Abstract
Transposable elements (TEs) comprise large fractions of many eukaryotic genomes and imperil host genome integrity. The host genome combats these challenges by encoding proteins that silence TE activity. Both the introduction of new TEs via horizontal transfer and TE sequence evolution requires constant innovation of host-encoded TE silencing machinery to keep pace with TEs. One form of host innovation is the adaptation of existing, single-copy host genes. Indeed, host suppressors of TE replication often harbor signatures of positive selection. Such signatures are especially evident in genes encoding the piwi-interacting-RNA pathway of gene silencing, for example, the female germline-restricted TE silencer, HP1D/Rhino. Host genomes can also innovate via gene duplication and divergence. However, the importance of gene family expansions, contractions, and gene turnover to host genome defense has been largely unexplored. Here, we functionally characterize Oxpecker, a young, tandem duplicate gene of HP1D/rhino. We demonstrate that Oxpecker supports female fertility in Drosophila melanogaster and silences several TE families that are incompletely silenced by HP1D/Rhino in the female germline. We further show that, like Oxpecker, at least ten additional, structurally diverse, HP1D/rhino-derived daughter and “granddaughter” genes emerged during a short 15-million year period of Drosophila evolution. These young paralogs are transcribed primarily in germline tissues, where the genetic conflict between host genomes and TEs plays out. Our findings suggest that gene family expansion is an underappreciated yet potent evolutionary mechanism of genome defense diversification.
Keywords: gene duplication, heterochromatin, genome defense, transposable elements, HP1 proteins
Introduction
Active and inactive copies of transposable elements (TEs) riddle eukaryotic genomes. TE copies make up ∼50% of the human genome (Lander et al. 2001; de Koning et al. 2011) and up to 85% of others (Schnable et al. 2009). TE activity imperils host fitness in many ways, for example, by interrupting exons, perturbing transcription, inducing alternative splicing, nucleating heterochromatin, or leading to ectopic recombination (Montgomery et al. 1987; Langley et al. 1988; Kazazian 2004; Slotkin and Martienssen 2007; Lee 2015). Despite imposing a fitness cost to the host, TE copy number can increase over evolutionary time if transposition occurs in the germline (Doolittle and Sapienza 1980; Orgel and Crick 1980). Here, TE copies have direct access to the next generation. On rare occasions, these germline insertion events are adaptive (Daborn et al. 2002; Schlenke and Begun 2004; Aminetzach et al. 2005; Feschotte 2008; Gonzalez et al. 2008; Guio et al. 2014; Kapusta and Feschotte 2014; Mateo et al. 2014). Most often, TE replication results in a fitness cost to the host. As a result, this relationship is akin to a host–pathogen interaction, driving rapid innovation on the part of both TEs as well as host machinery to curb TE proliferation.
The fate of individual TE copies and their evolutionary impact on host–TE genetic conflict depends on their site of insertion in host genomes. For instance, TE insertions into the gene-rich euchromatic genome compartment have a higher likelihood of disrupting host genes either directly or by ectopic exchange. Moreover, elevated recombination in the euchromatic compartment allows natural selection to become more efficient at removing even slightly deleterious TE insertions from the population (Charlesworth and Langley 1989). In contrast, TE insertions into the gene-poor heterochromatic compartment enjoy a lower likelihood of disrupting genes and so have higher likelihood of persistence. These forces all contribute to TE accumulation in heterochromatin (Charlesworth et al. 1994; Dimitri and Junakovic 1999; Blumenstiel et al. 2002; Hoskins et al. 2002; Maside et al. 2005).
Heterochromatin, however, is not exclusively a refuge for TEs. Instead, heterochromatin can be a potent source of TE silencing by virtue of its abundant repressive histone marks and the specialized proteins that propagate them (Elgin and Grewal 2003). Indeed, TEs embedded in constitutive heterochromatin near centromeres and telomeres are enriched for histone H3K9 di- and tri-methyl, marks diagnostic of chromatin inaccessible to transcriptional machinery (Filion et al. 2010; Ho et al. 2014). Furthermore, a heterochromatic insertion can have deleterious consequences on the fitness of the TE family (Grewal and Elgin 2007). For instance, host genomes can utilize the accumulation of some TEs in heterochromatin to suppress TE activity. In Drosophila and mice, clusters of degenerate TE “graveyards” and active transposons can seed the germline-specialized piRNA (“piwi-interacting-RNA”) pathway, which targets active TEs transcribed from these loci (Aravin et al. 2007; Brennecke et al. 2007; Zamore 2007). These “piRNA clusters” are concentrated at euchromatin–heterochromatin boundaries (Yamanaka et al. 2014) and produce piRNAs through an elaborate series of steps, which have been reviewed extensively (Iwasaki et al. 2015; Czech and Hannon 2016; Hirakata and Siomi 2016) and are briefly summarized below.
In Drosophila melanogaster, most primary piRNAs are generated from precursor transcripts produced from “dual-strand clusters”, in which piRNA precursors are transcribed off both genomic strands (“uni-strand” piRNA clusters also exist in Drosophila and produce piRNA precursor transcripts via a distinct mechanism) (Brennecke et al. 2007). Dual-strand clusters are epigenetically marked by the H3K9me3 epigenetic modification (Le Thomas et al. 2014) and recruit HP1D/Rhino, a female germline-specific HP1 paralog (Klattenhoff et al. 2009). HP1D/Rhino recruits the Cutoff protein via a linker protein Deadlock; together, these three proteins mediate transcription, block splicing, inhibit premature transcription termination and degradation of precursor transcripts and license them for further processing in cytoplasmic processing sites (Klattenhoff et al. 2009; Pane et al. 2011; Mohn et al. 2014; Sapetschnig and Miska 2014; Zhang et al. 2014). These piRNA precursors are then exported to the perinuclear cytoplasm (nuage), where they are cleaved into piRNA intermediates on mitochondrial surfaces and loaded onto PIWI proteins (Olivieri et al. 2010; Saito et al. 2010; Qi et al. 2011; Zhang et al. 2012; Murota et al. 2014; Han et al. 2015; Mohn et al. 2015), followed by trimming and methylation to produce mature PIWI–piRNA complexes. PIWI–piRNA complexes can directly mediate transposon silencing by heterochromatin nucleation (Klenov et al. 2011; Wang and Elgin 2011; Sienski et al. 2012; Huang et al. 2013; Le Thomas et al. 2013). Alternately, Aubergine–piRNA complexes can enter a feed–forward cycle of amplification to produce secondary piRNAs via the “ping-pong” pathway (Brennecke et al. 2007).
Host silencing mechanisms provide protection against TE proliferation and yet, evolutionary innovation of TEs recurrently evade this host machinery. Not only does the coding sequence across TEs within the same family change over time, but also entirely new TE families can frequently invade naïve genomes by horizontal transfer (Daniels et al. 1990; Jordan et al. 1999; Sanchez-Gracia et al. 2005; Bartolome et al. 2009; Gilbert et al. 2010; Lerat et al. 2011; Kofler et al. 2015). Closely related species can, therefore, encode non-overlapping TE families despite recent speciation. TE evasion of host machinery puts pressure back on the host to keep pace with TE innovation, and even to evolve novel recognition and silencing mechanisms. Consistent with this idea, several recent analyses have shown pervasive signatures of positive selection at host genome defense factors, including multiple genes that encode piRNA pathway factors (Obbard et al. 2006, 2011; Kolaczkowski et al. 2011; Bernhardt et al. 2012; Kelleher et al. 2012; Lee and Langley 2012; Simkin et al. 2013). Adaptive evolution at a subset of these genes may be driven by antagonism with infectious viruses as has been seen for components of the siRNA production machinery in insects (Marques and Carthew 2007; Obbard et al. 2009; Saleh et al. 2009; Nayak et al. 2010, 2013; Murray et al. 2013;; Schnettler et al. 2014; van Mierlo et al. 2014). Even if this were the case, several rapidly evolving host defense genes target TEs exclusively (Vermaak et al. 2005; Maheshwari et al. 2008; Satyaki et al. 2014), supporting the idea that TE innovation contributes to the directional selection of host defense factors.
Although directional selection of single-copy host gene orthologs is a potent mechanism of adaptation against TEs, it is not the only possible source of evolutionary innovation. Gene family expansion and turnover can also generate adaptive molecular novelty. Indeed, host genes involved in viral restriction show abundant evidence of both diversifying selection as well as gene turnover. Recurrent turnover of genes encoding antimicrobial peptides in Drosophila (Sackton et al. 2007; Lazzaro 2008), or TRIM and APOBEC proteins in mammals (Sawyer et al. 2007; Tareen et al. 2009; Munk et al. 2012) exemplify frequent lineage-specific expansions of host immune factors. One rare example of gene expansion that amplifies host defense repertoires against TEs comes from studies of the KRAB–ZNF family in mammals (Nowick et al. 2010; Thomas and Schneider 2011). Many of these proteins act as DNA-binding “adaptors” that target heterochromatin silencing machinery to individual TE copies (Wolf and Goff 2009; Jacobs et al. 2014). The pace of innovation of KRAB–ZNF proteins parallels the novelty of endogenous retroviruses in mammals (Thomas and Schneider 2011). This dynamic suggests a “tit-for-tat” cycle of gene duplication and adaptation to specifically repress newly originated TEs in mammalian genomes via epigenetic silencing (Jacobs et al. 2014). KRAB–ZNFs thus represent a powerful example of host gene innovation that is, at least in some cases, unambiguously driven by TE changes.
KRAB–ZNFs are found exclusively in mammals. We expect, however, that host gene family expansions that combat TE innovation are a common feature of eukaryotic genomes. We specifically investigated the hypothesis that gene family expansion is a potent evolutionary mechanism shaping host genome defense repertoires in Drosophila. We took advantage of the identification of a bona fide TE silencer—Heterochromatin Protein 1D (HP1D)/Rhino, which supports piRNA cluster transcription in D. melanogaster ovaries (Klattenhoff et al. 2009). HP1D/Rhino is a critical component of TE silencing in Drosophila females; mutant females are completely sterile (Volpe et al. 2001; Klattenhoff et al. 2009). De-repressed TEs wreak havoc on the female germline and the consequent loss of genome integrity results in embryo inviability (Klattenhoff et al. 2009). We previously showed that HP1D/Rhino evolves under strong positive selection, consistent with its role in a genetic conflict with TEs (Vermaak et al. 2005). We also previously discovered a few partial or complete HP1D/Rhino paralogs in a phylogenomic survey of the HP1 gene family in Drosophila (Levine et al. 2012; Levine and Malik 2013). We predict that this recurrent HP1D/Rhino gene duplication diversifies host defense.
To test this hypothesis, we investigated the function of one of these HP1D paralogs, Oxpecker (Oxp). Oxp is an evolutionarily young gene (∼10–15 million-years-old) in Drosophila. We show that Oxpecker mutant females have impaired fertility despite being wildtype for HP1D/Rhino. We also find that Oxp silences many TE families in a wildtype HP1D/Rhino genetic background. Using the newly available genome sequences in the melanogaster species group, we further discovered that the 15 million-year-old Oxpecker gene is one of many HP1D/Rhino-like paralogs to arise during this short snapshot of Drosophila evolution. We find evidence for at least ten independent gene duplication/divergence events. All assayed paralogs are expressed primarily in the germline where TEs can successfully proliferate. This combination of functional and phylogenomic analyses of the HP1D/Rhino gene family in Drosophila implicates recurrent gene duplication as a frequent source of novelty in host genome defense repertoires.
Results
Oxpecker Is a Recent Gene Duplicate of HP1D/Rhino
Like all canonical HP1 proteins, HP1D/Rhino has a tripartite structure, consisting of three domains (fig. 1A) (Canzio et al. 2014; Eissenberg and Elgin 2014). The N-terminal chromodomain typically recognizes histone modifications associated with silencing, for example, di- and tri-methyl H3K9 (Jacobs and Khorasanizadeh 2002; Nielsen et al. 2002). The middle hinge domain interacts with DNA and RNA (Meehan et al. 2003; Badugu et al. 2005), whereas the C-terminal chromoshadow self-dimerizes and interacts with other chromosomal proteins (Smothers and Henikoff 2000; Canzio et al. 2014). We previously carried out a phylogenomic analysis of all Drosophila HP1 proteins based on 12 published Drosophila species’ genome sequences (Levine et al. 2012). In this analysis, we found that the Oxpecker (“Oxp” from now on) gene was born via gene duplication from HP1D within the last 15 million years (fig. 1A). Like HP1D, Oxp is primarily expressed in D. melanogaster ovaries. We ruled out the possibility of an Oxp-HP1D fusion gene in D. melanogaster by RT-PCR analyses (supplementary fig. S1, Supplementary Material online). Oxp encodes a protein that contains only a chromodomain in D. melanogaster and other Drosophila genomes. In D. melanogaster, the Oxp and HP1D/Rhino chromodomains share 62% amino acid identity (supplementary fig. S2, Supplementary Material online). We hypothesize that the HP1D gene duplication event likely only spanned the chromodomain. However, it is also possible that a full-length HP1D/Rhino gene duplication was followed by the rapid degeneration of the hinge and chromoshadow domain in Oxp.
Fig. 1.
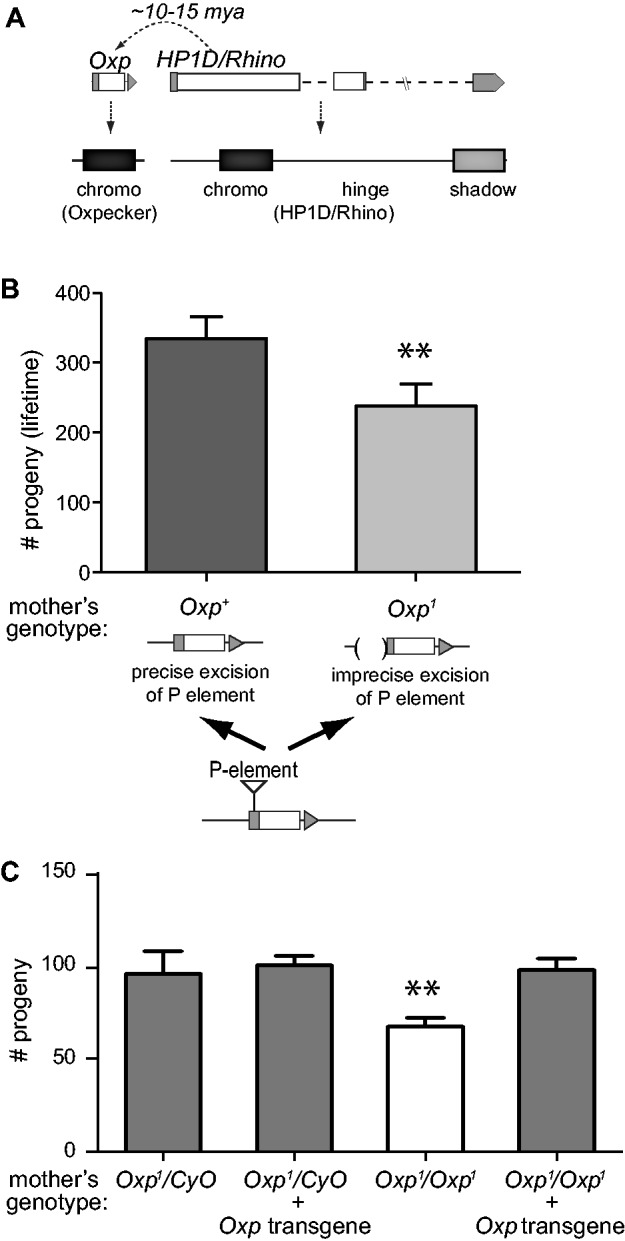
Oxpecker plays a role in female fertility. (A) Oxp is a chromodomain-only containing gene that is found immediately upstream of HP1D in the D. melanogaster genome. It derived from a HP1D/Rhino gene duplication ∼10–15 million years ago. (B) Isogenic females that possess an intact Oxp (Oxp+) or not (Oxp1) were crossed to wildtype males in egg-exhaustion experiments (see “Methods” section) that measured total progeny produced in their lifetime that developed to adulthood. Oxp deletion leads to a 20–25% drop in total progeny produced (“**” P < 0.05). (C) The effect of Oxp deletion on female fertility can be restored by an Oxp transgene. Compared with Oxp1/Oxp1female flies that exhibit compromised fertility, isogenic flies that express an Oxp transgene produce wildtype-like number of progeny.
We previously showed that HP1D/Rhino evolves under positive selection, which implicated its role in defense against TEs (Vermaak et al. 2005). This hypothesis was subsequently confirmed by functional studies that demonstrated HP1D/Rhino’s critical role in TE silencing via the piRNA pathway (Klattenhoff et al. 2009). We speculated that Oxp might participate in the same process as its parent gene HP1D/Rhino, that is, in genome defense. However, unlike its parent gene, we previously found that Oxp evolves under purifying selection (Levine et al. 2012). This dichotomy suggested an alternate possibility that Oxp could carry out a distinct function from HP1D, that is, a function unrelated to genome defense.
Oxp Supports Female Fertility and Silences Retrotransposons
To investigate its function, we characterized the biological consequences of creating a genetic lesion at the Oxp locus in a wildtype HP1D background. We mobilized a P-element near the Oxp start codon (see “Methods” section). This P-element mobilization gave rise to two types of strains. The first of these are “precise excision” stocks, in which the Oxp genomic sequence reverts to its intact form, leading to wildtype Oxp allele. The P-element mobilization also generated “imprecise deletion” stocks, in which a segment of the Oxp 5′ UTR and promoter sequence was deleted (fig. 1B). For subsequent analysis, we selected one “precise excision” control line and one “imprecise deletion” experimental line. We confirmed that the imprecise deletion removed 510 bp spanning the 5′ UTR and upstream sequence; using RT-PCR analyses, we confirmed this perturbed Oxp but not the HP1D/Rhino locus (supplementary fig. S3, Supplementary Material online). Using the precise deletion as our wildtype control (“Oxp+”), we confirmed that Oxp expression was virtually undetectable in the ovaries of the imprecise deletion (“Oxp1”) (supplementary fig. S3, Supplementary Material online). In contrast, there was no significant effect on transcription of the downstream parent gene, HP1D/Rhino (supplementary fig. S3, Supplementary Material online). Thus, our P-element mobilization strategy generated two near isogenic chromosomes that are both wildtype at the HP1D/Rhino locus but differ in their expression of the Oxp gene.
We next tested the consequences of this deletion on female fertility. We crossed Oxp+/Oxp+ or Oxp1/Oxp1 females to wildtype males (strain w1118) and counted the total progeny generated over their lifetime. We found that Oxp1/Oxp1 females generated ∼25% less adult progeny than the otherwise isogenic Oxp+/Oxp+ females (P < 0.02, fig. 1B). We also tested females homozygous or heterozygous for the imprecise excision (Oxp1/Oxp1 and Oxp1/CyO, respectively; CyO is a balancer second chromosome in D. melanogaster) crossed to wildtype males, counting all the adult progeny produced from eggs oviposited in a 3-day window. Again, we found a ∼25% reduction in progeny (P < 0.02, fig. 1C) for Oxp1/Oxp1 mothers. In contrast, Oxp1/CyO showed wildtype levels of progeny. Despite the reduction in fertility, we did not observe the characteristic “fused appendage” defects that are observed in eggs deposited by HP1D/Rhino homozygous females that led to its name (Volpe et al. 2001). We introduced an Oxp transgene driven by the native promoter from an ectopic location. This transgene restored Oxp expression partially in the Oxp1/Oxp1 strain (“rescue genotype”, supplementary fig. S4, Supplementary Material online). We found that this partial restoration of Oxp expression rescued fertility to wildtype levels (fig. 1C). These data demonstrate that Oxp function is non-redundant from HP1D/Rhino in the female germline.
To investigate the basis of Oxp’s fertility function, we first considered the well-characterized role of HP1D/Rhino in female fertility (Klattenhoff et al. 2009). HP1D/Rhino is a potent silencer of retrotransposons in the female germline. Indeed, massive TE derepression in HP1D-mutant females is associated with sterility. To test whether Oxp might support female fertility through a similar mechanism, we performed RNA-seq on the transcriptomes of ovaries dissected from Oxp+/Oxp+ and Oxp1/Oxp1 females, which are isogenic outside of the Oxp locus. We analyzed RNA-seq reads that mapped uniquely to coding genes and reads that mapped to repetitive elements (see “Methods” section). At a false discovery rate (fdr) of 0.01, we found that only 40 uniquely mapping coding genes were differentially expressed between the mutant and wildtype ovaries (supplementary fig. S5 and Table S1, Supplementary Material online). These differentially expressed euchromatic genes exhibited no significant bias of up- or down-regulation in the mutant genotype (24 and 16, respectively, binomial probability >0.1, supplementary fig. S5A, Supplementary Material online); none of the down-regulated genes encode a well-documented female fertility factor (supplementary table S1, Supplementary Material online). In wildtype ovaries, these 40 genes are expressed at low levels (supplementary fig. S5B, Supplementary Material online). We found that many of the significantly differentially expressed genes in Oxp1/Oxp1 ovaries were encoded on the fourth (“dot”) chromosome. Specifically, we observed a significant enrichment of Oxp-sensitive genes on the heterochromatic fourth chromosome (10% observed vs. <1% expected, P < 0.0001). In contrast, there was a dearth of such genes on the X chromosome (2.5% observed vs. 16% expected, P < 0.0001). This dichotomy suggests that the putatively indirect effects of Oxp on gene regulation strongly depend on the chromatin environment.
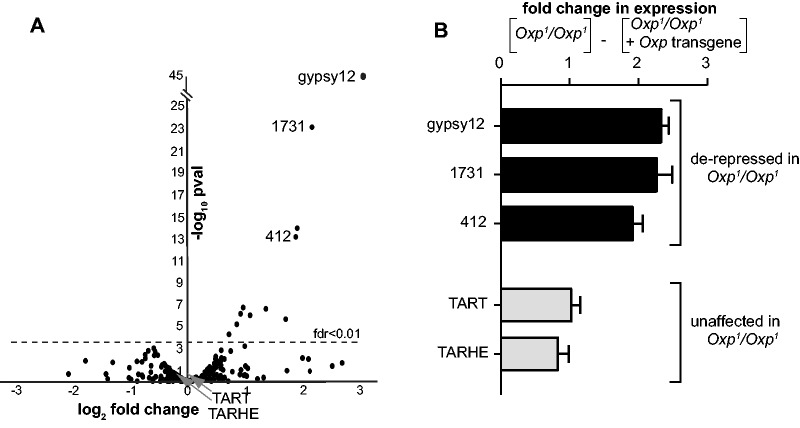
In addition to protein-coding genes, we found that 11 repetitive elements were de-repressed in Oxp1/Oxp1 ovaries at an fdr <0.01 (fig. 2A). In contrast to coding genes, we found that these differentially expressed repetitive elements were all overexpressed in Oxp mutants. These 11 elements represent nine TE families (supplementary table S2, Supplementary Material online). We confirmed that de-repression of these elements was specific to the Oxp locus. In our “rescue genotype” (Oxp transgene in Oxp1/Oxp1 genetic background), we were able to re-establish silencing in the ovaries of the three most significantly de-repressed elements (fig. 2B). These three, and indeed all nine oxp-silenced TE families, overlap with those silenced by HP1D/Rhino (Klattenhoff et al. 2009). The extent of TE overexpression, however, is comparatively modest in oxp mutants (Klattenhoff et al. 2009). Our findings suggest that although HP1D/Rhino plays a dominant role in the repression of TEs in the female germline, it is nevertheless insufficient for repressing a subset of D. melanogaster TEs targeted by the piRNA pathway. A previous study identified piRNAs that were present in the embryo relative to the ovary as a surrogate for germline versus somatic expression of piRNAs (Malone et al. 2009). We found that Oxp-silenced TEs are among those silenced by germline piRNAs, consistent with the model that Oxp-mediated TE repression occurs in the germline. Our findings support the hypothesis that gene duplication is an important evolutionary mechanism of genome defense diversification.
Fig. 2.
Oxpecker deletion results in the de-repression of retrotransposons in the female germline. (A) RNA-seq comparison between transcriptomes of Oxp+/Oxp+ and Oxp1/Oxp1ovaries reveal selected TEs (e.g., gypsy12, 1731, and 412 retrotransposons) that are upregulated upon Oxp deletion. However, the bulk of TEs are not affected by Oxp deletion, including the TART and TAHRE retrotransposons that are highly upregulated in HP1D homozygous mutant ovaries. (B) qPCR analyses of ovary-derived cDNA from either Oxp1/Oxp1flies or Oxp1/Oxp1flies with a “rescue” Oxp transgene confirms that TE overexpression (gypsy12, 1731, 412) in the imprecise deletion ovaries can be attributed to the loss of Oxp expression (“**” P < 0.05).
Recurrent Innovation by Gene Turnover in the HP1D/Rhino Subfamily
Our results show that Oxp supports TE silencing and fertility in the female germline in D. melanogaster. These findings also raised the possibility that other young duplications of HP1D could similarly expand the repertoire of TE silencing in Drosophila species beyond D. melanogaster and its close relatives. We took advantage of newly available genome sequences generated from eight Drosophila species (Drosophila modENCODE project (Chen et al. 2014)). These eight species shared a common ancestor around the time of Oxp’s birth, that is, 15 million years ago (Tamura et al. 2004). These eight genomes, combined with previously published genomes, lead to a total of 13 species of the melanogaster group (fig. 3A). Together, these species offer a unique opportunity to explore duplication, divergence, and degeneration of HP1D-like genes on a short evolutionary time scale.
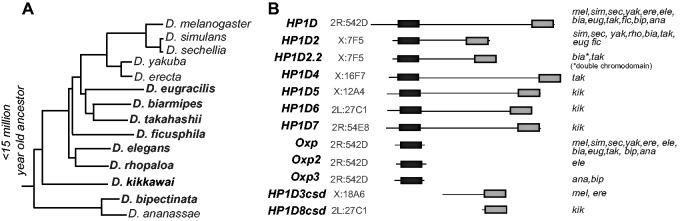
Fig. 3.
Many HP1D paralogs have arisen by gene duplication during Drosophila evolution. (A) A schematic of the melanogaster species group of Drosophila investigated in detail in this report. Species that were sequenced by the modENCODE project (Chen et al. 2014), which were previously not analyzed for HP1 duplications (Levine et al. 2012) are highlighted in boldface. (B) Schematic of all HP1D paralogs identified in the melanogaster group of Drosophila species, indicating the chromodomain (black) and chromoshadow (gray) domains. Putative orthology is assigned via shared syntenic position in the genome, with genome coordinates corresponding to the syntenic region of the D. melanogaster genome. Species in which a particular HP1D paralog is found are indicated on the right.
To define the HP1D-like gene complement in D. melanogaster and its relatives, we used the tBLASTn algorithm to query these genomes. We used the chromodomain and chromoshadow domains of HP1D/Rhino (“HP1D” from now on) and HP1D-like family members spanning 15 million years of Drosophila evolution (Levine et al. 2012). Our analysis identified previously undescribed orthologs of HP1D-like genes that were present in shared syntenic locations. In addition, we found many previously undescribed HP1D paralogs in non-syntenic locations (fig. 3B, supplementary table S3, Supplementary Material online). Using the Bayesian MCMC package in BEAST (see “Methods” section), we built a phylogenetic tree using the chromodomain of all known orthologs and paralogs across the entire Drosophila HP1 gene family. The HP1D orthologs and paralogs discovered via tBLASTn represent a monophyletic clade that is distinct from all other known HP1 genes in Drosophila (supplementary fig. S6, Supplementary Material online). This analysis confirms that these genes unambiguously belong to the HP1D clade.
We initially focused our analyses of the syntenic HP1D and Oxp orthologs in different Drosophila species. Every Drosophila species previously analyzed has been found to encode HP1D (Vermaak et al. 2005; Levine et al. 2012). To our surprise, we found that HP1D was not universally conserved in all queried genomes. For instance, we found that HP1D had degenerated along the lineage leading to a D. kikkawai ancestor (supplementary fig. S7, Supplementary Material online); this species represents the only currently sequenced genome from the species-rich montium subgroup. We were also unable to find HP1D in D. rhopaloa or D. takahashii due to a large gap (∼75 kb) in the publically available genome assemblies. We therefore sequenced the syntenic region to obtain HP1D and Oxp from both D. takahashii and D. rhopaloa genomic DNA. We discovered that D. takahashii encodes both Oxp and HP1D. In contrast, D. rhopaloa only encodes an intact HP1D but not Oxp (supplementary fig. S9, Supplementary Material online). Thus, we conclude that with one exception, HP1D is preserved in the melanogaster group of Drosophila species sampled. Oxp is almost equally well conserved; we found it to be absent only from D. kikkawai, D. rhopaloa, and D. ficusphila, in which it has recently degenerated (supplementary figs. S7–S9).
Using shared synteny, we found ten additional HP1D-related genes among the species we analyzed in addition to HP1D and Oxp (supplementary table S3, Supplementary Material online, and fig. 3B). We found that the full-length HP1D paralog, HP1D2, previously reported to be restricted to D. simulans and D. yakuba (Levine et al. 2012), was also present in D. rhopaloa, D. biarmipes, D. ficusphila, D. takahashii, and D. eugracilis in a shared syntenic location. We infer from this finding that HP1D2 originated over 15 million years ago, followed by subsequent loss in D. melanogaster and other species. Other “full-length” HP1D-related genes were only present in one or two closely-related species (fig. 3B). For instance, D. biarmipes and D. takahashii retain a tandem duplicate of HP1D2 (which we refer to as HP1D2.2). The HP1D2 chromodomain has internally duplicated in D. biarmipes, creating a unique HP1 gene that encodes two chromodomains and a single chromoshadow domain. D. takahashii also encodes an X chromosome-linked full-length HP1D-like gene (HP1D4) that is not found in any of the other sequenced species. D. kikkawai encodes a set of three full-length HP1D-like genes that are not found in other species: HP1D5, HP1D6, HP1D7 (fig. 3 and supplementary figs. S10–S12). Considering that D. kikkawai lacks the parental HP1D gene, it is very likely that at least one, and perhaps all three, of these younger paralogs have taken over the primary function of TE-silencing in this species.
Five of the HP1D-related genes were not full-length. In addition to Oxp, we found D. elegans encodes a tandem Oxpecker-like CD-only gene (Oxp2) while D. ananassae and D. bipectinata encode a third, independently derived Oxp3 (fig. 3). We also identified two genes that only encode the chromoshadow domain: the previously described HP1D3csd in D. melanogaster and D. erecta, and HP1D8csd found upstream of HP1D6 in D. kikkawai (fig. 3). The observed births and loss events of HP1D-like genes is consistent with recurrent HP1D-like gene family expansion and turnover.
Although the shared syntenic approach can be a powerful means to interrogate orthology, it can be misleading if multiple independent paralogs were born via gene-duplication in close proximity to the parental genes. We therefore carried out detailed phylogenetic analyses to ascertain orthology relationships. Although these analyses support the monophyletic relationship of all identified HP1D-like genes, unfortunately they failed to resolve many finer scale relationships (supplementary fig. S6, Supplementary Material online). The ∼60 aa chromodomain frequently had too little phylogenetic resolution (by a variety of distance and likelihood-based methods) to allow us to draw firm conclusions about orthology versus paralogy within the HP1D clade. For instance, we were unable to clearly establish common ancestry of the putative Oxp orthologs, which instead fall into two well-supported clades—one that encompasses the melanogaster subgroup and another exclusively the ananassae subgroup. It is possible that this bifurcation represents a burst of evolution immediately following the separation of these two lineages. But it is also likely that these two Oxp clades instead represent independent, tandem duplications of HP1D. Similarly, although the putative HP1D2 orthologs group together with presumed daughter genes HP1D2.2 and HP1D4, this phylogenetic grouping does not have strong bootstrap support. Overall, we attribute the poor resolution within HP1D to rapid divergence between these remarkably young genes. Thus, our assignment of orthology groups (fig. 3B) based on shared syntenic positions is not significantly improved by phylogenetic analyses. This limitation introduces ambiguity in orthology/paralogy only for tandem paralogs, such as Oxp and HP1D or HP1D2 and HP1D2.2. In all other cases, their shared syntenic location is a robust indicator of orthology.
HP1D-like Genes Are Germline-Expressed
HP1D and Oxp are both expressed in the female germline in D. melanogaster (Levine et al. 2012). This tissue-restricted expression pattern is consistent with their role in genome defense against TEs, which increase their copy number only if transposition occurs in the male or female germline. If the newly identified HP1D-related genes from non-model Drosophila species encode similar functions in genome defense as HP1D and Oxp, we predicted that they would also be expressed primarily in adult germline tissues.
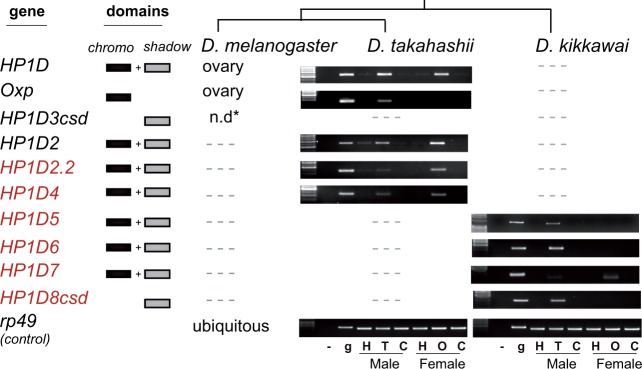
To test this prediction, we chose the two species that encode the largest subset of newly discovered HP1D-like genes—D. takahashii and D. kikkawai. In addition to HP1D and Oxp, D. takahashii encodes HP1D2, HP1D2.2, and HP1D4 (fig. 3). In D. takahashii, we found that HP1D, HP1D2, and HP1D4 were highly expressed in both ovaries and testes (fig. 4). In contrast, HP1D2.2 was most highly expressed in ovaries whereas Oxp was highly expressed only in testes. The additional testis expression of HP1D in D. takahashii suggests that this gene may encode an analogous TE silencing function in the male germline in this species even though such a function has not been elucidated in D. melanogaster. D. kikkawai encodes HP1D5, HP1D6, HP1D7, and HP1D8csd but neither HP1D nor Oxp (fig. 3 and supplementary fig. S7, Supplementary Material online). Intriguingly, we found that only D. kikkawai HP1D7 was expressed in ovaries where it might have replaced HP1D function (fig. 4). D. kikkawai HP1D5, HP1D6, and HP1D8csd are expressed primarily in testes (fig. 4). In contrast with the HP1D family members, the three founding HP1 genes, that is, HP1A, HP1B, and HP1C had ubiquitous expression in adult tissues (Vermaak et al. 2005; Levine et al. 2012). Based on their gonad-restricted expression patterns and strikingly dynamic evolution, we propose that the HP1D subfamily has recurrently undergone gene duplication to increase the repertoire of TE suppressors in the germline.
Fig. 4.
Expression analyses of HP1D paralogs in adult tissues of two Drosophila species. All HP1D paralogs analyzed in the RT-PCR analyses of adult tissues from either D. takahashii or D. kikkawai are indicated on the left, along with the previously measured expression pattern of their D. melanogaster orthologs. The rp49 gene is shown as a loading control on the bottom and as a quality measure of the cDNA versus genomic DNA (notice size difference). For adults, heads, germline tissue (testes or ovaries), and carcass were assayed. Note that many paralogs are specific to either species. When paralogs are not present, we indicate that as “—” [n.d.* refers to not determined because no adult expression was discovered (Levine et al. 2012)].
Discussion
Gene Duplications as a Genome Defense Strategy
HP1D is a critical component of piRNA defense against TEs in the female germline (Klattenhoff et al. 2009). We previously demonstrated that positive selection drives sequence evolution of HP1D orthologs across Drosophila evolution, suggesting that codon evolution might be one means for Drosophila host genomes to keep pace with TE sequence evolution and changing TE repertoires (Vermaak et al. 2005). In this report, we investigated HP1D gene duplication as a mechanism to diversify host genome defense against TEs. We first focused on a single HP1D duplication that gave rise to a tandem paralog encoding a chromodomain-only protein called Oxpecker. Despite its young age, we discovered that Oxp plays a significant role in female fertility. We found that Oxp augments HP1D silencing in the female germline of several TE families. Thus, for a subset of TEs, HP1D alone is insufficient to mediate complete suppression. We further demonstrate that at least 11 HP1D paralogs duplicated and diverged during a short 15-million-year snapshot of Drosophila evolution. The seven paralogs investigated further are all expressed in either or both male and female germlines—the putative battleground of host–TE evolutionary conflicts. Together with our genetic analysis of Oxp function, this pervasive history of HP1D-related gene duplication supports our hypothesis that HP1D gene family expansion has diversified genome defense repertoires in Drosophila genomes.
Other piRNA component gene amplifications have been documented in Drosophila, mosquito, and aphid genomes (Campbell et al. 2008; Lu et al. 2011; Lewis et al. 2016). Our findings especially complement previous studies that identified functional diversification of TUDOR domain proteins involved in piRNA processing in Drosophila. For instance, a recent study found that multiple genes encoding TUDOR domains (e.g., Vreteno) are essential for piRNA biogenesis (Handler et al. 2011; Zamparini et al. 2011). The TUDOR-domain containing genes may serve to make the ping-pong piRNA amplification cycle more robust, and thereby increase the efficiency of TE silencing by the piRNA machinery (Sato, Iwasaki, Shibuya et al. 2015; Sato, Iwasaki, Siomi et al. 2015; Wang et al. 2015). Unlike the HP1D gene family, however, the TUDOR-domain gene duplications appear to be quite ancient. Orthologs of these proteins are present in all sequenced Drosophila genomes, suggesting that the gene duplications predate the origin of the Drosophila genus, and have been strictly conserved since. Thus, they do not display the rapid “birth-and-death” dynamics we have seen in the HP1D gene family.
Oxpecker Silencing of TEs in the Germline
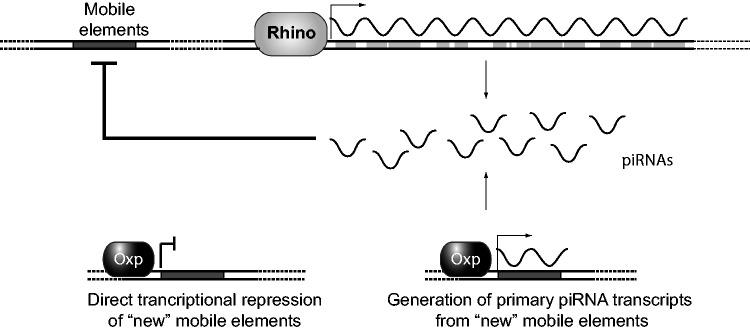
How does Oxp influence TE silencing in the female germline? We hypothesize two mechanisms by which Oxp could directly perform its TE-silencing function. First, Oxp could function analogously to HP1D, that is, supporting transcription of a primary piRNA transcript that is processed into TE-silencing piRNAs. This piRNA-related function of Oxp may be either dependent or independent of HP1D. In the latter scenario, Oxp may recruit HP1D to “new” genomic sites via an Oxp–HP1D protein–protein interaction, thereby expanding HP1D’s range of silencing (fig. 5). Alternatively, Oxp may act independently of the piRNA pathway, instead helping to recruit heterochromatin-silencing machinery to individual TEs or euchromatic genes via its chromodomain (fig. 5). Either of these mechanisms may allow Oxp to augment silencing of especially active TEs. We favor the latter because Oxpecker did not emerge as a strong candidate in screens for novel piRNA pathway members (Czech et al. 2013; Handler et al. 2013). Future work characterizing the genomic localization of Oxp (via ChIP-seq) and characterization of the piRNA repertoire (via piRNA-seq) in Oxp mutant ovaries will help distinguish between these possibilities.
Fig. 5.
Two possible mechanisms by which Oxp could augment protection against TEs in the female germline. Oxp might mediate binding and direct transcriptional repression of novel TE sites in the genome in cis (left). Alternatively, Oxp recruitment may generate piRNAs (either dependent or independent of subsequent HP1D/Rhino recruitment) that serve to silence TE copies in trans (right).
The evolutionary lability of TEs obscures the evolutionary force that drove oxp fixation 10 million years ago. However, the non-random subset of Oxp-silenced TE families offers insight into the initial selective force that may have driven oxp fixation nearly 15 million years ago. These nine families represent a diverse complement of TEs—long terminal repeat (LTR), non-LTR, and DNA elements. Nevertheless, copies within each family harbor high-percent identity and occur at a lower mean frequency in natural populations of D. melanogaster (supplementary table S4, Supplementary Material online, P < 0.001—Kelleher and Barbash 2013). These two features are also observed across the complement of TEs derepressed in piRNA pathway mutants (Kelleher and Barbash 2013). These two parameter estimates are consistent with Oxp silencing TEs that recently inserted into the genome (i.e., active or recently active elements). Oxp’s role in host defense might be to ensure more robust HP1D silencing of especially active TEs families. Thus, there is a net gain of TE-suppression activity by Oxp retention in a genome that already encodes HP1D. Similarly, the evolution of KRAB–ZNF genes in mammals to expand silencing to “new” TE families is largely the result of a “tit-for-tat” adaptive evolution (Thomas and Schneider 2011; Jacobs et al. 2014). Alternatively, Oxp may simply augment the pre-existing HP1D-generated piRNA pool, which is sufficient to repress all but the most active TE families in the host Drosophila genomes. This would be akin to the TUDOR-domain containing paralogs that increase the efficiency of the piRNA pathway response to active TE families by favoring the ping-pong amplification response (Sato, Iwasaki, Shibuya et al. 2015; Sato, Iwasaki, Siomi et al. 2015; Wang et al. 2015). This second possibility might help explain why Oxp evolves under purifying selection (Levine et al. 2012) unlike HP1D, which evolves under positive selection (Vermaak et al. 2005; Vermaak and Malik 2009).
We propose that the adaptation to combat recently encountered TE families also helps explain the observed pattern of recurrent HP1D gene duplication and loss. Gene duplications could result in functional redundancy that relaxes selective pressure for gene retention. For instance, Oxp birth may have led to a reduced repertoire of TE silencing by HP1D. Although HP1D has been largely conserved through this recurrent gene turnover, D. kikkawai represents an example of a genome in which HP1D was lost, likely due to its functional replacement by its female germline-restricted paralog, HP1D7. This hypothesis helps explain the dramatic gene turnover we observe in the HP1D gene family. Candidate replacement genes (e.g., in D. kikkawai) may support an equally efficient piRNA pathway-based mechanism of TE silencing, but one that leverages a recently refreshed set of genes. We propose that just like positive selection acting on codons in HP1D orthologs and other genome defense genes, the wholesale turnover of HP1D-like genes can be equally potent at amplifying the arsenal of host silencing machinery or making it more robust against newly encountered TEs. Future evolutionary analysis will determine if the dynamic gene turnover observed for the HP1D gene family here and previously in the KRAB–ZNF family (Nowick et al. 2010; Thomas and Schneider 2011) is a general feature of host defense genes across eukaryotes.
Materials and Methods
Oxpecker Mutant Construction and Confirmation
Bloomington stock 21086 encodes a mini-white-marked P-element (EPgy2) in the 5′ UTR of the Oxpecker gene. We crossed male flies homozygous for the insertion to virgin females heterozygous for Δ2-3 transposase (Δ2-3/CyO). Note that “CyO” refers to a chromosome II balancer. We crossed Δ2-3/21086 F1 males to virgin Bdx/CyO females. Next, we collected 85 F2 males from which the mini-white marked P-element had excised (white-eyed males). We backcrossed again to Bdx/CyO virgin females for long-term maintenance of each excision chromosome balanced over a CyO chromosome.
This white-eyed, chromosome II stock collection contained a both precise (clean) excision events and imprecise (deletion) events at the Oxpecker locus. We screened genomic DNA prepared from these stocks using primers that spanned the P-element insertion site (supplementary table S5, Supplementary Material online) and discovered 41 putatively precise excision and six imprecise excision chromosomes. We sequenced the PCR products of one putatively precise excision line and confirmed the genomic site was completely resurrected with no deletions upon P-element mobilization (supplementary fig. S3, Supplementary Material online). We also sequenced the PCR products of the five putatively imprecise excisions. One line harbored a large deletion of both Oxpecker and the upstream HP1D loci and four harbored smaller deletions ∼500 bp long. All four imprecise excisions deleted sequence upstream of the Oxpecker start codon. We chose the confirmed precise deletion line as the “wildtype” control going forward for subsequent functional analyses and an imprecise excision spanning the Oxp 5′ UTR and upstream flanking sequence—a 510-bp deletion upstream of the oxpecker start codon (supplementary fig. S3, Supplementary Material online). Based on our crossing scheme, we infer that these two lines are isogenic except for the 510-bp deletion.
Oxpecker is expressed primarily in D. melanogaster ovaries (Levine et al. 2012). We confirmed loss of Oxp expression in the imprecise excision line by PCR-amplifying the Oxp transcript from ovary-derived cDNA (supplementary fig. S3, Supplementary Material online). cDNA from the precise excision served as a control. Primers directed at the ubiquitously expressed rp49 locus confirmed equivalent cDNA concentrations across the genotypes (supplementary fig. S3, Supplementary Material online). Primer sequences are reported in supplementary table S5, Supplementary Material online.
Oxp Influence on Expression of Genes and Repetitive Elements in D. melanogaster
To investigate the genome-wide consequences of depleting Oxp expression, we conducted RNA-seq on ovaries dissected from 3 to 5-day-old females encoding either the precise and imprecise excision. We included three biological replicates per genotype. The FHCRC Shared Resources Genomics Core prepared six libraries using Illumina TruSeq Sample Prep Kit v2. We performed image analysis and base calling with Illumina’s RTA v1.13.sofware and de-multiplexed with Illumina’s CASAVA v1.8.2. We used TopHat v1.4.0 to align reads to BDGP5r66 and samtools v0.1.18 to convert files to sam format. To generate counts/gene and to cull genes with zero counts across all sample (our cut-off was at least 1 count/million in at least three samples), we used HTseq-count v0.5.3. This filtering left us with unique 7,547 genes (supplementary table S1, Supplementary Material online).
To investigate differences in expression across repetitive element classes, we first downloaded the dm3 repeat mask track as a GTF file from the UCSC Genome Browser website. We generated counts for each repeat using HTseq-count v0.5.3p9 (default “union” overlapped model). We removed repeats with <1 count/million in at least two samples. This filter left us with 469 repeat elements. We identified differentially expressed repeats using edgeR v2.6 (supplementary table S2, Supplementary Material online). For each TE, we extracted “mean pairwise identity” and “mean frequency” from Kelleher and Barbash (2013) (supplementary table S4, Supplementary Material online). Using a Mann–Whitney test, we compared the significantly differentially expressed set (identity: n = 7 and frequency: n = 9) with the global set (identity: n = 66, frequency: n = 88).
Oxpecker Transgene Construction and Rescue of TE Silencing
To determine if the functional consequences of the imprecise excision could be attributed to loss of Oxp expression alone, we engineered a native promoter-driven transgene rescue stock using the PhC31 technology. We cloned into the pattB vector the Oxp coding sequence, along with 3,150 and 375 bp 5′ and 3′ of the Oxp coding sequence, respectively. The Best Gene, Inc. (Chino Hills, CA) injected this plasmid into the 8622/attP2 line, which encodes a landing platform at cytolocation 68A4. We crossed a positive transformant line with the imprecise excision line, generating a genotype homozygous for the deletion (chromosome II) but heterozygous for the transgene (chromosomes III).
We performed qPCR on ovary-derived, poly-A selected cDNA prepared from the genotypes that were homozygous for the imprecise excision plus or minus a single copy of the transgene rescue construct. We chose the three most differentially expressed TEs in our RNA-seq dataset—Gypsy12 LTR, 1731, and 412—for downstream analysis. We extracted the bowtie alignments to each TE instance represented in the repeat mask GTF file (e.g., instance “Gypsy12_LTR_range = chr3RHet:640872-643144”). We designed qPCR primers to amplify the locus to which the majority of the reads mapped. The loci and primers can be found in supplementary table S5, Supplementary Material online. Two different primer pairs per TE, along with rp49 primers at the endogenous control locus, were used to amplify (SYBER green UPG with ROX, Life Technologies) each target. Each sample–primer pair combination was run in triplicate. We ran the reactions on an ABI 7900HT Real Time PCR Systems.
Fertility Assays
We assessed lifetime fertility of females homozygous for either the precise excision or the imprecise excision. In 15 replicate vials, we crossed two virgin females to two wildtype (w1118) males. Every 4 days, we transferred the parental generation onto new food for a total of 3 weeks. We counted adult progeny 8 days after first eclosion. We summed the data over the five time points per replicate and analyzed the data using a Mann–Whitney U-test (Prism 6).
To determine if the fertility defect detected in the above experiment could be attributed to the loss of Oxp expression alone, we repeated the fertility assay using females homozygous for the imprecise deletion with or without an ectopic Oxp transgene. We crossed three virgin females representing these two genotypes plus two additional controls (see fig. 1D) to two wildtype (genotype w1118) males per vial. We removed parents after 3 days and counted adult progeny on day 8 after the first eclosion. We analyzed the data again using a Mann–Whitney test (Prism 6). All fertility assays were conducted on standard cornmeal media at 25 °C on 12-h day/night cycles.
Bioinformatic Survey for HP1D Paralogs
Our previous phylogenomic analysis of the Heterochromatin Protein 1 gene family in Drosophila reported the discovery of nine genes that clustered phylogenetically within the HP1D/Rhino clade (Levine et al. 2012). These paralogs span the 60 million years of Drosophila evolution (Drosophila 12 Genomes et al. 2007). The analysis presented here takes advantage of eight recently sequenced Drosophila species related to D. melanogaster that span <15 million years: D. ficusphila, D eugracilis, D. biarmipes, D. takahashii, D. elegans, D. rhopaloa, D. kikkawai, and D. bipectinata (fig. 3A) (Chen et al. 2014). To discover new orthologs and paralogs derived from an HP1D/rhino or HP1D-like gene duplication, we first concatenated the known chromodomains or the chromoshadow domains (Levine et al. 2012) Using the tBLASTn algorithm (www.flybase.org), we searched for chromodomains and chromoshadow domains in the unannotated genomes of the eight Drosophila species.
For each species search, we initially retained hits to the genome scoring an e-value <0.1. We used the translated hit in a tBLASTn search of the D. melanogaster genome. Although we queried with exclusively HP1D/Rhino orthologs and paralogs, we identified all the previously characterized HP1 clades, including HP1A, HP1B, and HP1C in each species. Chromodomain-encoding, non-HP1 gene family members like Chromator or Polycomb, also came up in our gene lists for each species, indicating that our search for specifically HP1D-like genes was exhaustive. Those hits to HP1D-like genes were retained for synteny analysis. Specifically, we used the 5–10 kb region flanking each hit as the query in a BLAT search of the D. melanogaster genome. The location of the 10–20 kb region in D. melanogaster allowed us to classify each HP1D-like gene hit as either a potential ortholog of the previously known (shared syntenic) HP1D locus, of the HP1D-like loci reported in Levine et al. (2012), or a previously undescribed paralog (non-syntenic, supplementary table S4, Supplementary Material online).
A chromodomain and chromoshadow domain hit to a new, previously uncharacterized genomic location indicated the discovery of a new two-domain, “full HP1,” protein. For a subset of such hits, we computationally predicted an intronic sequence separating a chromodomain hit and a nearby chromoshadow domain hit. We confirmed presence or absence of a single coding gene in these locations by sequencing a PCR product amplified from cDNA that spanned the putative intron(s). Alignments of these transcripts to genomic DNA sequence are reported in supplementary figs. S10–S12, Supplementary Material online.
Phylogenetic Analyses
To uncover the phylogenetic relationships among our newly discovered orthologs and paralogs, we built phylogenetic trees restricted to the chromodomains. We perform tree-building using the Bayesian MCMC package BEAST v1.6.2 (Drummond and Rambaut 2007) using an uncorrelated log-normal relaxed clock (Drummond et al. 2006) and the SRD06 substitution model (Shapiro et al. 2006), which separates the evolutionary model for the third codon position from the first two. MCMC Chains ran until inspection of the traces and effective sample size of each parameter using the Tracer program v1.6 (http://tree.bio.ed.ac.uk/software/tracer) indicated acceptable mixing (ESS >200 for every parameter) and stationarity (as evaluated by the independent runs). Acceptable mixing occurred after running 20 million iterations. We present a maximum clade credibility tree (http://beast.bio.ed.ac.uk/summarizing-posterior-trees) in cladogram format for ease of presentation.
Expression Profiling of HP1 Paralogs in Adult Tissues
To determine if the newly discovered HP1D-like genes are germline-restricted, we conducted RT-PCR on a panel of adult male and female tissues dissected from D. takahashii and D. kikkawai. These two species share a common ancestor about 15 million years ago and encode a non-overlapping set of HP1D-like genes (also non-overlapping relative to D. melanogaster). We dissected ∼30 ovaries, 60 heads, and five “remaining carcasses” from adult females and stored the tissues in RNAlater (Qiagen) until total RNA extraction using the MirVana kit (Ambion). We performed the same procedure for males by dissecting 60 testis pairs, 60 heads, and five carcasses. We generated poly-A selected cDNA using SSIII reverse transcriptase (Invitrogen). Primers used for amplification of the seven loci can be found in supplementary table S5, Supplementary Material online.
Supplementary Material
Supplementary figures S1–S12 and Supplementary tables S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Danielle Vermaak for sharing results from her earlier analysis on Oxp and HP1D, and Ryan Basom for essential bioinformatics support of the RNA-seq analysis. We also thank Q. Helleu, T. Levin, L. Kursel, Grace Lee, A. Molaro, R. McLaughlin, J. Young, and S. Zanders for their comments and all members of the Malik and Peichel labs for valuable discussions. We especially thank the editor and four anonymous reviewers for their many helpful suggestions. This study was supported by an NIH K99/R00 Pathway to Independence Fellowship GM107351 to M.T.L. and grants from the Mathers Foundation and NIH R01 GM74108 to H.S.M. H.S.M. is an investigator of the Howard Hughes Medical Institute.
References
- Aminetzach YT, Macpherson JM, Petrov DA. 2005. Pesticide resistance via transposition-mediated adaptive gene truncation in Drosophila. Science 309:764–767. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316:744–747. [DOI] [PubMed] [Google Scholar]
- Badugu R, Yoo Y, Singh PB, Kellum R. 2005. Mutations in the heterochromatin protein 1 (HP1) hinge domain affect HP1 protein interactions and chromosomal distribution. Chromosoma 113:370–384. [DOI] [PubMed] [Google Scholar]
- Bartolome C, Bello X, Maside X. 2009. Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol. 10:R22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt SA, Simmons MP, Olson KE, Beaty BJ, Blair CD, Black WC. 2012. Rapid intraspecific evolution of miRNA and siRNA genes in the mosquito Aedes aegypti. PLoS One 7:e44198.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstiel JP, Hartl DL, Lozovsky ER. 2002. Patterns of insertion and deletion in contrasting chromatin domains. Mol Biol Evol. 19:2211–2225. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128:1089–1103. [DOI] [PubMed] [Google Scholar]
- Campbell CL, Black WCT, Hess AM, Foy BD. 2008. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genomics 9:425.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canzio D, Larson A, Narlikar GJ. 2014. Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol. 24:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Jarne P, Assimacopoulos S. 1994. The distribution of transposable elements within and between chromosomes in a population of Drosophila melanogaster. III. Element abundances in heterochromatin. Genet Res. 64:183–197. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Langley CH. 1989. The population genetics of Drosophila transposable elements. Annu Rev Genet. 23:251–287. [DOI] [PubMed] [Google Scholar]
- Chen ZX, Sturgill D, Qu J, Jiang H, Park S, Boley N, Suzuki AM, Fletcher AR, Plachetzki DC, FitzGerald PC, et al. 2014. Comparative validation of the D. melanogaster modENCODE transcriptome annotation. Genome Res. 24:1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. 2016. One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem Sci Available from: http://dx.doi.org/10.1016/j.tibs.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Preall JB, McGinn J, Hannon GJ. 2013. A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol Cell. 50:749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daborn PJ, Yen JL, Bogwitz MR, Le Goff G, Feil E, Jeffers S, Tijet N, Perry T, Heckel D, Batterham P, et al. 2002. A single p450 allele associated with insecticide resistance in Drosophila. Science 297:2253–2256. [DOI] [PubMed] [Google Scholar]
- Daniels SB, Peterson KR, Strausbaugh LD, Kidwell MG, Chovnick A. 1990. Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics 124:339–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 7:e1002384.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitri P, Junakovic N. 1999. Revising the selfish DNA hypothesis: new evidence on accumulation of transposable elements in heterochromatin. Trends Genet. 15:123–124. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Sapienza C. 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284:601–603. [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450:203–218. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 7:214.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Elgin SC. 2014. HP1a: a structural chromosomal protein regulating transcription. Trends Genet. 30:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin SC, Grewal SI. 2003. Heterochromatin: silence is golden. Curr Biol. 13:R895–R898. [DOI] [PubMed] [Google Scholar]
- Feschotte C. 2008. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 9:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143:212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Schaack S, Pace JK, 2nd, Brindley PJ, Feschotte C. 2010. A role for host-parasite interactions in the horizontal transfer of transposons across phyla. Nature 464:1347–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J, Lenkov K, Lipatov M, Macpherson JM, Petrov DA. 2008. High rate of recent transposable element-induced adaptation in Drosophila melanogaster. PLoS Biol. 6:e251.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC. 2007. Transcription and RNA interference in the formation of heterochromatin. Nature 447:399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guio L, Barron MG, Gonzalez J. 2014. The transposable element Bari-Jheh mediates oxidative stress response in Drosophila. Mol Ecol. 23:2020–2030. [DOI] [PubMed] [Google Scholar]
- Han BW, Wang W, Li C, Weng Z, Zamore PD. 2015. Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348:817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Meixner K, Pizka M, Lauss K, Schmied C, Gruber FS, Brennecke J. 2013. The genetic makeup of the Drosophila piRNA pathway. Mol Cell. 50:762–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Olivieri D, Novatchkova M, Gruber FS, Meixner K, Mechtler K, Stark A, Sachidanandam R, Brennecke J. 2011. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J. 30:3977–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakata S, Siomi MC. 2016. piRNA biogenesis in the germline: from transcription of piRNA genomic sources to piRNA maturation. Biochim Biophys Acta. 1859:82–92. [DOI] [PubMed] [Google Scholar]
- Ho JW, Jung YL, Liu T, Alver BH, Lee S, Ikegami K, Sohn KA, Minoda A, Tolstorukov MY, Appert A, et al. 2014. Comparative analysis of metazoan chromatin organization. Nature 512:449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Smith CD, Carlson JW, Carvalho AB, Halpern A, Kaminker JS, Kennedy C, Mungall CJ, Sullivan BA, Sutton GG, et al. 2002. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 3:RESEARCH0085.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XA, Yin H, Sweeney S, Raha D, Snyder M, Lin H. 2013. A major epigenetic programming mechanism guided by piRNAs. Dev Cell. 24:502–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki YW, Siomi MC, Siomi H. 2015. PIWI-Interacting RNA: its biogenesis and functions. Annu Rev Biochem. 84:405–433. [DOI] [PubMed] [Google Scholar]
- Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. 2014. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516:242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295:2080–2083. [DOI] [PubMed] [Google Scholar]
- Jordan IK, Matyunina LV, McDonald JF. 1999. Evidence for the recent horizontal transfer of long terminal repeat retrotransposon. Proc Natl Acad Sci U S A. 96:12621–12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A, Feschotte C. 2014. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet. 30:439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr. 2004. Mobile elements: drivers of genome evolution. Science 303:1626–1632. [DOI] [PubMed] [Google Scholar]
- Kelleher ES, Barbash DA. 2013. Analysis of piRNA-mediated silencing of active TEs in Drosophila melanogaster suggests limits on the evolution of host genome defense. Mol Biol Evol. 30:1816–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Edelman NB, Barbash DA. 2012. Drosophila interspecific hybrids phenocopy piRNA-pathway mutants. PLoS Biol. 10:e1001428.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. 2009. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138:1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Sokolova OA, Yakushev EY, Stolyarenko AD, Mikhaleva EA, Lavrov SA, Gvozdev VA. 2011. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc Natl Acad Sci U S A. 108:18760–18765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Hill T, Nolte V, Betancourt AJ, Schlotterer C. 2015. The recent invasion of natural Drosophila simulans populations by the P-element. Proc Natl Acad Sci U S A. 112:6659–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski B, Hupalo DN, Kern AD. 2011. Recurrent adaptation in RNA interference genes across the Drosophila phylogeny. Mol Biol Evol. 28:1033–1042. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. [DOI] [PubMed] [Google Scholar]
- Langley CH, Montgomery E, Hudson R, Kaplan N, Charlesworth B. 1988. On the role of unequal exchange in the containment of transposable element copy number. Genet Res. 52:223–235. [DOI] [PubMed] [Google Scholar]
- Lazzaro BP. 2008. Natural selection on the Drosophila antimicrobial immune system. Curr Opin Microbiol. 11:284–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Marinov GK, Aravin AA. 2014. A transgenerational process defines piRNA biogenesis in Drosophila virilis. Cell Rep. 8:1617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF. 2013. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 27:390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC. 2015. The role of piRNA-mediated epigenetic silencing in the population dynamics of transposable elements in Drosophila melanogaster. PLoS Genet. 11:e1005269.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC, Langley CH. 2012. Long-term and short-term evolutionary impacts of transposable elements on Drosophila. Genetics 192:1411–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E, Burlet N, Biemont C, Vieira C. 2011. Comparative analysis of transposable elements in the melanogaster subgroup sequenced genomes. Gene 473:100–109. [DOI] [PubMed] [Google Scholar]
- Levine MT, Malik HS. 2013. A rapidly evolving genomic toolkit for Drosophila heterochromatin. Fly 7:137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MT, McCoy C, Vermaak D, Lee YC, Hiatt MA, Matsen FA, Malik HS. 2012. Phylogenomic analysis reveals dynamic evolutionary history of the Drosophila heterochromatin protein 1 (HP1) gene family. PLoS Genet. 8:e1002729.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SH, Salmela H, Obbard DJ. 2016. Duplication and diversification of Dipteran Argonaute genes, and the evolutionary divergence of Piwi and Aubergine. Genome Biol Evol. 8:507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu HL, Tanguy S, Rispe C, Gauthier JP, Walsh T, Gordon K, Edwards O, Tagu D, Chang CC, Jaubert-Possamai S. 2011. Expansion of genes encoding piRNA-associated argonaute proteins in the pea aphid: diversification of expression profiles in different plastic morphs. PLoS One 6:e28051.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S, Wang J, Barbash DA. 2008. Recurrent positive selection of the Drosophila hybrid incompatibility gene Hmr. Mol Biol Evol. 25:2421–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137:522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JT, Carthew RW. 2007. A call to arms: coevolution of animal viruses and host innate immune responses. Trends Genet. 23:359–364. [DOI] [PubMed] [Google Scholar]
- Maside X, Assimacopoulos S, Charlesworth B. 2005. Fixation of transposable elements in the Drosophila melanogaster genome. Genet Res. 85:195–203. [DOI] [PubMed] [Google Scholar]
- Mateo L, Ullastres A, Gonzalez J. 2014. A transposable element insertion confers xenobiotic resistance in Drosophila. PLoS Genet. 10:e1004560.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan RR, Kao CF, Pennings S. 2003. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. EMBO J. 22:3164–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Handler D, Brennecke J. 2015. Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 348:812–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Sienski G, Handler D, Brennecke J. 2014. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 157:1364–1379. [DOI] [PubMed] [Google Scholar]
- Montgomery E, Charlesworth B, Langley CH. 1987. A test for the role of natural selection in the stabilization of transposable element copy number in a population of Drosophila melanogaster. Genet Res. 49:31–41. [DOI] [PubMed] [Google Scholar]
- Munk C, Willemsen A, Bravo IG. 2012. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol Biol. 12:71.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murota Y, Ishizu H, Nakagawa S, Iwasaki YW, Shibata S, Kamatani MK, Saito K, Okano H, Siomi H, Siomi MC. 2014. Yb integrates piRNA intermediates and processing factors into perinuclear bodies to enhance piRISC assembly. Cell Rep. 8:103–113. [DOI] [PubMed] [Google Scholar]
- Murray GG, Kosakovsky Pond SL, Obbard DJ. 2013. Suppressors of RNAi from plant viruses are subject to episodic positive selection. Proc Biol Sci. 280:20130965.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak A, Berry B, Tassetto M, Kunitomi M, Acevedo A, Deng C, Krutchinsky A, Gross J, Antoniewski C, Andino R. 2010. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat Struct Mol Biol. 17:547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak A, Tassetto M, Kunitomi M, Andino R. 2013. RNA interference-mediated intrinsic antiviral immunity in invertebrates. Curr Top Microbiol Immunol. 371:183–200. [DOI] [PubMed] [Google Scholar]
- Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416:103–107. [DOI] [PubMed] [Google Scholar]
- Nowick K, Hamilton AT, Zhang H, Stubbs L. 2010. Rapid sequence and expression divergence suggest selection for novel function in primate-specific KRAB-ZNF genes. Mol Biol Evol. 27:2606–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Gordon KH, Buck AH, Jiggins FM. 2009. The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci. 364:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Jiggins FM, Bradshaw NJ, Little TJ. 2011. Recent and recurrent selective sweeps of the antiviral RNAi gene Argonaute-2 in three species of Drosophila. Mol Biol Evol. 28:1043–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Jiggins FM, Halligan DL, Little TJ. 2006. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr Biol. 16:580–585. [DOI] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. 2010. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 29:3301–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel LE, Crick FH. 1980. Selfish DNA: the ultimate parasite. Nature 284:604–607. [DOI] [PubMed] [Google Scholar]
- Pane A, Jiang P, Zhao DY, Singh M, Schupbach T. 2011. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. EMBO J. 30:4601–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Watanabe T, Ku HY, Liu N, Zhong M, Lin H. 2011. The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J Biol Chem. 286:3789–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. 2007. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. 39:1461–1468. [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC. 2010. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 24:2493–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh MC, Tassetto M, van Rij RP, Goic B, Gausson V, Berry B, Jacquier C, Antoniewski C, Andino R. 2009. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature 458:346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gracia A, Maside X, Charlesworth B. 2005. High rate of horizontal transfer of transposable elements in Drosophila. Trends Genet. 21:200–203. [DOI] [PubMed] [Google Scholar]
- Sapetschnig A, Miska EA. 2014. Getting a grip on piRNA cluster transcription. Cell 157:1253–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Iwasaki YW, Shibuya A, Carninci P, Tsuchizawa Y, Ishizu H, Siomi MC, Siomi H. 2015. Krimper enforces an antisense bias on piRNA pools by binding AGO3 in the Drosophila germline. Mol Cell. 59:553–563. [DOI] [PubMed] [Google Scholar]
- Sato K, Iwasaki YW, Siomi H, Siomi MC. 2015. Tudor-domain containing proteins act to make the piRNA pathways more robust in Drosophila. Fly 9:86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyaki PR, Cuykendall TN, Wei KH, Brideau NJ, Kwak H, Aruna S, Ferree PM, Ji S, Barbash DA. 2014. The Hmr and Lhr hybrid incompatibility genes suppress a broad range of heterochromatic repeats. PLoS Genet. 10:e1004240.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. 2007. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog. 3:e197.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenke TA, Begun DJ. 2004. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc Natl Acad Sci U S A. 101:1626–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115. [DOI] [PubMed] [Google Scholar]
- Schnettler E, Tykalova H, Watson M, Sharma M, Sterken MG, Obbard DJ, Lewis SH, McFarlane M, Bell-Sakyi L, Barry G, et al. 2014. Induction and suppression of tick cell antiviral RNAi responses by tick-borne flaviviruses. Nucleic Acids Res. 42:9436–9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro B, Rambaut A, Drummond AJ. 2006. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol. 23:7–9. [DOI] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J. 2012. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151:964–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin A, Wong A, Poh YP, Theurkauf WE, Jensen JD. 2013. Recurrent and recent selective sweeps in the piRNA pathway. Evolution 67:1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. 2007. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 8:272–285. [DOI] [PubMed] [Google Scholar]
- Smothers JF, Henikoff S. 2000. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr Biol. 10:27–30. [DOI] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S. 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 21:36–44. [DOI] [PubMed] [Google Scholar]
- Tareen SU, Sawyer SL, Malik HS, Emerman M. 2009. An expanded clade of rodent Trim5 genes. Virology 385:473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Schneider S. 2011. Coevolution of retroelements and tandem zinc finger genes. Genome Res. 21:1800–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mierlo JT, Overheul GJ, Obadia B, van Cleef KW, Webster CL, Saleh MC, Obbard DJ, van Rij RP. 2014. Novel Drosophila viruses encode host-specific suppressors of RNAi. PLoS Pathog. 10:e1004256.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D, Henikoff S, Malik HS. 2005. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 1:96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D, Malik HS. 2009. Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu Rev Genet. 43:467–492. [DOI] [PubMed] [Google Scholar]
- Volpe AM, Horowitz H, Grafer CM, Jackson SM, Berg CA. 2001. Drosophila rhino encodes a female-specific chromo-domain protein that affects chromosome structure and egg polarity. Genetics 159:1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SH, Elgin SC. 2011. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci U S A. 108:21164–21169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Han BW, Tipping C, Ge DT, Zhang Z, Weng Z, Zamore PD. 2015. Slicing and Binding by Ago3 or Aub trigger Piwi-bound piRNA production by distinct mechanisms. Mol Cell. 59:819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP. 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458:1201–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Siomi MC, Siomi H. 2014. piRNA clusters and open chromatin structure. Mob DNA. 5:22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD. 2007. RNA silencing: genomic defence with a slice of pi. Nature 446:864–865. [DOI] [PubMed] [Google Scholar]
- Zamparini AL, Davis MY, Malone CD, Vieira E, Zavadil J, Sachidanandam R, Hannon GJ, Lehmann R. 2011. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development 138:4039–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, et al. 2012. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell 151:871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang J, Schultz N, Zhang F, Parhad SS, Tu S, Vreven T, Zamore PD, Weng Z, Theurkauf WE. 2014. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell 157:1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.