Abstract
The identification of genetic mechanisms underlying evolutionary change is critical to our understanding of natural diversity, but is presently limited by the lack of genetic and genomic resources for most species. Here, we present a new comparative genomic approach that can be applied to a broad taxonomic sampling of nonmodel species to investigate the genetic basis of evolutionary change. Using our analysis pipeline, we show that duplication and divergence of fgfr1a is correlated with the reduction of scales within fishes of the genus Phoxinellus. As a parallel genetic mechanism is observed in scale-reduction within independent lineages of cypriniforms, our finding exposes significant developmental constraint guiding morphological evolution. In addition, we identified fixed variation in fgf20a within Phoxinellus and demonstrated that combinatorial loss-of-function of fgfr1a and fgf20a within zebrafish phenocopies the evolved scalation pattern. Together, these findings reveal epistatic interactions between fgfr1a and fgf20a as a developmental mechanism regulating skeletal variation among fishes.
Keywords: comparative genomics, nonmodel organisms, parallelism, zebrafish, fgf signaling, epistasis
Introduction
Conventional approaches toward identifying the genetic basis for evolutionary change center on genetic linkage analysis to uncover associations between genetic and phenotypic variations (Hoekstra et al. 2006; Shapiro et al. 2006; Rebeiz et al. 2009; Hopkins and Rausher 2011; Pardo-Diaz et al. 2012; Wray 2013). However, linkage-based approaches require distinct populations that exhibit morphological divergence but are still interfertile. Unfortunately, the vast majority of species cannot be intercrossed. These requirements limit the populations that can be used in these approaches, and thus constrain the types of morphological change that can be investigated. Another method used to identify genetic basis of phenotypic differences is by association mapping whereby specific polymorphisms within a population are found to correlate with a particular trait. As this method relies on variation for the phenotype of interest within a population, in cases in which traits are fixed among species this method is no longer applicable. Thus, although the vast majority of phenotypic variation is between species, there are a limited number of cases in which genetic causation can be formally addressed and as such, reliance on these approaches may bias our understanding of evolutionary change to cases of recent and rapid evolutionary radiations.
With the recent explosion of sequencing data and technologies, comparative genomic approaches have become a powerful means to address evolutionary change within natural populations and to perform genetic comparisons between often distantly related species (Wray 2013). These approaches include genome-wide scans for signatures of selection (Vitti et al. 2013), isolation of large-effect mutations (Enard et al. 2009; Andersson et al. 2012; Baldwin et al. 2014), analysis of copy number alterations (Perry et al. 2007), and identification of species-specific noncoding changes (Pennacchio et al. 2006; Prabhakar et al. 2008; Visel et al. 2008; McLean et al. 2011; Wray 2013; Boyd et al. 2015). These approaches are an effective way to identify genetic contributions to a phenotype in species not amenable to genetic linkage or association analysis (Hiller et al. 2012; Wray 2013). In conjunction with functional analysis within model organisms, particular candidate loci underlying morphological variation can be formally tested (Pennacchio et al. 2006; Prabhakar et al. 2008; Enard et al. 2009; McLean et al. 2011; Wray 2013; Baldwin et al. 2014; Boyd et al. 2015). However, comparative genomics is predominantly limited to species with well-annotated genomes and/or extensive transcriptome data (Gayral et al. 2013). Despite the rising numbers in species with available genomic resources, the number and scope of investigations into natural variation remains limited.
To facilitate genomic analysis of a wide range of species and phenotypic diversity, independent of existing genomic resources, we devised an experimental method, Phylomapping, which enables identification of signals of selection and variance associated with the evolution of particular groups. Key in this analysis is the derivation of a shared, “ancestral” exome between species to enable a systematic analysis of genetic variation and patterns of selection. To examine the utility of this approach, we analyzed scale-loss within a lineage of cypriniform fishes for which no previous genomic or genetic resources were available. Because we have data on variance across orthologous genes in the genome, the method allowed us to systematically test for changes in gene function and selection to generate hypotheses about how character change evolved. We were able to identify key genes and genetic mechanisms underlying the evolution of scale reduction occurring in this group.
Results
Phylomapping Methodology
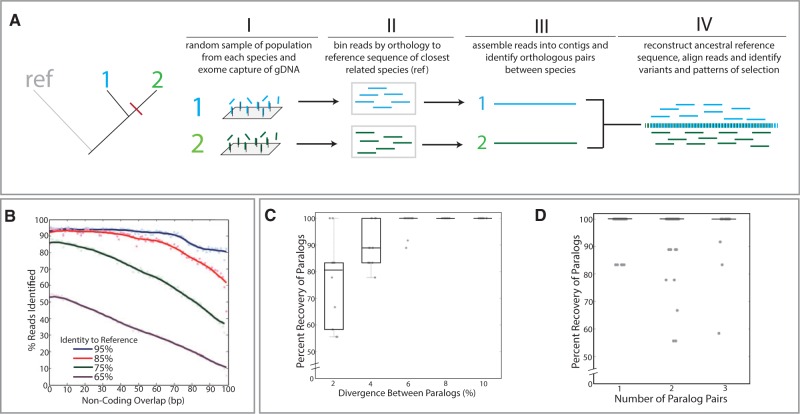
We set out to develop an approach to facilitate comparative genomic analysis in species that lack prior genetic resources. As a first step, pooled genomic DNA from populations of individual species is enriched for conserved orthologous protein-coding sequence through cross-hybridization to oligonucleotide “baits” designed against the known exome of a related species or a combination of species (fig. 1A-I). The use of genomic DNA through targeted sequence capture enables study of rare samples and the use of vast natural history collections (Mason et al. 2011) while being independent of the tissue type and developmental stage of sampled individuals that occurs with transcriptome data. After massively parallel sequencing, reads from the captured DNA are then grouped into bins of orthologous sequences by BLASTN (Altschul et al. 1990) against the reference exome used to design the capture array (fig. 1A-II). These read sets are assembled into exon-sized contigs by CAP3 (Huang 1999) (fig. 1A-III). To establish a shared ancestral reference exome, orthologous sequences are paired between species for reconstruction by “prequel” from the PHAST package (Hubisz et al. 2011) (fig. 1A-III and -IV). The imputed ancestral sequence is annotated by a BLASTX (Altschul et al. 1990) comparison with the orthologous reference exome. To facilitate recovery of reads flanking each exon that were previously not identified by exon-centered BLASTN, the raw reads are then realigned to this ancestral scaffold by Stampy (Lunter and Goodson 2011). The realignment of the raw reads to this shared scaffold then permits a broad comparative analysis of variation between species (fig. 1A-IV).
Fig. 1.
Phylomapping approach for analysis of character change in species lacking prior genetic resources. (A) Overview of approach. In this example, there are two species, “1” and “2,” with a common outgroup, “ref,” that has existing genetic resources from which to design the targeted capture array (I) and guide identification of reads (II). Reads are binned by orthology and assembled into contigs (III). Pairwise analysis between orthologous contigs enables reconstruction of a shared ancestral sequence that is then used to align and compare sequence variance (IV). (B–D) Test limits of approach through simulated assembly and annotation of reads from divergent genomes. (B) In silico 100-bp reads were generated at varying levels of divergence from the zebrafish reference genome and put into the phylomapping pipeline, utilizing the zebrafish reference genome to scaffold the analysis. Alignment and identification of reads from varying levels of divergence relative to the reference genome showing read recovery with high levels of variation and extending into noncoding sequence flanking the exons. Lines represent kernel regression. Simulated reconstruction of paralogs from mixed read data (supplementary fig. S1, Supplementary Material online) as a function of the divergence between paralogous sequences (C) and the number of paralogs present within the mixed read pools (D). Each dot represents an individual simulation.
To assess the limits of our approach, we asked how robust our pipeline was to variation between species and the reference genome used to guide exome reconstruction. Using the zebrafish genome, we artificially introduced variants into coding sequence at defined levels, created in silico reads, and then used the original zebrafish reference genome to scaffold the reconstruction of the variant exomes from the simulated reads. We were able to recover greater than 95% of reads containing even up to 15 single nucleotide polymorphisms (SNPs) per 100 bp of coding sequence (fig. 1B). Further, over 85% of reads with as many as 25 SNPs per 100 bp of coding sequence were able to be identified (fig. 1B). Although the analysis pipeline ignores noncoding sequence for the original identification of the reads, we were able to recover large amounts of noncoding sequence flanking each exon after realigning reads back to the ancestral scaffold (fig. 1B). Thus, this approach has the potential to recover and identify variation from species that lack a close relative with annotated genome resources.
Given variation in gene copy number between genomes, we next asked whether this approach can distinguish paralogs from variant haplotypes. The de novo assembly within this pipeline reliably distinguished paralogs from haplotypes as long as there was greater than 6% variation contained within the coding and flanking noncoding sequence (supplementary fig. S1, Supplementary Material online, and fig. 1C). Further, we found that paralog phasing is robust to multiple copy number (fig. 1D). This enables prediction and parsing of exons with increase in copy number between species and further expands the ability to assess species-specific variation.
Analysis of Skeletal Reduction in the Phoxinellus Lineage
As a case study of natural variation, we examined skeletal variation in fishes. Diversity in scale patterning in fishes is a common and defining feature of many species and genera, with over 13 independent cases of scale loss within Cypriniformes alone (Harris 2012). To explore the genetic basis of this natural diversity we selected a clade of minnows, Phoxinellus, where loss of scales is a defining, derived trait for the group (Freyhof et al. 2006) (fig. 2A–C). The Phoxinellus clade is thought to have diverged from a scaled fish lineage about 2.5 Ma (Freyhof et al. 2006). Fish of the genus Telestes are a close outgroup to Phoxinellus that have retained the ancestral scaled character state. Telestes or Phoxinellus species presently lack experimental or genetic tools making study of these species difficult. To enable analysis of the genetic mechanisms underlying the evolution of scale pattern divergence, we applied our Phylomapping pipeline to look at patterns of selection within these groups.
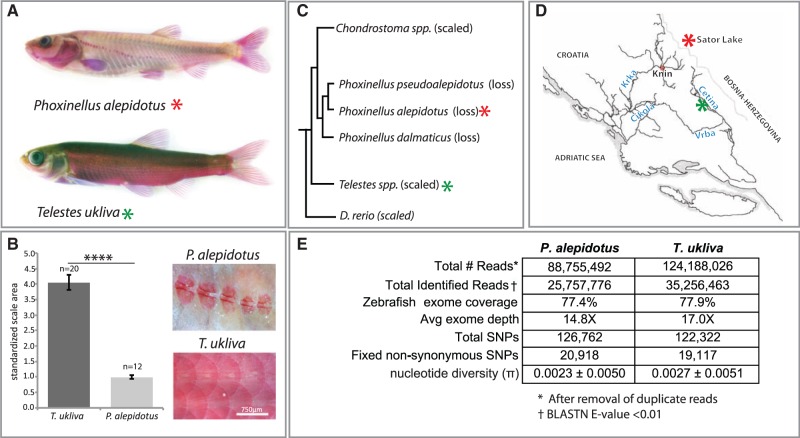
Fig. 2.
Comparative genomic analysis of scale reduction in natural populations. (A) Alizarin red stain of Phoxinellus alepidotus and Telestes ukliva highlighting loss of scales within Phoxinellus. (B) Average scale area relative to fish standard length. Data from three individuals of each species, with a total of n = 20 scales in T. ukliva and n = 12 scales in P. alepidotus. ****Two-tailed t-test P value < 0.0001. Error bars represent ±SEM. (C) Phylogenetic relationship between Phoxinellus and Telestes, with Danio rerio as the referenced outgroup used for analysis. Modified from (Perea et al. 2010) with addition of D. rerio. (D) Collection sites of P. alepidotus (red asterisk) and T. ukliva (green asterisk) used in this study. (E) Exome sequencing statistics from cross-species exome capture using zebrafish-targeted capture array. Single-end 100-bp Illumina HiSeq2000 reads were assembled using the Phylomapping pipeline.
During a field collection expedition in 2006, we collected Phoxinellus alepidotus from Sator Lake in Bosnia–Herzegovina and Telestes ukliva from locations in the Cetina river in Croatia (fig. 2D). Although Phoxinellus, Telestes and zebrafish are all cyprinids, they fall within different subfamilies and thus are quite distant relatives. Genomic DNA from 40 individuals of P. alepidotus and T. ukliva were pooled and hybridized to tiling arrays custom designed to cover the zebrafish exome (Zv9). Even given the evolutionary distance of approximately 50 My between zebrafish (Danioninae) and Phoxinellus/Telestes (Leuciscinae) lineages (Tang et al. 2010; Near et al. 2012), through use of the Phylomapping approach we were able to obtain genomic sequence from P. alepidotus and T. ukliva species pools orthologous to 77% of the zebrafish exome with an average depth of approximately 14–17× and 88% coverage of genes associated with development (fig. 2E and supplementary table S1, Supplementary Material online). The majority of zebrafish exons were represented equally by both P. alepidotus and T. ukliva reads, with only 1.3% of zebrafish coding bases covered by just one species. Roughly 10% of zebrafish genes had no exons covered by P. alepidotus or T. ukliva reads (supplementary table S2, Supplementary Material online). These genes are significantly enriched for highly variable gene classes, including zinc finger transcription factors, cadherins, and nucleosome complexes (supplementary table S2, Supplementary Material online). Analysis of SNP variation within T. ukliva and P. alepidotus permitted analysis of nucleotide diversity within the populations (fig. 2E). The observed level of nucleotide diversity (π = 0.0023 in P. alepidotus and π = 0.0027 in T. ukliva) is on the low end of the range seen in other fishes such as zebrafish (Whiteley et al. 2011) and is in-line with that of other Telestes species and Balkan freshwater cypriniforms (Stefani et al. 2004; Marešová et al. 2011; Palandačić et al. 2012). This may reflect the small endemic range of P. alepidotus and T. ukliva and suggests recovery of population diversity through the Phylomapping approach. We identified around 20,000 fixed nonsynonymous changes between P. alepidotus and T. ukliva (fig. 2E). This diversity is consistent with large amounts of coding variation within other species of Cyprinidae, such as zebrafish, where there may be as many as 13,000 fixed nonsynonymous variants even between strains (LaFave et al. 2014).
Analysis of SNP diversity among the assembled exomes of P. alepidotus and T. ukliva identified a large set of genes with potentially deleterious variants fixed within the populations. To identify the changes associated with the reduction and altered morphology of scales in P. alepidotus, we restricted our analysis to genes associated with developmental processes based on gene ontology (fig. 3A and supplementary table S1, Supplementary Material online). As a measure of selection and function of particular variants, we looked for the accumulation of mutations beyond a neutral rate of drift likelihood ratio test (LRT) (LRT; Pollard et al. 2010) along with functional effect prediction of fixed nucleotide variants (SIFT; Kumar et al. 2009). Out of 1,022 development-associated genes identified in P. alepidotus, 248 contained unique, fixed SNPs predicted to be deleterious (SIFT < 0.05), 215 genes were predicted to be under selection or drift (LRT, q value < 0.05), and 70 genes had both signatures (fig. 3A and supplementary table S1, Supplementary Material online). Within these gene classes, several candidates of the Fgf and Eda signaling pathways were present (fig. 3B)—both signaling pathways are integrally associated with development of integumentary appendages such as scales (Kere et al. 1996; Headon and Overbeek 1999; Kondo et al. 2001; Colosimo et al. 2005; Harris et al. 2008; Rohner et al. 2009). Although Eda and Edar in particular are commonly associated with variation in scale number (Kondo et al. 2001; Harris et al. 2008), we found no evidence of selection on either of these loci in P. alepidotus (fig. 3B and supplementary table S1, Supplementary Material online).
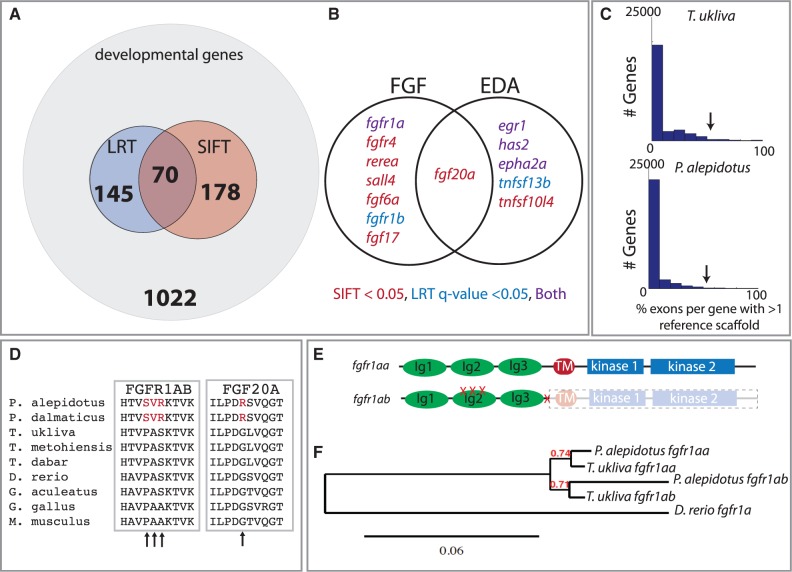
Fig. 3.
Selection on an FGF ligand-receptor pair within Phoxinellus underlying scale reduction. (A) Analysis of a subset of developmental genes as defined by select gene ontology terms (supplementary table S1, Supplementary Material online) with exons showing signatures of nonsense or deleterious SNPs (SIFT) and accelerated sequence evolution relative to neutral drift (LRT) (supplementary table S1, Supplementary Material online). (B) Genes within the EDA and FGF signaling pathways showing either LRT q value < 0.05 or SIFT < 0.05 (supplementary table S1, Supplementary Material online). (C) Histogram of the percentage of exons per gene showing a signature of duplication (supplementary table S3, Supplementary Material online); arrow indicates location of fgfr1a. (D) Multiple protein alignment of Fgfr1ab and Fgf20a. Arrows indicate amino acid changes within the Phoxinellus genus. (E) Location of Phoxinellus fgfr1ab nonsynonymous mutations and premature truncation mapped onto Fgfr1a protein (supplementary fig. S2, Supplementary Material online). (F) Reconstructed gene tree from cDNA of fgfr1aa and fgfr1ab transcripts isolated from Phoxinellus alepidotus skin and Telestes ukliva exome sequencing data.
Through our analysis, we detected a low level of duplication within both T. ukliva and P. alepidotus lineages (fig. 3C and supplementary table S3, Supplementary Material online). Of the 70 genes showing signatures of selection and predicted deleterious mutations, only four showed evidence of duplication (supplementary table S3, Supplementary Material online). Interestingly, one of these genes showing a shared duplication within the common ancestor of Phoxinellus and Telestes was fgfr1a (fig. 3C and supplementary table S3, Supplementary Material online). Reverse transcription polymerase chain reaction verified the presence of two independent paralogs of fgfr1a and indicated that both were expressed in skin (supplementary fig. S2, Supplementary Material online). Analysis of exon variation specifically within P. alepidotus indicated several fixed nonsynonymous changes in one fgfr1a paralog, fgfr1ab. The identified variants are predicted to be detrimental as they alter highly conserved residues within a hydrophobic binding interface between Fgf receptors and their ligands (Plotnikov et al. 2000) (fig. 3D and E). In this context, the P158S variant alters a specific hydrophobic contact (P169S; Plotnikov et al. 2000), suggesting a critical role of this change in regulating Fgfr1ab interaction with its ligands. These mutations are fixed and derived within the Phoxinellus genus, as they were not identified within T. ukliva or other closely related species of Telestes, T. metohiensis and T. dabar (Bogutskaya et al. 2012) (fig. 3D). Interestingly, identification of full-length fgfr1ab transcripts from P. alepidotus showed the presence of a truncated mature message with loss of the transmembrane and kinase domains (fig. 3E and supplementary fig. S2, Supplementary Material online). A splice acceptor mutation, consistent with this truncated mRNA, was observed in P. alepidotus but not T. ukliva sequencing reads. However, variance at this site within the P. alepidotus population suggests the fgfr1ab truncation is secondary to fixation of the binding interface mutations. A gene tree was constructed from these transcripts and the exome sequencing data (fig. 3F), revealing an extended branch length of P. alepidotus fgfr1ab, further supporting increased divergence of this paralog within this clade.
Identification of Key Modifiers of Skeletal Evolution
We extended our analysis to look for further clues to genetic regulators of scale evolution in this group. We identified a fixed, predicted function-altering, nonsynonymous mutation in the fgfr1a ligand fgf20a that is unique to P. alepidotus (fig. 3B and D). In mice, FGF20 is specifically upregulated downstream of EDA signaling (Lefebvre et al. 2012) and is sufficient to rescue phenotypes due to Eda deficiency during tooth development (Häärä et al. 2012). Further, Fgf20 is required for feather and scale development in chickens (Wells et al. 2012). The G80R SNP in fgf20a is fixed within the Phoxinellus genus and is not found within three different Telestes species (fig. 3D) suggesting that these mutations are unique to Phoxinellus and support a role of this ligand–receptor interaction in regulating scale patterning. However, no previous role was known for fgf20a in scalation as zebrafish mutants defective for fgf20a function do not have an apparent scale phenotype (Whitehead et al. 2005).
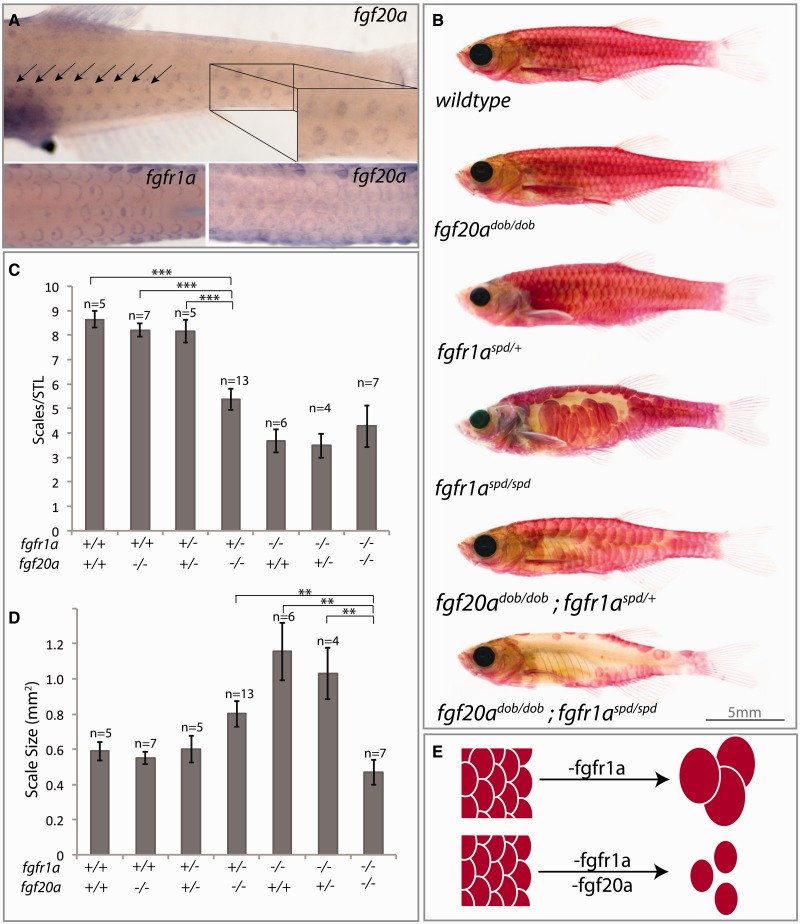
We asked whether the function of fgfr1a and fgf20a combine to affect the formation and phenotypic variation of scales. In zebrafish, fgf20a is expressed in the developing scale placodes and in the growing edges of scales in a domain overlapping with fgfr1a (fig. 4A). Zebrafish heterozygous for fgf20a/dob, or fgfr1a/spd loss-of-function mutations have normal scale numbers and morphology. However, a combined reduction in both ligand and receptor is sufficient to affect scale development, reducing the number of scales on the flank of each fish and limiting scale shape (fig. 4B–D). Although loss of fgfr1a alone leads to abnormally large, albeit fewer scales (fig. 4C and D), reduction of Fgf20a function in this context leads to scales that more closely resemble the small scale phenotype seen in P. alepidotus (fig. 2A and B). Thus, using a genomic approach centered on comparison between lineages with fixed morphological changes, we have identified a key role for Fgfr1ab–Fgf20a signaling in regulating the number and size of scales in zebrafish that reflects the phenotype observed in Phoxinellus (fig. 4E).
Fig. 4.
Epistatic interaction between Fgf20a and Fgfr1a regulates scale development and is sufficient to phenocopy Phoxinellus scale reduced phenotype. (A) Expression of fgfr1a and fgf20a by whole-mount in situ hybridization in 30-day-old zebrafish. (B) Representative alizarin red-stained zebrafish with loss-of-function mutations in fgf20a (dob) and/or fgfr1a (spd). (C) Average number of scales on lateral flank normalized to the standard length of each fish. ***P value < 0.001. (D) Average area of scales (mm2) in different genetic backgrounds. **P value < 0.01. Error bars represent ±SEM. (E) Model of epistatic interaction between fgfr1a and fgf20a regulating scale number and size.
Identification of Global Patterns of Selection within P. alepidotus
In addition to Phoxinellus, unrelated fish have independently lost their scales within the Adriatic drainage basin (Delic et al. 2005; Bogutskaya et al. 2012), suggesting a selective benefit for scale reduction within this environment. Scales are thought to protect against predators and, as a calcified component of the fish skeletal system, function in regulating calcium and phosphate homeostasis. However, the selective advantage for scale reduction in these fishes was unclear: The Krka drainage basin derives from limestone beds, potentially limiting the need for additional calcium storage by scales. Further, in Sator lake, where an unrelated and scale-reduced fish is found, other species of fish that could serve as predators were not identified (Delic et al. 2005).
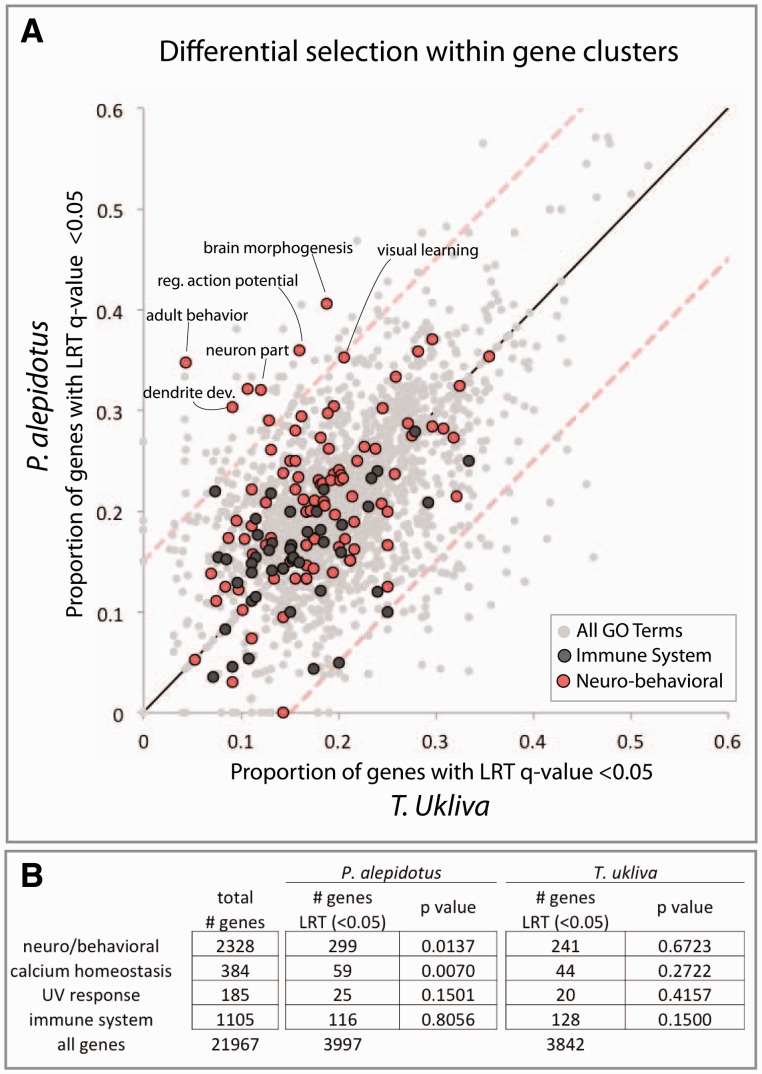
To explore the capability of our exome-wide data set to build hypotheses about selective causes for trait variance, we looked for accumulation of mutations beyond neutral drift by LRT (q value < 0.05) within gene ontology groups between P. alepidotus and T. ukliva (fig. 5A and supplementary table S4, Supplementary Material online). In support of a role for calcium in fish scale variation, we identified a P. alepidotus-specific enrichment for selection on biological processes related to calcium homeostasis (fig. 5B and supplementary table S5, Supplementary Material online). Unexpectedly, we also observed a P. alepidotus-specific enrichment for selection on gene classes associated with neurobiology and behavior (fig. 5A and B; supplementary table S5, Supplementary Material online). Although it is unknown what behavioral traits are associated with P. alepidotus, these patterns provide evidence for broad changes in addition to scale reduction during evolution of P. alepidotus.
Fig. 5.
Coordinated changes in Phoxinellus alepidotus point to multifactorial role of Fgfr1 signaling in trait evolution. (A) Proportion of genes within individual GO-terms with LRT q value < 0.05. Each dot represents a GO-term with a minimum of 20 genes. Mean represented by black line. Red-dashed lines represent ± 2 SD from the mean. (B) Enrichment for genes showing signature of accelerated sequence evolution relative to neutral drift (LRT q value < 0.05) within aggregate gene ontologies (supplementary table S4, Supplementary Material online).
Discussion
Here, we outline an analytic and experimental approach to investigate the genetic causes of evolution in species that do not have existing genetic resources or lend themselves to classical linkage analysis. We find that this method is robust to sequence variation and can be efficiently used to generate a platform for comparative genomic analysis even between species that are evolutionarily distantly related to each other. As zebrafish and Telestes/Phoxinellus share a common ancestor at least 50 Ma, there is a large evolutionary window in which this method can be applied. With the current coverage of annotated reference genomes and transcriptomes growing at a rapid rate, this approach should enable comparative analysis within a large number of species, from fish and mammals to plants and invertebrate groups.
Because the Phylomapping approach centers on conservation of coding regions to identify and compare genomic sequence between distantly related species, it is not powered to directly identify cis-regulatory mutations. However, signatures of selection within exons may reflect a hitchhiking effect of changes in linked noncoding sequence. Further, coding sequence has been used to estimate other indicators of regulatory change, such as codon bias (Chamary et al. 2006; Cutter et al. 2006), translation efficiency (Hudson et al. 2011, 2014), and pathway-level selection (Kosiol et al. 2008) that could serve as a proxy to detect signatures of regulatory evolution. Thus, genome-wide detection of regulatory variation may be indirectly detected within exome data assuming variation is sufficient within the sample populations. Incorporation of known or conserved noncoding elements in the DNA capture step may enable direct identification of putative enhancer and promoter regions modified during evolution.
Using our Phylomapping approach, we detail the role of Fgf signaling in scale reduction through duplication and divergence of fgfr1a within P. alepidotus. We have previously shown that independent duplication and divergence of fgfr1a is the cause of scale reduction in a domesticated variant of the common carp, Cyprinus carpio (Rohner et al. 2009). This suggests that there is a common mechanism underlying both transitions in morphology. Unlike the domesticated carp, where remaining scales are larger in size (Rohner et al. 2009), the scales in P. alepidotus remain small. Our finding of the combined effect of fgfr1a and fgf20a in regulating scale size in zebrafish suggests that these two genes are associated with the scalation phenotype seen in P. alepidotus. Although we previously have showed linkage of fgfr1a to scale loss in carp (Rohner et al. 2009), strong selection from domestication is likely not representative of the types of changes that would occur during natural selection. Our finding of a parallel processes by which scales are reduced in both natural and artificial selections suggests that there is significant constraint on the genetic changes that can cause viable variation in scalation within cyprinids. Given the essential roles for fgfr1a during development, gene duplication and subfunctionalization of the paralogs would release constraint on variation. Many studies have pointed to the important role of cis-regulatory regulation in evolution of morphology (Carroll 2008; Wittkopp and Kalay 2012). Although it is clear change in the regulation of genes during development is important mediator of evolution, gene duplication and functional divergence of gene function is quite common in most eukaryotic species and represents an important mechanism for evolutionary change that can encompass both regulatory and coding variance (Ohno 1970; Force et al. 1999; Hoekstra and Coyne 2007).
Analysis of synapomorphy, or shared derived characters, is a key element in understanding the potential causes of evolutionary transitions. As morphology can be frequently shaped to the varied life-history strategies of a species, it is tempting to ascribe selection directly on these physical adaptations for their behavioral and physiological advantages. However, it is likely that many species are “making fruitful use of available parts” (Gould and Lewontin 1979) and particular morphological states or changes do not necessarily impart direct selective advantage but rather can become fixed by alternate means. Genetic correlation of traits, in which traits are physically linked in the genome or are linked through the use of shared genetic mechanisms, are cases that reveal bias in phenotypes that can arise though selection. Such correlative evolution can lead to aberrant adaptive hypotheses, as putative adaptations may be simply linked to selection on other diverse traits under strong selection and thus are not independent entities.
Adaptive hypotheses for loss of scales and armor in fish have been many, ranging from calcium conservation in calcium poor environments (Giles 1983; Spence et al. 2013; Smith et al. 2014) to changes in buoyancy (Myhre and Klepaker 2009) and increased motility without armor (Bergstrom 2002). Most arguments involve relaxation of selective pressures by predation (Bell et al. 1993; Marchinko 2009) with growth advantages through the reduced metabolic demands of developing and maintaining scales across the body (Marchinko and Schluter 2007; Barrett et al. 2008). But to date support for these hypotheses has been inconsistent. Loss of fgfr1a in the zebrafish leads to increased boldness and aggression behavior (Norton et al. 2011), suggesting functional evidence for pleiotropy between the genes involved in scale development and behavior. Further, mirror carp that have lost a duplicated paralog of fgfr1a during artificial selection for scale-reduction also display an increase in boldness and are more easily captured than their scaled variety (Klefoth et al. 2012, 2013). As boldness can lead to increased fitness through enhanced foraging and ability to find mates, the behavioral aspects of loss of Fgfr1a function could provide a strong selective pressure for correlative reduction of scales within a permissive environment. Phoxinellus alepidotus from the field were easily collected with small aquarium nets (Delic et al. 2005; M.P.H. personal observation) in contrast to Telestes, suggesting potential parallels in these fish. However, detailed behavioral studies are needed to identify behavioral changes associated with scale reduction in these species to directly address the possibility of pleiotropy driving correlated changes in behavior and morphology.
Here, we detail an analytic and experimental approach to investigate the genetic causes of evolution in species that do not lend themselves to classical linkage analysis. We detail the role of Fgf signaling in scale development through duplication and divergence of fgfr1a and epistasis with Fgf20a. In addition to scale reduction, this data set provides multiple entry points in understanding changes in morphology, physiology, and behavior as a consequence of adaptation and speciation. Thus, this comparative genomic approach should enable broad expansion of genetic information to new species and improve our ability to understand natural diversity.
Materials and Methods
Sample Collection and Identification
Samples of Phoxinellus alepidotus and Telestes ukliva were collected at described localities during a field expedition in 2006. Communication with local authorities was made prior to collection with permission. Fish were caught through use of minnow-traps baited with bread, or simply by use of a small dip net. The majority of fish captured were measured, fin clipped, and released. Ten P. alepidotus and four T. ukliva were anesthetized in the field and preserved for analysis of morphology. Genomic DNA and trizol fixed tissue from P. Dalmaticus was kindly provided by Dr Joerg Freyhof.
The identity of P. alepidotus, P. dalmaticus, and T. ukliva were determined by meristic characters and were verified through Sanger sequencing of a portion of cytochrome B, with BLASTN identification against the NCBI nucleotide database. Primers for cytochrome b sequencing from Freyhof et al. (2006) (F′: TGACTTGAARAACCAYCGTTG, R′: GGCAAATAGGAARTATCATTC).
Zebrafish Husbandry and Mutants
Zebrafish wild-type and mutant lines used were housed and maintained in accordance with institutional IUCAC protocols. Mutant lines used are spiegeldanio (spd; fgfr1at3R705H) (Rohner et al. 2009), albino, and devoid of blastema (dob) (Whitehead et al. 2005). dob were kindly provided by Dr Kenneth Poss.
Genotyping
Fish were anesthetized with tricaine and the tail or pectoral fins were clipped with standard surgical scissors. fgf20adob and fgfr1aspd fish were genotyped by Sanger sequencing (dob primers-F′: TGCCTCTCTTAGGAAAAGCTG, R′:CATCTCTGGACGTCCCATCT; spd primers-F′: TGATGACCTCAGCTGGTCTTT, R′:AGTCTTACAGCTCATGTGTGCAT).
RNA Isolation and cDNA Preparation
Tissue for RNA extraction was collected in the field and stored in Trizol until RNA was extracted. RNA was transcribed to cDNA using Cloned AMV First-Strand Synthesis Kit (Invitrogen) and SuperScript III Reverse Transcription Kit (Invitrogen) using oligo dT primers.
Reconstruction of fgfr1aa and fgfr1ab Transcripts
Full-length fgfr1aa transcripts were isolated from P. alepidotus skin with the following primers: 5′-AGTGCGGATGTCCTAGCAGT-3′ and 5′-AAAGCGTGTTCTCCGTGAGT-3′.
For reconstruction of fgfr1ab, 3′-Rapid Amplification of cDNA Ends (RACE) was performed with SMARTer 5′/3′ RACE kit (Clontech). A fgfr1ab-specific 3′-RACE touchdown primer was designed from the exon under selection in P. alepidotus: 5′-ATAAGATGGAGAAGAAGCTCCACACGGTTTCCGT-3′. Nested primers for fgfr1ab: UPM-short (Clontech), 5′-CGTAGTCGAGTGTCCACCACAT-3′.
5′-Sequence of fgfr1ab was reconstructed with a universal fgfr1a exon 1 reverse primer (5′-ACCATGCTGCTGCTGATCT-3′) and paralog-specific fgfr1a primer designed from the identified exon under selection in P. alepidotus fgfr1ab (fgfr1aa : 5′-CTGGTCTCTCTTAAACTCTTTGCCG-3′, fgfr1ab: 5′-CTGGTCTCTCTTAAACTCTTTGCCA-3′).
Gene Tree Construction
Gene trees were constructed from using phylogeny.fr (Dereeper et al. 2008). Briefly, sequences were aligned by MUSCLE (Edgar 2004) and curated by Gblocks (Castresana 2000) before phylogenetic construction by PhyML (aLRT) (Guindon and Gascuel 2003). Tree rendering was performed with TreeDyn (Chevenet et al. 2006). Due to truncation of P. alepidotus fgfr1ab, only the first seven exons of fgfr1a were used to construct the gene tree.
Whole-Mount In Situ Hybridization
Whole-mount in situ hybridization was performed according to Harris et al. (2008) on 30-day-old albino zebrafish with a standard length of 10 mm. Primers for probe against fgf20a; f: GACCTGACGCATCTCAAAGG, r: GGGTGGTTTTGAGTTTGAGG. Primers for probe against fgfr1a; f: GCTTTGCTCAGGGACTCAAC, r: GCAGTCCATCAGGTCCGTAT.
Skeletal Stain and Morphometrics
Skeletal staining was performed using Alizarin Red in 1% KOH overnight and then cleared in 1% KOH (50 mg Alizarin Red per liter of 1% KOH). Scale area was measured using ImageJ. The scale area of five randomly selected scales was measured and normalized to the standard length of each fish, defined as the distance from the lower jaw to the end of the caudal peduncle.
Targeted Sequence Capture and Illumina Sequencing
Genomic DNA was extracted with the Qiagen DNAeasy Blood & Tissue DNA extraction kit from fin clips of 40 individuals from population samples of P. alepidotus and T. ukliva. Species-specific pools of 3–5 μg DNA were sheared according to manufacturer’s protocol (Covaris) to an average size of 200 bp, as verified on a 4% agarose gel. DNA libraries were blunt-ended, 5′ phosphorylated, A-tailed, and adapter ligated as previously described (Bowen et al. 2011). Phusion High-Fidelity DNA Polymerase (NEB) was used to amplify pre- and postcapture libraries.
To enrich DNA libraries for coding sequence, a custom Agilent Technologies 1 M SureSelect DNA capture array containing 974,016 probes covering all 41,131,682 bp of the zebrafish known protein-coding exome (Zv9) was designed to minimize overlap of bait oligos with noncoding sequence. The 60 mer oligos were tiled on average every 40 bases, with 20 bp of overlap between probes. Cross-species hybridization was performed at 60 °C to allow for potential mismatches between the zebrafish bait and the cross-species libraries. Postcapture libraries were amplified as previously described (Bowen et al. 2011).
As longer reads have a higher probability of identification by BLASTN, we performed 100-bp single-end Illumina HiSeq 2000 sequencing, with one lane each for P. alepidotus and T. ukliva. Leveraging the sequence information from the first run, a second custom capture array was designed to reflect the common ancestor of Phoxinellus and Telestes. Libraries of the same 40 individuals were constructed as previously described, only this time with 6-bp barcoded adapters. Exome capture was performed as above, and the DNA from P. alepidotus and T. ukliva pooled into one lane of Illumina HiSeq 2500 for 100-bp single-end sequencing.
Adapter sequence for P. alepidotus:
5′-/5Phos/GCCTAAAGATCGGAAGAGCGGTTCAGCAGGAATGCCGAG-3′
5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTTTAGGC*T-3′.
Adapter sequence for T. ukliva:
5′-/5Phos/TGGTCAAGATCGGAAGAGCGGTTCAGCAGGAATGCCGAG-3′
5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTTGACCA*T-3′.
Read Processing
Raw reads from sequencing were end quality trimmed (-Q 33 -t 33) and low-quality bases (-Q 33) were masked using the FASTX package (http://hannonlab.cshl.edu/fastx_toolkit, last accessed October 7, 2015). Identical reads were compressed using fastx_collapser (-Q 33).
Phylomapping Pipeline
Overview
Using the annotated reference sequence of a related species as bait, we perform targeted sequence capture to enrich DNA libraries for conserved, orthologous protein-coding genes in another species. After next-generation sequencing, reads are identified by BLASTN (E-value cutoff < 0.01, -outfmt 6; Altschul et al. 1990) and assembled into contigs by CAP3 (-o 25 -p 90, Huang 1999). Orthologous contigs are then identified between species through centroid clustering (k-means, SciPy) and are paired for the reconstruction of a shared ancestral sequence using prequel from the PHAST package (–subst-mod UNREST, –no-probs, –features; Hubisz et al. 2011). The ancestral sequence is annotated by BLASTX (E value < 0.001, -outfmt 6; Altschul et al. 1990) and used as a common reference scaffold for realignment of the original reads by Stampy (–substitutionrate = 0.09; Lunter and Goodson 2011). This shared genome serves as a scaffold for direct comparison of sequence variation within and between related species. Reads with greater than five mismatches to the ancestral sequence were removed as likely misalignments. See supplementary text, Supplementary Material online, for details.
Population Summary Statistics
Nucleotide diversity (π) was calculated and averaged from all coding sequence data using Popoolation (Kofler et al. 2011), with “–pool-size 80” and “–gtf” options.
Analysis of Variants
SAMtools (Li et al. 2009) was used to construct a shared mpileup file for P. alepidotus and T. ukliva. Variants were called from mpileup files using a custom python script. A minimum allelic depth of 3 was required for polymorphism identification. Heterozygous variants were called with an allele frequency greater than 25%, with homozygous variants having greater than 75% allele frequency. To increase confidence in identified variants for comparison between species, only variants with a read depth of at least three reads in each species were considered. Runs of greater than four consecutive SNPs between species were ignored as possible artifacts of the alignment and/or ancestral reference assembly. Variants were annotated using SnpEff (Cingolani et al. 2012). Fixed, nonsense and missense changes unique to P. alepidotus and T. ukliva were then isolated from the SnpEff file using custom python scripts.
PhyloP from the PHAST package (http://compgen.bscb.cornell.edu/phast/background.php) was used to identify exons under positive selection (Pollard et al. 2010). The consensus reference sequence for each species was reconstructed using the ancestral reference sequence as a template, and by amending bases in the imputed ancestral reference sequence to reflect fixed variants in a particular species as identified from the mpileup file. Neutral tree models were constructed using phyloFit (–tree “((Telestes,Phox), Zebrafish)”) with “–features” to restrict analysis to coding sequence (Siepel and Haussler 2004). MAFFT (Katoh et al. 2005) was used to align sequences of P. alepidotus, T. ukliva, and D. rerio. Tree models were constructed for each of 1,000 randomly selected exons, and the values averaged to generate a neutral tree-model. PhyloP (Pollard et al. 2010) was then run with –subtree for both Phoxinellus and Telestes, with –method LRT, –mode CONACC, and –features options using the BLASTX-generated GTF file. For multiple hypothesis testing, the P values were converted to false discovery rate (FDR) q values according to Storey (2002).
Statistical Analysis of Gene Ontology Enrichment
To identify trends of sequence evolution within P. alepidotus and T. ukliva (fig. 5B), genes were pooled from a custom grouping of related gene ontologies (supplementary table S5, Supplementary Material online). Statistical significance for enrichment of genes under selection within these broad gene ontology groupings was analyzed by bootstrap resampling of 20,000 permutations against the entire gene set within each species.
To identify individual gene ontology terms for which there is an increase in the number of genes under selection specifically in P. alepidotus (supplementary table S5, Supplementary Material online), bootstrap resampling was performed with 15,000 permutations against a null distribution based on the number of genes with q value < 0.05 within the same gene ontology category in T. ukliva. For multiple hypothesis testing, the exact P values were converted to FDR q values according to Storey (2002) using custom python scripts.
The imputed reference exome for T. ukliva and P. alepidotus and python scripts with phylomapping protocol used for the analysis are provided at www.fishyskeleton.com.
Supplementary Material
Supplementary text, figures S1–S3, and tables S1–S6 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank the support of Dr Christiane Nusslein-Volhard whose lab this work was initiated. Further, they also thank Dr Kenneth Poss for sharing fgf20a/dob mutant strains and Dr Jörg Feyhof for providing Phoxinellus dalmaticus samples. They especially appreciate the hospitality and assistance of the Curuvija Family and Zarko Jejina in Knin, Croatia that made this project possible. Further, the work was supported through advice and guidance from local wardens of the rivers within the Krka drainage basin and was completed with their permission. Finally, the authors specially thank Primoz Zupančič for sharing his advice and samples of Telestes species. They also acknowledge helpful comments from anonymous reviewers that helped to shape the manuscript. This work was supported in part by National Science Foundation (NSF) GRFP and DDIG (grant number DEB-1407092) to J.M.D., and John Simon Guggenheim Fellowship to M.P.H. as well as support from the Children’s Orthopaedic Surgery Foundation at Boston Children’s Hospital.
References
- Altschul S, Gish W, Miller W. 1990. Basic Local Alignment Search Tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Andersson LS, Larhammar M, Memic F, Wootz H, Schwochow D, Rubin C-J, Patra K, Arnason T, Wellbring L, Hjälm G, et al. 2012. Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature 488:642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin MW, Toda Y, Nakagita T, O’Connell MJ, Klasing KC, Misaka T, Edwards SV, Liberles SD. 2014. Evolution of sweet taste perception in hummingbirds by transformation of the ancestral umami receptor. Science 345:929–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RDH, Rogers SM, Schluter D. 2008. Natural selection on a major armor gene in threespine stickleback. Science 322:255–257. [DOI] [PubMed] [Google Scholar]
- Bell MA, Orti G, Walker JA, Koenings JP. 1993. Evolution of pelvic reduction in threespine stickleback fish: a test of competing hypotheses. Evolution 47:906–914. [DOI] [PubMed] [Google Scholar]
- Bergstrom CA. 2002. Fast-start swimming performance and reduction in lateral plate number in threespine stickleback. Can J Zool. 80:207–213. [Google Scholar]
- Bogutskaya NG, Zupančič P, Bogut I, Naseka AM. 2012. Two new freshwater fish species of the genus Telestes (Actinopterygii, Cyprinidae) from karst poljes in Eastern Herzegovina and Dubrovnik littoral (Bosnia and Herzegovina and Croatia). Zookeys 180:53–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen ME, Boyden ED, Holm IA, Campos-Xavier B, Bonafé L, Superti-Furga A, Ikegawa S, Cormier-Daire V, Bovée JV, Pansuriya TC, et al. 2011. Loss-of-function mutations in PTPN11 cause metachondromatosis, but not Ollier disease or Maffucci syndrome. PLoS Genet. 7:e1002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JL, Skove SL, Rouanet JP, Pilaz L-J, Bepler T, Gordân R, Wray GA, Silver DL. 2015. Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex. Curr Biol. 25:772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134:25–36. [DOI] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, Hurst LD. 2006. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 7:98–108. [DOI] [PubMed] [Google Scholar]
- Chevenet F, Brun C, Bañuls A-L, Jacq B, Christen R. 2006. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang L, Coon M. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Jr, Dickson M, Grimwood J, Schmutz J, Myers RM, Schluter D, Kingsley DM. 2005. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science 307:1928–1933. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Wasmuth JD, Blaxter ML. 2006. The evolution of biased codon and amino acid usage in nematode genomes. Mol Biol Evol. 23:2303–2315. [DOI] [PubMed] [Google Scholar]
- Delic A, Kucinic M, Maric D, Bucar M. 2005. New data about the distribution of Phoxinellus alepidotus (Heckel, 1841) and Aulopgye huegelii (Heckel, 1841). Nat Croat. 14:351–355. [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard J-F, Guindon S, Lefort V, Lescot M, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W, Gehre S, Hammerschmidt K, Hölter SM, Blass T, Somel M, Brückner MK, Schreiweis C, Winter C, Sohr R, et al. 2009. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell 137:961–971. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan Y, Postlethwait J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyhof J, Lieckfeldt D, Bogutskaya NG, Pitra C, Ludwig A. 2006. Phylogenetic position of the Dalmatian genus Phoxinellus and description of the newly proposed genus Delminichthys (Teleostei: Cyprinidae). Mol Phylogenet Evol. 38:416–425. [DOI] [PubMed] [Google Scholar]
- Gayral P, Melo-Ferreira J, Glémin S, Bierne N, Carneiro M, Nabholz B, Lourenco JM, Alves PC, Ballenghien M, Faivre N, et al. 2013. Reference-free population genomics from next-generation transcriptome data and the vertebrate-invertebrate gap. PLoS Genet. 9:e1003457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles N. 1983. The possible role of environmental calcium levels during the evolution of phenotypic diversity in Outer Hebridean populations of the three-spined stickleback, Gasterosteus aculeatus. J Zool. 199:535–544. [Google Scholar]
- Gould SJ, Lewontin RC. 1979. The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme. Proc R Soc Lond B Biol Sci. 205:581–598. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704. [DOI] [PubMed] [Google Scholar]
- Häärä O, Harjunmaa E, Lindfors PH, Huh S-H, Fliniaux I, Åberg T, Jernvall J, Ornitz DM, Mikkola ML, Thesleff I. 2012. Ectodysplasin regulates activator-inhibitor balance in murine tooth development through Fgf20 signaling. Development 139:3189–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MP. 2012. Comparative genetics of postembryonic development as a means to understand evolutionary change. J Appl Ichthyol. 28:306–315. [Google Scholar]
- Harris MP, Rohner N, Schwarz H, Perathoner S, Konstantinidis P, Nüsslein-Volhard C. 2008. Zebrafish eda and edar mutants reveal conserved and ancestral roles of ectodysplasin signaling in vertebrates. PLoS Genet. 4:e1000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Headon DJ, Overbeek PA. 1999. Involvement of a novel Tnf receptor homologue in hair follicle induction. Nat Genet. 22:370–374. [DOI] [PubMed] [Google Scholar]
- Hiller M, Schaar BT, Indjeian VB, Kingsley DM, Hagey LR, Bejerano G. 2012. A “forward genomics” approach links genotype to phenotype using independent phenotypic losses among related species. Cell Rep. 2:817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. 2007. The locus of evolution: evo devo and the genetics of adaptation. Evolution 61:995–1016. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. 2006. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science 313:101–104. [DOI] [PubMed] [Google Scholar]
- Hopkins R, Rausher MD. 2011. Identification of two genes causing reinforcement in the Texas wildflower Phlox drummondii. Nature 469:411–414. [DOI] [PubMed] [Google Scholar]
- Huang X. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Pollard KS, Siepel A. 2011. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief Bioinform. 12:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson NJ, Baker ML, Hart NS, Wynne JW, Gu Q, Huang Z, Zhang G, Ingham AB, Wang L, Reverter A. 2014. Sensory rewiring in an echolocator: genome-wide modification of retinogenic and auditory genes in the bat Myotis davidii. G3 4:1825–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson NJ, Gu Q, Nagaraj SH, Ding Y-S, Dalrymple BP, Reverter A. 2011. Eukaryotic evolutionary transitions are associated with extreme codon bias in functionally-related proteins. PLoS One 6:e25457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kere J, Srivastava A, Montonen O. 1996. X-linked anhidrotic (hypohidrotic) ectodermal dysplasia is caused by a mutation in a novel transmembrane protein. Nature 13:409–416. [DOI] [PubMed] [Google Scholar]
- Klefoth T, Pieterek T, Arlinghaus R. 2013. Impacts of domestication on angling vulnerability of common carp, Cyprinus carpio: the role of learning, foraging behaviour and food preferences. Fish Manag Ecol. 20:174–186. [Google Scholar]
- Klefoth T, Skov C, Krause J, Arlinghaus R. 2012. The role of ecological context and predation risk-stimuli in revealing the true picture about the genetic basis of boldness evolution in fish. Behav Ecol Sociobiol. 66:547–559. [Google Scholar]
- Kofler R, Orozco-terWengel P, De Maio N, Pandey RV, Nolte V, Futschik A, Kosiol C, Schlötterer C. 2011. PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One 6:e15925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Kuwahara Y, Kondo M, Naruse K, Mitani H, Wakamatsu Y, Ozato K, Asakawa S, Shimizu N, Shima A. 2001. The medaka rs-3 locus required for scale development encodes ectodysplasin-A receptor. Curr Biol. 11:1202–1206. [DOI] [PubMed] [Google Scholar]
- Kosiol C, Vinar T, da Fonseca RR, Hubisz MJ, Bustamante CD, Nielsen R, Siepel A. 2008. Patterns of positive selection in six Mammalian genomes. PLoS Genet. 4:e1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 4:1073–1081. [DOI] [PubMed] [Google Scholar]
- LaFave MC, Varshney GK, Vemulapalli M, Mullikin JC, Burgess SM. 2014. A defined zebrafish line for high-throughput genetics and genomics: NHGRI-1. Genetics 198:167–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Fliniaux I, Schneider P, Mikkola ML. 2012. Identification of ectodysplasin target genes reveals the involvement of chemokines in hair development. J Invest Dermatol. 132:1094–1102. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G, Goodson M. 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21:936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchinko KB. 2009. Predation’s role in repeated phenotypic and genetic divergence of armor in threespine sticklebacks. Evolution 63:127–138. [DOI] [PubMed] [Google Scholar]
- Marchinko KB, Schluter D. 2007. Parallel evolution by correlated response: lateral plate reduction in threespine stickleback. Evolution 61:1084–1090. [DOI] [PubMed] [Google Scholar]
- Marešová E, Delić A, Kostov V, Marić S, Mendel J, Sanda R. 2011. Genetic diversity of Sabanejewia balcanica (Actinopterygii: Cobitidae) in the western Balkans and comparison with other regions. Folia Zool. 60:335–342. [Google Scholar]
- Mason VC, Li G, Helgen KM, Murphy WJ. 2011. Efficient cross-species capture hybridization and next-generation sequencing of mitochondrial genomes from noninvasively sampled museum specimens. Genome Res. 21:1695–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Reno PL, Pollen AA, Bassan AI, Capellini TD, Guenther C, Indjeian VB, Lim X, Menke DB, Schaar BT, et al. 2011. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature 471:216–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhre F, Klepaker T. 2009. Body armour and lateral-plate reduction in freshwater three-spined stickleback Gasterosteus aculeatus: adaptations to a different buoyancy regime? J Fish Biol. 75:2062–2074. [DOI] [PubMed] [Google Scholar]
- Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, Wainwright PC, Friedman M, Smith WL. 2012. Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci U S A. 109:13698–13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton WHJ, Stumpenhorst K, Faus-Kessler T, Folchert A, Rohner N, Harris MP, Callebert J, Bally-Cuif L. 2011. Modulation of Fgfr1a signaling in zebrafish reveals a genetic basis for the aggression-boldness syndrome. J Neurosci. 31:13796–13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. New York: Springer-Verlag. [Google Scholar]
- Palandačić A, Matschiner M, Zupančič P, Snoj A. 2012. Fish migrate underground: the example of Delminichthys adspersus (Cyprinidae). Mol Ecol. 21:1658–1671. [DOI] [PubMed] [Google Scholar]
- Pardo-Diaz C, Salazar C, Baxter SW, Merot C, Figueiredo-Ready W, Joron M, McMillan WO, Jiggins CD. 2012. Adaptive introgression across species boundaries in Heliconius butterflies. PLoS Genet. 8:e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. 2006. In vivo enhancer analysis of human conserved non-coding sequences. Nature 444:499–502. [DOI] [PubMed] [Google Scholar]
- Perea S, Böhme M, Zupancic P, Freyhof J, Sanda R, Ozuluğ M, Abdoli A, Doadrio I. 2010. Phylogenetic relationships and biogeographical patterns in Circum-Mediterranean subfamily Leuciscinae (Teleostei, Cyprinidae) inferred from both mitochondrial and nuclear data. BMC Evol Biol. 10:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, et al. 2007. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 39:1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov AN, Hubbard SR, Schlessinger J, Mohammadi M. 2000. Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity. Cell 101:413–424. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. 2010. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar S, Visel A, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Morrison H, Fitzpatrick DR, Afzal V, et al. 2008. Human-specific gain of function in a developmental enhancer. Science 321:1346–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB. 2009. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 326:1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohner N, Bercsényi M, Orbán L, Kolanczyk ME, Linke D, Brand M, Nüsslein-Volhard C, Harris MP. 2009. Duplication of permits Fgf signaling to serve as a target for selection during domestication. Curr Biol. 19:1642–1647. [DOI] [PubMed] [Google Scholar]
- Shapiro MD, Bell MA, Kingsley DM. 2006. Parallel genetic origins of pelvic reduction in vertebrates. Proc Natl Acad Sci U S A. 103:13753–13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Haussler D. 2004. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol Biol Evol. 21:468–488. [DOI] [PubMed] [Google Scholar]
- Smith C, Spence R, Barber I, Przybylski M, Wootton RJ. 2014. The role of calcium and predation on plate morph evolution in the three-spined stickleback (Gasterosteus aculeatus). Ecol Evol. 4:3550–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence R, Wootton RJ, Barber I, Przybylski M, Smith C. 2013. Ecological causes of morphological evolution in the three-spined stickleback. Ecol Evol. 3:1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani F, Galli P, Zaccara S, Crosa G. 2004. Genetic variability and phylogeography of the cyprinid Telestes muticellus within the Italian peninsula as revealed by mitochondrial DNA. J Zool Syst Evol Res. 42:323–331. [Google Scholar]
- Storey JD. 2002. A direct approach to false discovery rates. J R Stat Soc B Stat Methodol. 64:479–498. [Google Scholar]
- Tang KL, Agnew MK, Hirt MV, Sado T, Schneider LM, Freyhof J, Sulaiman Z, Swartz E, Vidthayanon C, Miya M, et al. 2010. Systematics of the subfamily Danioninae (Teleostei: Cypriniformes: Cyprinidae). Mol Phylogenet Evol. 57:189–214. [DOI] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA. 2008. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 40:158–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitti JJ, Grossman SR, Sabeti PC. 2013. Detecting natural selection in genomic data. Annu Rev Genet. 47:97–120. [DOI] [PubMed] [Google Scholar]
- Wells KL, Hadad Y, Ben-Avraham D, Hillel J, Cahaner A, Headon DJ. 2012. Genome-wide SNP scan of pooled DNA reveals nonsense mutation in FGF20 in the scaleless line of featherless chickens. BMC Genomics 13:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead GG, Makino S, Lien C-L, Keating MT. 2005. Fgf20 is essential for initiating zebrafish fin regeneration. Science 310:1957–1960. [DOI] [PubMed] [Google Scholar]
- Whiteley AR, Bhat A, Martins EP, Mayden RL, Arunachalam M, Uusi-Heikkilä S, Ahmed ATA, Shrestha J, Clark M, Stemple D, et al. 2011. Population genomics of wild and laboratory zebrafish (Danio rerio). Mol Ecol. 20:4259–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G. 2012. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 13:59–69. [DOI] [PubMed] [Google Scholar]
- Wray GA. 2013. Genomics and the evolution of phenotypic traits. Annu Rev Ecol Evol Syst. 44:51–72 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.