Abstract
Background
Previously, we found that the expression of decoy receptor 3 (DcR3) in gliomas was significantly upregulated compared to normal brain tissues. However, the effect of DcR3-specific small interfering RNA (siRNA) on cell biological function of glioma cells remains incompletely understood.
Objective
The aim of this study was to explore the effect of DcR3 siRNA on cell growth and apoptosis of glioma cells and to investigate the potential downstream pathways affected by DcR3.
Methods
DcR3-specific siRNA was transfected into three glioma cell lines (U251MG, LN-308, and U87MG) using combiMAGnetofection method. MTS tetrazolium assay and fluorimetric resorufin viability assay were used to assess the growth of glioma cells. Then, apoptosis was examined using the Hoechst 33342/propidium iodide double-staining assay and fluorescent caspase-3/7 assay. Meanwhile, Western blot was performed to explore the probable pathway by which DcR3-specific siRNA acts in glioma cells. Also, microarray dataset analysis was applied to analyze the potential function of DcR3 in glioma.
Results
The DcR3-specific siRNA had a potent effect on cell growth and apoptosis of all three glioma cells tested, and the effects were time dependent. Among these three glioma cell lines, U251MG had the most significant effect with regard to growth inhibition and apoptosis induction. MTS assay showed that the proliferation rate at 72 and 96 hours after the transfection was 76.333%±5.131% (t=7.611, P=0.002) and 64.333%±5.859% (t=10.983, P<0.001), respectively. The viability rate of U251MG cells was 80.667%±2.309% (t=12.302, P<0.001) and 62.333%±2.082% (t=21.213, P<0.001) at 72 and 96 hours posttreatment, respectively. The caspase-3/7 activity of U251MG cells was 2.76 (t=−6.601, P=0.003) and 4.75 (t=−9.189, P=0.001) folds that of the mock control at 72 and 96 hours, respectively. The apoptosis rate was increased to 1.85 (t=−2.496, P=0.067) and 3.93 (t=−12.587, P<0.001) folds at 72 and 96 hours after transfection, respectively. Furthermore, the levels of phospho-ERK1/2 and phospho-AKT were significantly downregulated after DcR3 silencing.
Conclusion
The DcR3-specific siRNA could efficiently inhibit growth and induce apoptosis of cells via affecting ERK and AKT. Hence, DcR3-specific siRNA treatment could act as a supplementary targeted therapy strategy for gliomas.
Keywords: DcR3, glioma, siRNA, proliferation, viability, apoptosis
Introduction
Decoy receptor 3 (DcR3), also known as tumor necrosis factor receptor superfamily member 6B (TNFRSF6B), is located on chromosome 20q13.3.1–4 Some studies have published evidence on DcR3 overexpression in some malignant tumors such as colorectal carcinoma, gastric cancer, hepatocellular carcinoma, pancreatic head carcinoma, renal cell carcinoma, and bladder transitional cell carcinoma.5,6 Additionally, DcR3 was shown to correlate with biological functions of proliferation, apoptosis, invasion, and metastasis.5,6
Malignant glioma is one of the most common nosogenic and lethal malignant brain tumors worldwide. Malignant gliomas represent >50% of primary malignant brain cancers.7,8 Due to the difficulty of brain surgery and a high invasiveness and recurrence of glioma, it is still a challenging disease to treat.9 Thus, it is imperative to discover a new therapeutic strategy for the treatment of malignant glioma.
Until now, only three papers related to DcR3 in gliomas have been published. Arakawa et al10 concluded that the overexpression of DcR3 mRNA and protein might be correlated with gene amplification in glioblastomas. Roth et al11 found that DcR3 might be related to the progression and immune evasion of malignant gliomas. Hwang et al12 demonstrated that no change was found in DcR3 serum concentration before and after tumor removal in glioma patients. In our previous work, we also found that DcR3 was overexpressed in gliomas. The overexpression of DcR3 was related to tumorigenesis, differentiation, and proliferation of the glioma.13 We also verified that DcR3 neutralization antibody had an influence on inhibition of cell growth and induction of apoptosis in glioma cells.14 To further explore the role of DcR3 in malignant glioma cells, we herein transfected the designed and synthesized DcR3-specific small interfering RNA (siRNA) into three glioma cell lines. Subsequently, the function of DcR3 on growth and apoptosis of glioma cells was explored.
Materials and methods
Cell culture and transfection with DcR3-specific siRNA
Three malignant glioma cell lines (U251MG, LN-308, and U87MG) of humans were purchased from the American Type Culture Collection (ATCC, Rockville, MD, USA). These three cell lines were incubated in Dulbecco’s Modified Eagle Medium (DMEM). All cell lines were supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen Corp Grand Island, NY, USA) under a humidified 5% CO2 atmosphere with 2 mM glutamine and gentamicin at 37°C. Four DcR3-specific siRNAs (Table 1) were synthesized by GenePharma (ShangHai, People’s Republic of China) and merged into one siRNA pool. Five groups were designed in the current study: blank control, mock control, negative siRNA control, scramble siRNA, and DcR3-specific siRNA. Blank control groups were cultured with only serum-free DMEM medium without any transfection reagents or siRNA. In the mock control groups, only transfection reagent was added. Negative control groups were treated with negative siRNA (GenePharma). The sequences of DcR3 siRNAs were scrambled and rearranged as another control (Scramble siRNAs). CombiMAGnetofection (OZ Biosciences, Marseille cedex 9, France) was used for the transfection. All the in vitro experiments were performed in triplicate.
Table 1.
Sequence of DcR3-specific small interfering RNA
| Name of siRNA | Exon | Sequence | Location |
|---|---|---|---|
| DcR3 siRNA 103 | 1 | GCUCCAGCAAGGACCAUGATT | C103 |
| DcR3 siRNA 193 | 1 | GGCUGUACGCGGAGUGGCATT | C193 |
| DcR3 siRNA 355 | 1 | CGCAGUUCUGGAACUACCUTT | C355 |
| DcR3 siRNA 552 | 2 | GCCAGAACACGCAGUGCCATT | C552 |
Notes: Four siRNAs-targeting DcR3 sequences are shown. DcR3-specific siRNAs were synthesized by GenePharma in Shanghai and merged into a pool. All the sequences of siRNA were analyzed using BLAST to avoid nonspecific silencing of unrelated genes.
Abbreviations: siRNA, small interfering RNA; BLAST, Basic local alignment search tool.
Cell proliferation
A colorimetric tetrazolium (MTS) assay (CellTiter96 Aqueous One Solution Cell Proliferation Assay, G3580; Promega, Madison, WI, USA) was used to detect cell proliferation. A 100 µL volume of 2.5×103 cells per well was seeded in 96-well culture plates. Cells were placed in six parallel wells and then cultured at 37°C for 0, 24, 48, 72, and 96 hours, respectively. Then, 20 µL MTS reagent was added to each well and the plates cultivated at 37°C for 2 hours in a humidified environment with 5% CO2. Then, the absorbance at 490 nm was read using a 96-well microplate reader (Scientific Multiskan MK3, Thermo Finland). U251MG, LN-308, and U87MG cells were collected during the logarithmic growth phase. The results are shown by the ratio compared with mock control.
Cell viability
To confirm the results from the MTS assay, fluorimetric detection of resorufin (CellTiter-Blue Cell Viability Assay, G8080; Promega) was utilized to measure the cell viability. The procedure given in the manufacturer instructions was followed. An FL600 fluorescence plate reader (BioTek, Winooski, VT, USA) was used for fluorimetry analysis (excitation 560 nm/emission 590 nm). Fluorescence data were presented as the fluorescence of treated sample/mock control ×100%.
Caspase-3/7 activity detection
After the measurement of cell viability, caspase-3/7 activity was tested instantly using a synthetic rhodamine-labeled caspase-3/7 substrate (Apo-ONE® Homogeneous Caspase-3/7 Assay, G7790; Promega) on the same samples according to the manufacturer’s instructions. After incubation for 1 hour at room temperature, an FL600 fluorescence plate reader (BioTek) was used to measure the fluorescence of each well. Caspase-3/7 activity was obtained from the fluorescence of treated sample/mock control.
Evaluation of cell apoptosis
The evaluation of cell apoptosis and morphology was evaluated with Hoechst 33342 and propidium iodide (PI) (Sigma-Aldrich N.V., Bornem, Belgium) double fluorescent chromatin staining. Briefly, cells were washed in ice-cold phosphate-buffered saline (PBS) after treatment and then stained with Hoechst 33342 (1 mg/mL) and PI (1 mg/mL) for 15 minutes. Cells were observed under a fluorescence microscope. Cell apoptosis and morphology could be identified based on the chromatin condensation and nuclear fragmentation. The absolute number of Hoechst 33342-positive/PI-negative viable cells; PI-positive necrotic cells and debris signals; Hoechst 33342-positive/PI-negative early apoptotic cells with blue fragmentations in the cells; and Hoechst 33342-positive/PI-positive late apoptotic cells with red fragmentations in the cells could be identified. The collection of viable, apoptotic, and necrotic cells were counted under the 200× magnification in ten different fields by two persons independently, and the average result of these counts was then compared to the mock control.
Western blot analysis
After various treatments in six-well plates, the cells were washed with PBS and then lysed in a buffer of 10% glycerol, 5% sodium dodecyl sulfate (SDS), 5 mM ethylenediaminetetraacetic acid (pH 8), 80 mM Tris–HCl (pH 6.8), 0.2% bromophenolblue, 5% 2-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride. The lysates were centrifuged at 12,000× g for 10 minutes in a 4°C atmosphere and boiled for 5 minutes. Each sample with 25 µg of protein was run on SDS–polyacrylamide gel electrophoresis (8% SDS–acrylamide gel). Then, Hybond ECL nitrocellulose membranes (GE Healthcare Bio-sciences/Amersham, Diegem, Belgium) were used to separate proteins for 2 hours at 100 mA. The membrane was stained with DcR3 (ab11930; Abcam, Cambridge, UK), phospho-AKT (pS473; Invitrogen Corp.), phospho-ERK1/2 (pTpY185/187; Invitrogen Corp.), or β-actin (AC-15; Sigma-Aldrich) and subjected to chemiluminescence detection assay (GE Healthcare Bio-sciences/Amersham).14–18
Microarray datasets analysis
Considering some microarray datasets could also help us analyze the potential function of DcR3 in glioma, we then collected the microarray datasets of DcR3 in glioma from the European Bioinformatics Institute (EBI) Array Express (http://www.ebi.ac.uk/arrayexpress/) database, National Center of Biotechnology Information (NCBI), and the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) database up to May 24, 2016. The keywords for microarray screening were as follows: (“glioma” or “astrocytoma” or “oligodendroglioma” or “oligoastrocytoma” or “ependymoma” or “glioblastoma” or “gliomatosis cerebri” or “brain cancer” or “brain neoplasm” or “brain tumor” or “GBM”) and (DcR3 or “Decoy receptor 3” or TNFRSF6B or TR6 or M68).
Statistical analysis
SPSS 20.0 software (IBM Corporation, Armonk, NY, USA) was used for statistical analysis. Data on proliferation, cell viability, caspase-3/7 activity, and apoptosis were presented as mean ± standard deviation. Analysis of variance and Student’s t-test were used to examine statistical significance of alterations between groups. A P-value <0.05 was considered statistically significant.
Results
Cell growth inhibitory effect of DcR3-specific siRNA
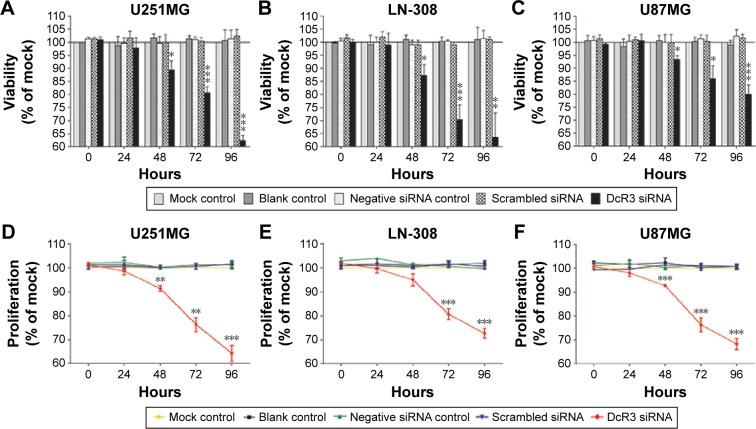
The MTS tetrazolium assay, Hoechst 33342/PI double-staining assay, and fluorimetric resorufin viability assay were applied to evaluate the growth of glioma cells. From the result of MTS tetrazolium assay, it was seen that a large cell growth inhibition was showed in all three cell lines, especially at 96 hours posttransfection. DcR3-specific siRNA exerted the most powerful effect on U251MG cells among the three cell lines. The proliferation rate at 72 and 96 hours after transfection was 76.333%±5.131% (t=7.611, P=0.002) and 64.333%±5.859% (t=10.983, P<0.001), respectively. Hoechst 33342/PI double-staining assay and fluorimetric resorufin viability assay were used to confirm these results, which could largely reflect the cell growth inhibitory effect of DcR3-specific siRNA. The viability rate of U251MG cells was 80.667%±2.309% (t=12.302, P<0.001) and 62.333%±2.082% (t=21.213, P<0.001) at 72 and 96 hours posttreatment, respectively. The other two cell lines had a less obvious effect on cell growth inhibition. The proliferation rate of LN-308 cells was 72.667%±3.512% (t=12.663, P<0.001) at 96 hours, and the viability rate was 63.667%±9.292% (t=6.919, P=0.002) as detected using MTS tetrazolium assay and fluorimetric resorufin viability assay, respectively (data from Hoechst 33342/PI double-staining assay not shown). Also, the proliferation rate of the third cell line U87MG was 68.333%±4.042% (t=12.962, P<0.001) as assessed by MTS tetrazolium assay at 96 hours, and the inhibitory effect on cell growth was slightly more sensitive than that detected by fluorimetric resorufin viability assay (viability rate: 80.000%±3.606%, t=9.584, P=0.001, Figure 1, Table 2). However, the suppressive role of DcR3-specific siRNA on growth of glioma cells showed the same trend as examined by three independent approaches.
Figure 1.
Effect of DcR3-specific siRNA on cell growth in glioma cell lines.
Notes: U251MG, LN-308, and U87MG cells (2.5×103 cells per well in 96-well plate) were cultured for 24 hours and then transfected with negative siRNA, scrambled siRNA, and DcR3-specific siRNA and incubated for another 96 hours. Cell viability (U251MG (A), LN-308 (B), and U87MG (C)) and cell proliferation (U251MG (D), LN-308 (E), and U87MG (F)) were assessed by MTS tetrazolium assay and fluorimetric resorufin viability assay, respectively. *P<0.05, **P<0.01, and ***P<0.001 compared to negative siRNA controls and scrambled siRNAs at the same time point.
Abbreviation: siRNA, small interfering RNA.
Table 2.
Cell growth inhibitory effect of DcR3-specific siRNA using different techniques and cell lines
| Time (hours) | U251MG
|
LN-308
|
U87MG
|
||||
|---|---|---|---|---|---|---|---|
| Scrambled siRNA (%) | DcR3 siRNA (%) | Scrambled siRNA (%) | DcR3 siRNA (%) | Scrambled siRNA (%) | DcR3 siRNA (%) | ||
| MTS tetrazolium assay | 0 | 100.333±1.528 | 101.333±1.528 | 101.000±2.646 | 102.000±1.000 | 99.667±0.577 | 100.667±1.528 |
| 24 | 100.667±1.154 | 98.667±2.517 | 101.000±1.732 | 99.667±3.215 | 99.667±0.577 | 98.000±2.646 | |
| 48 | 100.000±1.000 | 91.333±2.082 | 100.333±1.155 | 95.000±4.359 | 101.333±0.577 | 92.667±0.577 | |
| 72 | 100.667±2.082 | 76.333±5.131 | 101.667±2.309 | 80.667±4.042 | 101.000±2.000 | 76.333±5.132 | |
| 96 | 101.667±0.577 | 64.333±5.859 | 100.667±1.528 | 72.667±3.512 | 100.667±1.528 | 68.333±4.042 | |
| Fluorimetric resorufin viability assay | 0 | 101.333±0.577 | 101.000±1.000 | 101.667±1.155 | 100.000±1.000 | 101.333±1.528 | 99.333±0.577 |
| 24 | 101.667±2.517 | 97.667±4.041 | 102.000±2.000 | 99.000±4.359 | 101.000±1.000 | 100.667±2.517 | |
| 48 | 100.000±3.000 | 89.333±3.512 | 99.000±1.732 | 87.333±4.163 | 100.000±3.000 | 93.333±1.528 | |
| 72 | 100.333±1.528 | 80.667±2.309 | 99.000±1.000 | 70.333±5.686 | 100.333±2.517 | 86.000±5.000 | |
| 96 | 102.333±2.517 | 62.333±2.082 | 101.000±1.000 | 63.667±9.292 | 100.667±1.528 | 80.000±3.606 | |
Abbreviation: siRNA, small interfering RNA.
Apoptosis induced by DcR3-specific siRNA
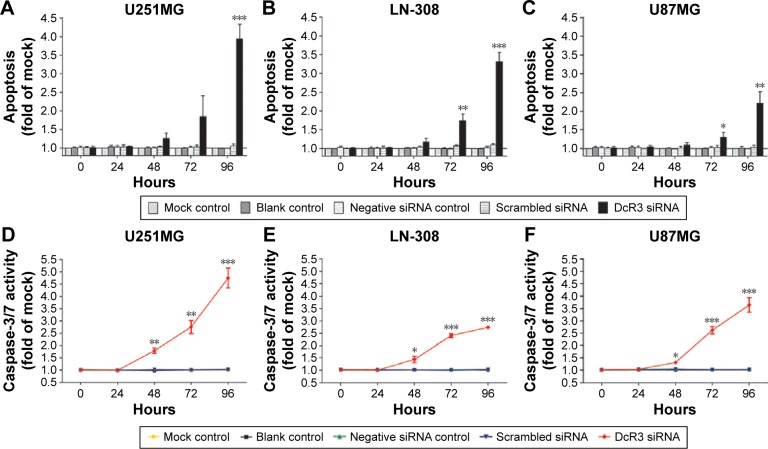
The caspase family is directly responsible for the dissolution of the protease system, and it occupies the central position in the cell apoptosis mechanism. So, caspase-3/7 could be used to reflect apoptosis. In this study, Hoechst 33342/PI double-staining assay was applied together with a fluorescent caspase-3/7 assay to validate whether DcR3-specific siRNA could affect apoptosis in glioma cells. Over time, the results showed that in all three glioma cell lines, caspase-3/7 activity was obviously enhanced with DcR3-specific siRNA, especially at 96 hours. Similarly, in all three cell lines, DcR3-specific siRNA had the most effective influence on U251MG cells. The caspase-3/7 activity of U251MG cells was 2.76 (t=−6.601, P=0.003) and 4.75 (t=−9.189, P=0.001) folds that of the mock control at 72 and 96 hours, respectively. The results from Hoechst 33342 and PI double fluorescent staining revealed that the apoptosis rate was increased to 1.85 (t=−2.496, P=0.067) and 3.93 (t=−12.587, P<0.001) folds at 72 and 96 hours after the transfection, respectively. The other two cell lines had a less obvious effect on cell apoptosis induction. In LN-308 cells, the caspase activity was 2.75 (t=−23.567, P<0.001) folds that of the mock control at 96 hours, and the apoptosis rate was increased to 3.31 (t=−15.027, P<0.001) folds that of the mock control. The caspase activity was 3.65 (t=−8.794, P=0.001) folds that of the mock control at 96 hours in U87MG cells, and the apoptosis rate was increased to 2.21 (t=−6.569, P=0.003) folds that of the mock control (Figures 2 and 3, Table 3). Furthermore, we also compared the functional change of DcR3-specific siRNA group to that of the other four groups (blank control, mock control, negative siRNA control, and scramble siRNA), and the results showed similar, statistically significant results among the four groups. Thus, we only showed the results of the comparison between the DcR3-specific siRNA group and mock control group.
Figure 2.
Effect of DcR3-specific siRNA on cell apoptosis in glioma cells.
Notes: U251MG, LN-308, and U87MG cells (2.5×103 cells per well in 96-well plate) were cultured for 24 hours and then transfected with negative siRNA, scrambled siRNA, and DcR3-specific siRNA and incubated for up to another 96 hours. Apoptosis (U251MG (A), LN-308 (B), and U87MG (C)) and caspase-3/7 activity (U251MG (D), LN-308 (E), and U87MG (F)) were assessed by the Hoechst 33342/PI double-staining assay and fluorescent caspase-3/7 assay, respectively. *P<0.05, **P<0.01, and ***P<0.001 compared to negative siRNA controls and scrambled siRNAs at the same time point.
Abbreviations: siRNA, small interfering RNA; PI, propidium iodide.
Figure 3.
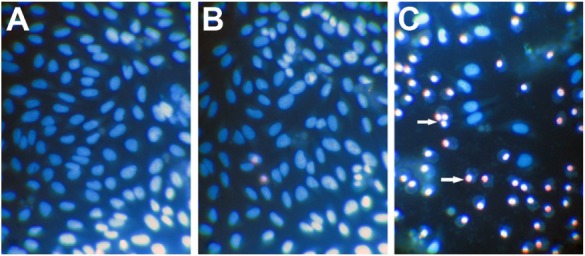
The evaluation of cell morphology and apoptosis.
Notes: DcR3-specific siRNA suppressed cell growth and induced apoptosis with Hoechst 33342/PI double fluorescent chromatin staining. U251MG cells (2.5×103 cells per well in 96-well plate) were cultured for 24 hours and then transfected with negative siRNA, scrambled siRNA, and DcR3-specific siRNA and incubated for up to another 96 hours. Hoechst 33342/PI double fluorescent chromatin staining was performed to examine the effect of DcR3-specific siRNA on the cell growth and apoptosis in U251MG cells. Hoechst 33342 positive/PI positive with red fragmentations in the cells were late apoptotic cells and Hoechst 33342 positive/PI negative with blue fragmentations in the cells were early apoptotic cells. (A) Mock control, (B) scrambled siRNA, (C) DcR3-specific siRNA. Arrows show apoptotic cells. Magnification, 400×.
Abbreviations: siRNA, small interfering RNA; PI, propidium iodide.
Table 3.
Cell apoptosis induced by DcR3-specific siRNA using different techniques and cell lines
| Time (hours) | U251MG
|
LN-308
|
U87MG
|
||||
|---|---|---|---|---|---|---|---|
| Scrambled siRNA (FC) | DcR3 siRNA (FC) | Scrambled siRNA (FC) | DcR3 siRNA (FC) | Scrambled siRNA (FC) | DcR3 siRNA (FC) | ||
| Fluorescent caspase-3/7 assay | 0 | 0.993±0.006 | 0.993±0.006 | 1.010±0.0173 | 1.020±0.0346 | 1.007±0.021 | 1.020±0.035 |
| 24 | 0.990±0.100 | 0.990±0.100 | 1.010±0.020 | 1.001±0.021 | 1.033±0.025 | 1.037±0.050 | |
| 48 | 1.023±0.035 | 1.790±0.182 | 1.023±0.035 | 1.433±0.219 | 1.043±0.047 | 1.307±0.104 | |
| 72 | 1.017±0.052 | 2.757±0.454 | 1.000±0.036 | 2.403±0.137 | 1.020±0.066 | 2.623±0.261 | |
| 96 | 1.033±0.068 | 4.750±0.697 | 1.040±0.066 | 2.747±0.107 | 1.027±0.045 | 3.647±0.514 | |
| Hoechst 33342/PI double-staining assay | 0 | 1.017±0.021 | 1.017±0.038 | 1.003±0.058 | 1.007±0.208 | 1.017±0.021 | 1.02±0.035 |
| 24 | 1.023±0.681 | 1.040±0.173 | 1.01±0.046 | 1.020±0.017 | 1.003±0.006 | 1.050±0.035 | |
| 48 | 1.033±0.252 | 1.263±0.142 | 1.033±1.025 | 1.170±0.106 | 1.013±0.045 | 1.097±0.061 | |
| 72 | 1.027±0.055 | 1.847±0.566 | 1.047±0.029 | 1.743±0.176 | 1.027±0.055 | 1.293±0.142 | |
| 96 | 1.057±0.577 | 3.937±0.392 | 1.087±0.035 | 3.313±0.254 | 1.037±0.050 | 2.213±0.306 | |
Abbreviations: siRNA, small interfering RNA; PI, propidium iodide; FC, fold change.
Contribution of DcR3-specific-siRNA on relevant cellular signaling
AKT (protein kinase B) and ERK (extracellular signal-regulated kinase) pathways are related to cell growth and apoptosis of glioma.19 To investigate the contribution of DcR3-specific siRNA in regulating cellular signaling, we tested the signaling of AKT and ERK pathways with Western blot. The phospho-AKT and phospho-ERK1/2 were downregulated by DcR3-specific siRNA at 96 hours posttreatment. No significant changes were observed in controls (Figure 4).
Figure 4.
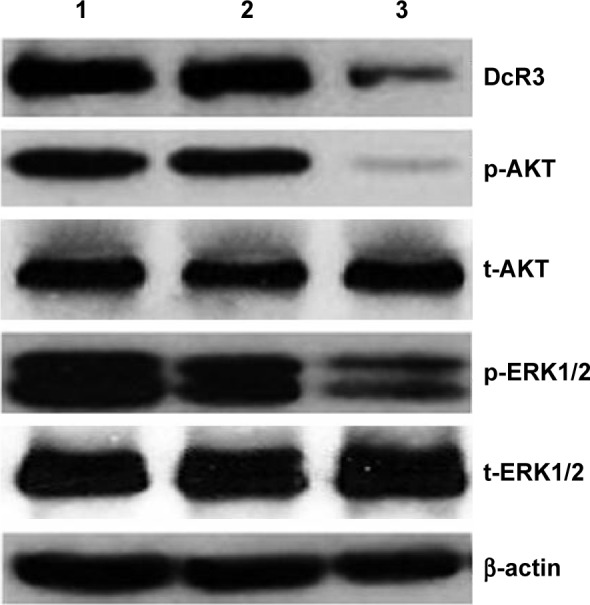
Effect of DcR3-specific siRNA on relevant downstream pathways in glioma U251MG cells.
Notes: The relevant downstream proteins were examined by Western blot. Antibodies included are as follows: DcR3, p-AKT. t-AKT, p-ERK1/2, t-ERK1/2, and β-actin. 1, Mock control; 2, scrambled siRNA; 3, DcR3-specific siRNA.
Abbreviation: siRNA, small interfering RNA.
Microarray datasets analysis
There were two eligible datasets available. Choi et al20 compared DcR3 expression between cyclophilin B (CypB)-depleted human glioblastoma multiforme (GBM) cells and the control group. The datasets showed that the expression level of DcR3 in CypB depletion (mean value 63.3243) was higher than that in the control group (mean value 57.1218). That study suggested that pharmacological inhibition of CypB induced several tumor suppressive responses, especially cell death and senescence in GBM cells; so, the CypB could be a novel molecular target for GBM clinical medication. Combining these experimental results and their profiling data, we concluded that the DcR3 could also be the therapeutic molecular target of glioma and that DcR3 might have a connection with CypB in glioma. Huang et al21 established that genes responded to focal adhesion kinase inhibitor Y15 alone and also to Y15 with temozolomide, which are essential for glioblastoma therapy, using microarray gene expression analysis. The dataset showed that the DcR3 expression in Y15 and temozolomide group had a tendency to decline compared with control group in both DBTRG and U87MG cells. The Y15 group and the Y15 and temozolomide combination group to treat glioblastoma were both associated with DcR3. The association also indicates that DcR3 might be a worthwhile candidate for further experimental study for the treatment strategies of glioblastoma.
Discussion
Previously, we found that DcR3 was expressed at a markedly higher level in glioma than in normal brain tissues. DcR3 was also positively correlated with tumorigenesis, differentiation, and proliferation of glioma.13 To investigate the function of DcR3 in glioma, we sequentially investigated the effect of anti-DcR3 neutralization monoclonal antibody on the malignant phenotype of glioma cells in vitro.14 We found that DcR3 neutralization antibody could function as a cell growth inhibitor and apoptosis inducer in glioma cells. In the study with DcR3 neutralization antibody, the most obvious response was observed in U251MG cells. The proliferation rate of U251MG cells at 96 hours was 70.667%±5.033%, and the caspase activity at 96 hours was 1.687 folds after treatment with a dose of 0.8 mg/L. To further explore the biological function and potential application of DcR3as a molecular target therapy in glioma, DcR3-specific siRNA was transfected into three cell lines (U251MG, LN-308, and U87MG) to detect the effect on cell proliferation, viability, apoptosis, and caspase-3/7 activity. As expected, DcR3-specific siRNA could influence cell growth inhibition and apoptosis induction of glioma cells. However, the effect was stronger compared to that produced by DcR3 neutralization antibody. Especially, the caspase activity of U251MG cells was 4.75 folds that of the mock control at 96 hours, much more powerful than the maximum effect from DcR3 neutralization antibody. Compared with anti-DcR3 antibody, which displays function at translation level, DcR3-specific siRNA could silence the expression of DcR3 at the transcription level and knockdown DcR3 more thoroughly. Until now, 19 siRNA-based molecular medicines have been used clinically to treat several diseases such as eye diseases, virus infection, cancer, and so on.22–24 But no siRNA-based molecular medicines have been used in glioma. The present research could provide the experimental evidence for clinical usage of siRNA-based molecular medicines in treating glioma patients.
Several studies have showed that DcR3 plays an important role in the malignancy of cancer. Zong et al25 reported that DcR3 may promote the lymph node metastasis of colorectal cancer and that DcR3 was related to a poor overall survival. Wang et al3 demonstrated that DcR3 expressed at a high level in pancreatic cancer and could act as an extracellular antiapoptotic molecule by binding to TRAIL and counteracting its death-promoting function. The expression of DcR3 was connected with resistance to chemotherapy and malignancy. Wu et al26 found that the tumor infiltration and lymphatic metastasis were closely related with the expression of DcR3 in breast cancer tissue. Jiang et al27 pointed out that DcR3 was related to the pathways of promoting proliferation, inhibiting apoptosis, and even inducing angiogenesis via vascular endothelial growth factor, and thus DcR3 could influence the survival of bladder urothelial carcinoma patients. We hypothesized from the results of the aforementioned studies that DcR3 might also be related with the invasiveness and treatment refractoriness of glioma. To further investigate the possible mechanisms between DcR3 and glioma, we searched the possible pathways in glioma. As reported, ERK and AKT pathways were related to cell biological function and cancer malignancy, which could also play a role proliferation, viability, and apoptosis in cancers.28 Until now, mechanisms of association between DcR3 and glioma have been explored. Toda et al29 demonstrated that DcR3 might enhance tumor progression and promote cell proliferation in human malignant fibrous histiocytoma via inhibiting FasL-induced apoptosis, thereby regulating cell invasion and migration by matrix metalloproteinase-2 activation via the PI3K/AKT pathway. In our research, Western blot was performed to explore the probable influence of DcR3-specific siRNA on AKT and ERK1/2 pathways. We also confirmed that DcR3 could influence glioma via the ERK and AKT pathways. Other mechanisms of DcR3 in cancer have also been explored. Roth et al11 found that DcR3 might be related to the progression and immune evasion of gliomas. Weissinger et al5 identified DcR3 as playing a role as a driver of adhesion, invasiveness, and migration in renal cell carcinoma via a signaling axis, including PI3K/AKT and NFATc1. Dang et al15 found that DcR3 was a target of miR-152 and that the overexpression of DcR3 could reduce the miR-152 level in hepatocellular carcinoma. But the function of miR-152 in glioma needs further investigation. From the clinical point of view, further research is needed to focus on the gene signal pathway in detail and the connection with clinical medication.
Conclusion
Together with the previous study regarding the effects of anti-DcR3 antibody on glioma, we demonstrated that DcR3 could lead to a reduction of growth and induction of apoptosis in glioma cells in vitro. This is the first report to confirm the effect of DcR3-specific siRNA on human glioma cells. Similar to the effect of anti-DcR3 antibody, DcR3 siRNA could exert more powerful effect on cell biological function, especially on activated caspase activity and apoptosis, via influencing ERK and AKT pathways. The effect of DcR3 on the growth and apoptosis of glioma cells could guide researchers to develop novel clinical medication directly targeting DcR3. The relationship between DcR3 and malignant glioma cells could lead to a breakthrough in glioblastoma treatment.
Acknowledgments
The study was supported partly by the fund of Guangxi Natural Scientific Research (No 2015GXNSFBA139130), Youth Science Foundation of Guangxi Medical University (GXMUYSF2014039), and Guangxi University Science and Technology Research Projects (201510598068). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Morishige T, Yoshioka Y, Inakura H, et al. Creation of a lysine-deficient LIGHT mutant with the capacity for site-specific PEGylation and low affinity for a decoy receptor. Biochem Biophys Res Commun. 2010;393(4):888–893. doi: 10.1016/j.bbrc.2010.02.119. [DOI] [PubMed] [Google Scholar]
- 2.Connor JP, Felder M, Kapur A, Onujiogu N. DcR3 binds to ovarian cancer via heparan sulfate proteoglycans and modulates tumor cells response to platinum with corresponding alteration in the expression of BRCA1. BMC Cancer. 2012;12:176. doi: 10.1186/1471-2407-12-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang W, Zhang M, Sun W, et al. Reduction of decoy receptor 3 enhances TRAIL-mediated apoptosis in pancreatic cancer. PLoS One. 2013;8(10):e74272. doi: 10.1371/journal.pone.0074272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang D, Fan X, Yin P, et al. Significance of decoy receptor 3 (Dcr3) and external-signal regulated kinase 1/2 (Erk1/2) in gastric cancer. BMC Immunol. 2012;13:28. doi: 10.1186/1471-2172-13-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weissinger D, Tagscherer KE, Macher-Goppinger S, Haferkamp A, Wagener N, Roth W. The soluble Decoy Receptor 3 is regulated by a PI3K-dependent mechanism and promotes migration and invasion in renal cell carcinoma. Mol Cancer. 2013;12(1):120. doi: 10.1186/1476-4598-12-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen G, Rong M, Luo D. TNFRSF6B neutralization antibody inhibits proliferation and induces apoptosis in hepatocellular carcinoma cell. Pathol Res Pract. 2010;206(9):631–641. doi: 10.1016/j.prp.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 7.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 8.Liu HL, Fan CH, Ting CY, Yeh CK. Combining microbubbles and ultrasound for drug delivery to brain tumors: current progress and overview. Theranostics. 2014;4(4):432–444. doi: 10.7150/thno.8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke J, Butowski N, Chang S. Recent advances in therapy for glioblastoma. Arch Neurol. 2010;67(3):279–283. doi: 10.1001/archneurol.2010.5. [DOI] [PubMed] [Google Scholar]
- 10.Arakawa Y, Tachibana O, Hasegawa M, Miyamori T, Yamashita J, Hayashi Y. Frequent gene amplification and overexpression of decoy receptor 3 in glioblastoma. Acta Neuropathol. 2005;109(3):294–298. doi: 10.1007/s00401-004-0956-6. [DOI] [PubMed] [Google Scholar]
- 11.Roth W, Isenmann S, Nakamura M, et al. Soluble decoy receptor 3 is expressed by malignant gliomas and suppresses CD95 ligand-induced apoptosis and chemotaxis. Cancer Res. 2001;61(6):2759–2765. [PubMed] [Google Scholar]
- 12.Hwang SL, Lin CL, Cheng CY, et al. Serum concentration of soluble decoy receptor 3 in glioma patients before and after surgery. Kaohsiung J Med Sci. 2004;20(3):124–127. doi: 10.1016/S1607-551X(09)70095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S, Chen G, Dang Y, Chen LH. Overexpression of DcR3 and its significance on tumor cell differentiation and proliferation in glioma. ScientificWorldJournal. 2014;2014:605236. doi: 10.1155/2014/605236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruan Y, Huang S, He D, Gopaul R, Li Z, Chen G. Effect of TNFRSF6B neutralization antibody on cell growth suppression and apoptosis induction in glioma cells. Neoplasma. 2015;62(4):574–581. doi: 10.4149/neo_2015_069. [DOI] [PubMed] [Google Scholar]
- 15.Dang YW, Zeng J, He RQ, Rong MH, Luo DZ, Chen G. Effects of miR-152 on cell growth inhibition, motility suppression and apoptosis induction in hepatocellular carcinoma cells. Asian Pac J Cancer Prev. 2014;15(12):4969–4976. doi: 10.7314/apjcp.2014.15.12.4969. [DOI] [PubMed] [Google Scholar]
- 16.Huang S, He R, Rong M, Dang Y, Chen G. Synergistic effect of MiR-146a mimic and cetuximab on hepatocellular carcinoma cells. Biomed Res Int. 2014;2014:384121. doi: 10.1155/2014/384121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rong M, Chen G, Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13:21. doi: 10.1186/1471-2407-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen G, Noor A, Kronenberger P, Teugels E, Umelo IA, De Greve J. Synergistic effect of afatinib with su11274 in non-small cell lung cancer cells resistant to gefitinib or erlotinib. PLoS One. 2013;8(3):e59708. doi: 10.1371/journal.pone.0059708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aroui S, Dardevet L, Ajmia WB, et al. A novel platinum-maurocalcine conjugate induces apoptosis of human glioblastoma cells by acting through the ROS-ERK/AKT-p53 Pathway. Mol Pharm. 2015;12(12):4336–4348. doi: 10.1021/acs.molpharmaceut.5b00531. [DOI] [PubMed] [Google Scholar]
- 20.Choi JW, Schroeder MA, Sarkaria JN, Bram RJ. Cyclophilin B supports Myc and mutant p53-dependent survival of glioblastoma multiforme cells. Cancer Res. 2014;74(2):484–496. doi: 10.1158/0008-5472.CAN-13-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang G, Ho B, Conroy J, Liu S, Qiang H, Golubovskaya V. The microarray gene profiling analysis of glioblastoma cancer cells reveals genes affected by FAK inhibitor Y15 and combination of Y15 and temozolomide. Anticancer Agents Med Chem. 2014;14(1):9–17. doi: 10.2174/18715206113139990141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Duan F, Lin L, Huang Q, Wu K. A new approach of delivering siRNA to the cornea and its application for inhibiting herpes simplex keratitis. Curr Mol Med. 2014;14(9):1215–1225. doi: 10.2174/1566524014666141021145909. [DOI] [PubMed] [Google Scholar]
- 23.Kong J, Liu X, Jia J, et al. Pokemon siRNA delivery mediated by RGD-modified HBV core protein suppressed the growth of hepatocellular carcinoma. Hum Gene Ther Methods. 2015;26(5):175–180. doi: 10.1089/hgtb.2015.093. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Stamatoyannopoulos G, Song CZ. Down-regulation of CXCR4 by inducible small interfering RNA inhibits breast cancer cell invasion in vitro. Cancer Res. 2003;63(16):4801–4804. [PubMed] [Google Scholar]
- 25.Zong L, Chen P, Wang DX. Death decoy receptor overexpression and increased malignancy risk in colorectal cancer. World J Gastroenterol. 2014;20(15):4440–4445. doi: 10.3748/wjg.v20.i15.4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Q, Zheng Y, Chen D, Li X, Lu C, Zhang Z. Aberrant expression of decoy receptor 3 in human breast cancer: relevance to lymphangiogenesis. J Surg Res. 2014;188(2):459–465. doi: 10.1016/j.jss.2014.01.058. [DOI] [PubMed] [Google Scholar]
- 27.Jiang YQ, Zhong TF, Dang YW, et al. Overexpression and clinicopathological contribution of DcR3 in bladder urothelial carcinoma tissues. Asian Pac J Cancer Prev. 2014;15(21):9137–9142. doi: 10.7314/apjcp.2014.15.21.9137. [DOI] [PubMed] [Google Scholar]
- 28.Yeon JS, Sun LH, Kyoung KE, Min HJ, Whan KY, SunSik B. Reactive oxygen species and PI3K/Akt signaling in cancer. Free Radic Biol Med. 2014;75:S34–S35. doi: 10.1016/j.freeradbiomed.2014.10.773. [DOI] [PubMed] [Google Scholar]
- 29.Toda M, Kawamoto T, Ueha T, et al. ‘Decoy’ and ‘non-decoy’ functions of DcR3 promote malignant potential in human malignant fibrous histiocytoma cells. Int J Oncol. 2013;43(3):703–712. doi: 10.3892/ijo.2013.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]