Abstract
Resilience of Mycobacterium tuberculosis (MTB) emerges from its ability to effectively counteract immunological, environmental, and antitubercular challenges. Here, we demonstrate that MTB can tolerate drug treatment by adopting a tolerant state that can be deciphered through systems analysis of its transcriptional responses. Specifically, we demonstrate how treatment with the antitubercular drug bedaquiline activates a regulatory network that coordinates multiple resistance mechanisms to push MTB into a tolerant state. Disruption of this network, by knocking out its predicted transcription factors, Rv0324 and Rv0880, significantly increased bedaquiline killing and enabled the discovery of a second drug pretomanid that potentiated killing by bedaquiline. We demonstrate that the synergistic effect of this combination emerges, in part, through disruption of the tolerance network. We discuss how this network strategy also predicts drug combinations with antagonistic interactions, potentially accelerating the discovery of new effective combination drug regimens for TB.
INTRODUCTION
Tuberculosis (TB) still claims more human lives each year than any other bacterial infection1. With millions dying each year, growing levels of drug-resistance, and unacceptably long and complex treatments, we need transformative changes in the way that TB drugs are developed. Increasingly, researchers studying TB are using systems biology approaches and omics technologies in their research. These approaches typically generate large genomic, transcriptomic or proteomic data sets from Mycobacterium tuberculosis (MTB), host animal models or human samples and seek to identify regulatory networks, metabolic pathways, and signaling programs associated with TB infection. In this study, we explored the feasibility of using these systems biology tools for TB drug development.
To test the potential of using a systems-scale approach for TB drug discovery and development, we focused on the antitubercular drug, bedaquiline. Bedaquiline is a new class of antitubercular drug and received FDA approval in 2012 for the treatment of multidrug-resistant TB2. Bedaquiline specifically inhibits the F1F0-ATP synthase of MTB, but kills relatively slowly compared to other frontline drugs, such as isoniazid3-6. The slow bactericidal activity of bedaquiline led us to speculate: (i) regulatory mechanisms push MTB into a tolerant state that resists bedaquiline killing, and (ii) a regulatory network model could capture this state and predict key regulators responsible for the transition. Furthermore, if these hypotheses were true, then inhibiting the key regulators of a resistant, or tolerant, state would render MTB more susceptible to bactericidal activity of bedaquiline.
We herein describe the use of systems-scale models to reveal drug tolerant states of MTB. Our results demonstrate that inhibiting regulators that control the tolerance response can increase bedaquiline killing. Moreover, we exploit these findings and drug transcriptome data to predict rationally and explain mechanistically bedaquiline and pretomanid as an effective drug combination. We also discuss how our approach can detect antagonistic drug combinations, thus, helping to reduce the combinatorial space when evaluating new TB drug regimens. Overall, we highlight the capability of network models to improve and advance the field of TB drug discovery.
RESULTS
Regulatory networks used to model the conditional response of MTB
To discover regulatory mechanisms directing MTB into drug specific tolerant states, we used existing environment and gene regulatory influence network (EGRIN)7,8 and probabilistic regulation of metabolism (PROM)9 models of MTB. While full description of the algorithms used to construct these models are beyond the scope of this work, readers are encouraged to refer to the original papers10,11 and Methods section for more detail.
Transcriptional response of MTB to bedaquiline
Insights into specific, conditionally-active regulatory networks from the MTB EGRIN and PROM models in response to bedaquiline treatment may provide clues into the regulatory mechanisms that control the response and drive MTB into a state that opposes bedaquiline killing. We identified such bedaquiline-response regulons by examining the global transcriptional response of MTB to bedaquiline and mapping differentially expressed genes onto the MTB EGRIN and PROM models. We focused on the gene expression changes at 48 to 96 hours after bedaquiline treatment. During the first 96 hours, bedaquiline shows very little bactericidal activity, even at 30× the minimal inhibitory concentration (MIC)3. Bedaquiline-mediated killing is observed after 96 hours, thus we determined 48 and 96 hours would be most representative of an established MTB bedaquiline-tolerant state. The genome wide gene expression changes at 48 and 96 hours clustered tightly in principal component analysis, were combined for further analysis, and referred to as ‘48-96 hours’. At this stage after bedaquiline treatment, 1121 genes were significantly differentially expressed when treated with 15 μM bedaquiline, compared to untreated samples using a moderated t-test (-1 ≤ log2 fold-change ≤ 1 and Benjamini-Hochberg, BH, multiple hypothesis adjusted P-value < 0.01, Figure S1 and Table S1). First, we explored expression changes of the dosR regulon and genes belonging to the ATP synthase operon, as they were previously reported to be upregulated early after bedaquiline treatment (30 and 180 min)3. However, at 48-96 hours, the expression profiles of the dosR regulon and ATP synthase operon were not significantly upregulated, compared to the untreated profiles (Figure S2). Therefore, we took an unbiased, data-driven approach to interpret the systems level transcriptome changes. Of the 1121 differentially expressed genes, a large majority (897 out of 1121) were downregulated and there was an overall gene enrichment for the TubercuList category12, ‘information pathways’ (BH adjusted P-value = 8.9 × 10−4). Similarly, 86 of the 89 differentially expressed genes associated with the ‘information pathway’ TubercuList category12 were downregulated and include many components of the large and small ribosomal subunits. The downregulation of ribosomal subunits was reported previously in response to bedaquiline treatment3 and suggests that bedaquiline-treatment is associated with a decrease in protein synthesis. While decreased protein synthesis is typically a reflection of slower growth, it is difficult to interpret from these data whether this is a cause or consequence of adopting a tolerant state.
Identification of MTB bedaquiline response regulons
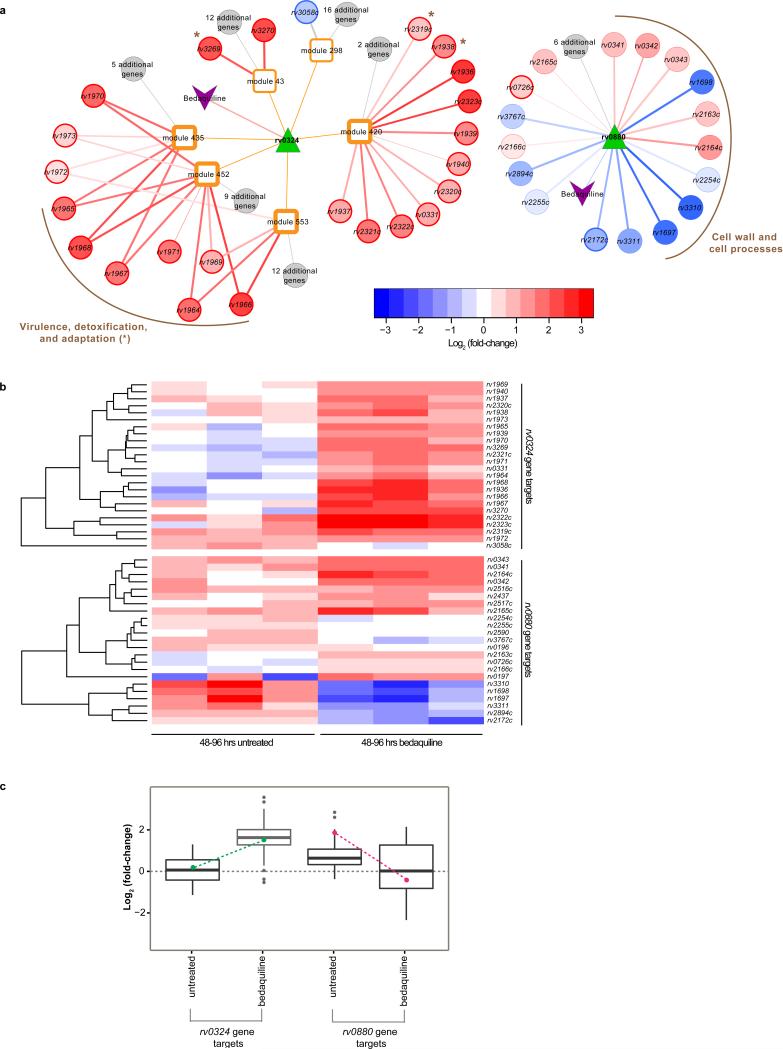
To identify bedaquiline response regulons from the MTB EGRIN and PROM models, we integrated the 1121 differentially expressed genes with each network model. Mapping the bedaquiline treated differentially expressed genes onto the MTB EGRIN model, we detected significant enrichment (BH adjusted P-value < 0.01) in 15 unique modules of co-regulated genes. For these 15 modules, we identified their predicted regulators and prioritized Rv0324 based on further examination of its predicted regulatory target genes (see Table S2 for full results). Rv0324, an ArsR-type TF, is predicted to regulate 4 modules significantly enriched in bedaquiline specific genes (BH adjusted P-value < 0.01) (Figure 1a). In the modules regulated by Rv0324, 22 out of the 23 target genes are upregulated in response to bedaquiline treatment (Figure 1b). This is remarkable considering 80% of the differentially expressed genes were downregulated in response to bedaquiline treatment at 48-96 hours. Moreover, Rv0324 itself has a significant increase in expression (>1.5 absolute log2 fold-change and BH adjusted P-value < 0.01) upon bedaquiline treatment (Figure 1c). This suggests bedaquiline treatment leads to the upregulation of a number of Rv0324 target genes, which may play a part in promoting a bedaquiline-tolerant state. In addition, the genes of the Rv0324 bedaquiline-response regulon are significantly enriched in the TubercuList category12, ‘virulence, detoxification, and adaptation’ (BH adjusted P-value = 3.5 × 10−8, Figure 1a). Many of the Rv0324 target genes that belong to this category are part of the cell wall-localized mce3 operon. While the exact role of the mce3 operon is unknown, the operon has been associated with antitubercular resistance and the mce3 deletion mutant is less virulent than wild type in mice13,14.
Figure 1. Bedaquiline response networks regulated by rv0324 and rv0880.
(a) Graphic representation of linkages between bedaquiline treatment, rv0324, and modules of co-regulated genes (left); and a network view of the rv0880 bedaquiline-response regulon (right). The color scale of regulatory targets represents log2 fold-change values. The edge width is scaled based on Benjamini Hochberg adjusted P-values from t-test comparison of log2 fold-change values of bedaquiline treated and untreated controls. Darkened borders indicate genes that were experimentally validated as functional targets of rv0324 or rv0880. Darkened borders of modules 420, 435, 452 and 553 indicate significant enrichment in bedaquiline-response genes (Benjamini Hochberg adjusted P-value < 0.01). The genes of the rv0324 regulon belonging to the TubercuList category12, ‘virulence, detoxification, and adaptation’ (Benjamini Hochberg adjusted P-value = 3.5 × 10−8) are grouped together as are the genes of the rv0880 regulon from the ‘cell wall and cell processes’ TubercuList category (Benjamini Hochberg adjusted P-value = 1.9 × 10−3). (b) The heat maps show the log2 fold-change of rv0324 regulatory targets (top) and rv0880 (bottom) regulatory targets for untreated and 48-96 hours of bedaquiline treatment from three biological samples. The color scale represents log2 fold-change values. (c) The box plots show average log2 fold-change for rv0324 and rv0880 gene targets for untreated and 48-96 hours of bedaquiline treatment from three biological samples. The box plots are overlayed with log2 fold-change values for rv0324 (green points) and rv0880 (magenta points).
A bedaquiline-specific PROM model was constructed by overlaying the bedaquiline treated transcriptome data using the iMAT approach15,16. The resulting iMAT-PROM model represents the metabolic state of MTB when exposed to bedaquiline and was used to simulate the phenotypic outcome of 104 TF knockout events. The model predicts that bedaquiline treatment leads to the drug-specific essentiality of 29 metabolic genes, which participate in processes including energy metabolism, as well as biosynthesis and degradation of key metabolites including amino acids, nucleotides, cofactors, and lipids. Out of the TF perturbations simulated, MTBPROM 2.0 identified Rv0880 as a knockout that would exploit the drug-specific vulnerabilities, yielding a strong defect when knocked out in the presence of bedaquiline, but not in its absence (see Table S3 for full results). Rv0880, under standard culture conditions, modulates the expression of 23 genes17, 17 of which were significantly differentially expressed when treated with bedaquiline (Figure 1B). The Rv0880 bedaquiline-response regulon also contains a notable number of upregulated genes (8 out of 17, Figure 1b). Nonetheless, Rv0880 has a significantly decreased expression (>2.0 absolute log2 fold-change and BH adjusted P-value < 0.01) after 48-96 hours of bedaquiline treatment (Figure 1c). Presumably, the bedaquiline-induced repression of Rv0880 leads to mixed expression changes of its regulatory targets. The regulatory targets of Rv0880 are enriched in the TubercuList category12, ‘cell wall and cell processes’ (BH adjusted P-value = 1.9 × 10−3, Figure 1a), which could contribute to a bedaquiline-tolerant state in complex ways.
Transcription factor knockout mutants, ΔRv0324 and ΔRv0880, are hypersensitive to bedaquiline
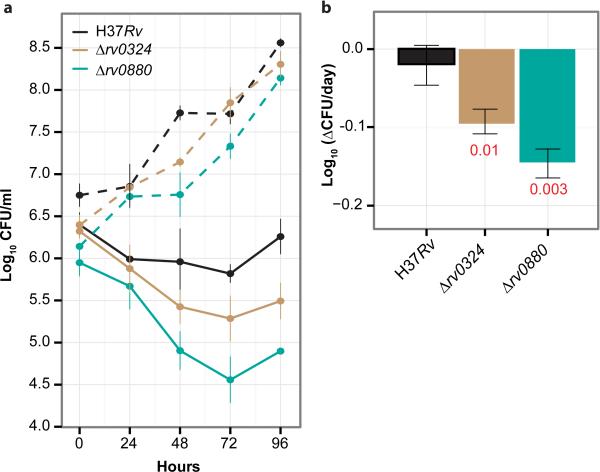
We hypothesized that strains lacking TFs controlling the bedaquiline-responsive regulons should be hypersensitive to bedaquiline and have altered kill kinetics. To test this hypothesis, we created Rv0324 and Rv0880 knockout mutants of MTB H37Rv using specialized transducing phages18. CFUs were monitored over a period of 96 hours (4 days) for H37Rv, ΔRv0324, and ΔRv0880 MTB strains treated with 15 μM bedaquiline (Figure 2a). There were fewer surviving cells from bedaquiline treatment for the ΔRv0324 and ΔRv0880 strains compared to H37Rv, with a 6 and 22-fold greater survival deficiency by 96 hours, respectively (Figure 2a). Over the 4 days of treatment, the ΔRv0324 and ΔRv0880 strains had significantly lower growth rates with 15 μM bedaquiline treatment compared to wildtype, indicating improved bedaquiline bactericidal activity with the TF knockout mutants (Figure 2b). Similar trends were also observed for 1.5 μM bedaquiline (Figure S3), but to a lesser extent as expected by a reduced drug dosage.
Figure 2. Killing of Δrv0324 and Δrv0880 MTB strains by bedaquiline.
(a) CFUs over a period of 168 hours (7 days) for untreated (dashed lines) and 15 μM bedaquiline (solid lines) liquid cultures of H37Rv wild type (black), Δrv0324 (brown) and Δrv0880 (teal). (b) First-order rate kinetics were calculated by linear regression of CFUs from H37Rv wild type and Δrv0324 and Δrv0880 strains obtained over 7 days of 15 μM bedaquiline treatment. Statistical significance of rate kinetics between wild type and mutant strains were calculated using t-test, Benjamini Hochberg adjusted P-values are numbers in red. Error bars show the standard deviation from three biological samples (a) and the standard error of the linear regression (b). Representative results from two experiments are presented.
Sensitivity of ΔRv0324 and ΔRv0880 to other antitubercular drugs
While the growth rates of ΔRv0324 and ΔRv0880 are comparable to wild type (dashed lines in Figure 2a), it is possible that even subtle changes in growth rate may affect the TF knockout mutants sensitivity to antitubercular drugs in a non-specific manner. To evaluate drug sensitivity, time-kill assays were performed for the MTB wild type H37Rv, ΔRv0324 and ΔRv0880 strains with three antitubercular drugs with distinct mechanisms of action: capreomycin, pretomanid and rifampin (Figure S4). Compared to wild type, ΔRv0324 and ΔRv0880 strains showed no significant shifts in the time-kill profiles for the antitubercular drugs tested. These time-kill profiles demonstrate that the hypersensitivity of the TF knockout mutant strains is specific to bedaquiline. We also evaluated drug sensitivity of the TF knockout strains by measuring their MIC values for bedaquiline, capreomycin, isoniazid and pretomanid (Table S4). The MIC values of the TF knockout mutant strains were consistent with MTB wild type H37Rv MIC values. Interestingly, there was no shift in the MIC of bedaquiline with the TF knockout mutant strains, despite their significant hypersensitivity to bedaquiline in time-kill assays and evidence that disruption of the tolerance network accelerated the dynamics of killing by bedaquiline. Importantly, these results illustrate how tolerance may be invisible to single time point MIC testing.
Upregulated, and not downregulated bedaquiline response regulons push MTB into a tolerant state
The Rv0324 and Rv0880 regulons contain a disproportionate number of target genes that are upregulated when MTB is treated with bedaquiline. To test if the induction of target genes is important to tolerate bedaquiline killing, we constructed knockout mutants of Rv1049 and Rv0238, which were also predicted by EGRIN and PROM models to control bedaquiline-specific regulons, respectively. However, all target genes belonging to the Rv1049 regulon and more than 85% of Rv0238 regulon genes are downregulated by bedaquiline (Figure S5). The bedaquiline susceptibility of the ΔRv1049, ΔRv0238 and wild type strains was tested by CFU counts over 7 days of treatment. We observed no significant difference between the bactericidal activity of bedaquiline for the wild type and mutant strains (Figure S6). These results suggested that induction and not repression of genes in bedaquiline-response regulons may be particularly important to the formation of an MTB tolerant state.
Rational drug combinations to potentiate bedaquiline bactericidal activity
The MTB EGRIN and PROM network models facilitated the discovery of key regulators of bedaquiline-tolerance regulons. We experimentally validated the importance of these regulons by demonstrating that deletion of their regulators renders MTB hypersensitive to bedaquiline treatment. Based on these promising results, we sought to identify drug(s) that could have a similar effect as the knockout strains, thus, potentiating the bactericidal activity of bedaquiline. To do this, we used transcriptome data from MTB exposed to 2×, 4×, and 8× MIC levels of current antitubercular drugs as well as test compounds with demonstrated antitubercular activity (GEO accession number GSE71200)19. Altogether, we mined an expression dataset of 36 antitubercular agents to identify drugs that significantly change the expression of Rv0324 or Rv0880 (see Table S5 for full results). From this analysis, we hypothesized that pretomanid20 (formerly PA-824) could synergize with bedaquiline through inhibition of the Rv0880 bedaquiline-response regulon.
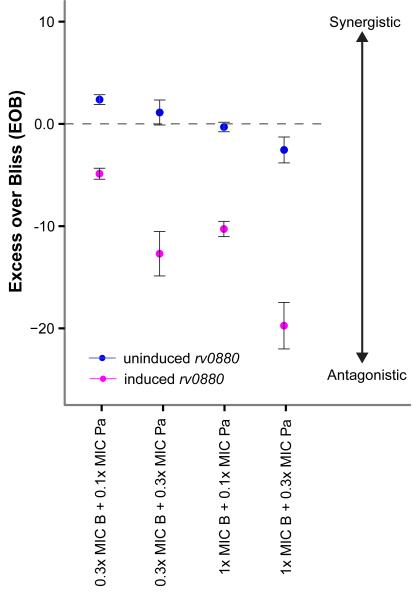
To experimentally evaluate the drug combination of bedaquiline and pretomanid (BPa), we tested the activity of single and combinatorial treatments with the two drugs using an inducible Rv0880 overexpression strain. The inducible overexpression strain was constructed by introducing a plasmid-borne FLAG-tagged version of Rv0880 under the control of the mycobacterial tetracycline-inducible promoter17,21. We tested the Rv0880 overexpression strain treated with 0.3× and 1× MIC of bedaquiline alone and in combination with 0.1× and 0.3× MIC of pretomanid. The dosage of bedaquiline was calibrated to the increased sensitivity (4-fold) of the Rv0880 overexpressing strain to bedaquiline (Table S2). We assayed the bacterial counts after 7 days of drug treatment for the Rv0880 overexpressor strain when un-induced (DMSO added) or induced with anhydrotetracycline (Figure S7). The BPa combination activity was estimated using Excess over Bliss (EOB), which determines whether the combined effect of two drugs is greater or smaller than the naïve (independent) combination of their individual effects22. The EOB estimates were used to determine synergism or antagonism of the tested BPa combination concentrations with un-induced or induced overexpression of Rv0880 (Figure 3). Our analysis demonstrates that the BPa combinations were additive to mildly synergistic at most of the sub-MIC concentrations tested with un-induced Rv0880 overexpression. In contrast, synergism was eliminated and BPa combinations demonstrated strong antagonistic behavior at all concentrations tested when Rv0880 overexpression was induced. The Rv0880-induced samples reveal the effectiveness of BPa is compromised through overexpression of the Rv0880 regulon.
Figure 3. Influence of Rv0880 overexpression on Excess over Bliss estimation of bedaquiline and pretomanid drug combination.
The Excess over Bliss (EOB) estimation for all bedaquiline and pretomanid (BPa) combinations tested with induced (magenta) and un-induced (blue) rv0880 overexpression cultures. Error bars represent the standard deviation of EOB, estimated from three biological samples. The dashed gray line is at EOB = 0 (additive) and represents a line over and below which BPa combinations are considered synergistic and antagonistic, respectively. This experiment was repeated twice with reproducible results. Bedaquiline, B. Pretomanid, Pa.
DISCUSSION
For decades, researchers have known that TB treatment must be delivered in regimens containing multiple drugs to prevent the development of drug resistance. Traditionally, novel drug combinations have been evaluated for improved efficacy and treatment-shortening potential by replacing a single drug within a regimen, or adding a new drug to an existing regimen in essentially a trial and error approach. However, the number of possible testing arrangements is extremely large and such efforts have yielded few promising therapies. Our results suggest that new drug combinations can be rationally predicted using a regulatory network model to uncover the adaptive consequences that result from perturbing MTB with a drug. Such a systems approach to drug combinations could advance TB drug discovery and facilitate the delivery of new and better TB treatments to patients.
Deletion of Rv0324 or Rv0880 significantly increases the killing efficacy of bedaquiline. Traditionally, these transcription factors would be dismissed as drug targets as their activity is not essential to MTB growth. They might also be rejected to potentiate bedaquiline as their regulatory targets are not enriched in the genes associated with intermediate metabolism and respiration that are most directly linked to the proposed mechanisms of action of bedaquiline: ATP depletion3,4 and uncoupling of respiration-driven ATP synthesis23. Recent studies have demonstrated increased bedaquiline killing for mycobacterium knockout mutants of cytochrome bd24,25, a component of the respiratory chain that establishes the proton motive force and energy necessary for ATP synthesis. Given its activity related to the proposed mode of action of bedaquiline and in vivo essentiality24, cytochrome bd is an obvious target26,27. In contrast, our systems approach was able to reveal novel, non-traditional targets with comparable potentiation of bedaquiline activity.
Based on the results, we propose the link between inhibition of Rv0324 or Rv0880 and hypersensitivity to bedaquiline is due to an MTB state that affords tolerance to bedaquiline. This bedaquiline-tolerant state is transcriptionally driven and does not rely on genetic mutations to trigger conventional antibiotic resistance mechanisms, such as conformational changes in drug target that decrease binding affinity of a drug28. Instead, the collective regulation of target genes must be important for the tolerant effect. In turn, the tolerant state maintains a higher cell population and affords more opportunity for MTB to develop a permanent resistance strategy. Our results also suggest a predominant upregulation of target genes is critical for the formation of a bedaquiline-tolerant state. This is also important to the distinction between tolerance and persistence. Both mechanisms transiently reduce MTB sensitivity to drugs and are stimulated by stress conditions. However, we propose that tolerance in MTB emerges through transcriptional induction of specific regulatory networks, while it is thought that subpopulations of MTB persisters enter a non-replicating or dormant state via various proposed mechanisms29-33. Importantly, both tolerance and persistence states of MTB need greater understanding and more study at the systems and single-cell levels.
It is likely that the bedaquiline-tolerant state is generated by an accompanying stress that MTB has encountered in its evolutionary history. The conditional grouping of genes within modules by the MTB EGRIN model provides an opportunity to understand the natural environmental stress context in which the Rv0324 regulon is active. From the transcriptome profiles used to construct the MTB EGRIN model, the expression changes of Rv0324 were correlated with co-regulated genes within modules 420, 435, and 452 under nutrient-limited conditions (module 420: R = 0.97, P-value = 1.4 × 10−5; module 435: R = 0.95, P-value = 9.8 × 10−5; module 452: R = 0.90, P-value = 0.003, Figure S7). This finding has important implications since the availability of nutrients (e.g. glucose, fatty acids) has been shown to affect bedaquiline bactericidal activity3. A greater bactericidal activity has been observed for bedaquiline in MTB-infected macrophages34, which are abundant in fatty acids. It is possible that the drug could be locally concentrated in the macrophage, resulting in greater bacterial killing. Alternatively, our network analysis predicts that the Rv0324 regulon is conditionally activated under standard and nutrient-limited conditions and we propose that under these conditions Rv0324 is able to generate a tolerant state, resulting in reduced bedaquiline killing. Therefore, it is also conceivable that MTB residing in lipid-rich human macrophages are incapable of transitioning to a tolerant state via the Rv0324 regulon. As such, we see potential in conditionally or chemically inhibiting the Rv0324 regulon, thereby reengineering MTB into a state that is more susceptible to killing by bedaquiline. We can use this network strategy to uncover TFs and regulons associated with other drug-specific tolerant states and rationally formulate strategies for reengineering MTB susceptibility, depending on the genetic background of the strain and drug-responsiveness of a patient.
The systems-scale understanding of how MTB adapts and responds to bedaquiline treatment enabled rational prediction of bedaquiline and pretomanid (BPa) as an effective drug combination. While this combination has been previously tested by others35,36, they were assayed in multi-agent screening studies. In contrast, we were able to rationally select the combination using in silico methods, and went on to demonstrate that mechanistically BPa acts, at least in part, through inhibition of the Rv0880 regulon. BPa is the core for two out of three clinical trials in TB Alliance's novel regimen development pipeline37. A Phase 2b clinical trial is currently recruiting for testing of the BPaZ regimen that is comprised of bedaquiline, pretomanid, and pyrazinamide38. This regimen shows potential to treat both drug-sensitive and drug-resistant forms of TB in as little as three months39. In addition, a phase 3 clinical trial is being conducted in XDR-TB patients with bedaquiline, pretomanid, and linezolid that aims to treat all forms of active TB40. Our findings elucidate a potential mechanism for why these drug combinations are so effective. We demonstrate that exploiting the transcriptional mechanism by which MTB responds to bedaquiline has led to the building blocks of new, faster, and better TB regimens.
The network strategy described here should also be able to predict antagonistic drug combinations, thereby, reducing the number of combinations to experimentally evaluate. For instance, even a small number of compounds results in a very large number of combinations; a collection of 1,000 compounds yields ~500,000 pairwise combinations, and many more higher-order combinations. Our approach is able to rationally reduce this huge combinatorial space by de-prioritizing ineffective combinations. As described in results, exploring a gene expression dataset of 36 antitubercular drugs led to the identification of pretomanid as significantly decreasing the expression of Rv0880. We went on to experimentally validate the synergy of bedaquiline and pretomanid at sub-MIC concentrations in the un-induced Rv0880 overexpression strain. The same transcriptome dataset also identified 5 other antitubercular agents that significantly increase the expression of Rv0324, including capreomycin and moxifloxacin (Table S1). We would expect these 5 antitubercular agents to enhance the tolerance to bedaquiline, thereby predicting antagonistic drug combinations. This is just one example of novel drug interaction predictions and biological insights that can be drawn from network analysis of transcriptional responses to drug treatment.
We have demonstrated a tolerance mechanism in MTB that can be deciphered and disrupted to potentiate antitubercular drugs in a rational manner. Moreover, the approach can predict drug combinations with antagonistic interactions, all based on network responses to single drug treatments. Given the expansive combinatorial space of double and triple drug therapy, this network-based strategy could accelerate the discovery of new drug combinations for treating TB, by rank prioritizing combinations with desired synergistic effects, tailored to the strain of MTB and drug responsiveness of a patient.
METHODS
The approach used in this study includes both computational and biological methods. Algorithms developed for the EGRIN model and the PROM model were implemented in the R programming language41 and MATLAB42, respectively. Plots were generated using R41, regulatory network diagrams generated using Cytoscape43, and images prepared using Adobe Illustrator CS5.
Strains
To investigate the growth properties of Mycobacterium tuberculosis (MTB) in the absence of different transcription factors (TFs), we used strains with the TF gene of interest deleted using allelic exchange substrates introduced by specialized transducing phages (phages provided by the labs of William Jacobs Jr. and Michelle Larsen, Albert Einstein College of Medicine), as previously described18. To measure the growth consequences of overexpressing TFs, we used strains containing an anhydrotetracycline (ATc)- inducible expression vector of the TF gene, as described previously17,44.
Culturing conditions
MTB strains were cultured in Middlebrook 7H9 with the ADC supplement (Difco), 0.05% Tween80 at 37°C under aerobic conditions with constant agitation to mid-log phase as described previously17,44. Strains containing the ATc-inducible expression vector were grown with the addition of 50 μg/mL hygromycin B to maintain the plasmid. Growth was monitored by OD600 and colony forming units (CFUs). Cultures were diluted to approximately 106 CFU/mL (OD600 ~ 0.002) and exposed to drug. For experiments featuring the Rv0880 overexpression strain, the gene overexpression was induced concurrently with drug treatment for the approximate duration of one cell doubling (18 h) using an ATc concentration 100 ng/mL culture.
For the bedaquiline time-kill assays, 0.75× and 7.5× MIC of bedaquiline (measured MIC of bedaquiline was 2 μM) were added to cultures of the wild type MTB H37Rv strain and of each TF knockout mutant strains. Time-kill assays were also similarly performed with 3x MIC of capreomycin (1.5 μg/mL), pretomanid (0.5 μg/mL) and rifampicin (0.6 μg/mL). Samples were taken for 4-7 days from each strain and antitubercular concentration to assess CFUs.
For the combinatorial drug exposure experiments, two concentrations of bedaquiline (0.3×, 1× MIC) and pretomanid (0.1×, 0.3× MIC) were tested in all four combined permutations (e.g. 0.3× MIC bedaquiline + 0.1× MIC pretomanid, 0.3× MIC bedaquiline + 0.3× MIC pretomanid, 1× MIC bedaquiline + 0.1× MIC pretomanid, and 1× MIC bedaquiline + 0.3× MIC pretomanid), and were compared with response to treating with bedaquiline alone, at the two concentrations (0.1× MIC and 0.3× MIC). For each dosing scheme, the drugs were supplemented into cultures of the wild type MTB H37Rv strain and of each TF mutant strain. Samples were taken for 7 days from each strain and bedaquiline concentration to assess CFUs.
Determination of minimum inhibitory concentration (MIC)
Twofold dilutions of antitubercular agents were prepared in Middlebrook 7H9 medium in a volume of 20 μL in 96-well culture plates. Approximately 180 μL of 106 CFU/mL MTB strains was added, yielding a final testing volume of 200 μL. The plates were incubated at 37°C; on the seventh day, 100 μL of culture was taken from each well and mixed with an equal volume of Bac Titer-Glo® reagent (Promega, Madison, WI, USA). The assay was performed according to the manufacturer's instructions. Luminescence was measured on a POLARstar Omega (BMG Lab Tech, Cary, NC) microplate reader at an integration time of 500 milliseconds.
RNA isolation
RNA was isolated as described previously17,44. Briefly, cell pellets in Trizol were transferred to a tube containing Lysing Matrix B (QBiogene, Inc.), and vigorously shaken at max speed for 30 s in a FastPrep 120 homogenizer (QBiogene) three times, with cooling on ice between shakes. This mixture was centrifuged at max speed for 1 min and the supernatant was transferred to a tube containing 300 μL chloroform and Heavy Phase Lock Gel (Eppendorf North America, Inc.), inverted for 2 minutes, and centrifuged at max speed for 5 min. RNA in the aqueous phase was then precipitated with 300 μL isopropanol and 300 μL high salt solution (0.8 M Na citrate, 1.2 M NaCl). RNA was purified using an RNeasy kit following manufacturer's recommendations (Qiagen) with one on-column DNase treatment (Qiagen). Total RNA yield was quantified using a Nanodrop (Thermo Scientific).
Microarray analysis
RNA was converted to Cy dye-labeled cDNA probes as described previously44. Sets of fluorescent probes were then hybridized to Agilent (Array ID ‘MTB.tiled.3.2013’) tiling arrays consisting of 135,000 probes spaced at approximately 100 bp intervals around the MTB H37Rv genome. These arrays provide 105,000 data points for each expression profile covering approximately 13,000 sense, antisense, and intergenic genome features. For background we compared the expression levels of these probes to a set of 30,000 randomers of equivalent GC distribution. Arrays were scanned and spots were quantified using a PerkinElmer ScanArray Express. Subsequent preprocessing by robust multichip average (RMA) normalization45 was carried out using Arraystar software. Array data are available at NCBI-GEO, accession number GSE72459.
EGRIN modeling
We integrated the MTB bedaquiline response transcriptional profiling gene expression data with the MTB EGRIN model as described in Peterson et al7. Briefly, the EGRIN model was constructed through semi-supervised biclustering of 2325 gene expression profiles of MTB, guided by biologically informative priors and de novo cis-regulatory GRE detection for module assignment11. We demonstrated that the MTB EGRIN model accurately predicts regulatory interactions in a network of 240 modules and validated the model with the DNA binding sites and transcriptional targets from overexpressing >150 MTB transcription factors (TFs)17,21. Thus, the EGRIN model serves as a powerful framework to formulate hypotheses of MTB regulatory interactions that respond to various environmental conditions, including the treatment of new antitubercular drugs, such as bedaquiline.
Using the differentially expressed genes at 48 and 96 hours (combined and referred to as ‘48-96 hours’) of 15 μM bedaquiline treatment compared to non-treated controls, we identified unique modules of co-regulated genes within the MTB EGRIN model that were significantly enriched (Benjamini Hochberg corrected hypergeometric P-value < 0.01). For these modules, we identified the predicted regulatory TFs and the functional enrichment of targets. See Table S2 for full results and details.
iMAT-PROM modeling
We also explored MTB's conditional response using an updated regulatory-metabolic model for MTB based on the PROM framework9. The updated model features a regulatory network for 104 TFs based on their ChIP-Seq (chromosome immunoprecipitation followed by sequencing) interactions upon TF overexpression21, interfaced with a refined genome-scale metabolic model of 810 genes. MTBPROM 2.0 model was used to generate knockout and overexpression phenotype predictions for 104 TFs, when exposed to a range of environmental and genetic perturbations, and it improved predictive accuracy of TF perturbation phenotypes over other MTB regulatory-metabolic models when tested against experimental gene essentiality and overexpression growth rate data17. This supports the PROM model as being amenable for use in predicting the phenotype of TF-perturbed strains under novel antitubercular drug (i.e., bedaquiline) treatment.
To model the MTB metabolic response to bedaquiline, we overlayed the bedaquiline treatment transcriptome data onto a genome-scale metabolic model of MTB described in Ma et al46 using the iMAT approach15,16 implemented by the COBRA toolbox in MATLAB47,48. We then simulated the effect of overexpressing and knocking out TFs that would synergize with the metabolic changes induced by bedaquiline by applying the PROM framework46, interfacing regulatory information from TF binding and transcriptional response of TF overexpression17,21. Using the bedaquiline-specific PROM model to simulate the effect of TF knockouts upon treatment with bedaquiline, we identified TF knockouts predicted to yield growth defect relative to wild type in the presence of bedaquiline but not in the absence of drug. See Table S1 for full results and details.
Excess over Bliss as a measurement for synergy
The Bliss Independence model49 predicts that if bedaquiline (B) and pretomanid (Pa), with experimentally measured fractional inhibitions fB and fPa, have an additive effect, then the expected fractional inhibition fBPa, induced by their combination should be:
Excess over Bliss (EOB) was determined by computing the difference in fractional inhibition induced by compound combination, fz, and the expected fractional inhibition, fBPa
A compound pair for which EOB ≈ 0 has an additive behavior, whereas a compound pair with positive or negative EOB values has synergistic or antagonistic behavior, respectively. Propagation of errors using standard deviation of fractional inhibitions was used to compute the error bars of EOB.
Supplementary Material
Acknowledgements
We thank members of the Baliga and Sherman labs for critical discussions; Tige Rustad, Jessica Winkler and Samuel Hobbs for generating knockout and overexpressing strains; Zac Simon, Megha Sarvothama and Reiling Liao for technical help. Funding was provided by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health [U19 AI10676, U19 AI111276 and ISBpilot-10135]; and National Institute of General Medical Sciences of the National Institutes of Health [P50GM076547].
Footnotes
Author Contributions E.J.R.P led the design and drafted the manuscript. E.J.R.P. and S.M. generated results and analyzed data. S.M. helped design the study and helped draft online methods. D.R.S. helped designed the study, discussed results and commented on the manuscript. N.S.B. conceived of the study, discussed results and drafted the manuscript.
Author Information Microarray data have been deposited in the GEO repository under accession number GSE72459. The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
REFERENCES
- 1.Phillips L. Infectious disease: TB's revenge. Nature. 2013;493:14–16. doi: 10.1038/493014a. doi:10.1038/493014a. [DOI] [PubMed] [Google Scholar]
- 2.Diacon AH, et al. The diarylquinoline TMC207 for multidrug-resistant tuberculosis. N Engl J Med. 2009;360:2397–2405. doi: 10.1056/NEJMoa0808427. doi:10.1056/NEJMoa0808427. [DOI] [PubMed] [Google Scholar]
- 3.Koul A, et al. Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nature communications. 2014;5:3369. doi: 10.1038/ncomms4369. doi:10.1038/ncomms4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koul A, et al. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat Chem Biol. 2007;3:323–324. doi: 10.1038/nchembio884. doi:10.1038/nchembio884. [DOI] [PubMed] [Google Scholar]
- 5.Koul A, et al. Diarylquinolines are bactericidal for dormant mycobacteria as a result of disturbed ATP homeostasis. J Biol Chem. 2008;283:25273–25280. doi: 10.1074/jbc.M803899200. doi:10.1074/jbc.M803899200. [DOI] [PubMed] [Google Scholar]
- 6.Andries K, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. doi:10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 7.Peterson EJ, et al. A high-resolution network model for global gene regulation in Mycobacterium tuberculosis. Nucleic Acids Res. 2015;42:11291–11303. doi: 10.1093/nar/gku777. doi:10.1093/nar/gku777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turkarslan S PE, Rustad TR, Minch KJ, Reiss DJ, Morrison R, Ma S, Price ND, Sherman DR, Baliga NS. A comprehensive map of genome-wide gene regulation in Mycobacterium tuberculosis. Sci Data. 2015;2:150010. doi: 10.1038/sdata.2015.10. doi:10.1038/sdata.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma S, et al. Integrated Modeling of Gene Regulatory and Metabolic Networks in Mycobacterium tuberculosis. PLoS Comput Biol. 2015;11:e1004543. doi: 10.1371/journal.pcbi.1004543. doi:10.1371/journal.pcbi.1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandrasekaran S, Price ND. Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2010;107:17845–17850. doi: 10.1073/pnas.1005139107. doi:10.1073/pnas.1005139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reiss DJ, Baliga NS, Bonneau R. Integrated biclustering of heterogeneous genome-wide datasets for the inference of global regulatory networks. BMC bioinformatics. 2006;7:280. doi: 10.1186/1471-2105-7-280. doi:10.1186/1471-2105-7-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lew JM, Kapopoulou A, Jones LM, Cole ST. TubercuList--10 years after. Tuberculosis (Edinb) 2011;91:1–7. doi: 10.1016/j.tube.2010.09.008. doi:10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Forrellad MA, et al. Virulence factors of the Mycobacterium tuberculosis complex. Virulence. 2013;4:3–66. doi: 10.4161/viru.22329. doi:10.4161/viru.22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Senaratne RH, et al. Mycobacterium tuberculosis strains disrupted in mce3 and mce4 operons are attenuated in mice. J Med Microbiol. 2008;57:164–170. doi: 10.1099/jmm.0.47454-0. doi:10.1099/jmm.0.47454-0. [DOI] [PubMed] [Google Scholar]
- 15.Shlomi T, Cabili MN, Herrgard MJ, Palsson BO, Ruppin E. Network-based prediction of human tissue-specific metabolism. Nature biotechnology. 2008;26:1003–1010. doi: 10.1038/nbt.1487. doi:10.1038/nbt.1487. [DOI] [PubMed] [Google Scholar]
- 16.Zur H, Ruppin E, Shlomi T. iMAT: an integrative metabolic analysis tool. Bioinformatics. 2010;26:3140–3142. doi: 10.1093/bioinformatics/btq602. doi:10.1093/bioinformatics/btq602. [DOI] [PubMed] [Google Scholar]
- 17.Rustad TR, et al. Mapping and manipulating the Mycobacterium tuberculosis transcriptome using a transcription factor overexpression-derived regulatory network. Genome Biol. 2014;15:502. doi: 10.1186/s13059-014-0502-3. doi:10.1186/PREACCEPT-1701638048134699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tufariello JM, et al. Enhanced specialized transduction using recombineering in Mycobacterium tuberculosis. MBio. 2014;5:e01179–01114. doi: 10.1128/mBio.01179-14. doi:10.1128/mBio.01179-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ioerger TR, et al. Identification of new drug targets and resistance mechanisms in Mycobacterium tuberculosis. PLoS One. 2013;8:e75245. doi: 10.1371/journal.pone.0075245. doi:10.1371/journal.pone.0075245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stover CK, et al. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature. 2000;405:962–966. doi: 10.1038/35016103. doi:10.1038/35016103. [DOI] [PubMed] [Google Scholar]
- 21.Minch KJ, et al. The DNA-binding network of Mycobacterium tuberculosis. Nature communications. 2015;6:5829. doi: 10.1038/ncomms6829. doi:10.1038/ncomms6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bansal M, et al. A community computational challenge to predict the activity of pairs of compounds. Nat Biotechnol. 2014;32:1213–1222. doi: 10.1038/nbt.3052. doi:10.1038/nbt.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hards K, et al. Bactericidal mode of action of bedaquiline. J Antimicrob Chemother. 2015;70:2028–2037. doi: 10.1093/jac/dkv054. doi:10.1093/jac/dkv054. [DOI] [PubMed] [Google Scholar]
- 24.Berney M, Hartman TE, Jacobs WR., Jr. A Mycobacterium tuberculosis cytochrome bd oxidase mutant is hypersensitive to bedaquiline. mBio. 2014;5:e01275–01214. doi: 10.1128/mBio.01275-14. doi:10.1128/mBio.01275-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu P, et al. The cytochrome bd-type quinol oxidase is important for survival of Mycobacterium smegmatis under peroxide and antibiotic-induced stress. Sci Rep. 2015;5:10333. doi: 10.1038/srep10333. doi:10.1038/srep10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pethe K, et al. Discovery of Q203, a potent clinical candidate for the treatment of tuberculosis. Nat Med. 2013;19:1157–1160. doi: 10.1038/nm.3262. doi:10.1038/nm.3262. [DOI] [PubMed] [Google Scholar]
- 27.Boshoff HI, et al. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J Biol Chem. 2004;279:40174–40184. doi: 10.1074/jbc.M406796200. doi:10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- 28.Telenti A, et al. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 29.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. Journal of bacteriology. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. doi:10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 31.Smith PA, Romesberg FE. Combating bacteria and drug resistance by inhibiting mechanisms of persistence and adaptation. Nat Chem Biol. 2007;3:549–556. doi: 10.1038/nchembio.2007.27. doi:10.1038/nchembio.2007.27. [DOI] [PubMed] [Google Scholar]
- 32.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic acids research. 2005;33:966–976. doi: 10.1093/nar/gki201. doi:10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sala A, Bordes P, Genevaux P. Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins (Basel) 2014;6:1002–1020. doi: 10.3390/toxins6031002. doi:10.3390/toxins6031002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dhillon J, Andries K, Phillips PP, Mitchison DA. Bactericidal activity of the diarylquinoline TMC207 against Mycobacterium tuberculosis outside and within cells. Tuberculosis. 2010;90:301–305. doi: 10.1016/j.tube.2010.07.004. doi:10.1016/j.tube.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Diacon AH, et al. 14-day bactericidal activity of PA-824, bedaquiline, pyrazinamide, and moxifloxacin combinations: a randomised trial. Lancet. 2012;380:986–993. doi: 10.1016/S0140-6736(12)61080-0. doi:10.1016/S0140-6736(12)61080-0. [DOI] [PubMed] [Google Scholar]
- 36.Tasneen R, et al. Sterilizing activity of novel TMC207- and PA-824-containing regimens in a murine model of tuberculosis. Antimicrob Agents Chemother. 2011;55:5485–5492. doi: 10.1128/AAC.05293-11. doi:10.1128/AAC.05293-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Developing Regimens. 2015 < http://www.tballiance.org/pipeline/developing-regimens-novel.php>.
- 38.A Phase 2 Open Label Partially Randomized Trial to Evaluate the Efficacy, Safety and Tolerability of Combinations of Bedaquiline, Moxifloxacin, PA-824 and Pyrazinamide in Adult Subjects With Drug-Sensitive or Multi Drug-Resistant Pulmonary Tuberculosis. 2015 (NC-005), < https://clinicaltrials.gov/ct2/show/NCT02193776>.
- 39.Diacon AH, et al. Bactericidal activity of pyrazinamide and clofazimine alone and in combinations with pretomanid and bedaquiline. Am J Respir Crit Care Med. 2015;191:943–953. doi: 10.1164/rccm.201410-1801OC. doi:10.1164/rccm.201410-1801OC. [DOI] [PubMed] [Google Scholar]
- 40.A Phase 3 Study Assessing the Safety and Efficacy of Bedaquiline Plus PA-824 Plus Linezolid in Subjects With Drug Resistant Pulmonary Tuberculosis. 2015 < https://clinicaltrials.gov/ct2/results?term=Nix-TB&Search=Search>.
- 41.Team RC. R: A language and environment for statistical computing. Vienna, Austria: 2013. [Google Scholar]
- 42.Rogers DJ, Tanimoto TT. A Computer Program for Classifying Plants. Science. 1960;132:1115–1118. doi: 10.1126/science.132.3434.1115. doi:10.1126/science.132.3434.1115. [DOI] [PubMed] [Google Scholar]
- 43.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rustad TR, Harrell MI, Liao R, Sherman DR. The enduring hypoxic response of Mycobacterium tuberculosis. PloS one. 2008;3:e1502. doi: 10.1371/journal.pone.0001502. doi:10.1371/journal.pone.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. doi:10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 46.Ma S, Minch KJ, Rustad TR, Hobbs S, Zhou S, Sherman DR, Price ND. Integrated modeling of gene regulatory and metabolic networks in Mycobacterium tuberculosis. accepted at PLoS Computational Biology. 2015 doi: 10.1371/journal.pcbi.1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Becker SA, et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protoc. 2007;2:727–738. doi: 10.1038/nprot.2007.99. doi:10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- 48.Schellenberger J, et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. doi:10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borisy AA, et al. Systematic discovery of multicomponent therapeutics. Proc Natl Acad Sci U S A. 2003;100:7977–7982. doi: 10.1073/pnas.1337088100. doi:10.1073/pnas.1337088100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.