Abstract
Introduction
Ovarian cancer (OvCa) is among the most common types of cancer and is the leading cause of death from gynecological malignancies in western countries. Cancer biomarkers have a potential for improving the management of OvCa patients at every point from screening and detection, diagnosis, prognosis, follow up, response to therapy and outcome.
Areas covered
The literature search has indicated a number of candidate biomarkers have recently emerged that could facilitate the molecular definition of OvCa, providing information about prognosis and predicting response to therapy. These potentially promising biomarkers include immune cells and their products, tumor-derived exosomes, nucleic acids and epigenetic biomarkers.
Expert commentary
Although most of the biomarkers available today require prospective validation, the development of noninvasive liquid biopsy-based monitoring promises to improve their utility for evaluations of prognosis, response to therapy and outcome in OvCa.
Keywords: Ovarian carcinoma, serum, tissue, immune biomarkers, exosomes, microRNAs
1. INTRODUCTION
Malignant ovarian neoplasms are a heterogeneous group of tumors that include primary epithelial and non-epithelial ovarian cancers, with the former having the highest mortality rates of all gynecological malignancies with a median age at diagnosis of 65 years [1]. According to International Federation of Gynecology and Obstetrics (FIGO), most patients with ovarian carcinoma (OvCa) present with advanced stage, due to the lack of specific symptoms and adequate diagnostic tools [2]. The etiology of OvCa is unknown, but multiple genetic and environmental factors have been identified as potential contributors to its pathogenesis. Based on recently obtained morphological and molecular data, a new paradigm of the OvCa pathogenesis has emerged, which divides the epithelial tumor into two groups. Type I tumors are linked to ovarian precursor lesions such as cystadenofibroma, borderline tumors and endometriosis and include low-grade serous carcinoma, endometrioid, mucinous and clear cell carcinomas. These tumors are characterized by slow growth, presentation as low stage tumors and contain multiple genetic mutations. In contrast, type II OvCa are linked to precursors arising from the fallopian tube epithelium and consist of high-grade serous and endometrioid carcinomas, carcinosarcomas and undifferentiated carcinomas. Type II tumors are highly aggressive, are in advanced stage at diagnosis and have poor prognosis. Type I and type II tumors are also genomically distinct. Type I tumors are frequently associated with specific mutations in oncogenes such as KRAS, BRAF, PTEN and ARID1A. In the vast majority of type II tumors, TP53 is mutated, and a high level of chromosomal disruption is evident [3,4]. Standard treatment for advanced stage OvCa is tumor-debulking surgery and adjuvant platinum-based chemotherapy. However, after complete surgery and chemotherapy, the risk of relapse is approximately 85%, with substantial variations based on the well-known clinicopathological features. The five-year survival of patients with advanced disease is only 10–30% [5]. In the past two decades, several new drugs have become available for patients with OvCa. However, in individual patients, it is difficult to determine the effectiveness of a given treatment due to a lack of objective measures that define efficacy. These clinical findings have emphasized the need to identify novel biomarkers for OvCa that can be used to measure responses to therapy, prognosis and overall outcome. Recently, a number of candidate biomarkers have been evaluated, but the lack of the understanding of the OvCa etiology as well as the disease heterogeneity, make it unlikely that a single biomarker will satisfy criteria formulated for acceptance of cancer biomarkers [6].
2. DEFINITION OF A CANCER BIOMARKER
The so called REMARK criteria have been formulated and are listed in the literature as guidelines for biomarker studies [7,8]. Briefly, a new biomarker has to be validated in prospective (and not retrospective) studies of adequate size and statistical power. These studies should include a unique cohort of patients in whom the biomarker correlates with disease activity and the known (if any) molecular factors predictive of survival. The biomarker should be able to discriminate between pathologic and physiologic conditions even if they are similar. The biomarker should have a defined molecular mechanism of biological activity, and the data in support of its validity have to be based on thorough specimen collection, assay results confirming specificity, sensitivity, reproducibility, robustness as well as statistical rigor and on stringent patient follow-up. Cancer biomarkers may be discovered using molecular, cellular, and imaging methodologies. They should be detectable in biological samples that are easily obtainable, for example; serum, plasma, whole blood, ascites, urine and tissues accessible for sampling. Biomarkers can be normal endogenous products that are produced at a greater rate in cancer cells or the products of newly switched on genes that remained inactive in normal cells. Biomarkers may include intracellular molecules or proteins in tissues or may be released into the circulation and body fluids. In addition, the assays for biomarkers to be used clinically should be simple, inexpensive and lend themselves readily to high through-put technologies. These are by no means trivial requirements, and they emphasize the difficulties that are associated with the field of biomarker discovery.
3. CA125 AND HE4 BIOMARKERS FOR OVCA
Today, the most frequently used biomarker for OvCa is CA 125 (MUC 16). CA125 is useful for monitoring responses to chemotherapy, detecting disease recurrence and for differentiation of malignant from non-malignant pelvic masses. In the past decade, major efforts have been made to improve the performance of CA125 in differential diagnosis of pelvic masses and in screening for OvCa [9]. However, the positive predictive value is low for CA125, and it is only effective when used in combination with other diagnostic tests (Table 1). Moreover, CA125 can be elevated in a number of conditions unrelated to OvCa [9], and 20% of OvCa have little or no expression of CA125. Nevertheless, according to the current guidelines, CA125 remains the only serum marker accepted for diagnosis and follow up of OvCa.
Table 1.
Clinically-used serum biomarkers for detection of ovarian carcinomaa
| Biomarkers | Sensitivity | Specificity | References |
|---|---|---|---|
| CA125 | 92% | 62% | [17] |
| RM1 (risk of malignancy index) CA125 + ultrasoundb |
96% | 82% | [10,17] |
| HE4 | 91% | 63% | [18] |
| ROMA (risk of ovarian malignancy algorithm) CA125 + HE4b |
95% | 77% | [17, 18] |
|
| |||
| OVA1 | 96% | 40% (pre)c | [19] |
| 5 different markers: (↑CA125; ↑β macroglobulin ↓apolipoprotein A1; ↓prealbumin ↓transferrin) | 85% | 28% (post)c | |
The sensitivity and specificity values are for differentiating benign diseases from OvCa.
The menopausal status influences risk. The values shown are for all patients without consideration of the menopausal status.
Pre vs. post refer to pre-menopause and post-menopause
To improve the diagnostic value of CA125 in OvCa, especially in the presence of a suspicious pelvic mass or symptoms, the risk malignancy index (RMI) has been developed [10]. The RMI combines CA125 results with ultrasound findings and the menopausal status to calculate the index score that helps in predicting the risk of OvCa in women with adnexal mass (Table 1). The RMI remains the most accurate tool for stratifying patients into high and low risk groups.
Another commonly used serum biomarker for OvCa is human epididymis protein 4 (HE4), recently approved by the FDA for monitoring disease progression or recurrence in OvCa. This protein was initially identified in the epithelium of the distal epididymis and then discovered to be a protease inhibitor involved in sperm maturation [11]. It appears to be useful in differential diagnosis of adnexal masses and in early diagnosis of OvCa (Table 1). Various factors aside from malignancy including age, smoking and chronic renal disease may influence serum HE4 levels and, thus, the interpretation of the results of HE4 assays is not straightforward [12,13]. On the other hand, the HE4 serum level, in contrast to that for CA125, is not affected by menstrual cycle, endometriosis and oral contraceptives [14,15]. Therefore, HE4 seems to be a potentially more specific detection marker than CA125 for early diagnosis and has been cleared by the FDA for monitoring disease progression and recurrence [16].
Promising HE4 results in OvCa diagnosis, especially in combination with CA125, have led to the development of the Risk of Ovarian Malignancy Algorithm (ROMA) [17]. It combines HE4, CA 125 and the menopausal status to enhance sensitivity and specificity of OvCa detection (Table 1). As neither CA125 nor HE4 measurements alone meet the requirements for sensitivity or specificity that should be attributes of the biomarkers used for screening or monitoring responses to therapy in OvCa, ROMA provides a greater power for diagnosis of OvCa than either of the two markers alone (Table 1) [18]. Whether ROMA is better than the RMI in distinguishing OvCa from benign diseases remains unclear [18]. Furthermore, a survival advantage of early detection of OvCa or OvCa recurrence using HE4 or CA125 markers is yet to be established.
In 2009, the FDA approved another test for clinical testing of OvCa named OVA1 [19]. It evaluates serum concentrations of five different markers (Table 1), and appears to have not only high levels of sensitivity but also high negative predictive value [19].
In view of the paucity of reliable biomarkers in OvCa and inadequate diagnostic or prognostic performance of those that are available, the identification of novel biomarkers for early detection, prognosis, prediction of treatment efficacy and overall outcome in OvCa is an unmet need. This review considers some of the most promising recently described biomarkers and their potential role in the detection and prognosis of OvCa.
4. EMERGING SERUM BIOMARKERS
Blood contains a wide range of proteins that reflect the physiological conditions in tissues and thus can be used as a biological marker of impending or ongoing disease. Blood collection is a minimally invasive procedure, and sampling can be carried out in large cohorts of patients. Recently, rapid advances in high-throughput technologies has allowed for new and exciting opportunities for serum biomarkers discovery. More than 30 serum markers have been evaluated alone and in combination with CA125 by different investigators. Some of the most promising include: osteopontin, mesothelin, kallikrein(s), macrophage colony-stimulating factor (M-CSF), and soluble epidermal growth factor receptor (EGFR). Much of this effort has utilized proteomics analyses to monitor and identify proteins that regulate functions and cellular interactions responsible for cancer cell growth. For example, in 2002, Petricoin et al. used an independent set of 116 serum samples: 50 from women with OvCa and 66 from normal controls or those with benign tumors to search for markers that would best distinguish the two cohorts. The results showed that the used algorithm identified a cluster of proteins which completely segregated OvCa sera from non-cancer specimens [20]. In 2014, Cheng et al. analyzed serum samples from patients with OvCa and from healthy controls. Proteomic analyses identified 1200 serum proteins, among which 57 were upregulated in the sera collected from cancer patients. Retinol binding protein 4 (RBP4), which is an adipokine secreted by adipose tissue and liver, was found to be highly elevated in sera of OvCa patients compared to healthy individuals [21]. These examples suggest that proteomics profiles of OvCa patients’ sera are distinct from those of sera of healthy controls.
Among proteins found to be consistently elevated in OvCa sera is osteopontin, a multifunctional cytokine that plays an important role in many physiological and pathological processes, including cell proliferation, survival, drug resistance, invasion, inflammation, immune response, stem cell behavior and tumorigenesis [22,23]. Osteopontin is overexpressed in many different tumor types, including OvCa [24]. Preoperative plasma levels of osteopontin were significantly higher in patients with OvCa compared to those with benign ovarian tumors and to healthy women’s plasma. Thus, preoperative osteopontin levels could be a potentially useful biomarker for detecting OvCa, especially when used together with CA125 [25].
Mesothelin, a cell surface protein on OvCa cells, is a potential target for antibody-based therapy due to its high expression levels in OvCa. It is also a new target for chimeric antigen receptor (CAR) T cell adoptive therapy [26]. Mesothelin plays a role in cancer progression; it may aid in the peritoneal implantation and metastasis of tumor through its interaction with CA125; it promotes cancer cell proliferation and survival via the NF-kB signaling pathway; it also promotes resistance to chemotherapy, including paclitaxel, platinum and cyclophosphamide. In patients with OvCa, high expression of mesothelin in both tissues and serum indicate poor prognosis [27]. Ibrahim et al evaluated diagnostic accuracy of serum mesothelin levels in 110 patients with ovarian masses relative to serum CA125 levels (Table 2). Mesothelin seemed to have the same sensitivity, but higher specificity than CA125 and was a significant predictor of early stage OvCa [28].
Table 2.
Emerging serum biomarkers in ovarian carcinomaa
| Marker | Sensitivity | Specificity | References |
|---|---|---|---|
| Osteopontin | 77% | 90% | [22, 24] |
| Osteopontin & CA125 | 87% | 88% | [25] |
| Mesothelin | 97% | 98% | [28] |
| Mesothelin & CA125 | 97% | 98% | [28] |
| sEGFRb | 64% | 65% | [31] |
| sEGFR & CA125 | 68% | 100% | [32] |
| M-CSFb | 70% | 94% | [35] |
| M-CSF & CA125 | 86% | 86% | [35] |
| sFRAb | 59% | 98% | [36] |
The sensitivity and specificity values are for differentiating benign disease for OvCa
- sEGFR: soluble epidermal growth factor receptor;
- M-CSF: macrophage colony stimulating factor;
- s-FRA: soluble folate receptor alpha
The epidermal growth factor receptor (EGFR) is overexpressed in 30–98% of EOC, and signaling cascades it activates impact on tumor cell proliferation, migration, invasion and resistance to chemotherapy [29]. While EGFR emerges as an attractive therapeutic target in OAC, soluble EGFR (sEGFR) has been of interest as a potential biomarker for EOC diagnosis and/or resistance to platinum therapy [30]. Circulating sEGFR is a 90–110kDisoform of EGFR readily detectable by immunoassays. Its concentrations are lower in OvCa patients compared to healthy women or women with benign ovarian tumors or other gynecological conditions [31,32]. This suggests that sEGFR alone or in combination with CA125 may be a useful biomarker for risk assessment, early detection and diagnosis of OvCa (Table 2). Also, sEGFR in combination with CA125 can differentiate benign from malignant OvCa [32]. A recent study of sEGFR levels in late-stage patients suggested that OvCa patients with increased sEGFR levels had poor PFS [33].
The macrophage colony-stimulating factor (M-CSF), a cytokine also known as colony stimulating factor-1 (CSF-1), regulates growth and differentiation of myeloid cells. I t is a chemoattractant for monocytes, and is strongly expressed in OvCa specimens of patients with advanced disease. Detectable in sera by immunoassays, it has been used alone or in combinations with HE4 or CA125 as a screening assay for OvCa [34]. In a recent study of 110 OvCa patients, detection sensitivity of M-CSF was somewhat greater than that of HE4 or CA125 and improved upon its combination with these two markers [35]. Its specificity was very high at 94% (Table 2). Thus, M-CSF emerges as a potentially useful screening test for OvCa.
Another promising serum biomarker for OvCa diagnosis is the soluble form of the folate receptor alpha (sFRA). It is a GPI-anchored glycoprotein encoded by the FOLR1 gene, which can be shed from the cell surface into the circulation [36]. Kurosaki et al. investigated serum levels of sFRA in 231 patients and found high sFRA in sera of EOC patients, but not in patients with benign or borderline gynecological disease or with OvCa metastasizing from colorectal cancers. Serum levels of sFRA were highly correlated to the clinical stage, tumor grade and histological tumor type and demonstrated greater accuracy for the detection of OvCa than did serum CA125 (Table 2). In both early and advanced stage patients, high sFRA levels were significantly associated with shorter PFS [36].
A number of other potentially promising diagnostic markers for EOC are currently under investigation, including vascular endothelial growth factor (VEGF), interleukin-10 (IL-10), metalloproteinases (MMP-2, MMP-7, MMP-9) or the tissue inhibitor of metalloproteinase-1 (TIMP-1).
5. TISSUE BIOMARKERS
Immunohistochemistry (IHC) with antibodies specific for OVCa antigens helped to identify those that are overexpressed or aberrantly expressed in tumor tissues. Human leukocyte antigen-G (HLA-G) is a non-classical MHC class I molecule. Its major biological role is suppression of ongoing immune responses. Measurements of HLA-G protein or mRNA levels have shown promise in detection and prognosis of OvCa [37,38]. HLA-G expression can be associated with either favorable or unfavorable clinical outcome. On the one hand, high expression levels of HLA-G such as seen in aggressive types of OvCa, i.e., high-grade serous carcinomas, tend to suppress anti-tumor immune responses promoting tumor escape. On the other hand, HLA-G expression emerges as potential marker of tumor susceptibility to chemotherapy and associates with longer progression-free survival [38]. In high-grade epithelial OvCa, HLA-G expression is an independent prognostic factor and a predictor of platinum sensitivity [38], which outweighs its tumor immunoprotective role.
A growing body of evidence suggests that reproductive hormones can affect tumor progression, proliferation and metastasis through paracrine, autocrine or intracrine mechanisms [39]. Hydroxysteroid (17B) dehydrogenase type 12 (HSD17B12) is a multifunctional isoenzyme functional in the conversion of estron to estradiol and elongation of long-chain fatty acids, in particular the conversion of palmitic to archadonic acid, the precursor of sterols and the inflammatory mediator, prostaglandin E2 (PGE2). In the normal ovary HSD17B is detected in granulosa cells of developing follicles, but not in the surface epithelium. In contrast, epithelial OvCa is positive for HSD17B, and its expression in OvCa is associated with poor prognosis [40]. The clinical implications attributed to HSD17B12 overexpression in OvCa suggest that it could serve not only as biomarker but also as candidate for T-cell based immunotherapy [41].
Proteomics have introduced another powerful approach to the detection of proteins that have promise as biomarkers in OvCa such as kallikrein-related peptidase (KLK). Tissue kallikrein (KLK1) and kallikrein-related peptidases (KLK2-15) are aberrantly expressed in OvCa tissues, in particular, in the more metastatic type II-tumors. KLKs play a role in many aspects of pathophysiology including hydrolysis of growth factors, proteases, cell membrane bound receptors, adhesion proteins, and cytokines, regulating intracellular signaling pathways and their downstream events. High KLK levels are associated with poor prognosis of OvCa patients, suggesting that KLK might be a useful as a marker of disease progression [42]. Two kallikrein serine protease family members (KLK6 and KLK7) were found to be significantly overexpressed relative to normal tissue controls in most of the OvCa cell lines. Overexpression of KLK6 and KLK7 mRNA was specific for ovarian cancer, in particular for serous and papillary serous cancer subtypes. In situ hybridization and IHC further confirmed significantly elevated levels of KLK6 and KLK7 mRNA and proteins, respectively, in tissue epithelia relative to a lack of expression in the neighboring stroma [43].
6. IMMUNOLOGICAL BIOMARKERS
The idea of the immune system as a valuable parameter in the development and prognosis of ovarian cancer has been neglected for a long time [44]. Newer data indicate that the host immune system strongly influences outcome in patients with OvCa. Studies of immune cells infiltrating OvCa have indicated that accumulations of lymphocytes, regulatory T cells, macrophages, dendritic cells (DC), myeloid derived suppressor cells (MDSC) and natural killer (NK) cells might determine biologic behavior of the tumor as well as cancer therapy and prognosis. Interestingly, the absolute lymphocyte count (ALC) in peripheral blood was found to be associated with survival in OvCa independent of tumor-infiltrating lymphocytes (TILs) in OvCa [45]. T lymphocytes represent a major population of TILs. Their diversity makes it difficult to evaluate the significance of these cells in the tumor development, since individual subtypes of T cells could have opposite effects on tumor progression. High density of tumor-infiltrating CD3+ T cells has been associated with favorable prognosis in various types of cancer, including OvCa [46–48]. Among specific subtypes of TILs, cytotoxic T cells (CD8+) have also been associated with better survival in OvCa patients [49]. TIL infiltration patterns in OvCa can range from CD8+T cells alone to complex lymphoid follicle-like structures that include CD8+T, CD4+T and CD20+ B cells [50]. Cooperation between tumor-infiltrating B and T cells leads to increased patient survival, presumably because B cells, which serve as APC and secrete polarizing cytokines, enhance antitumor immunity [51]. Analyses of TILs in high-grade serous OvCa further showed that expression by intraepithelial CD8+ T cells of CD103 (an αE integrin) strongly correlated with increased disease-specific survival [52]. In aggregate, new insights into anti-tumor immunity in OvCa indicate that patient survival rates increase as the complexity and size of the TIL compartment increases, suggesting that cooperative interactions between various infiltrating immune cells are important for survival.
Another subset of CD4+ T cells variably present within the tumor environment are the CD4+CD25+regulatory T cells (Treg). They usually represent a smaller fraction of tumor infiltrating lymphocytes, but may have a significant influence on the tumor development [53,54]. It has been shown that the frequency of Treg is higher in tumors than in normal tissues due to an active recruitment of Treg by factors present in the tumor microenvironment [55]. Increased percentages of Treg are also seen in the peripheral circulation of patients with cancer [56,57]. Their potential to inhibit Th1 responses by interfering with anti-tumor functions of effector T cells could contribute to poor outcome, as seems to be the case in some human solid tumors, including OvCa [55]. Moreover, high percentages of Treg in OvCa contribute to an immunosuppressive microenvironment that curbs anti- tumor immunity [55].
Macrophages represent a major population of tumor–infiltrating leukocytes. Their number is often increased in tumors compared to healthy tissues [44]. It is well known that M1 macrophages, predominantly situated within tumor epithelial islets, have anti-tumor effects, whereas M2 cells, located in the stroma, promote tumor growth. Emerging evidence suggests that tumor-associated macrophages (TAM) display a unique activation profile in OvCa and are able to create an immunosuppressive microenvironment, allowing tumors to evade immune detection and promoting tumor progression. Studies investigating the presence of TAMs in OvCa demonstrate a significant increase in their number in malignant ovarian tumors compared to benign and borderline tumors [58,59]. However, the quantity of CD68+ tumor–infiltrating macrophages does not appear to influence outcome [60,61]. More detailed studies using specific M2-associated markers in OvCa indicated that TAM can predict the patient’s prognosis in certain subsets of OvCa. In addition to CD68+ cells, Lan et al. also studied expression of CD163, an M2 marker, in 110 advanced stage OvCa and demonstrated that both PFS and overall survival (OS) were significantly reduced in patients whose tumors contained high numbers of CD163+ cells [62]. Serum levels of CD163 have also been shown to predict poor prognosis in patients with OvCa [63].
Dendritic cells (DC) are professional antigen presenting cells that regulate immune system. The two major DC subset are the classic DC and the plasmacytoid DC, and both have been studied in OvCa.[64]. Unlike other TILs, the density of DC at the tumor site is lower than that in the corresponding normal tissues. The results of several studies indicated that DC from OvCa patients are functionally defective or immunosuppressive and that prognosis of OvCa is inversely related to the presence of tumor infiltrating DC [65,66].
Myeloid-derived suppressor cells (MDSC) are a heterogeneous population of immature myeloid cells with profound immunosuppressive abilities that contribute to immune dysfunction and tumor progression. MDSC accumulate in OvCa, inhibit T cells responses and correlate with more rapid recurrence in OvCa patients [67,68]. Obermajer et al showed that MDSC accumulated in the ascites of patients with advanced OvCa, and that they suppressed T cell proliferation ex vivo [69]. These findings show that MDSC, representing a specific innate immune population, may serve as a potential prognostic marker with the potential to predict time to relapse in OvCa.
Extensive research of the last two decades suggest that tumors are inflammatory organs, in which the tumor microenvironment (TME) has been co-opted to support tumor growth [70]. In this context, inflammatory chemokines assume major roles in cancer. OvCas are known to produce a variety of chemokines which impact on the TME, including cancer cells and immune cells. These factors that in principle could have protected the individual against the developing tumor are being used by the tumor cells for their own propagation, motility and spread. A recent study of OvCa progression has shown that two phenotypically distinct monocyte subsets were present in the peritoneum at different stages of tumor progression. These two monocytes population suppressed activities of naïve CD8+ and CD4+ T cells. CCR2, a chemokine which specifically mediates monocyte chemotaxis, was a critical factor in recruiting these suppressive cells to the ovarian tumor microenvironment [71]. Moreover, CCR2-expressing MDSC limited the efficacy of immune therapy by down regulating the migration of CD8+ T cells to the tumor site [72]. Another chemokine produced by OvCa cells as well as associated macrophages is CCL22, which mediates trafficking of Treg to the tumor. This specific recruitment of Treg represents a mechanism by which tumors may foster immune privilege [55].
7. TUMOR-DERIVED EXOSOMES (TEX)
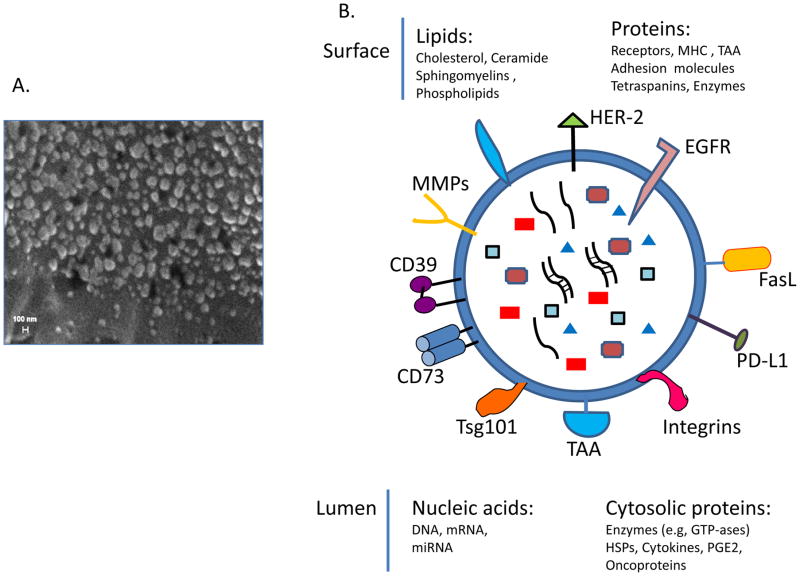
One of the biggest challenges in identification of biomarkers of pathological mechanisms operating in the ovary during OvCa progression is the inaccessibility of the diseased tissue. Many secreted molecules and factors, such as cytokines or chemokines, are readily detectable in body fluids, including ascites, and can serve as important biomarkers of the inflammatory processes. However, secreted biomolecules originating from non-circulating OvCa cells are often present in very low concentrations and thus are difficult to detect. In recent years, a better understanding of cell-to-cell signaling through secreted extracellular vesicles (EVs) has indicated that EVs may be an important source of biomarkers in many different diseases [73]. All cells secrete EVs, which vary in size from large apoptotic bodies (1,000–5,000 nm), smaller microvesicles (MVs, 200–1,000nm) and the smallest, exosomes (30–150nm). Tumor cells produce masses of EVs, which are found in all body fluids. Exosomes represent information-containing packages, which transfer their content to target cells, inducing changes in the phenotype and functions of the target cells [74]. The molecular profile of exosomes found in body fluids resembles that seen on the surface membrane of cells from which they originate (Figure 1). Exosome cargos include proteins, such as enzymes, cytokines, growth factors, lipids and nucleic acids, including mRNA or microRNA and ncRNA [75–77]. It has been shown that tumor cells are avid producers of exosomes, which play a role in many aspects of tumor progression including: degradation of the extracellular matrix [78]; angiogenesis [79]; suppression of immune cell functions [56,80,81] and drug resistance [82].
Figure 1.
Exosomes in plasma of OvCa patients. A. A scanning electron micrograph image of gold-coated exosomes isolated from plasma of a patient with OvCa.. Reproduced with permission from Szajnik M, Derbis M, Lach M et al. Exosomes in Plasma of Patients with Ovarian Carcinoma: Potential Biomarkers of Tumor Progression and Response to Therapy. Gynecol Obstet (Sunnyvale) 2013 Suppl 4:3. doi:10.4172/2161-0932.S4-003 B. A diagram of a tumor-derived exosome illustrating the variety of molecules in the exosome cargo. Reproduced from Whiteside TL. Tumor-derived exosomes and their role in tumor progression. Adv Clin Chem. 2016;74:103-41. doi: 10.1016/bs.acc.2015.12.005. Epub 2016 Apr 7. with permission from Elsevier.
OvCa cells release large quantities of exosomes which have a molecular composition in part resembling that of cell surface membranes in the parent tumor cells. The OvCa patients’ plasma contains higher levels of exosomal proteins compared to those found in plasma of patients with benign tumors or normal controls (NC) [83]. Exosomes isolated from OvCa patients’ plasma carry TGFβ-B1 and other OvCa-associated antigens, e.g., melanoma-associated antigen MAGE 3/6, previously shown to be present in OvCas [84] which distinguish OvCa patients from benign tumors or NCs [83]. High protein levels of exosome fractions are seen in newly diagnosed patients [83]. Moreover, in advanced stages of OvCa, the protein content of isolated exosomes is significantly higher than that in early stages [83]. Thus, the protein levels in isolated exosome fractions could differentiate OvCa patients with early from those with advanced disease. The exosome protein levels variably changed during/after chemotherapy [83]. Preliminary correlations between the changes in exosomal protein levels and clinical data suggested that the protein content of exosomes in plasma might be useful in predicting responses to therapy and prognosis in OvCa patients [83]. A study by Luciani et al. suggested that tumor-derived exosomes could be responsible for removing cytotoxic drugs from tumor cells thus reducing the anti-tumor potential of chemotherapy [85].
Also, Safaei et al. showed that cisplatin-resistant OvCa patients secreted higher quantities of microvesicles which carried the cisplatin export transporters, MRP2, ATP7A, and ATP7B [86]. Moreover, enhanced microvesicles production and resistance to cisplatin were associated with higher expression of genes whose products are known to be responsible for membrane fusion, vesicle trafficking and export of drugs by microvesicles [86]. In recent years, it has become increasingly clear that exosomes obtained from plasma of OvCa patients contained significant levels of mRNA and miRNA, providing an explanation for the ability of TEX to transfer genetic material between the cells [87]. miRNA and other molecular features of MV represent a unique combination representative of the cancer cells from which they were derived [88]. Thus, their presence in cancer-derived exosomes may serve as a novel source of disease-related information and possibly as unique and specific cancer biomarkers that may prove useful for screening and diagnosis [75].
8. CANCER STEM CELL BIOMARKERS
Cancer stem cells (CSC) are a small population of malignant cells within a tumor (typically 0,01–1% of cells) that have increased tumorigenicity and differentiating capacity and unlimited self-renewal. Thus, CSC are capable of recreating the original tumor. The concept of OvCa as a stem-cell disease has been gaining credence in recent years. It profiles CSC as important drivers of carcinogenesis, metastasis, tumor recurrence and drug resistance. In this context, CSC are seen as predictors of poor prognosis. [89,90]. Tumors identified as being stem cell-like generally have a high tumor grade and significantly worse prognosis [91,92]. However, there exists a significant controversy in regard to markers that define CSC in human OvCa and prognostic significance of these markers [93]. For example, a number of cell surface markers including CD133 (prominin), CD44 (hyaluronic acid and receptor), CD24 (mucin-like cell surface glycoprotein marker), ALDH (aldehyde dehydrogenase), CD117 (mast/stem cell growth factor receptor), EpCAM (tumor associated calcium signal transducer 1) and ABCG2 (ATP-binding cassette sub-family G member 2), are used singly or in combination to identify and isolate CSC from human cancers. Among these markers, the membrane glycoprotein prominin (CD133) is most widely used but the least understood in terms of its functions [94]. Nevertheless, it has been shown that increased expression of CD133 is a negative prognostic factor in OvCa [95]. OvCa patients with combined expression of CD133 and ALDH within the tumor had significantly poorer outcome. Single ALDH expression in OvCa correlated with poor prognosis in some studies but not others [96,97] [98]. Steffensen et al reported that early stage OvCa patients whose tumor contained >20% of CD44+ CSC, had a shorter PFS compared to patients with tumors with <20% of these cells [99]. Similarly, CD24 expression of has been correlated with poor prognosis [100]. Zhang et al found that CSC express a type I receptor tyrosine kinase-like orphan receptor (ROR1) and that patients with high ROR1 expression levels had high rates of relapse and short median survival. [101]. Recently, an expression profile analysis of 145 serous OvCas identified a stem-like subtype characterized by the 51-gene signature. This signature was significantly enriched in type-II tumors and it was prognostic within high-stage serous OvCa, identifying a small subset of high stage tumors with better prognosis. This signature remained a significant independent predictor of relapse even after adjusting for common clinical factors [102].
9. EPIGENETIC BIOMARKERS
Aberrant DNA methylation occurs frequently in human tumors and is considered to be one of the earliest molecular changes in carcinogenesis [103–105]. Thus, epigenetic candidate gene studies promise to reveal new biomarkers specially for early cancer diagnosis. Indeed, already numerous studies of differential methylation have identified potential biomarkers not only for diagnosis, but also for progression and response to therapy [106]. In OvCa, two opposite epigenetic phenomena take place; namely, a global decrease in DNA methylation of heterochromatin leading to demethylation of several oncogenes and a specific CpG island hypermethylation associated with the promoters of tumor suppressor genes (TSG) [107]. The epigenetic silencing of TSGs in OvCa is best described for BRCA1. Studies have demonstrated that hypermethylation of the BRCA1 promoter occurs in 10–15% of sporadic cases of OvCa leading to a loss of BRCA 1expression [108,109]. It is associated with the serous histotype [108,110] and aggressive malignancy [111]. Accordingly, patients with hypermethylation of the BRCA1 promoter have a significantly shorter PFS and OS compared with patients harboring mutated or wild-type BRCA1 [112]. Additionally, BRCA1 methylation has been shown to be associated with sensitivity to platinum treatment in a pilot study of 35 OvCa cases [113]. Another protein responsible for DNA repair, MutL homolog 1 (MLH1) is encoded by the MLH1 gene which is also hypermethylated in OvCa, leading to resistance to platinum based therapy in patients. This platinum resistance is reversed upon restoration of MLH1 expression [114]. Gifford et al. compared the DNA methylation patterns of MLH1 in plasma DNA from OvCa patients prior to and following treatment (at relapse) with platinum-based therapy [115]. They found that promoter MLH1 methylation during treatment was significantly correlated with poor outcome. However, expression of MLH1 prior to treatment was not able to predict outcome [116]. These results suggest that MLH1 methylation may serve as a marker for enrichment of inherently resistant cells under therapy and could help in determining response to platinum treatment.
Montavon et al. found that the gene encoding homeobox protein A9 (HOXA9) is methylated in 75/79 (97.5%) of high-grade serous ovarian carcinoma and only in 1/12 (8%) of normal ovarian surface epithelium [117]. Furthermore, methylation of both HOXA9 and EN1 in combination with CA-125 levels was able to discriminate high-grade serous OvCa from normal ovarian epithelium with a sensitivity of 100% and a specificity of 91.7%, suggesting that HOXA9 methylation could serve as a potential biomarker for the detection of OvCa. Tam et al. investigated methylation of HIC1 in a large number of OvCas, including borderline and benign ovarian tumors, as well as normal ovarian tissues, and found that HIC1 was methylated in >50% of OvCa cancer tissues, >40% of borderline tumors, >20% of benign tumors and 10% of normal ovarian tissues [118], suggesting that HIC1 methylation correlates with the presence of invasive OvCa. Another TSG in OvCa, secreted protein/acidic/cysteine-rich (SPARC), is involved in cell adhesion, motility and ECM interactions, and is downregulated in OvCa through hypermethylation of its promoter. Socha et al. recently demonstrated that the SPARC promoter is hypermethylated full in 68.2% OvCa and partially in 22.7% OvCa, respectively. In addition, SPARC expression was inversely correlated with tumor grade [119]. It has been shown that genes involved in the TGF-β and Wnt pathways are often dysregulated through methylation in numerous cancers including melanoma, gastric and prostate carcinomas. Kang et al. observed TGFBI methylation in 23/38 (60.5%) of OvCa cases, 5/18 (27.8%) borderline ovarian tumors and 0/38 normal ovarian tissues [120]. However in this cohort, TGFBI methylation was not associated with any clinical outcome. Matsumura et al. identified depressed TGF-beta pathway activity by methylation of several genes involved in 39 ovarian cancer cell lines and OvCa tissues [121].
In a recent study Chou et al showed that FBXO32, the gene encoding F-box-only protein 32, is a TGF-beta/SMAD4 target gene and a regulator of apoptosis. This gene was downregulated through methylation in 96 OvCa cases but remained unmethylated and at normal levels in healthy ovarian surface epithelium [122]. Methylation was significantly associated with tumor stage and poor PFS. Re-expression of FBXO32 restored sensitivity to cisplatin in vitro, suggesting that methylation of FBXO32 may serve as a prognostic marker for OvCa.
In contrast to the downregulated TGF-β pathway, the Wnt pathway is often activated in cancers – but again through the hypermethylation of specific regulators. For example, the gene encoding secreted frizzled-related protein, SFRP5, a Wnt antagonist, was found to be hypermethylated in 44.4% of examined OvCa and only in 1.3% of benign cases [123]. Also, expression of SFRP5 sensitized OvCa cells to cisplatin and taxol in vitro, and patients with methylated SFRP5 had worse clinical responses [124]. Dai et al. demonstrated that methylation at 7 loci (FZD4, DVL1, NFATC3, ROCK1, LRP5, AXIN1, and NKD1) in a panel of 137 Wnt pathway genes in a screening cohort of 111 and a validation cohort of 48 serous and endometrioid OvCa was associated with poor progression-free survival. Hypermethylation of DVL1 and NFATC3 significantly correlated with poor response, with patients with progressive or stable disease harboring increased methylation versus patients with partial or complete response [125]. These studies demonstrate that epigenetic regulation of the Wnt pathway may serve as a biomarker for prognosis and/or treatment response in OvCa.
A novel biomarker candidate could be opioid-binding protein/cell adhesion molecule (OPCML), an immunoglobulin domain-containing GPI-anchored cell adhesion molecule encoded by the gene OPCML which behaves like a tumor suppressor. It is methylated and inactivated in a high proportion (33–83%) of ovarian tumors [126,127]. These studies also indicate that methylation of OPCML may be an early event, as higher methylation frequencies were present in borderline and early stage tumors compared with late-stage tumors, showing its potential to serve as a specific biomarker of surgically curable ovarian tumors.
Taken together, since aberrant methylation is one of the earliest molecular alterations during tumorigenesis it represents a promising strategy for the early detection of OvCa. However, the studies so far suggest that methylation in a panel of biomarkers, rather than individual gene methylation, will be necessary to achieve a suitable diagnostic assay with high specificity [128]. Indeed, studies that have examined combinations of gene methylation as OvCa biomarkers have achieved the greatest degree of sensitivity and specificity [125,129].
10. NUCLEIC ACIDS AS BIOMARKERS
Since most ovarian-cancer patients are diagnosed with advanced-stage disease, identification of an early tumor is essential to improve prognosis and therapy. Micro-RNAs as well as cell-free DNA promise to be good candidates for early diagnostic biomarkers because they are abundant in blood and can be easily detected. Since the first discovery of microRNA (miR) dysregulation in cancer in 2005 (i.e., deletion of miR-15a and miR-16-1 in B-CLL), extensive research has demonstrated that miRs are involved in the initiation and progression of human cancers, including OvCa [130]. MicroRNAs are small (19–25 nucleotides) non-coding RNAs that suppress the translation of target mRNAs by binding to their 3’ untranslated regions. Thereby, they act as critical regulators of cellular processes such as proliferation, differentiation, apoptosis and development [131]. Numerous studies have shown that expression of individual miRs or specific miR signatures can be linked to the diagnosis and prognosis of many cancer types [132].
Interestingly, circulatory miRs are resistant to degradation and remain stable, despite high levels of RNAses in the blood. This startling stability has been explained by the association of miRs with protein complexes such as Argo2 or high-density lipoprotein (HDL) [133,134]. Around 10% of circulating miRs are packed in circulating extracellular microvesicles such as exosomes [135]. In this context, Gercel-Taylor showed that miR signatures of exosomes released from tumors in the bloodstream were distinct from signatures of patients with benign disease and could be strongly correlated with the tumor stage [87]. Twelve miRNAs were present at a higher proportion in malignant cells (e.g., mir-155, -29), while 31 were present at elevated levels exclusively in exosomes (e.g., miR-203, -205). A recent miRNA profiling performed by Vaksman et al identified 99 miRNAs with high expression levels in exosomes from OvCa effusion supernatants [136]. Some of these miRNAs showed significant associations with effusion sites (peritoneum versus pleura) and the FIGO stage. In univariate survival analysis, high levels of miRNAs 21, 23b and 29a were associated with poor PFS whereas high expression of miRNA 21 correlated with poor OS. In experimental mouse models, OvCa exosomes affect both tumor cells and cells in the tumor microenvironment and induce more aggressive disease. Table 3 lists the circulating miRNAs that could be potentially useful for early diagnosis of OvCa.
Table 3.
Potential diagnostic micro RNAs for OvCa
| Elevated miR | Source | Reference | Decreased miR | Source | Reference |
|---|---|---|---|---|---|
| miR-21,-29a, -92, -93, -126 | serum | [135] | let-7b, miR-26a, -132, -145 | serum | [167] |
| miR-16, -21, -191 | plasma | [168] | mi-99b, -127, -155 | serum | [135] |
| miR-205 | plasma | [169] | let-7f | serum | [169] |
| miR-30c-1-p | whole blood | [170] | miR-181a-3p, -342-3p, -450-5p | whole blood | [170] |
| miR-21,-141, -200a | serum exosomes | [87] | miR-145 | serum | [171] |
| miR200a/b/c | serum | [172] | |||
| miR-92 | serum | [173] | |||
| miR-205, -483-5p | serum | [169] | |||
| miR-221 | serum | [174] | |||
Additionally, some miR signatures can be used to differentiate the histological subtypes of OvCa [137–139]. The Cancer Genome Atlas (TCGA) study, identified two clinically relevant subtypes, one mesenchymal and one epithelial, based on the specific miR signatures [140]. The mesenchymal subtype was associated with poor prognosis and included the miRNA regulatory network consisting of miR-29c, miR-101, miR-128, miR-141, miR-200, and miR-506 [141]. Various miRs promise a potential utility also as predictive markers of prognosis and clinical response to chemotherapy (Table 4). Several miRs were identified as oncogenes upregulated in tumors and were associated with poor overall survival. In contrast, other miRs were shown to act as tumor suppressors, being downregulated in ovarian cancers. Table 4 summarizes potential prognostic miRNAs which were significant in multivariate analysis.
Table 4.
Potential prognostic micro RNAs for OvCa
| Elevated miR | Prognosis | Reference | Decreased miR | Prognosis | Reference |
|---|---|---|---|---|---|
| miR-21 | poor | [175] | miR-31 | good | [143] |
| miR-25 | poor | [176] | miR-100 | good | [177] |
| miR-29b | poor | [178] | miR-150 | good | [179] |
| miR-203 | poor | [180] | miR-187 | good | [181] |
| miR-221 | poor | [174] | miR-200a | good | [182] |
| miR-484 | good | [143] | miR-200c | good | [183] |
| miR-520d-3p | good | [184] | miR-221/222 ratio | good | [185] |
| miR-335 | good | [186] | |||
| miR-410, miR-645 | good | [187] | |||
Recent data show that microRNAs could also be used to predict the clinical response to chemotherapy. For example Leskelä et al [142] demonstrated that the miR-200 family showed a significant association with response to paclitaxel–carboplatin. Patients who achieved complete response had tumors with significantly higher miR-200c expression levels than patients unresponsive to paclitaxel–carboplatin. In addition, higher expression of miR-200c was associated with lower relapse/progression rates. Lower expression of miR-31 significantly correlated with chemoresistance and poor prognosis in OvCa patients [143]. Vecchione et al analyzed miR signatures in 198 serous OvCa samples and identified that expression of miR-217, miR-484 and miR-617 predicted chemoresistance of these tumors [144].
Similar to miRNAs, circulating cell-free DNA (cfDNA) could be used for the early detection and monitoring of OvCa. Circulating cfDNAs are short (70–200 bp) or long (up to 21 kb) fragments of double-stranded DNA, which are released mostly by apoptotic or necrotic cells. Genome-wide sequencing of plasma DNA showed that circulating tumor DNA represents the tumor genome and reflects the clonality of tumor cells [145]. The total cfDNA level in blood samples of a patient is higher than in healthy controls, particularly in advanced stages of disease. OvCa cfDNA can be detected already in early stages [146] and high pre-operative cfDNA plasma levels were significantly associated with decreased patient survival and constituted an independent predictor of death from OvCa [147–149]. Interestingly, OvCa patients have significantly different levels of circulating cell-free mitochondrial DNA, but not of circulating nuclear DNA [150]. The concentration of tumor-specific DNA in plasma increases, as one can suspect, with the tumor burden and decreases following chemotherapy. Kamat et al showed that in mice treated with a combination of cytotoxic chemotherapy and anti-angiogenic agents plasma DNA levels were significantly lower than in untreated mice [151]. Thus, tumor-specific cfDNA might serve as useful biomarker of therapeutic response. Indeed, recent papers showed that the exome-wide analysis of circulating tumor DNA could represent a non-invasive liquid biopsy, complementing or even in future replacing the current invasive biopsy approaches.
Overall, the above studies indicate that cell-free nucleic acids, such as miRNAs and cfDNAs, are likely to play an increasingly important role as non-invasive markers of OvCa detection/prognosis. However, the broad heterogeneity of results, based on differences in the samples analyzed and technologies used, suggests that additional studies are needed to define predictive and reliable nucleic acids signatures that can find a clinico-pathological application.
11. BIOMARKERS PREDICTING RESPONSES TO BIOTHERAPIES IN OVCA
An improved understanding of the molecular basis of OvCa has encouraged testing of newer therapies, including pharmacological or biological agents, such as poly (ADP-ribose) polymerase inhibitors (PARPi) or an anti-angiogenic antibody (Ab), bevacizumab, respectively [152,153]. More recently, clinical trials with immune checkpoint inhibitors, ipilimumab (anti-CTLA-4 Ab), nivolumab (anti-PD-1 Ab) or avelumab (anti-PD-L1 Ab) have been offered to patients with cancer, including patients with OvCa either as monotherapies or in combination with ParPi or with chemotherapy [154]. While biological therapies are promising, only a proportion of patients achieve long-term responses, and there exists an urgent need for the establishment of biomarkers that would predict effectiveness of these therapies.
PARP inhibition (PARPi) has offered new opportunities for treatment of BRCA1- and BRCA2-related breast and ovarian cancers. One of the PARP inhibitors, olaparib, was recently approved by the FDA to treat BRCA1 and BRCA2 germline-mutated OvCa patients, and other PARPi drugs are undergoing active phase I and II trials [155]. However, BRCA1 and 2 germline mutations only account for approximately 15% of the OvCa patients. Recent studies showed that PARPi sensitivity is not restricted to familial BRCA-mutated cancers and that all tumors incapable of the error-free DSB repair, i.e., tumors with a defect in the HR repair pathway, can benefit from PARPi treatment [152]. Zhang et al. identified the copy number deletion of RAD50 as a candidate marker for survival and response to PARPi in BRCA-wildtype OvCa tumors [156]. Lee et al. developed a multiparameter flow cytometry assay to measure γH2AX and MRE11, indicators of double-strand breaks and DBS-repair, in PBMCs [157]. Other studies identified gene BRCA-like signatures [158] or LOH patterns [159] or mutations in eight ADAMTS genes [160] as potential markers. None of these potentially useful assays for surrogate biomarkers has been validated, and prospective studies will be required to confirm their role in predicting responses to PARPi-based therapy.
Bevacizumab (BV) is a humanized monoclonal Ab targeting vascular endothelial growth factor (VEGF). BV therapy has been initially reported to be effective for OvCa. However, more recent data show that the BV treatment does not prolong OS of many patients with either primary or recurrent OvCa. Given the high cost associated with the treatment, patients who receive BV treatment should be carefully selected. To this end, Backen et al, suggested that circulating Ang1 and Tie2 proteins (in combination) could serve as potential predictive biomarkers of improved PFS in BV-treated patients [161]. Other potential candidates for predictive biomarkers are VEGFR-1 and neuropilin-1 found in the plasma or cancer tissue [153] as well as plasma cell-free DNA [162].
Numerous clinical trials have reported an outstanding anti-tumor efficacy of immune checkpoint inhibitors in non-small lung cancer, melanoma or renal cell cancer [163]. However, in OvCa response rates to these inhibitors were relatively low, ranging from 6–20% [154]. Thus, predictive biomarkers of anti-tumor responses are urgently needed. Several studies have described that expression levels of PD-L1 on tumor cells serves as a correlate of response to anti-PD-1 Ab therapy [164]. However, the predictive validity of PD-L1 expression has not been confirmed in more recent clinical trials in patients with solid tumors. The numbers of TILS and the proportions of T cells positive for PD-L1 and/or PD-1 expression were reported to correlate to therapeutic responses. In OvCa, two clinical trials, one with nivolumab, the other with avelumab, showed no correlation between PD-L1 expression levels and anti-tumor response. The high frequency of somatic gene mutations was linked to therapeutic responses in various solid tumors [165]. In OvCa, mutations of BRCA1/2 genes were reported to be closely related to tumor-infiltrating lymphocyte (TIL) frequency and PD-L1 expression levels on tumor cells [166]. Thus, OvCa patients with BRCA-mutated tumors might be good candidates for PD-1/PD-L1 inhibitors.
12. EXPERT COMMENTARY
Recent technological advances have made it possible to in part define molecular, epigenetic or immunologic signatures of human OvCas. These signatures of tumor cells or the tumor microenvironment (TME) derived from genomic, proteomic or immune-based analyses have provided a complex map of factors with potential diagnostic or prognostic significance. However, few of these factors have been validated and so far, their correlations to disease, its progression or response to therapy remain incomplete. Perhaps the most important conclusion to date is that no single biomarker or a single signature emerges as the universally informative adjunct to clinical evaluations. Rather, the definition of a profile of markers obtained for each OvCa at the time of diagnosis seem to offer the most promising approach. Subsequent linking of the biomarker profile to outcome will be necessary to establish its prognostic value. Qualitative or quantitative changes taking place in the composition of this profile during therapy and disease progression or regression monitored over time are expected to inform about responses to therapy. It remains to be determined which of the biomarkers discussed above should be included in this profile. The availability of non-invasive biomarkers, i.e., serum markers or “liquid biopsies,” would make serial monitoring easy and less costly. In the era of personalized medicine, it is expected that individual profiles of biomarkers will be compiled for each patient and used in combination with clinicopathologic endpoints to predict prognosis, select appropriate treatment and monitor responses to therapy.
13. FIVE YEAR VIEW
The large number of potentially useful biomarkers for OvCa will be subject to intense evaluations in the near future. The development and validation of blood tests that can accurately detect OvCa at an early stage when the disease is asymptomatic are of utmost importance. Today, there are no screening tests that are sufficiently sensitive and reliable to meet the challenge. Given the large variety of serum and tissue candidate biomarkers, including “liquid biopsy” sampling, the next five years are likely to see optimization and validation of a new crop of screening assays for OvCa detection. One such candidate biomarker might be a combination of osteopontin and CA125. Assays that require minimal quantities of biological fluids and are noninvasive will be of special interest. Among a wide variety of molecular, genetic or immunological assays already available, those that best meet requirements for accurate early diagnosis of OvCa and its differentiation from benign diseases will be selected and validated. These biomarkers will make it possible to screen individuals at risk of OvCa, e.g., OvCa patients’ first-degree relatives, close relatives diagnosed with OvCa at an early age or those with the history of early-age breast cancer. A rapid development of prognostic biomarkers for evaluating OvCa responses to therapy or outcome will result in an improved selection of therapies for OvCa patients. In this respect, biomarkers of sensitivity/resistance to chemotherapies as well as biomarkers for responses to immunotherapies will become available and will be validated in prospective clinical trials. The developmental efforts will focus on high throughput technologies performed in real time at a minimal cost. Because monitoring using a single serum or tissue biomarker is not likely to be applicable to different clinical situations, panels of biomarkers will be used for following responses to therapies or predicting outcome.
At the time when immunotherapy is increasingly frequently used alone or in combination with conventional therapies to treat OvCa, immunologic biomarkers of response are likely to gain in importance. The next five years will see rapid growth in the development and prospective validation of new biomarkers that include OvCa-reactive immune cells in situ or in the peripheral circulation. In conjunction with other molecular and genetic markers, immunologic profiling will provide much needed metrics of the role the host immune system plays in OvCa.
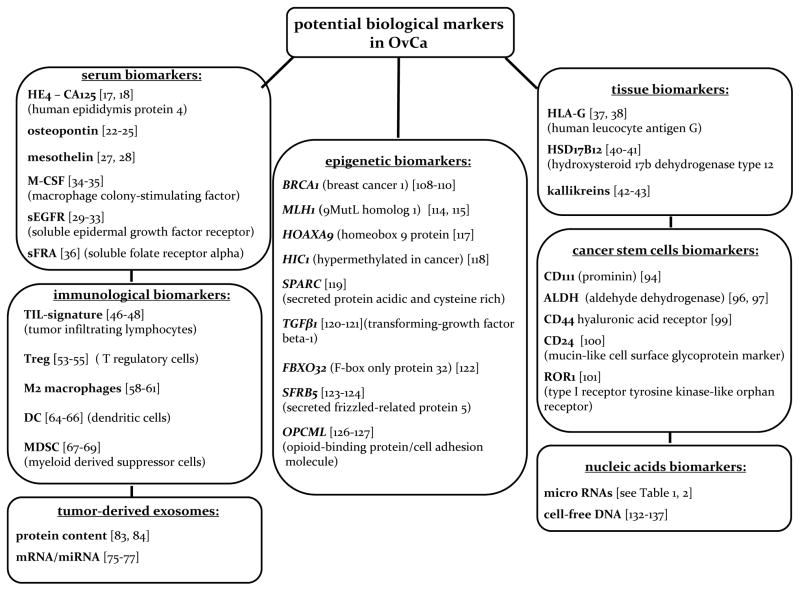
Figure 2.
A diagram listing various categories of potential biological markers that are various categories of being developed for OvCa diagnosis, prognosis and response to therapies.
14. KEY ISSUES.
There exists an unmet need for OvCa biomarkers that could serve as surrogates of response to therapy, prognosis or outcome.
A search for novel, reliable biomarkers for early OvCa detection, prognosis, treatment efficacy or outcome is an immediate and urgent current goal.
Currently, emerging serum or tissue biomarkers that measure anti-tumor immunological responses of the host offer special promise and deserve attention.
Tumor-derived exosomes potentially serving as a “liquid tumor biopsy” are being considered as future OvCa biomarkers.
Profiling of OvCa stem cells and quantitation of circulating tumor cells (CTC) are potentially promising future biomarkers.
Numerous epigenetic biomarkers are available and await validation.
Nucleic acid, including circulating DNAs and miRNAs, are in the limelight, with considerable progress made in establishing their clinical significance.
None of these potential OvCa biomarkers have been validated in prospective clinical trials so far.
The next five years will determine which of these biomarkers will be included in a panel of potentially non-invasive biomarkers for evaluating patients with OvCa.
Footnotes
Declaration of interest
This study was supported in part by the Foundation for Polish Science (Parent Bridge Program/2011/186) grant to M Szajnik and M Czystowska-Kuzmicz and by the NIH grant R01 CA-16862 to TL Whiteside. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as:
* of interest
** of considerable interest
- 1.Poole EM, Merritt MA, Jordan SJ, et al. Hormonal and reproductive risk factors for epithelial ovarian cancer by tumor aggressiveness. Cancer Epidemiol Biomarkers Prev. 2013;22(3):429–437. doi: 10.1158/1055-9965.EPI-12-1183-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Escayola C, Ferron G, Romeo M, Torrent JJ, Querleu D. The impact of pleural disease on the management of advanced ovarian cancer. Gynecol Oncol. 2015;138(1):216–220. doi: 10.1016/j.ygyno.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Vang R, Shih Ie M, Kurman RJ. Fallopian tube precursors of ovarian low- and high-grade serous neoplasms. Histopathology. 2013;62(1):44–58. doi: 10.1111/his.12046. [DOI] [PubMed] [Google Scholar]
- 4.Kurman RJ, Vang R, Junge J, et al. Papillary tubal hyperplasia: the putative precursor of ovarian atypical proliferative (borderline) serous tumors, noninvasive implants, and endosalpingiosis. Am J Surg Pathol. 2011;35(11):1605–1614. doi: 10.1097/PAS.0b013e318229449f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasey PA, Jayson GC, Gordon A, et al. Phase III randomized trial of docetaxel-carboplatin versus paclitaxel-carboplatin as first-line chemotherapy for ovarian carcinoma. J Natl Cancer Inst. 2004;96(22):1682–1691. doi: 10.1093/jnci/djh323. [DOI] [PubMed] [Google Scholar]
- 6.Sundar S, Neal RD, Kehoe S. Diagnosis of ovarian cancer. BMJ. 2015;351:h4443. doi: 10.1136/bmj.h4443. [DOI] [PubMed] [Google Scholar]
- 7**.McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23(36):9067–9072. doi: 10.1200/JCO.2004.01.0454. This is an important paper which defines the parameters of what can be considered a reliable biomarker. It is an obligatory text for all investigators dealing with biomarkers. [DOI] [PubMed] [Google Scholar]
- 8.McShane LM, Hayes DF. Publication of tumor marker research results: the necessity for complete and transparent reporting. J Clin Oncol. 2012;30(34):4223–4232. doi: 10.1200/JCO.2012.42.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9*.Visintin I, Feng Z, Longton G, et al. Diagnostic markers for early detection of ovarian cancer. Clin Cancer Res. 2008;14(4):1065–1072. doi: 10.1158/1078-0432.CCR-07-1569. Early detection is a significant unmet need in OvCa. This paper describes what is available today in this field. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs I, Oram D, Fairbanks J, et al. A risk of malignancy index incorporating CA 125, ultrasound and menopausal status for the accurate preoperative diagnosis of ovarian cancer. Br J Obstet Gynaecol. 1990;97(10):922–929. doi: 10.1111/j.1471-0528.1990.tb02448.x. [DOI] [PubMed] [Google Scholar]
- 11.Clauss A, Ng V, Liu J, et al. Overexpression of elafin in ovarian carcinoma is driven by genomic gains and activation of the nuclear factor kappaB pathway and is associated with poor overall survival. Neoplasia. 2010;12(2):161–172. doi: 10.1593/neo.91542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anastasi E, Granato T, Falzarano R, et al. The use of HE4, CA125 and CA72-4 biomarkers for differential diagnosis between ovarian endometrioma and epithelial ovarian cancer. J Ovarian Res. 2013;6(1):44. doi: 10.1186/1757-2215-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hallamaa M, Suvitie P, Huhtinen K, et al. Serum HE4 concentration is not dependent on menstrual cycle or hormonal treatment among endometriosis patients and healthy premenopausal women. Gynecol Oncol. 2012;125(3):667–672. doi: 10.1016/j.ygyno.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 14.Hallamaa M, Huhtinen K, Suvitie P, Perheentupa A. Serum concentrations of HE4 change little during in vitro fertilization. Acta Obstet Gynecol Scand. 2014;93(7):640–646. doi: 10.1111/aogs.12393. [DOI] [PubMed] [Google Scholar]
- 15.Moore RG, Miller MC, Steinhoff MM, et al. Serum HE4 levels are less frequently elevated than CA125 in women with benign gynecologic disorders. Am J Obstet Gynecol. 2012;206(4):351, e351–358. doi: 10.1016/j.ajog.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granato T, Porpora MG, Longo F, et al. HE4 in the differential diagnosis of ovarian masses. Clin Chim Acta. 2015;446:147–155. doi: 10.1016/j.cca.2015.03.047. [DOI] [PubMed] [Google Scholar]
- 17.Moore RG, McMeekin DS, Brown AK, et al. A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecol Oncol. 2009;112(1):40–46. doi: 10.1016/j.ygyno.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlsen MA, Sandhu N, Hogdall C, et al. Evaluation of HE4, CA125, risk of ovarian malignancy algorithm (ROMA) and risk of malignancy index (RMI) as diagnostic tools of epithelial ovarian cancer in patients with a pelvic mass. Gynecol Oncol. 2012;127(2):379–383. doi: 10.1016/j.ygyno.2012.07.106. [DOI] [PubMed] [Google Scholar]
- 19.Bast RC, Jr, Skates S, Lokshin A, Moore RG. Differential diagnosis of a pelvic mass: improved algorithms and novel biomarkers. Int J Gynecol Cancer. 2012;22(Suppl 1):S5–8. doi: 10.1097/IGC.0b013e318251c97d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petricoin EF, Ardekani AM, Hitt BA, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359(9306):572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Y, Liu C, Zhang N, Wang S, Zhang Z. Proteomics analysis for finding serum markers of ovarian cancer. Biomed Res Int. 2014;2014:179040. doi: 10.1155/2014/179040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmed M, Behera R, Chakraborty G, et al. Osteopontin: a potentially important therapeutic target in cancer. Expert Opin Ther Targets. 2011;15(9):1113–1126. doi: 10.1517/14728222.2011.594438. [DOI] [PubMed] [Google Scholar]
- 23.Shevde LA, Samant RS. Role of osteopontin in the pathophysiology of cancer. Matrix Biol. 2014;37:131–141. doi: 10.1016/j.matbio.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moszynski R, Szubert S, Szpurek D, Michalak S, Sajdak S. Role of osteopontin in differential diagnosis of ovarian tumors. J Obstet Gynaecol Res. 2013;39(11):1518–1525. doi: 10.1111/jog.12097. [DOI] [PubMed] [Google Scholar]
- 25.Nakae M, Iwamoto I, Fujino T, et al. Preoperative plasma osteopontin level as a biomarker complementary to carbohydrate antigen 125 in predicting ovarian cancer. J Obstet Gynaecol Res. 2006;32(3):309–314. doi: 10.1111/j.1447-0756.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- 26.Morello A, Sadelain M, Adusumilli PS. Mesothelin-Targeted CARs: Driving T Cells to Solid Tumors. Cancer Discov. 2016;6(2):133–146. doi: 10.1158/2159-8290.CD-15-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng WF, Huang CY, Chang MC, et al. High mesothelin correlates with chemoresistance and poor survival in epithelial ovarian carcinoma. Br J Cancer. 2009;100(7):1144–1153. doi: 10.1038/sj.bjc.6604964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28*.Ibrahim M, Bahaa A, Ibrahim A, et al. Evaluation of serum mesothelin in malignant and benign ovarian masses. Arch Gynecol Obstet. 2014;290(1):107–113. doi: 10.1007/s00404-014-3147-2. This paper reviews the role of serum mesothelin as a marker of OvCa progression and the potential target for therapies. [DOI] [PubMed] [Google Scholar]
- 29.Gui T, Shen K. The epidermal growth factor receptor as a therapeutic target in epithelial ovarian cancer. Cancer Epidemiol. 2012;36(5):490–496. doi: 10.1016/j.canep.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Granados ML, Hudson LG, Samudio-Ruiz SL. Contributions of the Epidermal Growth Factor Receptor to Acquisition of Platinum Resistance in Ovarian Cancer Cells. PLoS One. 2015;10(9):e0136893. doi: 10.1371/journal.pone.0136893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baron AT, Cora EM, Lafky JM, et al. Soluble epidermal growth factor receptor (sEGFR/sErbB1) as a potential risk, screening, and diagnostic serum biomarker of epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2003;12(2):103–113. [PubMed] [Google Scholar]
- 32.Baron AT, Boardman CH, Lafky JM, et al. Soluble epidermal growth factor receptor (sEGFR) [corrected] and cancer antigen 125 (CA125) as screening and diagnostic tests for epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(2):306–318. doi: 10.1158/1055-9965.EPI-04-0423. [DOI] [PubMed] [Google Scholar]
- 33.Tas F, Karabulut S, Serilmez M, Ciftci R, Duranyildiz D. Increased serum level of epidermal growth factor receptor (EGFR) is associated with poor progression-free survival in patients with epithelial ovarian cancer. Cancer Chemother Pharmacol. 2014;73(3):631–637. doi: 10.1007/s00280-014-2396-x. [DOI] [PubMed] [Google Scholar]
- 34.Skates SJ, Horick N, Yu Y, et al. Preoperative sensitivity and specificity for early-stage ovarian cancer when combining cancer antigen CA-125II, CA 15-3, CA 72-4, and macrophage colony-stimulating factor using mixtures of multivariate normal distributions. J Clin Oncol. 2004;22(20):4059–4066. doi: 10.1200/JCO.2004.03.091. [DOI] [PubMed] [Google Scholar]
- 35.Bedkowska GE, Lawicki S, Gacuta E, Pawlowski P, Szmitkowski M. M-CSF in a new biomarker panel with HE4 and CA 125 in the diagnostics of epithelial ovarian cancer patients. J Ovarian Res. 2015;8:27. doi: 10.1186/s13048-015-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurosaki A, Hasegawa K, Kato T, et al. Serum folate receptor alpha as a biomarker for ovarian cancer: Implications for diagnosis, prognosis and predicting its local tumor expression. Int J Cancer. 2016;138(8):1994–2002. doi: 10.1002/ijc.29937. [DOI] [PubMed] [Google Scholar]
- 37.Sheu JJ, Shih Ie M. Clinical and biological significance of HLA-G expression in ovarian cancer. Semin Cancer Biol. 2007;17(6):436–443. doi: 10.1016/j.semcancer.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rutten MJ, Dijk F, Savci-Heijink CD, et al. HLA-G expression is an independent predictor for improved survival in high grade ovarian carcinomas. J Immunol Res. 2014;2014:274584. doi: 10.1155/2014/274584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeon SY, Hwang KA, Choi KC. Effect of steroid hormones, estrogen and progesterone, on epithelial mesenchymal transition in ovarian cancer development. J Steroid Biochem Mol Biol. 2016;158:1–8. doi: 10.1016/j.jsbmb.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 40**.Szajnik M, Szczepanski MJ, Elishaev E, et al. 17beta Hydroxysteroid dehydrogenase type 12 (HSD17B12) is a marker of poor prognosis in ovarian carcinoma. Gynecol Oncol. 2012;127(3):587–594. doi: 10.1016/j.ygyno.2012.08.010. This is a report featuring HSD17B12 as a potentially useful prognostic biomarker in OvCa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Visus C, Ito D, Dhir R, et al. Identification of Hydroxysteroid (17beta) dehydrogenase type 12 (HSD17B12) as a CD8+ T-cell-defined human tumor antigen of human carcinomas. Cancer Immunol Immunother. 2011;60(7):919–929. doi: 10.1007/s00262-011-1001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong Y, Loessner D, Irving-Rodgers H, et al. Metastasis of ovarian cancer is mediated by kallikrein related peptidases. Clin Exp Metastasis. 2014;31(1):135–147. doi: 10.1007/s10585-013-9615-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamir A, Jag U, Sarojini S, et al. Kallikrein family proteases KLK6 and KLK7 are potential early detection and diagnostic biomarkers for serous and papillary serous ovarian cancer subtypes. J Ovarian Res. 2014;7:109. doi: 10.1186/s13048-014-0109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sickert D, Aust DE, Langer S, et al. Characterization of macrophage subpopulations in colon cancer using tissue microarrays. Histopathology. 2005;46(5):515–521. doi: 10.1111/j.1365-2559.2005.02129.x. [DOI] [PubMed] [Google Scholar]
- 45.Milne K, Alexander C, Webb JR, et al. Absolute lymphocyte count is associated with survival in ovarian cancer independent of tumor-infiltrating lymphocytes. J Transl Med. 2012;10:33. doi: 10.1186/1479-5876-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giannakakis A, Karapetsas A, Dangaj D, et al. Overexpression of SMARCE1 is associated with CD8+ T-cell infiltration in early stage ovarian cancer. Int J Biochem Cell Biol. 2014;53:389–398. doi: 10.1016/j.biocel.2014.05.031. [DOI] [PubMed] [Google Scholar]
- 47**.Nelson BH. The impact of T-cell immunity on ovarian cancer outcomes. Immunol Rev. 2008;222:101–116. doi: 10.1111/j.1600-065X.2008.00614.x. This review comprehensively discusses the potential role of T cell immunity in regulating OvCa progression and outcome. [DOI] [PubMed] [Google Scholar]
- 48.Tomsova M, Melichar B, Sedlakova I, Steiner I. Prognostic significance of CD3+ tumor-infiltrating lymphocytes in ovarian carcinoma. Gynecol Oncol. 2008;108(2):415–420. doi: 10.1016/j.ygyno.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 49.Sato E, Olson SH, Ahn J, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci U S A. 2005;102(51):18538–18543. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nielsen JS, Nelson BH. Tumor-infiltrating B cells and T cells: Working together to promote patient survival. Oncoimmunology. 2012;1(9):1623–1625. doi: 10.4161/onci.21650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51**.Nielsen JS, Sahota RA, Milne K, et al. CD20+ tumor-infiltrating lymphocytes have an atypical CD27- memory phenotype and together with CD8+ T cells promote favorable prognosis in ovarian cancer. Clin Cancer Res. 2012;18(12):3281–3292. doi: 10.1158/1078-0432.CCR-12-0234. This paper is a must for all who wish to consider the role of the host immune system in regulating OvCa progression and outcome. [DOI] [PubMed] [Google Scholar]
- 52.Webb JR, Milne K, Watson P, Deleeuw RJ, Nelson BH. Tumor-infiltrating lymphocytes expressing the tissue resident memory marker CD103 are associated with increased survival in high-grade serous ovarian cancer. Clin Cancer Res. 2014;20(2):434–444. doi: 10.1158/1078-0432.CCR-13-1877. [DOI] [PubMed] [Google Scholar]
- 53.Sakaguchi S, Sakaguchi N, Shimizu J, et al. Immunologic tolerance maintained by CD25+ CD4+ regulatory T cells: their common role in controlling autoimmunity, tumor immunity, and transplantation tolerance. Immunol Rev. 2001;182:18–32. doi: 10.1034/j.1600-065x.2001.1820102.x. [DOI] [PubMed] [Google Scholar]
- 54.Bates GJ, Fox SB, Han C, et al. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J Clin Oncol. 2006;24(34):5373–5380. doi: 10.1200/JCO.2006.05.9584. [DOI] [PubMed] [Google Scholar]
- 55**.Curiel TJ, Coukos G, Zou L, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10(9):942–949. doi: 10.1038/nm1093. This is one of the first and most widely quoted manuscripts which demonstrates the role of Treg in OvCa progression and patient survival. [DOI] [PubMed] [Google Scholar]
- 56.Szajnik M, Czystowska M, Szczepanski MJ, Mandapathil M, Whiteside TL. Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg) PLoS One. 2010;5(7):e11469. doi: 10.1371/journal.pone.0011469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bergmann C, Strauss L, Wang Y, et al. T regulatory type 1 cells in squamous cell carcinoma of the head and neck: mechanisms of suppression and expansion in advanced disease. Clin Cancer Res. 2008;14(12):3706–3715. doi: 10.1158/1078-0432.CCR-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kawamura K, Komohara Y, Takaishi K, Katabuchi H, Takeya M. Detection of M2 macrophages and colony-stimulating factor 1 expression in serous and mucinous ovarian epithelial tumors. Pathol Int. 2009;59(5):300–305. doi: 10.1111/j.1440-1827.2009.02369.x. [DOI] [PubMed] [Google Scholar]
- 59.Wang H, Rosen DG, Wang H, et al. Insulin-like growth factor-binding protein 2 and 5 are differentially regulated in ovarian cancer of different histologic types. Mod Pathol. 2006;19(9):1149–1156. doi: 10.1038/modpathol.3800637. [DOI] [PubMed] [Google Scholar]
- 60.Klimp AH, Hollema H, Kempinga C, et al. Expression of cyclooxygenase-2 and inducible nitric oxide synthase in human ovarian tumors and tumor-associated macrophages. Cancer Res. 2001;61(19):7305–7309. [PubMed] [Google Scholar]
- 61.Le Page C, Marineau A, Bonza PK, et al. BTN3A2 expression in epithelial ovarian cancer is associated with higher tumor infiltrating T cells and a better prognosis. PLoS One. 2012;7(6):e38541. doi: 10.1371/journal.pone.0038541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lan C, Huang X, Lin S, et al. Expression of M2-polarized macrophages is associated with poor prognosis for advanced epithelial ovarian cancer. Technol Cancer Res Treat. 2013;12(3):259–267. doi: 10.7785/tcrt.2012.500312. [DOI] [PubMed] [Google Scholar]
- 63.No JH, Moon JM, Kim K, Kim YB. Prognostic significance of serum soluble CD163 level in patients with epithelial ovarian cancer. Gynecol Obstet Invest. 2013;75(4):263–267. doi: 10.1159/000349892. [DOI] [PubMed] [Google Scholar]
- 64**.Wilke CM, Kryczek I, Zou W. Antigen-presenting cell (APC) subsets in ovarian cancer. Int Rev Immunol. 2011;30(2–3):120–126. doi: 10.3109/08830185.2011.567362. This is an informative review of antigen presentation in the environment of OvCa and of the effects OvCa exerts on immune responses. [DOI] [PubMed] [Google Scholar]
- 65**.Zhang Z, Huang J, Zhang C, et al. Infiltration of dendritic cells and T lymphocytes predicts favorable outcome in epithelial ovarian cancer. Cancer Gene Ther. 2015;22(4):198–206. doi: 10.1038/cgt.2015.7. This paper reports that attributes of OvCa-associated infiltrates of T cells and DC play an important role in outcome of OvCa. [DOI] [PubMed] [Google Scholar]
- 66.Labidi-Galy SI, Sisirak V, Meeus P, et al. Quantitative and functional alterations of plasmacytoid dendritic cells contribute to immune tolerance in ovarian cancer. Cancer Res. 2011;71(16):5423–5434. doi: 10.1158/0008-5472.CAN-11-0367. [DOI] [PubMed] [Google Scholar]
- 67.Khan AN, Kolomeyevskaya N, Singel KL, et al. Targeting myeloid cells in the tumor microenvironment enhances vaccine efficacy in murine epithelial ovarian cancer. Oncotarget. 2015;6(13):11310–11326. doi: 10.18632/oncotarget.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kryczek I, Zou L, Rodriguez P, et al. B7-H4 expression identifies a novel suppressive macrophage population in human ovarian carcinoma. J Exp Med. 2006;203(4):871–881. doi: 10.1084/jem.20050930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Obermajer N, Muthuswamy R, Odunsi K, Edwards RP, Kalinski P. PGE(2)-induced CXCL12 production and CXCR4 expression controls the accumulation of human MDSCs in ovarian cancer environment. Cancer Res. 2011;71(24):7463–7470. doi: 10.1158/0008-5472.CAN-11-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Balkwill FR, Capasso M, Hagemann T. The tumor microenvironment at a glance. J Cell Sci. 2012;125(Pt 23):5591–5596. doi: 10.1242/jcs.116392. [DOI] [PubMed] [Google Scholar]
- 71.Hart KM, Bak SP, Alonso A, Berwin B. Phenotypic and functional delineation of murine CX(3)CR1 monocyte-derived cells in ovarian cancer. Neoplasia. 2009;11(6):564–573. doi: 10.1593/neo.09228. 561 p following 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72*.Lesokhin AM, Hohl TM, Kitano S, et al. Monocytic CCR2(+) myeloid-derived suppressor cells promote immune escape by limiting activated CD8 T-cell infiltration into the tumor microenvironment. Cancer Res. 2012;72(4):876–886. doi: 10.1158/0008-5472.CAN-11-1792. This paper illustrates one of the mechanisms responsible for blocking of T cell infiltration into the tumor tissues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ohno S, Ishikawa A, Kuroda M. Roles of exosomes and microvesicles in disease pathogenesis. Adv Drug Deliv Rev. 2013;65(3):398–401. doi: 10.1016/j.addr.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 74.Schifferli JA. Microvesicles are messengers. Semin Immunopathol. 2011;33(5):393–394. doi: 10.1007/s00281-011-0276-6. [DOI] [PubMed] [Google Scholar]
- 75.Rak J. Microparticles in cancer. Semin Thromb Hemost. 2010;36(8):888–906. doi: 10.1055/s-0030-1267043. [DOI] [PubMed] [Google Scholar]
- 76.Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.D'Souza-Schorey C, Clancy JW. Tumor-derived microvesicles: shedding light on novel microenvironment modulators and prospective cancer biomarkers. Genes Dev. 2012;26(12):1287–1299. doi: 10.1101/gad.192351.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Graves LE, Ariztia EV, Navari JR, et al. Proinvasive properties of ovarian cancer ascites-derived membrane vesicles. Cancer Res. 2004;64(19):7045–7049. doi: 10.1158/0008-5472.CAN-04-1800. [DOI] [PubMed] [Google Scholar]
- 79**.Valenti R, Huber V, Iero M, et al. Tumor-released microvesicles as vehicles of immunosuppression. Cancer Res. 2007;67(7):2912–2915. doi: 10.1158/0008-5472.CAN-07-0520. This is an early report calling attention to potential effects of tumor-derived vesicles exert on the host immune system. [DOI] [PubMed] [Google Scholar]
- 80.Castellana D, Zobairi F, Martinez MC, et al. Membrane microvesicles as actors in the establishment of a favorable prostatic tumoral niche: a role for activated fibroblasts and CX3CL1-CX3CR1 axis. Cancer Res. 2009;69(3):785–793. doi: 10.1158/0008-5472.CAN-08-1946. [DOI] [PubMed] [Google Scholar]
- 81.Czystowska M, Han J, Szczepanski MJ, et al. IRX-2, a novel immunotherapeutic, protects human T cells from tumor-induced cell death. Cell Death Differ. 2009;16(5):708–718. doi: 10.1038/cdd.2008.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao L, Liu W, Xiao J, Cao B. The role of exosomes and “exosomal shuttle microRNA” in tumorigenesis and drug resistance. Cancer Lett. 2015;356(2 Pt B):339–346. doi: 10.1016/j.canlet.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 83*.Szajnik M, Derbis M, Lach M, et al. Exosomes in Plasma of Patients with Ovarian Carcinoma: Potential Biomarkers of Tumor Progression and Response to Therapy. Gynecol Obstet (Sunnyvale) 2013;(Suppl 4):3. doi: 10.4172/2161-0932.S4-003. This is the first report showing that exosomes isolated from plasma of OvCa patients might influence responses to chemotherapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wieckowski EU, Visus C, Szajnik M, et al. Tumor-derived microvesicles promote regulatory T cell expansion and induce apoptosis in tumor-reactive activated CD8+ T lymphocytes. J Immunol. 2009;183(6):3720–3730. doi: 10.4049/jimmunol.0900970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luciani F, Spada M, De Milito A, et al. Effect of proton pump inhibitor pretreatment on resistance of solid tumors to cytotoxic drugs. J Natl Cancer Inst. 2004;96(22):1702–1713. doi: 10.1093/jnci/djh305. [DOI] [PubMed] [Google Scholar]
- 86.Safaei R, Larson BJ, Cheng TC, et al. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol Cancer Ther. 2005;4(10):1595–1604. doi: 10.1158/1535-7163.MCT-05-0102. [DOI] [PubMed] [Google Scholar]
- 87*.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. This paper introduces the concept of exosomes in plasma of OvCa patients serving as “liquid tumor biopsy.”. [DOI] [PubMed] [Google Scholar]
- 88.Mause SF, Weber C. Microparticles: protagonists of a novel communication network for intercellular information exchange. Circ Res. 2010;107(9):1047–1057. doi: 10.1161/CIRCRESAHA.110.226456. [DOI] [PubMed] [Google Scholar]
- 89.Pantic I. Cancer stem cell hypotheses: impact on modern molecular physiology and pharmacology research. J Biosci. 2011;36(5):957–961. doi: 10.1007/s12038-011-9155-5. [DOI] [PubMed] [Google Scholar]
- 90.Tirino V, Desiderio V, Paino F, et al. Cancer stem cells in solid tumors: an overview and new approaches for their isolation and characterization. FASEB J. 2013;27(1):13–24. doi: 10.1096/fj.12-218222. [DOI] [PubMed] [Google Scholar]
- 91.Ben-Porath I, Thomson MW, Carey VJ, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40(5):499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shats I, Gatza ML, Chang JT, et al. Using a stem cell-based signature to guide therapeutic selection in cancer. Cancer Res. 2011;71(5):1772–1780. doi: 10.1158/0008-5472.CAN-10-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Burgos-Ojeda D, Rueda BR, Buckanovich RJ. Ovarian cancer stem cell markers: prognostic and therapeutic implications. Cancer Lett. 2012;322(1):1–7. doi: 10.1016/j.canlet.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tirino V, Desiderio V, d'Aquino R, et al. Detection and characterization of CD133+ cancer stem cells in human solid tumours. PLoS One. 2008;3(10):e3469. doi: 10.1371/journal.pone.0003469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang J, Guo X, Chang DY, et al. CD133 expression associated with poor prognosis in ovarian cancer. Mod Pathol. 2012;25(3):456–464. doi: 10.1038/modpathol.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96*.Landen CN, Jr, Goodman B, Katre AA, et al. Targeting aldehyde dehydrogenase cancer stem cells in ovarian cancer. Mol Cancer Ther. 2010;9(12):3186–3199. doi: 10.1158/1535-7163.MCT-10-0563. This is one example of several papers that describe the detection and clinical significance of cancer stem cells in OvCa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deng S, Yang X, Lassus H, et al. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS One. 2010;5(4):e10277. doi: 10.1371/journal.pone.0010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chang B, Liu G, Xue F, et al. ALDH1 expression correlates with favorable prognosis in ovarian cancers. Mod Pathol. 2009;22(6):817–823. doi: 10.1038/modpathol.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Steffensen KD, Alvero AB, Yang Y, et al. Prevalence of epithelial ovarian cancer stem cells correlates with recurrence in early-stage ovarian cancer. J Oncol. 2011;2011:620523. doi: 10.1155/2011/620523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kristiansen G, Denkert C, Schluns K, et al. CD24 is expressed in ovarian cancer and is a new independent prognostic marker of patient survival. Am J Pathol. 2002;161(4):1215–1221. doi: 10.1016/S0002-9440(10)64398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang S, Cui B, Lai H, et al. Ovarian cancer stem cells express ROR1, which can be targeted for anti-cancer-stem-cell therapy. Proc Natl Acad Sci U S A. 2014;111(48):17266–17271. doi: 10.1073/pnas.1419599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schwede M, Spentzos D, Bentink S, et al. Stem cell-like gene expression in ovarian cancer predicts type II subtype and prognosis. PLoS One. 2013;8(3):e57799. doi: 10.1371/journal.pone.0057799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Balch C, Fang F, Matei DE, Huang TH, Nephew KP. Minireview: epigenetic changes in ovarian cancer. Endocrinology. 2009;150(9):4003–4011. doi: 10.1210/en.2009-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Widschwendter M, Jiang G, Woods C, et al. DNA hypomethylation and ovarian cancer biology. Cancer Res. 2004;64(13):4472–4480. doi: 10.1158/0008-5472.CAN-04-0238. [DOI] [PubMed] [Google Scholar]
- 105.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6(2):107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 106*.Wei SH, Balch C, Paik HH, et al. Prognostic DNA methylation biomarkers in ovarian cancer. Clin Cancer Res. 2006;12(9):2788–2794. doi: 10.1158/1078-0432.CCR-05-1551\. A paper that shows the importance of DNA methylation in OvCa and its potential role as a biomarker. [DOI] [PubMed] [Google Scholar]
- 107.Shih Ie M, Kurman RJ. Ovarian tumorigenesis: a proposed model based on morphological and molecular genetic analysis. Am J Pathol. 2004;164(5):1511–1518. doi: 10.1016/s0002-9440(10)63708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Baldwin RL, Nemeth E, Tran H, et al. BRCA1 promoter region hypermethylation in ovarian carcinoma: a population-based study. Cancer Res. 2000;60(19):5329–5333. [PubMed] [Google Scholar]
- 109.Press JZ, De Luca A, Boyd N, et al. Ovarian carcinomas with genetic and epigenetic BRCA1 loss have distinct molecular abnormalities. BMC Cancer. 2008;8:17. doi: 10.1186/1471-2407-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang HJ, Liu VW, Wang Y, Tsang PC, Ngan HY. Differential DNA methylation profiles in gynecological cancers and correlation with clinico-pathological data. BMC Cancer. 2006;6:212. doi: 10.1186/1471-2407-6-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wiley A, Katsaros D, Chen H, et al. Aberrant promoter methylation of multiple genes in malignant ovarian tumors and in ovarian tumors with low malignant potential. Cancer. 2006;107(2):299–308. doi: 10.1002/cncr.21992. [DOI] [PubMed] [Google Scholar]
- 112.Chiang JW, Karlan BY, Cass L, Baldwin RL. BRCA1 promoter methylation predicts adverse ovarian cancer prognosis. Gynecol Oncol. 2006;101(3):403–410. doi: 10.1016/j.ygyno.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 113.Chaudhry P, Srinivasan R, Patel FD. Utility of gene promoter methylation in prediction of response to platinum-based chemotherapy in epithelial ovarian cancer (EOC) Cancer Invest. 2009;27(8):877–884. doi: 10.1080/07357900902849699. [DOI] [PubMed] [Google Scholar]
- 114.Strathdee G, MacKean MJ, Illand M, Brown R. A role for methylation of the hMLH1 promoter in loss of hMLH1 expression and drug resistance in ovarian cancer. Oncogene. 1999;18(14):2335–2341. doi: 10.1038/sj.onc.1202540. [DOI] [PubMed] [Google Scholar]
- 115.Gifford G, Paul J, Vasey PA, Kaye SB, Brown R. The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin Cancer Res. 2004;10(13):4420–4426. doi: 10.1158/1078-0432.CCR-03-0732. [DOI] [PubMed] [Google Scholar]
- 116.Samimi G, Fink D, Varki NM, et al. Analysis of MLH1 and MSH2 expression in ovarian cancer before and after platinum drug-based chemotherapy. Clin Cancer Res. 2000;6(4):1415–1421. [PubMed] [Google Scholar]
- 117.Montavon C, Gloss BS, Warton K, et al. Prognostic and diagnostic significance of DNA methylation patterns in high grade serous ovarian cancer. Gynecol Oncol. 2012;124(3):582–588. doi: 10.1016/j.ygyno.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 118.Tam KF, Liu VW, Liu SS, et al. Methylation profile in benign, borderline and malignant ovarian tumors. J Cancer Res Clin Oncol. 2007;133(5):331–341. doi: 10.1007/s00432-006-0178-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Socha MJ, Said N, Dai Y, et al. Aberrant promoter methylation of SPARC in ovarian cancer. Neoplasia. 2009;11(2):126–135. doi: 10.1593/neo.81146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kang S, Dong SM, Park NH. Frequent promoter hypermethylation of TGFBI in epithelial ovarian cancer. Gynecol Oncol. 2010;118(1):58–63. doi: 10.1016/j.ygyno.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 121.Matsumura N, Huang Z, Mori S, et al. Epigenetic suppression of the TGF-beta pathway revealed by transcriptome profiling in ovarian cancer. Genome Res. 2011;21(1):74–82. doi: 10.1101/gr.108803.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chou JL, Su HY, Chen LY, et al. Promoter hypermethylation of FBXO32, a novel TGF-beta/SMAD4 target gene and tumor suppressor, is associated with poor prognosis in human ovarian cancer. Lab Invest. 2010;90(3):414–425. doi: 10.1038/labinvest.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Su HY, Lai HC, Lin YW, et al. An epigenetic marker panel for screening and prognostic prediction of ovarian cancer. Int J Cancer. 2009;124(2):387–393. doi: 10.1002/ijc.23957. [DOI] [PubMed] [Google Scholar]
- 124.Su HY, Lai HC, Lin YW, et al. Epigenetic silencing of SFRP5 is related to malignant phenotype and chemoresistance of ovarian cancer through Wnt signaling pathway. Int J Cancer. 2010;127(3):555–567. doi: 10.1002/ijc.25083. [DOI] [PubMed] [Google Scholar]
- 125.Dai W, Teodoridis JM, Zeller C, et al. Systematic CpG islands methylation profiling of genes in the wnt pathway in epithelial ovarian cancer identifies biomarkers of progression-free survival. Clin Cancer Res. 2011;17(12):4052–4062. doi: 10.1158/1078-0432.CCR-10-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sellar GC, Watt KP, Rabiasz GJ, et al. OPCML at 11q25 is epigenetically inactivated and has tumor-suppressor function in epithelial ovarian cancer. Nat Genet. 2003;34(3):337–343. doi: 10.1038/ng1183. [DOI] [PubMed] [Google Scholar]
- 127.Czekierdowski A, Czekierdowska S, Szymanski M, et al. Opioid-binding protein/cell adhesion molecule-like (OPCML) gene and promoter methylation status in women with ovarian cancer. Neuro Endocrinol Lett. 2006;27(5):609–613. [PubMed] [Google Scholar]
- 128.Clarke-Pearson DL. Clinical practice. Screening for ovarian cancer. N Engl J Med. 2009;361(2):170–177. doi: 10.1056/NEJMcp0901926. [DOI] [PubMed] [Google Scholar]
- 129.Liggett TE, Melnikov A, Yi Q, et al. Distinctive DNA methylation patterns of cell-free plasma DNA in women with malignant ovarian tumors. Gynecol Oncol. 2011;120(1):113–120. doi: 10.1016/j.ygyno.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130*.Kinose Y, Sawada K, Nakamura K, Kimura T. The role of microRNAs in ovarian cancer. Biomed Res Int. 2014;2014:249393. doi: 10.1155/2014/249393. This is one of multiple recent papers describing the potential microRNAs as noninvasive biomarkers of OvCa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ruan K, Fang X, Ouyang G. MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett. 2009;285(2):116–126. doi: 10.1016/j.canlet.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 132.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Resnick KE, Alder H, Hagan JP, et al. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112(1):55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 136.Vaksman O, Trope C, Davidson B, Reich R. Exosome-derived miRNAs and ovarian carcinoma progression. Carcinogenesis. 2014;35(9):2113–2120. doi: 10.1093/carcin/bgu130. [DOI] [PubMed] [Google Scholar]
- 137.Calura E, Fruscio R, Paracchini L, et al. MiRNA landscape in stage I epithelial ovarian cancer defines the histotype specificities. Clin Cancer Res. 2013;19(15):4114–4123. doi: 10.1158/1078-0432.CCR-13-0360. [DOI] [PubMed] [Google Scholar]
- 138.Nam EJ, Yoon H, Kim SW, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14(9):2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 139.Vang S, Wu HT, Fischer A, et al. Identification of ovarian cancer metastatic miRNAs. PLoS One. 2013;8(3):e58226. doi: 10.1371/journal.pone.0058226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yang D, Sun Y, Hu L, et al. Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell. 2013;23(2):186–199. doi: 10.1016/j.ccr.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Leskela S, Leandro-Garcia LJ, Mendiola M, et al. The miR-200 family controls beta-tubulin III expression and is associated with paclitaxel-based treatment response and progression-free survival in ovarian cancer patients. Endocr Relat Cancer. 2011;18(1):85–95. doi: 10.1677/ERC-10-0148. [DOI] [PubMed] [Google Scholar]
- 143.Mitamura T, Watari H, Wang L, et al. Downregulation of miRNA-31 induces taxane resistance in ovarian cancer cells through increase of receptor tyrosine kinase MET. Oncogenesis. 2013;2:e40. doi: 10.1038/oncsis.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Vecchione A, Belletti B, Lovat F, et al. A microRNA signature defines chemoresistance in ovarian cancer through modulation of angiogenesis. Proc Natl Acad Sci U S A. 2013;110(24):9845–9850. doi: 10.1073/pnas.1305472110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 146.Zhang Q, Hu G, Yang Q, et al. A multiplex methylation-specific PCR assay for the detection of early-stage ovarian cancer using cell-free serum DNA. Gynecol Oncol. 2013;130(1):132–139. doi: 10.1016/j.ygyno.2013.04.048. [DOI] [PubMed] [Google Scholar]
- 147.Kamat AA, Sood AK, Dang D, et al. Quantification of total plasma cell-free DNA in ovarian cancer using real-time PCR. Ann N Y Acad Sci. 2006;1075:230–234. doi: 10.1196/annals.1368.031. [DOI] [PubMed] [Google Scholar]
- 148.Kamat AA, Baldwin M, Urbauer D, et al. Plasma cell-free DNA in ovarian cancer: an independent prognostic biomarker. Cancer. 2010;116(8):1918–1925. doi: 10.1002/cncr.24997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.No JH, Kim K, Park KH, Kim YB. Cell-free DNA level as a prognostic biomarker for epithelial ovarian cancer. Anticancer Res. 2012;32(8):3467–3471. [PubMed] [Google Scholar]
- 150.Zachariah RR, Schmid S, Buerki N, et al. Levels of circulating cell-free nuclear and mitochondrial DNA in benign and malignant ovarian tumors. Obstet Gynecol. 2008;112(4):843–850. doi: 10.1097/AOG.0b013e3181867bc0. [DOI] [PubMed] [Google Scholar]
- 151.Kamat AA, Bischoff FZ, Dang D, et al. Circulating cell-free DNA: a novel biomarker for response to therapy in ovarian carcinoma. Cancer Biol Ther. 2006;5(10):1369–1374. doi: 10.4161/cbt.5.10.3240. [DOI] [PubMed] [Google Scholar]
- 152.Ledermann JA, El-Khouly F. PARP inhibitors in ovarian cancer: Clinical evidence for informed treatment decisions. Br J Cancer. 2015;113(Suppl 1):S10–16. doi: 10.1038/bjc.2015.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lambrechts D, Lenz HJ, de Haas S, Carmeliet P, Scherer SJ. Markers of response for the antiangiogenic agent bevacizumab. J Clin Oncol. 2013;31(9):1219–1230. doi: 10.1200/JCO.2012.46.2762. [DOI] [PubMed] [Google Scholar]
- 154.Hamanishi J, Mandai M, Konishi I. Immune checkpoint inhibition in ovarian cancer. Int Immunol. 2016 doi: 10.1093/intimm/dxw020. [DOI] [PubMed] [Google Scholar]
- 155.Gunderson CC, Moore KN. Olaparib: an oral PARP-1 and PARP-2 inhibitor with promising activity in ovarian cancer. Future Oncol. 2015;11(5):747–757. doi: 10.2217/fon.14.313. [DOI] [PubMed] [Google Scholar]
- 156.Zhang M, Liu G, Xue F, et al. Copy number deletion of RAD50 as predictive marker of BRCAness and PARP inhibitor response in BRCA wild type ovarian cancer. Gynecol Oncol. 2016;141(1):57–64. doi: 10.1016/j.ygyno.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lee JM, Gordon N, Trepel JB, et al. Development of a multiparameter flow cytometric assay as a potential biomarker for homologous recombination deficiency in women with high-grade serous ovarian cancer. J Transl Med. 2015;13:239. doi: 10.1186/s12967-015-0604-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Konstantinopoulos PA, Spentzos D, Karlan BY, et al. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J Clin Oncol. 2010;28(22):3555–3561. doi: 10.1200/JCO.2009.27.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Abkevich V, Timms KM, Hennessy BT, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer. 2012;107(10):1776–1782. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Liu Y, Yasukawa M, Chen K, et al. Association of Somatic Mutations of ADAMTS Genes With Chemotherapy Sensitivity and Survival in High-Grade Serous Ovarian Carcinoma. JAMA Oncol. 2015;1(4):486–494. doi: 10.1001/jamaoncol.2015.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Backen A, Renehan AG, Clamp AR, et al. The combination of circulating Ang1 and Tie2 levels predicts progression-free survival advantage in bevacizumab-treated patients with ovarian cancer. Clin Cancer Res. 2014;20(17):4549–4558. doi: 10.1158/1078-0432.CCR-13-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Steffensen KD, Madsen CV, Andersen RF, et al. Prognostic importance of cell-free DNA in chemotherapy resistant ovarian cancer treated with bevacizumab. Eur J Cancer. 2014;50(15):2611–2618. doi: 10.1016/j.ejca.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 163.Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Strickland KC, Howitt BE, Shukla SA, et al. Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chung YW, Bae HS, Song JY, et al. Detection of microRNA as novel biomarkers of epithelial ovarian cancer from the serum of ovarian cancer patients. Int J Gynecol Cancer. 2013;23(4):673–679. doi: 10.1097/IGC.0b013e31828c166d. [DOI] [PubMed] [Google Scholar]
- 168.Suryawanshi S, Vlad AM, Lin HM, et al. Plasma microRNAs as novel biomarkers for endometriosis and endometriosis-associated ovarian cancer. Clin Cancer Res. 2013;19(5):1213–1224. doi: 10.1158/1078-0432.CCR-12-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zheng H, Zhang L, Zhao Y, et al. Plasma miRNAs as diagnostic and prognostic biomarkers for ovarian cancer. PLoS One. 2013;8(11):e77853. doi: 10.1371/journal.pone.0077853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hausler SF, Keller A, Chandran PA, et al. Whole blood-derived miRNA profiles as potential new tools for ovarian cancer screening. Br J Cancer. 2010;103(5):693–700. doi: 10.1038/sj.bjc.6605833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wu H, Xiao Z, Wang K, Liu W, Hao Q. MiR-145 is downregulated in human ovarian cancer and modulates cell growth and invasion by targeting p70S6K1 and MUC1. Biochem Biophys Res Commun. 2013;441(4):693–700. doi: 10.1016/j.bbrc.2013.10.053. [DOI] [PubMed] [Google Scholar]
- 172.Kan CW, Hahn MA, Gard GB, et al. Elevated levels of circulating microRNA-200 family members correlate with serous epithelial ovarian cancer. BMC Cancer. 2012;12:627. doi: 10.1186/1471-2407-12-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Guo F, Tian J, Lin Y, et al. Serum microRNA-92 expression in patients with ovarian epithelial carcinoma. J Int Med Res. 2013;41(5):1456–1461. doi: 10.1177/0300060513487652. [DOI] [PubMed] [Google Scholar]
- 174.Hong F, Li Y, Xu Y, Zhu L. Prognostic significance of serum microRNA-221 expression in human epithelial ovarian cancer. J Int Med Res. 2013;41(1):64–71. doi: 10.1177/0300060513475759. [DOI] [PubMed] [Google Scholar]
- 175.Xu YZ, Xi QH, Ge WL, Zhang XQ. Identification of serum microRNA-21 as a biomarker for early detection and prognosis in human epithelial ovarian cancer. Asian Pac J Cancer Prev. 2013;14(2):1057–1060. doi: 10.7314/apjcp.2013.14.2.1057. [DOI] [PubMed] [Google Scholar]
- 176.Wang X, Meng X, Li H, et al. MicroRNA-25 expression level is an independent prognostic factor in epithelial ovarian cancer. Clin Transl Oncol. 2014;16(11):954–958. doi: 10.1007/s12094-014-1178-6. [DOI] [PubMed] [Google Scholar]
- 177.Peng DX, Luo M, Qiu LW, He YL, Wang XF. Prognostic implications of microRNA-100 and its functional roles in human epithelial ovarian cancer. Oncol Rep. 2012;27(4):1238–1244. doi: 10.3892/or.2012.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Flavin R, Smyth P, Barrett C, et al. miR-29b expression is associated with disease-free survival in patients with ovarian serous carcinoma. Int J Gynecol Cancer. 2009;19(4):641–647. doi: 10.1111/IGC.0b013e3181a48cf9. [DOI] [PubMed] [Google Scholar]
- 179.Jin M, Yang Z, Ye W, Xu H, Hua X. MicroRNA-150 predicts a favorable prognosis in patients with epithelial ovarian cancer, and inhibits cell invasion and metastasis by suppressing transcriptional repressor ZEB1. PLoS One. 2014;9(8):e103965. doi: 10.1371/journal.pone.0103965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wang S, Zhao X, Wang J, et al. Upregulation of microRNA-203 is associated with advanced tumor progression and poor prognosis in epithelial ovarian cancer. Med Oncol. 2013;30(3):681. doi: 10.1007/s12032-013-0681-x. [DOI] [PubMed] [Google Scholar]
- 181.Chao A, Lin CY, Lee YS, et al. Regulation of ovarian cancer progression by microRNA-187 through targeting Disabled homolog-2. Oncogene. 2012;31(6):764–775. doi: 10.1038/onc.2011.269. [DOI] [PubMed] [Google Scholar]
- 182.Hu X, Macdonald DM, Huettner PC, et al. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol. 2009;114(3):457–464. doi: 10.1016/j.ygyno.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 183.Marchini S, Cavalieri D, Fruscio R, et al. Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: a retrospective study of two independent tumour tissue collections. Lancet Oncol. 2011;12(3):273–285. doi: 10.1016/S1470-2045(11)70012-2. [DOI] [PubMed] [Google Scholar]
- 184.Nishimura M, Jung EJ, Shah MY, et al. Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer Discov. 2013;3(11):1302–1315. doi: 10.1158/2159-8290.CD-13-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wurz K, Garcia RL, Goff BA, et al. MiR-221 and MiR-222 alterations in sporadic ovarian carcinoma: Relationship to CDKN1B, CDKNIC and overall survival. Genes Chromosomes Cancer. 2010;49(7):577–584. doi: 10.1002/gcc.20768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Cao J, Cai J, Huang D, et al. miR-335 represents an independent prognostic marker in epithelial ovarian cancer. Am J Clin Pathol. 2014;141(3):437–442. doi: 10.1309/AJCPLYTZGB54ISZC. [DOI] [PubMed] [Google Scholar]
- 187.Shih KK, Qin LX, Tanner EJ, et al. A microRNA survival signature (MiSS) for advanced ovarian cancer. Gynecol Oncol. 2011;121(3):444–450. doi: 10.1016/j.ygyno.2011.01.025. [DOI] [PubMed] [Google Scholar]