Abstract
Widespread sequencing efforts are revealing unprecedented amount of genomic variation in populations. Such information is routinely used to derive consensus reference sequences and to infer positions subject to natural selection. Here, we present a new molecular evolutionary method for estimating neutral evolutionary probabilities (EPs) of each amino acid, or nucleotide state at a genomic position without using intraspecific polymorphism data. Because EPs are derived independently of population-level information, they serve as null expectations that can be used to evaluate selective forces on alleles at both polymorphic and monomorphic positions in populations. We applied this method to coding sequences in the human genome and produced a comprehensive evolutionary variome reference for all human proteins. We found that EPs accurately predict neutral and disease-associated alleles. Through an analysis of discordance between allelic EPs and their observed population frequencies, we discovered thousands of novel candidate sites for nonneutral evolution in human proteins. Many of these were validated in a joint analysis of disease-associated variants and population data. The EP method is also directly applicable to the analysis of noncoding sequences and genomic analyses of nonmodel species.
Keywords: phylomedicine, evolution, disease, adaptation, neutrality
Introduction
According to the neutral theory of molecular evolution, intraspecific variation is a transient phase of interspecific evolution (Kimura 1983). Therefore, patterns of genome conservation and divergence observed across species should predict the frequency and fate of genomic variation within a species. Based on this principle, we developed a new method to estimate the neutral evolutionary probability (EP) of amino acid state at a protein position (or each possible nucleotide state at a genomic position) in a given species using only the interspecific evolutionary history of the position. In the new method, population-level information on observed alleles is not needed when deriving EPs. This independence enables EPs to serve as null expectations when evaluating variation in one or more populations. Collectively, EPs over all positions constitute a multistate evolutionary variome (eVar), which is unaffected by the patterns of population sampling, vagaries of genetic drift within populations, and local changes in natural selection. Therefore, this method can be used to evaluate alleles at both polymorphic and monomorphic positions in a population, including all known and unknown variants.
New Approaches
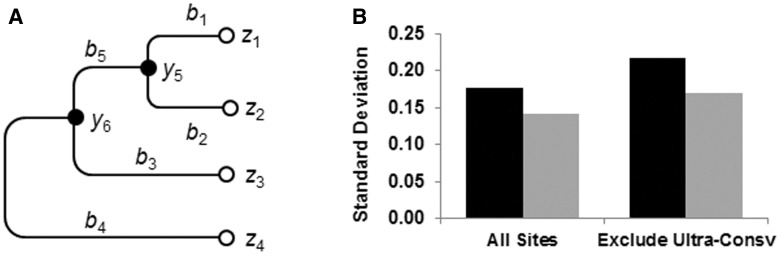
The new approach aims to estimate the EP of observing an amino acid residue at a protein position (focal position) in a given species using a multispecies sequence alignment and phylogenetic relationships among sequences, independent of population-level information on the frequency of alleles at the focal position. To accomplish this, we use a Bayesian framework to calculate the posterior probability (PP) for each possible residue state at a given position in a species (species 1 in fig. 1), where the multispecies alignment is modified to replace the residue at the focal position with a missing data symbol (see Materials and Methods). Then, we compute multiple PP values by progressively pruning evolutionary lineages that are sister groups to species 1. For the example in figure 1A, we compute PP0 using the whole data set, PP1 after pruning sister group containing species 2, and PP2 after additional pruning of the sister group containing species 3. Then, we obtain the EP as an average of PPi values weighted by the evolutionary time depth of the closest relative of species 1 (human in our case) in the corresponding evolutionary tree used. The iterative pruning and the normalization by evolutionary time is intended to ameliorate the effects caused by incomplete species sampling and evolutionary extinction of species. We tested the effect of decreasing species sampling by sequentially removing the closest species to humans and their corresponding sequences from the data set. In this analysis, EPs showed a 23% lower variance than PPs (fig. 1B), and thus are more robust to bias due to incomplete species coverage than PPs. For a given protein, EPs were estimated for each position independently, which collectively represent a multistate eVar. Although we described the new approach using amino acid sequences as an example, it can be directly applied for the analysis of nucleotide sequences, which would produce EPs for different nucleotide bases when using a nucleotide sequence alignment and phylogenetic relationships.
Fig. 1.
EP calculation. (A) A simple tree of four sequences. Leaf nodes are marked by open circles, where the observed amino acid residues are denoted by z1,…, z4. Residues at ancestral nodes are denoted by y5 and y6. Along each branch, a branch length (b1,…, b5) is displayed. (B) Standard deviations of PPs (black bars) and EPs (gray bars) when species and their corresponding sequences were progressively pruned from the data set. Data were derived from 100 randomly selected human protein sequences and the evolutionary lineages of human and 45 species (Kumar et al. 2012).
Results and Discussion
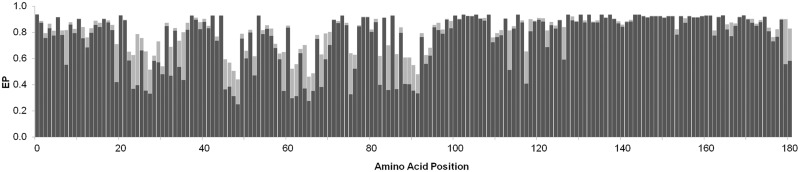
We applied the new method to estimate eVar for all proteins in the human genome, which were compiled based on the National Center for Biotechnology Information (NCBI) RefSeq annotations of the human genome build GRCh37 (also known as hg19). For genes producing multiple isoforms of proteins, the longest isoform was used (see Materials and Methods). We used a phylogenetic tree that included 46 vertebrate species spanning over 500 My (Kumar et al. 2012). For each position in a given human protein, EP was calculated for each of the 20 possible amino acid residues (alleles) using the protein sequence alignment. Figure 2 shows eVar for a small protein (NP_000064, CD3G). In eVar, evolutionarily likely alleles will have high EP values and evolutionarily unlikely alleles will have low EP values. Conserved positions tend to have one dominant allele with high EP and variable positions tend to have multiple alleles with EP > 0.05.
Fig. 2.
eVar for a small protein (NP_000064, CD3G). At each position, the black bar represents the allele with the highest EP and the gray bar represents the allele with the second highest EP. All other alleles contribute EPs (aggregated and represented by white space) such that the total EP is 1.0.
Proteome-Wide EP Estimates
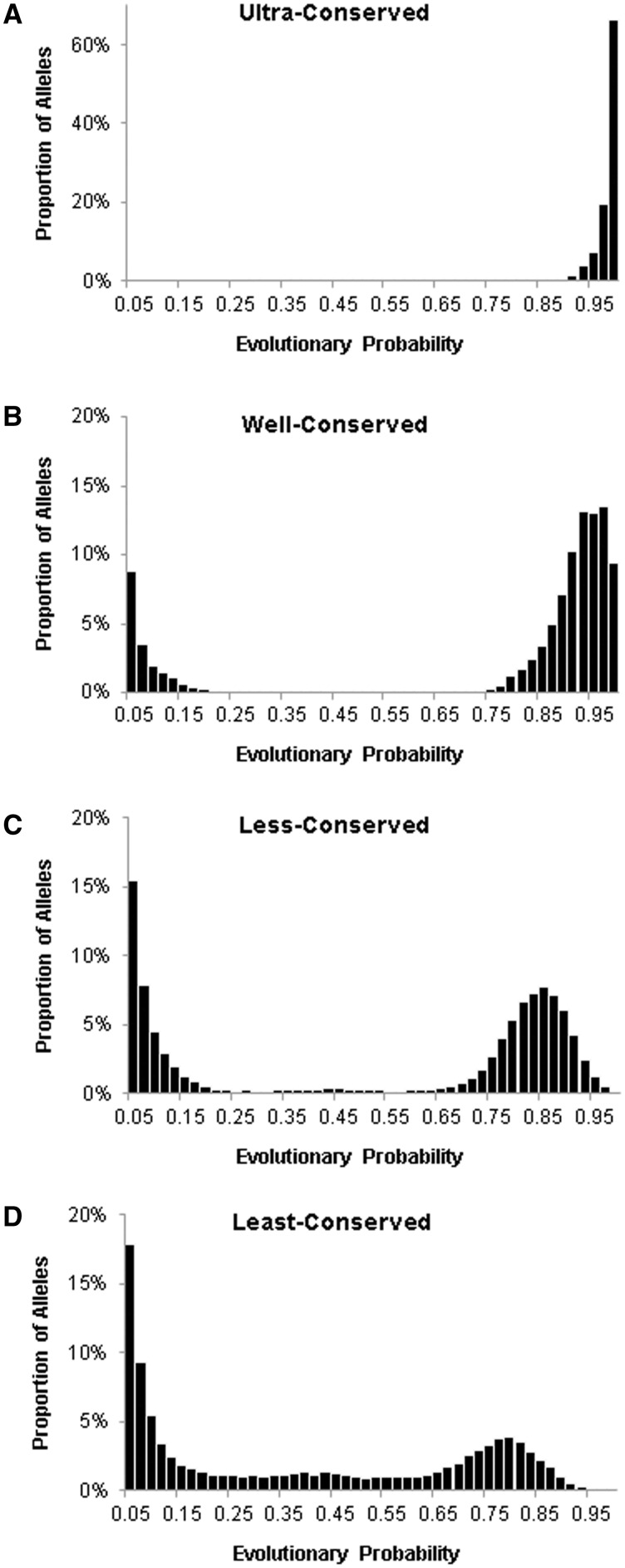
For the human protein collection, EPs of alleles were calculated for 10,575,180 amino acid positions in 18,390 protein sequences. Collectively, these estimates constitute the proteomic eVar for human. Consistent with the expectation that most new mutations are deleterious, 94.4% of alleles in eVar had EPs lower than 0.05. For alleles with EP ≥ 0.05, the allelic EP distribution was right skewed at ultraconserved positions (fig. 3A), because these positions are constrained to allow only one amino acid. In contrast, the distribution is left skewed at least conserved positions, which have permitted multiple amino acids over the long-term evolutionary history (fig. 3D). A scarcity of alleles with intermediate EPs evokes the U-shaped theoretical distribution of allele frequencies of a population under the neutral models (Crow 2005; Haegeman and Weitz 2012).
Fig. 3.
Distributions of allelic EPs at positions over a spectrum of conservation level, from highly conserved to highly variable. A total of 10,575,180 amino acid positions in 18,390 proteins were grouped into ultraconserved (A), well conserved (B), less conserved (C), or least conserved (D). Only alleles with EP>0.05 are included.
Comparing EPs Versus Population Frequencies
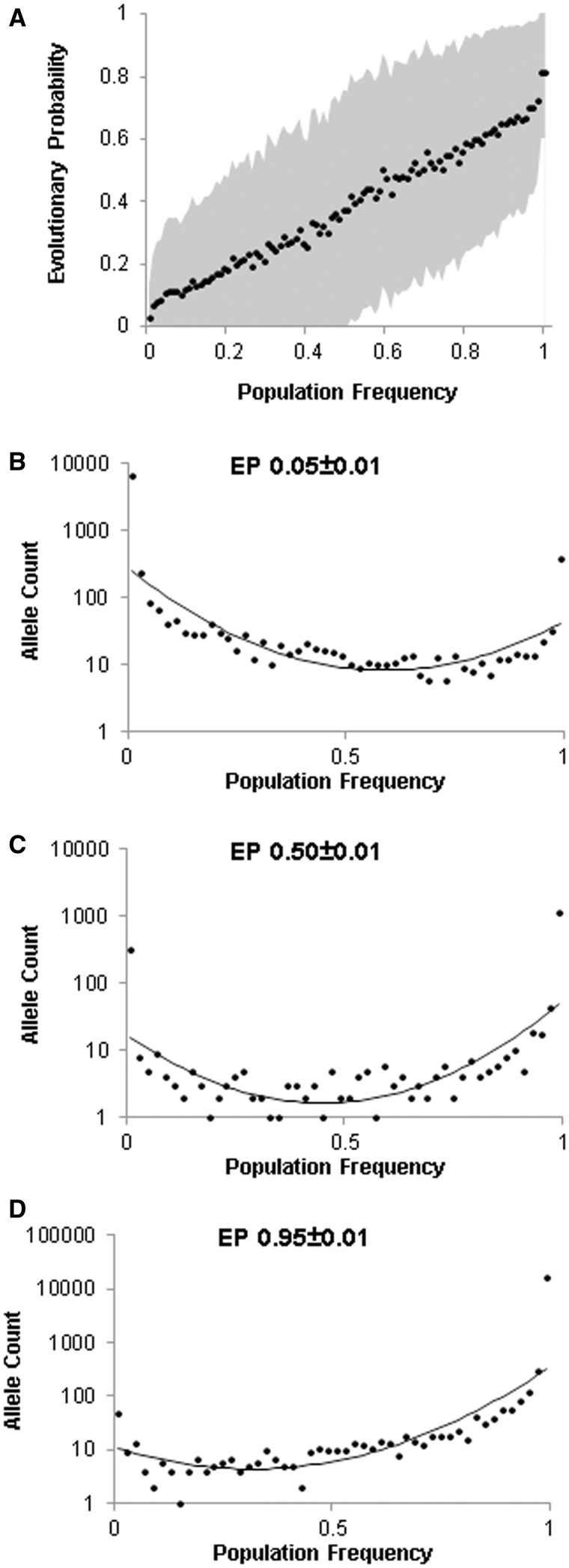
We first constructed a consensus sequence for each human protein based on the reference genome (GRCh37) and the 1000 Genomes (1KG) data (phase 3). This consensus sequence consists of 10,324,216 monomorphic positions (allele frequency = 100%) and major alleles at 250,964 polymorphic positions. At 96% of all positions, human consensus alleles had the highest EPs. At polymorphic positions, allelic EPs were strongly correlated with observed population frequencies (Pearson correlation coefficient = 0.99, P << 0.01; fig. 4A). These observations confirm the neutral theory (Kimura 1983) prediction that relates long- and short-term evolutionary patterns.
Fig. 4.
Relationship of allelic EPs with their observed population frequencies in the 1000 Genomes (1KG) data. (A) A scatter plot showing the relationship of population frequencies of alleles and their average EPs. A total of 250,964 nSNVs were binned into population frequencies in increments of 1% and plotted against average allelic EPs (black circle) and their standard deviation (shaded area). The observed correlation is high for the running average (Pearson correlation coefficient [PCC] = 0.99, P << 0.01) and for individual EPs (PCC = 0.90, P << 0.01). A similarly strong relationship was observed when EPs were binned first and their average allele frequencies were considered (PCC = 0.90, P << 0.01). Distributions of population frequencies of alleles with similar evolutionary probabilities: (B) EP = 0.05 ± 0.01, (C) EP = 0.50 ± 0.01, and (D) EP = 0.95 ± 0.01. Fitted lines of second order of polynomial regression were shown.
However, we observed high dispersion in EP distributions, where alleles with similar population frequencies showed a wide range of EPs (shaded area in fig. 4A). This is because the frequencies of alleles segregating today are a product of their time of origin, the intensity of natural selection, and genetic drift. Ultimately, large variances can be attributed to genetic drift that has driven most standing alleles toward loss or fixation in the population (Kimura 1983). In general, low EP alleles showed lower allele frequencies (fig. 4B), whereas high EP alleles had rather high frequencies (fig. 4D).
EPs for Disease-Associated Alleles
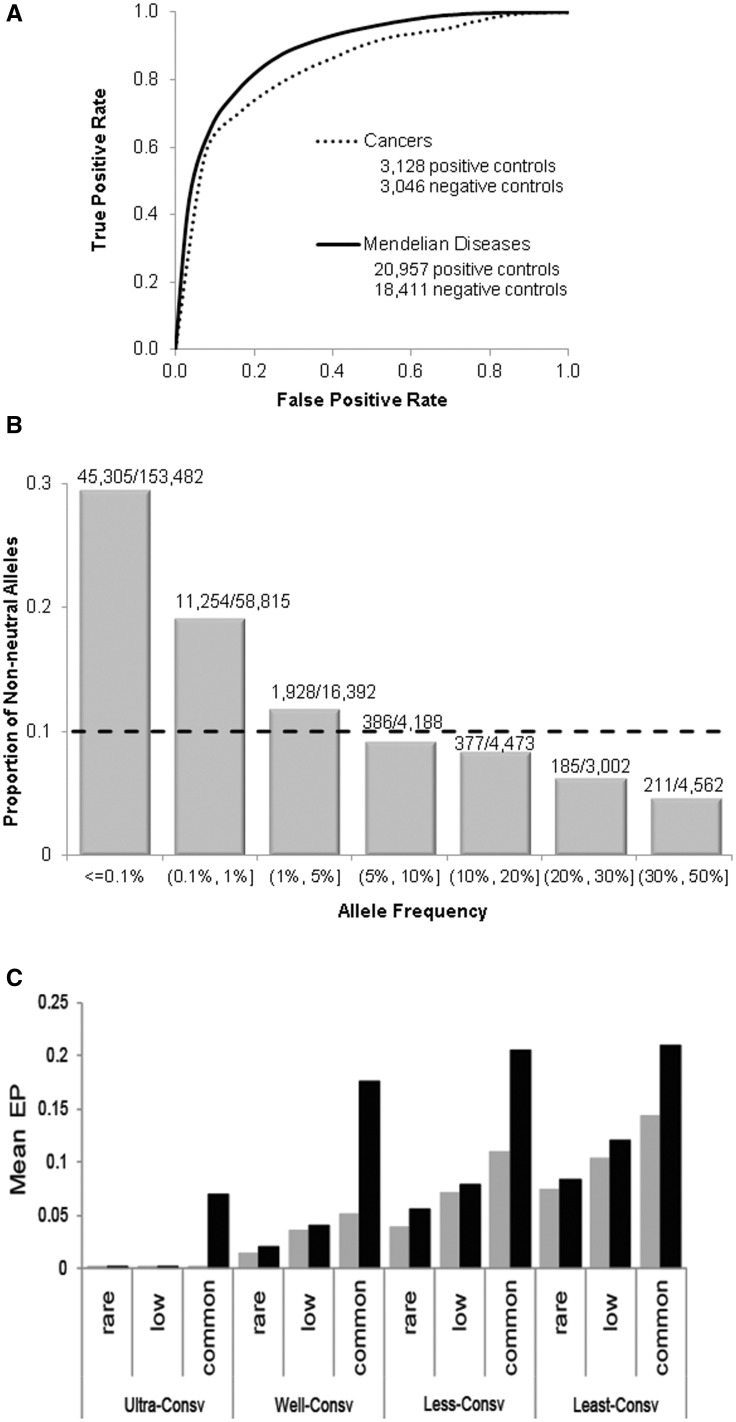
We also tested the efficacy of EPs to discriminate between neutral and disease-associated alleles, a vast majority of which are expected to be under negative selection (Dudley et al. 2012). We made use of two existing sets of benchmark data. The first benchmark data set (HumVar; Adzhubei et al. 2010) contains 20,957 deleterious single nucleotide polymorphisms (SNPs) associated with diseases (positive controls) and 18,411 common population polymorphisms (negative controls). EPs showed a quickly rising receiver operating characteristic (ROC) curve that contrasts the false positive and true positive diagnosis rates at different EP thresholds (fig. 5A). This trend shows that EPs will afford high rates of correct diagnosis of disease alleles at low rates of false positive diagnosis. The area under the curve (AUC) value for EPs was high (0.89) and similar to that for classical and recently developed methods, including SIFT (0.88), PolyPhen-2 (0.89), and CADD (0.84) for the same collection of variants (Ng and Henikoff 2001; Adzhubei et al. 2010; Kircher et al. 2014). Thus, EPs perform comparably with other mutation diagnosis methods that require the use of disease-associated variants and neutral population polymorphisms to build predictive models. Dependencies on the use of such training data in building predictive models are known to cause problems for such classifiers, as their performances decline when applied to variants not represented in the training data sets (Dorfman et al. 2010; Cline and Karchin 2011).
Fig. 5.
Application of EPs to diagnose disease-associated variants. (A) ROC curves displaying the true positive rates at different levels of false positive rates when using allelic EPs to predict disease-associated alleles. Positive (disease) and negative (nondisease) control variants in two benchmark data sets were used, one (Adzhubei et al. 2010) for diagnosing alleles associated with Mendelian diseases (solid line) and the other (Capriotti and Altman 2011) for cancer-associated alleles (broken line). (B) Predicted proportion of nonneutral alleles in the 1000 Genomes data. EP threshold of 0.0022 corresponds to a false positive rate of 10% (dotted line, panel A). Results are shown separately for polymorphisms with low and high frequencies. The numbers of alleles predicted to be deleterious and the total number of alleles are shown above each bar. (C) Average EP of alleles occurring in homozygous genotypes (black bars) and those occurring only as heterozygotes (gray bars). Alleles are grouped by their positional conservation into ultra, well, less, and least conserved categories and minor allele frequency (MAF) in the 1KG data set (rare: MAF < 1%; low: MAF 1–5%; common: MAF > 5%).
Our second benchmark data set (CNO; Capriotti and Altman 2011) consisted of 3,128 cancer driver mutations (positive controls) and 3,046 passenger mutations (negative controls). Again, EP shows a quickly rising ROC curve and a high AUC (0.84; fig. 5A). Therefore, we expect allelic EPs to be useful in prioritizing variants in biological and clinical investigations, even when sufficient population variation data are not available to train good predictive models.
Putatively Deleterious Population Variation
The ROC curve in figure 5A shows that an EP value of 0.0022 produced a 10% rate of false positive diagnosis for the HumVar benchmark data. Using this threshold, we identified putatively deleterious variation reported in the 1KG population survey (1000 Genomes Project Consortium et al. 2012) (fig. 5B). The proportion of deleterious alleles was higher than the false positive rate only for alleles that occur with population frequency of 1% or lower, with the highest percentage (30%) of deleterious alleles seen in the collection of private variants. This overall trend may be explained by the action of negative selection, which would prevent deleterious alleles from rising to high frequencies.
The effect of negative selection can be directly observed by comparing EPs of minor alleles found in heterozygous and homozygous genotypes in individuals. In 1KG data, we found 31,835 SNPs that have at least one homozygous individual and 213,079 SNPs that exist only in heterozygous states. Overall, alleles found only in heterozygotes are 25 times less evolutionarily likely than those found as homozygotes (average EP = 0.001 vs. 0.025, P << 0.01). Furthermore, this pattern varied when we stratified SNPs by their positional conservation and population frequency (fig. 5C). The greatest difference is observed for common alleles (population frequency > 5%) at ultraconserved positions. EPs for homozygous and heterozygous alleles were quite similar at less conserved positions or least conserved positions, because they are under lower functional and, thus, evolutionary constraints.
EPs Predict Potentially Adaptive Alleles
Analysis of EPs also revealed many alleles present at unexpectedly high frequency in humans. These constitute signs of potential nonneutral (e.g., adaptive) evolution. We found 36,691 evolutionarily unlikely alleles (EP < 0.05) that occur at 100% frequency in the 1KG data, that is, these evolutionarily unlikely alleles appear to be fixed in modern humans. Although this set represents a miniscule proportion of all positions analyzed (0.4%), it likely contains many candidates for adaptive evolution. We reasoned that if the fixation of an evolutionarily unlikely allele at a position was due to adaptation (including functional compensation), then mutations that revert back to an evolutionarily likely (high EP) allele would be detrimental. To search for such cases, we examined EPs of 54,034 missense mutations in the HGMD database (Stenson et al. 2009), which are implicated in Mendelian disorders and absent from 1KG data. We found ten positions where a mutation from a low EP allele (<0.05) to a high EP allele (>0.50) was associated with a Mendelian disease (table 1).
Table 1.
Evolutionarily Unlikely Alleles Occurring with High Frequency in Humans, But associated with Diseases and Other Traits.
| Gene | Variant | Major Allele |
Disease/Trait | |
|---|---|---|---|---|
| EP | Frequency | |||
| Monomorphic | ||||
| PRSS1 | N29T | 0.004 | 100% | Hereditary pancreatitis |
| F8 | L69V | 0.006 | 100% | Hemophilia A |
| FOXI1 | P239L | 0.007 | 100% | Pendred syndrome |
| CYP21A2 | M240K | 0.007 | 100% | 21-hydroxylase deficiency |
| CYP2A6 | K194E | 0.008 | 100% | Altered activity |
| PKD1 | L2696R | 0.009 | 100% | Polycystic kidney disease 1 |
| USP26a | L165S | 0.010 | 100% | Azoospermia/oligozoospermia |
| RHCE | Q233E | 0.011 | 100% | Rhesus blood group variant |
| CRB1 | G959S | 0.019 | 100% | Retinitis pigmentosa |
| MLL2 | P4353L | 0.037 | 100% | Kabuki syndrome |
| Polymorphic | ||||
| STAT2a | M594I | 0.011 | 95% | Height |
| APOEa | C130R | 0.007 | 85% | Alzheimer’s disease biomarkers |
| ICAM1a | K469E | 0.046 | 65% | Soluble ICAM-1 |
| KNG1a | I581T | 0.003 | 58% | Activated partial thromboplastin time |
| CFHa | V62I | 0.005 | 57% | Serum myeloperoxidase levels |
| COL11A1a | L1335P | 0.002 | 53% | Glaucoma (primary open angle) |
Note.—Allele frequencies are from the 1000 genomes data.
aDenotes genes that have been shown to be under positive selection in past studies analyzing human population data and comparison with other species (Nielsen et al. 2005; Zhang et al. 2010; Grossman et al. 2013; Li et al. 2014).
One of these genes (USP26) was previously identified as experiencing positive selection in a study that analyzed the ratio of nonsynonymous to synonymous differences in the genomes of humans and chimpanzees (Nielsen et al. 2005). This gene is involved in spermatogenesis, and mutation to an evolutionarily likely allele at position 165 causes Azoospermia (Asadpor et al. 2013). For the other nine alleles, genomic scanning that compared humans and other species failed to infer nonneutral evolution (Nielsen et al. 2005; Zhang et al. 2010), either due to the low statistical power of traditional methods in multispecies analysis (Arbiza et al. 2006) or because these adaptive variants are fixed in the human population and are, thus, not detectable in population scans.
EPs for Coding Alleles Implicated in Complex Diseases
We also looked for evidence of nonneutral evolution in the collection of variants that have been strongly associated with complex diseases or quantitative traits in genome-wide association studies (GWAS). We found 376 positions harboring nonsynonymous SNPs (nSNPs) in the GRASP 2.0 database, which aggregates more than 6 million SNP–phenotype associations from 2,082 GWAS studies (Leslie et al. 2014). The same 376 nSNPs were also found in the 18,152 high confidence variants available from the NHGRI GWAS catalog. A substantial proportion of these positions (13%) contained low EP alleles that have high population frequency (>50%) in the 1KG data. These low EP, high-frequency alleles are potentially involved in nonneutral evolution, including adaptive evolution. In fact, six of these positions (table 1) are located in genes that are already implicated as targets of positive selection in previous population genomic analyses (Li et al. 2014). This interpretation is supported by the observation of low EP alleles at a vast majority of positions that have been implicated in ongoing adaptive change in comprehensive population genomic analyses (Grossman et al. 2013) (table 2). These results demonstrate the usefulness of EP to complement traditional methods in the discovery of alleles that deviate from neutral expectations.
Table 2.
Putative Positively Selected Alleles and Their Diagnosis.
| Protein | Varianta | Population | Evolutionarily Unexpected High-Frequency Allele |
|||||
|---|---|---|---|---|---|---|---|---|
| Amino Acid | Frequency (%) | EP | SIFT | PolyPhen-2 | EvoD | |||
| NP_995322 | T111A | CEU | T | 100 | 0.001 | Non-N | — | — |
| NP_057264 | F374L | CEU | F | 98 | 0.004 | Non-N | Non-N | — |
| NP_006336 | M50V | CHB, JPT | V | 96 | 0.006 | N | N | N |
| NP_071731 | V370A | CHB, JPT | A | 95 | 0.002 | Non-N | N | Non-N |
| NP_057645 | M140R | YRI | M | 92 | 0.002 | Non-N | — | — |
| NP_002033 | M26V | CHB, JPT | M | 90 | 0.005 | N | — | — |
| NP_003259 | F616L | YRI | F | 89 | 0.237 | N | — | — |
| NP_002215 | L2436V | YRI | L | 85 | 0.002 | N | — | — |
| NP_006585 | L480S | YRI | S | 84 | 0.003 | N | N | Non-N |
| NP_057565 | V324M | YRI | M | 84 | 0.031 | N | N | N |
| NP_001136235 | D435A | CHB, JPT | A | 83 | 0.002 | N | Non-N | Non-N |
| NP_006579 | D5E | CHB, JPT | D | 82 | 0.005 | N | — | — |
| NP_002199 | R482Q | YRI | Q | 81 | 0.006 | N | N | N |
| NP_861445 | P267L | CHB, JPT | L | 79 | 0.089 | N | N | N |
| NP_055861 | I2587V | CEU | I | 74 | 0.013 | N | — | — |
| NP_062818 | S112A | YRI | S | 67 | 0.005 | N | — | — |
| NP_001122087 | L367V | YRI | V | 64 | 0.022 | Non-N | N | N |
| NP_073594 | T324P | CEU | P | 63 | 0.009 | N | N | N |
| NP_002199 | V1019A | YRI | A | 62 | 0.046 | N | N | N |
| NP_060224 | Q454R | CEU | Q | 62 | 0.111 | N | — | — |
| NP_057457 | A179T | CEU | T | 59 | 0.002 | N | N | Non-N |
| NP_114157 | R2141W | YRI | W | 57 | 0.023 | Non-N | — | N |
| NP_061139 | R390Q | YRI | Q | 54 | 0.040 | N | N | N |
| NP_004160 | L474F | YRI | F | 43 | 0.007 | Non-N | N | Non-N |
| NP_001009894 | V238L | CEU | V | 42 | 0.110 | N | — | — |
| NP_060328 | A111G | CHB, JPT | G | 40 | 0.001 | N | N | Non-N |
Note.—Alleles potentially involved in positive selection, reported in a comprehensive scan of 1000 Genome data by Grossman et al. (2013) are shown.
aEach variant is shown in the format of reference allele as defined in hg19 RefSeq annotation followed by position and alternative allele. For each evolutionary unexpected allele reported to be under positive selection, its population frequency, EP, and neutrality predictions from SIFT (Ng and Henikoff 2001), PolyPhen-2 (Adzhubei et al. 2010), and EvoD (Kumar et al. 2012) are shown. N, neutral; Non-N, nonneutral; —, no result because the mutant high-frequency population-specific allele is found in the reference genome as the consensus allele. Allele frequencies are from the corresponding populations in the 1000 Genomes data.
EP Versus Other Mutation Diagnosis Methods
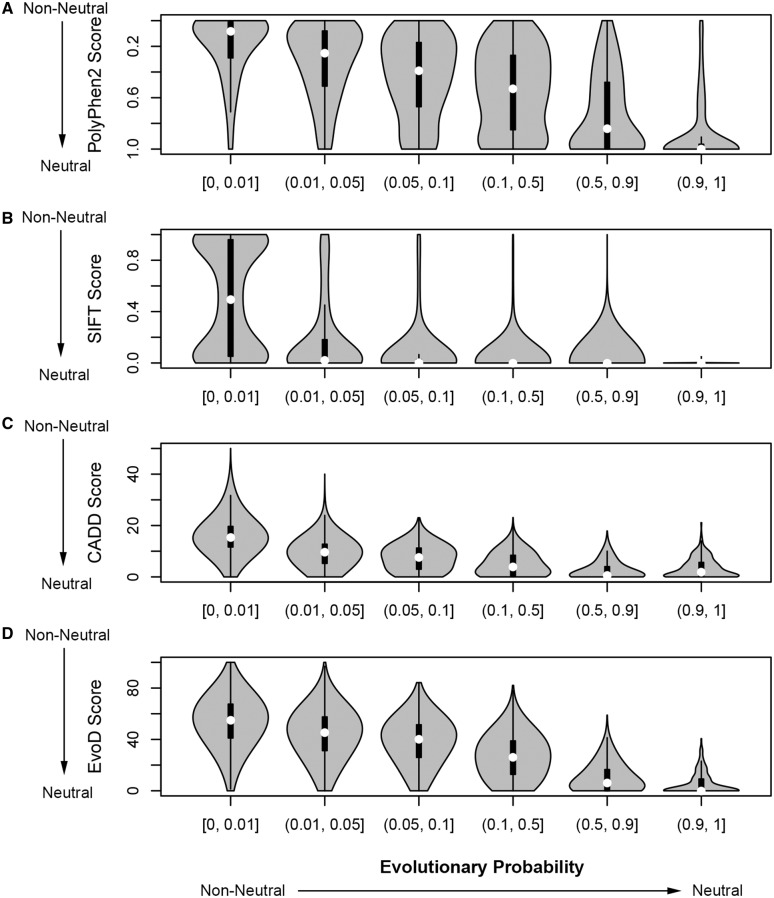
Several methods are available to estimate impact scores for alternative alleles (Sunyaev 2012); these scores are used to diagnose alleles as functionally neutral or nonneutral. Using 100,000 randomly chosen human population polymorphisms from the 1KG data, we explored the relationship of EP scores with impact scores produced by SIFT, PolyPhen-2, EvoD, and CADD (Ng and Henikoff 2001; Adzhubei et al. 2010; Kumar et al. 2012; Kircher et al. 2014). In all cases, EPs showed a strong positive correlation (fig. 6), primarily because all the approaches use similar evolutionary information. However, many differences were also revealed. In particular, many minor alleles with low EPs (EP < 0.05; nonneutral) receive “neutral” impact scores with current methods. These included potentially nonneutral alleles that are candidates for positive selection identified by comprehensive scan of the 1KG data (table 2). Although most (85%) of them have EP < 0.05, SIFT, PolyPhen-2, and EvoD deemed a vast majority to be either neutral or undiagnosable (73%, 92%, and 77%, respectively). These methods failed to diagnose these variants because they all use intraspecific variation data along with interspecies information during their calculation. This is also the reason why putatively adaptive alleles fixed in human population, as well as many other known high-frequency alleles shown in table 1, also received neutral diagnosis via these methods but were found to be nonneutral in EP calculations.
Fig. 6.
Relationship of EPs of human population polymorphisms with their impact scores produced by PolyPhen-2, SIFT, CADD, and EvoD. Using the 1KG data (1000 Genomes Project Consortium et al. 2012), 100,000 population polymorphisms were randomly selected. EPs and impact scores were calculated for minor alleles. Variants were grouped according to their EPs and violin plots produced to display the spread of impact scores from (A) SIFT, (B) PolyPhen-2, (C) CADD, and (D) EvoD. A violin plot displays the distribution of the impact scores for each group of variants; the white circle shows the median score and the black box shows the interquartile range.
In conclusion, our new method produces an eVar for a given genome using interspecific evolutionary history, which will prioritize function-impacting variants and identify positions that have undergone adaptive and other nonneutral evolution. This method can be applied to any species to generate reference variant sets for any part of its genome as long as multispecies sequence alignments can be assembled. The eVar for human proteins will be publicly available on the myPEG server (www.mypeg.info).
Materials and Methods
Computation of EP
To estimate the EP of observing an amino acid residue at a sequence position in a given species, we employ a Bayesian framework, a multispecies alignment consisting of orthologous sequences, and the phylogenetic tree relating the species in the alignment. For a simple data set consisting of four aligned orthologous sequences, let zi represents the residue at the focal position in the contemporary sequence Si, such that z = (z1, z2, z3, z4), and the ancestral states are given by the vector y = (y5, y6) (fig. 1A). We aim to estimate the relative likelihood of different residues at node 1. The PP for each possible residue is given by
| (1) |
where z* is the vector of observed residues at the given position with the constraint that the residue in species 1 at that position is unknown, that is, z* = (“?”, z2, z3, z4). And, b* is the vector of branch lengths (b1,…, b5) that is computed by using a maximum-likelihood method for the protein-specific amino acid alignment where the amino acid at the focal position is replaced by a missing data symbol. Under a time-reversible model of amino acid substitution, the evolutionary change of state can be assumed to start from any node of the tree, say y5. Then, the first component in equation (1) is given by
| (2) |
where is the probability of change from residues y5 to z1. is the frequency of residue y5 in the sequence data in the altered alignment as specified above. The second component in equation (1) is given by
| (3) |
And, the denominator in equation (1) is given by
| (4) |
where z1, y5, and y6 are allowed to take all possible amino acid states. Using the probabilities for all possible combinations of amino acid states z1, y5 and y6, the probabilities of all sets of allele (e.g., z1 = Ala, y5, y6) are summed to get the PP of any amino acid state (e.g., Ala) at z1:
| (5) |
For computing transition probabilities, we used a time-reversible model of substitution with equal probability of change from one amino acid to another (uniform prior at a given position) in order to avoid imposing a specific model of substitution on individual positions. This is important because it is becoming clear that the instantaneous substitution matrix for different positions is not the same (Lartillot and Philippe 2004). So, it is better to be conservative and not use standard, global substitution matrices when predicting the variome by molecular evolutionary analyses. Analyses using an equal evolutionary rate across positions in proteins produced PP estimates similar to those obtained when using a gamma distribution of rates across sites with 5 discrete categories.
To estimate EP, we compute multiple PP values by progressively pruning evolutionary lineages that are sister groups to the species of interest (species 1 here). For the example in figure 1A, we compute PP0 using the whole data set, PP1 after pruning S2 that previously induced ancestral node 5, and PP2 after additionally pruning S3 that previously induced ancestral node 6. Then, we obtain the EP as an average of PPi values weighted by the evolutionary time depth of the closest relative of species 1 (human) in the evolutionary tree used (Ti). Specifically,
| (6) |
where T(w) is the evolutionary divergence time between species 1 and species w, and PP(w) is computed by retaining only those species in the tree that share a common ancestor with humans starting with the ancestral node w and earlier.
The above method can be directly applied for the analysis of nucleotide sequences, which would produce EPs for different nucleotide bases by using a nucleotide sequence alignment and their phylogenetic relationships. More generally, one can apply this method for any sequence alignment where the evolutionary tree relating the sequences assumed a priori or inferred from the alignment. If evolutionary divergence times for nodes in the tree are not available, then one could employ a simple average instead of weighted average in equation (6) or use alternative measures of node depth (e.g., relative divergence times obtained using the RelTime method; Tamura et al. 2012) or linearized tree (Takezaki et al. 1995).
EP Analysis of Human Proteins
A total of 18,621 protein-coding genes were defined in the NCBI RefSeq database in the human genome build GRCh37. For each protein, the alignments of orthologous amino acid sequences in 46 vertebrate species were downloaded from the UCSC Genome Browser. Due to alternative splicing, multiple isoforms of proteins were found for 5,026 genes. In these cases, the longest isoform and its alignments were used, such that each gene and each position had only one representation in the eVar. Because the multiple alignments were derived based on sequence similarity and synteny (Miller et al. 2007), they consist of homologous exons. In a given species, if an exon homologous to a human exon was not found, these positions were represented as missing data (or “gaps”). We excluded 231 sequences/genes because the UCSC human sequences in the downloaded alignments were different from the RefSeq canonical sequences (similarity <98%). This resulted in 18,390 human proteins with alignments of orthologous sequences, to which we applied the new EP method. Here, divergence times were obtained from the Timetree resource (Kumar and Hedges 2011).
We set EP to 1 for the observed human allele and 0 for all other alleles at positions where only human sequences identified an amino acid residue (i.e., missing or insertion/deletion for the rest of the species in the UCSC alignments). Changes in the first position in a protein, early termination via gain of stop codons, and extension via loss of stop codons were not considered. To expedite calculations, we compared the EP estimates obtained position-by-position following the above procedure with those obtained simultaneously for all positions in a protein. This approximation involves generating PPs without replacing the site-specific bases by a missing symbol, which reduced the number of calculations per protein from the length of a protein to only one. The results were either identical or extremely similar (differ in the value in the third decimal place). Therefore, we have reported EPs generated using the faster method. In total, the eVar consists of alleles and their EPs at 10,575,180 amino acid positions in 18,390 protein sequences.
Conservation Categories
Using the same set of multiple alignments of orthologous sequences from 46 species, we estimated the evolutionary rate (r) of a protein position as the absolute substitution rate, reported as the number of substitutions per site per billion years (Kumar et al. 2012). A position is regarded as ultraconserved if r = 0, well conserved if 0 < r ≤ 1, less conserved if 1 < r ≤ 2, or least conserved if r > 2.
EP Analysis of Variants
We analyzed EPs for a wide collection of variants in human proteins, including population polymorphisms, variants associated with Mendelian diseases, complex diseases or somatic cancers, and variants under positive selection. Population polymorphisms were retrieved from the 1000 Genomes Project phase 3 data (1KG) (1000 Genomes Project Consortium et al. 2012) that contained 250,964 nSNPs and their overall population frequencies. From the HGMD database, we retrieved 60,502 missense mutations implicated in heritable diseases. We removed 6,468 mutations that contained potential annotation errors or are present in the 1KG data. The remaining 54,034 mutations were regarded as associated with Mendelian diseases. Variants associated with complex diseases were downloaded from the GRASP 2.0 database, which aggregates more than 6 million SNP–phenotype associations from 2,082 GWAS studies (Leslie et al. 2014). Among these associations, 376 unique missense variants were identified based on their chromosomal locations and primary protein isoforms, as well as presence in the 1KG data. Positions that were previously reported to be under positive selection were collected from studies that compared human with other species (Nielsen et al. 2005; Zhang et al. 2010) or scanned 1000 Genome data using population statistics (Grossman et al. 2013; Li et al. 2014).
To evaluate the accuracy of EP and other methods in diagnosing disease-associated variants, we used two existing sets of benchmark data. The first benchmark data set (HumVar; Adzhubei et al. 2010) consisted of 20,957 deleterious SNPs associated with diseases (positive controls), and 18,411 common population polymorphisms (negative controls). The second benchmark data set (CNO; Capriotti and Altman 2011) consisted of 3,128 cancer driver mutations (positive controls) and 3,046 passenger mutations (negative controls). PolyPhen-2, SIFT, and CCAD predictions were obtained from the dbNSFP database (Liu et al. 2013).
To cross-reference different sets of variants with the corresponding EPs, variants were first mapped to chromosomal locations in the human genome build GRCh37/hg19 and then to amino acid positions in the longest isoform of proteins as defined in NCBI RefSeq. Affected codons were identified, based on which wild-type and mutant nucleotides were translated into amino acid alleles. Unique variants were identified based on their chromosomal locations, RefSeq protein IDs, amino acid positions, wild-type and mutant alleles.
Acknowledgments
We thank S. Miura, J. Hey, R. Patel, K. Xu, G. Gibson, H. Rowe, and R. Kanda for comments. This work was supported by research grants from the National Institutes of Health (DK098242-03 to Joel Dudley and HG-002096-12 to S.K.).
References
- 1000 Genomes Project Consortium., Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. 2010. A method and server for predicting damaging missense mutations. Nat Methods. 7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbiza L, Dopazo J, Dopazo H. 2006. Positive selection, relaxation, and acceleration in the evolution of the human and chimp genome. PLoS Comput Biol. 2:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadpor U, Totonchi M, Sabbaghian M, Hoseinifar H, Akhound MR, Zari Moradi S, Haratian K, Sadighi Gilani MA, Gourabi H, Mohseni Meybodi A. 2013. Ubiquitin-specific protease (USP26) gene alterations associated with male infertility and recurrent pregnancy loss (RPL) in Iranian infertile patients. J Assist Reprod Genet. 30:923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E, Altman RB. 2011. A new disease-specific machine learning approach for the prediction of cancer-causing missense variants. Genomics 98:310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Karchin R. 2011. Using bioinformatics to predict the functional impact of SNVs. Bioinformatics 27:441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF. 2005. An introduction to population genetics theory. Caldwell (NJ): Blackburn Press. [Google Scholar]
- Dorfman R, Nalpathamkalam T, Taylor C, Gonska T, Keenan K, Yuan XW, Corey M, Tsui LC, Zielenski J, Durie P. 2010. Do common in silico tools predict the clinical consequences of amino-acid substitutions in the CFTR gene? Clin Genet . 77:464–473. [DOI] [PubMed] [Google Scholar]
- Dudley JT, Kim Y, Liu L, Markov GJ, Gerold K, Chen R, Butte AJ, Kumar S. 2012. Human genomic disease variants: a neutral evolutionary explanation. Genome Res. 22:1383–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, Park DJ, Griesemer D, Karlsson EK, Wong SH, et al. 2013. Identifying recent adaptations in large-scale genomic data. Cell 152:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman B, Weitz JS. 2012. A neutral theory of genome evolution and the frequency distribution of genes. BMC Genomics 13:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1983. The neutral theory of molecular evolution. Cambridge: Cambridge University Press. [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. 2011. TimeTree2: species divergence times on the iPhone. Bioinformatics 27:2023–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Sanderford M, Gray VE, Ye J, Liu L. 2012. Evolutionary diagnosis method for variants in personal exomes. Nat Methods. 9:855–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 21:1095–1109. [DOI] [PubMed] [Google Scholar]
- Leslie R, O’Donnell CJ, Johnson AD. 2014. GRASP: analysis of genotype-phenotype results from 1390 genome-wide association studies and corresponding open access database. Bioinformatics 30:i185–i194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MJ, Wang LY, Xia Z, Wong MP, Sham PC, Wang J. 2014. dbPSHP: a database of recent positive selection across human populations. Nucleic Acids Res. 42:D910–D916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E. 2013. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 34:E2393–E2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W, Rosenbloom K, Hardison RC, Hou M, Taylor J, Raney B, Burhans R, King DC, Baertsch R, Blankenberg D, et al. 2007. 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res. 17:1797–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. 2001. Predicting deleterious amino acid substitutions. Genome Res. 11:863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Bustamante C, Clark AG, Glanowski S, Sackton TB, Hubisz MJ, Fledel-Alon A, Tanenbaum DM, Civello D, White TJ, et al. 2005. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 3:e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN. 2009. The Human Gene Mutation Database: 2008 update. Genome Med. 1:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev SR. 2012. Inferring causality and functional significance of human coding DNA variants. Hum Mol Genet. 21:R10–R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezaki N, Rzhetsky A, Nei M. 1995. Phylogenetic test of the molecular clock and linearized trees. Mol Biol Evol. 12:823–833. [DOI] [PubMed] [Google Scholar]
- Tamura K, Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, Kumar S. 2012. Estimating divergence times in large molecular phylogenies. Proc Natl Acad Sci U S A. 109:19333–19338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Pei Z, Krawczak M, Ball EV, Mort M, Kehrer-Sawatzki H, Cooper DN. 2010. Triangulation of the human, chimpanzee, and Neanderthal genome sequences identifies potentially compensated mutations. Hum Mutat. 31:1286–1293 [DOI] [PubMed] [Google Scholar]