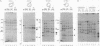Abstract
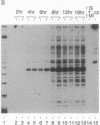
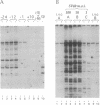
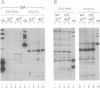
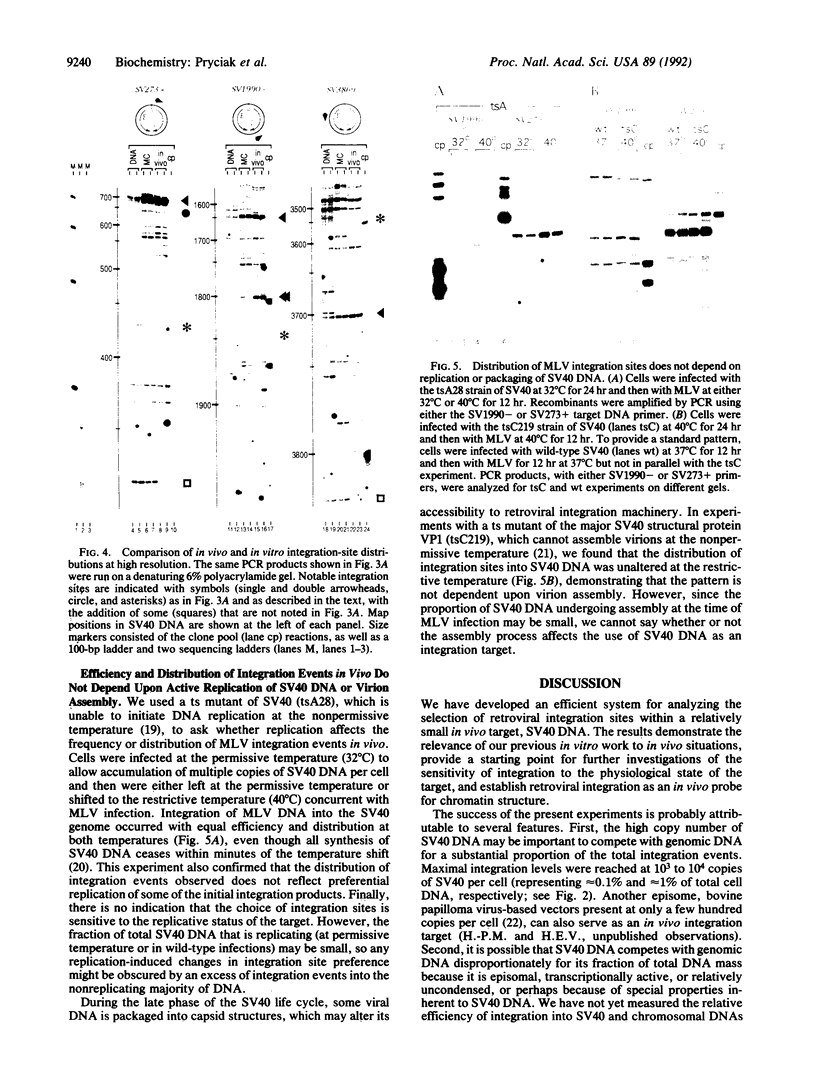
We present a method for studying multiple retroviral integration events into a small DNA target in vivo. Episomal simian virus 40 (SV40) genomes established by infection of CV-1 cells served as integration targets during subsequent infection with murine leukemia virus (MLV). Using a PCR-based assay for the abundance and distribution of integration events, nonrandom integration of MLV DNA into SV40 DNA is detectable as early as 4 hr and reaches a maximum level by 8 hr after MLV infection. The level of integration but not the distribution of integration sites is sensitive to the stage in the SV40 life cycle at which MLV infection is performed. Using a temperature-sensitive tumor (T) antigen mutant SV40 strain, we observed that active replication of the target DNA is not required for efficient integration in vivo. The distribution of integration sites in vivo is closely approximately by in vitro reactions with isolated SV40 minichromosomes as integration targets. However, the degree of bias between the most and least favored sites is greater in vivo than in vitro.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambrose C., Lowman H., Rajadhyaksha A., Blasquez V., Bina M. Location of nucleosomes in simian virus 40 chromatin. J Mol Biol. 1990 Aug 20;214(4):875–884. doi: 10.1016/0022-2836(90)90342-J. [DOI] [PubMed] [Google Scholar]
- Bina M., Blasquez V., Ng S. C., Beecher S. SV40 morphogenesis. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):565–569. doi: 10.1101/sqb.1983.047.01.066. [DOI] [PubMed] [Google Scholar]
- Brown P. O., Bowerman B., Varmus H. E., Bishop J. M. Correct integration of retroviral DNA in vitro. Cell. 1987 May 8;49(3):347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- Chinsky J., Soeiro R. Studies with aphidicolin on the Fv-1 host restriction of Friend murine leukemia virus. J Virol. 1982 Jul;43(1):182–190. doi: 10.1128/jvi.43.1.182-190.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J. Y., Avila J., Martin R. G. Viral DNA synthesis in cells infected by temperature-sensitive mutants of simian virus 40. J Virol. 1974 Jul;14(1):116–124. doi: 10.1128/jvi.14.1.116-124.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Calladine C. R. Sequence-specific positioning of core histones on an 860 base-pair DNA. Experiment and theory. J Mol Biol. 1987 May 5;195(1):143–173. doi: 10.1016/0022-2836(87)90333-0. [DOI] [PubMed] [Google Scholar]
- Drew H. R., McCall M. J. Structural analysis of a reconstituted DNA containing three histone octamers and histone H5. J Mol Biol. 1987 Oct 5;197(3):485–511. doi: 10.1016/0022-2836(87)90560-2. [DOI] [PubMed] [Google Scholar]
- Gershey E. L. Simian virus 40-host cell interaction during lytic infection. J Virol. 1979 Apr;30(1):76–83. doi: 10.1128/jvi.30.1.76-83.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J. B., Defendi V. Simian virus 40 gene A regulation of cellular DNA synthesis. I. In permissive cells. J Virol. 1979 May;30(2):590–599. doi: 10.1128/jvi.30.2.590-599.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T. W., Taylor J. M. Effect of aphidicolin on avian sarcoma virus replication. J Virol. 1982 Nov;44(2):493–498. doi: 10.1128/jvi.44.2.493-498.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura Y., Lee Y. M., Coffin J. M. Nonrandom integration of retroviral DNA in vitro: effect of CpG methylation. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5532–5536. doi: 10.1073/pnas.89.12.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt A. D., Rose R. B., Varmus H. E. Both substrate and target oligonucleotide sequences affect in vitro integration mediated by human immunodeficiency virus type 1 integrase protein produced in Saccharomyces cerevisiae. J Virol. 1992 Apr;66(4):2359–2368. doi: 10.1128/jvi.66.4.2359-2368.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeber G., Tevethia M. J., Schwedes J. F., Tegtmeyer P. Temperature-sensitive mutants identify crucial structural regions of simian virus 40 large T antigen. J Virol. 1989 Oct;63(10):4426–4430. doi: 10.1128/jvi.63.10.4426-4430.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. D., Buttimore C. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol Cell Biol. 1986 Aug;6(8):2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooslehner K., Karls U., Harbers K. Retroviral integration sites in transgenic Mov mice frequently map in the vicinity of transcribed DNA regions. J Virol. 1990 Jun;64(6):3056–3058. doi: 10.1128/jvi.64.6.3056-3058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan J. E., Blankenship J. W., Matthews H. R. Polyamines and acetylpolyamines increase the stability and alter the conformation of nucleosome core particles. Biochemistry. 1987 Jun 16;26(12):3643–3649. doi: 10.1021/bi00386a058. [DOI] [PubMed] [Google Scholar]
- Oudet P., Weiss E., Regnier E. Preparation of simian virus 40 minichromosomes. Methods Enzymol. 1989;170:14–25. doi: 10.1016/0076-6879(89)70040-9. [DOI] [PubMed] [Google Scholar]
- Pages J., Manteuil S., Stehelin D., Fiszman M., Marx M., Girard M. Relationship between replication of simian virus 40 DNA and specific events of the host cell cycle. J Virol. 1973 Jul;12(1):99–107. doi: 10.1128/jvi.12.1.99-107.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryciak P. M., Sil A., Varmus H. E. Retroviral integration into minichromosomes in vitro. EMBO J. 1992 Jan;11(1):291–303. doi: 10.1002/j.1460-2075.1992.tb05052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryciak P. M., Varmus H. E. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell. 1992 May 29;69(5):769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- Richard-Foy H., Hager G. L. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987 Aug;6(8):2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohdewohld H., Weiher H., Reik W., Jaenisch R., Breindl M. Retrovirus integration and chromatin structure: Moloney murine leukemia proviral integration sites map near DNase I-hypersensitive sites. J Virol. 1987 Feb;61(2):336–343. doi: 10.1128/jvi.61.2.336-343.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeyer S. B., Hansen L. J., Chalker D. L. Integration specificity of retrotransposons and retroviruses. Annu Rev Genet. 1990;24:491–518. doi: 10.1146/annurev.ge.24.120190.002423. [DOI] [PubMed] [Google Scholar]
- Scherdin U., Rhodes K., Breindl M. Transcriptionally active genome regions are preferred targets for retrovirus integration. J Virol. 1990 Feb;64(2):907–912. doi: 10.1128/jvi.64.2.907-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C. C., Stoye J. P., Coffin J. M. Highly preferred targets for retrovirus integration. Cell. 1988 May 20;53(4):531–537. doi: 10.1016/0092-8674(88)90569-7. [DOI] [PubMed] [Google Scholar]
- Shimotohno K., Temin H. M. No apparent nucleotide sequence specificity in cellular DNA juxtaposed to retrovirus proviruses. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7357–7361. doi: 10.1073/pnas.77.12.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C., Hoffman J., Goff S. P., Baltimore D. Intramolecular integration within Moloney murine leukemia virus DNA. J Virol. 1981 Oct;40(1):164–172. doi: 10.1128/jvi.40.1.164-172.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E., Padgett T., Heasley S., Simon G., Bishop J. M. Cellular functions are required for the synthesis and integration of avian sarcoma virus-specific DNA. Cell. 1977 Jun;11(2):307–319. doi: 10.1016/0092-8674(77)90047-2. [DOI] [PubMed] [Google Scholar]
- Vijaya S., Steffen D. L., Robinson H. L. Acceptor sites for retroviral integrations map near DNase I-hypersensitive sites in chromatin. J Virol. 1986 Nov;60(2):683–692. doi: 10.1128/jvi.60.2.683-692.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]