Abstract
To maintain a particular cell fate, a unique set of genes should be expressed while another set is repressed. One way to repress gene expression is through Polycomb group (PcG) proteins that compact chromatin into a silent configuration. In addition to cell fate maintenance, PcG proteins also maintain normal cell physiology, for example cell cycle. In the absence of PcG, ectopic activation of the PcG-repressed genes leads to developmental defects and malignant tumors. Little is known about the molecular nature of ectopic gene expression; especially what differentiates expression of a given gene in the orthotopic tissue (orthotopic expression) and the ectopic expression of the same gene due to PcG mutations. Here we present that ectopic gene expression in PcG mutant cells specifically requires dBRWD3, a negative regulator of HIRA/Yemanuclein (YEM)-mediated histone variant H3.3 deposition. dBRWD3 mutations suppress both the ectopic gene expression and aberrant tissue overgrowth in PcG mutants through a YEM-dependent mechanism. Our findings identified dBRWD3 as a critical regulator that is uniquely required for ectopic gene expression and aberrant tissue overgrowth caused by PcG mutations.
Author Summary
Genetic information is stored in our genomic DNA, and different cells retrieve distinct sets of information from our genome. While it is important to activate genomic regions encoding proteins that are essential for a given cell type, it is equally important to silence genomic regions encoding proteins that are potentially harmful to this type of cells. One of the gene silencing mechanisms frequently used during and after development is mediated by the Polycomb group (PcG) proteins. If this guardian function does not perform correctly due to PcG mutations, genes that are normally silenced—such as oncogenes—are expressed aberrantly. Due to the activation of oncogenes and the loss of other PcG functions, PcG mutant cells often begin to display hallmarks of cancer, such as proliferating beyond control, acquiring stem-cell-like properties, and migrating to distant sites. If the transcriptional mechanisms underlying aberrant gene expression in PcG-mutant cancer cells differ from gene expression in normal cells, we may be able to selectively inhibit the growth of cancer cells without affecting their normal counterparts. Here we show that the difference between these two types of gene expression resides in their sensitivity to dBRWD3, a negative regulator of the deposition of histone H3 variant H3.3. Our results indicate that the inactivation of dBRWD3 or promotion of H3.3 deposition may selectively suppress ectopic gene expression and tumorigenesis driven by mutations in PcG.
Introduction
The eukaryotic genome is packaged in a macromolecular complex termed chromatin. Chromatin is composed of DNA, RNA, histones, and non-histone proteins. The nucleosome, the basic unit of chromatin, consists of a histone octamer containing two copies of histones (H3, H2A, H2B, and H4) and 147 base pairs of DNA wrapped around the octamer [1]. Variants of H2A, H2B, and H3 differ from the canonical histones by a few amino acids [2]. Moreover, canonical histones are encoded by multiple repeated sequence arrays and expressed during the S-phase, while the variants are encoded by single-copy genes and expressed in the interphase [2,3]. Once the histone variants are incorporated into nucleosomes, they confer distinct physical and biochemical properties to DNA templates and thus regulate DNA replication, repair and gene transcription [3,4]. The deposition of histone variants is mediated by specific chaperone complexes. For example, H3.3 deposition, which often occurs in actively transcribed regions, is mediated by a histone chaperone named histone repressor A (HIRA) and its associated chaperone Yemanuclein (YEM) [5–10]. We previously showed that HIRA/YEM activity is negatively regulated by dBRWD3 (Bromodomain and WD repeat-containing protein 3) [11], which adds a second layer of complex regulation to H3.3 deposition. Dendritic arborization of peripheral neurons and photoreceptor development are disrupted in dBRWD3 mutants. These phenotypes are effectively suppressed by mutations in yem or H3.3, indicating that dBRWD3 functions largely through restricting YEM-dependent H3.3 deposition [11]. However, it remains unknown where in the genome this regulation of H3.3 deposition takes place and how it affects transcription.
Distinct patterns of transcriptional activation and inactivation of the genome contribute to the diversity of cell types in multicellular organisms. To inactivate transcription, Polycomb group (PcG) proteins bind to specific genomic regions and modify histones posttranslationally [12,13]. PcG proteins are grouped into two evolutionarily conserved complexes, PRC1 and PRC2. In Drosophila, the PRC1 complex consists of Polycomb, Posterior sex combs, Sex combs extra (Sce, the Drosophila homolog of human RING1), Polyhomeotic proximal, Polyhomeotic distal with an accessory molecule, and Sex comb on midleg (Scm) [14]. The PRC2 complex is composed of Enhancer of zeste (E(z)), Suppressor of zeste 12, and extra sex combs [13]. Functionally, PRC1 adds a monoubiquitin moiety onto histone H2AK119 (H2AK118 in Drosophila), whereas PRC2 catalyzes the trimethylation of H3K27 (H3K27me3). The combined activities of PRC1 and PRC2 repress transcription by compacting chromatin [15]. PcG proteins may also silence gene expression through a compaction-independent mechanism, such as by blocking transcription initiation [16–18].
Misregulation of transcription within typically inactive genomic regions leads to the disorganization of tissues and organisms [19]. For instance, loss of PcG function causes the ectopic expression of Homeotic (Hox) genes specific to posterior segments and thus disrupts the anterior-to-posterior body plan in embryos [20,21]. In Drosophila, loss of PRC1 leads to ectopic expression of Unpaired 1–3, driving aberrant cell proliferation and tissue overgrowth by activating the JAK-STAT pathway [22]. In humans, loss of PRC1 function has been shown to promote tumorigenesis [23,24]. Reduced expression of the PRC1 subunit CBX7 has been implicated in bladder, breast, colon, glioma, lung, pancreatic, and thyroid carcinomas [25–31], and CBX7 knockout mice develop lung and liver carcinoma [29]. Loss of PRC2 function also causes tumor formation. For example, the tumor-driving H3.3K27M mutation in pediatric diffuse intrinsic pontine gliomas (DIPGs) results in the inactivation of the PRC2 complex, causing ectopic expression of LIN28B, PLAG1, and PLAGL1, and leading to the de-differentiation and hyperproliferation of tumor cells [32–34]. A second mutation in the PRC2 complex genes in patients with neurofibromatosis increases the likelihood of developing malignant peripheral nerve sheath tumors [35,36].
Currently, no therapeutic strategies have been developed for tumorigenesis caused by ectopic gene expression. This is mainly because little is known about how ectopic gene expression is initiated within de-repressed genomic regions, and how it differs from conventional transcription initiation. Here we show that dBRWD3 is specifically required for ectopic gene expression and tissue overgrowth caused by PcG mutations. dBRWD3 sustains PcG mutation-induced ectopic gene transcription by regulating H3.3 deposition, which in turn affects the way RNA polymerase II occupies transcription start sites. Thus, our results suggest that human BRWD3 could be a potential therapeutic target for PcG mutation-driven tumors.
Results
dBRWD3 mutations suppress ectopic antennapedia expression caused by mutations in the PRC1 subunits, Scm and Sce
In the process of investigating how dBRWD3 might affect gene expression, we unexpectedly found that the dBRWD3 mutations suppress the lethality of Scm mutants. Similar to other PcG mosaic mutants [22], ScmD1 mosaic mutant flies died in the pupal stage. Interestingly, a significant portion of the mosaic ScmD1, dBRWD3s5349 double mutants survived to the adult stage, so did the mosaic ScmD1, dBRWD3PX2 double mutants (Table 1). To explore the relationship between dBRWD3 and PcG genes, we examined the genetic interaction between dBRWD3 and Posterior sex comb (Psc), another PcG gene. We found that knockdown of Psc was semi-lethal, whereas simultaneous knockdown of Psc and dBRWD3 was fully viable (Table 2). Taken together, these results suggest a role for dBRWD3 as a suppressor of PcG genes.
Table 1. The genetic interaction between Scm and dBRWD3.
| Genotype | Eclosure rate |
|---|---|
| wild type | 95.16% (n = 186) |
| ScmD1 | 0% (n = 240) |
| ScmD1, dBRWD3s5349 | 11.2% (n = 125) |
| ScmD1, dBRWD3PX2 | 10.5% (n = 228) |
| dBRWD3s5349 | 9.56% (n = 168) |
Table 2. The genetic interaction between Psc and dBRWD3.
| Genotype | Eclosure rate |
|---|---|
| wild type | 100% (n = 150) |
| Psc-dsRNA | 19.2% (n = 166) |
| Psc-dsRNA, dBRWD3-dsRNA | 100% (n = 136) |
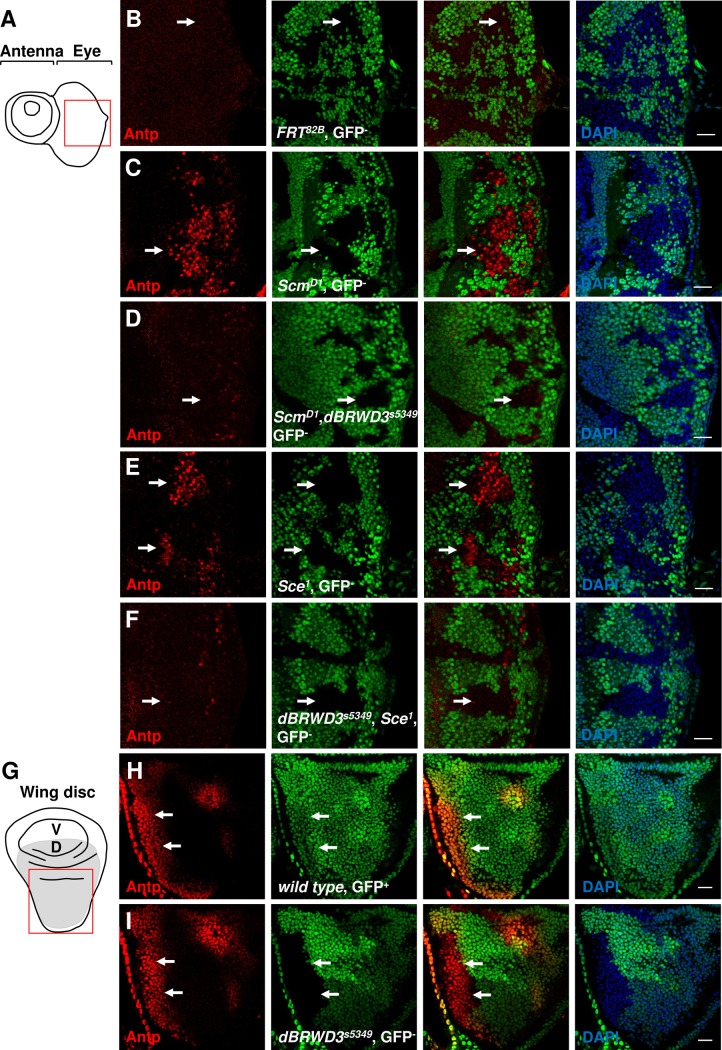
Since ectopic gene expression underlies many phenotypes of PcG mutations, we then investigated whether the dBRWD3 mutations also suppresses ectopic gene expression. While the second thoracic segment-specific Hox gene, antennapedia (Antp), was repressed in wild-type eye clones (Fig 1A and 1B, arrow), it was ectopically expressed in Scm mutant eye clones located in the posterior region (Fig 1C, arrow). This ectopic Antp expression was dramatically reduced in ScmD1, dBRWD3s5349 or ScmD1, dBRWD3PX2 double-mutant eye clones (Figs 1D, S1A, S1B and S1C). Similarly, Antp is ectopically expressed in Sce1 mutant eye clones (Fig 1E, arrows) but not in dBRWD3s5349, Sce1 double-mutant clones (Figs 1F and S1D). Interestingly, dBRWD3 is dispensable for the orthotopic expression of Antp in wings (Figs 1G, 1H, 1I and S1E). Overall, these observations reveal that dBRWD3 is involved in the ectopic expression of Antp caused by PcG mutations.
Fig 1. dBRWD3s5349 mutation suppresses ectopic expression of Antp in Scm D1 or Sce1 mutant eye clones.
(A) A schematic illustration of a 3rd instar eye imaginal disc. The red square indicates the region examined in the following experiments. (B-F) Antennapedia (Antp) levels (arrows) in wild-type clones (B), ScmD1 mutant clones (C), ScmD1, dBRWD3s5349 double-mutant clones (D), Sce1 mutant clones (E), and dBRWD3s5349 Sce1 double-mutant clones (F) generated in the 3rd instar eye imaginal discs by ey-flp and marked by the absence of GFP. Scale bars indicate 50μm. (G) A schematic illustration of a 3rd instar wing imaginal disc. V and D stand for ventral and dorsal (marked by grey) compartments, respectively. (H) Antp levels (arrows) in a wild-type wing. (I) Antp levels (arrowheads) in dBRWD3s5349 mutant wing disc clones generated by hs-flp and marked by the absence of GFP. Scale bars indicate 50μm.
Knockdown of dBRWD3 suppresses ectopic ultrabithorax expression in the PRC1 and PRC2 depleted brains
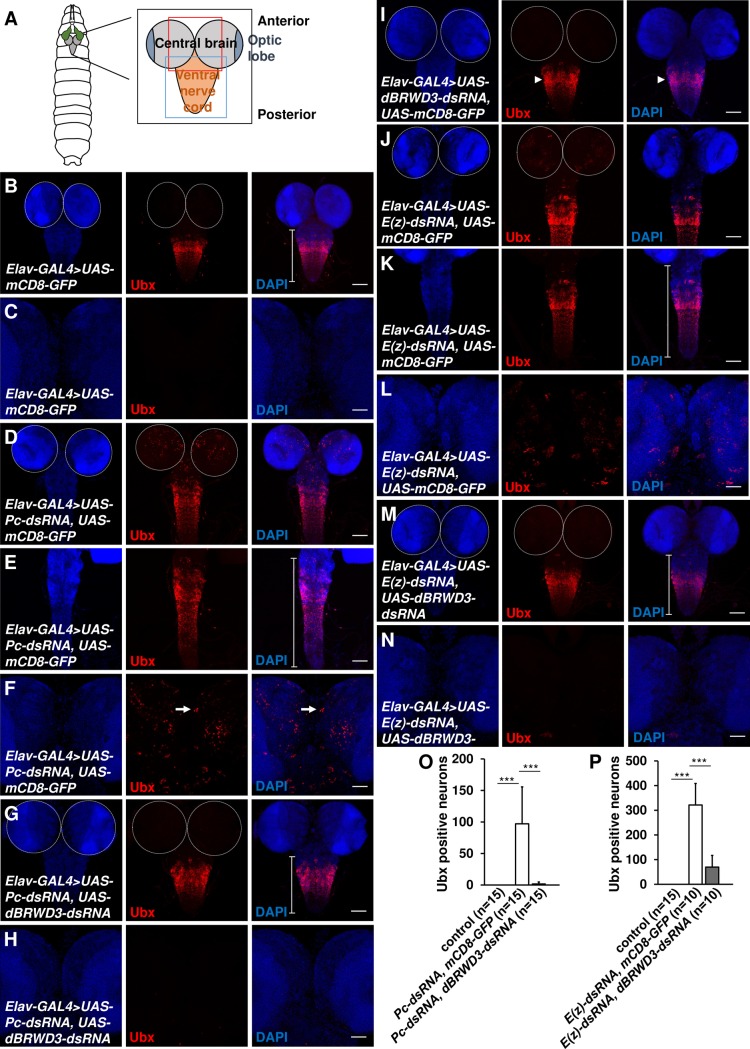
To determine whether dBRWD3 suppresses ectopic gene expression other than Antp in the eyes, we knocked down Pc in the central nervous system by Elav-GAL4, reducing the level of Pc mRNA to 5% (S2A Fig). Ubx is ectopically expressed in the Pc-depleted brains but not in the control (Fig 2A, 2B, 2C, 2D, 2F and 2O). In addition, the Pc depleted ventral nerve cord was thinner and more elongated compared to the control (Fig 2E, bracket). We found that both ectopic expression of Ubx and elongation of ventral nerve cords were suppressed by depletion of dBRWD3 (Fig 2G and 2H). On the other hand, orthotopic expression of Ubx in the ventral nerve cord was not affected in the dBRWD3, Pc double knockdown (Fig 2G) or in dBRWD3 knockdown animals (Figs 2I, S2B, S2C and S2D, arrowhead). Thus, ectopic expression of Ubx depends on dBRWD3 whereas orthotopic expression of Ubx does not.
Fig 2. Knockdown of dBRWD3 suppresses ectopic expression of Ubx in Pc or E(z) depleted brains and condensation failure of Pc or E(z) depleted ventral nerve cords.
(A) A schematic diagram of a 3rd instar central nervous system. The black, red and blue squares indicate the regions examined for the central nervous system, central brain, and ventral nerve cord, respectively. (B and C) Ubx protein levels in the ventral nerve cord (B) and brains (C) of flies expressing mCD8-GFP under the control of Elav-GAL4. The average number of Ubx positive neurons per brain is 0, n = 15. The average length of the ventral nerve cord is 395.6μm, n = 21. (D-F) Ubx protein levels in the ventral nerve cords (D and E) and brains (D and F) of flies expressing UAS-Pc-dsRNA, UAS-mCD8-GFP under the control of Elav-GAL4 to deplete Pc expression. The average number of Ubx positive neurons per brain is 97.1, n = 15, p<0.00001 vs. GAL4 control by Student's t-test. The average length of ventral nerve cord is 468.5μm, n = 49, p<0.001 vs. GAL4 control by Student's t-test. (G and H) Ubx protein levels in the ventral nerve cord (G) and brains (H) of Pc, dBRWD3 doubly depleted flies. The average Ubx positive neurons per brain is 1.9, n = 15, p<0.0001 vs. knockdown of Pc by Student's t-test. The average length of ventral nerve cords is 389.4μm, n = 45, p<0.00001 vs. knockdown of Pc by Student's t-test. (I) Ubx protein (arrowhead) levels in the ventral nerve cord of dBRWD3 depleted flies. (J-L) Ubx protein levels in the ventral nerve cords (J and K) and brains (J and L) of flies expressing UAS-E(z)-dsRNA and UAS-mCD8-GFP by Elav-GAL4. The average number of Ubx positive neurons per brain is 321.6, n = 10, p<0.0001 vs. GAL4 control by Student's t-test. The average length of ventral nerve cords is 456.5μm, n = 39, p<0.01 vs. GAL4 control by Student's t-test. (M and N) Ubx protein levels in the ventral nerve cord (M) and brains (N) of E(z), dBRWD3 doubly depleted flies. The average number of Ubx positive neurons per brain is 69.9, n = 10, p<0.0001 vs. knockdown of E(z) by Student's t-test. The average length of the ventral nerve cords is 393.9μm, n = 40, p<0.001 vs. knockdown of E(z) by Student's t-test. (O and P) Suppression of Pc (O) or E(z) (P) depletion-induced ectopic expression of Antp by dBRWD3 depletion. The number of Antp positive neurons in Pc, dBRWD3 (O), E(z), dBRWD3 (P) doubly depleted brains. *** indicates p<0.0001 by Student's t-test. Brackets indicate the length of the ventral nerve cord. Scale bars indicate 100μm in B, D, G, I, J, and M and 50μm in C, E, F, H, K, L and N.
Depleting E(z), which encodes the H3K27 methyltransferase in PRC2, caused ectopic expression of Ubx in brains (Figs 2J, 2L and S2E) and condensation failure in ventral nerve cords (Fig 2K, bracket). The ectopic expression of Ubx and condensation failure of ventral nerve cords were also suppressed by knockdown of dBRWD3 (Fig 2M, 2N and 2P). By contrast, orthotopic expression of Ubx was not affected in dBRWD3, E(z)-doubly depleted ventral nerve cords (S2F, S2G and S2H Fig). Taken together, our data indicates that ectopic Hox gene expression depends on dBRWD3 whereas orthotopic Hox gene expression does not.
dBRWD3s5349 suppresses ectopic expression of unpaired in Scm or Sce mutants
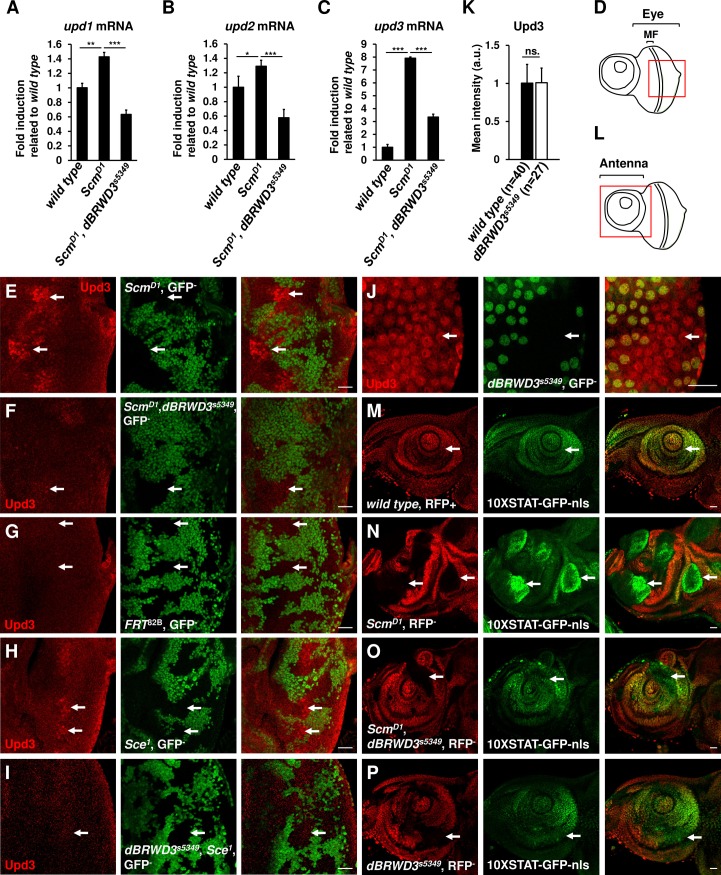
In addition to ectopic expression of Hox genes, loss of Ph, Psc, or Pc induces ectopic expression of unpaired (upd) 1–3, and therefore activation of the JAK-STAT pathway that leads to overgrowth of tumor-like tissues [22,37–39]. By RT-qPCR, we detected mild increases of upd1 and upd2 mRNAs (Fig 3A and 3B) and a strong induction of upd3 mRNA (Fig 3C) in the mosaic ScmD1 mutant brain-eye complex. This upregulation of upd1-3 was prevented or significantly weakened in the mosaic ScmD1, dBRWD3s5349 double mutants compared with mosaic ScmD1 mutants (Fig 3A, 3B and 3C). Consistently, our immunofluorescent micrographs showed that Upd3 accumulated in ScmD1 mutant clones adjacent to the morphogenetic furrow (Figs 3D, 3E and S3A, arrows), but not in ScmD1, dBRWD3s5349 double-mutant clones (Figs 3F and S3A) or wild-type clones (Fig 3G). Similarly, we detected accumulation of Upd3 in Sce1 (Figs 3H and S3B, arrows) and SceKO (S4 Fig) mutant clones, but not in the dBRWD3s5349, Sce1 double-mutant clones (Figs 3I and S3B, arrow). On the other hand, we found that orthotopic upd3 expression in the posterior end of the 2nd instar eye disc was not altered in dBRWD3s5349 mutant clones (Fig 3J and 3K, arrows), indicating that the regulation of upd3 by dBRWD3 is specific to ectopic expression.
Fig 3. dBRWD3 suppresses ectopic expression of upd and activation of the JAK-STAT pathway.
(A-C) The upd1 (A), upd2 (B), and upd3 (C) mRNA levels of mosaic eye brain complexes isolated from wild type, ScmD1, and ScmD1, dBRWD3s5349 as indicated. Data are shown as means ± S.D. *, **, *** indicate P<0.01, 0.001, 0.0001 respectively by Student's t-test, n = 4. (D) A schematic illustration of a 3rd instar eye imaginal disc. MF stands for morphogenic furrow. The red square indicates the region examined in the following experiments. (E-I) Upd3 levels (arrows) in ScmD1 mutant clones (E), ScmD1, dBRWD3s5349 double-mutant clones (F), wild-type clones (G), Sce1 mutant clones (H), and dBRWD3s5349, Sce1 double-mutant clones (I) generated in 3rd instar eye imaginal discs by ey-flp and marked by the absence of GFP. Scale bars indicate 50μm. (J) Orthotopic Upd3 levels (arrows) in dBRWD3s5349 mutant clones generated in 2nd instar larval imaginal eye discs marked by the absence of GFP. Scale bars indicate 50μm. (K) Quantification analyses of orthotopic Upd3 levels (arrows) in dBRWD3s5349 mutant clones. ns. indicates not significant. (L) A schematic illustration of a 3rd instar antennal imaginal disc. The red square indicates the region examined in the following experiments. (M) GFP levels (arrows) of the 10XSTAT-nls-GFP reporter in a wild-type disc. (N-P) GFP levels (arrows) of the 10XSTAT-nls-GFP reporter in ScmD1 mutant clones (N), ScmD1, dBRWD3s5349 double-mutant clones (O), and dBRWD3s5349 mutant clones (P) generated in 3rd instar antennal imaginal discs by ey-flp and marked by the absence of the ubi promoter driven mof-RFP. Scale bars indicate 50μm.
We also used the 10XSTAT-GFP reporter to determine whether the JAK-STAT pathway, which is activated by Upd1-3, is affected by dBRWD3 in PcG mutant cells [40,41]. In contrast to the weak and uniform expression observed in wild-type antennal discs (Fig 3L and 3M, arrows), the GFP signal was much higher in ScmD1 mutant clones (Fig 3N), likely due to up-regulation of upd1-3. It remained unchanged in ScmD1, dBRWD3s5349 double mutants (Fig 3O, arrow). The STAT activity in the antennal disc was not affected in dBRWD3s5349 single mutants (Fig 3P, arrow). Given these results, we propose that dBRWD3s5349 suppresses ectopic activation of the JAK-STAT pathway caused by ScmD1 or Sce1 mutations.
dBRWD3s5349 suppresses tissue overgrowth caused by the loss of PcG function
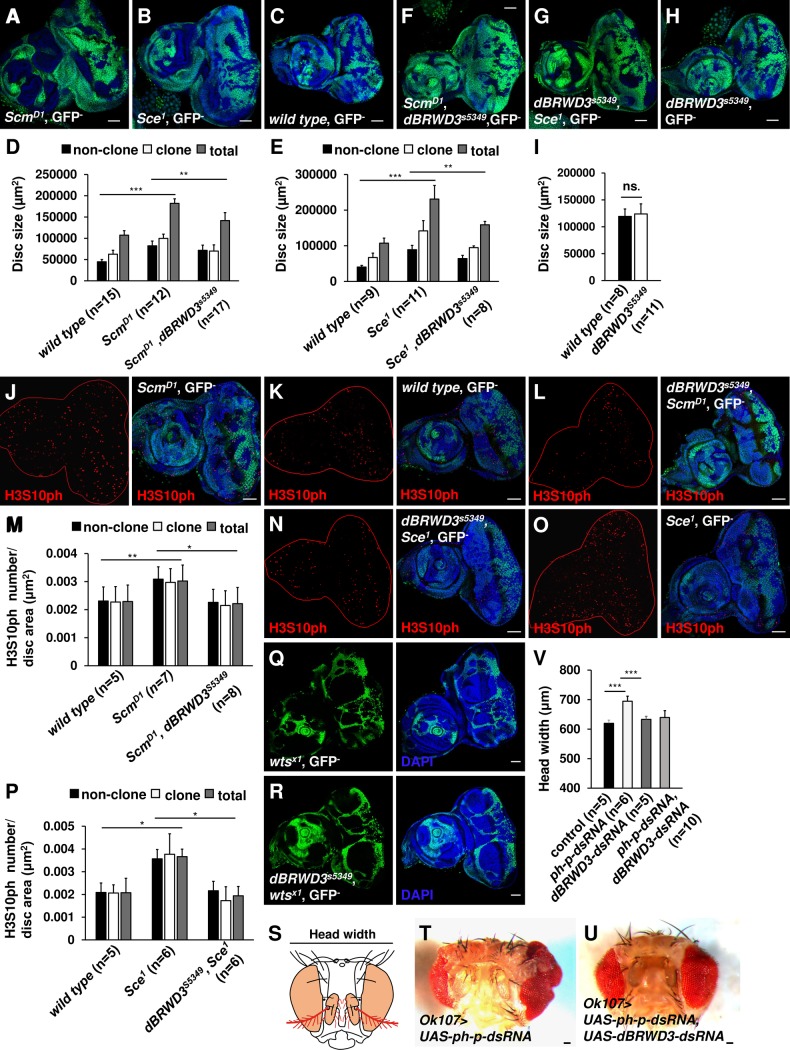
By ectopically expressing Upd1, Upd2, and Upd3, mutations in PRC1 and PRC2 complexes cause cell autonomous and non-autonomous proliferations [22,37]. Consistently, we found that ScmD1 (Fig 4A) or Sce1 (Fig 4B) mutant clones were larger than wild-type clones (Fig 4C, 4D and 4E). Since Upd3 is a diffusible ligand stimulating non-cell-autonomous proliferation, we found the non-clonal area of mosaic ScmD1 (Fig 4D) or Sce1 (Fig 4E) eye antennal discs were also larger. Overall, mosaic ScmD1 or Sce1 mutant discs were 1.5- (Fig 4D) or 1.8-fold in size (Fig 4E) compared to wild-type respectively. This tissue overgrowth could be suppressed by dBRWD3s5349 (Fig 4D, 4E, 4F and 4G). As a control, the mosaic dBRWD3s5349 alone did not reduce the disc size (Fig 4H and 4I).
Fig 4. dBRWD3s5349 suppresses tissue overgrowth caused by PcG mutations.
(A-C) ScmD1(A), Sce1(B), and wild-type (C) mosaic clones were generated in 3rd instar eye antennal discs by ey-flp and marked by the absence of GFP. Scale bars indicate 50μm. (D and E) Bar chart presentations of clonal, non-clonal, and total eye antennal disc area of wild type, ScmD1, and ScmD1, dBRWD3s5349 (D), and wild type, Sce1, and dBRWD3s5349, Sce1 (E). Data are shown as means ± S.D. **, *** indicate P<0.001, 0.0001 respectively by Student's t-test. ns. indicates not significant. (F-H) ScmD1, dBRWD3s5349 (F), dBRWD3s5349, Sce1 (G), or dBRWD3s5349 (H) mosaic clones were generated in 3rd instar eye antennal discs by ey-flp and marked by the absence of GFP. Scale bars indicate 50μm. (I) Bar chart presentations of clonal, non-clonal, and total eye antennal disc areas of wild type and dBRWD3s5349. Data are shown as means ± S.D. ns. indicates not significant by Student's t-test. (J-L) H3S10ph levels (red) in ScmD1 (J), wild type (K), ScmD1, dBRWD3s5349 (L), mosaic mutant eye discs. Scale bars indicate 50μm. (M) The clonal, non-clonal, and total density of H3S10ph positive cells of wild type, ScmD1, and ScmD1, dBRWD3s5349 mosaic eye antennal discs. Data are shown as means ± S.D. *, ** indicate p<0.01, 0.001 respectively by Student's t-test. (N and O) H3S10ph levels (red) in dBRWD3s5349, Sce1 (N), and Sce1 (O) mosaic mutant eye discs. Scale bars indicate 50μm. (P) The clonal, non-clonal, and total density of H3S10ph positive cells of wild type, Sce1, and dBRWD3s5349, Sce1 mosaic eye antennal discs. Data are shown as means ± S.D. * indicates p<0.01 by Student's t-test. (Q and R) wtsx1 (Q), dBRWD3s5349, wtsx1 (R) mosaic clones were generated in 3rd instar eye antennal discs by ey-flp and marked by the absence of GFP. Scale bars indicate 50μm. (S) A schematic illustration of an adult head. Colored areas indicate OK107-GAL4 expression domains. (T) Eye phenotype produced by knockdown of polyhomeotic proximal (ph-p) by the OK-107 GAL4 driver. The scale bar indicates 50μm. (U) An adult eye that resulted from double knockdown of ph-p and dBRWD3. Scale bars indicate 50μm. (V) Head width of ph-p-depleted, dBRWD3-depleted, and dBRWD3, ph-p-doubly-depleted eyes. *** indicates p<0.0001 by Student's t-test.
Quantitatively, the numbers of clonal, non-clonal, and total mitotic cells marked by phosphorylation of H3S10 (H3S10ph) were increased in mosaic ScmD1 mutant eye-antennal disc (Fig 4J). It was reduced to a wild-type level in mosaic ScmD1, dBRWD3s5349 mutant eye-antennal discs (Fig 4K, 4L and 4M). A similar suppression of proliferation was found in mosaic dBRWD3s5349, Sce1 mutant eye-antennal discs (Fig 4N) as opposes to Sce1 mutants (Fig 4O and 4P). The elongated Pc and E(z) depleted ventral nerve cords and control ventral nerve cord had comparable mitotic indices and undetectable expression of upd1, upd2 and upd3, indicating that the elongation of the ventral nerve cord was not caused by excessive proliferation.
To determine whether the dBRWD3 mutation also suppresses other types of oncogenic tissue overgrowth, we sampled tissue overgrowth caused by the warts (wts) mutation that activates the hippo pathway [42,43]. We found that dBRWD3s5349 did not suppress the expression of the hippo pathway target gene, expanded (ex) (S5A and S5B Fig) and tissue overgrowth (Fig 4Q and 4R). Thus, the dBRWD3 mutation appears to suppress the oncogenic tissue overgrowth specifically related to PcG mutations. To examine the growth-inhibition effect of the dBRWD3 mutation beyond the developmental stage, we generated overgrown eyes and surrounding tissues by knockdown of ph-p (Fig 4S and 4T). In dBRWD3 and ph-p double-knockdown eyes, the tissue overgrowth phenotype was suppressed (Fig 4U and 4V). From these data, we infer that the tissue overgrowth induced by PcG gene depletion requires dBRWD3.
H3.3 accumulation correlates with suppression of ectopic gene expression mediated by loss of dBRWD3
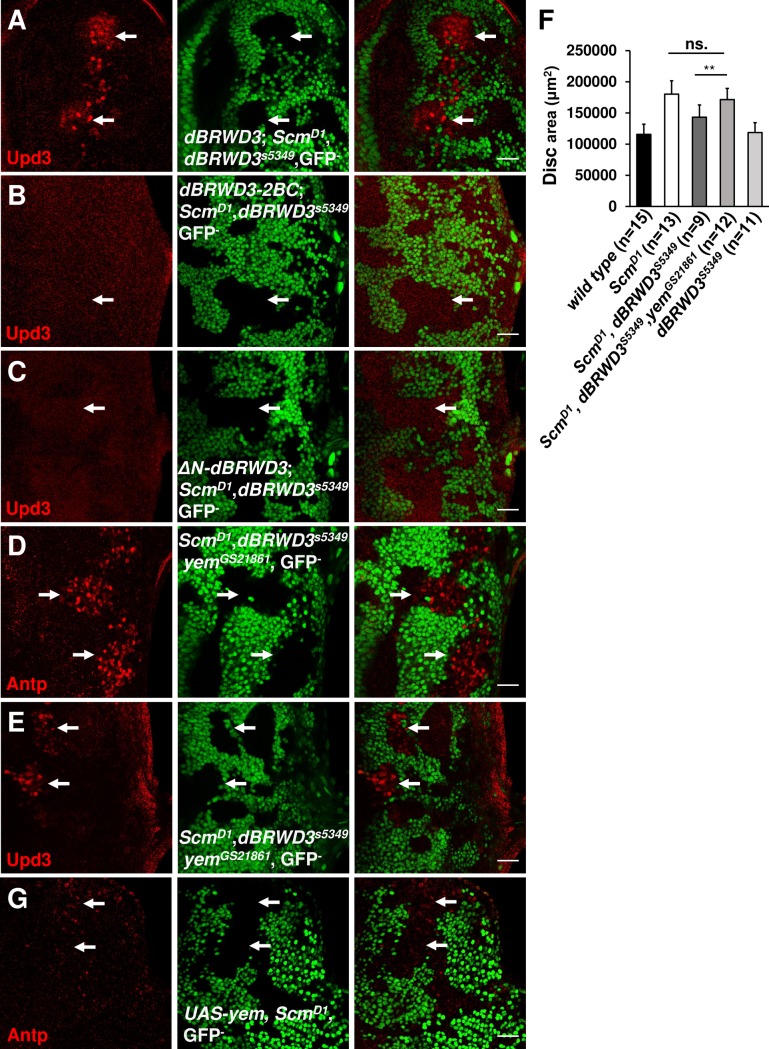
dBRWD3 contains bromodomain I and II (BRDI and BRDII) that were predicted to be acetylated histone-binding domains (S6A Fig) [44]. To investigate the function of these bromodomains, we complemented ScmD1, dBRWD3s5349 double-mutant cells with wild-type dBRWD3, dBRWD3-N1287A, and dBRWD3-N1451A, in which the conserved asparagines in the BC loops of BRDI and BRDII were mutated into alanines. The wild-type dBRWD3-RFP, dBRWD3-N1287A-RFP, and dBRWD3-N1451A-RFP could restore the ectopic upd3 expression in ScmD1, dBRWD3s5349 double-mutant cells (Figs 5A, S6B and S6C). However, when both the BRDI and BRDII were disrupted, the dBRWD3-N1287A, N1451A-RFP (designated as the dBRWD3-2BC-RFP) could not restore the ectopic expression of upd3 (Fig 5B). This failure to complement is not related to expression levels because dBRWD3-2BC-RFP was expressed more than wild-type dBRWD3-RFP (S7 Fig). Therefore, the BRDI and BRDII of dBRWD3 are functionally redundant in supporting ectopic gene expression. We also complemented ScmD1, dBRWD3s5349 double-mutant cells with a HLH motif-deleted, ΔN-dBRWD3, which no longer interacts with the DNA damage binding protein 1 (DDB1) and cannot be recruited to cullin4/DDB1 organized E3 ligase [11]. The ΔN-dBRWD3-RFP also failed to restore the ectopic upd3 expression in ScmD1, dBRWD3s5349 double-mutant cells (Fig 5C). Together, these results suggest that the activities of dBRWD3 binding to acetylated histones and cullin 4/DDB1 organized E3 ligase are essential for maintaining ectopic gene expression.
Fig 5. dBRWD3 regulates ectopic gene expression and clone size in a YEM-dependent manner.
(A-C) upd3 levels (arrows) in ScmD1, dBRWD3s5349 double-mutant clones complemented with wild type dBRWD3-RFP (A), dBRWD3-2BC-RFP (B) or ΔN-dBRWD3-RFP (C). Scale bars indicate 50μm. (D and E) Antp (D) and upd3 (E) levels (arrows) in ScmD1, dBRWD3s5349, yemGS21861 mosaic mutant clones in eye discs. Scale bars indicate 50μm. (F) Disc sizes of mosaic ScmD1, ScmD1 dBRWD3s5349 double-mutant, and ScmD1, dBRWD3s5349, yemGS21861 triple-mutant eye antennal discs. Data are shown as means ± S.D. ** indicates p<0.001 by Student's t-test. ns. indicates not significant. (G) Antp protein levels in mosaic ScmD1 mutant eye antennal discs expressing yem-Flag under the control of GMR-GAL4. Scale bars indicate 50μm.
Previously, we demonstrated that dBRWD3 limits HIRA/YEM-mediated H3.3 deposition [11]. However, it is not clear whether the BRDI, BRDII, and HLH motif of dBRWD3 are important for dBRWD3 regulation of H3.3. When we complemented dBRWD3s5349 mutant cells with ΔN-dBRWD3-RFP or dBRWD3-2BC-RFP, the H3.3 levels in the dBRWD3s5349 mutant cells remained higher than those in wild-type cells (S8A and S8B Fig, arrows). By contrast, dBRWD3-N1287A and dBRWD3-N1451A reduced the H3.3-dendra2 to a normal level (S8C and S8D Fig, arrows), indicating a negative correlation between accumulation of H3.3 and ectopic gene expression. Indeed, the negative correlation was also observed in the dBRWD3 knockdown brains, where the endogenous H3.3 levels were higher than in control brains (S8E Fig). When we co-immuno-stained the mosaic discs with ant-Antp antibody, the ectopic anti-Antp signals were strongly reduced along with accumulated H3.3 in ScmD1, dBRWD3s5349 double-mutant clones (S9B Fig, arrows).
dBRWD3s5349 suppresses ScmD1 through a yemanuclein (yem)-dependent mechanism
We next examined whether the increased H3.3 deposition suppresses ectopic gene expression. To this end, we introduced a yem mutation to reduce the dBRWD3 mutation-induced H3.3 deposition (S9C Fig) [11]. In ScmD1, dBRWD3s5349, yemGS21861 triple-mutant clones, the ectopic expression of Antp was restored and coincided with the reduction of H3.3 (Figs 5D, S9C and S10A), suggesting that the dBRWD3 mutation suppresses the Scm mutation through a YEM-dependent mechanism. Moreover, upd3 was ectopically expressed in this triple mutant clone (Figs 5E and S10B). Consistently, the size of the ScmD1, dBRWD3s5349, yemGS21861 triple-mutant eye-antennal disc was larger than that in the ScmD1, dBRWD3s5349 double-mutant (Fig 5F). To further substantiate the role for H3.3 in ectopic gene expression, we investigated whether ectopic gene expression could be suppressed by YEM-induced H3.3 deposition (S11A, S11B and S11C Fig) without any mutation in dBRWD3. YEM over-expression effectively suppressed the ectopic expression of Antp (Figs 5G and S10C). Taken together, these data indicate that dBRWD3 supports ectopic gene expression and tissue overgrowth mediated by PcG mutations by limiting HIRA/YEM-mediated H3.3 deposition.
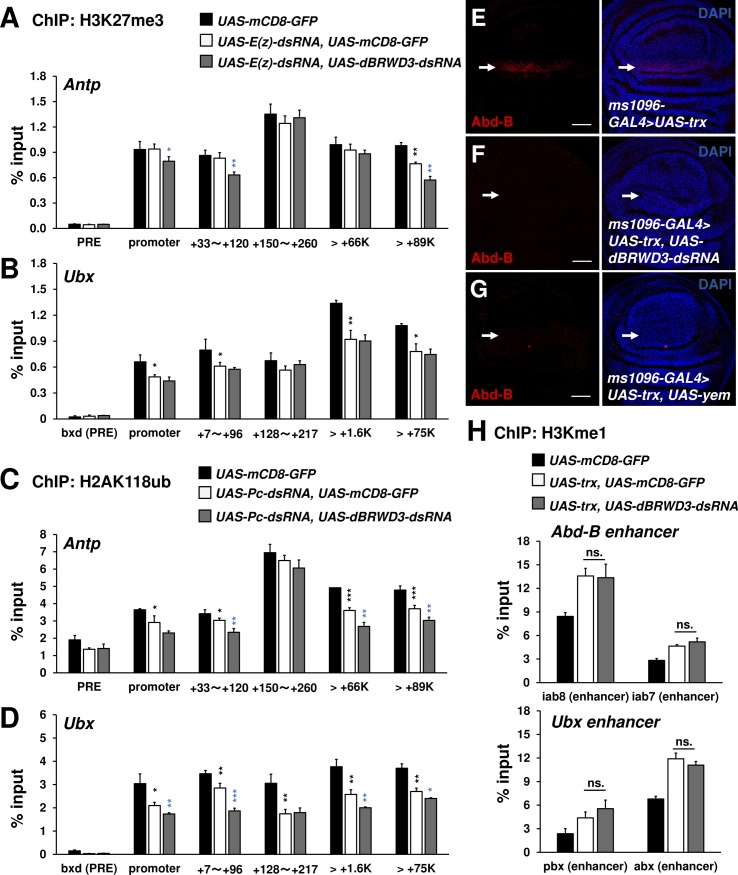
dBRWD3 is epistatic to trithorax during ectopic gene expression through its role in maintaining PolII at the proximal region
To understand how the dBRWD3 mutation suppresses ectopic gene expression, we investigated whether dBRWD3 is required for the removal of pre-existing H3K27me3 and H2A118ub at Antp and Ubx loci upon depletion of E(z) and Pc. The ChIP-qPCR analysis revealed a reduction of H3K27me3 in the distal region of Antp and at Ubx when E(z) was depleted (Fig 6A and 6B). Similarly, H2A118ub levels at Antp and Ubx were also decreased in the Pc depleted brains (Fig 6C and 6D). When dBRWD3 was depleted by RNAi together with E(z) or Pc, the H3K27me3 or H2A118ub levels at Antp and Ubx loci remained low or became lower (Fig 6A, 6B, 6C and 6D), indicating that knockdown of dBRWD3 promotes or does not affect the removal of the pre-existing H3K27me3 and H2A118ub. Moreover, dBRWD3 is not required for the removal of pre-existing H3K27me3 in the E(z) depleted wings at the global level, as revealed by the equally reduced H3K27me3 immunostaining signals in the E(z) knockdown, and E(z), dBRWD3 double-knockdown wings (S12A, S12B and S12C Fig). H3K27me3 levels were not changed in the Pc knockdown, and Pc, dBRWD3 double-knockdown wings compared with the control (S12D, S12E, S12F Fig). Similarly, dBRWD3 was not required for the removal of pre-existing H2AK118ub in the Sce mutant clones at the global level (S12G and S12H Fig). trithorax (trx) encodes an H3K4 monomethyltransferase [45] and antagonizes PcG activity by binding to PRE sites, the enhancer cis-elements targeted by PcG proteins. In different cellular contexts, ectopic gene expression might or might not depend on trx [46,47]. To investigate the requirement of trx in ectopic expression of Antp in eyes, we generated ScmD1, trxE2 double-mutant eye clones and found that the ectopic expression of Antp was suppressed (S13 Fig), indicating that trx, like dBRWD3, is required for ectopic Antp expression. We next investigated whether dBRWD3 and trx function in a linear pathway or in parallel. We found that over-expression of trx in the eye disc proper, peripodial epithelium of the eye disc, and wing disc was sufficient to induce ectopic expression of Antp or Abd-B (Figs 6E, S14A, S14B and S14D), but not in a dBRWD3 knockdown background (Figs 6F, S14C and S14E). In addition, Trx-induced Ubx ectopic expression in wing discs was strongly suppressed by knockdown of dBRWD3, albeit incompletely (S14F and S14G Fig). It seems that H3.3 deposition underlies the suppression of this ectopic gene expression, since Trx-induced Abd-B ectopic expression was also completely suppressed by YEM over-expression (Fig 6G).
Fig 6. dBRWD3 is epistatic to trx in the ectopic expression of Ubx and Abd-B.
(A and B) A ChIP-qPCR analysis of H3K27me3 levels at the enhancers, promoters, and transcription start sites of Antp (A) and Ubx (B) in Elav-GAL4 control, E(z) depleted, and E(z), dBRWD3 doubly depleted brains. Black asterisks indicate control versus E(z) depletion. Blue asterisks indicate E(z) depletion versus E(z), dBRWD3 double depletion. (C and D) A ChIP-qPCR analysis of H2AK118ub levels at the enhancers, promoters, and transcription start sites of Antp (C) and Ubx (D) in Elav-GAL4 control, Pc depleted, and Pc, dBRWD3 doubly depleted brains. Black asterisks indicate control versus Pc depletion. Blue asterisks indicates Pc depletion versus Pc, dBRWD3 double depletion. (E-G) trx was overexpressed under the control of ms-1096-GAL4. The TRX-induced Abd-B expression (arrows) in wild-type (E), dBRWD3 depletion (F), and yem over-expression (G) backgrounds. Scale bars indicate 20μm. (H) A ChIP-qPCR analysis of H3K4me1 levels at Ubx and Abd-B enhancers in the UAS-mCD8-GFP control, trx over-expression, and trx over-expression, dBRWD3 depleted wings as indicated. ns. indicates not significant. ChIP-qPCR Data are shown as means ± S.D from 4 technical replicates. *, **, *** indicate P<0.05, 0.01, 0.001 respectively by Student's t-test.
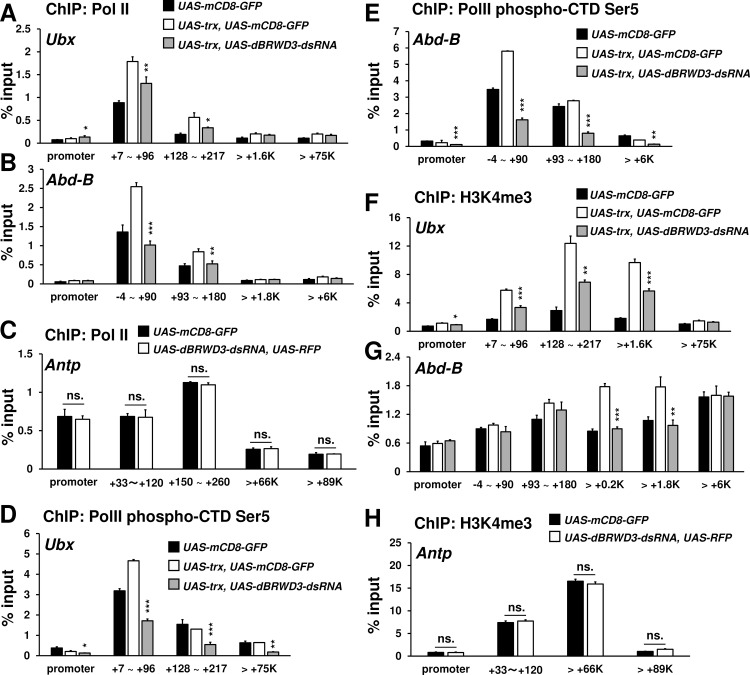
To understand how dBRWD3 affects Trx-induced ectopic gene expression, we used ChIP to determine the levels of H3K4me1 and PolII over Ubx and Abd-B loci. Compared to the control, Trx increased H3K4me1 levels at the enhancer regions of Ubx and Abd-B irrespective of dBRWD3 depletion (Fig 6H), which was later found to have no effects on Trx-induced monomethylation of H3K4 on a global scale (S15A, S15B, S15C, S15D, and S15E Fig). These data indicate that dBRWD3 is epistatic to trx with respect to ectopic gene expression. By contrast, Trx-induced PolII levels at the transcription start sites and 5' ends of Ubx and Abd-B were significantly reduced when dBRWD3 was depleted by RNAi (Fig 7A and 7B), while the PolII levels for orthotopically expressing Antp were not affected (Fig 7C). When trx is overexpressed in the wing imaginal discs, RNA PolII increased on the Antp promoter, which is likely contributed by the purely orthotopic Antp expression and the Trx-induced ectopic Antp expression. The additional knockdown of dBRWD3 restored the PolII occupancy to a level similar to the orthotopic Antp expression control (S16 Fig), indicating that knockdown of dBRWD3 suppressed only the Trx-induced increase of PolII occupancy but not the PolII occupancy of orthotopically expressing Antp. Similarly, PolII phospho-CTD Ser5 levels around the transcription start sites of Ubx and Abd-B were reduced upon knockdown of dBRWD3 (Fig 7D and 7E). We also detected higher levels of H3K4me3, a marker for active chromatins, at the transcription start sites and 5' ends of Ubx and Abd-B upon trx over-expression in a dBRWD3-dependent manner (Fig 7F and 7G). Nevertheless, the levels of H3K4me3 at the orthotopically expressed Antp were not sensitive to dBRWD3 depletion (Fig 7H). These observations suggest that, in ectopic gene expression, dBRWD3 is required for the activation of chromatin specifically at transcription start sites but not in enhancer regions.
Fig 7. dBRWD3 is required for maintaining PolII and H3K4me3 levels at the 5' end of Ubx and Abd-B.
(A and B) A representative ChIP-qPCR analysis of PolII levels at Ubx (A) and Abd-B (B) in the UAS-mCD8-GFP control, in wings over-expressing trx, or in wings with concurrent trx over-expression and dBRWD3 depletion. Black asterisks indicate trx over-expression versus trx over-expression and dBRWD3 depletion. (C) A representative ChIP-qPCR analysis of PolII levels at the promoters and transcription start sites of Antp in the UAS-mCD8-GFP control and dBRWD3-depleted wings. ns. indicates not significant. (D and E) Similar to (A and B), a ChIP-qPCR analysis of PolII phospho-CTD Ser5 levels at Ubx (D) and Abd-B (E). Black asterisks indicate trx over-expression versus trx over-expression and dBRWD3 depletion. (F and G) Similar to (A and B), a representative ChIP-qPCR analysis of H3K4me3 levels at Ubx (F) and Abd-B (G). Black asterisks indicate trx over-expression versus trx over-expression and dBRWD3 depletion. (H) Similar to (C), a representative ChIP-qPCR analysis of H3K4me3 levels at the promoters and transcription start sites of Antp in the UAS-mCD8-GFP control and dBRWD3-depleted wings. ns. indicates not significant. ChIP-qPCR Data are shown as means ± S.D from 4 technical replicates. *, **, *** indicate P<0.05, 0.01, 0.001 respectively by Student's t-test.
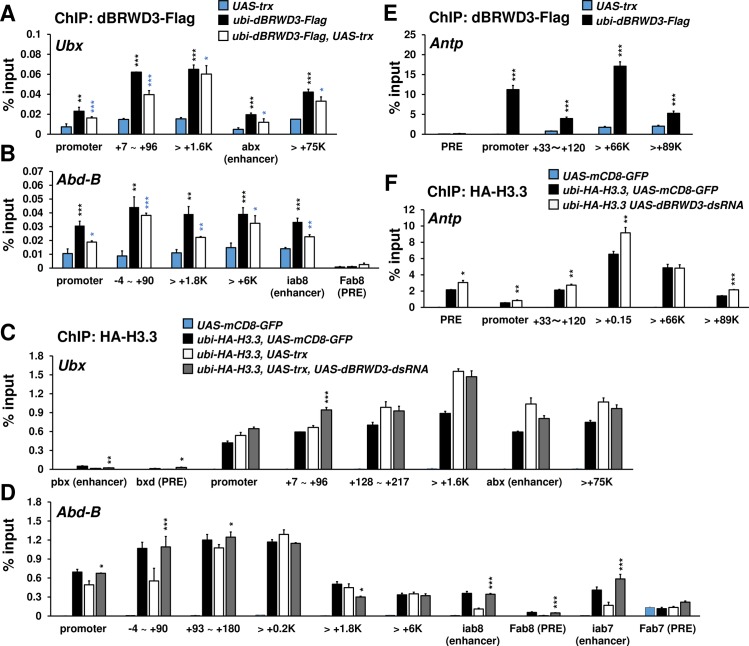
To examine whether the reduction of PolII and H3K4me3 are directly caused by H3.3 deposition, we examined dBRWD3 and H3.3 levels across Ubx and Abd-B loci. ChIP revealed that dBRWD3 was present at these regions and Trx over-expression moderately reduced the occupancy of dBRWD3 (Fig 8A and 8B). Although dBRWD3 was recruited to the promoters, 5' and 3' regions of these ectopically expressed loci, it predominantly reduced H3.3 levels at the transcription start sites (Fig 8C and 8D). These results imply that dBRWD3 maintains PolII levels of ectopically expressed genes by limiting H3.3 deposition at the transcription start sites. Moreover, dBRWD3 was also present at Antp locus (Fig 8E) and limited H3.3 levels at the promoters and the transcription start site of Antp (Fig 8F). Nevertheless, PolII and H3K4me3 levels at Antp were not affected by knockdown of dBRWD3 (Fig 7C and 7H), indicating that the sensitivity toward H3.3 but not H3.3 levels at the promoter and transcription start site per se distinguishes ectopic gene expression from orthotopic gene expression.
Fig 8. dBRWD3 negatively regulates H3.3 at the promoter and 5' regions of Ubx and Abd-B in the trx over-expressing wing discs.
(A and B) A ChIP-qPCR analysis of dBRWD3-Flag levels at Ubx (A) and Abd-B (B). Black asterisks indicate negative control versus dBRWD3-Flag. Blue asterisks indicate negative control versus dBRWD3-Flag and trx over-expression. (C and D) A representative ChIP-qPCR analysis of HA-H3.3 levels at Ubx (C) and Abd-B (D) in the UAS-mCD8-GFP control, the trx over-expressing, and the trx over-expressing, dBRWD3 depleted wings as indicated. Black asterisks indicate trx over-expression versus trx over-expression and dBRWD3 depletion. (E) A ChIP-qPCR analysis of dBRWD3-Flag levels at Antp. Black asterisks indicate negative control versus dBRWD3-Flag. (F) A representative ChIP-qPCR analysis of HA-H3.3 levels at Antp. Black asterisks indicate control versus dBRWD3 depletion. ChIP-qPCR Data are shown as means ± S.D from 4 technical replicates. *, **, *** indicate P<0.05, 0.01, 0.001 respectively by Student's t-test.
Loss of TFII-D or TFII-H activities suppresses ectopic gene expression
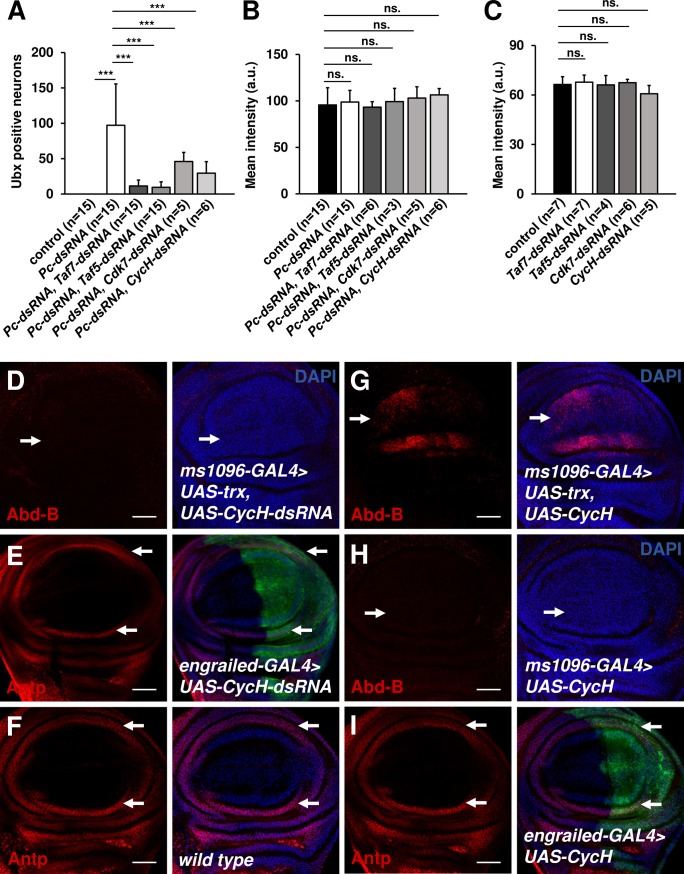
Since in ectopic gene expression PolII occupancy appears to be more sensitive to H3.3 deposition at transcription start sites, we speculated that the initiation of ectopic transcription is more vulnerable to perturbation. To test this hypothesis, we reduced the activities of the general transcriptional factor TFII-D by knocking down various TATA box-binding protein (TBP)-associated factors (TAF). Although TAFs are essential factors, animals with 65% reduction of Taf5 or 40% reduction of Taf7 in the central nervous system can grow to the adult stage without discernible defects (S17A and S17B Fig). Ubx-expressing neurons in Pc, Taf5 or Pc, Taf7 double-knockdown brains exhibited 88% or 90% reduction in number relative to Pc depleted brains, respectively (Fig 9A). Next, we investigated whether ectopic expression is more sensitive to general transcriptional factor TFII-H subunits, Cdk7 and CycH, which phosphorylate PolII CTD serine 5. Similarly, Pc, Cdk7 or Pc, CycH double depletion by RNAi significantly reduced the number of neurons ectopically expressing Ubx in brains (Figs 9A, S17C and S17D). By contrast, orthotopic expression of Ubx in ventral nerve cords was not affected by partial depletion of Taf5, Taf7, Cdk7 or CycH (Fig 9B and 9C). The Trx-induced ectopic expression of Abd-B was also sensitive to knockdown of CycH (Figs 9D and S18A), whereas the orthotopic expression of Antp was not (Figs 9E, 9F and S18B). Interestingly, the ectopic expression domain of Abd-B extended to the ventral compartment by over-expressing CycH (Figs 9G and S18A). As a control, over-expression of CycH alone did not induce ectopic expression of Abd-B (Figs 9H and S18A) or affect the orthotopic express of Antp (Figs 9I and S18C). Collectively, we propose that ectopic gene expression involves more sensitive coordination between H3.3 deposition, TFII-D, and TFII-H activities than is required for orthotopic gene expression.
Fig 9. Ectopic expression of Antp is more sensitive to partial knockdown of Taf5, Taf7, Cdk7, and CycH.
(A) Suppression of Pc depletion-induced ectopic expression of Ubx by depletion of Taf5, Taf7, Cdk7, and CycH. Quantification of Ubx positive neuron number in the doubly depleted brains as indicated. (B) The orthotopic expression of Ubx in the doubly depleted ventral nerve cords as indicated. (C) The orthotopic expression of Ubx in control, Taf5, Taf7, Cdk7, and CycH depleted ventral nerve cords. *** indicates p<0.0001 by Student's t-test. ns. indicates not significant. (D) The Trx-induced Abd-B expression (arrows) in the CycH knockdown wings. (E and F) The orthotopic expression of Antp in the CycH knockdown (E), and the control (F) wings. (G and H) The Abd-B expression in wings over-expressing both trx and CycH (G) or CycH alone (H) under the control of ms1096-GAL4. (I) The orthotopic expression of Antp in the CycH over-expression wings. Scale bars indicate 20μm.
Discussion
In this study, we provide evidence that loss of dBRWD3 suppresses ectopic gene expression and the tissue proliferation caused by the loss of PcG function, but not orthotopic gene expression. Loss of dBRWD3 also suppresses the ectopic gene expression induced by Trx. This suppression is related to enhanced H3.3 deposition at transcription start sites and reduced H3K4me3, activated PolII, and total PolII levels around the 5' regions of the ectopically expressed genes.
Excessive histone H3.3 deposition negatively affects ectopic gene transcription
A genome-wide H3.3 ChIP study revealed that H3.3 is enriched in enhancers, promoters, and gene bodies of actively transcribed genes [48]. This correlation suggests that H3.3 deposition may promote gene transcription, a concept that has been supported by the fact that H3.3s are more likely to possess marks associated with active gene expression, including trimethylation at lysine 4 (H3.3K4me3) and acetylation at several lysine residues [49,50]. However, an elegant study demonstrated that gene transcription remains normal when H3.3 is replaced by H3.3K4A, casting doubt regarding the importance of H3.3K4me3 [51]. Moreover, the extent to which H3.3 deposition truly promotes gene transcription is difficult to determine from genetic studies because knockout of H3.3 concurrently leads to up-regulation of one set of genes and down-regulation of another[52]. In the case of trans-retinoid acid induced expression of Cyp26A1 in embryonic stem cells, H3.3 is actively deposited to the enhancer before induction. Upon induction, H3.3 is depleted from the enhancer but deposited into the promoter. Knockdown of H3.3 reduces the binding of RAR and Tip60 to the enhancer region, indicating that deposition of H3.3 at enhancer regions facilitates the activation of inducible genes [53]. However, the role of H3.3 at promoter and gene body in transcription remain unclear.
It is even less clear how H3.3 affects ectopic gene expression, a pathological condition frequently associated with various cancers in humans [54,55]. In this study, we provide several lines of evidence that show that the regulation of H3.3 by dRBWD3 is required for the ectopic gene expression observed in PcG mutants or upon over-expression of trx. Firstly, only dBRWD3 transgenes that are able to reduce H3.3 levels in dBRWD3 mutant cells are capable of restoring ectopic expression of upd3 in Scm, dBRWD3 double-mutant cells. Secondly, the loss of yem, which prevents the aberrant incorporation of H3.3, also restores the ectopic expression of upd3 in Scm, dBRWD3, yem triple-mutant clones. Conversely, the over-deposited H3.3 induced by YEM is sufficient to suppress the ectopic Antp expression and Trx-induced ectopic Abd-B expression.
In ectopic gene expression, H3.3 deposition at the enhancers is variably regulated by dBRWD3. Nevertheless, through a not entirely clear mechanism, the H3K4me1 induced by Trx at enhancers is insensitive to dBRWD3. By contrast, the dBRWD3 depletion-enhanced H3.3 deposition at transcription start sites interferes with H3K4me3 and PolII enrichment at the same regions as well as the 5' ends of the gene bodies. Taken together, these observations suggest that H3.3 deposition at these regions disrupts transcription by interfering with trimethylation of H3K4 as well as PolII engagement or activation. This notion is supported by a recent finding that ectopic gene expression persists longer in Hira mutants, in which H3.3 deposition is reduced at the promoter and 5' regions [56]. Different from the known role of H3K4me3 on the promoters, our data shows that H3K4me3 is enriched on the gene bodies of ectopically or orthotopically expressed genes rather than the promoters. This is most likely due to the bias associated with the selected PCR amplicons.
dBRWD3 regulates the deposition of H3.3 more prominently at the promoters and transcription start sites in both ectopic and orthotopic gene expression. However, dBRWD3 regulates the PolII occupancy and H3K4me3 levels only in ectopic gene expression. Due to a not-yet-defined “robustness” of transcription, orthotopic gene expression is rendered insensitive to the increase of H3.3. Our data suggest that the same robustness of orthotopic gene expression closely cooperates with TFII-D and TFII-H since orthotopic gene expression remains intact under the suboptimal TFII-D or TFII-H activities. Further studies are needed to understand molecular nature of the robustness.
In addition to their negative effects on ectopic gene expression, H3.3 deposition and dBRWD3 may also interact with PcG in different contexts. For example, it has been reported that H3.3 deposition directs PRC2 to bivalent promoters in ES cells [57]. In addition, loss of dBRWD3 up-regulates Pc, pho, and tna but down-regulates phol, Jarid2 and the trithorax group genes ash1 and Iswi [11]. The regulation of ash1, which encodes H3K4 monomethylase, is particularly interesting because an independent transcriptome analysis of adult heads in dBRWD3 hypomorphic mutants also confirmed a lower level of ash1 mRNA. However, functional studies revealed that ash1 depletion by RNAi rescues rather than exacerbates the rough eye phenotype caused by dBRWD3 depletion. In addition, the H3K4me1 levels in dBRWD3 mutant cells are similar to those in wild-type cells. Further studies are needed to determine the significance of dBRWD3 regulation of ash1 mRNA. Finally, knockdown of dBRWD3 causes further reduction of H3K27me3 or H2AK118ub in the E(z) or Pc depleted brains, suggesting that H3.3 deposition may accelerate the removal of these repressive marks. Further investigations are warranted to understand whether the accelerated removal of repressive marks is mainly contributed by the nucleosome turnover associated with H3.3 deposition or involves activation of demethylases and deubiquitylase.
Biological implications of differences between ectopic and orthotopic gene expression
The H3K4me3 levels at the promoter and 5' ends of genes correlate well with active transcription. In fact, it is both a cause and a consequence of active transcription. As a cause, H3K4me3 recruits the TFII-D subunit TAF3, a general transcription factor involved in PolII engagement and transcription initiation [58]. Based on such a scenario, we propose that dBRWD3 increases H3K4me3 levels to the extent required for promoters to recruit TFII-D in ectopic gene expression. Consistently, partial knockdown of Taf5 or Taf7 affects ectopic but not orthotopic gene expression. In other words, ectopic and orthotopic gene expression may require different levels of TFII-D activities.
During active transcription, H3K4me3 is established by hSet1A/B, which is recruited to actively transcribed gene regions by CTD Ser5-phosphorylated PolII [59]. The phosphorylation of PolII’s CTD Ser5 is mediated by CDK7/CycH and is preferentially required for ectopic gene expression. Consistent with this idea, we demonstrated that partial knockdown of Cdk7 or CycH indeed affected ectopic gene expression without a discernible effect on orthotopic gene expression, perhaps because it was more dependent on the phosphorylation of PolII CTD Ser5. A complement study in RING1A, RING1B double knockout embryonic stem cells revealed that de-repressed loci displaying higher levels of PolII phospho-CTD Ser5 are ectopically expressed at higher levels [60], supporting our findings that phosphorylation of PolII CTD Ser5 plays an unique role in ectopic gene expression that is not shared with orthotopic gene expression. Based in part on these findings, we propose that dBRWD3 could play a preferential role in ectopic gene expression by facilitating the phosphorylation of PolII CTD Ser5.
PcG mutations and reduced expression of PcG proteins contribute to tumorigenesis in several human malignancies [25–31,34,35,61–64]. Hence, understanding the regulation of ectopic gene expression will have important medical implications. It has been shown that the ectopic expression of upd1, upd2, and upd3 also underlies tissue overgrowth in Drosophila PcG mutants, suggesting an evolutionarily conserved role for PcG in tumor suppression from insects to humans. Based on our results, we speculate that inhibition of the BRWD3, TFII-D, and TFII-H complex, for example by the CDK7 inhibitor THZ1 [65–67], might preferentially suppress a broad spectrum of tumors driven by PcG mutations.
In summary, we found that ectopic gene expression differs from orthotopic gene expression in their sensitivities to dBRWD3. Inactivation of dBRWD3 selectively suppresses ectopic gene expression and tissue overgrowth induced by loss of PcG function.
Materials and Methods
Constructs
p-ENTR-dBRWD3-N1287A-3XFlag, p-ENTR-dBRWD3-N1451A-3XFlag, p-ENTR-dBRWD3-N1287A, N1451A-3XFlag (p-ENTR-dBRWD3-2BC-3XFlag) were generated with the Thermo Scientific Phusion Site-Directed Mutagenesis kit using the previously described p-ENTR-dBRWD3-3XFlag as a template. p-ENTR-HA-yem was generated by PCR from the cDNA clone RE33235, Drosophila Genetic Resource Center. p-ENTR-dBRWD3-N1287A-3XFlag, p-ENTR-dBRWD3-N1451A-3XFlag, and pENTR- dBRWD3-2BC-3XFlag were recombined into the pUWR vector (DGRC Gateway collection) to generate pUWR-dBRWD3-N1287A-3XFlag-RFP, pUWR-dBRWD3-N1451A-3XFlag-RFP, pUWR-dBRWD3-2BC-3XFlag-RFP. p-ENTR-HA-yem was recombined into the pTWF vector (DGRC Gateway collection) to generate pTWF-HA-yem.
Fly strains and genetics
Flies were raised in standard conditions at 25°C except as otherwise mentioned. The dBRWD3s5349, and yemGS21861 were described earlier [11,68]. SceKO and ScmD1, trxE2 were kindly provided by Dr. Muller [39,47]. hs-H3.3-GFP was a gift from Dr. Kami Ahmad [69]. GMR-GAL4 (stock number 9146), Elav-GAL4 (stock number 458), ScmD1 (stock number 24158), Sce1 (stock number 24618), wtsx1 (stock number 44251), ex-LacZ (stock number 11067), UAS-mCD8-GFP (stock number 5146), UAS-Taf5-shRNA (stock number 35367), and UAS-Taf7-shRNA (stock number 55216) were obtained from the Bloomington stock center. UAS-trx (stock number 12194) and OK107-GAL4, were obtained from the Drosophila Genetic Resource Center, Kyoto. The ScmD1, dBRWD3s5349 double mutant, ScmD1, dBRWD3PX2 double mutant, dBRWD3s5349, Sce1 double mutant, ScmD1, dBRWD3s5349, yemGS21861 triple mutant, and wtsx1, dBRWD3s5349 double mutant were generated by recombination. UAS-Psc-dsRNA (NIG3886R-4) UAS-ph-p-dsRNA (NIG18412R-1), UAS-E(z)-dsRNA (NIG6502R-3), UAS-Pc-dsRNA (NIG32443R-1), UAS-CycH-dsRNA (NIG7405R-1), and UAS-Cdk7-dsRNA (NIG3319R-1) were obtained from the fly stocks of the National Institute of Genetics, Kyoto, Japan (NIG-FLY). UAS-dBRWD3-dsRNA (VDRC40209) was obtained from the Vienna Drosophila RNAi Center (VDRC). The transgenic flies ubi-dBRWD3-N1287A-3XFlag-RFP, ubi-dBRWD3-N1451A-3XFlag-RFP, ubi-dBRWD3-N1287A, N1451A-3XFlag-RFP (ubi-dBRWD3-2BC-3XFlag-RFP), and pTWF-HA-yem were generated by microinjection for germ-line transformation. The transgenic flies ubi-H3.3-dendra2, ubi-dBRWD3-3XFlag-RFP, ubi-delta-N-dBRWD3-3XFlag-RFP, and 10XSTAT-nlsGFP were described previously [11,41].
Clonal analysis
Genotypes for mosaic mutant clones in eyes:
ey-flp/+; FRT82B /FRT82B ubi-nlsGFP (Figs 1B, 3A, 3B, 3C, 3G, 4C and 4K)
ey-flp/+; FRT82B ScmD1/FRT82B ubi-nlsGFP (Figs 1C, 3A, 3B, 3C, 3E, 4A and 4J)
ey-flp/+; FRT82B ScmD1, dBRWD3s5349/FRT82B ubi-nlsGFP (Figs 1D, 3A, 3B, 3C, 3F, 4F and 4L)
ey-flp/+; FRT82B Sce1/FRT82B ubi-nlsGFP (Figs 1E, 3H, 4B and 4O)
ey-flp/+; FRT82B dBRWD3s5349, Sce1/FRT82B ubi-nlsGFP (Figs 1F, 3I, 4G and 4N)
hs-flp/+; FRT82B /FRT82B ubi-nlsGFP (Fig 1H)
hs-flp/+; FRT82B dBRWD3s5349/FRT82B ubi-nlsGFP (Fig 1I)
ey-flp/+; FRT82B dBRWD3s5349/FRT82B ubi-nlsGFP (Figs 3J and 4H)
ey-flp/+; 10XSTAT-nlsGFP/+; FRT82B ScmD1/FRT82B ubi-mof-RFP (Fig 3N)
ey-flp/+; 10XSTAT-nlsGFP/+; FRT82B ScmD1, dBRWD3s5349/FRT82B ubi-mof-RFP (Fig 3O)
ey-flp/+; 10XSTAT-nlsGFP/+; FRT82B dBRWD3s5349/FRT82B ubi-mof-RFP (Fig 3P)
ey-flp/+; FRT82B wtsx1/FRT82B ubi-nlsGFP (Fig 4Q)
ey-flp/+; FRT82B dBRWD3s5349, wtsx1/FRT82B ubi-nlsGFP (Fig 4R)
ey-flp/+; ubi-dBRWD3-RFP/+; FRT82B ScmD1, dBRWD3s5349/FRT82B ubi-nlsGFP (Fig 5A)
ey-flp/+; ubi-dBRWD3-2BC-RFP/+; FRT82B ScmD1, dBRWD3s5349/FRT82B ubi-nlsGFP (Fig 5B)
ey-flp/+; ubi-△N-dBRWD3-RFP/+; FRT82B ScmD1, dBRWD3s5349/FRT82B ubi-nlsGFP (Fig 5C)
ey-flp/+; FRT82B ScmD1, dBRWD3s5349, yemGS21861/FRT82B ubi-nlsGFP (Fig 5D and 5E)
ey-flp/GMR-GAL4; UAS-HA-yem-Flag, FRT82B ScmD1/FRT82B ubi-nlsGFP (Fig 5G)
RNA extraction, reverse transcription and RT-PCR
Total RNA was isolated from instar larval mosaic eye brain complexes using TRIzol reagent (Invitrogen). Following the manufacturer’s protocol, cDNA was synthesized using oligo(dT) and SuperScript reverse transcriptase (Invitrogen). OmicsGreen qPCR 5X Master Mix (Omics Bio) was used for real-time quantitative PCR on a CFX96 connect Real-Time PCR System (Bio-Rad). RPL32 was used as an endogenous loading control.
Immunostaining and antibodies
3rd instar larval eye imaginal discs were dissected in PBS and fixed for 17 minutes in 4% formaldehyde, followed by three 10-min washes in PBS supplemented with 0.3% Triton-X-100 (PBT) and 30-min blocking in PBT containing 5% normal donkey serum (NDS). After blocking, discs were incubated with primary antibody either overnight at 4°C or 2 hours at room temperature in PBT containing 5% NDS. After incubation with primary antibody, discs were washed three times in PBT before incubating with secondary antibody in PBT containing 5% NDS for one hour at room temperature. After three subsequent washes, discs were mounted with glycerol. Primary antibodies used in this study include mouse anti-H2AK118ub (1:100, Millipore, E6C5), rabbit anti-H3K27me3 (1:100, Millipore), rabbit anti-H3K4me1 (1:100, Active Motif), rabbit anti-H3S10ph (1:500, Millipore), mouse anti-Ubx (1:20, DSHB, Ubx), mouse anti-Antp (1:20, DSHB, 8C11), rabbit anti-upd3 (1:750), and mouse anti-β-Galactosidase (1:1000, Sigma, GAL-50). Secondary antibodies include goat anti-mouse Cy3 (1:1000, Jackson ImmunoResearch), goat anti-mouse Cy5 (1:1000, Jackson ImmunoResearch), and goat anti-rabbit Cy3 (1:1000, Jackson ImmunoResearch).
Chromatin immunoprecipitation
Chromatin immunoprecipitation was done with a ChIP-IT High Sensitivity (HS) Kit (Active Motif) following the instructions provided by the manufacturer. Briefly, 300 pairs of brain lobes (leaving out the attached ventral nerve cords) or wing imaginal discs of 3rd instar larvae were collected. The collected tissues were fixed with complete tissue fixation solution (28μl 37% formaldehyde in 970μl PBS) at room temperature for 15 minutes. Fixation was stopped with the stop solution at room temperature for 5 minutes. The fixed tissues were washed with ice-old PBS wash buffer and then immersed in the tissues with the chromatin prep buffer. The fixed tissues were sonicated using the UP50H Ultrasonic Processor (Hielscher-Ultrasound Technology), with 30% amplitude and 20 pulse cycles of 30 seconds on followed by 30 seconds off. 6 μg sheared chromatin was incubated with 1μg antibodies overnight at 4°C. 30 μl Protein G agarose beads were added to each IP reaction. The mixture was rotated at 4°C for 3 hours. The ChIP reactions were loaded into columns and washed. The ChIP DNA was obtained by eluting the columns with elution buffer AM4. The elute was treated with Protease K at 55°C for 30 minutes, 80°C for two hours, followed by column clean-up. OmicsGreen qPCR 5X Master Mix (Omics Bio) was used for real-time quantitative PCR on a CFX96 connect Real-Time PCR System (Bio-Rad) to measure the amount of ChIP DNA and input DNA containing indicated sequences from enhancers, promoters, and 5' transcription regions of Antp, Ubx, and Abd-B. Primary antibodies used include rabbit anti-H2AK118ub (Cell signaling, D27C4), mouse anti-H3K27me3 (Abcam, mAbcam 6002), rabbit anti-H3K4me1 (Abcam, ab8895), rabbit anti-H3K4me3 (Abcam, ab8580), mouse anti-PolII (Abcam, 4H8), and rabbit anti-PolII phospho-CTD Ser5 (Abcam, ab5131), rabbit anti-HA (Cell Signaling, C29F4) and mouse anti-Flag (Sigma, M2)
Image quantification
All confocal images were obtained by LSM 700 laser scanning confocal microscope (Carl Zeiss). For quantitative analysis of protein levels, the antibody staining conditions, laser power, and pinhole sizes were kept identical among groups. Pixel number, pixel intensity, and area were provided by the built-in software in LSM 700. The areas of clones (marked by the absence of GFP) and non-clones (marked by GFP) were calculated by the total GFP positive and GFP negative areas respectively. Antp-positive regions in ventral nerve cords were manually marked. The Antp expression areas and lengths were calculated by the built-in software according to the marked regions. H3S10ph-positive mitotic cells and Antp-positive brain cells were manually counted.
Supporting Information
(DOCX)
(A) Antp levels (arrows) in ScmD1, dBRWD3PX2 double-mutant clones generated in the 3rd instar eye imaginal discs by ey-flp and marked by the absence of GFP. Scale bars indicate 50μm. (B) A schematic illustration of how protein levels are calculated from a confocal image of a mosaic imaginal disc. (C and D) Quantifications of Antp expression. The ectopic expression of Antp in ScmD1, ScmD1, dBRWD3PX2 and ScmD1, dBRWD3s5349 mutant clones (C), Sce1 and dBRWD3s5349, Sce1 mutant clones (D). (E)The orthotopic expression of Antp in wild type and dBRWD3s5349 mutant clones. a.u. indicates arbitrary unit. Data are shown as means ± S.D. *, **, *** indicate P< 0.01, 0.001, 0.0001, respectively, by Student's t-test. ns. indicates not significant.
(TIF)
(A) The knockdown efficiency of Pc-dsRNA driven by Elav-GAL4. (B-D) The mean intensity (B), expression area (C), and length of expression domain (D) of Antp in control, Pc depleted, dBRWD3 depleted, and Pc, dBRWD3 doubly depleted ventral nervous cords. (E) The knockdown efficiency of E(z)-dsRNA driven by Elav-GAL4. (F-H) The mean intensity (F), expression area (G), and length of expression domain (H) in the control, E(z) depleted, dBRWD3 depleted, and E(z), dBRWD3 doubly depleted ventral nervous cords. a.u. indicates arbitrary unit. Data are shown as means ± S.D. ns. indicates P>0.05 by Student's t-test. *, *** indicate P< 0.01, 0.0001, respectively, by Student's t-test.
(TIF)
(A and B) Quantification of Upd3 ectopic expression in ScmD1 and ScmD1, dBRWD3s5349 mutant clones (A) or Sce1 and dBRWD3s5349, Sce1 mutant clones (B). Data are shown as means ± S.D. *, **, *** indicate P< 0.01, 0.001, 0.0001, respectively, by Student's t-test. ns. indicates not significant. a.u. indicates arbitrary unit.
(TIF)
Scale bar indicates 50μm.
(TIF)
(A and B) expanded-LacZ (ex-LacZ) expression (arrows) in wtsx1 mutant clones (A), and dBRWD3s5349, wtsx1 double-mutant clones (B). Scale bars indicate 50μm.
(TIF)
(A) A diagram illustrating the molecular structure of dBRWD3. (B and C) upd3 levels (arrows) in ScmD1, dBRWD3s5349 double-mutant clones complemented with ubiquitously expressed dBRWD3-N1287A-RFP (B) or dBRWD3-N1451A-RFP (C). Scale bars indicate 50μm.
(TIF)
Data represent the mRNA levels of indicated dBRWD3 transgenes plus endogenous dBRWD3 and are shown in means ± S.D., n = 4. *, **, *** indicate P< 0.01, 0.001, 0.0001 respectively in comparison to that in Canton-S by student's t-test. a.u. indicates arbitrary unit.
(TIF)
(A-D) H3.3-dendra2 driven by a ubi-promoter was expressed in dBRWD3s5349 mosaic mutant eye discs. dBRWD3s5349 mutant clones were marked by the absence of RFP (arrows), with concomitant expression of ΔN-dBRWD3-RFP (A), dBRWD3-2BC-RFP (B), dBRWD3-N1287A-RFP (C), and dBRWD3-N1451A-RFP (D). Scale bars indicate 50μm. (E) The levels of endogenous H3.3 in dBRWD3-depleted brains and ventral nerve cords.
(TIF)
(A) H3.3-dendra2 and Antp levels in wild type eye discs. (B-D) H3.3-dendra2 and Antp levels in ScmD1, dBRWD3s5349, double-mutant clones (B), ScmD1, dBRWD3s5349, yemGS21861 triple-mutant clones (C), and ScmD1 single-mutant clones (D) that were generated in the eye disc and marked by the absence of RFP (arrows). Scale bars indicate 50μm.
(TIF)
(A and B) The mean intensity of Antp (A) and Upd3 (B) staining in wild type and ScmD1 dBRWD3s5349, yemGS21861 triple-mutant clones. *** indicates p<0.0001 by Student's t-test. (C) The mean intensity of Antp staining in ScmD1 and ScmD1 UAS-yem. *** indicates p<0.0001 by Student's t-test.
(TIF)
(A and B) Heat-shock inducible H3.3-GFP was expressed in wild-type (A) and yem-expressing (B) salivary glands. (C) The quantification of H3.3-GFP levels in (A) and (B).
(TIF)
(A) A schematic illustration of the expression domain of ms1096-GAL4. Expression in the dorsal compartment (dark grey) is higher than in the ventral compartment (light grey). (B-F) Immunofluorescence studies of H3K27me3 levels in the E(z), dBRWD3 doubly depleted (B), E(z) depleted (C), Pc, dBRWD3 doubly depleted (D), Pc depleted (E), and control (F) wing discs. (G and H) H2AK118ub levels in SceKO (G) and dBRWD3s5349, SceKO (H) mutant clones. Scale bars indicate 50μm.
(TIF)
The ectopic expression of Antp in ScmD1, trxE2 double-mutant clones marked by the absence of GFP (arrows). Scale bars indicate 50μm.
(TIF)
(A) A schematic illustration of the disc proper and the peripodial epithelium of the eye disc. (B and C) Antp levels in the trx-expressing (B) and trx-expressing, dBRWD3-depleted (C) peripodial epithelia. Scale bars indicate 50μm. (D-E) trx was overexpressed under the control of GMR-GAL4. The TRX-induced Antp expression (arrows) in wild-type (D), dBRWD3 depletion (E) backgrounds. (F-G) trx was overexpressed under the control of ms-1096-GAL4. The TRX-induced Ubx expression (arrows) in wild-type (F), dBRWD3 depletion (G) backgrounds. Scale bars indicate 20μm.
(TIF)
(A) A schematic illustration of the apterous-GAL4 expression region in the 3rd wing imaginal discs. D indicates the dorsal compartment. (B-D) Immunofluorescence studies of H3K4me1 levels in the dorsal compartment of the apterous-GAL4 control (B), trx-expressing (C), and trx-expressing, dBRWD3 depleted (D) wing discs marked by GFP. Scale bars indicate 50μm. (E) A quantitative analysis of H3K4me1 levels in control, trx over-expression, and trx over-expression, dBRWD3-depleted wings. *** indicates p<0.0001 by Student's t-test. ns. indicates not significant.
(TIF)
A ChIP-qPCR analysis of PolII levels at the promoters and transcription start sites of Antp in the UAS-mCD8-GFP control, in wings over-expressing trx, or in wings with concurrent trx over-expression and dBRWD3 depletion. ns. indicates not significant. ChIP-qPCR Data are shown as means ± S.D from 4 technical replicates. *, ** indicate P<0.05, 0.01 respectively by Student's t-test.
(TIF)
(A-D) A quantitative analysis of the knockdown efficiencies of Taf5 (A), Taf7 (B), Cdk7 (C), and CycH (D) RNAi. *** indicates p<0.0001 by Student's t-test.
(TIF)
(A) A quantitative analysis of Abd-B expression in the control, Trx-over-expressing, Trx-over-expressing plus CycH knockdown, Trx and CycH-over-expressing, and CycH-over-expressing wings. (B and C) quantitative analyses of orthotopic expression of Antp in the CycH knockdown (B) and the CycH-over-expressing (C) wings. *** indicates p<0.0001 by Student's t-test. ns. indicates not significant.
(TIF)
Acknowledgments
We thank Dr. Chih-Chiang Chan, Dr. Chun-Liang Pan, Dr. Tso-Pang Yao, Dr. Cheng-Ting Chien for constructive comments. We thank Taiwan Fly Core, the fly core facility at National Taiwan University, and the sixth common core lab at the National Taiwan University Hospital for their technical support. We also thank Dr. Katherine Olsson Carter for critical reading of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants (NSC101-2321- B-002- 082-) to JTW and (NSC102-2633- B-029- 002-) and (MOST104-2632-B-029 -001) to YCT from the Ministry of Science and Technology of Taiwan (https://www.most.gov.tw/en/public), and (NTU-ERP-105R89215) to JTW from National Taiwan University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705. [DOI] [PubMed] [Google Scholar]
- 2.Maze I, Noh KM, Soshnev AA, Allis CD (2014) Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat Rev Genet 15: 259–271. 10.1038/nrg3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zink LM, Hake SB (2016) Histone variants: nuclear function and disease. Curr Opin Genet Dev 37: 82–89. 10.1016/j.gde.2015.12.002 [DOI] [PubMed] [Google Scholar]
- 4.Gaume X, Torres-Padilla ME (2015) Regulation of Reprogramming and Cellular Plasticity through Histone Exchange and Histone Variant Incorporation. Cold Spring Harb Symp Quant Biol. [DOI] [PubMed] [Google Scholar]
- 5.Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, et al. (2011) Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell 44: 928–941. 10.1016/j.molcel.2011.12.006 [DOI] [PubMed] [Google Scholar]
- 6.Schneiderman JI, Orsi GA, Hughes KT, Loppin B, Ahmad K (2012) Nucleosome-depleted chromatin gaps recruit assembly factors for the H3.3 histone variant. Proc Natl Acad Sci U S A 109: 19721–19726. 10.1073/pnas.1206629109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loppin B, Bonnefoy E, Anselme C, Laurencon A, Karr TL, et al. (2005) The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437: 1386–1390. [DOI] [PubMed] [Google Scholar]
- 8.Orsi GA, Algazeery A, Meyer RE, Capri M, Sapey-Triomphe LM, et al. (2013) Drosophila Yemanuclein and HIRA cooperate for de novo assembly of H3.3-containing nucleosomes in the male pronucleus. PLoS Genet 9: e1003285 10.1371/journal.pgen.1003285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M (2014) Hira-mediated h3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev Cell 30: 268–279. 10.1016/j.devcel.2014.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inoue A, Zhang Y (2014) Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat Struct Mol Biol 21: 609–616. 10.1038/nsmb.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen WY, Shih HT, Liu KY, Shih ZS, Chen LK, et al. (2015) Intellectual disability-associated dBRWD3 regulates gene expression through inhibition of HIRA/YEM-mediated chromatin deposition of histone H3.3. EMBO Rep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G (2007) Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745. [DOI] [PubMed] [Google Scholar]
- 13.Geisler SJ, Paro R (2015) Trithorax and Polycomb group-dependent regulation: a tale of opposing activities. Development 142: 2876–2887. 10.1242/dev.120030 [DOI] [PubMed] [Google Scholar]
- 14.Kang H, McElroy KA, Jung YL, Alekseyenko AA, Zee BM, et al. (2015) Sex comb on midleg (Scm) is a functional link between PcG-repressive complexes in Drosophila. Genes Dev 29: 1136–1150. 10.1101/gad.260562.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon JA, Kingston RE (2013) Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 49: 808–824. 10.1016/j.molcel.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dellino GI, Schwartz YB, Farkas G, McCabe D, Elgin SC, et al. (2004) Polycomb silencing blocks transcription initiation. Mol Cell 13: 887–893. [DOI] [PubMed] [Google Scholar]
- 17.King IF, Francis NJ, Kingston RE (2002) Native and recombinant polycomb group complexes establish a selective block to template accessibility to repress transcription in vitro. Mol Cell Biol 22: 7919–7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehmann L, Ferrari R, Vashisht AA, Wohlschlegel JA, Kurdistani SK, et al. (2012) Polycomb repressive complex 1 (PRC1) disassembles RNA polymerase II preinitiation complexes. J Biol Chem 287: 35784–35794. 10.1074/jbc.M112.397430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aloia L, Di Stefano B, Di Croce L (2013) Polycomb complexes in stem cells and embryonic development. Development 140: 2525–2534. 10.1242/dev.091553 [DOI] [PubMed] [Google Scholar]
- 20.Struhl G (1981) A gene product required for correct initiation of segmental determination in Drosophila. Nature 293: 36–41. [DOI] [PubMed] [Google Scholar]
- 21.Struhl G, Brower D (1982) Early role of the esc+ gene product in the determination of segments in Drosophila. Cell 31: 285–292. [DOI] [PubMed] [Google Scholar]
- 22.Classen AK, Bunker BD, Harvey KF, Vaccari T, Bilder D (2009) A tumor suppressor activity of Drosophila Polycomb genes mediated by JAK-STAT signaling. Nat Genet 41: 1150–1155. 10.1038/ng.445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauvageau M, Sauvageau G (2010) Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 7: 299–313. 10.1016/j.stem.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pallante P, Forzati F, Federico A, Arra C, Fusco A (2015) Polycomb protein family member CBX7 plays a critical role in cancer progression. Am J Cancer Res 5: 1594–1601. [PMC free article] [PubMed] [Google Scholar]
- 25.Hinz S, Kempkensteffen C, Christoph F, Krause H, Schrader M, et al. (2008) Expression parameters of the polycomb group proteins BMI1, SUZ12, RING1 and CBX7 in urothelial carcinoma of the bladder and their prognostic relevance. Tumour Biol 29: 323–329. 10.1159/000170879 [DOI] [PubMed] [Google Scholar]
- 26.Mansueto G, Forzati F, Ferraro A, Pallante P, Bianco M, et al. (2010) Identification of a New Pathway for Tumor Progression: MicroRNA-181b Up-Regulation and CBX7 Down-Regulation by HMGA1 Protein. Genes Cancer 1: 210–224. 10.1177/1947601910366860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pallante P, Terracciano L, Carafa V, Schneider S, Zlobec I, et al. (2010) The loss of the CBX7 gene expression represents an adverse prognostic marker for survival of colon carcinoma patients. Eur J Cancer 46: 2304–2313. 10.1016/j.ejca.2010.05.011 [DOI] [PubMed] [Google Scholar]
- 28.Li G, Warden C, Zou Z, Neman J, Krueger JS, et al. (2013) Altered expression of polycomb group genes in glioblastoma multiforme. PLoS One 8: e80970 10.1371/journal.pone.0080970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forzati F, Federico A, Pallante P, Abbate A, Esposito F, et al. (2012) CBX7 is a tumor suppressor in mice and humans. J Clin Invest 122: 612–623. 10.1172/JCI58620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karamitopoulou E, Pallante P, Zlobec I, Tornillo L, Carafa V, et al. (2010) Loss of the CBX7 protein expression correlates with a more aggressive phenotype in pancreatic cancer. Eur J Cancer 46: 1438–1444. 10.1016/j.ejca.2010.01.033 [DOI] [PubMed] [Google Scholar]
- 31.Pallante P, Federico A, Berlingieri MT, Bianco M, Ferraro A, et al. (2008) Loss of the CBX7 gene expression correlates with a highly malignant phenotype in thyroid cancer. Cancer Res 68: 6770–6778. 10.1158/0008-5472.CAN-08-0695 [DOI] [PubMed] [Google Scholar]
- 32.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, et al. (2013) Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340: 857–861. 10.1126/science.1232245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan KM, Fang D, Gan H, Hashizume R, Yu C, et al. (2013) The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 27: 985–990. 10.1101/gad.217778.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Funato K, Major T, Lewis PW, Allis CD, Tabar V (2014) Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science 346: 1529–1533. 10.1126/science.1253799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang M, Wang Y, Jones S, Sausen M, McMahon K, et al. (2014) Somatic mutations of SUZ12 in malignant peripheral nerve sheath tumors. Nat Genet 46: 1170–1172. 10.1038/ng.3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Raedt T, Beert E, Pasmant E, Luscan A, Brems H, et al. (2014) PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 514: 247–251. 10.1038/nature13561 [DOI] [PubMed] [Google Scholar]
- 37.Martinez AM, Schuettengruber B, Sakr S, Janic A, Gonzalez C, et al. (2009) Polyhomeotic has a tumor suppressor activity mediated by repression of Notch signaling. Nat Genet 41: 1076–1082. 10.1038/ng.414 [DOI] [PubMed] [Google Scholar]
- 38.Feng S, Huang J, Wang J (2011) Loss of the Polycomb group gene polyhomeotic induces non-autonomous cell overproliferation. EMBO Rep 12: 157–163. 10.1038/embor.2010.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutierrez L, Oktaba K, Scheuermann JC, Gambetta MC, Ly-Hartig N, et al. (2012) The role of the histone H2A ubiquitinase Sce in Polycomb repression. Development 139: 117–127. 10.1242/dev.074450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, et al. (2007) GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns 7: 323–331. [DOI] [PubMed] [Google Scholar]
- 41.Tsai YC, Grimm S, Chao JL, Wang SC, Hofmeyer K, et al. (2015) Optomotor-blind negatively regulates Drosophila eye development by blocking Jak/STAT signaling. PLoS One 10: e0120236 10.1371/journal.pone.0120236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, et al. (2002) salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110: 467–478. [DOI] [PubMed] [Google Scholar]
- 43.Wu S, Huang J, Dong J, Pan D (2003) hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114: 445–456. [DOI] [PubMed] [Google Scholar]
- 44.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, et al. (2012) Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149: 214–231. 10.1016/j.cell.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tie F, Banerjee R, Saiakhova AR, Howard B, Monteith KE, et al. (2014) Trithorax monomethylates histone H3K4 and interacts directly with CBP to promote H3K27 acetylation and antagonize Polycomb silencing. Development 141: 1129–1139. 10.1242/dev.102392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, et al. (2010) Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet 6: e1000805 10.1371/journal.pgen.1000805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klymenko T, Muller J (2004) The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep 5: 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, et al. (2010) Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140: 678–691. 10.1016/j.cell.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKittrick E, Gafken PR, Ahmad K, Henikoff S (2004) Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci U S A 101: 1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, et al. (2006) Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J Biol Chem 281: 559–568. [DOI] [PubMed] [Google Scholar]
- 51.Hodl M, Basler K (2009) Transcription in the absence of histone H3.3. Curr Biol 19: 1221–1226. 10.1016/j.cub.2009.05.048 [DOI] [PubMed] [Google Scholar]
- 52.Bush KM, Yuen BT, Barrilleaux BL, Riggs JW, O'Geen H, et al. (2013) Endogenous mammalian histone H3.3 exhibits chromatin-related functions during development. Epigenetics Chromatin 6: 7 10.1186/1756-8935-6-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen P, Zhao J, Wang Y, Wang M, Long H, et al. (2013) H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev 27: 2109–2124. 10.1101/gad.222174.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang W, Qin JJ, Voruganti S, Nag S, Zhou J, et al. (2015) Polycomb Group (PcG) Proteins and Human Cancers: Multifaceted Functions and Therapeutic Implications. Med Res Rev 35: 1220–1267. 10.1002/med.21358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koppens M, van Lohuizen M (2016) Context-dependent actions of Polycomb repressors in cancer. Oncogene 35: 1341–1352. 10.1038/onc.2015.195 [DOI] [PubMed] [Google Scholar]
- 56.Sadasivam DA, Huang DH (2016) Maintenance of Tissue Pluripotency by Epigenetic Factors Acting at Multiple Levels. PLoS Genet 12: e1005897 10.1371/journal.pgen.1005897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Banaszynski LA, Wen D, Dewell S, Whitcomb SJ, Lin M, et al. (2013) Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155: 107–120. 10.1016/j.cell.2013.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, et al. (2013) H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 152: 1021–1036. 10.1016/j.cell.2013.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee JH, Skalnik DG (2008) Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol 28: 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, et al. (2012) Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell 10: 157–170. 10.1016/j.stem.2011.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang W, Yuasa T, Tsuchiya N, Ma Z, Maita S, et al. (2009) The novel tumor-suppressor Mel-18 in prostate cancer: its functional polymorphism, expression and clinical significance. Int J Cancer 125: 2836–2843. 10.1002/ijc.24721 [DOI] [PubMed] [Google Scholar]
- 62.Sanchez-Beato M, Sanchez E, Gonzalez-Carrero J, Morente M, Diez A, et al. (2006) Variability in the expression of polycomb proteins in different normal and tumoral tissues. A pilot study using tissue microarrays. Mod Pathol 19: 684–694. [DOI] [PubMed] [Google Scholar]
- 63.Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, et al. (2010) Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet 42: 665–667. 10.1038/ng.620 [DOI] [PubMed] [Google Scholar]
- 64.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, et al. (2010) Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet 42: 722–726. 10.1038/ng.621 [DOI] [PubMed] [Google Scholar]
- 65.Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, et al. (2014) Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511: 616–620. 10.1038/nature13393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Christensen CL, Kwiatkowski N, Abraham BJ, Carretero J, Al-Shahrour F, et al. (2014) Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell 26: 909–922. 10.1016/j.ccell.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, et al. (2014) CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 159: 1126–1139. 10.1016/j.cell.2014.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.D'Costa A, Reifegerste R, Sierra S, Moses K (2006) The Drosophila ramshackle gene encodes a chromatin-associated protein required for cell morphology in the developing eye. Mech Dev 123: 591–604. [DOI] [PubMed] [Google Scholar]
- 69.Schwartz BE, Ahmad K (2005) Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev 19: 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(A) Antp levels (arrows) in ScmD1, dBRWD3PX2 double-mutant clones generated in the 3rd instar eye imaginal discs by ey-flp and marked by the absence of GFP. Scale bars indicate 50μm. (B) A schematic illustration of how protein levels are calculated from a confocal image of a mosaic imaginal disc. (C and D) Quantifications of Antp expression. The ectopic expression of Antp in ScmD1, ScmD1, dBRWD3PX2 and ScmD1, dBRWD3s5349 mutant clones (C), Sce1 and dBRWD3s5349, Sce1 mutant clones (D). (E)The orthotopic expression of Antp in wild type and dBRWD3s5349 mutant clones. a.u. indicates arbitrary unit. Data are shown as means ± S.D. *, **, *** indicate P< 0.01, 0.001, 0.0001, respectively, by Student's t-test. ns. indicates not significant.
(TIF)
(A) The knockdown efficiency of Pc-dsRNA driven by Elav-GAL4. (B-D) The mean intensity (B), expression area (C), and length of expression domain (D) of Antp in control, Pc depleted, dBRWD3 depleted, and Pc, dBRWD3 doubly depleted ventral nervous cords. (E) The knockdown efficiency of E(z)-dsRNA driven by Elav-GAL4. (F-H) The mean intensity (F), expression area (G), and length of expression domain (H) in the control, E(z) depleted, dBRWD3 depleted, and E(z), dBRWD3 doubly depleted ventral nervous cords. a.u. indicates arbitrary unit. Data are shown as means ± S.D. ns. indicates P>0.05 by Student's t-test. *, *** indicate P< 0.01, 0.0001, respectively, by Student's t-test.
(TIF)
(A and B) Quantification of Upd3 ectopic expression in ScmD1 and ScmD1, dBRWD3s5349 mutant clones (A) or Sce1 and dBRWD3s5349, Sce1 mutant clones (B). Data are shown as means ± S.D. *, **, *** indicate P< 0.01, 0.001, 0.0001, respectively, by Student's t-test. ns. indicates not significant. a.u. indicates arbitrary unit.
(TIF)
Scale bar indicates 50μm.
(TIF)
(A and B) expanded-LacZ (ex-LacZ) expression (arrows) in wtsx1 mutant clones (A), and dBRWD3s5349, wtsx1 double-mutant clones (B). Scale bars indicate 50μm.
(TIF)
(A) A diagram illustrating the molecular structure of dBRWD3. (B and C) upd3 levels (arrows) in ScmD1, dBRWD3s5349 double-mutant clones complemented with ubiquitously expressed dBRWD3-N1287A-RFP (B) or dBRWD3-N1451A-RFP (C). Scale bars indicate 50μm.
(TIF)
Data represent the mRNA levels of indicated dBRWD3 transgenes plus endogenous dBRWD3 and are shown in means ± S.D., n = 4. *, **, *** indicate P< 0.01, 0.001, 0.0001 respectively in comparison to that in Canton-S by student's t-test. a.u. indicates arbitrary unit.
(TIF)
(A-D) H3.3-dendra2 driven by a ubi-promoter was expressed in dBRWD3s5349 mosaic mutant eye discs. dBRWD3s5349 mutant clones were marked by the absence of RFP (arrows), with concomitant expression of ΔN-dBRWD3-RFP (A), dBRWD3-2BC-RFP (B), dBRWD3-N1287A-RFP (C), and dBRWD3-N1451A-RFP (D). Scale bars indicate 50μm. (E) The levels of endogenous H3.3 in dBRWD3-depleted brains and ventral nerve cords.
(TIF)
(A) H3.3-dendra2 and Antp levels in wild type eye discs. (B-D) H3.3-dendra2 and Antp levels in ScmD1, dBRWD3s5349, double-mutant clones (B), ScmD1, dBRWD3s5349, yemGS21861 triple-mutant clones (C), and ScmD1 single-mutant clones (D) that were generated in the eye disc and marked by the absence of RFP (arrows). Scale bars indicate 50μm.
(TIF)
(A and B) The mean intensity of Antp (A) and Upd3 (B) staining in wild type and ScmD1 dBRWD3s5349, yemGS21861 triple-mutant clones. *** indicates p<0.0001 by Student's t-test. (C) The mean intensity of Antp staining in ScmD1 and ScmD1 UAS-yem. *** indicates p<0.0001 by Student's t-test.
(TIF)
(A and B) Heat-shock inducible H3.3-GFP was expressed in wild-type (A) and yem-expressing (B) salivary glands. (C) The quantification of H3.3-GFP levels in (A) and (B).
(TIF)
(A) A schematic illustration of the expression domain of ms1096-GAL4. Expression in the dorsal compartment (dark grey) is higher than in the ventral compartment (light grey). (B-F) Immunofluorescence studies of H3K27me3 levels in the E(z), dBRWD3 doubly depleted (B), E(z) depleted (C), Pc, dBRWD3 doubly depleted (D), Pc depleted (E), and control (F) wing discs. (G and H) H2AK118ub levels in SceKO (G) and dBRWD3s5349, SceKO (H) mutant clones. Scale bars indicate 50μm.
(TIF)
The ectopic expression of Antp in ScmD1, trxE2 double-mutant clones marked by the absence of GFP (arrows). Scale bars indicate 50μm.
(TIF)
(A) A schematic illustration of the disc proper and the peripodial epithelium of the eye disc. (B and C) Antp levels in the trx-expressing (B) and trx-expressing, dBRWD3-depleted (C) peripodial epithelia. Scale bars indicate 50μm. (D-E) trx was overexpressed under the control of GMR-GAL4. The TRX-induced Antp expression (arrows) in wild-type (D), dBRWD3 depletion (E) backgrounds. (F-G) trx was overexpressed under the control of ms-1096-GAL4. The TRX-induced Ubx expression (arrows) in wild-type (F), dBRWD3 depletion (G) backgrounds. Scale bars indicate 20μm.
(TIF)
(A) A schematic illustration of the apterous-GAL4 expression region in the 3rd wing imaginal discs. D indicates the dorsal compartment. (B-D) Immunofluorescence studies of H3K4me1 levels in the dorsal compartment of the apterous-GAL4 control (B), trx-expressing (C), and trx-expressing, dBRWD3 depleted (D) wing discs marked by GFP. Scale bars indicate 50μm. (E) A quantitative analysis of H3K4me1 levels in control, trx over-expression, and trx over-expression, dBRWD3-depleted wings. *** indicates p<0.0001 by Student's t-test. ns. indicates not significant.
(TIF)
A ChIP-qPCR analysis of PolII levels at the promoters and transcription start sites of Antp in the UAS-mCD8-GFP control, in wings over-expressing trx, or in wings with concurrent trx over-expression and dBRWD3 depletion. ns. indicates not significant. ChIP-qPCR Data are shown as means ± S.D from 4 technical replicates. *, ** indicate P<0.05, 0.01 respectively by Student's t-test.
(TIF)
(A-D) A quantitative analysis of the knockdown efficiencies of Taf5 (A), Taf7 (B), Cdk7 (C), and CycH (D) RNAi. *** indicates p<0.0001 by Student's t-test.
(TIF)
(A) A quantitative analysis of Abd-B expression in the control, Trx-over-expressing, Trx-over-expressing plus CycH knockdown, Trx and CycH-over-expressing, and CycH-over-expressing wings. (B and C) quantitative analyses of orthotopic expression of Antp in the CycH knockdown (B) and the CycH-over-expressing (C) wings. *** indicates p<0.0001 by Student's t-test. ns. indicates not significant.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.