Abstract
Coral reefs are one of the most spectrally diverse environments, both in terms of habitat and animal color. Species identity, sex, and camouflage are drivers of the phenotypic diversity seen in coral reef fishes, but how the phenotypic diversity is reflected in the genotype remains to be answered. The labrids are a large, polyphyletic family of coral reef fishes that display a diverse range of colors, including developmental color morphs and extensive behavioral ecologies. Here, we assess the opsin sequence and expression diversity among labrids from the Great Barrier Reef, Australia. We found that labrids express a diverse palette of visual opsins, with gene duplications in both RH2 and LWS genes. The majority of opsins expressed were within the mid-to-long wavelength sensitive classes (RH2 and LWS). Three of the labrid species expressed SWS1 (ultra-violet sensitive) opsins with the majority expressing the violet-sensitive SWS2B gene and none expressing SWS2A. We used knowledge about spectral tuning sites to calculate approximate spectral sensitivities (λmax) for individual species’ visual pigments, which corresponded well with previously published λmax values for closely related species (SWS1: 356–370 nm; SWS2B: 421–451 nm; RH2B: 452–492 nm; RH2A: 516–528 nm; LWS1: 554–555 nm; LWS2: 561–562 nm). In contrast to the phenotypic diversity displayed via color patterns and feeding ecology, there was little amino acid diversity within the known opsin sequence tuning sites. However, gene duplications and differential expression provide alternative mechanisms for tuning visual pigments, resulting in variable visual sensitivities among labrid species.
Keywords: visual ecology, RNA-Seq, opsins, reef fish, Labridae
Introduction
Coral reefs are one of the most spectrally diverse environments, both in terms of habitat and animal color (McFarland and Munz 1975; Goda and Fujii 1995; Marshall et al. 2003a). Reef fish show the greatest diversity of color and patterns within this particular habitat but the function of many of these colors and patterns is largely unknown. Reasons for their color have been speculated about since the time of Darwin and Wallace, and are likely to be related to species identity, sex, and camouflage (Darwin 1859; Wallace 1877; Cott 1940; Lorenz 1962; Thresher 1984; Marshall 2000; Siebeck 2004; Cheney, Grutter, et al. 2009).
In order to understand the behavioral and ecological functions of the colors and patterns displayed by coral reef fish, or indeed any animal, we need to analyze them using information on the visual systems that view them (Lythgoe 1979; Cuthill et al. 1999; Marshall et al. 2006). Coral reefs may contain several thousand species of fish, many of which are phylogenetically distinct, having arisen tens of millions of years ago (Near et al. 2012). In this article, the visual systems of the labrids are investigated; a polyphyletic group of colorful fishes with a well-resolved phylogeny that are found in both tropical and temperate waters (Westneat and Alfaro 2005; Cowman et al. 2009; Price et al. 2011). The labrids are a large group of both wrasses and parrotfish including more than 500 species in 60 genera, with a huge size range encompassing, for example, the pygmy possum wrasse, Wetmorella tanakai (4 cm total length) to the humphead parrotfish, Bulbometapon muricatum (130 cm total length) (Parenti and Randall 2000). Their feeding, reproductive and behavioral ecologies are diverse, and they include variable color morphs at different developmental stages (Randall et al. 1997; Allen et al. 2003). Additionally, several species possess specific “labrid complex-colors,” described by spectra with multiple peaks in wavelengths, allowing simultaneous communication and camouflage (Marshall et al. 2003a).
Little is known about the visual ecology of labrids. One of the reasons for this is that microspectrophotometry (MSP), a technique commonly used to determine the spectral sensitivities of fishes, is difficult to perform on labrids. The labrid retinal pigment epithelium (RPE), as in other fishes, is the site for enzymatic activities of the visual cycle including the isomerization of all-trans retinal to 11-cis retinal (Hubbard and Wald 1952). In labrids, the RPE is particularly thick, highly pigmented, and in close contact to the cone cell outer segments, causing damage to these parts of the cell when the pigment is removed (Kawamura and Tamura 1973; Fineran and Nicol 1974; Nicol 1975; Ali and Anctil 1976). From two successful studies of labrid visual ecology, one using MSP and extraction spectrophotometry (ESP) and the other using electroretinograms (ERG), we know that labrids are likely to have at least dichromatic color vision (Barry and Hawryshyn 1999; Losey and George 2003). However, these studies do not provide a complete picture—in Bodianus bilunnulatus no MSP recordings were taken from single cones, despite labrids having two different classes of single cones in addition to double or twin cones as is evident in both wholemounted and sectioned labrid retinas (Kawamura and Tamura 1973; Fineran and Nicol 1974; Nicol 1975; Ali and Anctil 1976).
Visual sensitivities in many animals can now be described to a first approximation with molecular techniques, and are determined by the absorption of light by visual pigments contained within retinal photoreceptors. Visual pigments are composed of an opsin protein bound to a chromophore that typically, but not always, is 11-cis-retinal (Wald 1968). Variation in spectral sensitivities can therefore arise from changes in opsin sequence, opsin expression, the type of the retinal chromophore (11-cis-retinal [A1] or 11-cis-3,4,didehydroretinal [A2]), and filtering mechanisms such as transmission properties of the cornea and lens (Loew 1976; Bowmaker 1995; Carleton and Kocher 2001; Siebeck and Marshall 2001; Shand et al. 2008; Yokoyama 2008). Additional diversity can arise from opsin gene duplications or gene losses within a family or even genus (Hofmann and Carleton 2009; Rennison et al. 2012; Meredith et al. 2013; Cortesi et al. 2015). These mechanisms have been shown to play a role in concert or individually, and may even occur within a single aquatic group (i.e., Lake Malawi cichlids [Hofmann et al. 2009; Hofmann et al. 2010]).
With a dichromatic visual system (Barry and Hawryshyn 1999), labrids might construct basic color vision, and be able to discriminate particular colors from the background (Lythgoe 1979; Marshall et al. 2003b). However from previous studies on the visual systems of other reef fish and retinal anatomy, it is likely that labrids are at least trichromatic with spectrally distinct single and double or twin cones (Losey et al. 2003; Hart et al. 2004; Pignatelli et al. 2010; Cheney et al. 2013). We might expect labrid visual sensitivities to vary between species as they have diverse ecologies and related anatomy (Loew and Lythgoe 1978; Levine and MacNichol 1979; Marshall 1988; Ferry-Graham et al. 2002; Wainwright et al. 2004; Schmitz and Wainwright 2011). Spectral variation could relate to the aquatic light environment, which changes with depth and water quality (Lythgoe 1979; Bowmaker et al. 1994; Meredith et al. 2013), or could relate to the fishes’ behavioral ecology, for example, their diet or sexual selection. (Ferry-Graham et al. 2002; Losey et al. 2003; Marshall et al. 2003b, 2015; Cronin et al. 2014).
In addition to the human-visible light spectrum (400–700 nm), several reef fish species have the potential to see ultra-violet (UV) light (<400 nm) with implications for private communication channels between individuals of the same species (Siebeck et al. 2010). In species with UV vision, the cornea and lens necessarily allow the transmission of UV wavelengths of light. In many species of labrids, UV light is filtered out by the lens and cornea by yellow carotenoid-based pigments that in fact allow little light below 500 nm to pass. Some authors have suggested this may be to prevent tissue damage via UV radiation as the retina is highly susceptible to photoxidative damage (Douglas and Marshall 1999); whereas others suggest reducing UV light may reduce chromatic aberration (Thorpe et al. 1993; Douglas and Marshall 1999; Siebeck and Marshall 2000); although UV vision may be more related to function (Douglas and Thorpe 1992). The distribution of the pigment may be uneven across the cornea, with densely pigmented areas often found toward the periphery, and a clear “window” toards the central area. Therefore, the presence of a short-wavelength absorbing pigment in the cornea does not necessarily mean that no UV light can be transmitted, if the lens is UV transmitting (Douglas and Marshall 1999; Siebeck and Marshall 2000). Where seeing UV light is important for intraspecific communication or identifying prey, such as zooplankton, both the lens and cornea should be UV-transmitting (Siebeck and Marshall 2001) and some wrasse species are know to be potential planktivores and may reflect UV from body colors (Marshall 2000). Therefore it is important to consider not only the spectral sensitivities of the photoreceptors but also the optical properties of the whole eye when studying animal visual systems.
In this study of labrid visual sensitivities, we hypothesized that there would be spectral sensitivity diversity within the labrids that is related to their ecology and ethology (table 1). To test this hypothesis, we utilized next-generation sequencing (NextGen RNA-Seq) of retinal transcriptomes of 12 different labrid species and took a molecular approach to examining visual system diversity (fig. 1). We also used opsin sequences and known data on spectral tuning sites to estimate spectral sensitivities (λmax) for each opsin expressed in the individual species. We then related our results to labrid behavioral ecology to assess to what extent visual sensitivity variation is correlated with ecology. Comparative opsin expression data within fish families have been used previously to study the visual system diversity of both the highly diverse freshwater cichlids (Cichlidae) and their closest relatives within the coral reefs, the damselfish (Pomacentridae) (Carleton et al. 2000, 2008; Carleton and Kocher 2001; Hofmann et al. 2009, 2010, 2012; O'Quin et al. 2010; Smith et al. 2011).
Table 1.
A Brief Summary of Published Ecological and Visual Data on the Labrids in This Study, (Randall et al. 1997; Siebeck and Marshall 2001, 2007; Allen et al. 2003).
| Species | Lens T50 and Type | Cornea T50 and Type | Diet | Depth (m) | Opsins |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SWS1 | SWS2B | SWS2A | RH2B | RH2A | LWS | RH1 | |||||
| Gomphosus varius | 433 (I) | 348–445 (III) | Invertebrates | 1–30 | — | 1 | — | — | 2 | 1 | 1 |
| Thalassoma lunare | 431 (I) | 386–489 (III) | Fish eggs, small fishes, invertebrates | 1–20 | — | 1 | — | — | 1 | 1 | 1 |
| Coris gaimard | 430 (I) | 347–393 (III) | Invertebrates | 2–78 | — | 1 | — | — | 2 | 1 | 1 |
| Halichoeres ornatissimus | 426 (I) | 392–513 (III) | Invertebrates | 4–15 | — | 1 | — | — | 1 | 1 | 1 |
| Halichoeres chrysus | — | — | Invertebrates | 2–70 | — | 1 | — | — | 1 | 1 | 1 |
| Halichoeres chloropterus | — | — | Invertebrates | ≤ 10 | — | 1 | — | — | 1 | 1 | 1 |
| Labroides dimidiatus | 414 (I) | 361 (I) | Zooplankton and parasitic crustaceans | ≤ 40 | — | 1 | — | — | 1 | 1 | 1 |
| Cirrhilabrus punctatus | 378 (I) | n/a (IV) | Invertebrates | 1–50 | 1 | — | — | — | 4 | 1 | 1 |
| Chlorurus sordidus | 425 (I) | 388 (II) | Algae and sand | ≤ 40 | — | 1 | — | — | 1 | 2 | 1 |
| Epibulus insidiator | 426 (I) | 398–495 (III) | Invertebrates and small fishes | 1–42 | — | 1 | — | — | 2 | 2 | 1 |
| Choerodon fasciatus | 393–403 (I) | 387–500 (III) | Invertebrates | 5–35 | 1 | 1 | — | — | 1 | 1 | 1 |
| Bodianus mesothorax | 374 (I) | 378–453 | Zooplankton and parasitic crustaceans | 5–40 rarely >20 | 1 | 1 | — | 1 | 2 | 1 | 1 |
Note.—Also noted is the number of opsin genes identified in this work.
Fig. 1.
Phylogeny of the Labridae modified to show the species in this analysis (modified from [Cowman et al. 2009; Kazancioglu et al. 2009; Price et al. 2011]). Where known, branches show the different feeding ecology of each species, and numbers at nodes show divergence estimates in time (Ma). Filled circles represent the opsins that were expressed in each of the species sampled in this study. From L-R: SWS1, SWS2B, RH2B, RH2A, RH2Ab, LWS1, LWS2.
Results
Assemblies
The RNAseq data obtained by multiplexing eight retinal samples in a lane provided sufficient data to assemble and quantify opsin transcripts. From the 12 labrid samples, we obtained an average of 25.5 ± 8 million paired end reads. Approximately 80% of the pairs passed trimming, and after assembly produced an average of 63,000 transcripts. Because the opsins are some of the most highly expressed genes in the retina, this was sufficient data to construct complete opsin sequences for at least three cone opsins and the rod opsin per species and quantify their expression. This multiplexing approach makes RNAseq a new and affordable way to rapidly survey visual systems and opsin diversity in new groups of organisms.
Phylogenetic Trees
BLAST output using the opsin gene sequences from Oreochromis niloticus (Spady et al. 2006) identified putative opsins from all of the labrid species. All labrids expressed an RH1 opsin and at least three cone opsins from the known opsin classes (SWS1, SWS2B, RH2A or RH2B and LWS) with the exception of SWS2A, which was not identified in any of the species studied. For the majority of opsin genes, we obtained full-length sequences (supplementary table S1, Supplementary Material online), though there were a few exceptions. For one species, Cirrhilabrus punctatus, there were four versions of the RH2 opsin that were highly expressed, with good E-values with highly similar coding sequences (supplementary table S1, Supplementary Material online). An additional four putative RH2 opsin sequences were also found in this species, although these had low E-values and shorter lengths (726–757 bp) than known RH2 opsins and so were excluded from further analysis. There were similar results for other genes such as an SWS1 gene in Chlorurus sordidus, and putative RH2B transcripts in Chl. sordidus, Halichoeres chrysus, C. punctatus, and Choerodon fasciatus (three potential genes). These transcripts had low E-values, or were of a shortened length than expected and so were discounted from the analysis.
Protein distance trees confirmed the identity of the opsins as they grouped with well-known opsin classes defined by the other fishes. Nucleotide trees determined by jModelTest were consistent with the protein trees (fig. 2; supplementary figs. S1–S4, Supplementary Material online) and both trees grouped the labrid opsins together in a group that was distinct and separate from the other fishes (i.e., Danio rerio and O. niloticus). Labrid opsin phylogenetic trees agreed with previously published phylogenies based on other genetic information (Cowman et al. 2009; Price et al. 2011); figs. 1 and 2). The phylogenetic trees suggested potential gene duplications for Epibulus insidiator and Chl. sordidus in the LWS opsin class, whereas E. insidiator, Gomphosus varius, and Coris gaimard all showed potential gene duplications in the RH2A opsin class (fig. 2).
Fig. 2.
Maximum likelihood trees showing phylogenetic relationships between (A) RH1, RH2A, and RH2B, (B) SWS1 and SWS2B, and (C) LWS opsin genes of the labrids used in this study. Trees were created using Phyml based on 100 bootstrap samples. Filled circles represent a 95% confidence at that node.
Gene Expression
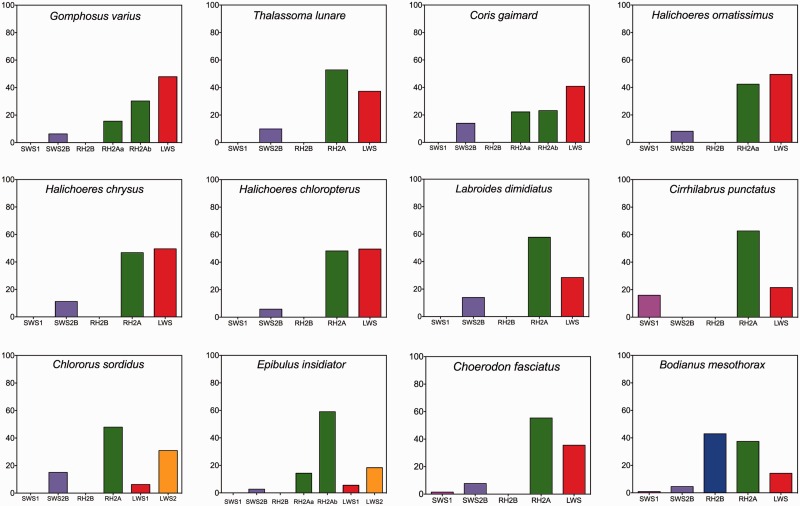
The predominant labrid visual system is based on expression of three cone opsin genes: SWS2B, RH2A, and LWS. The labrids expressed mainly LWS and RH2A opsins with SWS1 and SWS2B opsins making up a much smaller percentage of the overall opsin expression. In all labrids except G. varius and Halichoeres ornatissimus, RH2A was preferentially expressed over LWS. In these two species, LWS was preferentially expressed (fig. 3). There were some variations on this three-gene system. Epibulus insidiator and Chl. sordidus expressed two unique LWS copies (fig. 3). Two RH2A genes were expressed in E. insidiator, G. varius, and Co. gaimard. Bodianus mesothorax was the only labrid to express an RH2B gene and express it at a similar level to RH2A opsin expression in other species. Only three species significantly expressed a copy of a full length SWS1 opsin (B. mesothorax, C. punctatus, Cho. fasciatus) although expression levels were lower than SWS2B for B. mesothorax and Cho. fasciatus. While transcripts suggested C. punctatus expressed an SWS2B gene in addition to SWS1, expression levels were so low that sequencing failed to obtain a full-length gene sequence. Therefore, C. punctatus was the only wrasse to show significant SWS1 opsin expression rather than the SWS2B opsin class.
Fig. 3.
Opsin expression levels (% of expressed cone opsins) for each species of labrid in this study. Opsins are sorted into the common vertebrate opsin classes. Where putative gene duplications are suggested in E. insidiator, Co. gaimard, G. varius, and Chl. sordidus, relative percentage expression levels for each gene are plotted separately. SWS2A was omitted from the graph, as it is not expressed in any members of the labrids in this study. For each species, n = 1.
Spectral Sensitivities
Between species, there was little variation at the known tuning sites (supplementary figs. S5–S8, Supplementary Material online), resulting in similar estimations of spectral sensitivities across the family. All estimated spectral sensitivities were well within the ranges reported for other reef fishes (fig. 4; Losey et al. 2003; Hart et al. 2004; Cheney, Skogh, et al. 2009; Cheney et al. 2013). Estimates for λmax are reported below as ranges for all species analyzed; and in most cases, are the same for individual species as there was low amino acid variation between species at critical tuning sites (fig. 4; supplementary figs. S5–S8, Supplementary Material online).
Fig. 4.
Ranges for the spectral sensitivities of the labrid family, as estimated from opsin amino acid sequences (nm; bold bars). Solid filled rectangles represent the different opsin classes: from L-R = SWS1, SWS2B, RH2B, RH2A, LWS1, LWS2. Lightly shaded regions show the range of spectral sensitivities from MSP and ERG recordings of single cones from a range of coral reef fishes. Readings from double cones and twin cones in that same study range from 457 to 548 nm and the dotted rectangular outline represents these values (Losey et al. 2003).
The rod pigment, RH1 has an estimated λmax of between 486 and 499 nm which is within the range of MSP measurements made on other reef fish (RH1 = λmax of 477–502 nm; (Losey et al. 2003). For the cone pigments, only three species of labrid (B. mesothorax, C. punctatus, and Cho. fasciatus) expressed low levels of the SWS1 visual pigment (usually UV-sensitive) with an estimated λmax of between 356 and 370 nm, well within the UV range (<400 nm). The SWS2B opsin gene expressed by all other species (except C. punctatus) was calculated to have visual pigment with an estimated λmax of between 421 and 451 nm. RH2A and RH2B pigments were separated by a difference at amino acid 122 (RH2A = 122 E; RH2B = 122Q) that shifts the RH2B visual pigment from B. mesothorax into the blue-sensitive region with an estimated λmax between 452 and 492 nm (Yokoyama et al. 1999). RH2A visual pigments were estimated to have a λmax of between 516 and 528 nm. We calculated narrow spectral sensitivities for the LWS visual pigments for all labrids. The majority of labrids expressed one LWS opsin (LWS2) with an estimated visual pigment λmax of 561–562 nm. Interestingly, the estimates for the LWS2 visual pigment λmax fall outside the range of λmax recorded from double cones from a range of Hawaiian coral reef fishes: 457–548 nm (Losey et al. 2003).
In E. insidiator and Chl. sordidus where two LWS genes were expressed; the two pigments (LWS1 and LWS2) are estimated to be spectrally distinct, with LWS1 estimated to have a spectral sensitivity of between 554 and 555 nm compared with the LWS2 pigment that is estimated to have a spectral sensitivity of between 561 and 562 nm (fig. 4). The significant difference at a known tuning site between LWS1 and LWS2 (bovine rhodopsin site A164S) in both E. insidiator and Chl. sordidus strongly suggests a gene duplication event within the labrid family (bovine rhodopsin site A164S; supplementary fig. S7, Supplementary Material online).
It is important to note that estimating λmax from sequence data alone may be inaccurate due to a lack of knowledge of all tuning sites within the opsin sequences. Additionally, experimental mutations at particular sites may cause different changes in λmax depending on the animal model or in vitro background (Yokoyama 2012). As such, we have presented λmax data as ranges for individual labrid species in order to generate hypotheses about the role of spectral sensitivities.
Phylogenetic Analysis by Maximum Likelihood (PAML)
As only three species expressed SWS1, and one expressed a full length RH2B gene, PAML analysis was not carried out on these opsins. Of the other expressed opsins, when comparing codeml models 1a and 2, all opsins were under positive selection (SWS2B: P = 9.62 e − 10; RH2A: P = 5.40 e−4; LWS: P = 4.43 e−13; RH1: P = 3.37 e−4; supplementary tables S2 and S3, Supplementary Material online). Similarly, when comparing codeml models M8 and M8a, all expressed opsins were under positive selection (SWS2B: P = 1.94 e−9; RH2A: P = 3.81 e−4; LWS: P = 1.22 e−12; RH1: P = 4.89 e−3; supplementary tables S2 and S3, Supplementary Material online). The PAML model 1/2a identified several sites under positive selection in RH1 (three), RH2A (three), LWS (7), and SWS2B (9), whereas model 8/8a identified more sites than model 1/2a (RH1 = 6; RH2A = 13; LWS = 17, and SWS2B = 19; table 2). Although both the PAML model 1/2a and 8/8a comparisons identified several sites within each opsin that were under positive selection (P ≥ 0.95), only one in RH1 (labrid and bovine site 292) and one in SWS2B (bovine site 116/labrid site 122) agreed with known tuning sites as described previously (table 2; supplementary figs. S5–S8, Supplementary Material online) (Yokoyama 2008).
Table 2.
Number of Sites with Amino Acid (AA) Differences within the Transmembrane and Retinal Binding Pocket Regions for the Opsin Classes and Number of Positively Selected Sites Identified Using PAML.
| Sites with Amino Acid Differencesa |
Sites under Selection: P > 0.95a |
Significant PAML Result (P > 0.95) at Known Tuning Sites for Specific Opsina |
||||||
|---|---|---|---|---|---|---|---|---|
| Total | Transmembrane Regions | Retinal Binding Pockets | Known Tuning Sites for Specific Opsin | PAML Model 1/2a | PAML Model 8/8a | PAML Model 1/2a | PAML Model 8/8a | |
| RH1 | 34 | 22 | 3 | 2 (292 and 299) | 3 | 6 | 1 (292) | 1 (292) |
| RH2A | 65 | 36 | 3 | 0 | 3 | 13 | 0 | 0 |
| LWS | 97 | 48 | 13 | 1 (177) | 7 | 17 | 0 | 0 |
| SWS2B | 106 | 55 | 15 | 4 (52, 115, 122, and 275) | 9 | 19 | 0 | 1 (122) |
Note.—These results show that SWS2B has the highest number of sites with amino acid differences between the labrid species, followed by LWS, then the RH2A and RH1 opsins. The PAML models find similar results (Sites under selection: P > 0.95) although there were only two known tuning sites that were under positive selection as determined by the PAML models (where P > 0.95).
aAA sites based on labrid sequence numbers. Please refer to amino acid alignments (supplementary figs. S5–S8, Supplementary Material online) for corresponding bovine rhodopsin (RH1) amino acid site numbers.
Discussion
All labrids we studied express at least three classes of visual opsins: one SWS opsin, one RH2 opsin, and one LWS opsin. A comparison of opsin expression data across labrid genera suggests that labrid genomes contain SWS1, SWS2B, RH2B, RH2A, and LWS opsin genes, with species varying in which genes are expressed. This includes likely duplications of the RH2A and LWS opsins in several lineages (figs. 1 and 2). Our phylogenetic trees of opsins fit with previously published phylogenies of the labrid family—see figure 1 (Kazancioglu et al. 2009; Price et al. 2011). Our results also suggest that older lineages (Hypsigenyines, Odacines, Cheilines, Labrines, and Scarines) express multiple visual opsin classes (SWS1, SWS2B, RH2A, RH2B, and at least one LWS gene) whereas more recent lineages (Pseudocheilines, Novaculines, Pseudolabrines, Labricthyines, and Julidines) express predominantly three opsin classes—SWS2B, RH2A, and LWS.
The molecular sequences of each opsin class are relatively similar between labrid species, and only differ by a few amino acid substitutions within the critical transmembrane and retinal binding pocket regions (table 2). At known tuning sites there are few differences between species, with 2 (RH1), 0 (RH2A), 1 (LWS), and 4 (SWS2B) amino acid substitutions at known tuning sites between the 12 species studied (table 2). The overall sequence differences between species are much higher than those observed in cichlids, where expressed opsins were found to be essentially identical, with no differences at known tuning sites (Carleton and Kocher 2001). This result is consistent with the older age of the labrid genera. The greater difference in opsin sequence diversity in labrids compared with cichlids may predict greater visual pigment diversity within this family, as a change in the opsin sequence at novel spectral tuning sites can have dramatic effects on the spectral sensitivity of the visual pigment within the photoreceptor (Carleton and Kocher 2001; O'Quin et al. 2010; Hofmann et al. 2012). This result would be very different to that found in cichlids, where gene expression was shown to drive spectral sensitivity diversity (Carleton and Kocher 2001). We also found diversity in both opsin gene duplications and opsin gene expression, suggesting visual pigment spectral sensitivity diversity should be higher in the labrids than that seen in other fish, including the well-studied cichlids.
PAML
To identify whether the diversity seen between opsin sequences is being positively selected for, we ran PAML analyses on the labrid opsins. Both the PAML model 1/2a and 8/8a comparisons identified several sites within each gene that were under positive selection (P ≥ 0.95), but there was little overlap between PAML-identified sites that were undergoing positive selection and known spectral tuning sites, the transmembrane region or the retinal binding pocket regions (thought to be important in tuning vertebrate opsin spectral sensitivity (table 2). In fact, there were two such sites (bovine/labrid site 292 in RH1 and bovine 116/labrid 112 in SWS2B; table 2) where PAML-identified sites overlapped with known tuning sites. Of particular interest was the lack in finding many of the known tuning sites (e.g., bovine sites: 164, 261, 269, and 292 in LWS) that have a significant effect on the spectral tuning properties of those opsins. Similar to results in another reef fish family, the Pomacentrids (damselfish), many of the positively selected sites identified by PAML were found in the transmembrane regions and not the retinal binding pocket where it is assumed that variations in amino acids will have a greater effect on spectral tuning (Chang et al. 1995; Hofmann et al. 2012). In this study, many of the PAML-selected sites were found close to or adjacent the retinal binding pocket (supplementary figs. S5–S8, Supplementary Material online) suggesting either the tuning sites are shifted in labrids or the binding pocket regions are highly conserved and differences in amino acids outside this pocket have little effect on spectral tuning. In fact, changes in amino acids in the transmembrane region may have indirect effects on spectral tuning, or may be important for other aspects of visual pigment function and not spectral tuning (Schott et al. 2014). Therefore, at least for coral reef fish visual pigments, PAML appears to miss several spectrally important amino acid sites, and so is unlikely to provide definitive information on spectral tuning, until we know more about the functions of specific amino acids sites within labrid visual pigments.
Gene Duplications
Our study found putative gene duplications in both the LWS and RH2 genes. The RH2A gene duplication we found in the labrids may have occurred up to 59 Ma, based on phylogenetic data from previous studies (node where Labrines, Scarines, and Cheilines branch away from the Pseudocheilines, Novaculines, Pseudolabrines, Labricthyines, and Julidines [Cowman et al. 2009]). Based on our phylogenies, the new RH2A gene duplication in labrids (RH2Aa and RH2Ab) is likely to have been lost by some groups, while being retained or even reduplicated in others (fig. 2). We found that RH2Aa and Rh2Ab nucleotide sequences differed by 10%, 6%, and 10% in E. insidiator, G. varius, and Co. gaimard, respectively, and amino acid sequence differences were similar: 10%, 5%, and 9%. These differences were found throughout the length of the sequences, suggesting misassembly is unlikely (supplementary fig. S6, Supplementary Material online, for amino acid alignments). All RH2A genes were situated far enough apart on our phylogenetic trees (fig. 2), to support them being real genes and not alternate alleles.
Duplications of the RH2 gene have occurred several times in fish and are probably more common than we think. For example, the zebrafish D. rerio has four separate RH2 genes whereas the Pacific bluefin tuna (Thunnus orientalis) has five RH2 genes, the most reported of any fish yet (Chinen et al. 2003; Rennison et al. 2012; Nakamura et al. 2013). Similar gene duplication events have happened within other fish lineages, including coral reef fishes where several duplications of the SWS2 opsin have occurred (Cortesi et al. 2015). Despite the knowledge that many fish, and indeed other animals express several versions of the same opsin gene, there is still little understanding of the function of expressed opsin gene duplications (Marshall et al. 2015).
The RH2Aa and RH2Ab genes in this study have phylogenetically distinct sequences, although they show no differences at known tuning sites (supplementary fig. S6, Supplementary Material online). These results could suggest that there are additional tuning sites within these genes that we do not know about. Or, if expression of RH2Aa and RH2Ab produces visual pigments that are spectrally sensitive to similar wavelengths (through future protein expression analyses), these pigments may be found in different photoreceptors across the retina, contributing to inter-retinal variation in opsin expression. For example, RH2Aa may be found in single cones or one member of a double cone, whereas RH2Ab may be coexpressed with the LWS gene or found in the other member of a double cone. Many animals, including fish have been shown to have differential expression of opsins, or cone cells with differing spectral sensitivities in particular areas of the retina, suggesting that the retina is organized to optimize the visual system (for review, see [Temple 2011]). To understand whether the RH2A genes that we have found in the labrids are spectrally distinct, or expressed in different photoreceptors across the retina, site-directed mutagenesis of expressed proteins and in situ hybridization would need to be carried out to test potential spectral tuning sites.
Slightly less common among fishes is duplication in the LWS gene, although this may be because the longer wavelengths are of greater visual importance to the labrids compared with Pseudochromids where SWS duplication occurs (Cortesi et al. 2015). Only E. insidiator and Chl. sordidus expressed two LWS opsin genes, LWS1 and LWS2. From the existing labrid phylogenies, it is likely that this LWS gene duplication occurred around 53 Ma when the Scarines (Chl. sordidus) and Cheilines (E. insidiator) split from the Labrines (fig. 1). Individuals in the Hypsigenyines (an older order of labrids) do not have this gene duplication, suggesting it is a derived feature of the Cheilines and Scarines.
Interestingly, the LWS2 gene duplicate in E. insidiator and Chl. sordidus suggests these species could be specializing in longer wavelength vision (Losey et al. 2003; Marshall et al. 2003b). Previous research into labrid RPE has identified a unique red pigment within the RPE, not found in other fish families, that strongly absorbs light at wavelengths less than 600 nm (Fineran and Nicol 1974). This compound has been suggested to act as a filter on labrid-specific long single cones, suggesting longer wavelength vision may be possible in this family than for other coral reef fishes (Fineran and Nicol 1974; Nicol 1975; Ali and Anctil 1976; Lythgoe 1979; Marshall et al. 2003b). Recent work has also confirmed the presence of fluorescent body patterns (with reflectance spectra peaks >600 nm) that may be involved in territoriality and intraspecific communication in labrids (Michiels et al. 2008). Long wavelength vision may also be useful in the detection of algae or other chlorophyll containing substances, for example in foraging, as they have reflectance spectra peaks greater than 600 nm.
There are three known cone cell types in labrids (long single cones, short single cones, and unequal double cones [Fineran and Nicol 1974]). In vertebrates, LWS opsins are normally expressed within double cone cells, along with a proportion of the RH2A opsins (Cheng and Flamarique 2007; Palacios et al. 2010; Novales Flamarique et al. 2012). A LWS gene duplication in labrids suggests either that each member of the double cone is expressing a different LWS opsin or that they are both expressed within the same double cone cell member, with an RH2A opsin. For example, LWS1 may be found in one member of the labrid double cone, whereas the other member expresses an LWS2 opsin. Alternatively, LWS1 and LWS2 could be coexpressed in specific single, double, or twin cones across the retina, either with RH2A or another opsin gene (Dalton et al. 2014). Coexpression of two spectrally unique visual pigments within the same photoreceptor would help to broaden the sensitivity of a particular cone, by increasing the range of wavelengths that the individual cone is sensitive to, thereby establishing broadband sensitive photoreceptors (Temple et al. 2010; Dalton et al. 2014). Opsin coexpression may also allow for better contrast detection, which may be useful for fish trying to identify potential prey or even the early detection of predators through the water column (Dalton et al. 2014). Confirmation of these hypotheses awaits in situ labeling or other more cell-specific techniques. Clearly, the Labridae show diversity within both the gene sequence and in gene duplications, which may, in turn lead to spectral sensitivity diversity within the family, as not all labrids express the same complement of opsins.
Gene Expression Diversity
There was considerable diversity within the labrids in terms of both the opsins expressed within the retinal tissue, and the relative levels of each opsin that were expressed by different species (fig. 3). The majority of species expressed predominantly LWS and RH2 opsins with only a small percentage of SWS opsin expressed (mean LWS + RH2% expression: 90.2% ± 4.26 SD; mean SWS % expression: 9.77% ± 4.26 SD). Most species expressed greater levels of RH2 than LWS, with exceptions being Halichoeres spp. and Chl. sordidus, the latter with duplication in the LWS gene. Of the three species that expressed SWS1, only one, C. punctatus expressed relatively high levels of the gene (15% of expressed opsins cf. 0.9% and 1.42% for B. mesothorax and Cho. fasciatus, respectively), suggesting functionality of UV-vision may not be present in the other two species. As in previous work on cichlids (Carleton and Kocher 2001; Carleton 2009; Hofmann et al. 2009; O'Quin et al. 2010), we find considerable variation in relative opsin expression, which, in turn leads to diversity in relative visual pigment levels across the retina, suggesting visual diversity stems from gene expression diversity. The lack of a full-length SWS2B gene in C. punctatus and RH2B gene in several species, and the low levels of expression of the partial-length gene could suggest presence of an expressed copy of both SWS2B and RH2B in a different ontogenetic phase, (i.e. larval phases) or expression in only a few cone cells across the retina, suggesting subsequent in situ hybridization experiments would be interesting in this species. Next-Gen sequencing using a multiplexing approach may have biased our results to only highly expressed opsins within the retina, so subsequent investigations into the species in this study and other labrids may find full-length SWS2B and RH2B genes if less barcoding is used.
Spectral Sensitivities
From the diversity we found in gene sequence, gene expression, and the highly positive selection suggested by PAML models, we expected spectral sensitivities for the labrids to be diverse too. Our estimates of labrid spectral sensitivity are broad; however, there appears to be little variation between species (fig. 4), with all species fitting into similar λmax ranges. There were few amino acid substitutions at key tuning sites; LWS amino acid sequences were the same for all species except in E. insidiator and Chl. sordidus where the S164A substitution is predicted to shift the visual pigment λmax to shorter wavelengths (−7 nm). The SWS1 sequence was also identical between species at known tuning sites, except in B. mesothorax where the substitution Y265W is likely to shift the visual pigment λmax to longer wavelengths (+10 nm). The most variation was seen in the SWS2B amino acid sequence, with many substitutions seen over four main tuning sites. E. insidiator is predicted to have a longer wavelength SWS2B pigment than the other labrids in the study as it did not contain the substitutions S97C (−17 nm, A109G (−2 nm), or I116T (−7 nm); supplementary table S4, Supplementary Material online) (Chang et al. 1995; Yokoyama 2008).
Our estimates of labrid spectral sensitivities fit well with, and extend the two known previous studies of labrid visual ecology, suggesting our methods provide reasonable estimates of λmax for different photoreceptor types in the retina (Barry and Hawryshyn 1999; Losey et al. 2003).
Although our λmax estimations for the visual pigments in labrids are close to previous work, our calculations for the λmax of all opsins in the labrids were based on predictions from only two fish species, O. niloticus and Oryzias latipes. Neither of these fish species is a coral reef fish, as there is a lack of information on both opsin sequence and corresponding λmax in reef fishes. We calculated λmax estimations based on amino acid substitutions being additive, for simplicity. There is literature to suggest that substitutions at key tuning sites may not always be additive (Asenjo et al. 1994; Hauser et al. 2014) and so these estimates should be taken as a first approximation, until MSP or behavioral work can confirm spectral sensitivities for labrids.
UV Vision
Both our estimates of λmax and gene expression data identified three species with potential for UV vision: C. punctatus, Cho. fasciatus, and B. mesothorax. Two of these species, Cho. fasciatus and B. mesothorax expressed SWS1 at such low levels that it was barely measurable (fig. 3). One of the reasons for low SWS1 expression in labrids as a family could be individual variation (we only sampled single individuals), or for the one species where we subsampled the retina, inadvertently sampling from an area of the retina with a low density of SWS cone cells. Additionally, as the expression levels of this opsin class were so low, it may be that there is no functional significance for UV sensitivity at this particular life stage, although UV cones may be present in the juvenile retina. In other reef fish, such as damselfish (Pomacentridae), potential UV vision is only likely in the adults, with juvenile fish possessing the violet sensitive SWS2B visual pigment, whereas adults are UV sensitive (McFarland and Loew 1994; Braun et al. 2014).
UV vision in reef fish may be implied based on transmission by the ocular media (Siebeck and Marshall 2000, 2001). For most reef fish, there has been little research on UV vision, the pomacentrids being the best-studied family from MSP, transmission, body patterns, and behavior (Losey and George 2003; Losey et al. 2003; Siebeck 2004; Siebeck and Marshall 2007; Siebeck et al. 2010). There are many suggested functions for UV vision in fishes and at least for B. mesothorax and C. punctatus, planktivory appears to be the most obvious (Schmitz and Wainwright 2011). UV vision has been hypothesized to increase the contrast between UV-absorbing zooplankton, and the scattered UV background of a coral reef, therefore increasing the prey detection ability of the zooplanktivore (Bowmaker and Kunz 1987; Douglas and Hawryshyn 1990; Loew et al. 1993; Browman et al. 1994; McFarland and Loew 1994). Choerodon fasciatus is not known to be planktivorous as an adult (table 1) and so expression of SWS1 was particularly interesting, as piscivorous coral reef predators are not thought to be UV-sensitive (Siebeck and Marshall 2001). SWS1 expression therefore may be nonfunctional in the adult, but used in the juvenile—when zooplanktivory may be an important factor for foraging, similar to the situation in rainbow trout (Cheng and Flamarique 2007). Alternatively, expression may be upregulated should long-term ecology require it, for example in salmonids, SWS1 expression is turned off in the adults but the functionality is available, should conditions change (Loew et al. 1993; Browman et al. 1994)
Relating spectral sensitivities to aspects of labrid ecology may provide greater insight into why they have the visual systems that they do. For example, studies on damselfish have suggested that UV vision may be beneficial for discriminating individuals using UV-reflecting body patterns (Siebeck 2004; Siebeck et al. 2010). Although Cho. fasciatus has several UV reflective patches on its body, including its blue teeth, the transmission of the cornea and lens (T50 lens = 397 nm; T50 cornea = 387–500 nm; table 1; fig. 5) and the low expression of the SWS1 gene suggest it is unlikely that UV vision is of considerable importance to this species, at least in adult phase. Instead, it may be that the orange stripes (peak reflectance ∼600 nm; fig. 5) may be more relevant for signaling (Braun et al. 2014). For any ecological comparisons, however, we would need to incorporate data from several individuals of each species for robust quantitative and statistical comparisons, and therefore utilize techniques such as quantitative polymerase chain reaction.
Fig. 5.
(A) Spectral reflectance of specific color patches on Cho. fasciatus. Wavelengths less than 400 nm (marked by the solid vertical black line) are considered to be UV. (B) Putative visual pigment absorption curves for Cho. fasciatus based on the median calculated λmax from specific cone opsin sequences (from L-R: SWS1, SWS2B, RH2A, LWS). Curves are based on the A1 visual pigment template (Bowmaker et al. 1994). The dotted line represents the lens transmission (T50 = 397 nm) whereas the dashed line represents the cornea transmission (T50 = 387–500 nm; (Siebeck and Marshall 2007).
Important future steps include modeling reflectance spectra of labrid color patterns though the eyes of the fish themselves in the putative color space of these animals (Vorobyev and Osorio 1998), thus highlighting any potential communication or camouflage patterns. In, addition, a greater understanding of the visual acuity of the species in question would highlight which of the beautiful and intricate labrid patterns are visible to conspecifics, and which are visible to interspecifics (Marshall 2000; Marshall et al. 2003b).
As such, there remains much to learn about both the spectral sensitivities and opsin expression in labrids, as in other coral reef fish families. Future work will concentrate on linking spectral sensitivities of individual photoreceptors with opsin sequences, and potential additional tuning sites within them using protein expression. Additionally, MSP, in situ hybridization, and further RNA-Seq will allow us to explore various theories on the evolution of visual systems, such as the idea of neutral drift or relaxed adaptation in which several spectral sensitivity combinations might perform equally well (Marshall et al. 2015).
Conclusions
In conclusion, labrid visual system diversity is likely to be influenced by multiple mechanisms: opsin sequence variation, changes in gene expression between species, and gene losses and duplications over a number of as yet poorly understood timescales. As labrids are one of the reef fish groups that undergo sex-change, it would be particularly interesting to examine this in context of any retinal changes. These mechanisms are likely to shape the possible visual pigments that can be utilized by labrids to function and perform their many visual tasks. Such diversity warrants further investigation.
Materials and Methods
Species Chosen
We examined the retinal transcriptomes of 12 species of labrid fish. These were chosen from across the labrid phylogeny (Cowman et al. 2009; Price et al. 2011; fig. 1). The majority were wrasses and included C. punctatus, Co. gaimard, E. insidiator, G. varius, Halichoeres chloropterus, H. chrysus, H. ornatissimus, Labroides dimidiatus, and Thalassoma lunare. We also included B. mesothorax, Cho. fasciatus, and a parrotfish, Chl. sordidus. These species include a variety of foraging styles and habitats (table 1). To ensure good transcriptome assemblies we included only one individual per species.
Fish were collected either at Lizard Island Research Station (14° 40′ 8″ S, 145° 27′ 34″ E) or from nearby locations on the Great Barrier Reef via Cairns Marine. Fish were euthanized according to ethics (The University of Queensland Animal Ethics Council Approval No. QBI/192/13/ARC) using a 2 mg / ml solution of clove oil in methanol and seawater. Retinas were dissected out and preserved in RNAlater within 15 min of euthanasia to prevent RNA degradation. RNA was isolated from one or two whole retina for all species except E. insidiator, whose large retina was cut in half. Total RNA was extracted with an RNeasy kit (Qiagen) and quantified and quality checked on an Agilent 2100 Bioanalyzer. RNAseq libraries were made using the TruSeq RNA Sample kit Preparation Kit v.2 (Illumina, San Diego) by the Genomics Facility of the Queensland Brain Institute and sequenced on an Illumina HiSeq 2000 (chemistry v.3). Individual species were barcoded and multiplexed eight to a lane to generate 100 bp paired end sequences.
The demultiplexed data was checked with FastQC version 0.10.1 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/, last accessed October 17 2015). Data were trimmed using Trimmomatic version 0.32 (Bolger et al. 2014) to remove overrepresented sequences and to retain sequences with a minimum quality score of 20 and a minimum length of 80 bp. Only paired sequences were included in the final assembly, which was completed using Trinity version r20140413 (Grabherr et al. 2011; Haas et al. 2013) with a minimum coverage of two to join contigs.
To test the pipeline and confirm the parameters used in trimming and assembling the data, we also assembled paired end reads from a retinal transcriptome of the cichlid fish, Metriaclima zebra which was sequenced to a comparable depth by the Broad Institute (Brawand et al. 2014). For this species, we had previously mapped the reads to the M. zebra genome and counted the reads with Cufflinks (Schulte et al. 2014). This enabled us to compare the transcript sequences as well as their expression level obtained using a de novo approach based on Trinity with a genome guided approach based on the Tuxedo suite (Trapnell et al. 2010, 2012).
Phylogenetic Trees
To accurately group new labrid opsin sequences into ancient vertebrate visual opsin classes, we constructed nucleotide trees using opsin sequence information from various vertebrate species. First, labrid opsin genes were identified from the resulting FASTA files, by querying transcripts with the cichlid opsin genes of O. niloticus (Spady et al. 2006). Using TBLASTx, we identified transcripts that were rod and cone opsins as well as other opsins including vertebrate ancient opsins, melanopsins, and pinopsins. Both full and some partial sequences were returned, with partial sequences typically having few reads. To ensure accurate results in the rest of the analysis, we only used full-length sequences in further analyses. Amino acid sequences were derived from nucleotide sequences using DNAtranslate at the Swiss Institute of Bioinformatics (ExPASy DNA Translate Tool, (Artimo et al. 2012).
Each transcript was then assigned to a particular opsin class based on the phylogenetic relationships of their coding sequences with opsin genes from Ory. latipes, O. niloticus, M. zebra, and Lucania goodei, sourced from GenBank (Benson et al. 2005). We used nucleotide or amino acid sequences to construct maximum likelihood trees using PhyML using the web-based bioinformatics interface Mobyle (Néron et al. 2009). To construct the most likely tree, we used DNA models that were analyzed using jModelTest 2.1.4 (Posada 2008; Darriba et al. 2012). The best tree was then selected using Aikaike Information Criteria (Akaike 1973). Bootstrap analyses (100 bootstrap sets) quantified the significance of the nodes (P ≥ 0.95; fig. 2). These final trees allowed us to be confident that the genes we had found were real, and allowed us to identify potential gene duplications (fig. 2). All labrid sequences were deposited in GenBank (supplementary table S5, Supplementary Material online).
PAML
To look for evidence of selection on the opsin sequences, we first constructed nucleotide trees using jModelTest (Guindon and Gascuel 2003; Darriba et al. 2012) from sequences aligned using the Clustal Omega alignment tool from the EMBL-EBI; and subsequently ran the codeml program from PAML 4.8 on these labrid sequences (Yang 2007; Goujon et al. 2010; Sievers et al. 2011; McWilliam et al. 2013). (supplementary figs. S1–S4, Supplementary Material online). Only labrid sequences were used in the PAML analysis, and no other outgroups. The codeml program calculates the ratio of nonsynonymous (dN) to synonymous (dS) substitution rates using the codon-based model of sequence evolution (Goldman and Yang 1994). Site specific models M1a versus M2a and M8a versus M8 were used that allow dN/dS ratios to vary among codon sites of the proteins. M1a and M8a were neutral models that do not allow for positive selection while M2a and M8 added an additional class of sites where dN/dS could be greater than 1 and so positively selected. This enabled us to determine whether any of the opsin genes were under selection and if so, which amino acid sites selection was acting on. We performed likelihood ratio tests (LRT) by comparing twice the difference in the log-likelihoods (2Δℓ) between models M1a and M2a and also between models M8a and M8. These comparisons were made against a χ2 2 with two degrees of freedom for M1a versus M2a (critical values of 5.99 (P > 0.95) and 9.21 (P > 0.99)) and a χ1 2 with critical values of 2.71 (P > 0.95) and 5.41 (P > 0.99) for M8a versus M8 (Yang 2007). If LRT were significant, sites inferred to be under positive selection were identified using the Bayes Empirical Bayes method, which is employed by the codeml program under models M2a and M8 (Yang et al. 2005).
Gene Expression
To estimate gene expression, reads were mapped back to the assembled transcripts using RSEM, part of the Trinity package (Li and Dewey 2011). Some of the transcripts were longer than expected for opsin genes (2–5 kb cf. 1.5–2 kb). The segments beyond those that aligned with the opsins blasted to nonopsin genes in Genbank and were therefore likely chimeric. These transcripts were trimmed to lengths comparable to the other labrid cone opsins (1.5–2 kb) and then replaced prior to read mapping. This included the SWS2B gene for Co. gaimard, L. dimidiatus, H. chrysus, and T. lunare and the LWS gene for G. varius and H. ornatissimus. The read counts for each of the opsin genes were extracted from the RSEM output (quantified as fragments per kilobase of transcript per million reads). Cone opsin read counts were normalized by dividing by the sum of all the cone opsin genes to get the fraction of each opsin expressed. Specific opsin expression levels were compared with total opsin expression within the retina as this difference reflects potential differences in color sensitivity (Fuller et al. 2003, 2004; Shand et al. 2008). Opsins are some of the most highly expressed genes, and expressed at comparable levels to housekeeping genes such as beta-actin.
Spectral Sensitivities
In order to estimate spectral sensitivities from the opsin sequences, we aligned the labrid sequences in each opsin class with bovine rhodopsin (GenBank Accession No.: NP_001014890.1). This allowed us to identify specific spectral tuning sites as outlined by Yokoyama (2008). For the LWS and RH2 genes, these sites are particularly well studied and we know that the individual amino acid site changes are approximately additive (Chang et al. 1995; Yokoyama and Radlwimmer 2001; Yokoyama and Takenaka 2004; Yokoyama and Tada 2010). For the SWS1, SWS2A, and SWS2B visual pigments, some of the tuning sites have been characterized, however the sites in these pigments are thought to act epistatically rather than additively making estimates of spectral sensitivities more difficult (Cowing et al. 2002; Shi and Yokoyama 2003; Yokoyama et al. 2007; Goujon et al. 2010).
To estimate putative spectral sensitivities, we used two well-studied fishes, Ory. latipes and O. niloticus with known spectral sensitivities that were estimated using absorption spectroscopy on expressed opsin proteins (Matsumoto et al. 2006; Spady et al. 2006). These two fish were chosen as the closest related species for which there was both λmax data and opsin sequence data. In preliminary analyses, two other species of fish were included in estimates (M. zebra and L. goodei), however the opsin sequences of these fish introduced new sites and only served to complicate λmax estimates, and so were omitted from final calculations. For each of the opsin sequences, if the labrid sequence differed from either Ory. latipes or O. niloticus sequences at known tuning sites, the change in λmax was estimated by applying approximate site effects (Takahashi and Ebrey 2003; Yokoyama et al. 2007; Yokoyama 2008). This provided us with two estimates of spectral sensitivity, inside which the labrid spectral sensitivity should lie. We then compared these estimates to known values for spectral sensitivities within the labrid family, and other reef fish from previously published work (Barry and Hawryshyn 1999; Losey et al. 2003). To simplify estimates, all sequence changes were applied additively. We calculated all spectral sensitivities assuming an A1 template as this has been shown to be the most likely retinal pigment found in reef fishes (Wald 1936; Toyama et al. 2008).
Transmission Properties of the Eye
To complete this first analysis of the labrid visual systems, we included previously published information on the light transmission properties of the eye (lens T50 and cornea transmission curves) to our opsin expression findings for specific species (Siebeck and Marshall 2000; table 1). In animals where UV light is important for their behavioral ecology, that is, feeding or individual recognition, UV transmissible lenses and corneas are necessary. Previous work has theorized that UV vision is likely when the lens and cornea transmit light <400 nm. However, often there has not been additional data on the spectral sensitivity of the photoreceptors to confirm functional UV vision. In this article, we hypothesize that fishes with UV transmissible lenses and corneas will express the UV-sensitive opsin class (SWS1) for functional UV vision to be a possibility. These fishes are: B. mesothorax, C. punctatus, and Cho. fasciatus.
Supplementary Material
Supplementary figures S1–S8 and tables S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Janette Edson, Queensland Brain Institute Genomics Facility, for sequencing the libraries and to Ian Misner (funded by the Department of Biology and the University of Maryland’s Bioinformatics Core) for help with putting together the Trinity assembly pipeline. The authors acknowledge the University of Maryland supercomputing resources (http://www.it.umd.edu/hpcc) made available in conducting the research reported in this article. The authors also thanks to Cairns Marine, the directors of Lizard Island Research Station and many field volunteers and to Dr Yakir L. Gagnon who wrote the MATLAB code to generate figure 4. They also like to thank three anonymous reviewers for their helpful comments on earlier versions of the manuscript. This work was supported by a Joan Allsop Travel Award through The University of Queensland Graduate School (2014 to G.A.C.P.); a The University of Queensland International Travel Award (2013 to K.L.C.); a University of Maryland Summer Research and Scholarship Award (2014 to K.L.C.); the National Institutes of Health (Grant 1R01EY024639 to K.L.C.); and the Australian Research Council (Discovery Grant DP110105389 to N.J.M.).
References
- Akaike H. 1973. Information theory and an extension of the maximum likelihood principle. In: Parzen E, Tanabe K, Kitagawa G, editors. Selected papers of Hirotugu Akaike. New York: Springer; p. 199–213. [Google Scholar]
- Ali MA, Anctil M. 1976. Retinas of fishes: an atlas. New York: Springer-Verlag. [Google Scholar]
- Allen G, Steene R, Humann P, Deloach N. 2003. Reef fish identification: tropical pacific. Jackonsville (FL): New World Publications, Inc. [Google Scholar]
- Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, et al. 2012. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40:W597–W603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asenjo AB, Rim J, Oprian DD. 1994. Molecular determinants of human red/green color discrimination. Neuron 12:1131–1138. [DOI] [PubMed] [Google Scholar]
- Barry KL, Hawryshyn CW. 1999. Spectral sensitivity of the Hawaiian saddle wrasse, Thalassoma duperrey, and implications for visually mediated behaviour on coral reefs. Environ Biol Fish. 56:429–442. [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. 2005. GenBank. Nucleic Acids Res. 33:D34–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowmaker JK. 1995. The visual pigments of fish. Prog Retinal Eye Res. 15:1–31. [Google Scholar]
- Bowmaker JK, Govardovskii VI, Shukolyukov SA, Zueva LV, Hunt DM, Sideleva VG, Smirnova OG. 1994. Visual pigments and the photic environment: the cottoid fish of Lake Baikal. Vision Res. 34:591–605. [DOI] [PubMed] [Google Scholar]
- Bowmaker JK, Kunz YW. 1987. Ultraviolet receptors, tetrachromatic colour vision and retinal mosaics in the brown trout (Salmo trutta): age-dependent changes. Vision Res. 27:2101–2108. [DOI] [PubMed] [Google Scholar]
- Braun C, Michiels NK, Siebeck UE, Sprenger D. 2014. Signalling function of long wavelength colours during agonistic male-male interactions in the wrasse Coris julis. MEPS 504:277–286. [Google Scholar]
- Brawand D, Wagner CE, Li YI, Malinsky M, Keller I, Fan S, Simakov O, Ng AY, Lim ZW, Bezault E, et al. 2014. The genomic substrate for adaptive radiation in African cichlid fish. Nature 513:375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browman HI, Novales Flamarique I, Hawryshyn CW. 1994. Ultraviolet photoreception contributes to prey search behaviour in two species of zooplanktivorous fishes. J Exp Biol. 186:187–198. [DOI] [PubMed] [Google Scholar]
- Carleton K. 2009. Cichlid fish visual systems: mechanisms of spectral tuning. Int Zool. 4:75–86. [DOI] [PubMed] [Google Scholar]
- Carleton KL, Hárosi FI, Kocher TD. 2000. Visual pigments of African cichlid fishes: evidence for ultraviolet vision from microspectrophotometry and DNA sequences. Vision Res. 40:879–890. [DOI] [PubMed] [Google Scholar]
- Carleton KL, Kocher TD. 2001. Cone opsin genes of African cichlid fishes: tuning spectral sensitivity by differential gene expression. Mol Biol Evol. 18:1540–1550. [DOI] [PubMed] [Google Scholar]
- Carleton KL, Spady TC, Streelman JT, Kidd MR, McFarland WN, Loew ER. 2008. Visual sensitivities tuned by heterochronic shifts in opsin gene expression. BMC Biol. 6:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang BS, Crandall KA, Carulli JP, Hartl DL. 1995. Opsin phylogeny and evolution: a model for blue shifts in wavelength regulation. Mol Phylogenet Evol. 4:31–43. [DOI] [PubMed] [Google Scholar]
- Cheney KL, Grutter AS, Blomberg SP, Marshall NJ. 2009. Blue and yellow signal cleaning behavior in coral reef fishes. Curr Biol. 19:1283–1287. [DOI] [PubMed] [Google Scholar]
- Cheney KL, Newport C, McClure EC, Marshall NJ. 2013. Colour vision and response bias in a coral reef fish. J Exp Biol. 216:2967–2973. [DOI] [PubMed] [Google Scholar]
- Cheney KL, Skogh C, Hart NS, Marshall NJ. 2009. Mimicry, colour forms and spectral sensitivity of the bluestriped fangblenny, Plagiotremus rhinorhynchos. Proc R Soc Lond B Biol Sci. 276:1565–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CL, Flamarique IN. 2007. Chromatic organization of cone photoreceptors in the retina of rainbow trout: single cones irreversibly switch from UV (SWS1) to blue (SWS2) light sensitive opsin during natural development. J Exp Biol. 210:4123–4135. [DOI] [PubMed] [Google Scholar]
- Chinen A, Hamaoka T, Yamada Y, Kawamura S. 2003. Gene duplication and spectral diversification of cone visual pigments of zebrafish. Genetics 163:663–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortesi F, Musilová Z, Stieb SM, Hart NS, Siebeck UE, Malmstrøm M, Tørresen OK, Jentoft S, Cheney KL, Marshall NJ, et al. 2015. Ancestral duplications and highly dynamic opsin gene evolution in percomorph fishes. Proc Natl Acad Sci U S A. 112:1493–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cott HB. 1940. Adaptive coloration in animals. London: Methuen. [Google Scholar]
- Cowing JA, Poopalasundaram S, Wilkie SE, Robinson PR, Bowmaker JK, Hunt DM. 2002. The molecular mechanism for the spectral shifts between vertebrate ultraviolet- and violet-sensitive cone visual pigments. Biochem J. 367:129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman PF, Bellwood DR, van Herwerden L. 2009. Dating the evolutionary origins of wrasse lineages (Labridae) and the rise of trophic novelty on coral reefs. Mol Phylogenet Evol. 52:621–631. [DOI] [PubMed] [Google Scholar]
- Cronin TW, Johnsen S, Marshall NJ, Warrant EJ. 2014. Visual ecology. Princetown University Press. [Google Scholar]
- Cuthill IC, Bennett ATD, Partridge JC, Maier EJ. 1999. Plumage reflectance and the objective assessment of avian sexual dichromatism. Am Nat. 153:183–200. [DOI] [PubMed] [Google Scholar]
- Dalton BE, Loew ER, Cronin TW, Carleton KL. 2014. Spectral tuning by opsin coexpression in retinal regions that view different parts of the visual field. Proc R Soc Lond B Biol Sci. 281, 20141980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. 1859. On the origin of species by means of natural selection. London: Murray. [Google Scholar]
- Douglas RH, Hawryshyn CW. 1990. Behavioural studies of fish vision: an analysis of visual capabilities. In: Douglas RH, Djamgoz MBA, editors. The visual system of fish. London: Chapman and Hall Ltd; p. 373–418. [Google Scholar]
- Douglas RH, Marshall NJ. 1999. A review of vertebrate and invertebrate ocular filters. In: Archer SN, Djamgoz MBA, Loew ER, Partridge JC, Vallerga S, editors. Adaptive mechanisms in the ecology of vision. Netherlands: Springer; p. 95–162. [Google Scholar]
- Douglas RH, Thorpe A. 1992. Short-wave absorbing pigments in the ocular lenses of deep-sea teleosts. J Mar Biol UK. 72:93–93. [Google Scholar]
- Ferry-Graham LA, Wainwright PC, Westneat MW, Bellwood DR. 2002. Mechanisms of benthic prey capture in wrasses (Labridae). Mar Biol. 141:819–830. [Google Scholar]
- Fineran BA, Nicol JAC. 1974. Studies on the eyes of New Zealand parrot-fishes (Labridae). Proc R Soc Lond B Biol Sci. 186:217–247. [DOI] [PubMed] [Google Scholar]
- Fuller RC, Carleton KL, Fadool JM, Spady TC, Travis J. 2004. Population variation in opsin expression in the bluefin killifish, Lucania goodei: a real-time PCR study. J Comp Physiol A. 190:147–154. [DOI] [PubMed] [Google Scholar]
- Fuller RC, Fleishman LJ, Leal M, Travis J, Loew E. 2003. Intraspecific variation in retinal cone distribution in the bluefin killifish, Lucania goodei. J Comp Physiol A. 189:609–616. [DOI] [PubMed] [Google Scholar]
- Goda M, Fujii R. 1995. Blue chromatophores in two speces of Callionymid fish. Zool Sci. 12:811–813. [Google Scholar]
- Goldman N, Yang Z. 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 11:725–736. [DOI] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38:W695–W699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst Biol. 52:696–704. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart NS, Lisney TJ, Marshall NJ, Collin SP. 2004. Multiple cone visual pigments and the potential for trichromatic colour vision in two species of elasmobranch. J Exp Biol. 207:4587–4594. [DOI] [PubMed] [Google Scholar]
- Hauser FE, van Hazel I, Chang BS. 2014. Spectral tuning in vertebrate short wavelength-sensitive 1 (SWS1) visual pigments: can wavelength sensitivity be inferred from sequence data? J Exp Zool B Mol Dev Evol. 322:529–539. [DOI] [PubMed] [Google Scholar]
- Hofmann CM, Carleton KL. 2009. Gene duplication and differential gene expression play an important role in the diversification of visual pigments in fish. Integr Comp Biol. 49:630–643. [DOI] [PubMed] [Google Scholar]
- Hofmann CM, Marshall NJ, Abdilleh K, Patel Z, Siebeck UE, Carleton KL. 2012. Opsin evolution in damselfish: convergence, reversal, and parallel evolution across tuning sites. J Mol Evol. 75:79–91. [DOI] [PubMed] [Google Scholar]
- Hofmann CM, O'Quin KE, Justin Marshall N, Carleton KL. 2010. The relationship between lens transmission and opsin gene expression in cichlids from Lake Malawi. Vision Res. 50:357–363. [DOI] [PubMed] [Google Scholar]
- Hofmann CM, O'Quin KE, Marshall NJ, Cronin TW, Seehausen O, Carleton KL. 2009. The eyes have it: regulatory and structural changes both underlie cichlid visual pigment diversity. PLoS Biol. 7:e1000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard R, Wald G. 1952. Cis-trans isomers of vitamin A and retinene in the rhodopsin system. J Gen Physiol. 36:269–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura G, Tamura T. 1973. Morphological studies on the retina of two teleosts Scomber tapeinocephalus and Halichoeres poecilopterus. Bull Jpn Soc Sci Fish. 39:715–726. [Google Scholar]
- Kazancioglu E, Near TJ, Hanel R, Wainwright PC. 2009. Influence of sexual selection and feeding functional morphology on diversification rate of parrotfishes (Scaridae). Proc R Soc Lond B Biol Sci. 276:3439–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JS, MacNichol EF. 1979. Visual pigments in teleost fishes: effects of habitat, microhabitat, and behavior on visual system evolution. Sens Processes. 3:95–131. [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loew ER. 1976. Vitamin A1/A2-based visual pigment mixtures in cones of the rudd. Vision Res. 16:891–896. [DOI] [PubMed] [Google Scholar]
- Loew ER, Lythgoe JN. 1978. The ecology of cone pigments in teleost fishes. Vision Res. 18:715–722. [DOI] [PubMed] [Google Scholar]
- Loew ER, McFarland WN, Mills EL, Hunter D. 1993. A chromatic action spectrum for planktonic predation by juvenile yellow perch, Perca flavescens. Can J Zool. 71:384–386. [Google Scholar]
- Lorenz K. 1962. The function of colour in coral reef fishes. Proc R Inst Gr Br. 39:282–296. [Google Scholar]
- Losey GS, George SL., Jr 2003. Crypsis and communication functions of UV-visible coloration in two coral reef damselfish, Dascyllus aruanus and D.reticulatus . Anim Behav. 66:299–307. [Google Scholar]
- Losey GS, McFarland WN, Loew ER, Zamzow JP, Nelson PA, Marshall NJ. 2003. Visual biology of Hawaiian coral reef fishes. I. Ocular transmission and visual pigments. Copeia 3:433–454. [Google Scholar]
- Lythgoe JN. 1979. The ecology of vision. New York: Oxford University Press. [Google Scholar]
- Marshall J, Carleton KL, Cronin T. 2015. Colour vision in marine organisms. Curr Opin Neurobiol. 34C:86–94. [DOI] [PubMed] [Google Scholar]
- Marshall J, Vorobyev M, Siebeck UE. 2006. What does a reef fish see when it sees a reef fish? Eating ‘Nemo'©. In: Kapoor B, Ladich F, Collin SP, editors. Communication in fishes. Enfield: Science Publisher Inc; p. 393–422. [Google Scholar]
- Marshall NJ. 1988. A unique colour and polarization vision system in mantis shrimps. Nature 333:557–560. [DOI] [PubMed] [Google Scholar]
- Marshall NJ. 2000. Communication and camouflage with the same ‘bright' colours in reef fishes. Philos Trans R Soc Lond B Biol Sci. 355:1243–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NJ, Jennings K, McFarland WN, Loew ER, Losey GS. 2003a. Visual biology of Hawaiian coral reef fishes. II. Colours of Hawaiian coral reef fish. Copeia 3:455–466. [Google Scholar]
- Marshall NJ, Jennings K, McFarland WN, Loew ER, Losey GS. 2003b. Visual biology of Hawaiian coral reef fishes. III. Environmental light and an integrated approach to the ecology of reef fish vision. Copeia 3:467–480. [Google Scholar]
- Matsumoto Y, Fukamachi S, Mitani H, Kawamura S. 2006. Functional characterization of visual opsin repertoire in Medaka (Oryzias latipes). Gene 371:268–278. [DOI] [PubMed] [Google Scholar]
- McFarland WN, Loew ER. 1994. Ultraviolet visual pigments in marine fishes of the family Pomacentridae. Vision Res. 34:1393–1396. [DOI] [PubMed] [Google Scholar]
- McFarland WN, Munz FW. 1975. Part II: the photic environment of clear tropical seas during the day. Vision Res. 15:1063–1070. [DOI] [PubMed] [Google Scholar]
- McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. 2013. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 41:W597–W600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith RW, Gatesy J, Emerling CA, York VM, Springer MS. 2013. Rod monochromacy and the coevolution of cetacean retinal opsins. PLoS Genet. 9.4:e1003432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels NK, Anthes N, Hart NS, Herler J, Meixner AJ, Schleifenbaum F, Schulte G, Siebeck UE, Sprenger D, Wucherer MF. 2008. Red fluorescence in reef fish: a novel signalling mechanism? BMC Ecol. 8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Mori K, Saitoh K, Oshima K, Mekuchi M, Sugaya T, Shigenobu Y, Ojima N, Muta S, Fujiwara A, et al. 2013. Evolutionary changes of multiple visual pigment genes in the complete genome of Pacific bluefin tuna. Proc Natl Acad Sci U S A. 110:11061–11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, Wainwright PC, Friedman M, Smith WL. 2012. Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci U S A. 109:13698–13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Néron B, Ménager H, Maufrais C, Joly N, Maupetit J, Letort S, Carrere S, Tuffery P, Letondal C. 2009. Mobyle: a new full web bioinformatics framework. Bioinformatics 25:3005–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol JAC. 1975. Studies on the eyes of fishes: structure and ultrastructure. In: Ali MA, editor. Vision in fishes. New York: Plenum Press; p. 579–607. [Google Scholar]
- Novales Flamarique I, Cheng C, Bergstrom C, Reimchen T. 2012. Pronounced heritable variation and limited phenotypic plasticity in visual pigments and opsin expression of threespine stickleback photoreceptors. J Exp Biol. 216:656–667. [DOI] [PubMed] [Google Scholar]
- O'Quin KE, Hofmann CM, Hofmann HA, Carleton KL. 2010. Parallel evolution of opsin gene expression in African cichlid fishes. Mol Biol Evol. 27:2839–2854. [DOI] [PubMed] [Google Scholar]
- Palacios AG, Bozinovic F, Vielma A, Arrese CA, Hunt DM, Peichl L. 2010. Retinal photoreceptor arrangement, SWS1 and LWS opsin sequence, and electroretinography in the South American marsupial Thylamys elegans (Waterhouse, 1839). J Comp Neurol. 518:1589–1602. [DOI] [PubMed] [Google Scholar]
- Parenti P, Randall JE. 2000. An annotated checklist of the species of the labroid fish families Labridae and Scaridae. Ichthyol Bull JLB Smith Inst. 68:1–97. [Google Scholar]
- Pignatelli V, Champ C, Marshall J, Vorobyev M. 2010. Double cones are used for colour discrimination in the reef fish, Rhinecanthus aculeatus. Biol Lett. 6:537–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25:1253–1256. [DOI] [PubMed] [Google Scholar]
- Price SA, Holzman R, Near TJ, Wainwright PC. 2011. Coral reefs promote the evolution of morphological diversity and ecological novelty in labrid fishes. Ecol Lett. 14:462–469. [DOI] [PubMed] [Google Scholar]
- Randall JE, Allen GR, Steene RC. 1997. Fishes of the Great Barrier Reef and Coral Sea. Hawaii: University of Hawai'i Press. [Google Scholar]
- Rennison DJ, Owens GL, Taylor JS. 2012. Opsin gene duplication and divergence in ray-finned fish. Mol Phylogenet Evol. 62:986–1008. [DOI] [PubMed] [Google Scholar]
- Schmitz L, Wainwright PC. 2011. Nocturnality constrains morphological and functional diversity in the eyes of reef fishes. BMC Evol Biol. 11:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott RK, Refvik SP, Hauser FE, López-Fernández H, Chang BSW. 2014. Divergent positive selection in rhodopsin from lake and riverine cichlid fishes. Mol Biol Evol. 31:1149–1165. [DOI] [PubMed] [Google Scholar]
- Schulte JE, O'Brien CS, Conte MA, O'Quin KE, Carleton KL. 2014. Interspecific variation in Rx1 expression controls opsin expression and causes visual system diversity in African cichlid fishes. Mol Biol Evol. 31:2297–2308. [DOI] [PubMed] [Google Scholar]
- Shand J, Davies WL, Thomas N, Balmer L, Cowing JA, Pointer M, Carvalho LS, Trezise AEO, Collin SP, Beazley LD, et al. 2008. The influence of ontogeny and light environment on the expression of visual pigment opsins in the retina of the black bream, Acanthopagrus butcheri. J Exp Biol. 211:1495–1503. [DOI] [PubMed] [Google Scholar]
- Shi Y, Yokoyama S. 2003. Molecular analysis of the evolutionary significance of ultraviolet vision in vertebrates. Proc Natl Acad Sci U S A. 100:8308–8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebeck UE. 2004. Communication in coral reef fish: the role of ultraviolet colour patterns in damselfish territorial behaviour. Anim Behav. 68:273–282. [Google Scholar]
- Siebeck UE, Marshall NJ. 2000. Transmission of ocular media in labrid fishes. Philos Trans R Soc Lond B Biol Sci. 355:1257–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebeck UE, Marshall NJ. 2001. Ocular media transmission of coral reef fish—can coral reef fish see ultraviolet light? Vision Res. 41:133–149. [DOI] [PubMed] [Google Scholar]
- Siebeck UE, Marshall NJ. 2007. Potential ultraviolet vision in pre-settlement larvae and settled reef fish–a comparison across 23 families. Vision Res. 47:2337–2352. [DOI] [PubMed] [Google Scholar]
- Siebeck UE, Parker AN, Sprenger D, Mäthger LM, Wallis G. 2010. A species of reef fish that uses ultraviolet patterns for covert face recognition. Curr Biol. 20:407–410. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AR, D'Annunzio L, Smith AE, Sharma A, Hofmann CM, Marshall NJ, Carleton KL. 2011. Intraspecific cone opsin expression variation in the cichlids of Lake Malawi. Mol Ecol. 20:299–310. [DOI] [PubMed] [Google Scholar]
- Spady TC, Parry JWL, Robinson PR, Hunt DM, Bowmaker JK, Carleton KL. 2006. Evolution of the cichlid visual palette through ontogenetic subfunctionalization of the opsin gene arrays. Mol Biol Evol. 23:1538–1547. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Ebrey TG. 2003. Molecular basis of spectral tuning in the newt short wavelength sensitive visual pigment. Biochemistry 42:6025–6034. [DOI] [PubMed] [Google Scholar]
- Temple S, Hart NS, Marshall NJ, Collin SP. 2010. A spitting image: specializations in archerfish eyes for vision at the interface between air and water. Proc R Soc Lond B Biol Sci. 277:2607–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple SE. 2011. Why different regions of the retina have different spectral sensitivities: a review of mechanisms and functional significance of intraretinal variability in spectral sensitivity in vertebrates. Vis Neurosci. 28:281–293. [DOI] [PubMed] [Google Scholar]
- Thorpe A, Douglas RH, Truscott RJW. 1993. Spectral transmission and short-wave absorbing pigments in the fish lens—I. Phylogenetic distribution and identity. Vision Res. 33:289–300. [DOI] [PubMed] [Google Scholar]
- Thresher RE. 1984. Reproduction in reef fishes. Neptune City: TFH Publications. [Google Scholar]
- Toyama M, Hironaka M, Yamahama Y, Horiguchi H, Tsukada O, Uto N, Ueno Y, Tokunaga F, Seno K, Hariyama T. 2008. Presence of rhodopsin and porphyropsin in the eyes of 164 fishes, representing marine, diadromous, coastal and freshwater species-a qualitative and comparative study. Photochem Photobiol. 84:996–1002. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobyev M, Osorio D. 1998. Receptor noise as a determinant of colour thresholds. Proc R Soc Lond B Biol Sci. 265:351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright PC, Bellwood DR, Westneat MW, Grubich JR, Hoey AS. 2004. A functional morphospace for the skull of labrid fishes: patterns of diversity in a complex biomechanical system. Biol J Linn Soc. 82:1–25. [Google Scholar]
- Wald G. 1936. Pigments of the retina: II. Sea robin, sea bass, and scup. J Gen Physiol. 20:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald G. 1968. The molecular basis of visual excitation. Nature 219:800–807. [DOI] [PubMed] [Google Scholar]
- Wallace AR. 1877. The colors of animals and plants. Am Nat. 11:641–662. [Google Scholar]
- Westneat MW, Alfaro ME. 2005. Phylogenetic relationships and evolutionary history of the reef fish family Labridae. Mol Phylogenet Evol. 36:370–390. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielsen R. 2005. Bayes-Empirical-Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 22:1107–1118. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. 2008. Evolution of dim-light and color vision pigments. Ann Rev Genomics Hum Genet. 9:259–282. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. 2012. Synthesis of experimental molecular biology and evolutionary biology: an example from the world of vision. Bioscience 62:939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Radlwimmer FB. 2001. The molecular genetics and evolution of red and green color vision in vertebrates. Genetics 158:1697–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Tada T. 2010. Evolutionary dynamics of rhodopsin Type 2 opsins in vertebrates. Mol Biol Evol. 27:133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Takenaka N. 2004. The molecular basis of adaptive evolution of squirrelfish rhodopsins. Mol Biol Evol. 21:2071–2078. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Takenaka N, Blow N. 2007. A novel spectral tuning in the short wavelength-sensitive (SWS1 and SWS2) pigments of bluefin killifish (Lucania goodei). Gene 396:196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Zhang H, Radlwimmer FB, Blow NS. 1999. Adaptive evolution of color vision of the Comoran coelacanth (Latimeria chalumnae). Proc Natl Acad Sci U S A. 96:6279–6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.