (P = 0.024). Across all three treatments, expression of these 21 genes showed a minimum at intermediate egg-laying rates instead of a linear relationship (supplementary file S3, Supplementary Material online).
Fig. 3.
Expression of all 21 mating-associated genes across all four conditions. Illustrated are mean and standard errors of log2-values of normalized counts. Genes are named according to Drosophila melanogaster orthologs/homologs if present and grouped by functions.
Table 2.
List of Genes Downregulated by Mating.
| Gene ID | Fold Change |
Drosophila melanogaster (o, ortholog; h, homolog) |
||||
|---|---|---|---|---|---|---|
| VQ18 versus MQ18 | VQ18 versus SQ18 | Name | Human Homologs | Function | ||
| Cobs_05812 | 1.5 | 1.5 | NLaz | o | APOD | Carbohydrate and triglyceride homeostasis |
| Cobs_09609 | 1.5 | 1.3 | Ac76E | o | ADCY2, ADCY4, ADCY7 | Negative regulation of growth |
| Cobs_03273 | 2.1 | 1.6 | Trehalase | h | TREA | Carbohydrate metabolism |
| Cobs_15800 | 2.2 | 1.5 | Maltase B1 | h | RBAT | Carbohydrate metabolism |
| Cobs_02493 | 2.1 | 1.5 | Glucose dehydrogenase | h | — | Carbohydrate metabolism |
| Cobs_06870 | 1.7 | 1.8 | Obstructor-E | o | — | Carbohydrate metabolism |
| Cobs_08335 | 1.6 | 1.4 | — | — | DAK | Carbohydrate metabolism |
| Cobs_10061 | 2.0 | 1.5 | cathD | o | CATD and others | Lysosomal proteolysis |
| Cobs_08331 | 1.5 | 1.4 | CG4678 | o | CBPD, CBPM | Proteolysis |
| Cobs_15592 | 2.5 | 1.7 | CG3108 | o | CBPA4 and other CBPs | Proteolysis |
| Cobs_16232 | 2.0 | 2.0 | CG34316 | h | — | Hemolymph JH binding |
| Cobs_01124 | 1.4 | 1.5 | CG8888 | o | BDH, DHI2 | Metabolism |
| Cobs_15266 | 1.5 | 1.6 | Wings up A | o | — | Muscle development and contraction |
| Cobs_05213 | 1.4 | 1.5 | PFTAIRE-interacting factor 1A | o | — | Regulation of transcription |
| Cobs_01240 | 1.3 | 1.2 | PRL-1 | o | PRL1, PRL2, PRL3 | Protein dephosphorylation |
| Cobs_03852 | 1.4 | 1.4 | CG32082 | o | BAIAP2, BI2L1, BI2L2 | Membrane organization |
| Cobs_05552 | 1.6 | 1.3 | CG31121 | o | — | Transport |
| Cobs_01171 | 1.6 | 1.4 | Aspartyl β-hydroxylase | o | ASPH | Protein modification |
| Cobs_14086 | 1.5 | 1.6 | Drop dead | h | — | Digestion, oogenesis |
| Cobs_09826 | 1.7 | 1.7 | — | — | — | — |
| Cobs_12991 | 2.1 | 1.5 | — | — | — | — |
Note.—Corresponding D. melanogaster orthologs, human homologs and fold changes of both VQ18–MQ18 and VQ18–SQ18 comparisons are indicated.
Comparison of Genes Differentially Expressed with Age in Mated Queens and Fruit Fly Females
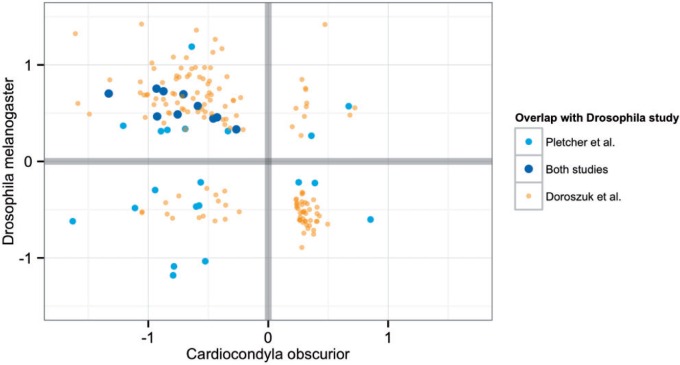
We performed a quantitative comparison of age-related gene expression changes in C. obscurior and D. melanogaster females by including expression differences between young and aged females (about 65% survival; Pletcher et al. 2002) as well as between young and extremely old females (10% survival; Doroszuk et al. 2012). The D. melanogaster studies resembled each other regarding the lists of genes upregulated in older individuals (table 3). A significant number of these upregulated genes were downregulated in aged C. obscurior queens. Ten genes were consistently upregulated in flies and downregulated in ant queens (fig. 4). Analysis of GO terms suggested that cellular ketone, carbohydrate, and organic acid metabolic processes are oppositely regulated in aging ant and fly females (table 4). Furthermore, transcripts, which contribute to the development and contraction of muscles, were less abundant in MQ18, but more highly abundant in extreme old flies. Genes that were increasingly expressed with age in C. obscurior, but downregulated in very old D. melanogaster, play a role in cell division and reproduction.
Table 3.
Overlap of Genes Found to Be Upregulated (+) or Downregulated (−) with Age in Cardiocondyla obscurior and Drosophila melanogaster Females (Pletcher et al. 2002; Doroszuk et al. 2012).
| MQ18 versus MQ4 |
Pletcher et al. |
|||||
|---|---|---|---|---|---|---|
| + | − | + | − | |||
| DEGs | 160 | 336 | 176 | 134 | ||
| Pletcher et al. (65 % survival) | + | 176 | 2 (1) | 16 (0.026) | ||
| − | 134 | 3 (1) | 10 (0.2) | |||
| Doroszuk et al. (10 % survival) | + | 648 | 12 (1) | 84 (2.6e-15) | 50 (1.0e-12) | 22 (0.018) |
| − | 1,233 | 43 (0.009) | 16 (1) | 19 (1) | 7 (1) | |
Note.—Number of common genes are shown with statistical significance as extracted from Fisher’s exact tests and FDR-correction in parentheses. Significant overlaps are italicized.
Fig. 4.
Relation of expression changes (log2 fold changes = lfc) of genes showing age-regulated transcription in Cardiocondyla obscurior and Drosophila melanogaster. Ten genes were consistently upregulated in aged (Pletcher et al. 2002) and very old flies (Doroszuk et al. 2012) and downregulated in ant queens: ref(2)P, emp, IscU, P5cr-2, CAP, CCHa2, CG3168, CG13124, CG9701, CG11796. For these genes, the mean lfc of both analyses is given.
Table 4.
Overlap of Significant GO Terms in Genes Upregulated (+) or Downregulated (−) with Age in Cardiocondyla obscurior (CO) and Drosophila melanogaster (DM).
| Biological Process | GO Code | CO | DM | CO | DM | |
|---|---|---|---|---|---|---|
| Gene expression changes in opposite directions | ||||||
| Metabolism | ||||||
| Oxidation reduction | GO:0055114 | − | + | 39 (306) | 42/115 (620) | |
| Cellular ketone metabolism | GO:0042180 | − | + | 19 (195) | 21/51 (290) | |
| Carbohydrate metabolism | GO:0005975 | − | + | 16 (193) | 24/73 (393) | |
| Carbohydrate catabolism | GO:0016052 | − | + | 10 (39) | 21 (83) | |
| Cellular carbohydrate metabolism | GO:0044262 | − | + | 11 (109) | 12 (182) | |
| Monosaccharide metabolism | GO:0005996 | − | + | 8 (55) | 10 (97) | |
| Hexose metabolism | GO:0019318 | − | + | 8 (46) | 10 (85) | |
| Glucose metabolism | GO:0006006 | − | + | 8 (33) | 9 (61) | |
| Organic acid metabolism | GO:0006082 | − | + | 19 (181) | 21/50 (270) | |
| Oxoacid metabolism | GO:0043436 | |||||
| Carboxylic acid metabolism | GO:0019752 | |||||
| Cellular amino acid metabolism | GO:0006520 | − | + | 16 (145) | 32 (199) | |
| Amine metabolism | GO:0009308 | − | + | 13 (117) | 28 (151) | |
| Cellular amine metabolism | GO:0044106 | − | + | 18 (188) | 60 (332) | |
| Aromatic amino acid family metabolism | GO:0009072 | − | + | 5 (14) | 8 (19) | |
| Alcohol metabolism | GO:0006066 | − | + | 16 (103) | 16 (173) | |
| Alcohol catabolism | GO:0046164 | − | + | 9 (26) | 5 (51) | |
| DNA metabolism | GO:0006259 | + | − | 9 (159) | 110 (221) | |
| DNA packaging | GO:0006323 | + | − | 5 (42) | 28 (65) | |
| Development/Reproduction | ||||||
| Anatomical structure formation | GO:0048646 | + | − | 12 (203) | 16/63 (369) | |
| Myofibril assembly | GO:0030239 | − | −/+ | 4 (11) | 3/8 (14) | |
| Actomyosin structure organization | GO:0031032 | − | + | 5 (23) | 10 (38) | |
| Mesoderm development | GO:0007498 | − | + | 8 (59) | 21 (91) | |
| Cell differentiation | GO:0030154 | + | − | 26 (636) | 158 (1,060) | |
| Anterior/posterior axis specification | GO:0009948 | + | − | 8 (84) | 28 (147) | |
| Reproduction | GO:0000003 | + | − | 21 (408) | 126 (848) | |
| Reproductive process | GO:0022414 | |||||
| Female gamete generation | GO:0007292 | + | − | 15 (286) | 96 (546) | |
| Cell cycle | GO:0007049 | + | − | 22 (349) | 173 (616) | |
| Cell cycle checkpoint | GO:0000075 | + | − | 5 (20) | 17 (24) | |
| Cell cycle phase | GO:0022403 | + | − | 16 (270) | 136 (496) | |
| M phase | GO:0000279 | + | − | 15 (255) | 126 (478) | |
| Meiotic M phase | GO:0051327 | + | − | 7 (72) | 49 (200) | |
| Mitotic M phase | GO:0000087 | + | − | 8 (92) | 49 (148) | |
| Cell division | GO:0051301 | + | − | 13 (136) | 61 (205) | |
| Asymmetric cell division | GO:0008356 | + | − | 6 (47) | 15 (58) | |
| Chromosome segregation | GO:0007059 | + | − | 9 (73) | 54 (117) | |
| Organelle fission | GO:0048285 | + | − | 8 (97) | 59 (153) | |
| Other processes | ||||||
| Muscle system process | GO:0003012 | − | + | 5 (10) | 7 (13) | |
| Muscle contraction | GO:0006936 | |||||
| RNA localization | GO:0006403 | − | + | 6 (80) | 39 (125) | |
| Macromolecular complex assembly | GO:0065003 | + | − | 10 (177) | 75 (246) | |
| Regulation of programmed cell death | GO:0043067 | + | − | 6 (75) | 22 (111) | |
| Negative regulation of biological process | GO:0048519 | + | − | 17 (369) | 116 (604) | |
| Negative regulation of cellular process | GO:0048523 | + | − | 15 (323) | 97 (538) | |
| Gene expression changes in same direction | ||||||
| Cell fate determination | GO:0001709 | + | + | 8 (89) | 22 (129) | |
| Neurogenesis | GO:0022008 | + | + | 16 (347) | 76 (499) | |
| Generation of neurons | GO:0048699 | + | + | 15 (332) | 69 (477) | |
| Anatomical structure homeostasis | GO:0060249 | + | + | 4 (31) | 10 (43) | |
Note.—The number of annotated genes for each GO category contained in the lists of DEGs is given in the left-most columns together with whole-genome annotations for both organisms in parentheses. Underlined, both D. melanogaster studies (first number corresponds to Pletcher et al., second to Doroszuk et al.); italic, Doroszuk et al. only.
Genes expressed in the same direction did not overlap significantly, but showed a significant enrichment in cell differentiation (P = 0.004). In addition, cell fate determination, neurogenesis, and anatomical structure homeostasis were identified as processes upregulated with age in both species.
Cross-Species Comparison of Mating- and Reproduction-Associated Transcriptomic Changes
We compared our data with several previous studies in D. melanogaster and honeybees, Apis mellifera, which focused on short-term gene expression changes linked to mating or reproduction (supplementary file S4, Supplementary Material online). Though we examined queens only several weeks after mating, we found significant overlap of the DEGs in these studies with our DEGs in VQ18 versus SQ18, but not with the other two contrasts (table 5). Genes with higher expression in VQ18 were significantly enriched for genes downregulated by sperm, but surprisingly also for genes upregulated by accessory gland proteins in female fruit flies. Likewise, genes downregulated in the brains of mated honeybee queens and genes downregulated in reproductive honeybee workers were overrepresented in the list of genes with higher expression in VQ18. A GO term enrichment analysis revealed that the expression of genes involved in muscle development and contraction is consistently reduced by mating in C. obscurior queens and D. melanogaster females and by the onset of reproduction in A. mellifera workers. Furthermore, significantly more genes were found to be upregulated in SQ18 and brains of incompletely mated honeybee queens (intermediate) compared with virgin individuals than expected by chance.
Table 5.
Overlap of Genes Found to be Differentially Expressed due to Mating and/or the Onset of Reproduction in Cardiocondyla obscurior, Drosophila melanogaster, and Apis mellifera (−, downregulated; +, upregulated).
| VQ18 versus SQ18 |
VQ18 versus MQ18 |
SQ18 versus MQ18 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| + | − | + | − | + | − | ||||
| Drosophila melanogaster orthologs (6,959) | DEGs | 186 | 74 | 19 | 1 | 4 | 1 | ||
| Whole Female (McGraw 2004) | Sperm | + | 139 | 3 (0.9) | 1 | 0 | 0 | 0 | 0 |
| − | 152 | 16 (3.8e-5) | 2 (0.8) | 2 (0.2) | 0 | 0 | 0 | ||
| Acps | + | 41 | 5 (0.027) | 1 | 0 | 0 | 0 | 0 | |
| − | 29 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Nonsperm/Non-Acps | + | 317 | 13 (0.2) | 2 (0.3) | 0 | 0 | 0 | 0 | |
| − | 196 | 11 (0.4) | 0 | 0 | 0 | 0 | 0 | ||
| Apis mellifera othologs (7,948) | DEGs | 215 | 80 | 27 | 3 | 4 | 1 | ||
| Queen Brain (Kocher et al. 2008) | Virgin versus Mated | + | 42 | 1 | 0 | 0 | 0 | 0 | 0 |
| − | 30 | 1 | 2 (0.1) | 0 | 0 | 0 | 0 | ||
| Virgin versus Intermediate | + | 66 | 3 (0.4) | 1 | 0 | 0 | 0 | 0 | |
| − | 336 | 2 (1) | 12 (0.001) | 0 | 0 | 0 | 0 | ||
| Intermediate versus Mated | + | 261 | 1 | 4 (0.4) | 0 | 0 | 0 | 0 | |
| − | 100 | 3 (0.7) | 1 | 0 | 0 | 0 | 0 | ||
| Queen Ovary (Kocher et al. 2008) | Virgin versus Mated | + | 74 | 2 (0.8) | 0 | 1 | 0 | 0 | 0 |
| − | 11 | 0 | 1 | 0 | 0 | 0 | 0 | ||
| Virgin versus Intermediate | + | 119 | 2 (1) | 2 (0.5) | 1 | 0 | 0 | 0 | |
| − | 26 | 1 | 0 | 0 | 0 | 0 | 0 | ||
| Intermediate versus Mated | + | 18 | 0 | 0 | 0 | 0 | 0 | 0 | |
| − | 19 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Queen Brain (Kocher et al. 2010) | Virgin versus Mated | + | 84 | 9 (0.003) | 1 | 1 | 0 | 0 | 0 |
| − | 45 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Worker Brain (Grozinger et al. 2007) | Sterile versus Reproductive | + | 21 | 1 | 0 | 0 | 0 | 0 | 0 |
| − | 59 | 4 (0.2) | 0 | 0 | 0 | 0 | 0 | ||
| Whole Worker (Cardoen et al. 2011) | Sterile versus Reproductive | + | 410 | 39 (8.9e-11) | 2 (1) | 3 (0.3) | 0 | 0 | 0 |
| − | 632 | 5 (1) | 12 (0.1) | 1 | 0 | 1 | 0 | ||
| Worker Ovary (Wang et al. 2012) | Low pollen versus high pollen strain | + | 616 | 25 (0.1) | 2 (1) | 0 | 0 | 0 | 0 |
| − | 1,140 | 21 (1) | 18 (0.1) | 3 (1) | 0 | 2 (0.2) | 0 | ||
Note.—Fisher’s exact tests were performed on comparisons revealing at least two common genes, resulting P values after FDR-correction are shown in parenthesis. Significant overlaps are italicized.
Genes Related to Aging in D. melanogaster
We screened the C. obscurior genome for 136 genes with documented effects on longevity in D. melanogaster and found 95 orthologs. Eight of them showed differential expression with age or mating status or both (table 6). Neural Lazarillo (NLaz) was identified to be less expressed in both SQ18 and MQ18 types. Nlaz and Sirt6 were oppositely expressed in ant queens and fly females with regard to age, whereas rutabaga and Muscle LIM protein at 84B were regulated in the same direction.
Table 6.
Drosophila melanogaster Aging Candidate Genes Showing Differential Expression (−, downregulated; +, upregulated) with Age and/or Mating in Cardiocondyla obscurior (CO).
| Gene ID | Name | Longevity Effect | Age |
Mating |
||
|---|---|---|---|---|---|---|
| DM | CO | DM | CO | AM | ||
| Age-specific changes in opposite directions | ||||||
| Cobs_05812 | NLaz | Pro | − | + (4) | VQ18 > SQ18 and MQ18 | Virgin > Mated (3) |
| Cobs_12553 | Sirt6 | Pro | + | − (4) | Intermediate < Mated (2) | |
| Age-specific changes in same direction | ||||||
| Cobs_05934 | Muscle LIM protein at 84B | Pro | − | − (1,5) | VQ18 > SQ18 | |
| Cobs_00579 | rutabaga | Pro | − | − (4) | ||
| Other pattern | ||||||
| Cobs_15768 | InR | Anti | + | |||
| Cobs_03800 | Myospheroid | Anti | + (4) | VQ18 > SQ18 | ||
| Cobs_01798 | Parkin | Pro | VQ18 < SQ18 | Virgin < Intermediate (2, brain) | ||
| Cobs_04768 | CG3776 | Anti+pro | VQ18 < SQ18 | Virgin > Intermediate and Mated (2, ovary) | ||
Note.—The following references provided information about age- and mating-related expression in D. melanogaster (DM) and Apis mellifera (AM): (1) Pletcher et al. (2002), (2) Kocher et al. (2008), (3) Kocher et al. (2010), (4) Doroszuk et al. (2012), (5) Zhou et al. (2014).
Discussion
Opposite Gene Expression Changes in Aging Queens and Female Flies
Female reproductives of social insects appear to suffer less “mortality costs” from mating and reproduction than females of solitary insects (Partridge et al. 1987; Trevitt and Partridge 1991; Eady et al. 2007). On the contrary, mating extends the lifespan of Cardiocondyla ant queens (Schrempf et al. 2005) and their longevity increases with egg laying rate (Heinze and Schrempf 2012; Heinze et al. 2013; A. Schrempf, unpublished data). Evolutionary theories of aging explain the long lifespan of social insect queens and the absence of the fecundity/longevity trade-off from their low extrinsic mortality, as queens live in the relative safety of well-protected, often subterraneous, nests (Keller and Genoud 1997; Carey 2001). Proximately, this suggests an alternative regulation of the conserved pathways that mediate the interplay among mating, egg laying, and aging. Our study provides support for this hypothesis at the transcriptome level: Age-related changes in gene expression had opposite directions in two taxa with opposite life histories.
Transcriptional changes of aged female (Pletcher et al. 2002; Doroszuk et al. 2012) and male (Zou et al. 2000; Girardot et al. 2006) flies reflect their decline in reproductive capacity (e.g., Tatar et al. 1996). In contrast, the observation that C. obscurior queens increase their reproductive efforts with age and show reproductive senescence only immediately before they die, if at all (Heinze and Schrempf 2012), is consistent with the differences between the transcriptomes of young and older C. obscurior queens found in this study.
Furthermore, aged and extremely old female D. melanogaster exhibit a higher expression of genes involved in cellular ketone, carbohydrate, and organic acid metabolism than young female flies, whereas these genes were downregulated in older relative to young C. obscurior queens. The decline of muscle formation and contraction in aging C. obscurior queens is consistent with the adaptation to a stationary mode of life and might contribute to save energy. Together with the downregulation of metabolism genes, it might also delay the accumulation of physiological damage. Reproductives of several social insects have lower levels and activities of oxidant enzymes than nonreproductives, which might indicate a reduced generation of oxygen radicals (Parker et al. 2004; Corona et al. 2005; Schneider et al. 2011), perhaps because of reduced metabolism.
Common Gene Expression Changes in Aging Queens and Female Flies
Aging is largely regarded as the result of wear and tear. At the same time, the overlap of gene expression changes found during aging and during development in mammals suggests that aging is a regulated process under genetic control (de Magalhães 2012). From this point of view, genes showing age-specific expression changes in the same direction across taxa might be universal regulators of aging. Here, we identified the downregulation of rut and Mlp84B and the upregulation of genes involved in cell fate determination, neurogenesis, and anatomical structure homeostasis as common signatures of aging. This result supports the idea that developmental processes might continue beyond maturity and become detrimental later in life when selection is relaxed (de Magalhães and Church 2005). In Caenorhabditis elegans, age-related expression changes are controlled by three transcription factors, which are not affected by the accumulation of damage (Budovskaya et al. 2008). Extending this theory to our study, selection for late reproduction in social insect queens might specifically prevent the drift or cessation of developmental programs, which optimize reproduction, such as insulin signaling.
Gene Expression Changes Associated with the Lifespan-Prolonging Effect of Mating
A comparison of the transcriptomes among 18-week-old egg-laying virgin queens, mated queens, and queens mated with sterilized males yielded additional insight into the effects of mating and reproduction. Cardiocondyla obscurior queens that are unmated (or sham-mated) and lay only few eggs are tolerated in the colony and receive the same treatment from workers as more fecund, mated queens (Schrempf et al. 2005, 2011). Nevertheless, both mated and sham-mated queens live significantly longer than virgin queens do. Consistent with this phenotypic similarity, MQ18 and SQ18 differed in their transcriptomic profiles in only five genes. Interestingly, the transcriptomic profile of VQ18 was more similar to the profile of MQ18 than to the profile of SQ18 considering the number of gene expression differences. Similar to our results, Kocher et al. (2008) found more DEGs between unmated and “intermediate” queens than between unmated and mated, egg-laying queens. Furthermore, significant overlap of genes downregulated in SQ18 compared with VQ18 with genes downregulated by sperm in D. melanogaster (McGraw et al. 2004) and by mating in brains of honey bees (Kocher et al. 2008, 2010) indicates that short- and long-term consequences of the mating event are similar, even across taxa. However, our analysis revealed that genes downregulated by sham-mating in ant queens contained a significant part of genes upregulated by the transfer of accessory gland proteins during mating in flies (McGraw et al. 2004).
The differential regulation of conserved, public mechanisms may relate to lifespan regulation in these different biological contexts, although presumably with opposing consequences. This corresponds to the contrasting effect of mating on longevity in these taxa. Out of the 257 genes with higher expression in VQ18 compared with SQ18, 21 genes had also significant higher expression than in MQ18. Given that sham-mated queens live as long as mated queens and at the same time display low fecundity similar to that of virgin queens, these genes are particularly interesting because they might be correlated with the different speed of aging. The five carbohydrate-degrading and proteolytic enzymes Trehalase, Maltase B1, cathD, CG4678, and CG3108 point to a reduced need of these energy resources in mated queens. The differential expression of Trehalase, Maltase B1, NLaz, obstructor-E, Glucose dehydrogenase and a homolog of human DAK give further support that mating has an effect on carbohydrate metabolism and homeostasis. In addition, NLaz, Adenylyl cyclase 76E (Ac76E), and a JH binding protein (homolog to CG34316) indicate an involvement of the Insulin/IGF signaling pathway (IIS) and JH. Noticeably, our data do not hint at a major role of vitellogenin in regulating fecundity or longevity in C. obscurior. So far, we identified Cobs_01486 as the only gene in the genome of this species possessing the vitellogenin domain (pfam01347). This gene is orthologous to the honeybee “vitellogenin-like” GB52464 and was moderately downregulated in MQ18 compared with MQ4 (fold change = 0.7).
Candidate Genes
The lipocalin NLaz, which is homologous to vertebrate Apolipoprotein D (APOD), was shown to promote metabolic homeostasis and tolerance to certain types of stress by repressing Insulin/IGF signaling (IIS) in the fly model (Hull-Thompson et al. 2009). Consequently, flies overexpressing NLaz have an extended lifespan at the expense of reduced growth (Hull-Thompson et al. 2009; Ruiz et al. 2011). Experiments with female flies predict that NLaz decreases food intake, decreases fat storage with age, increases locomotor activity, and enhances mating behavior. Ant queens depend on extensive energy intake for the continuous production of eggs. It is therefore not surprising that this gene is less expressed in older, more fertile queens than in younger queens. Furthermore, NLaz expression was reduced in mated C. obscurior and A. mellifera queens (Kocher et al. 2010) relative to virgin queens, indicating a regulatory function of postmating behavior and metabolism. Consistent with the expression pattern of NLaz, the differential regulation of Ac76E—a direct transcriptional target of foxo (Mattila et al. 2009)—indicates that IIS activity is lower in short-lived virgin queens. Corona et al. (2007) hypothesized that a reduction of IIS in the head of bee queens contributes to their longer lifespan compared with workers. In contrast, Insulin-like receptor (InR) was shown to be important for ovary development and reproduction in dipterans and ants (Tatar et al. 2001; Okada et al. 2010; Lu and Pietrantonio 2011). Our results, including the upregulation of InR in older, more fertile queens, point to the involvement of IIS but do not suggest a general reversal of the traditional relationship between nutrition and IIS as proposed for the honeybee (Corona et al 2007). Instead, we found that lifespan differences are accompanied by the differential expression of carbohydrate-metabolizing enzymes. This suggests that mating triggers a change in metabolism to allow a long life and maximize the reproductive output at the same time.
Conclusions
Our study reveals a number of genes that change expression with age and as a function of reproductive status. The important commonalities and differences in age-related expression changes between C. obscurior queens and D. melanogaster females may be of broad interest in the community of aging researchers working on diverse organisms.
The comparison among virgin, sham-mated, and mated queens shows how the effects of mating and fecundity on queen longevity can be separated and suggest a number of promising candidates for further in depth studies on the complex regulation of fundamental life-history traits in social insects.
Materials and Methods
The Study Organism
Cardiocondyla obscurior is a tropical tramp species (Heinze et al. 2006), which nests in cavities of dead twigs and leaves (Seifert 2002 ). Its successful establishment around the globe through human activities is possible because of several specific life-history traits, such as the continuous production of sexuals, the presence of multiple queens per nest, mating in the nest, and colony propagation by budding. These traits also facilitate rearing and maintenance of Cardiocondyla colonies in the lab. Its small colonies contain on average 20 female workers, several reproductive queens, and a single wingless male, which monopolizes mating with any newly produced queen by killing younger rival males reared in the colony (Kinomura and Yamauchi 1987; Stuart et al. 1987; Heinze and Delabie 2005).
Experimental Design and Sampling
We established 73 experimental colonies from laboratory stock colonies derived from the genome reference population in Bahia, Brazil (Schrader et al. 2014). Each nest contained 20 workers, 10 larvae, and a single queen pupa, which was assigned to one of three treatments: Mated (MQ), sham-mated (SQ), and virgin (VQ). MQ colonies were set up with an additional male pupa about to eclose simultaneously with the queen, whereas VQ did not have contact to males. For the SQ treatments, we sterilized the added male prior to its introduction to the nest by exposure to X-rays (120 G; 2.95 ± 0.12 G/min; Schrempf et al. 2005). Colonies with males that died within 1 week after irradiation were excluded from further analysis. Sterilized males transfer only unviable sperm and consequently SQ can only produce male offspring from unfertilized eggs (Schrempf et al. 2005). From these three treatments (MQ, SQ, and VQ), individuals were sampled after 18 weeks (MQ18, SQ18, and VQ18), corresponding to the age when 50% of virgin queens had died in a previous experiment (Schrempf et al. 2005). In addition, mated queens which were set up as the mated queens described above were sampled after 4 weeks (=MQ4; Schrempf et al. 2015) to assess age-related changes under normal circumstances. These young queens started to lay eggs 1 week after emergence and consistently increased their egg-laying rate within the 3 weeks before sampling.
The colonies were reared in Petri dishes with plaster and fed three times per week with chopped cockroaches and diluted honey according to standard protocols ad libitum. All eggs were counted and removed twice per week in the first month and subsequently once per week. The number of workers and larvae was standardized by adding or removing individuals to 20 workers and 10 larvae per colony. Developing male and queen pupae were removed to avoid replacement or (additional) mating of the focal queens. All queens were individually snap-frozen in liquid nitrogen and stored at −80 °C until further processing.
We monitored survival and reproductive output of queens. Mating type had no significant effect on the survival of queens until 18 weeks, but mean egg-laying rates differed significantly between all three queen types (details in supplementary file S5A, Supplementary Material online).
Library Preparation and Sequencing
Individual queens were transferred into RLT Plus buffer (QIAGEN) and homogenized by using Lysing Matrix Tubes and a FastPrep bead shaker (MP Biomedicals). Subsequently, total RNA was extracted following the RNeasy Plus Micro Kit protocol (QIAGEN). We measured RNA content and quality with an Agilent 2100 Bioanalyzer, which indicated yields of 20–200 ng of total RNA per queen. To obtain sufficient RNA to prepare a sequencing library, whole RNA was amplified after conversion into cDNA (NuGEN Ovation RNA-Seq System V2). After sonic fragmentation, and adapter ligation and incorporation of multiplex barcodes (NuGEN Encore Rapid Library Systems), the 28 samples were randomly distributed across different lanes of a flow cell and sequenced on an Illumina HiSeq1000 platform.
RNA-Seq Analysis
On average, 24 million 100-bp single reads were generated per sample. Quality of raw reads (phred scores > 30) was assessed by FastQC version 0.10.1 (Andrews 2010). Adapter residuals were trimmed with Cutadapt version 1.2.1 (Martin 2011). Using Bowtie2 version 2.1.0 (Langmead and Salzberg 2012) in combination with the splice junction mapper TopHat version 2.0.8 (Trapnell et al. 2009), the sequences were mapped with default settings against the C. obscurior reference genome Cobs1.4 (Schrader et al. 2014, mapping statistics in supplementary file S5B, Supplementary Material online). Subsequently, HTSeq-count version 0.5.4 (Anders et al. 2014) was used for counting reads. Normalization of raw counts and the tests for differential gene expression were performed with DESeq2 version 1.6.2 (Love et al. 2014 ) in R version 3.1.2 (R Core Team 2014). We tested MQ18 against MQ4 and then contrasted the three treatments MQ18, SQ18, and VQ18. Raw P values were adjusted for multiple testing (Benjamini and Hochberg 1995). A principal component analysis was conducted on expression values of the top 500 genes with the highest variance across all samples after variance stabilization in DESeq2. Centroids were added by means of the package Vegan version 2.2-1 (Oksanen et al. 2015). Area-proportional Venn diagrams were generated with EulerAPE (Micallef and Rodgers 2014).
Functional Annotation
We inferred orthology by applying a reciprocal BLASTp between all 17,552 predicted C. obscurior genes and the D. melanogaster (dmel-all-translation-r5.56.fasta) and A. mellifera (amel_OGSv1.1_pep.fa) genomes by means of the BLAST+ toolkit (Camacho et al. 2009). This resulted in 6,959 fruit fly and 7,948 honeybee orthologs, corresponding to 68% and 72% of the annotated fly or bee genes, respectively. For the remaining genes, the most similar homolog was defined as the best hit of the one-way protein BLAST against the fruit fly or honeybee genome on condition that the e value was smaller than 1 e-05. Functional annotation of genes was obtained by loading all 6,959 genes with reciprocal orthologous relationships to fly genes as background into DAVID (Huang et al. 2008). A modified Fisher’s exact test was used for testing of enrichment (EASE < 0.05) for GO terms (Ashburner et al. 2000) and KEGG pathways (Kanehisa and Goto 2000) in the sets of DEGs relative to the background. As the lists of DEGs between VQ18 and MQ18 and of the common genes between the contrasts VQ18-MQ18 and VQ18-SQ18 did not reveal a functional enrichment considering orthologs, we repeated the test by including homologs.
Cross-Species Comparisons
We compared our results with similar studies in insects that addressed mating- and aging-related transcriptome changes. We compared our data with two independent studies to determine overlap in age-related changes between C. obscurior queens and D. melanogaster females: A study of young and old virgin female D. melanogaster (90% vs. 10% survival) by Doroszuk et al. (2012) and a study that compared the transcriptomes of 7- and 23-day-old mated fly females (=aged females with about 65% survival) from Pletcher et al. (2002). We did not identify any other data sets that were suitable to compare age-related transcriptome changes in insect females.
We assessed six further data sets for similarities to our transcriptomic comparisons among mated, sham-mated, and virgin C. obscurior queens. Three of these data sets focused on mating-induced changes in gene expression of D. melanogaster females (McGraw et al. 2004) and A. mellifera queens (Kocher et al. 2008, 2010). The other three comparative data sets were derived from studies of reproduction- or ovary-status-associated gene expression changes in A. mellifera workers (Grozinger et al. 2007; Cardoen et al. 2011; Wang et al. 2012). We excluded another study (Zhou et al. 2014) because it did not report on a sufficient number of orthologs of our genes (<50%) to allow for a meaningful comparative analysis (see supplementary file S4, Supplementary Material online). We also screened our lists of DEGs for the presence of putative D. melanogaster aging candidates retrieved from the GenAge database (Tacutu et al. 2012).
The given identifiers were converted to the current gene annotations that we used for determining orthologs (as described above). Normalized log-transformed expression values from microarrays of two studies on flies (Pletcher et al. 2002; Doroszuk et al. 2012) were analyzed with limma version 3.22.4 (Ritchie et al 2015). The definition of DEGs was based on an false discovery rate (FDR) < 0.05 for all data sets, only Grozinger et al. (2007) applied a 97.5% confidence level cutoff. As for the functional enrichment analyses, we restricted the comparison to the set of unambiguous orthologs. Up- and downregulated genes were analyzed separately. To perform quantitative comparisons we generated contingency tables containing the number of DEGs found in both studies, the number of DEGs not found in the other study in each case and the number of all remaining genes. A one-sided Fisher’s exact test then revealed whether more genes overlapped than expected by chance.
Data Accessibility
Raw sequencing data have been deposited in SRA under the BioProject accession numbers PRJNA293450 (MQ18, SQ18, VQ18: SRR2177525–SRR2177544) and PRJNA284224 (MQ4: SRR2033894–SRR2033897, SRR2033903–SRR2033905).
Supplementary Material
Supplementary files S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft (He1623/31). Collection of colonies of C. obscurior was possible through permit SISBIO 20324–1. The study was also supported by Regensburger Universitätsstiftung and NIA (R21AG046837) to O.R. The authors thank Alexandra Schrempf, Abel Bernadou, and two anonymous referees for valuable comments on the manuscript. This research utilized Queen Mary’s MidPlus computational facilities, supported by QMUL Research-IT and funded by EPSRC grant EP/K000128/1.
References
- Anders S, Pyl PT, Huber W. 2014. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC. Available from: http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/. [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene Ontology: tool for the unification of biology. Nat Genet. 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Methodol. 57:289–300. [Google Scholar]
- Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, Kim SK. 2008. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell 134:291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoen D, Wenseleers T, Ernst UR, Danneels EL, Laget D, De Graaf DC, Schoofs L, Verleyen P. 2011. Genome-wide analysis of alternative reproductive phenotypes in honeybee workers. Mol Ecol. 20:4070–4084. [DOI] [PubMed] [Google Scholar]
- Carey JR. 2001. Demographic mechanisms for the evolution of long life in social insects. Exp Gerontol. 36:713–722. [DOI] [PubMed] [Google Scholar]
- Corona M, Hughes KA, Weaver DB, Robinson GE. 2005. Gene expression patterns associated with queen honey bee longevity. Mech Ageing Dev. 126:1230–1238. [DOI] [PubMed] [Google Scholar]
- Corona M, Velarde RA, Remolina S, Moran-Lauter A, Wang Y, Hughes KA, Robinson GE. 2007. Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. Proc Natl Acad Sci U S A. 104:7128–7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhães JP. 2012. Programmatic features of aging originating in development: aging mechanisms beyond molecular damage? FASEB J. 26:4821–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhães JP, Church GM. 2005. Genomes optimize reproduction: aging as a consequence of the developmental program. Physiology 20:252–259. [DOI] [PubMed] [Google Scholar]
- Doroszuk A, Jonker MJ, Pul N, Breit TM, Zwaan BJ. 2012. Transcriptome analysis of a long-lived natural Drosophila variant: a prominent role of stress-and reproduction-genes in lifespan extension. BMC Genomics 13:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eady PE, Hamilton L, Lyons RE. 2007. Copulation, genital damage and early death in Callosobruchus maculatus. Proc R Soc Lond B Biol Sci. 274:247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. 2000. Oxidants, oxidative stress, and the biology of aging. Nature 408:239–247. [DOI] [PubMed] [Google Scholar]
- Flatt T. 2011. Survival costs of reproduction in Drosophila. Exp Gerontol. 46:369–375. [DOI] [PubMed] [Google Scholar]
- Flatt T, Kawecki TJ. 2007. Juvenile hormone as a regulator of the trade-off between reproduction and life span in Drosophila melanogaster. Evolution 61:1980–1991. [DOI] [PubMed] [Google Scholar]
- Flatt T, Tu MP, Tatar M. 2005. Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. Bioessays 27:999–1010. [DOI] [PubMed] [Google Scholar]
- Girardot F, Lasbleiz C, Monnier V, Tricoire H. 2006. Specific age related signatures in Drosophila body parts transcriptome. BMC Genomics 7:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozinger CM, Fan Y, Hoover SE, Winston ML. 2007. Genome-wide analysis reveals differences in brain gene expression patterns associated with caste and reproductive status in honey bees (Apis mellifera). Mol Ecol. 16:4837–4848. [DOI] [PubMed] [Google Scholar]
- Haddad LS, Kelbert L, Hulbert AJ. 2007. Extended longevity of queen honey bees compared to workers is associated with peroxidation-resistant membranes. Exp Gerontol. 42:601–609. [DOI] [PubMed] [Google Scholar]
- Havukainen H, Münch D, Baumann A, Zhong S, Halskau Ø, Krogsgaard M, Amdam GV. 2013. Vitellogenin recognizes cell damage through membrane binding and shields living cells from reactive oxygen species. J Biol Chem. 288:28369–28381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinze J, Cremer S, Eckl N, Schrempf A. 2006. Stealthy invaders: the biology of Cardiocondyla tramp ants. Insectes Soc. 53:1–7. [Google Scholar]
- Heinze J, Delabie J. 2005. Population structure of the male-polymorphic ant Cardiocondyla obscurior. Stud Neotrop Fauna Environ. 40:187–190. [Google Scholar]
- Heinze J, Frohschammer S, Bernadou A. 2013. Queen life-span and total reproductive success are positively associated in the ant Cardiocondyla cf. kagutsuchi. Behav Ecol Sociobiol. 67:1555–1562. [Google Scholar]
- Heinze J, Hölldobler B. 1993. Fighting for a harem of queens: physiology of reproduction in Cardiocondyla male ants. Proc Natl Acad Sci U S A. 90:8412–8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinze J, Schrempf A. 2008. Aging and reproduction in social insects—a mini-review. Gerontology 54:160–167. [DOI] [PubMed] [Google Scholar]
- Heinze J, Schrempf A. 2012. Terminal investment: individual reproduction of ant queens increases with age. PLoS One 7:e35201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2008. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Hughes KA, Reynolds RM. 2005. Evolutionary and mechanistic theories of aging. Annu Rev Entomol. 50:421–445. [DOI] [PubMed] [Google Scholar]
- Hull-Thompson J, Muffat J, Sanchez D, Walker DW, Benzer S, Ganfornina MD, Jasper H. 2009. Control of metabolic homeostasis by stress signaling is mediated by the lipocalin NLaz. PLoS Genet. 5:e1000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemielity S, Chapuisat M, Parker JD, Keller L. 2005. Long live the queen: studying aging in social insects. Age 27:241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemielity S, Kimura M, Parker KM, Parker JD, Cao X, Aviv A, Keller L. 2007. Short telomeres in short-lived males: what are the molecular and evolutionary causes? Aging Cell 6:225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller L. 1998. Queen lifespan and colony characteristics in ants and termites. Insectes Soc. 45:235–246. [Google Scholar]
- Keller L, Genoud M. 1997. Extraordinary lifespans in ants: a test of evolutionary theories of ageing. Nature 389:958–960. [Google Scholar]
- Kinomura K, Yamauchi K. 1987. Fighting and mating behaviors of dimorphic males in the ant. J Ethol. 5:75–81. [Google Scholar]
- Kocher SD, Richard FJ, Tarpy DR, Grozinger CM. 2008. Genomic analysis of post-mating changes in the honey bee queen (Apis mellifera). BMC Genomics 9:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher SD, Tarpy DR, Grozinger CM. 2010. The effects of mating and instrumental insemination on queen honey bee flight behaviour and gene expression. Insect Mol Biol. 19:153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu HL, Pietrantonio PV. 2011. Insect insulin receptors: insights from sequence and caste expression analyses of two cloned hymenopteran insulin receptor cDNAs from the fire ant. Insect Mol Biol. 20:637–649. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17:10. [Google Scholar]
- Mattila J, Bremer A, Ahonen L, Kostiainen R, Puig O. 2009. Drosophila FoxO regulates organism size and stress resistance through an adenylate cyclase. Mol Cell Biol. 29:5357–5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw LA, Gibson G, Clark AG, Wolfner MF. 2004. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr Biol. 14:1509–1514. [DOI] [PubMed] [Google Scholar]
- Micallef L, Rodgers P. 2014. eulerAPE: drawing area-proportional 3-Venn diagrams using ellipses. PLoS One 9:e101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michod RE, Viossat Y, Solari CA, Hurand M, Nedelcu AM. 2006. Life-history evolution and the origin of multicellularity. J Theor Biol. 239:257–272. [DOI] [PubMed] [Google Scholar]
- Okada Y, Miyazaki S, Miyakawa H, Ishikawa A, Tsuji K, Miura T. 2010. Ovarian development and insulin-signaling pathways during reproductive differentiation in the queenless ponerine ant Diacamma sp. J Insect Physiol. 56:288–295. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Guillaume Blanchet F, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, MHH Stevens, Wagner H. 2015. vegan: Community Ecology Package. R package version 2.2-1. Available from: http://CRAN.R-project.org/package=vegan. [Google Scholar]
- Parker JD, Parker KM, Sohal BH, Sohal RS, Keller L. 2004. Decreased expression of Cu–Zn superoxide dismutase 1 in ants with extreme lifespan. Proc Natl Acad Sci U S A. 101:3486–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Gems D, Withers DJ. 2005. Sex and death: what is the connection? Cell 120:461–472. [DOI] [PubMed] [Google Scholar]
- Partridge L, Green A, Fowler K. 1987. Effects of egg-production and of exposure to males on female survival in Drosophila melanogaster. J Insect Physiol. 33:745–749. [Google Scholar]
- Pletcher SD, Macdonald SJ, Marguerie R, Certa U, Stearns SC, Goldstein DB, Partridge L. 2002. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr Biol. 12:712–723. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2014. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; Available from: http://www.R-project.org/. [Google Scholar]
- Remolina SC, Hughes KA. 2008. Evolution and mechanisms of long life and high fertility in queen honey bees. Age 30:177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MR. 1990. Evolutionary biology of aging. New York: Oxford University Press. [Google Scholar]
- Ruiz M, Sanchez D, Canal I, Acebes A, Ganfornina MD. 2011. Sex-dependent modulation of longevity by two Drosophila homologues of human Apolipoprotein D, GLaz and NLaz. Exp Gerontol. 46:579–589. [DOI] [PubMed] [Google Scholar]
- Schmidt CM., Jarvis JU, Bennett NC. 2013. The long-lived queen: reproduction and longevity in female eusocial Damaraland mole-rats (Fukomys damarensis): short communication. Afr Zool. 48:193–196. [Google Scholar]
- Schneider SA, Schrader C, Wagner AE, Boesch-Saadatmandi C, Liebig J, Rimbach G, Roeder T. 2011. Stress resistance and longevity are not directly linked to levels of enzymatic antioxidants in the ponerine ant Harpegnathos saltator. PLoS One 6:e14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader L, Kim JW, Ence D, Zimin A, Klein A, Wyschetzki K, Weichselgartner T, Kemena C, Stökl J, Schultner E, et al. 2014. Transposable element islands facilitate adaptation to novel environments in an invasive species. Nat Commun. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrempf A, Cremer S, Heinze J. 2011. Social influence on age and reproduction: reduced lifespan and fecundity in multi-queen ant colonies. J Evol Biol. 24:1455–1461. [DOI] [PubMed] [Google Scholar]
- Schrempf A, Heinze J, Cremer S. 2005. Sexual cooperation: mating increases longevity in ant queens. Curr Biol. 15:267–270. [DOI] [PubMed] [Google Scholar]
- Schrempf A, Wyschetzki K, Klein A, Schrader L, Oettler J, Heinze J. 2015. Mating with an allopatric male triggers immune response and decreases longevity of ant queens. Mol Ecol. 24:3618–3627. [DOI] [PubMed] [Google Scholar]
- Seehuus SC, Norberg K, Gimsa U, Krekling T, Amdam GV. 2006. Reproductive protein protects functionally sterile honey bee workers from oxidative stress. Proc Natl Acad Sci U S A. 103:962–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert B. 2002. The ant genus Cardiocondyla (Insecta: Hymenoptera: Formicidae)—a taxonomic revision of the C. elegans, C. bulgarica, C. batesii, C. nuda, C. shuckardi, C. stambuloffii, C. wroughtonii, C. emeryi, and C. minutior species groups. Ann Nat Hist Mus Wien Ser B Bot Zool. 104:203–338. [Google Scholar]
- Stearns SC. 1989. Trade-offs in life-history evolution. Funct Ecol. 3:259–268. [Google Scholar]
- Stuart RJ, Francoeur A, Loiselle R. 1987. Lethal fighting among dimorphic males of the ant, Cardiocondyla wroughtonii. Naturwissenschaften 74:548–549. [Google Scholar]
- Tabatabaie V, Atzmon G, Rajpathak SN, Freeman R, Barzilai N, Crandall J. 2011. Exceptional longevity is associated with decreased reproduction. Aging (Albany NY) 3:1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacutu R, Craig T, Budovsky A, Wuttke D, Lehmann G, Taranukha D, Costa J, Fraifeld VE, de Magalhães JP. 2012. Human ageing genomic resources: integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. 2001. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292:107–110. [DOI] [PubMed] [Google Scholar]
- Tatar M, Promislow DE, Khazaeli AA, Curtsinger JW. 1996. Age-specific patterns of genetic variance in Drosophila melanogaster. II. Fecundity and its genetic covariance with age-specific mortality . Genetics 143:849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevitt S, Partridge L. 1991. A cost of receiving sperm in the female fruitfly Drosophila melanogaster. J Insect Physiol. 37:471–475. [Google Scholar]
- Wang Y, Kocher SD, Linksvayer TA, Grozinger CM, Page RE, Amdam GV. 2012. Regulation of behaviorally associated gene networks in worker honey bee ovaries. J Exp Biol. 215:124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorp RG, Kirkwood TB. 1998. Human longevity at the cost of reproductive success. Nature 396:743–746. [DOI] [PubMed] [Google Scholar]
- Zhou S, Mackay TF, Anholt RR. 2014. Transcriptional and epigenetic responses to mating and aging in Drosophila melanogaster. BMC Genomics 15:927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Meadows S, Sharp L, Jan LY, Jan YN. 2000. Genome-wide study of aging and oxidative stress response in Drosophila melanogaster. Proc Natl Acad Sci U S A. 97:13726–13731 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data have been deposited in SRA under the BioProject accession numbers PRJNA293450 (MQ18, SQ18, VQ18: SRR2177525–SRR2177544) and PRJNA284224 (MQ4: SRR2033894–SRR2033897, SRR2033903–SRR2033905).