Abstract
Background
Pancreatic cancer is one of the most lethal cancers worldwide, due to the lack of efficient therapy and difficulty in early diagnosis. ULBPs have been shown to behave as important protectors with prognostic significance in various cancers.
Materials and methods
Immunohistochemistry and enzyme-linked immunosorbent assays were used to explore the expression of ULBPs in cancer tissue and in serum, while survival analysis was used to evaluate the subsequent clinical value of ULBPs.
Results
Statistics showed that high expression of membrane ULBP1 was a good biomarker of overall survival (18 months vs 13 months), and a high level of soluble ULBP2 was deemed an independent poor indicator for both overall survival (P<0.001) and disease-free survival (P<0.001).
Conclusion
ULBP1 provides additional information for early diagnosis, and soluble ULBP2 can be used as a novel tumor marker to evaluate the risk of pancreatic cancer patients.
Keywords: pancreatic cancer, NKG2D ligands, prognostic marker, cancer immunotherapy
Introduction
Pancreatic cancer (PC) has a high mortality rate, due to the lack of methods for early diagnosis and effective therapy, even though many researchers have provided insights into its etiology.1 PC was listed as seventh in the ranking of cancer deaths worldwide, both for men and women, and in 2012 a total of 338,000 people were diagnosed with PC.2 Siegel et al estimated a total of 48,960 patients with PC and 40,560 deaths from this disease in the US, for the year 2015.2 A 2010 cancer survey in the People’s Republic of China showed that PC ranked sixth for males and seventh for females in the top-ten cancer-mortality ranking list.3 Presently, surgery is the best treatment for PC,4 but unfortunately most PC patients are diagnosed at advanced stages and can thus no longer fully benefit from surgery. Recently, research has been carried out into adjuvant therapies for treating PC, which has been related to the immune behavior of cancer cells and the host.5 Therefore, studying immunotherapy was considered to be a promising means to cure cancer indefinitely. Here, we studied the NKG2D immunosignaling pathway in PC. ULBPs, also called retinoic acid early transcripts, are ligands of the NKG2D receptor. The NKG2D receptor and its ligands form an important pathway of immunological defense.6 The NKG2D receptor is a C-type lectin-like transmembrane-activating receptor, which has been reported to be expressed on many cytotoxic lymphocytes, including NK cells, NKT cells, CD8+ T cells, γβ T cells, and certain subsets of human CD4+ T cells.7 Immunological surveillance was achieved when NKG2D, present on the surface of immune cells, came into contact with ULBPs, which were expressed in various malignancies and virus-infected tissues or when cells suffered from other injuries, such as DNA damage. Two isoforms of the NKG2D receptor (the long and the short form) are expressed as the result of alternative splicing. However, only the long isoform exists in humans. Two molecules of long NKG2D bind as a homodimer with one molecule of DAP10, whereas the short homodimer can interact with both DAP10 and DAP12. In addition, the function and composition of the NKG2D-signaling pathway are different in various types of cells. KLRK1 is an NKG2D-encoding gene distantly related to those encoding the remaining NKG2 receptors. NKG2D is found in both humans and mice, but its ligands vary extraordinarily.8 In humans, MICA/B and ULBPs have been identified as NKG2D ligands. ULBP has six isoforms (ULBP1–6), whose encoding genes are all located on chromosome 6. Their structures are not all the same; ULBP1, -2, -3, and -6 are attached to the cell membrane via glycosylphosphatidylinositol anchors, and ULBP4 and -5 are attached via cytoplasmic tails. These ligands are readily found in malignant diseases, but rarely in normal tissues.9,10 Furthermore, many in vivo and in vitro studies have clearly shown that the NKG2D-signaling pathway participates in the antitumor process. Numerous experiments have been carried out in an attempt to explain the origin of soluble ULBP ligands.11–13 Some researchers found that distinct MMPs, eg, MMP9 and MMP14, participated in the production of soluble ligands and that ERP5 was needed for the MMP-dependent cleavage of MICA.14–16 Moreover, some studies have indicated that several of the ADAM (a disintegrin and metalloproteinase) domains have the function of cleaving the extracellular domain of membrane ligands.17,18 Soluble ULBP (s-ULBP) cannot trigger the following cascade, and it reduces anticancer immunity by decreasing the number of membrane NKG2D receptors, thus contrasting the effect of membrane ULBP (m-ULBP).19–21 Interestingly, s-ULBP also has its own function: clinical analyses have claimed that s-ULBP1 is a prognosis indicator of cervical cancer and that s-ULBP2 is a prognosis indicator of ovarian cancer and melanoma.9,22,23 Many in vivo and in vitro experiments have confirmed that the NKG2D-signaling pathway contributes to therapeutic antitumor immunity.24 Recently, experts have declared that effective chemotherapeutics, eg, gemcitabine and valproic acid, enhance immunosurveillance by increasing the expression of MICA/B.25 In addition, the murine model experiment demonstrated that combination therapy of doxorubicin and IFNα upregulated the NKG2D ligands Mult1 and Rae1, inhibiting tumor growth. All these experiments demonstrated that NKG2D signaling is important in antitumor immunotherapy. However, no studies have explored the function or expression of ULBPs, which are ligands of NKG2D, in PC. For the purpose of promoting the development of diagnosis and treatment of PC, our research group collected the clinical data of patients with PC and detected the expression of m-ULBP1–3 via immunohistochemistry (IHC) and levels of s-ULBP1–3 by enzyme-linked immunosorbent assay (ELISA). We then analyzed the differences in expression of these ligands and their clinical value by means of survival analysis.
Materials and methods
Patients and samples
Collectively, volunteers with PC were from the same general surgery-treatment group of the Affiliated Provincial Hospital of Anhui Medical University (Hefei, People’s Republic of China), and healthy subjects were from our research group and physical examination center. Our research was approved by the ethics committee and the human research review committee of the Affiliated Provincial Hospital of Anhui Medical University, and all participants signed the informed consent form. Cancer tissues and adjacent noncancerous tissues were collected from 80 PC patients who had been diagnosed by postoperative pathology between 2009 and 2012. Venous blood was drawn from these patients before the operation and prior to radiotherapy and chemotherapy. Blood samples were taken from a total of 30 volunteers to act as the control group. These blood samples were centrifuged at 2,000 rpm for 10 minutes and the supernatants kept at −80º C until required. Patients were followed and clinicopathological information recorded from the time of diagnosis. Information regarding age, gender, tumor location, histological grade, tumor diameter, carbohydrate antigen (CA)-19-9, and TNM staging (tumor, node, metastasis) was obtained for screening of the data related to prognosis.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the ethics committee and the human research review committee of the Affiliated Provincial Hospital of Anhui Medical University, and with the 1964 Helsinki declaration and its subsequent amendments or comparable ethical standards.
Immunohistochemistry
Preliminary experiments were carried out to determine the best antibody concentration for each ligand. The following three steps were then carried out: 1) deparaffinization – cancerous and paracancerous tissue sections (4 μm thick) were deparaffinized with xylene, rehydrated through graded alcohol, and then washed in phosphate-buffered saline (PBS); 2) antigenicity recovery – sections were immersed in citrate buffer (pH 6) and heated using a microwave for 10 minutes and washed with PBS, followed by immersion of sections in 0.3% hydrogen peroxide for 20 minutes to block endogenous peroxidase activity; 3) reaction and staining – subsequently, hydrophobic circles were drawn and slides reacted overnight with 1:100 diluted primary antibodies, namely rabbit anti-ULBP1, anti-ULBP2, and anti-ULBP3 antibody (Novus Biologicals, Littleton, CO, USA) at 4ºC. Biotinylated secondary antibody (1:500) was incubated for 30 minutes at 37°C, and a diaminobenzidine-detection kit was used to visualize the immunostaining. Sections were counterstained with hematoxylin, then dehydrated and mounted in orderly fashion. For the negative-control groups, the steps were the same, apart from the fact that primary antibodies were replaced by PBS. Staining of ULBPs was assessed semiquantitatively on two scores: 1) staining intensity (no evidence of staining, 0; weak staining, 1+; moderate staining, 2+; and strong positive staining in most cells, 3+), and 2) the percentage of positively stained pancreatic cells (less than 10% of positively stained cells, 1+; 10%–30% of positively-stained cells, 2+; >30% of positively stained cells, 3+). An overall protein-expression score was calculated by multiplying the intensity and positivity scores. The IHC staining scores were then dichotomized into low expression (less than/equal to mean score of cancer specimens) and high expression (greater than mean score of cancer specimens). Two physicians of the pathology department were invited to complete the evaluation. A consensus was reached after discussion when there were different scores.
ELISA analysis
The frozen supernatants described in the “Patients and samples” section were taken and completely thawed at room temperature. Commercial ELISA kits (R&D Systems Inc., Minneapolis, MN, USA) were used to detect levels of s-ULBP. Preliminary tests were carried out to find the appropriate concentration of serum for s-ULBP (s-ULBP1, 1:5; s-ULBP2, 2:5; s-ULBP3, 1:5). All procedures were strictly followed according to the specifications. Briefly, standard serial dilutions and serum samples were added to each well in 96-well flat-bottomed plates including anti-ULBPs monoclonal antibody. A horseradish peroxidase-conjugated reagent was added, and color was developed using the tetramethylbenzidine system. Optical density (OD) was read at 450 nm using a microtiter plate reader within 15 minutes. Serum was replaced by sample diluent in the control group; other steps were the same as the experimental groups. The standard curve was generated by plotting the average OD (450 nm) obtained for each of the six standard concentrations on the vertical (y) axis vs the corresponding concentration on the horizontal (x) axis. Finally, the concentration of every sample was calculated by its OD value. According to the specifications of the ELISA kits, neither the interassay nor intra-assay coefficient of variation was more than 15%.
Statistical analysis
Statistical analyses were performed using SPSS version 18.0 (SPSS Inc., Chicago, IL, USA). Results are expressed as means ± standard deviation. The statistical significance of the differences in IHC staining of ULBPs in the different groups was calculated by the χ2 test. Student’s t-test was used to calculate the results of the ELISA. Overall and disease-free survival curves were generated by the Kaplan–Meier method, and the difference between the survival curves was calculated by the log-rank test. In addition, receiver-operating characteristic analysis was used to evaluate the potential value of s-ULBP. Each concentration had its own corresponding sensitivity and specificity. Youden’s index = sensitivity + specificity – 1. Therefore, each concentration had its own Youden’s index. The concentration of ULBP used as the cutoff point was that in which the Youden’s index was minimum. Finally, the Cox regression model was applied to multivariate analysis to ascertain the independent significance of relevant clinical covariates. In all cases, the test was two-sided, and a P-value <0.05 was considered statistically significant.
Results
Immunohistochemical staining
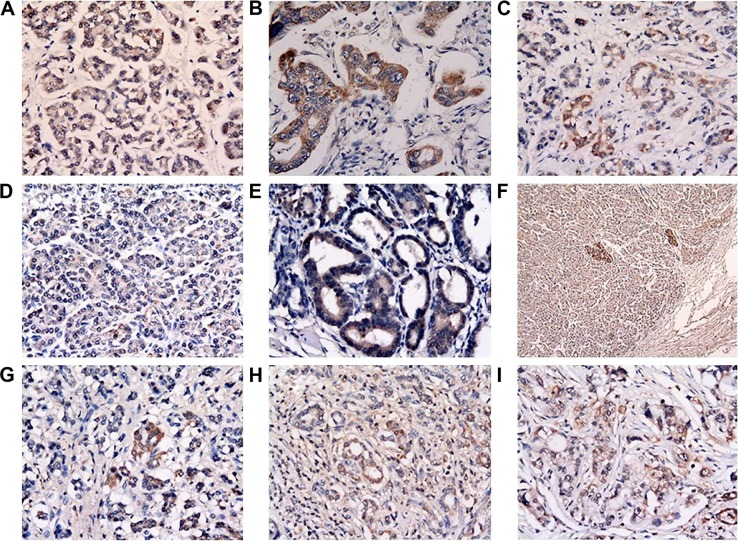
Hematoxylin and eosin staining was used to confirm the malignancy of the slides. We found that not all of the ULBPs were expressed on the membrane of PC cells, but they were also stained in the cytoplasm. Furthermore, m-ULBP was more likely to be found in pancreatic islet cells and cells around small vessels than other cells, as shown in Figure 1. The positive expression of ULBP1 and ULBP3 in terminal stage PC was lower than in adjacent noncancerous tissues (m-ULBP1, χ2=15.87, P<0.001; m-ULBP3, χ2=12.76, P<0.001), but m-ULBP1 was upregulated in early stage PC cells compared with normal cells (χ2=3.92, P=0.048), as shown in Table 1 and Figure 1.
Figure 1.
Expression of m-ULBP1–3 in pancreatic cancer and adjacent noncancerous tissues.
Notes: (A–C) Expression of m-ULBP1 in normal cells, early stage tumor cells, and terminal cells, respectively. (D, E) Expression of m-ULBP2 in normal (D) and tumor (E) cells. (F) Expression of m-ULBP2 in islet cells. (G–I) Expression of m-ULBP3 in normal cells, early stage tumor cells, and terminal stage tumor cells, respectively. All pictures taken at 400× magnification, except F (100×).
Abbreviations: m-ULBP, membrane ULBP; s-ULBP, soluble ULBP.
Table 1.
Expression of ULBPs in different TNM stages of pancreatic cancer (PC) compared with adjacent noncancerous tissues
| Group | PC (n) | Control (n) | Pearson χ2 | P-value |
|---|---|---|---|---|
| All (ULBPl) | 0.25 | |||
| High | 25 | 32 | ||
| Low | 55 | 48 | 1.34 | |
| I–II (ULBPl) | 0.048 | |||
| High | 21 | 32 | ||
| Low | 14 | 48 | 3.92 | |
| III–IV (ULBP1) | <0.001 | |||
| High | 4 | 32 | ||
| Low | 41 | 48 | 15.87 | |
| AII (ULBP2) | 0.69 | |||
| High | 15 | 17 | ||
| Low | 65 | 63 | 0.16 | |
| I–II (ULBP2) | 0.60 | |||
| High | 9 | 17 | ||
| Low | 26 | 63 | 0.28 | |
| III–IV (ULBP2) | 0.27 | |||
| High | 6 | 17 | ||
| Low | 39 | 63 | 1.2 | |
| AII (ULBP3) | 0.001 | |||
| High | 18 | 38 | ||
| Low | 62 | 42 | 10.99 | |
| I–II (ULBP3) | 0.11 | |||
| High | 11 | 38 | ||
| Low | 24 | 82 | 2.57 | |
| III–IV (ULBP3) | <0.001 | |||
| High | 7 | 38 | ||
| Low | 38 | 42 | 12.76 |
ELISA detection
Serum from 80 PC patients before surgery was used for the experimental group, and serum from healthy subjects was used for the no-treatment control group. Levels of s-ULBP1 and s-ULBP2 were significantly higher in PC patients compared to the healthy volunteers (s-ULBP1 29.42±1.43 pg/mL vs 19.23±1.51 pg/mL; s-ULBP2 132.14±13.24 pg/mL vs 34.12±6.23 pg/mL), while s-ULBP3 was weakly expressed in these patients and volunteers (Figure 2).
Figure 2.
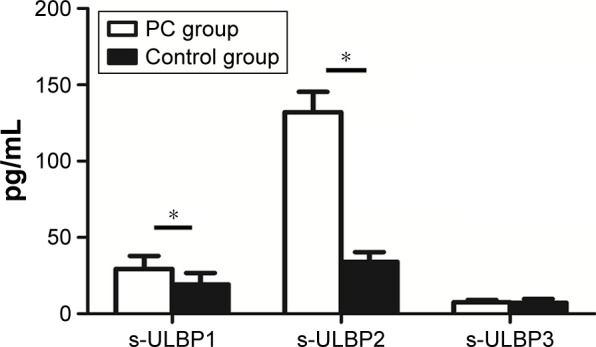
Expression of s-ULBPs in the serum of pancreatic cancer (PC) patients and healthy volunteers (means of 80 patients and 30 volunteers).
Note: *P<0.001.
Abbreviation: s-ULBP, soluble ULBP.
Survival analyses
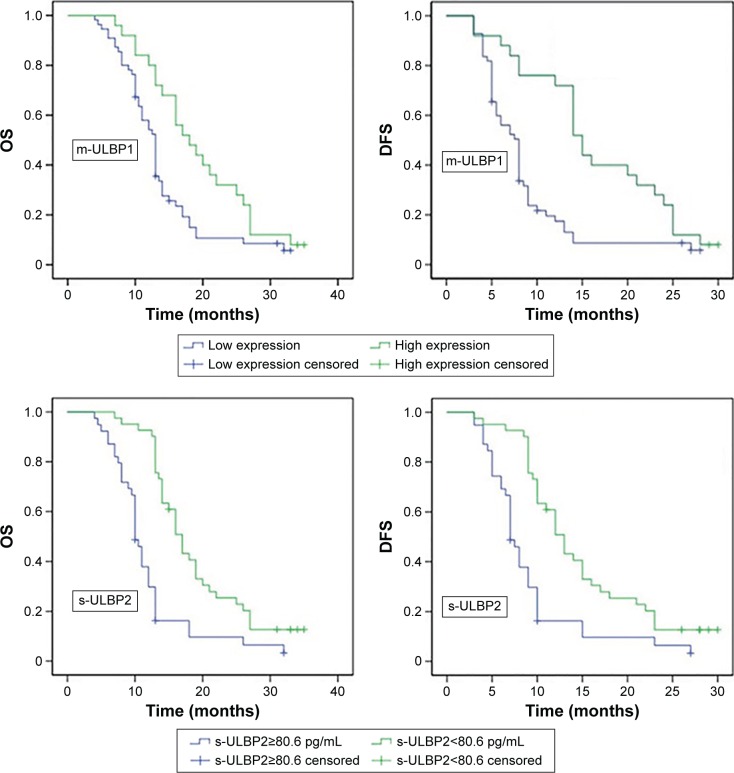
The results of the Spearman’s correlation test indicated that the m-ULBP1 level of PC patients was correlated with the TNM stage of PC: stage I–II patients had higher m-ULBP1 expression than stage III–IV patients (r=−0.31, P<0.001). However, neither m-ULBP2 nor m-ULBP3 were related to the TNM stage of patients, as indicated in Table 2. Moreover, s-ULBP was calculated to have no relation to the TNM stage of patients (Table 2). Subsequently, receiver-operating characteristic curves were drawn to evaluate the diagnosis accuracy and reliability of serum s-ULBP1 and s-ULBP2. The optimal cutoff values of 24.4 pg/mL and 80.6 pg/mL were defined for s-ULBP1 (sensitivity 0.69%, specificity 0.73%) and s-ULBP2 (sensitivity 0.64%, specificity 0.87%), respectively. In addition, the area under the curve was 0.746 for s-ULBP1 and 0.795 for s-ULBP2 (Figure 3). s-ULBP3 was not included in the survival analysis, due to its weak concentration in patients (7.31±0.26 pg/mL). Kaplan–Meier curves (Figure 4) and log-rank tests were taken to analyze overall survival (OS) and disease-free survival (DFS) between the different groups. Results showed that patients with higher m-ULBP1 expression (χ2=10.77, P=0.001) and/or lower concentration of s-ULBP2 (χ2=19.77, P<0.001) had longer OS. Similarly, DFS was also prolonged in these patients (m-ULBP1, χ2=11.19, P=0.001; s-ULBP2, χ2=14.42, P<0.001; Table 3). Univariate analysis revealed that CA19-9, histological grade, TNM, m-ULBP1, and s-ULBP2 were significantly related to OS and DFS. However, there may also have been a correlation between these indices. Therefore, multivariate analysis was carried out to eliminate any interference between these factors. Results (Table 4) indicated that high m-ULBP1 expression was an independent biomarker of good prognosis for OS (hazard ratio [HR] 0.32, P=0.001), and a low s-ULBP2 level was an independent indicator of good prognosis for both OS (HR 3.118, P<0.001) and DFS (HR 3.014, P<0.001). Additionally, CA19-9, histological grade, and TNM stage were also independent prognostic markers for OS and DFS.
Table 2.
Correlation between TNM stage of pancreatic cancer and ULBP ligands
| Ligands | TNM stage I–II (n) | TNM stage III–IV (n) | P-value |
|---|---|---|---|
| m-ULBP1 | <0.001 | ||
| Low | 14 | 41 | |
| High | 21 | 4 | |
| m-ULBP2 | 0.41 | ||
| Low | 26 | 39 | |
| High | 9 | 6 | |
| m-ULBP3 | 0.09 | ||
| Low | 24 | 38 | |
| High | 11 | 7 | |
| s-ULBP1 (mean ±SD) | 28.78±2.12 | 29.91±1.94 | 0.70 |
| s-ULBP2 (mean ±SD) | 137.67±22.84 | 127.83±15.62 | 0.72 |
| s-ULBP3 (mean ±SD) | 6.73±0.47 | 7.56±0.29 | 0.07 |
Abbreviations: TNM, tumor, node, metastasis; m-ULBP, membrane ULBP; s-ULBP, soluble ULBP; SD, standard deviation.
Figure 3.

Receiver-operating characteristic (ROC) curve of soluble ULBP2 (s-ULBP2).
Note: The area under the curve was 0.795.
Figure 4.
Kaplan–Meier curves for overall survival (OS) and disease free survival (DFS) of pancreatic cancer patients.
Note: Patients were divided into two groups according to s-ULBP1 expression and s-ULBP2 cutoff 80.6 pg/mL, calculated by receiver-operating characteristic analysis.
Abbreviation: s-ULBP, soluble ULBP.
Table 3.
Univariate survival analysis of factors associated with OS and DFS
| Variables | Patients (n) | OS
|
DFS
|
||||
|---|---|---|---|---|---|---|---|
| MST (months) |
χ2 | Log-rank P-value |
MST (months) |
χ2 | Log-rank P-value |
||
| Age (years) | 0.978 | 0.96 | |||||
| <60 | 31 | 14 | 0.001 | 9 | 0.003 | ||
| ≥60 | 49 | 13 | 10 | ||||
| Sex | 0.167 | 0.173 | |||||
| Men | 42 | 13 | 1.908 | 9 | 1.858 | ||
| Women | 38 | 15 | 11 | ||||
| Tumor diameter (mm) | 0.768 | 0.212 | |||||
| <30 | 44 | 13 | 0.087 | 13 | 1.559 | ||
| ≥30 | 36 | 13 | 9 | ||||
| CA19-9 (U/mL) | 0.003 | <0.001 | |||||
| <37 | 21 | 14 | 8.937 | 12 | 12.49 | ||
| ≥37 | 59 | 9.5 | 7 | ||||
| Histological grade | 0.01 | 0.012 | |||||
| Poor–moderate | 41 | 13 | 6.726 | 8 | 6.34 | ||
| Good | 39 | 17 | 12 | ||||
| TNM stage | 0.02 | 0.013 | |||||
| I–II | 35 | 14 | 5.378 | 10 | 6.151 | ||
| III–IV | 45 | 13 | 9 | ||||
| m-ULBP1 | 0.01 | 0.001 | |||||
| Low | 55 | 13 | 6.68 | 8 | 11.19 | ||
| High | 25 | 18 | 15 | ||||
| m-ULBP2 | 0.393 | 0.542 | |||||
| Low | 65 | 13 | 0.729 | 9 | 0.371 | ||
| High | 15 | 17 | 9 | ||||
| m-ULBP3 | 0.944 | 0.098 | |||||
| Low | 62 | 14 | 0.005 | 8.5 | 2.731 | ||
| High | 18 | 12.5 | 9.5 | ||||
| s-ULBP1 (pg/mL) | 0.49 | 0.189 | |||||
| ≥24.4 | 37 | 13 | 0.477 | 7 | 1.725 | ||
| <24.4 | 43 | 15 | 11 | ||||
| s-ULBP2 (pg/mL) | <0.001 | <0.001 | |||||
| ≥80.6 | 39 | 10 | 19.767 | 7 | 14.42 | ||
| <80.6 | 41 | 17 | 13 | ||||
Abbreviations: MST, median survival time; OS, overall survival; DFS, disease-free survival; CA, carbohydrate antigen; TNM, tumor, node, metastasis; m-ULBP, membrane ULBP; s-ULBP, soluble ULBP.
Table 4.
Multivariate analysis of factors associated with OS and DFS
| Variables | OS
|
DFS
|
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| CA19-9 (<37 U/mL vs =37 U/mL) | 1.43 | 0.159–0.49 | <0.001 | 1.417 | 0.256–0.887 | 0.019 |
| Histological grade (poor–moderate vs good) | 0.248 | 0.285–0.818 | 0.007 | 0.189 | 0.293–0.817 | 0.006 |
| TNM (I–II vs III–IV) | 2.03 | 1.407–5.413 | 0.008 | 2.34 | 1.218–4.315 | 0.002 |
| m-ULBP1 (low vs high) | 0.42 | 0.165–0.619 | 0.001 | 0.613 | 0.312–1.326 | 0.232 |
| s-ULBP2 (<80.6 pg/mL vs =80.6 pg/mL) | 3.118 | 2.325–7.293 | <0.001 | 3.014 | 2.086–6.262 | <0.001 |
Abbreviations: OS, overall survival; DFS, disease-free survival; HR, hazard ratio; CI, confidence interval; CA, carbohydrate antigen; TNM, tumor, node, metastasis; m-ULBP, membrane ULBP; s-ULBP, soluble ULBP.
Discussion
Many results were obtained from our experiments, namely: 1) expression of ULBP1-3 was heterogeneous and the presence of m-ULBP1 and m-ULBP3 crucial to strengthen immunoprotection against PC, similar to the function of m-MICA; 2) s-ULBP1 and s-ULBP2 were detected at higher levels in the serum of PC patients; and 3) high expression of m-ULBP1 in tissues and low levels of s-ULBP2 in blood were good prognostic indicators for OS of PC patients, the latter also implying an increased DFS.
The NKG2D/NKG2D ligands system contributes to both innate and acquired immune responses.26 Various experiments have shown that NKG2D ligands expression is heterogeneous and can be influenced by stress, posttranscriptional factors, and other stimuli, such as DNA damage. To date, the mechanism of action of this signaling pathway has not been fully elucidated, but for NK cells in humans, the molecular cascade is as follows: 1) DAP10 is phosphorylated when membranous NKG2D recognizes and combines with its ligands (MICA/B or ULBPs present on the surface of cells); 2) in cytoplasm, PI3K or Grb2–Vav1–Sos1 is recruited by DAP10; and 3) NK cells secrete cytokines and cytotoxic granules consisting of IFNγ, GM-CSF, and MIP1β when another costimulatory molecule (such as 2B4 or NKP46) is activated. The results of the IHC indicated that expression of the m-ULBPs was varied.27 Expression of m-ULBP1 was higher in stage I–II PC tissue, but the staining scores of m-ULBP1 and m-ULBP3 were higher in paracancerous tissues than in PC tissues in terminal stages. Expression of ULBPs was affected by the various factors depicted, but which factors were responsible for regulating these ligands in PC is unclear.28,29 Several experiments have demonstrated that DNA damage could induce transcription of ULBPs.30 Global genomic analyses for human PC revealed that a core set of 12 cellular signaling pathways, which accelerate the process of carcinogenesis and genes in the Wnt/Notch- and Hedgehog-signaling pathways, were 100% altered to different degrees. Therefore, we speculated that DNA damage could participate in the regulation of ULBP expression in PC. Moreover, the expression level of m-ULBP1 was considered to be useful in evaluating the risk of patients and screening suitable resectable PCs in patients, on the basis of our finding that m-ULBP1 was highly upregulated in early stage patients compared with terminal patients. The loss of expression of ULBP1 in the terminal stage may help tumor cells escape from immunosurveillance.31 The mechanism by which ULBP is released from the cell membrane is complicated, but has been shown to include such molecules as ADAM, MMP14, and ERP5. Some studies have demonstrated that m-ULBP1–3 could be transferred from donor cells to NK cells only after receptor–ligand interaction and granule mobilization, and that NK cells with acquired NKG2D ligands are targets for autologous NK cells, which can also sequentially transfer the acquired molecule.32 Therefore, the reason that m-ULBP was poorly expressed in the terminal stage after being upregulated in the early stage of PC requires further investigation. Before our study, Duan et al showed that MICA expression was correlated with the TNM stage of PC.32 Our experiments demonstrated that m-ULBP1 is correlated with the TNM stage of PC. Consequently, we concluded that both MICA and m-ULBP1 could be used to evaluate the risk of PC patients, but determining which is better and whether there is any synergistic effect of evaluation between the two ligands needs further investigation.33 The mechanism of soluble ULBP generation has been explained, and ULBPs can also be secreted into the serum in the form of exosomes. The ELISA results showed that the levels of s-ULBP1 and s-ULBP2 were higher in PC patients than in healthy volunteers, but the levels of s-ULBP3 were weakly detected in both groups. Both univariate and multivariate survival analysis demonstrated that a low level of s-ULBP2 in the serum was a good prognostic indicator for OS and DFS.34 These results could be explained by the fact that soluble NKG2D ligands could downregulate the expression of the membrane NKG2D receptor to impair the function of NK cells and T cells and the release of soluble ligands, representing a form of immunoevasion strategy for tumor cells.35 Furthermore, ligand expression is heterogeneous between different tumor types and between individual cells within a tumor entity.36 Moreover, the level of s-MICA was related to the TNM of PC and cancer metastasis, with a lower level of s-MICA being associated with longer OS. Taken together, the results of our experiments confirm the idea that m-ULBPs (membrane NKG2D ligands) are involved in the antitumor immune response, and high levels of s-ULBPs (soluble NKG2D ligands) are correlated with a poor prognosis of patients.37 This is supported by the fact that s-ULBP2 was shown to be a novel serum marker for identification of PC.19 Similarly, Cho et al showed that m-ULBP1–5 were all expressed in cervical cancer tissue and that m-ULBP1 was an independent predictor of good prognosis regarding both DFS and OS.19 Yamaguchi et al demonstrated that s-ULBP2 was useful in the diagnosis of squamous cell carcinoma and could be considered a prognostic indicator for patients with advanced non-small-cell lung cancer.38 However, not all experiments were consistent with our results.21,39 In contrast to our study, some researchers demonstrated that high expression of m-ULBP2 was associated with poor prognosis in ovarian cancer and that soluble Mult1, a kind of high-affinity NKG2D ligand, caused NK-cell activation and tumor rejection.
In summary, the expression and effects of NKG2D ligands differ according to the type of tumor cell, due to the heterogeneity of their generation and sensitivity to the microenvironment of the tumor. However, NKG2D-receptor signaling was shown to be one of the most important pathways of immunotherapy. Therefore, investigating ways in which to specifically upregulate or downregulate the members of this signaling pathway after studying their function in a certain tumor will be the main focus of our future work.
Acknowledgments
This study was funded by the National Natural Science Foundation of China (81071985) and Anhui Province Science and Technology Projects Foundation (10080703038).
Footnotes
Disclosure
The authors report no conflicts of interest in this work. There are no financial relationships with any commercial entities that produce health care-related products and/or services relevant to this article.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Zhang S, Zhao P, Zeng H, Zou X. Report of cancer incidence and mortality in China, 2010. Ann Transl Med. 2014;2:61. doi: 10.3978/j.issn.2305-5839.2014.04.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iovanna J, Mallmann MC, Gonçalves A, Turrini O, Dagorn JC. Current knowledge on pancreatic cancer. Front Oncol. 2012;2:6. doi: 10.3389/fonc.2012.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makkouk A, Weiner GJ. Cancer immunotherapy and breaking immune tolerance: new approaches to an old challenge. Cancer Res. 2015;75:5–10. doi: 10.1158/0008-5472.CAN-14-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.González S, López-Soto A, Suarez-Alvarez B, López-Vázquez A, López-Larrea C. NKG2D ligands: key targets of the immune response. Trends Immunol. 2008;29:397–403. doi: 10.1016/j.it.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Vivier E, Raulet DH, Moretta A, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosman D, Müllberg J, Sutherland CL, et al. ULBPs, novel MHC class I related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14:123–133. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 9.Wennerberg E, Pfefferle A, Ekblad L, et al. Human anaplastic thyroid carcinoma cells are sensitive to NK cell-mediated lysis via ULBP2/5/6 and chemoattract NK cells. Clin Cancer Res. 2014;20:5733–5744. doi: 10.1158/1078-0432.CCR-14-0291. [DOI] [PubMed] [Google Scholar]
- 10.de Kruijf EM, Sajet A, van Nes JG, et al. NKG2D ligand tumor expression and association with clinical outcome in early breast cancer patients: an observational study. BMC Cancer. 2012;12:24. doi: 10.1186/1471-2407-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chitadze G, Lettau M, Bhat J, et al. Shedding of endogenous MHC class I-related chain molecules A and B from different human tumor entities: heterogeneous involvement of the “a disintegrin and metalloproteases” 10 and 17. Int J Cancer. 2013;133:1557–1566. doi: 10.1002/ijc.28174. [DOI] [PubMed] [Google Scholar]
- 12.Liu G, Atteridge CL, Wang X, Lundgren AD, Wu JD. Cutting edge: the membrane type matrix metalloproteinase MMP14 mediates constitutive shedding of MHC class I chain-related molecule A independent of A disintegrin and metalloproteinases. J Immunol. 2010;184:3346–3350. doi: 10.4049/jimmunol.0903789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaiser BK, Yim D, Chow IT, et al. Disulphide-isomerase-enabled shedding of tumour-associated NKG2D ligands. Nature. 2007;447:482–486. doi: 10.1038/nature05768. [DOI] [PubMed] [Google Scholar]
- 14.Kohga K, Takehara T, Tatsumi T, et al. Anticancer chemotherapy inhibits MHC class I-related chain A ectodomain shedding by downregulating ADAM10 expression in hepatocellular carcinoma. Cancer Res. 2009;69:8050–8057. doi: 10.1158/0008-5472.CAN-09-0789. [DOI] [PubMed] [Google Scholar]
- 15.Sun D, Wang X, Zhang H, Deng L, Zhang Y. MMP9 mediates MICA shedding in human osteosarcomas. Cell Biol Int. 2011;35:569–574. doi: 10.1042/CBI20100431. [DOI] [PubMed] [Google Scholar]
- 16.Boutet P, Agüera-González S, Atkinson S, et al. Cutting edge: the metalloproteinase ADAM17/TNF-alpha-converting enzyme regulates proteolytic shedding of the MHC class I-related chain B protein. J Immunol. 2009;182:49–53. doi: 10.4049/jimmunol.182.1.49. [DOI] [PubMed] [Google Scholar]
- 17.Song H, Kim J, Cosman D, Choi I. Soluble ULBP suppresses natural killer cell activity via down-regulating NKG2D expression. Cell Immunol. 2006;239:22–30. doi: 10.1016/j.cellimm.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Ashiru O, Boutet P, Fernández-Messina L, et al. Natural killer cell cytotoxicity is suppressed by exposure to the human NKG2D ligand MICA*008 that is shed by tumor cells in exosomes. Cancer Res. 2010;70:481–489. doi: 10.1158/0008-5472.CAN-09-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho H, Chung JY, Kim S, et al. MICA/B and ULBP1 NKG2D ligands are independent predictors of good prognosis in cervical cancer. BMC Cancer. 2014;14:957. doi: 10.1186/1471-2407-14-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paschen A, Sucker A, Hill B, et al. Differential clinical significance of individual NKG2D ligands in melanoma: soluble ULBP2 as an indicator of poor prognosis superior to S100B. Clin Cancer Res. 2009;15:5208–5215. doi: 10.1158/1078-0432.CCR-09-0886. [DOI] [PubMed] [Google Scholar]
- 21.McGilvray RW, Eagle RA, Rolland P, Jafferji I, Trowsdale J, Durrant LG. ULBP2 and RAET1E NKG2D ligands are independent predictors of poor prognosis in ovarian cancer patients. Int J Cancer. 2010;127:1412–1420. doi: 10.1002/ijc.25156. [DOI] [PubMed] [Google Scholar]
- 22.Guerra N, Tan YX, Joncker NT, et al. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 2008;28:571–580. doi: 10.1016/j.immuni.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu G, Lu S, Wang X, et al. Perturbation of NK cell peripheral homeostasis accelerates prostate carcinoma metastasis. J Clin Invest. 2013;123:4410–4422. doi: 10.1172/JCI69369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi P, Yin T, Zhou F, Cui P, Gou S, Wang C. Valproic acid sensitizes pancreatic cancer cells to natural killer cell-mediated lysis by upregulating MICA and MICB via the PI3K/Akt signaling pathway. BMC Cancer. 2014;14:370. doi: 10.1186/1471-2407-14-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang WJ, Qin SH, Zhang JW, Jiang YY, Zhang JN, Zhao L. Combination doxorubicin and interferon-α therapy stimulates immunogenicity of murine pancreatic cancer Panc02 cells via up-regulation of NKG2D ligands and MHC class I. Asian Pac J Cancer Prev. 2014;15:9667–9672. doi: 10.7314/apjcp.2014.15.22.9667. [DOI] [PubMed] [Google Scholar]
- 26.López-Soto A, Huergo-Zapico L, Acebes-Huerta A, Villa-Alvarez M, Gonzalez S. NKG2D signaling in cancer immunosurveillance. Int J Cancer. 2015;136:1741–1750. doi: 10.1002/ijc.28775. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Basher F, Wu JD. NKG2D ligands in tumor immunity: two sides of a coin. Front Immunol. 2015;6:97. doi: 10.3389/fimmu.2015.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raulet DH, Gasser S, Gowen BG, Deng W, Jung H. Regulation of ligands for the NKG2D activating receptor. Annu Rev Immunol. 2013;31:413–441. doi: 10.1146/annurev-immunol-032712-095951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Textor S, Fiegler N, Arnold A, Porgador A, Hofmann TG, Cerwenka A. Human NK cells are alerted to induction of p53 in cancer cells by upregulation of the NKG2D ligands ULBP1 and ULBP2. Cancer Res. 2011;71:5998–6009. doi: 10.1158/0008-5472.CAN-10-3211. [DOI] [PubMed] [Google Scholar]
- 30.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.López-Cobo S, Romera-Cárdenas G, García-Cuesta EM, Reyburn HT, Valés-Gómez M. Transfer of the human NKG2D ligands UL16 binding proteins (ULBP) 1–3 is related to lytic granule release and leads to ligand retransfer and killing of ULBP-recipient natural killer cells. Immunology. 2015;146:70–80. doi: 10.1111/imm.12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duan X, Deng L, Chen X, et al. Clinical significance of the immunostimulatory MHC class I chain-related molecule A and NKG2D receptor on NK cells in pancreatic cancer. Med Oncol. 2011;28:466–474. doi: 10.1007/s12032-010-9480-9. [DOI] [PubMed] [Google Scholar]
- 33.Chitadze G, Bhat J, Lettau M, Janssen O, Kabelitz D. Generation of soluble NKG2D ligands: proteolytic cleavage, exosome secretion and functional implications. Scand J Immunol. 2013;78:120–129. doi: 10.1111/sji.12072. [DOI] [PubMed] [Google Scholar]
- 34.Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 2002;419:734–738. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 35.Mincheva-Nilsson L, Baranov V. Cancer exosomes and NKG2D receptor-ligand interactions: impairing NKG2D-mediated cytotoxicity and anti-tumour immune surveillance. Semin Cancer Biol. 2014;28:24–30. doi: 10.1016/j.semcancer.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 36.Märten A, von Lilienfeld-Toal M, Büchler MW, Schmidt J. Soluble MIC is elevated in the serum of patients with pancreatic carcinoma diminishing γδ T cell cytotoxicity. Int J Cancer. 2006;119:2359–2365. doi: 10.1002/ijc.22186. [DOI] [PubMed] [Google Scholar]
- 37.Chang YT, Wu CC, Shyr YM, et al. Secretome-based identification of ULBP2 as a novel serum marker for pancreatic cancer detection. PLoS One. 2011;6:e20029. doi: 10.1371/journal.pone.0020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamaguchi K, Chikumi H, Shimizu A, et al. Diagnostic and prognostic impact of serum-soluble UL16-binding protein 2 in lung cancer patients. Cancer Sci. 2012;103:1405–1413. doi: 10.1111/j.1349-7006.2012.02330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng W, Gowen BG, Zhang L, et al. Antitumor immunity: a shed NKG2D ligand that promotes natural killer cell activation and tumor rejection. Science. 2015;348:136–139. doi: 10.1126/science.1258867. [DOI] [PMC free article] [PubMed] [Google Scholar]