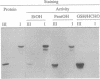
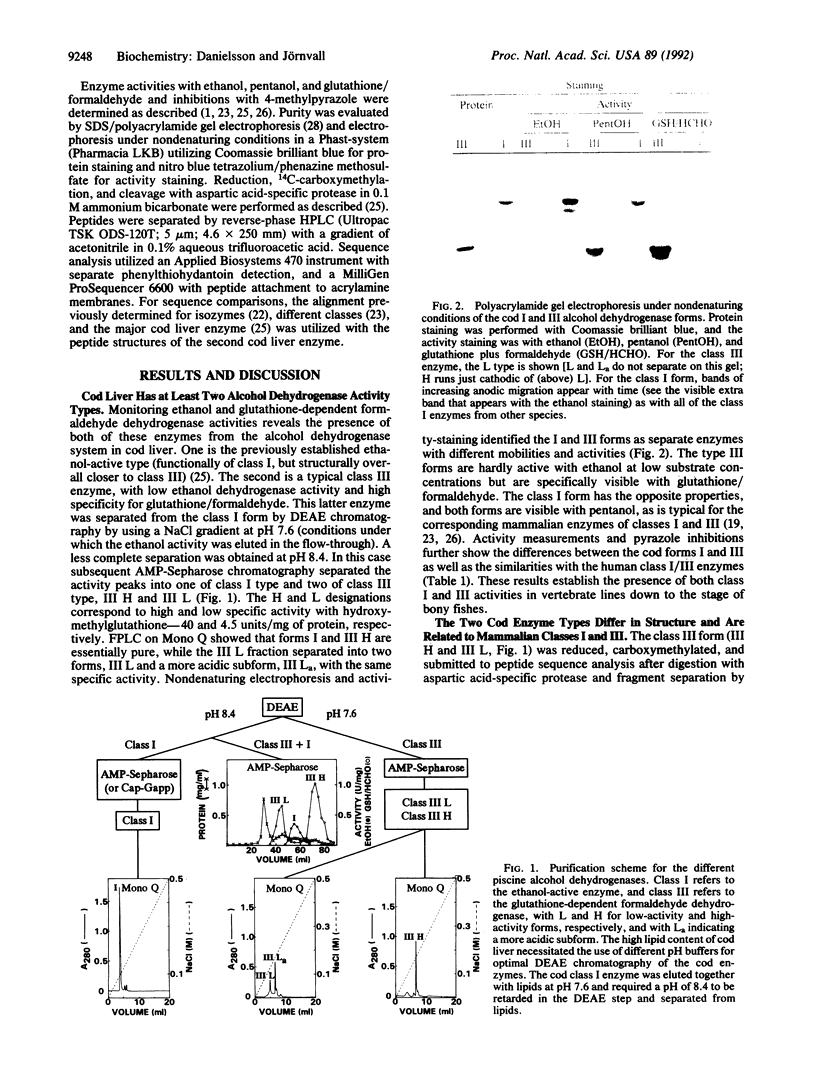
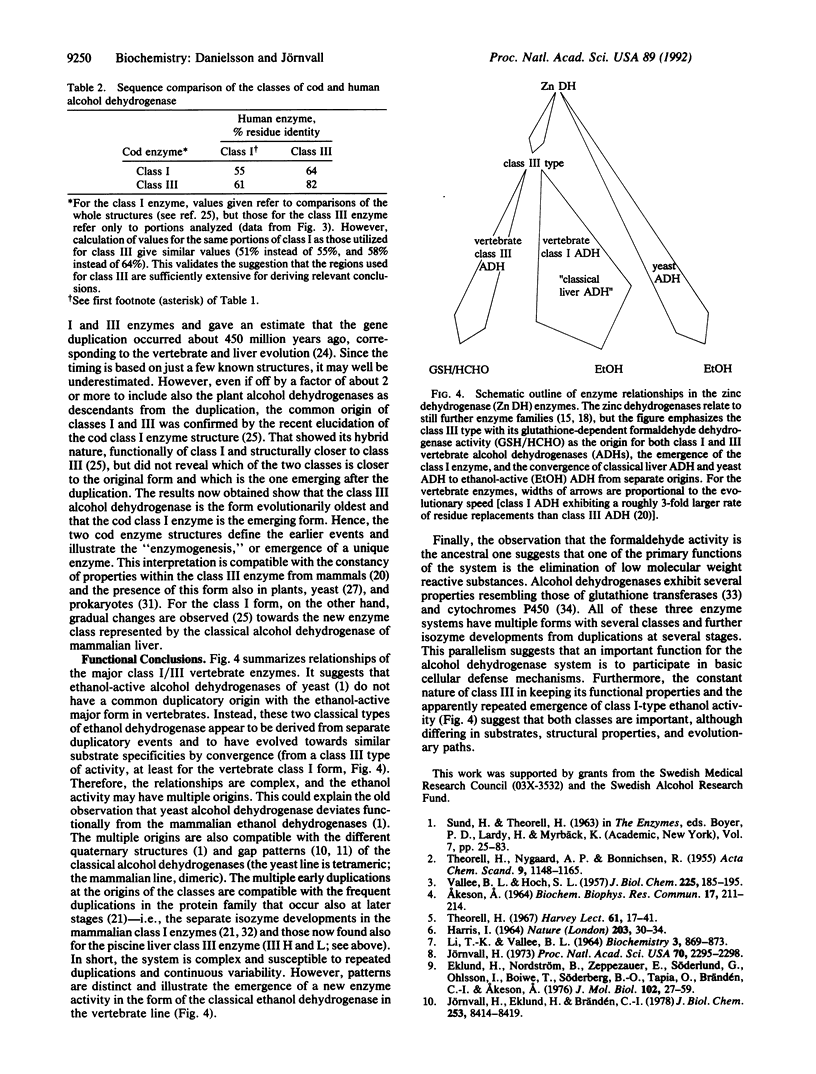
Abstract
Analysis of the activity and structure of lower vertebrate alcohol dehydrogenases reveals that relationships between the classical liver and yeast enzymes need not be continuous. Both the ethanol activity of class I-type alcohol dehydrogenase (alcohol:NAD+ oxidoreductase, EC 1.1.1.1) and the glutathione-dependent formaldehyde activity of the class III-type enzyme [formaldehyde:NAD+ oxidoreductase (glutathione-formylating), EC 1.2.1.1] are present in liver down to at least the stage of bony fishes (cod liver: ethanol activity, 3.4 units/mg of protein in one enzyme; formaldehyde activity, 4.5 units/mg in the major form of another enzyme). Structural analysis of the latter protein reveals it to be a typical class III enzyme, with limited variation from the mammalian form and therefore with stable activity and structure throughout much of the vertebrate lineage. In contrast, the classical alcohol dehydrogenase (the class I enzyme) appears to be the emerging form, first in activity and later also in structure. The class I activity is present already in the piscine line, whereas the overall structural-type enzyme is not observed until amphibians and still more recent vertebrates. Consequently, the class I/III duplicatory origin appears to have arisen from a functional class III form, not a class I form. Therefore, ethanol dehydrogenases from organisms existing before this duplication have origins separate from those leading to the "classical" liver alcohol dehydrogenases. The latter now often occur in isozyme forms from further gene duplications and have a high rate of evolutionary change. The pattern is, however, not simple and we presently find in cod the first evidence for isozymes also within a class III alcohol dehydrogenase. Overall, the results indicate that both of these classes of vertebrate alcohol dehydrogenase are important and suggest a protective metabolic function for the whole enzyme system.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akeson A. On the zinc content of horse liver alcohol dehydrogenase. Biochem Biophys Res Commun. 1964 Oct 14;17(3):211–214. doi: 10.1016/0006-291x(64)90385-7. [DOI] [PubMed] [Google Scholar]
- Baker M. E. A common ancestor for human placental 17 beta-hydroxysteroid dehydrogenase, Streptomyces coelicolor actIII protein, and Drosophila melanogaster alcohol dehydrogenase. FASEB J. 1990 Feb 1;4(2):222–226. doi: 10.1096/fasebj.4.2.2153594. [DOI] [PubMed] [Google Scholar]
- Cederlund E., Peralba J. M., Parés X., Jörnvall H. Amphibian alcohol dehydrogenase, the major frog liver enzyme. Relationships to other forms and assessment of an early gene duplication separating vertebrate class I and class III alcohol dehydrogenases. Biochemistry. 1991 Mar 19;30(11):2811–2816. doi: 10.1021/bi00225a011. [DOI] [PubMed] [Google Scholar]
- Danielsson O., Eklund H., Jörnvall H. The major piscine liver alcohol dehydrogenase has class-mixed properties in relation to mammalian alcohol dehydrogenases of classes I and III. Biochemistry. 1992 Apr 21;31(15):3751–3759. doi: 10.1021/bi00130a004. [DOI] [PubMed] [Google Scholar]
- Eklund H., Horjales E., Vallee B. L., Jörnvall H. Computer-graphics interpretations of residue exchanges between the alpha, beta and gamma subunits of human-liver alcohol dehydrogenase class I isozymes. Eur J Biochem. 1987 Sep 1;167(2):185–193. doi: 10.1111/j.1432-1033.1987.tb13322.x. [DOI] [PubMed] [Google Scholar]
- Eklund H., Müller-Wille P., Horjales E., Futer O., Holmquist B., Vallee B. L., Hög J. O., Kaiser R., Jörnvall H. Comparison of three classes of human liver alcohol dehydrogenase. Emphasis on different substrate binding pockets. Eur J Biochem. 1990 Oct 24;193(2):303–310. doi: 10.1111/j.1432-1033.1990.tb19337.x. [DOI] [PubMed] [Google Scholar]
- Eklund H., Nordström B., Zeppezauer E., Söderlund G., Ohlsson I., Boiwe T., Söderberg B. O., Tapia O., Brändén C. I., Akeson A. Three-dimensional structure of horse liver alcohol dehydrogenase at 2-4 A resolution. J Mol Biol. 1976 Mar 25;102(1):27–59. doi: 10.1016/0022-2836(76)90072-3. [DOI] [PubMed] [Google Scholar]
- Gonzalez F. J., Nebert D. W. Evolution of the P450 gene superfamily: animal-plant 'warfare', molecular drive and human genetic differences in drug oxidation. Trends Genet. 1990 Jun;6(6):182–186. doi: 10.1016/0168-9525(90)90174-5. [DOI] [PubMed] [Google Scholar]
- Gutheil W. G., Holmquist B., Vallee B. L. Purification, characterization, and partial sequence of the glutathione-dependent formaldehyde dehydrogenase from Escherichia coli: a class III alcohol dehydrogenase. Biochemistry. 1992 Jan 21;31(2):475–481. doi: 10.1021/bi00117a025. [DOI] [PubMed] [Google Scholar]
- HARRIS I. STRUCTURE AND CATALYTIC ACTIVITY OF ALCOHOL DEHYDROGENASES. Nature. 1964 Jul 4;203:30–34. doi: 10.1038/203030a0. [DOI] [PubMed] [Google Scholar]
- Jeck R., Woenckhaus C., Harris J. J., Runswick M. J. Identification of the amino acid residue modified in Bacillus stearothermophilus alcohol dehydrogenase by the NAD+ analogue 4-(3-bromoacetylpyridinio)butyldiphosphoadenosine. Eur J Biochem. 1979 Jan 2;93(1):57–64. doi: 10.1111/j.1432-1033.1979.tb12794.x. [DOI] [PubMed] [Google Scholar]
- Jörnvall H. Differences between alcohol dehydrogenases. Structural properties and evolutionary aspects. Eur J Biochem. 1977 Feb;72(3):443–452. doi: 10.1111/j.1432-1033.1977.tb11268.x. [DOI] [PubMed] [Google Scholar]
- Jörnvall H., Eklund H., Brändén C. I. Subunit conformation of yeast alcohol dehydrogenase. J Biol Chem. 1978 Dec 10;253(23):8414–8419. [PubMed] [Google Scholar]
- Jörnvall H. Partial similarities between yeast and liver alcohol dehydrogenases. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2295–2298. doi: 10.1073/pnas.70.8.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörnvall H., Persson M., Jeffery J. Alcohol and polyol dehydrogenases are both divided into two protein types, and structural properties cross-relate the different enzyme activities within each type. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4226–4230. doi: 10.1073/pnas.78.7.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser R., Holmquist B., Vallee B. L., Jörnvall H. Characteristics of mammalian class III alcohol dehydrogenases, an enzyme less variable than the traditional liver enzyme of class I. Biochemistry. 1989 Oct 17;28(21):8432–8438. doi: 10.1021/bi00447a024. [DOI] [PubMed] [Google Scholar]
- Koivusalo M., Baumann M., Uotila L. Evidence for the identity of glutathione-dependent formaldehyde dehydrogenase and class III alcohol dehydrogenase. FEBS Lett. 1989 Oct 23;257(1):105–109. doi: 10.1016/0014-5793(89)81797-1. [DOI] [PubMed] [Google Scholar]
- Krook M., Marekov L., Jörnvall H. Purification and structural characterization of placental NAD(+)-linked 15-hydroxyprostaglandin dehydrogenase. The primary structure reveals the enzyme to belong to the short-chain alcohol dehydrogenase family. Biochemistry. 1990 Jan 23;29(3):738–743. doi: 10.1021/bi00455a021. [DOI] [PubMed] [Google Scholar]
- LI T. K., VALLEE B. L. ACTIVE-CENTER PEPTIDES OF LIVER-ALCOHOL DEHYDROGENASE. I. THE SEQUENCE SURROUNDING THE ACTIVE CYSTEINYL RESIDUES. Biochemistry. 1964 Jun;3:869–873. doi: 10.1021/bi00894a025. [DOI] [PubMed] [Google Scholar]
- Mannervik B., Alin P., Guthenberg C., Jensson H., Tahir M. K., Warholm M., Jörnvall H. Identification of three classes of cytosolic glutathione transferase common to several mammalian species: correlation between structural data and enzymatic properties. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7202–7206. doi: 10.1073/pnas.82.21.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson I., Axiö-Fredriksson U. B., Degerman M., Olsson B. Fast horizontal electrophoresis. I. Isoelectric focusing and polyacrylamide gel electrophoresis using PhastSystem. Electrophoresis. 1988 Jan;9(1):16–22. doi: 10.1002/elps.1150090104. [DOI] [PubMed] [Google Scholar]
- Park D. H., Plapp B. V. Isoenzymes of horse liver alcohol dehydrogenase active on ethanol and steroids. cDNA cloning, expression, and comparison of active sites. J Biol Chem. 1991 Jul 15;266(20):13296–13302. [PubMed] [Google Scholar]
- Persson B., Krook M., Jörnvall H. Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur J Biochem. 1991 Sep 1;200(2):537–543. doi: 10.1111/j.1432-1033.1991.tb16215.x. [DOI] [PubMed] [Google Scholar]
- Schwartz M. F., Jörnvall H. Structural analyses of mutant and wild-type alcohol dehydrogenases from drosophila melanogaster. Eur J Biochem. 1976 Sep;68(1):159–168. doi: 10.1111/j.1432-1033.1976.tb10774.x. [DOI] [PubMed] [Google Scholar]
- Thatcher D. R. The complete amino acid sequence of three alcohol dehydrogenase alleloenzymes (AdhN-11, AdhS and AdhUF) from the fruitfly Drosophila melanogaster. Biochem J. 1980 Jun 1;187(3):875–883. doi: 10.1042/bj1870875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theorell H. Function and structure of liver alcohol dehydrogenase. Harvey Lect. 1967;61:17–41. [PubMed] [Google Scholar]
- VALLEE B. L., HOCH F. L. Zinc in horse liver alcohol dehvdrogenase. J Biol Chem. 1957 Mar;225(1):185–195. [PubMed] [Google Scholar]
- Vallee B. L., Bazzone T. J. Isozymes of human liver alcohol dehydrogenase. Isozymes Curr Top Biol Med Res. 1983;8:219–244. [PubMed] [Google Scholar]