Abstract
We present Insight as an integrated database and analysis platform for epilepsy self-management research as part of the national Managing Epilepsy Well Network. Insight is the only available informatics platform for accessing and analyzing integrated data from multiple epilepsy self-management research studies with several new data management features and user-friendly functionalities. The features of Insight include, (1) use of Common Data Elements defined by members of the research community and an epilepsy domain ontology for data integration and querying, (2) visualization tools to support real time exploration of data distribution across research studies, and (3) an interactive visual query interface for provenance-enabled research cohort identification. The Insight platform contains data from five completed epilepsy self-management research studies covering various categories of data, including depression, quality of life, seizure frequency, and socioeconomic information. The data represents over 400 epilepsy patients with 7,552 data points. The Insight data exploration and cohort identification query interface has been developed using Ruby on Rails Web technology and open source Web Ontology Language Application Programming Interface to support ontology-based reasoning. We have developed an efficient ontology management module that automatically updates the ontology mappings each time a new version of the Epilepsy and Seizure Ontology is released. The Insight platform features a Role-based Access Control module to authenticate and effectively manage user access to different research studies. User access to Insight is managed by the Managing Epilepsy Well Network database steering committee consisting of representatives of all current collaborating centers of the Managing Epilepsy Well Network. New research studies are being continuously added to the Insight database and the size as well as the unique coverage of the dataset allows investigators to conduct aggregate data analysis that will inform the next generation of epilepsy self-management studies.
1. Introduction
Persons with chronic health conditions can significantly benefit from self-management techniques, which consist of understanding their health conditions and adopting a set of behaviors to manage their conditions (1-5). Epilepsy is one of the most common serious neurological disorders, affecting an estimated 50 million persons worldwide (6), with 200,000 new cases reported each year (7). Development and adoption of self-management techniques have been strongly recommended for persons with epilepsy to positively influence their prognosis and ability to manage the symptoms of epilepsy (8, 9). Patients with epilepsy experience repeated seizures that manifest as physical or behavioral changes that disrupt normal activities (10). Repeated epilepsy seizures have a negative impact on quality of life, education, and employment, and also increase risk of early mortality (11). In addition, epilepsy as a chronic condition imposes a high economic burden on patients, their families, and society; in the US the total cost per year for medical expenditure and informal care for patients with epilepsy is estimated to be $9.6 billion (12). Some studies have described the burden associated with non-adherence to anti-seizure drugs, which can result in negative outcome for persons with epilepsy (13). Therefore, there is clear need to develop and use approaches that improve the ability of people with epilepsy to control their seizures and improve their health.
Epilepsy self-management includes a set of behavioral practices that are likely to help patients better control their seizures and that may positively impact their symptoms as well as prognosis (8, 14). These self-management techniques are often categorized into three areas of: (1) treatment management (e.g., medication adherence), (2) seizure management (e.g., keeping track of seizures), and (3) lifestyle management (e.g., getting regular sleep, engaging in safe physical activity) (7). Self-management techniques can reduce healthcare utilization by lowering the number of inpatient hospitalizations, inpatient days and visits to the Emergency Department (13). Since 2007, the U.S. Centers for Disease Control and Prevention’s (CDC) Prevention Research Centers (PRC) has funded the Managing Epilepsy Well (MEW) Network, a thematic research network whose mission is to advance the science of epilepsy self-management. CDC MEW Network programs are designed to enhance the quality of life of people with epilepsy (7). A key feature of MEW Network self-management approaches has been to incorporate consideration of comorbidity such as depression and to address the pervasive problem of cognitive impairment among individuals with epilepsy. The MEW Network currently comprises eight PRCs (connected with accredited schools of public health or schools of medicine with a preventive medicine residency program) that collaborate on epilepsy self-management research.
1.1 Motivation for integrated analysis of MEW Network data
The MEW Network was founded on principles of community-based participatory research, Each collaborating site develops its own capacity to conduct independent research with community partners to respond to variations in the needs and interests specific to these communities. Since 2007, each site has collected, securely stored, analyzed, and published their study findings. However, pooled data from both completed and ongoing MEW self-management studies represent valuable, untapped information relevant to the epilepsy self-management research community. Integrative analysis of these datasets will enable epilepsy researchers to gain new insights into various aspects of epilepsy self-management that may ultimately help people with epilepsy and their families. A MEW database can harness the power of aggregated data to better understand the different factors that impact people with epilepsy, for example identifying the association between seizure frequency and quality of life or depression. Indeed, effective secondary use of healthcare data for advancing research, a critical aspect of biomedical informatics, maximizes the value of existing data (15, 16). An integrated database containing common data collected by the MEW Network sites will potentially allow researchers to propose new studies on self-management techniques across subpopulations, geographical locations, and healthcare settings with sufficient statistical power.
There are many study-specific data repositories that store data using various approaches, including relational databases, MS Excel spreadsheet, and paper forms (17). These data repositories are conceptually similar to “data silos” with limited or no support for data sharing, integration, and secondary analysis. In addition, there is limited terminological standardization across different epilepsy research studies, which impedes the integrated analysis of data across different studies. Similar to initiatives such as the International Epilepsy Electrophysiology Portal (IEEG-Portal), which aims to create a database for epilepsy electrophysiological data (18), there is a clear need to develop an integrated and scalable database for epilepsy self-management research data. To meet this need, a MEW Network database workgroup was established in 2014 to explore the feasibility of an integrated epilepsy self-management database. The goals of the workgroup included establishment of an analytical platform that would eventually enable members of the MEW Network to interactively create cohorts of patients for secondary data analysis.
The consortium of researchers involved in this initiative included representatives from all the MEW Network collaborating centers. Since September 2014, a Steering Committee representing research data stakeholders, has developed a standardized process for sharing de-identified study data across the MEW Network through Data User Agreements (DUA) and provides continued oversight and guidance for the expansion, management and use of the MEW self-management study data for research purposes. The MEW Network is currently coordinated by Dartmouth College. While the MEW Network currently includes 8 collaborating centers, a total of 11 centers have been active during the extant tenure of the MEW Network, including previously and currently funded centers. These centers are: Emory University; the University of Texas Health Center at Houston; the University of Michigan; the Dartmouth Institute at the Geisel School of Medicine; New York University; the University of Arizona; the Morehouse College School of Medicine; the University of Illinois at Chicago; the University of Minnesota; the University of Washington at Seattle; and Case Western Reserve University (CWRU).
The development of the Insight MEW Network data analytics platform (hereafter called Insight) is led by CWRU. Data contribution from participating sites is purely voluntary and not required under the respective CDC PRC cooperative agreements. As of November 2015, data from five research studies have been integrated into Insight. The five research studies have a total of over 400 de-identified participants with more than 7,552 data points corresponding to 16 MEW Common Data Elements (CDEs) recommended for inclusion in study designs by the MEW Network to standardize terminology in the integrated database. Although Insight contains only de-identified data from the research studies, the data is stored using standard best practices in terms of data security, data accessibility, and maintenance of user access log data.
1.2 Aims of the Insight database
Insight is being developed as a national resource for the epilepsy self-management research community. To the best of our knowledge there is no existing epilepsy self-management data analysis platform similar to Insight, which together with the integrated dataset features a rich set of data exploration and query functions for easier data analysis by epilepsy researchers. The four primary goals of Insight are: (1) integrate data using MEW CDEs as standard terminology, (2) enable users to interactively explore the data using visualization functions, (3) provide an intuitive and flexible provenance-enabled query environment for users to perform cohort identification; and (4) serve as the basis of a proposed epilepsy self-management registry. Insight uses an epilepsy domain ontology called Epilepsy and Seizure Ontology (EpSO) (19) to support advanced data exploration, query composition, and execution strategies through use of ontology reasoning for “query unfolding” and for reconciling semantic heterogeneity. These techniques are described in detail in the next section.
2. Material and methods
Data from a MEW Network research study are added into Insight in accordance with a standardized protocol. After a DUA is completed between the Insight team and a specific MEW Network collaborating center, the study data is transferred to the Insight team either as Microsoft Excel or Comma Separated Value (CSV) files together with study protocols describing the provenance metadata (e.g., inclusion and exclusion criteria), and the study-specific data dictionary. No data is or will be used to identify any study participant. Using the data dictionary, we map the study data to the MEW CDEs and invoke the data transformation pipeline for integration and loading of data into the database. The data transformation pipeline is implemented as a flexible Extract Transform Load (ETL) workflow, which can be easily modified to incorporate the mappings between MEW CDEs and the data dictionaries of different research studies. Although there has been extensive work in data integration techniques (20, 21), there are no existing techniques that can completely automate the generation of correct mappings between two terminologies; therefore we manually generated mappings between the MEW CDEs and study-specific data dictionaries.
The MEW CDEs are the core component of the ETL workflow and are based on data elements recommended for epilepsy surveillance studies by the Institute of Medicine (IOM) (22), the CDEs developed by the National Institute of Neurological Disorders and Stroke (NINDS) (23), and terms defined in the Behavioral Risk Factor Surveillance System (BRFSS) Questionnaire (24). The MEW Network database initiative is using an incremental approach to define epilepsy self-management CDEs with 16 CDEs selected as Tier-1 variables (mainly demographic variables) and 15 additional CDEs approved as Tier-2 variables (mainly clinical outcome variables). The Insight database schema and the ETL workflow are designed to be easily extensible to incorporate additional CDEs as they are recommended for adoption by the MEW database Steering Committee. In addition to data transformation, the ETL workflow annotates the appropriate subset of study data with ontology classes defined in EpSO. This “semantic annotation” is used to support ontology-based reasoning during data exploration and query execution, which significantly improves the quality of query results (described in the following section).
The integrated database created by the ETL workflow is the first step towards supporting cross-cohort and multi-study data analysis. At present, most biomedical databases follow a cumbersome approach for data retrieval, which involves request for data by researchers to a database manager, retrieval of study cohort data or variable values by the manager, and data analysis by researchers to evaluate their hypothesis. However, if the researchers are not satisfied with the retrieved data, they have to resubmit their requests to the database manager and this process may involve multiple iterations. Clearly, there are several limitations to this approach, including significant time delay between data request and subsequent analysis, limited support for data exploration, and lack of tools to dynamically modify search criteria during cohort identification.
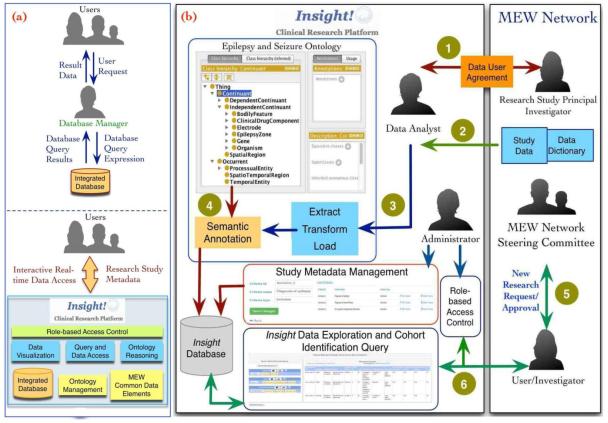
Insight has been designed to support an alternate approach that allows researchers to directly access data in the integrated database through an intuitive visual interface (Figure 1 (a) illustrates the differences in the two approaches). In the following section, we describe the implementation of the integrated database, the ontology-driven data exploration, and features of the visual query interface (Figure 1 (b) illustrates the information flow in Insight).
Figure 1.
(a): Traditional approaches to biomedical database require a database manager to retrieve data for secondary analysis, however Insight allows users to directly retrieve relevant data. Figure 1(b): Information and data flow implemented by the MEW Network database group is shown with steps covering completion of a Data User Agreement (DUA) to access by users. The complete workflow consists of 6 steps (marked with a numeric circle in the figure). Step 1 involves completion of a data user agreement between the MEW Network center and CWRU followed by mapping of CDEs to data dictionary terms of the new research study in Step 2. Step 3 consists of using the study-specific ETL to integrate the new study data into Insight. In Step 4, the data is annotated with EpSO before storage in the database. Once the MEW Network coordination committee approves a new research study in Step 5, an investigator can access the integrated data using the Insight user interface in Step 6.
2.1 Implementation of the integrated database
The integrated data is stored in a relational database using MySQL (version 5.6.16). The tables store: (a) configuration details (e.g., values corresponding to the MEW CDEs, and user profile information), and (b) study data (e.g., visit information, and epilepsy seizure details). The configuration details include: the inclusion/exclusion criteria for each research study; each study protocol; the mappings from the ontology classes to the study values; the ontology class hierarchy (used to support the multi-level drop down menus in the visual query widgets and query unfolding during query execution); and the mappings between de-identified study identifier and study name. The user credentials and user groups with well-defined access privileges (as determined by the MEW database Steering Committee) allows the implementation of a flexible and easy to maintain Role-based Access Control (RBAC) functionality in Insight.
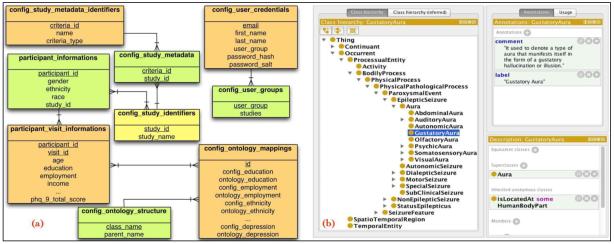
In contrast to a naïve approach of assigning access privileges to each individual user, which is difficult to modify and maintain with large number of users, granting access to user groups allows systematic maintenance of access privileges for all user assigned to a specific user groups. The research study data are stored in 2 tables with unique identifier of the participant (generated by Insight to uniquely identify participants), the source study of the participant, and demographic information. The data collected at different time points in a clinical trial corresponding to the research design of individual studies (e.g., baseline, interim, and follow-up visits) are stored in a separate table. Figure 2(a) shows the current entity-relationship diagram of the Insight database and Figure 2(b) shows a screenshot of the epilepsy ontology class hierarchy used to support query composition and execution.
Figure 2.
(a): The Entity-Relationship (ER) diagram used for the Insight database is shown consisting of tables to store configuration information and study data. Figure 2(b): A screenshot of the Epilepsy and Seizure Ontology (EpSO) class hierarchy is shown with details of various types of epileptic seizures, including Aura, Autonomic Seizures, Dialeptic Seizures, and Motor Seizures.
2.2 Research studies and data elements
The current version of the Insight database integrates data from five completed MEW Network research studies (as of November 2015) with a total of over 400 participants. The five MEW Network research studies in the Insight database are: (a) Targeted Self-Management for Epilepsy and Mental Illness (TIME) study, (b) WebEase (Epilepsy Awareness, Support, and Education) study (25), (c) F.O.C.U.S. on Epilepsy: pilot and randomized control trial (RCT) studies, and (d) Home-Based Self-management and Cognitive Training Changes Lives (HOBSCOTCH) study (26). The TIME study tested an intervention consisting of in-person group sessions for adults with epilepsy and co-morbid serious mental illness, such as severe depression, bipolar disorder or schizophrenia. The on-line WebEase Program, which was tested in a RCT, uses principles of the trans-theoretical model of behavioral change, social behavioral theory and motivational interviewing, to provide tailored and responsive communications, that adapt activities and resources to meet the user’ readiness to change, self-management needs and confidence in behavior change. WebEase guides the adoption of favorable medication adherence, stress reduction, and sleep management behaviors by the adult with epilepsy (27). The FOCUS study tested an intervention in adults with epilepsy and member of their social support network. The intervention goal was teach self-management techniques via a workshop, telephonic coaching sessions, and workbooks leveraging the patient’s social support (25). The HOBSCOTCH program targets memory and cognitive problems in adults with epilepsy. The HOBSCOTCH is a phone-delivered program, which was tested in a small pilot RCT (28). All studies integrated into the database used a repeated measures research design, with data points collected at discrete time intervals. For this report, only baseline data points from the studies are featured.
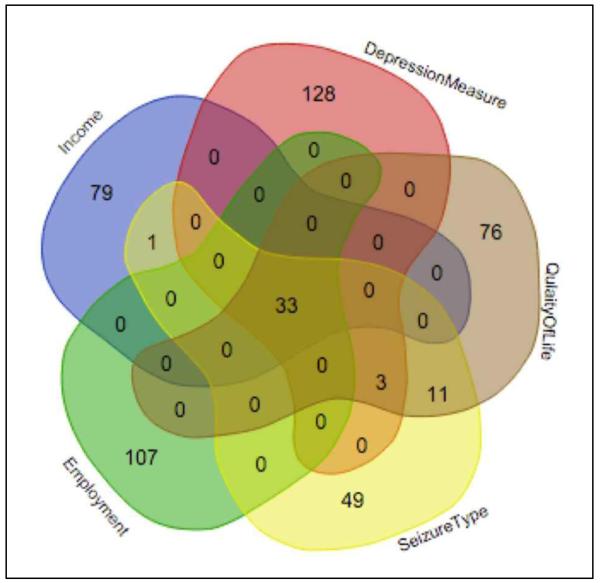
The data from these research studies are mapped and transformed to the MEW CDEs by the study-specific ETL workflow. The Tier-1 MEW CDEs can be divided into five categories: (a) demographics, (b) seizure details, (c) quality of life, (d) depression, and (e) other health measures. Table 1 shows the data categories, the variables corresponding to each data category, the number of data points, and the data value corresponding to each variable. Table 1 also shows the distribution of participants across the five studies (the FOCUS Pilot and RCT values are aggregated into single set of values). Figure 3 is a Venn diagram showing the distribution of the number of patients corresponding to quality of life, depression, income, employment status, and seizure frequency values. Figure 3 shows that there is overlap between some of the measures, including Income, Quality of Life, and Depression measures (PHQ-9). In the next section, we describe the role of the epilepsy domain ontology in data integration, query composition, and query execution features of Insight.
Table 1.
The distribution of data points corresponding to five data categories and their Managing Epilepsy Well Common Data Elements in the Insight database
| Data Category | Study Variables | Number of Data Points |
MEW Network Studies | Total Number of Participants |
|||
|---|---|---|---|---|---|---|---|
| WebEase Study |
FOCUS Study (Pilot and RCT studies) |
TIME Study |
HOBSCOTCH Study |
||||
|
Socioeconomic -Demography |
Gender | 430 | |||||
| Male | 39 (26%) | 69 (38%) | 18 (41%) | 18 (31%) | 144 (33%) | ||
| Female | 109 (74%) | 111 (63%) | 26 (59%) | 40 (69%) | 286 (67%) | ||
| Race | 372 | ||||||
| White | 132 (89%) | 140 (78%) | 16 (36%) | - | 288 (77%) | ||
| Black/African- American |
2 (1%) | 22 (12%) | 25 (57%) | - | 49 (13%) | ||
| Asian | 3 (2%) | 0 | 0 | - | 3 (1%) | ||
| American Indian or Alaska Native |
0 | 1 (1%) | 3 (7%) | - | 4 (1%) | ||
| Other* | 11 (8%) | 17 (9%) | 0 | - | 28 (8%) | ||
| Ethnicity | 372 | ||||||
| Hispanic or Latino |
7 (5%) | 9 (5%) | 3 (7%) | - | 19 (5%) | ||
| Not Hispanic or Latino |
141 (95%) | 170 (94%) | 41 (93%) | - | 352 (95%) | ||
| Other | 0 | 1 (1%) | 0 | - | 1 (1%) | ||
| Education | 430 | ||||||
| Never attended school or only attended kindergarten |
0 | 0 | 0 | 0 | 0 | ||
| Grades 1 through 8 (Elementary) |
0 | 1 (1%) | 0 | 0 | 1 (1%) | ||
| Grades 9 through 11 (Some high school) |
1 (1%) | 3 (1%) | 11 (25%) | 0 | 15 (3%) | ||
| Grade 12 or GED (High school graduate) |
19 (13%) | 33 (18%) | 8 (18%) | 27 (47%) | 87 (20%) | ||
| College 1 year to 3 years (Some college or technical school) |
68 (46%) | 61 (34%) | 21 (48%) | 0 | 150 (35%) | ||
| College 4 years or more (College graduate) |
58 (39%) | 75 (42%) | 4 (9%) | 30 (52%) | 167 (39%) | ||
| Other | 2 (1%) | 7 (4%) | 0 | 1 (1%) | 10 (2%) | ||
| Income | 224 | ||||||
| Less than $25K | - | 81 (45%) | 42 (95%) | - | 123 (55%) | ||
| $25,000–$49,999 | - | 16 (9%) | 2 (5%) | - | 18 (8%) | ||
| $50,000 or greater |
- | 7 (4%) | 0 | - | 7 (3%) | ||
| Other | - | 76 (42%) | 0 | - | 76 (34%) | ||
| Employment | 430 | ||||||
| Self-employed | 1 (1%) | 1 (1%) | 0 | 0 | 2 (1%) | ||
| Employed for wages |
76 (51%) | 68 (38%) | 3 (7%) | 21 (36%) | 168 (39%) | ||
| Retired | 3 (2%) | 17 (9%) | 5 (11%) | 0 | 25 (6%) | ||
| Student | 5 (3%) | 6 (3%) | 1 (2%) | 0 | 12 (3%) | ||
| Homemaker | 3 (2%) | 8 (4%) | 4 (9%) | 0 | 15 (3%) | ||
| Do not work/Disability |
4 (3%) | 48 (27%) | 19 (43%) | 0 | 71 (17%) | ||
| Out of work for less than 1 year |
0 | 8 (4%) | 2 (5%) | 0 | 10 (2%) | ||
| Out of work for 1 year or more |
0 | 19 (11%) | 10 (23%) | 37 (64%) | 66 (15%) | ||
| Other | 56 (38%) | 5 (3%) | 0 | 0 | 61 (14%) | ||
|
Relationship
Status |
192 | ||||||
| Married | 69 (47%) | - | 4 (9%) | - | 73 (38%) | ||
| Member of an unmarried couple |
12 (8%) | - | 2 (5%) | - | 14 (7%) | ||
| Never married | 36 (24%) | - | 21 (48%) | - | 57 (30%) | ||
| Divorced | 22 (15%) | - | 14 (32%) | - | 36 (19%) | ||
| Widowed | 3 (2%) | - | 1 (2%) | - | 4 (2%) | ||
| Separated | 1 (1%) | - | 2 (4%) | - | 3 (2%) | ||
| Refused/ Unknown/Other |
5 (3%) | - | 0 | - | 5 (2%) | ||
| Seizure Details | Type of Seizure | 250 | |||||
| Partial Seizure | 65 (44%) | - | 7 (16%) | 49 (85%) | 121 (48%) | ||
| Generalized Seizure |
76 (51%) | - | 25 (57%) | 9 (15%) | 110 (44%) | ||
| Other | 7 (5%) | - | 12 (27%) | 0 | 19 (8%) | ||
|
Health
Measures |
Health Status | 44 | |||||
| Excellent | - | - | 1 (2%) | - | 1 (2%) | ||
| Very good | - | - | 4 (9%) | - | 4 (9%) | ||
| Good | - | - | 9 (20%) | - | 9 (20%) | ||
| Fair | - | - | 20 (46%) | - | 20 (46%) | ||
| Poor | - | - | 7 (16%) | - | 7 (16%) | ||
| Don't know/not sure |
- | - | 3 (7%) | - | 3 (7%) | ||
|
Quality of Life
Measure |
QOLIE-10 | 2182 | |||||
| Average total value (Baseline) |
2.59 | 3.4 | 2.9 | - | 3.0 | ||
|
Depression
Measure |
PHQ-9 | 2626 | |||||
| Average total value (Baseline) |
- | 8.2 | 10.9 | 9.3 | 9.1 | ||
Other includes additional or non-standard categories, missing data or results that are not interpretable QOLIE-10: 10-item quality of life measure, lower scores indicate better quality PHQ-9: Patient Health Questionnaire, lower scores indicate less depression
Figure 3.
The Venn diagram shows the data points corresponding to 437 participants in different categories of MEW CDE. The data categories are Income, Employment, Seizure Type, Depression Measure (PHQ-9), and Quality of Life Measure (QOLIE-10) (the diagram was created using tool available at: http://bioinformatics.psb.ugent.be/webtools/Venn)
2.3 Role of the Epilepsy and Seizure Ontology
The Epilepsy and Seizure Ontology (EpSO) is an epilepsy domain ontology that has been developed by an interdisciplinary team of clinical researchers and computer scientists to support various data management tasks in epilepsy research (19). A domain ontology is a formal knowledge representation structure that models domain information for consistent and correct interpretation of data by software applications (29). Domain ontologies, such as the Gene Ontology (GO) (30) and Human Phenotype Ontology (HPO) (31), play a key role in reconciling data heterogeneity, harmonizing data from different sources, and ensure the “completeness” of query results using ontology reasoning (32). EpSO is being developed as a domain ontology using the well-known four-dimensional classification of epilepsy and seizures with information about seizures, anatomical locations, etiology, and related medical conditions (19). In addition, EpSO also models terms describing medication, genes (with known association to epilepsy syndromes), electrophysiological signal features, and the MEW CDEs. We use various types of ontology modeling constructs to represent domain-specific information in EpSO. For example, ontology class-level restrictions are used to model information about preferred medication, paroxysmal events, and signal features associated with different epilepsy syndromes.
Using the description logic-based Web Ontology Language (OWL2) features (33), EpSO can support identification of equivalent terms with different syntactic labels (e.g., using synonym information and commonly used acronyms), development of user friendly visual interface (e.g., using textual description of ontology terms), and improvement in quality of query results through “query unfolding” technique using reasoning. The query unfolding strategy uses the ontology class hierarchy together with OWL2 subsumption reasoning to expand a user query expression by including all the appropriate subclasses of a query term in the query expression. This enables the query execution module to retrieve results corresponding to query term and all its sub classes. For example, if a cohort identification query includes “Aura” as a type of seizure, the Insight query execution module automatically expands the query expression to include all its sub classes such as “Auditory Aura”, “Gustatory Aura”, etc. Many biomedical database query execution approaches rely only on exact string matching between the query term and the data values, and therefore query result is often incomplete. This limitation is addressed in Insight through use of EpSO class hierarchy during query execution.
Given the key role of EpSO in Insight, the ontology management module supports: (a) automated updates of EpSO OWL file as soon as a new version of the ontology is released; and (b) semantic annotation of research study data as part of the ETL workflow (described earlier). The automated update feature uses a remote Web service invocation to retrieve the ontology OWL file, which is parsed using the open source OWL Application Programming Interface (OWLAPI) (34). The OWLAPI was developed to allow users to programmatically create and modify OWL ontologies with support for parsing, validation of OWL2 profiles, and use of reasoners. The parsed ontology classes together with the class structure information are used to update the Insight database. In addition to query unfolding, Insight uses the EpSO class hierarchy to support the data exploration and query composition functionalities also, which are described in the following section.
2.4 Data exploration, visualization, and query composition
Insight allows users to interactively explore the different study variables stored in the database and perform queries to create research cohorts across different MEW research studies for subsequent analysis. This module was implemented using the Ruby on Rails (RoR) Web application framework that uses Model View Controller (MVC) architecture. The MVC architecture facilitates the logical separation of different components of Insight for easier maintenance and extension with new functionalities. Using agile software engineering principles, we developed Insight over multiple iterations with user-guided prioritization of functionalities and frequent user feedback. Agile development technique allowed evolutionary development of Insight with easier incorporation of requested changes and higher user satisfaction through early availability of working software (35). Insight can be accessed at http://mew.meds.cwru.edu. After logging into Insight, the user accesses the data exploration and query interface.
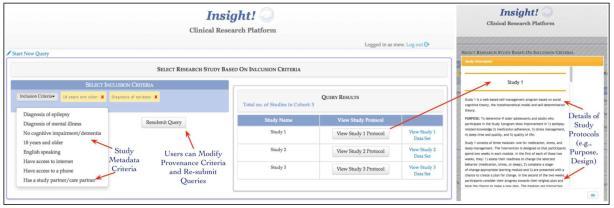
Based on user recommendations, the data access and query process is divided into two phases. In the first phase, users can select the most appropriate research study for data exploration and cohort identification by filtering research studies based on their provenance metadata (e.g., inclusion and/or exclusion criteria). The study metadata is manually extracted from the protocol description of each research study, categorized into specific categories, and made available to the user as “provenance metadata widgets”. The provenance widget consists of visual interactive query composition features. Provenance information describes the history or lineage of data, which enables accurate interpretation of data in the correct context (36). Therefore, the provenance of the research studies allows users to select research studies that meet the requirements of their research hypothesis and create research cohorts with comparable data points. After a user selects the provenance values in the study, metadata query widgets and the resulting research studies are displayed in the “Query Result” window with links provided for the user to view the complete study protocol (Figure 4). Users can also download all the data corresponding to a particular research study in the “Query Result” window without completing the second phase of querying. In addition, users can also modify their provenance metadata values in the query widgets to refine their research study search queries.
Figure 4.
A screenshot of the first phase of querying using provenance metadata of the research studies using a query widget with editable values. The users can also review a detailed description of study protocol of a research study.
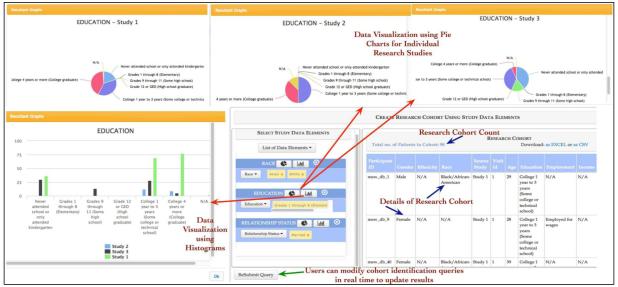
In the second phase, users can create a research cohort from the research studies selected in the first phase by selecting an appropriate MEW CDE from a “drop-down” menu. Each MEW CDE is implemented as an individual query widget and users can select one or more values using either a drop-down menu or input interface for numeric values (as shown in Figure 5). The values selected by the user in each MEW CDE query widget are displayed as “editable tags” and these tags can be easily deleted to modify the cohort query.
Figure 5.
The data exploration features of Insight allows users to view the distribution of data values corresponding to a MEW CDE in real time and helps them to compose appropriate cohort identification queries. The cohort identification query is composed using a set of visual query widgets with editable values and the details of the resulting research cohort is also available for viewing.
To help the users to select the most relevant values corresponding to a MEW CDE, Insight features intuitive data visualization functionality. The data visualization functionality allows users to explore the distribution of data values across multiple research studies by selecting an option from different graphic visualization options such as histogram or pie chart. These visualization features allow users to select the most appropriate MEW CDE values that lead to inclusion of greater or smaller number of participants in their research cohorts. For example, the visualization feature may show that most of the study participants in the Insight database have “college or technical school” qualification corresponding to “education” CDE. Therefore, users can select “college or technical school” to ensure that their research cohort has maximum number of participants. Figure 5 shows histogram and pie charts corresponding to three MEW studies for the “education” MEW CDE. Figure 5 shows the research cohort corresponding to the cohort identification query composed by the user. The users can access the result cohort using features of the “result explorer”, which allows users to download the research cohort data as an MS Excel spreadsheet or CSV file for further analysis. In the following section, we discuss some of the planned extensions and challenges currently being addressed in the MEW Network database initiative.
3. Discussion
As the Insight system is expanded to include data from additional MEW Network studies, there is increasing interest in effectively using the integrated datasets to gain new insights into epilepsy neurological disorder and self-management techniques..
3.1 Application of the Insight platform in epilepsy self-management research
The integrated dataset together with the various features supported by the Insight platform can provide useful support to the MEW Network in advancing it’s mission and plan future research priorities. For example, preliminary descriptive data extracted from the database suggests that women with epilepsy make up the largest sub population in the MEW Network enrolled sample with mean value of 0.67 women and 0.33 men for data aggregated from four research studies (Table 1). Table 1 also describes the study-specific distribution of women and men. This finding is perhaps not surprising given that women may be more likely than men to seek help for chronic health conditions (37). However, the data on epilepsy does not suggest a clear gender susceptibility for most forms of epilepsy (38). Thus, men with epilepsy appear to be inadequately represented in the samples collected by various MEW Network studies. This insight gained from the integrated dataset can form the basis for devising and implementing strategies for a more gender-balanced sampling approach. This will provide truly representative data findings for people with epilepsy with respect to different self-management techniques.
3.2 Towards dynamic integration of data from ongoing MEW Network research studies
At present, the Insight ETL workflow is executed in a “batch mode” to integrate data from completed research studies. In contrast to completed studies, data from ongoing research studies need to be integrated into Insight as a continuous data feed. In future, we propose to use a Service Oriented Architecture (SOA) that consists of RESTful service endpoints hosted at each MEW Network collaborating center on a dedicated server to transfer de-identified study data. The service endpoint can be remotely queried by Insight using a secure connection for new data and allow easy transfer of data to Insight. We believe that this approach (based on “pull” instead of “push”) will allow integration of a continuous data feed from ongoing research studies with minimal additional effort by the researchers at different MEW centers.
The ETL workflow will be extended to process data from multiple research studies simultaneously. Although we do not expect the size of data from different research studies to be large, we will implement a parallelized ETL workflow using scalable Hadoop technology, for example MapReduce (39) deployed on a cluster-computing infrastructure to process data from multiple studies simultaneously. In addition, the ETL workflow will also be extended to incorporate the Tier-2 MEW CDEs, which have recently been approved by the MEW Network database steering committee. The new Tier-2 CDEs will also be incorporated in EpSO as ontology classes and these classes will be used for semantic annotation of the data.
3.3 Formal representation of research study provenance metadata
The current implementation of Insight does not structure the research study metadata into specific categories and only presents them as list of inclusion or exclusion criteria. There has been extensive work on formalization of research study protocols (40, 41), for example the Ontology for Clinical Research (OCRe) that models the details of study design and eligibility criteria (42). The OCRe project has also developed an annotation pipeline called ERGO, which is used for formal representation of eligibility criteria of research studies. We propose to extend this work together with standard representation model of provenance, such as the new PROV specifications developed by the World Wide Web Consortium (W3C) (43), to model the research study metadata in Insight. This formal representation of research study metadata will enable users to systematically select inclusion and exclusion criteria during the first phase of querying in Insight.
In addition, the formal representation of study provenance will also enable reasoning over metadata values from multiple research studies for cross-study cohort identification queries. We expect that our experience with formal modeling of study provenance will allow us to provide appropriate feedback to the MEW Network collaborating centers in terms of capturing relevant study metadata for subsequent secondary data analysis in Insight. The use of W3C PROV specifications to model study provenance will also make it easier for Insight to interface with existing biomedical datasets on the Web, such as the Linked Open Data (LOD) (44), which also use PROV to model provenance information. For example, Insight can retrieve data about epilepsy related genes (together with its available provenance information) from LOD.
Limitations
The MEW Network was not funded as a centralized entity with standardized procedures and standard preassigned data elements; therefore interpretation of results is limited by heterogeneity at the micro and macro-level. Investigators using Insight should read through the specific study protocols carefully to understand how samples may differ based upon enrollment criteria, study design and study implementation. Data harmonization procedures may obscure the finer-grained detail that a specific individual study has identified and results can differ from individual study reports because of the way missing or outlier data is handled or analyzed. However, the MEW database represents an innovative approach for using data that takes advantage of the strengths of larger samples to identify patterns or outcomes that would not be possible with smaller studies. Even for studies where efficacy findings are negative (such as the FOCUS RCT study), the collected data is still useful to evaluate other potentially important clinical questions about epilepsy self-management.
4. Conclusion
In this paper, we described the development of Insight, an integrated database and analytical platform for the MEW Network collaborating centers to facilitate cross-study cohort identification for aggregate analysis. This approach effectively leverages the valuable epilepsy self-management data being collected across the U.S. Insight is the first provenance-enabled epilepsy data integration and analysis system that allows users to explore and perform research cohort identification. In addition, Insight uses a novel epilepsy domain ontology called EpSO for semantic annotation, data integration, and query execution. The EpSO class structure is used for implementing a “query unfolding” strategy, which ensures completeness of the cohort identification query results. Insight also features an ontology management module to automatically update the database with each new release of EpSO and provides fine-granularity access control through a flexible RBAC feature.
We aim to develop Insight as a national informatics resource for the epilepsy self-management research community to allow advanced data analytics and research cohort identification queries. The Insight database is already being used to support analysis by MEW Network investigators who aim to understand the correlation between quality of life and other clinical variables among persons with epilepsy. As new MEW Network research studies are added to Insight, we believe the MEW Network researchers can evaluate novel research hypotheses with sufficient statistical power and data variety to advance our understanding of self-management techniques for epilepsy.
Highlights.
Insight is an ontology-driven integrated database to aggregate epilepsy self management research data from different sites
Provenance-aware cohort query identification for cross-study data analysis
Interactive user interface with features for data exploration and visualization to support cohort identification query
Managing Epilepsy Well (MEW) Network consists of 8 collaborating centers across the US that conduct independent studies in epilepsy self management techniques
The analysis of the integrated data set provides new insights to researchers involved in epilepsy self management techniques
Acknowledgement
The authors thank the members of the MEW Network database Steering Committee for their continuing support in developing Insight. We also thank Rosemarie Kobau and Matthew Zack from the CDC for reviewing our manuscript and providing valuable comments
Funding
This work is supported in part by the Centers for Disease Control and Prevention (CDC) (grant#U48DP005030 and U48DP001930), and the National Institutes of Biomedical Imaging and Bioengineering (NIBIB) Big Data to Knowledge (BD2K) grant (1U01EB020955).
Glossary of Terms
- Managing Epilepsy Well (MEW) Network
is a thematic research network whose mission is to advance the science of epilepsy self-management.
- Common Data Elements (CDE)
is a set of well-defined terms used consistently across multiple research studies to reduce data heterogeneity, improve data quality, and facilitate data integration.
- Ruby on Rails (RoR)
is a web development framework that uses Model View Controller (MVC) architecture together with Ruby language for rapid application development
- Web Ontology Language Application Programming Interface (OWLAPI)
was developed to allow users to programmatically create and modify OWL ontologies with support for parsing, validation of OWL2 profiles, and use of reasoners.
- Epilepsy and Seizure Ontology (EpSO)
is a domain ontology developed to formally model terms associated with the well-known four-dimensional classification of epilepsy consisting of seizure, anatomical location, etiology, and related medical conditions.
- Role-based Access Control (RBAC)
is an approach to manage accessibility of users to computational resources based on their role.
- U.S. Centers for Disease Control and Prevention’s (CDC)
is a federal organization in the United States focused on national public health through control and prevention of diseases.
- Prevention Research Centers (PRC)
is a network of research centers funded by the US CDC to study techniques to address risks of chronic illness such as obesity, cancer, and heart disease.
- Data User Agreements (DUA)
is an agreement for transfer and usage of data across different entities, for example research centers or universities.
Footnotes
Author Contributions:
SSS, PR, MS designed the overall architecture of Insight with user input from EW. PR, SSS, JV implemented the software platform with data collection and curation done by AB, EW, YB, SS, BCJ, and CT. All co-authors contributed to writing, reviewing, and editing the final manuscript.
Conflict of interest: None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Robinson E, DiIorio C, DePadilla L, McCarty F, Yeager K, Henry T, Schomer D, Shafer P. Psychosocial predictors of lifestyle management in adults with epilepsy. Epilepsy & Behavior. 2008;13(3):523–8. doi: 10.1016/j.yebeh.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DiIorio C, Shafer PO, Letz R, Henry TR, Schomer DL, Yeager K, Project EASE study group Project EASE: a study to test a psychosocial model of epilepsy medication managment. Epilepsy & Behavior. 2004;5(6):926–36. doi: 10.1016/j.yebeh.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 3.DiIorio C, Faherty B, Manteuffel B. Epilepsy self-management: Partial replication and extension. Research in Nursing & Health. 1994;17:167–74. doi: 10.1002/nur.4770170304. [DOI] [PubMed] [Google Scholar]
- 4.DiIorio C, Hennessy M, Manteuffel B. Epilepsy self-management: A test of a theoretical model. Nursing Research. 1996;45:211–7. doi: 10.1097/00006199-199607000-00004. [DOI] [PubMed] [Google Scholar]
- 5.Institute of Medicine (IOM) Report. Living Well with Chronic Illness: A Call for Public Health Action. 2012.
- 6.Dua T, de Boer HM, Prilipko LL, Saxena S. Epilepsy Care in the World: results of an ILAE/IBE/WHO Global Campaign Against Epilepsy survey. Epilepsia. 2006;47(7):1225–31. doi: 10.1111/j.1528-1167.2006.00595.x. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention [Retrieved on December 24, 2015]; Available from: http://www.cdc.gov/
- 8.DiIorio C, Henry M. Self-management in persons with epilepsy. Journal of Neuroscience Nursing. 1995;27(6):338–43. doi: 10.1097/01376517-199512000-00004. [DOI] [PubMed] [Google Scholar]
- 9.Kendall S, Thompson D, Couldridge L. The information needs of carers of adults diagnosed with epilepsy. Seizure. 2004;13(7):499–508. doi: 10.1016/j.seizure.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Hauser WA, Lee JR. Do seizures beget seizures? Progress in Brain Research. 2002;135:215–9. doi: 10.1016/s0079-6123(02)35021-0. [DOI] [PubMed] [Google Scholar]
- 11.Kobau R, Zahran H, Thurman DJ, Zack MM, Henry TR, Schachter SC, Price PH, Centers for Disease Control and Prevention (CDC) Epilepsy surveillance among adults--19 States, Behavioral Risk Factor Surveillance System, 2005. Morbidity and Mortality Weekly Report Surveillance Summary. 2008;57(6):1–20. [PubMed] [Google Scholar]
- 12.Yoon D, Frick KD, Carr DA, Austin JK. Economic impact of epilepsy in the United States. Epilepsia. 2009;50:2186–91. doi: 10.1111/j.1528-1167.2009.02159.x. [DOI] [PubMed] [Google Scholar]
- 13.Faught RE, Weiner JR, Guérin A, Cunnington MC, Duh MS. Impact of nonadherence to antiepileptic drugs on health care utilization and costs: findings from the RANSOM study. Epilepsia. 2009;50(3):501–9. doi: 10.1111/j.1528-1167.2008.01794.x. [DOI] [PubMed] [Google Scholar]
- 14.Cole KA, Gaspar PM. Implementation of an epilepsy self-management protocol. Journal of Neuroscience Nursing. 2015;47(1):3–9. doi: 10.1097/JNN.0000000000000105. [DOI] [PubMed] [Google Scholar]
- 15.Holdren JP, Lander E. PCAST Report. Washington, D.C.: 2010. Realizing the Full Potential of Health Information Technology to Improve Healthcare for Americans: The Path Forward. [Google Scholar]
- 16.Murphy S, Mendis ME, Berkowitz DA, Kohane I, Chueh H, editors. Integration of clinical and genetic data in the i2b2 architecture. AMIA Annu Symp Proc; 2006. [PMC free article] [PubMed] [Google Scholar]
- 17.Sahoo SS, Zhang GQ, Bamps Y, Fraser R, Stoll S, Lhatoo SD, Tatsuoka C, Sams J, Welter E, Sajatovic M. Managing information well: Toward an ontology-driven informatics platform for data sharing and secondary use in epilepsy self-management research centers. Health Informatics Journal. 2015:1–14. doi: 10.1177/1460458215572924. [DOI] [PubMed] [Google Scholar]
- 18.Wagenaar JB, Brinkmann BH, Ives Z, Worrell GA, Litt B. A Multimodal Platform for Cloud-based Collaborative Research. 6th International IEEE/EMBS Conference on Neural Engineering (NER); San Diego, CA. IEEE; 2013. pp. 1386–9. [Google Scholar]
- 19.Sahoo SS, Lhatoo SD, Gupta DK, Cui L, Zhao M, Jayapandian C, Bozorgi A, Zhang GQ. Epilepsy and seizure ontology: towards an epilepsy informatics infrastructure for clinical research and patient care. Journal of American Medical Informatics Association. 2014;21(1):82–9. doi: 10.1136/amiajnl-2013-001696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doan A, Halevy A, Ives Z. Principles of Data Integration. Morgan Kaufmann; Waltham, MA: 2012. [Google Scholar]
- 21.Mena E, Illarramendi A, Kashyap V, Sheth A. OBSERVER: An Approach for Query Processing in Global Information Systems based on Interoperation across Pre-existing Ontologies. Distributed and Parallel Databases (DAPD) 2000;8(2):223–71. [Google Scholar]
- 22.Hesdorffer DC, Begley CE. Surveillance of epilepsy and prevention of epilepsy and its sequelae: lessons from the Institute of Medicine report. Curr Opin Neurol. 2013;26(2):168–73. doi: 10.1097/WCO.0b013e32835ef2c7. [DOI] [PubMed] [Google Scholar]
- 23.Loring DW, Lowenstein DH, Barbaro NM, Fureman BE, Odenkirchen J, Jacobs MP, Austin JK, Dlugos DJ, French JA, Gaillard WD, Hermann BP, Hesdorffer DC, Roper SN, Van Cott AC, Grinnon S, Stout A. Common data elements in epilepsy research: development and implementation of the NINDS epilepsy CDE project. Epilepsia. 2011;52(6):1186–91. doi: 10.1111/j.1528-1167.2011.03018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.US Center for Disease Control and Prevention (CDC) [Retrieved on December 24, 2015];Behavioral Risk Factor Surveillance System (BRFSS) 2012. Available from: http://www.cdc.gov/brfss/about/brfss_today.htm.
- 25.DiIorio CK, Bamps YA, Edwards AL, Escoffery C, Thompson NJ, Begley CE, Shegog R, Clark NM, Selwa L, Stoll SC, Fraser RT, Ciechanowski P, Johnson EK, Kobau R, Price PH. Managing Epilepsy Well Network. The prevention research centers' managing epilepsy well network. Epilepsy & Behavior. 2010;19(3):218–24. doi: 10.1016/j.yebeh.2010.07.027. [DOI] [PubMed] [Google Scholar]
- 26.Caller TA, Ferguson RJ, Roth RM, Secore KL, Alexandre FP, Zhao W, Tosteson TD, Henegan PL, Birney KA, Jobst BC. A cognitive-behavioral intervention (HOBSCOTCH) improves quality of life and attention in epilepsy: a pilot study. Epilepsy and Behavior. 2016 doi: 10.1016/j.yebeh.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 27.DiIorio C, Escoffery C, Yeager KA, McCarty F, Henry TR, Koganti A, Reisinger E, Robinson E, Kobau R, Price P. WebEase: development of a Web-based epilepsy self-management intervention. Preventing chronic illness. 2009;6(1):A28. [PMC free article] [PubMed] [Google Scholar]
- 28.Caller T, Secore K, Ferguson R, Roth R, Alexandre F, Harrington J, Jobst B. A Pilot Study of a Self-Management Intervention for Cognitive Impairment in Epilepsy Neurology. 2014;82(10 Supplement S43.001) [Google Scholar]
- 29.Bodenreider O, Stevens R. Bio-ontologies: Current trends and future directions. Briefings in Bioinformatics. 2006;7(3):256–74. doi: 10.1093/bib/bbl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Köhler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, Black GC, Brown DL, Brudno M, Campbell J, FitzPatrick DR, Eppig JT, Jackson AP, Freson K, Girdea M, Helbig I, Hurst JA, Jähn J, Jackson LG, Kelly AM, Ledbetter DH, Mansour S, Martin CL, Moss C, Mumford A, Ouwehand WH, Park SM, Riggs ER, Scott RH, Sisodiya S, Van Vooren S, Wapner RJ, Wilkie AO, Wright CF, Vulto-van Silfhout AT, de Leeuw N, de Vries BB, Washingthon NL, Smith CL, Westerfield M, Schofield P, Ruef BJ, Gkoutos GV, Haendel M, Smedley D, Lewis SE, Robinson PN. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42(Database Issue):966–74. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sahoo SS, Bodenreider O, Rutter JL, Skinner KJ, Sheth AP. An ontology-driven semantic mashup of gene and biological pathway information: Application to the domain of nicotine dependence. Journal of Biomedical Informatics. 2008;41(5 (Semantic Mashup of Biomedical Data)):752–65. doi: 10.1016/j.jbi.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hitzler P, Krötzsch M, Parsia B, Patel-Schneider PF, Rudolph S. OWL 2 Web Ontology Language Primer. World Wide Web Consortium W3C; 2009. [Google Scholar]
- 34.Horridge M, Bechhofer S. The OWL API: A Java API for OWL Ontologies. Semantic Web Journal. 2011;2(1):11–21. [Google Scholar]
- 35.Beck K, Beedle M, Bennekum AV, Cockburn A, Cunningham W, Fowler M, Grenning J, et al. Manifesto for agile software development. 2001.
- 36.Sahoo SS, Nguyen V, Bodenreider O, Parikh P, Minning T, Sheth AP. A unified framework for managing provenance information in translational research. BMC Bioinformatics. 2011;12(461) doi: 10.1186/1471-2105-12-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Dea B, Glozier N, Purcell R, McGorry PD, Scott J, Feilds KL, Hermens DF, Buchanan J, Scott EM, Yung AR, Killacky E, Guastella AJ, Hickie IB. A cross-sectional exploration of the clinical characteristics of disengaged (NEET) young people in primary mental healthcare. BMJ Open. 2014;4(12) doi: 10.1136/bmjopen-2014-006378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perucca P, Camfield P, Camfield C. Does gender influence susceptibility and consequences of acquired epilepsies? Neurobiology of Disease. 2014;72(Pt. B):125–30. doi: 10.1016/j.nbd.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 39.Dean J, Ghemawat S. MapReduce: a flexible data processing tool. Communications of the ACM. 2010;53(1):72–7. [Google Scholar]
- 40.Weng C, Wu X, Luo Z, Boland MR, Theodoratos D, Johnson SB. EliXR: an approach to eligibility criteria extraction and representation. Journal of American Medical Informatics Association. 2011;18(Suppl 1):116–24. doi: 10.1136/amiajnl-2011-000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milian K, Teije A, editors. Towards automatic patient eligibility assessment: from free-text criteria to queries. 14th Conference on Artificial Intelligence in Medicine, AIME 2013; Murcia, Spain. 2013; Springer; [Google Scholar]
- 42.Sim I, Tu SW, Carini S, Lehmann HP, Pollock BH, Peleg M, Wittkowski KM. The Ontology of Clinical Research (OCRe): an informatics foundation for the science of clinical research. Journal of Biomedical Informatics. 2014;52:78–91. doi: 10.1016/j.jbi.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moreau L, Missier P. PROV Data Model (PROV-DM) World Wide Web Consortium W3C; 2013. [Google Scholar]
- 44.Bizer C, Heath T, Berners-Lee T. Linked Data - The Story So Far. International Journal on Semantic Web and Information Systems. 2009 Special Issue on Linked Data. [Google Scholar]