Abstract
The main familial focal epilepsies of childhood are autosomal dominant nocturnal frontal lobe epilepsy, familial temporal lobe epilepsy and familial focal epilepsy with variable foci. A frameshift mutation in the DEPDC5 (DEP domain containing protein 5) gene was identified in a family with focal epilepsy with variable foci, by linkage analysis and exome sequencing. Subsequent pyrosequencing of DEPDC5 in a cohort of 15 additional families with focal epilepsies revealed four nonsense and one missense mutations. Our findings provided evidence for frequent (37%) loss-of-function mutations in DEPDC5 associated with a broad spectrum of focal epilepsies. The implication of a DEP domain (Dishevelled, Egl-10 and Pleckstrin domain)-containing protein that may be involved in membrane trafficking and/or G-protein signaling, opens new avenues for research.
Epilepsy is a frequent neurological disorder characterized by spontaneous, recurrent seizures. Focal epileptic seizures are thought to originate within networks limited to one hemisphere. Several autosomal dominant, non-lesional, focal epilepsies have been described as specific age-related and electro-clinical syndromes: autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE [MIM 600513])1, familial temporal lobe epilepsy (FTLE [MIM 600512])2, including mesial and lateral forms (also termed autosomal dominant epilepsy with auditory features (ADEAF [MIM 604619]))3, and familial focal epilepsy with variable foci (FFEVF [MIM 604364]), characterized by focal seizures that are initiated in distinct cortical regions in different family members4. The only gene mutations currently identified are those linked to ADNFLE (CHRNA4, CHRNA2, CHRNB2 and KCNT1, encoding respectively the α4, α2, β2 subunits of the neuronal nicotinic acetylcholine receptor and a potassium channel subunit)5,6 and to ADEAF (LGI1 encoding epitempin)7. While no causal genes have yet been identified for FFEVF, linkage to 22q12 has been reported in several families4,8–11.
We have previously described the electro-clinical characteristics of 19 families with autosomal dominant focal epilepsies, subdivided into ADNFLE, FTLE and FFEVF12. Two large multiplex French families (N and S) with diagnosed FFEVF were selected from this cohort for linkage analysis using a high-density genome-wide scan with 10,000 SNPs. Both families mapped to the FFEVF locus on chromosome 22q12, with a maximum LOD score of 2.08 (family N) and 1.81 (family S). Haplotype reconstruction confirmed the segregation of a disease haplotype in each family (Supplementary Fig. 1-2).
We then sequenced the entire exome of patients N-III:4 and N-III:6 and sought rare coding variants in the 22q12 candidate region. In both patients we found the same single base deletion, c.1122delA, in exon 16 of the DEPDC5 (also known as KIAA0645) gene. This variant caused a frameshift, p.Leu374Phefs*30, introducing a premature stop codon 29 amino acids further (RefSeq NM_001242896). The c.1122delA variant fully segregated within the family (Fig. 1).
Figure 1.
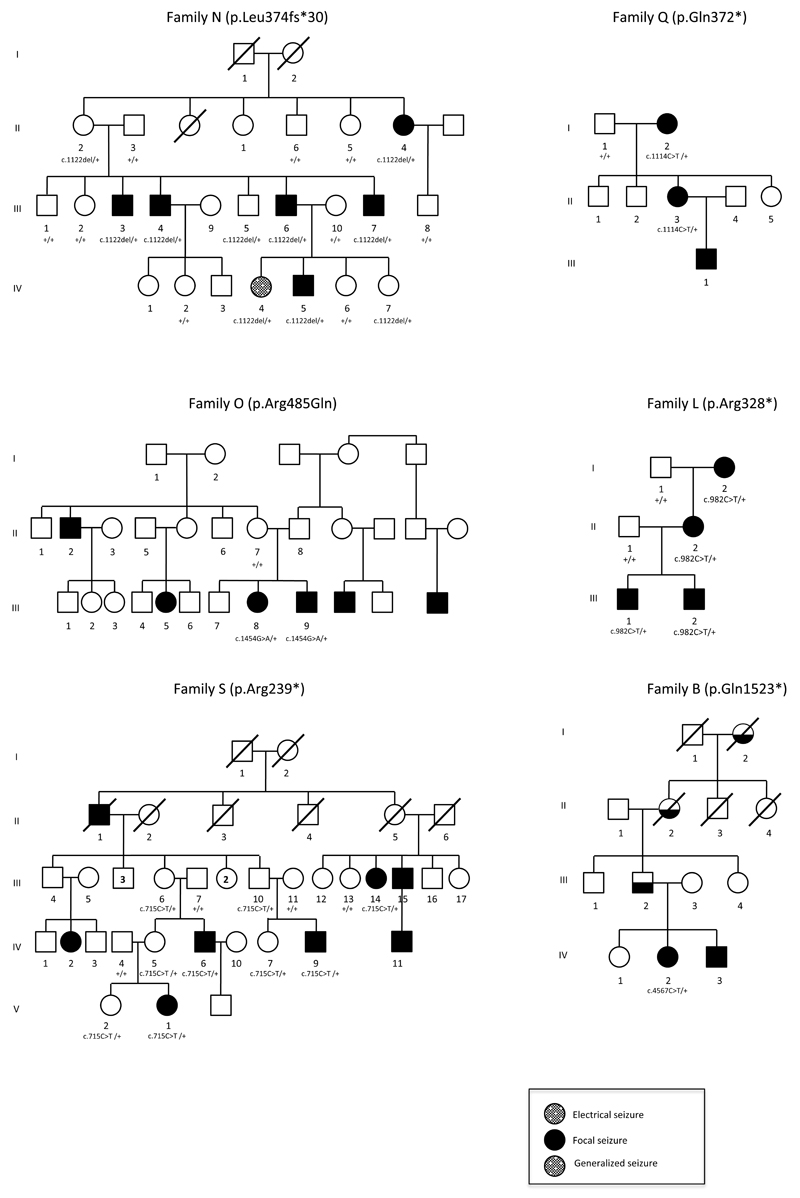
Pedigrees of families with segregation of DEPDC5 mutations: c.1122delA (p.Leu374Phefs*30) in family N, c.715C>T (p.Arg239*) in family S, c.982C>T (p.Arg328*) in family L, c.1114C>T (p.Gln372*) in family Q, c.1454G>A (p.Arg485Gln) in family O, c.4567C>T (p.Gln1523*) in family B. Individual N-IV:4 showed frontal spikes at the EEG, but no clinical seizure; ? indicates individuals born in the 1870s, with doubtful epilepsy histories. Diagonal lines indicate deceased individuals.
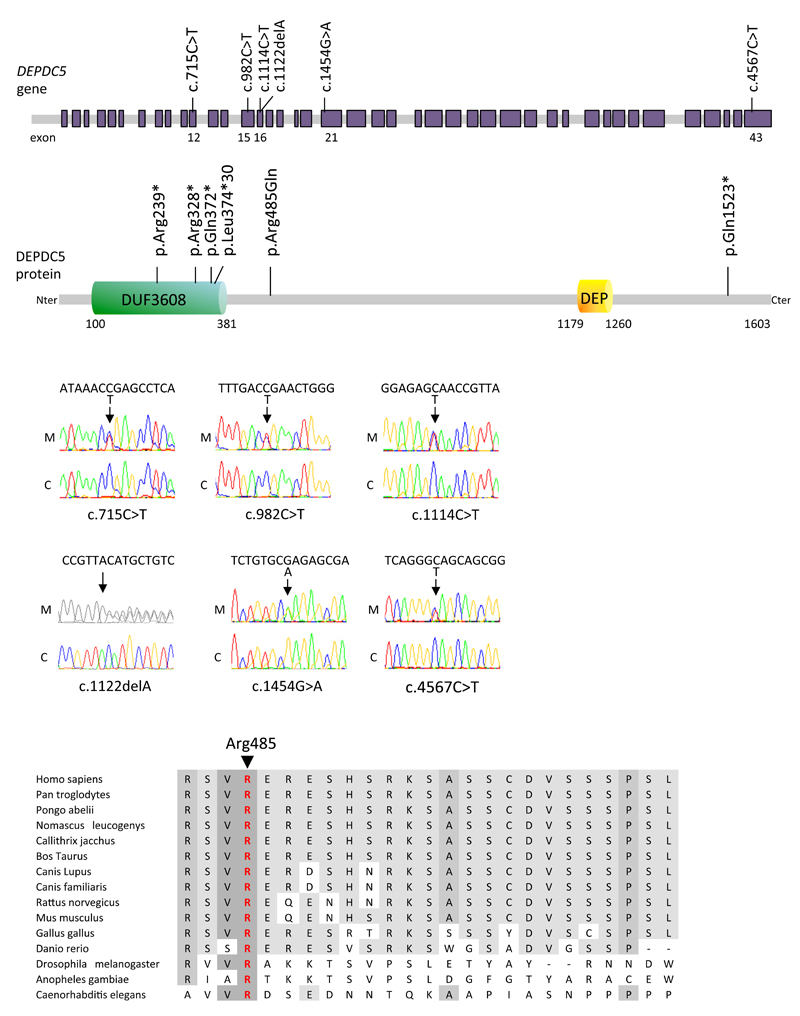
We next sought DEPDC5 mutations in 15 additional probands of these families with autosomal dominant focal epilepsies (ADNFLE= 7, FTLE= 4, FFEVF= 4)12, including family S linked to the 22q12 locus. Massively parallel pyrosequencing screening of all 43 coding exons and splice regions of DEPDC5 revealed four further nonsense mutations (p.Arg239*/family S, p.Arg328*/family L, p.Gln372*/family Q, p.Gln1523*/family B) and one missense mutation (p.Arg485Gln/family O) in 5 families (Fig. 2a, Supplementary Fig. 3, Table 1). All mutations were shown to segregate within the families, except for p.Gln1523* in family B, where additional family members were unavailable for analysis (Fig. 1). In family O, the father (O-II:8) may have transmitted the p.Arg485Gln mutation since it was not inherited from the mother (O-II:7). The arginine at position 485 is a highly conserved amino acid (Fig. 2b). Different prediction software tools – Mutation Taster, SIFT and Polyphen2 – predict that the arginine to glutamine at position 485 is disease causing, tolerated and possibly damaging, respectively. This substitution, p.Arg485Gln, was not detected in a cohort of 450 ethnically matched controls. None of the six DEPDC5 variants was present in the dbSNP135, 1000 Genomes Project database or the 6,503 exomes for which variants are currently listed in the NHLBI exome variant server (EVS) database. One nonsense (1/11,813) and one frameshift (1/11,775) mutation have been identified in the 6,503 exomes listed in the EVS database and no copy number variations disrupting the DEPDC5 coding sequence have been reported in the database of genomic variants. These mutations occurred significantly more often in our group of patients (5/32, p = 5.10−12 Fisher exact test).
Figure 2.
(a) Illustration of the exon-intron structure of DEPDC5 gene with the position of the mutations based on the human genomic sequence (RefSeq NM_001242896) and protein. DEPDC5 is a multidomain protein carrying an N-terminal domain of unknown function termed DUF3608 and a DEP domain at the C-terminal. Mutations p.Arg239*, p.Arg328*, p.Gln372*, p.Leu374Phefs*30 are clustered in the DUF3608 domain. (b) Multiple protein alignment showing conservation of the arginine residue in position 485 in orthologs of DEPDC5 across species. (c) Sequence chromatograms of one affected (S-IV:9) and one unaffected (S-III:11) member of family S, showing the presence of p.Arg239* mutation in DEPDC5 cDNA of S-IV:9 lymphoblasts pretreated with emetine, but not in untreated cells.
Table 1. Mutations identified in DEPDC5 gene.
| Family | Phenotype | Genomic (hg 19) | Coding (NM_001242896) | Exon | Protein | Allele frequency (EVS) |
|---|---|---|---|---|---|---|
| N | FFEVF | Chr22:g.32200188delA | c.1122delA | 16 | p.Leu374Phefs*30 | 0% |
| S | FFEVF | Chr22:g.32188751C>T | c.715C>T | 12 | p.Arg239* | 0% |
| L | FTLE | Chr22:g.32198725C>T | c.982C>T | 15 | p.Arg328* | 0% |
| Q | FFEVF | Chr22:g.32200180C>T | c.1114C>T | 16 | p.Gln372* | 0% |
| O | FTLE | Chr22:g.32210986G>A | c.1454G>A | 21 | p.Arg485Gln | 0% |
| B | ADNFLE | Chr22:g.32302238C>T | c.4567C>T | 43 | p.Gln1523* | 0% |
ADNFLE, autosomal dominant nocturnal frontal lobe epilepsy; FFEVF, familial focal epilepsy with variable foci; FTLE, familial temporal lobe
Four out of six mutations (p.Arg239*, p.Arg328*, p.Gln372*, p.Leu374Phefs*30) are predicted to be degraded by the nonsense mediated decay (NMD) system, while the nonsense mutation p.Gln1523*, located in the last exon of the gene, is not likely to be targeted by the NMD system. To confirm that the p.Arg239* mutation leads to NMD degradation, we treated cultured lymphoblasts from 3 mutation carriers (S-III:10, S-IV:7 and S-IV:9) and one spouse (S-III:11) with emetine, a NMD inhibitor. Sequencing of DEPDC5 cDNA revealed that the p.Arg239* mutation is only detected when NMD is inhibited (Fig. 2c), demonstrating that this nonsense mutation specifically leads to transcript degradation. DEPDC5 haploinsufficiency is likely to be the cause of the disease.
We identified mutations in one third of the families in our cohort (6/16, 37%) and, overall, 20 patients with epilepsy carried a DEPDC5 mutation. The age of onset ranged from 0 to 39 years (mean: 12.9 ±10.9 s.d.). DEPDC5 mutations were not limited to families with familial focal epilepsy with variable foci (FFEVF) phenotype but were also found in a family with two patients exhibiting a typical nocturnal frontal lobe epilepsy (family B)13 and another family including four patients with temporal lobe seizures (family L). A phenotype of FFEVF was attributed to family N because frontal seizures have been diagnosed in one patient (N-IV:5) and frontal electrical seizures in another (N-IV:4) while seizures in other family members originate in the temporal lobe. Penetrance was incomplete in families N, S and O, in agreement with reports of low penetrance of 62 %14 or 50 %4 in families with FFEVF. Seven asymptomatic obligate carriers (N-II:2, S-II:5, S-III:4, S-III:7, S-III:10, S-IV:5, O-II:8) and four asymptomatic at-risk individuals aged 8–42 (N-III:5, N-IV:7, S-IV:7, S-V:2) carried the mutations. Our data strongly support the causal role of DEPDC5 in a subset of our families with focal epilepsies. However, the observed reduced penetrance and variable expressivity (age at onset, seizure focus in various brain areas) suggest that other gene(s) or epigenetic and environmental factors modulate the phenotype.
Since five out of the six mutations introduce a premature stop mutation, a loss-of-function of DEPDC5presumably causes the epileptic phenotype. It will now be crucial to determine the function of DEPDC5 gene product and how its loss causes epilepsy. DEPDC5 transcript was first identified in human brain cDNA libraries15. It is strongly expressed at rather constant levels in both developing and adult human brain (Human Brain Transcriptome project), consistent with a role in epilepsy. We attempted to obtain clues from sequence analysis and database mining. The Reference Sequence (RefSeq) collection reports five putative isoforms of DEPDC5, of lengths 1572, 559, 1594, 1603 and 1503 amino acids. One of the mutations (p.Gln1523*) lies within an exon absent from isoform 2, suggesting this isoform is not involved in the pathology. Alignment with orthologs, using reciprocal best BLASTP hits, and phylogeny revealed a highly conserved protein present in metazoa and fungi (but absent from plant) (Supplementary Fig. 4), indicating a core role for this ancient eukaryotic gene. We note also that DEPDC5, and orthologs, occur as single gene copies even within genomes known to be ancient polyploids, such as D. rerio and S. cerevisiae. This, together with the dominance of epilepsy-causing mutations, suggests that gene dosage may be critical. Transmembrane prediction using Hidden Markov Models (TMHMM) analysis also shows that DEPDC5 has no transmembrane domain and no homology with known ion channels. The presence of a DEP domain (Dishevelled, Egl-10 and Pleckstrin domain), a globular protein motif of about 80 amino acids present in many proteins of G-protein signaling pathways, suggests this gene may be a G-protein substrate. DEPDC5 ortholog in yeast Yjr138p (also known as SEA1) is part of an evolutionary conserved multi-protein SEA complex involved in membrane trafficking16. DEPDC5 may also be involved in oncology: i) it was reported to be deleted in two glioblastomas17, ii) an intronic SNP (rs1012068) in the DEPDC5 locus has been associated with hepatocellular carcinoma risk18.
In conclusion, this work provides clear evidence that loss-of-function mutations of DEPDC5 are a major cause in a broad spectrum of autosomal dominant focal epilepsies. It shows that a single gene, DEPDC5, is a common genetic actor for epileptic syndromes with different brain localization and electro-clinical expression, including nocturnal frontal lobe epilepsy (ADNFLE), familial temporal lobe epilepsy (FTLE) and familial focal epilepsy with variable foci (FFEVF). Thus seizure initiation sites may be dissociated from underlying genetic mechanisms, in striking contrast to previous data, which link LGI1 mutations with a seizure focus localized in the lateral temporal lobe and mutations in nicotinic acetylcholine subunit genes with a seizure focus localized in the frontal lobe. Understanding of relations between DEPDC5 loss-of-function and the genesis of pathological neuronal synchronies in different epileptic networks and syndromes will provide new, clinically useful insights and knowledge that may be useful in genetic counseling. With no known homology between the DEPDC5 protein and other proteins, new pathogenic mechanisms seem likely. DEPDC5 is the second gene, after LGI1 found to be mutated in familial focal epilepsies but not encoding ion channel or transmitter receptor subunits. After channelopathies, we must consider alternative genetic pathways leading to epileptogenesis.
Online Methods
Families and patients
Sixteen unrelated families from Western Europe with non-lesional focal epilepsies were studied12. Mutations in CHRNA4, CHRNB2 and LGI1 have been excluded in all families. Local ethics committee (CCPPRB of Pitié-Salpêtrière Hospital, Paris, No. 69-03, 25/9/2003) approved this study. Informed consent was obtained from all participants or parents.
Genotyping and linkage analysis
A genome-wide screen was performed on family N and S with Illumina 6K panel microarrays. LOD scores were calculated with MERLIN software assuming an autosomal dominant trait with 70% penetrance, a disease frequency of 0.0001 and 0% phenocopy. Haplotypes were reconstructed according to hg19 annotations.
Exome sequencing
Exome sequencing was carried out in patients III-4 and III-6 of family N. Targeted exome sequencing, library preparation, capture, sequencing, and variant detection and annotation, were performed by IntegraGen (Evry, France). Exons of genomic DNA samples were captured using Agilent in-solution enrichment methodology with a biotinylated oligonucleotide probes library, followed by paired-end 75 bases massively parallel sequencing on Illumina HiSEQ2000. Sequence capture was performed according to the manufacturer’s instructions (Human All exon kit V4+UTRs, 70 Mb, Agilent). Briefly, 3 µg of each genomic DNA were fragmented by sonication and purified to yield fragments of 150-200 bp. Paired-end adaptor oligonucleotides from Illumina were ligated on repaired A-tailed fragments, then purified and enriched by 6 PCR cycles. Purified libraries (500 ng) were hybridized to the SureSelect oligo probe capture library for 24 hr. After hybridization, washing, and elution, the eluted fraction was PCR-amplified with 10 to 12 cycles, purified and quantified by qPCR to obtain sufficient DNA template for downstream applications. Each eluted-enriched DNA sample was then sequenced on an IlluminaHiSEQ2000 as paired-end 75 bases reads. Image analysis and base calling were performed using Illumina Real Time Analysis Pipeline version 1.14 with default parameters.
Bioinformatics analysis
Bioinformatics analysis of sequencing data was based on the Illumina pipeline (CASAVA 1.8). CASAVA aligns reads to the human reference genome (hg19) with the alignment algorithm ELANDv2 (performs multiseed and gapped alignments), then calls the SNPs based on the allele calls and read depth, and detects variants (SNPs & Indels). Only positions included in the bait coordinates were conserved. Genetic variation was annotated with the IntegraGen in-house pipeline, consisting of gene annotation (RefSeq), detection of known polymorphisms (dbSNP135, 1000 Genomes Project database) and characterization of mutations as exonic, intronic, silent or nonsense.
Screening of a cohort by pyrosequencing
All 43 exons and intron-exon junctions of DEPDC5 (except exon 2 screened by Sanger sequencing) were analyzed by universal tailed amplicon sequencing (454 Sequencing Technology, Roche). This approach used the 454 GS Junior system and involved an initial PCR amplification with primer sets designed to amplify exons of accession number NM_001242896, a second PCR aiming to incorporate multiplex identifier and 454 adaptors and finally an emulsion PCR step done according to the emPCR Amplification Method Manual.
Validation of exome and pyrosequencing variants by Sanger sequencing
Mutations found with exome and pyrosequencing were validated by Sanger sequencing using the Big-Dye® Terminator kit on an ABI Prism 3730 DNA Analyzer (Applied Biosystems). Segregation analysis of within-family variants was done using the same primers as for pyrosequencing. Mutations interpretation was assessed with Alamut version 2.2 (Interactive Biosoftware, France). The effects of amino acid substitutions on protein function were predicted using Mutation Taster, SIFT and Polyphen2.
Cell culture and mRNA experiments
Lymphoblastic cells from three mutation carriers (S-III:10, S-IV:7, S-IV:9) and one spouse (S-III:11) from family S were treated (or not) overnight with 10 mg/ml emetine to inhibit nonsense-mediated decay (NMD). Total RNA was extracted with the QIAGEN RNeasy Mini kit and reverse-transcribed with the ThermoScript™ RT-PCR System (Invitrogen). DEPDC5 cDNA was amplified and sequenced using specific primers located in exons 12 and 15.
Phylogenetic tree
All amino acid sequences of orthologous genes were retrieved from NCBI and aligned using MUSCLE. Visual inspection of the alignment with SEAVIEW shows that a homology dispersed across several regions of the protein. Protein alignment was trimmed to 454 homologous sites using GBLOCK. The tree reconstruction was performed by maximum likelihood using the software PHYML with an LG matrix.
Supplementary Material
Acknowledgments
We thank the genotyping and sequencing platform of ICM for technical assistance, the DNA and cell bank of ICM for DNA extraction and cell culture, Philippe Couarch for technical assistance, Marion Gaussen for linkage analysis and Christel Depienne for helpful discussions. S.I received a grant of the French Government. This study was supported by the program “Investissements d’avenir” ANR-10-IAIHU-06.
Footnotes
URLs
1000 Genomes Project, http://www.1000genomes.org/
BLASTP, http://blast.ncbi.nlm.nih.gov/
Database of Genomic Variants, http://projects.tcag.ca/variation/
dbSNP, http://www.ncbi.nlm.nih.gov.gate2.inist.fr/projects/SNP/
Human Brain Transcriptome, http://hbatlas.org/
MERLIN, http://www.sph.umich.edu/csg/abecasis/merlin/
Mutation Taster, http://www.mutationtaster.org/
NHLBI Exome Variant Server, http://evs.gs.washington.edu/EVS/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
RefSeq, http://www.ncbi.nlm.nih.gov/refseq/rsg/
SIFT, http://sift.jcvi.org/
Author Contributions
S.I. performed experiments and analyzed data; F.P. performed phenotyping and collected clinical data; G.R. collected and extracted DNA samples; E.N. and E.M. participated in genetic experiments, G.A. carried out the bioinformatics analysis on DEPDC5 and statistical analysis; P.T, P.G., M.W, C.M. and E.H. performed phenotyping of families; R.M. and M.B. contributed to the writing of the manuscript; E.L. supervised the project and contributed to the writing of the manuscript; S.B. designed the study, supervised the data analysis and wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Scheffer IE, et al. Autosomal dominant nocturnal frontal lobe epilepsy. A distinctive clinical disorder. Brain. 1995;118:61–73. doi: 10.1093/brain/118.1.61. [DOI] [PubMed] [Google Scholar]
- 2.Berkovic SF, et al. Familial temporal lobe epilepsy: a common disorder identified in twins. Ann Neurol. 1996;40:227–35. doi: 10.1002/ana.410400214. [DOI] [PubMed] [Google Scholar]
- 3.Ottman R, et al. Localization of a gene for partial epilepsy to chromosome 10q. Nat Genet. 1995;10:56–60. doi: 10.1038/ng0595-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein KM, et al. Familial focal epilepsy with variable foci mapped to chromosome 22q12: expansion of the phenotypic spectrum. Epilepsia. 2012;53:e151–5. doi: 10.1111/j.1528-1167.2012.03585.x. [DOI] [PubMed] [Google Scholar]
- 5.Baulac S, Baulac M. Advances on the genetics of mendelian idiopathic epilepsies. Clin Lab Med. 2010;30:911–929. doi: 10.1016/j.cll.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Heron SE, et al. Missense mutations in the sodium-gated potassium channel gene KCNT1 cause severe autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet. 2012;44:1188–90. doi: 10.1038/ng.2440. [DOI] [PubMed] [Google Scholar]
- 7.Kalachikov S, et al. Mutations in LGI1 cause autosomal-dominant partial epilepsy with auditory features. Nat Genet. 2002;30:335–41. doi: 10.1038/ng832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkovic SF, et al. Familial partial epilepsy with variable foci: clinical features and linkage to chromosome 22q12. Epilepsia. 2004;45:1054–60. doi: 10.1111/j.0013-9580.2004.30502.x. [DOI] [PubMed] [Google Scholar]
- 9.Callenbach PM, et al. Familial partial epilepsy with variable foci in a Dutch family: clinical characteristics and confirmation of linkage to chromosome 22q. Epilepsia. 2003;44:1298–305. doi: 10.1046/j.1528-1157.2003.62302.x. [DOI] [PubMed] [Google Scholar]
- 10.Morales-Corraliza J, et al. Familial partial epilepsy with variable foci: a new family with suggestion of linkage to chromosome 22q12. Epilepsia. 2010;51:1910–4. doi: 10.1111/j.1528-1167.2010.02680.x. [DOI] [PubMed] [Google Scholar]
- 11.Xiong L, et al. Mapping of a gene determining familial partial epilepsy with variable foci to chromosome 22q11-q12. Am J Hum Genet. 1999;65:1698–710. doi: 10.1086/302649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Picard F, et al. Dominant partial epilepsies. A clinical, electrophysiological and genetic study of 19 European families. Brain. 2000;123:1247–62. doi: 10.1093/brain/123.6.1247. [DOI] [PubMed] [Google Scholar]
- 13.Thomas P, Picard F, Hirsch E, Chatel M, Marescaux C. Autosomal dominant nocturnal frontal lobe epilepsy. Rev Neurol (Paris) 1998;154:228–35. [PubMed] [Google Scholar]
- 14.Scheffer IE, et al. Familial partial epilepsy with variable foci: a new partial epilepsy syndrome with suggestion of linkage to chromosome 2. Ann Neurol. 1998;44:890–9. doi: 10.1002/ana.410440607. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa K, et al. Prediction of the coding sequences of unidentified human genes. X. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1998;5:169–76. doi: 10.1093/dnares/5.3.169. [DOI] [PubMed] [Google Scholar]
- 16.Dokudovskaya S, et al. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.006478. M110 006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seng TJ, et al. Complex chromosome 22 rearrangements in astrocytic tumors identified using microsatellite and chromosome 22 tile path array analysis. Genes Chromosomes Cancer. 2005;43:181–93. doi: 10.1002/gcc.20181. [DOI] [PubMed] [Google Scholar]
- 18.Miki D, et al. Variation in the DEPDC5 locus is associated with progression to hepatocellular carcinoma in chronic hepatitis C virus carriers. Nat Genet. 2011;43:797–800. doi: 10.1038/ng.876. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.