Abstract
Foods rich in resistant starch can help prevent various diseases, including diabetes, colon cancers, diarrhea, and chronic renal and hepatic diseases. Variations in starch biosynthesis enzymes could contribute to the high content of resistant starch in some cultivars of rice (Oryza sativa L.). Our previously published work indicated that the sbe3-rs gene in the rice mutant line, ‘Jiangtangdao1’ was a putative allele of the rice starch branching enzyme gene SBEIIb (previously known as SBE3); sbe3-rs might control the biosynthesis of the high resistant starch content in the rice line. Biomolecular analysis showed that the activity of SBEs was significantly lower in soluble extracts of immature seeds harvested from ‘Jiangtangdao1’ 15 days after flowering than in the extracts of the wild-type rice line ‘Huaqingdao’. We performed gene complementation assays by introducing the wild-type OsSBEIIb into the sbe3-rs mutant ‘Jiangtangdao1’. The genetically complemented lines demonstrated restored seed-related traits. The structures of endosperm amylopectin and the morphological and physicochemical properties of the starch granules in the transformants recovered to wild-type levels. This study provides evidence that sbe3-rs is a novel allele of OsSBEIIb, responsible for biosynthesis of high resistant starch in ‘Jiangtangdao1’.
Keywords: resistant starch, sbe3-rs, rice, starch branching enzyme, complementation assay, Jiangtangdao1
Introduction
Starch is a biologically and commercially important polymer of glucose synthesized and stored in the form of starch grains inside amyloplasts. Most of the starch in cereal products is digested rapidly in the upper gastrointestinal tract and is called “rapidly digested starch” (RDS). However, a proportion of the starch in cereals is resistant to enzymatic hydrolysis and hence escapes degradation in the stomach and the small intestine; this starch is termed “resistant starch” (RS; Englyst et al. 1982). RS is recognized as a significant contributor to gastrointestinal health. It is fermented by microorganisms in a large bowel to produce short chain fatty acids (SCFAs) including acetate, propionate, butyrate, isobutyrate, valerate, and isovalerate, which are thought to promote the optimal function of the viscera (Perera et al. 2010, Topping and Clifton 2001, Wong et al. 2006). RS can be classified into the following four groups: RS1, RS2, RS3, and RS4 (Perera et al. 2010). RS is considered to be beneficial for both prevention and management of diet-related illnesses, such as type-II diabetes, obesity, and colorectal cancer (Wolever 2003). RS has been paid attracting attention from geneticists and breeders for the last two decades. Many high-amylose cultivars of rice (Oryza sativa), maize (Zea mays), and barley (Hordeum vulgare) developed by mutation or biotechnological techniques have been found to contain high RS content (Bird et al. 2008, Hallström et al. 2011, Jiang et al. 2010).
Starch biosynthesis is a complicated process in plants. Three groups of enzymes are known to be involved in the synthesis pathways (Hennen-Bierwagen et al. 2008). There are starch synthases (SSs), which catalyze the formation of linear α-1,4 linked glucan chains, disbranching enzymes (DBEs) and pullulanase, which are important for the formation of semi-crystalline amylopectin (Fujita et al. 2009, Wattebled et al. 2005), and starch branching enzymes (SBEs), which play an essential role in starch biosynthesis, catalyzing chain transfer by cleavage of an α-1,4 linkage following the condensation of an α-1,6 linkage (Tanaka et al. 2004). SBE isoenzymes have been characterized in many important crops, such as potato tuber (Solanum tuberosum; Larsson et al. 1996), rice (Mizuno et al. 1993), maize (Boyer and Preiss 1978), barley (Sun et al. 1997), and wheat (Morell et al. 1997). There are two different classes of SBEs in cereals: SBEI and SBEII (which is further classified into SBEIIa, and SBEIIb). Rice has three SBE isoforms, SBEI, SBEIIa (previously known as SBE4), and SBEIIb (previously known as SBE3). SBEI plays a role in the formation of long chains of amylopectin, producing a range of short and intermediate chains with degree of polymerization (DP) ≤ 40. SBEIIb generates short chains; in vitro experiments with rice enzymes revealed that SBEIIb transferred chains of DP6 and DP7. Additionally, SBEIIa transferred chains of DP6–15, which partially, but not fully, supports the functions of SBEI and IIb (Zhu et al. 2012). Alterations in SBE activity affect the number and size distribution of amylopectin branches. Loss of activity of SBEI in maize endosperm, rice endosperm, or potato tuber did not alter starch content and led to minor differences in amylopectin structure and/or the properties of starch (Blauth et al. 2002, Satoh et al. 2003, Xia et al. 2011). In contrast, loss of SBEII resulted in significant changes, such as an increase in the apparent amylose content (AAC), as observed in several species, including maize (Stinard et al. 1993), potato (Jobling et al. 1999), pea (Bhattacharyya et al. 1990), rice (Mizuno et al. 1993), barley (Regina et al. 2010), and wheat (Regina et al. 2006).
In rice, allelic variations in genes related to starch synthesis affect phenotypic variations in grain quality traits (Mo et al. 2014). Mutations in SBEIIb and the resulting phenotypes have been reported in ‘japonica’ and ‘indica’ backgrounds. For example, an SNP causing a G/C transversion at the downstream end of the SBEIIb gene was significantly associated with multiple trait indices (Lu and Park 2012). In a previous study, we located a putative gene, sbe3-rs, in mutant ‘Jiangtangdao1’, which contained a missense mutation, Leu-599-Pro, in SBEIIb (Yang et al. 2012). This locus explained 60.4% of the RS variation in the F2 population of a cross between ‘Jiangtangdao1’ and ‘Milyang23’.
To further determine whether or not the high RS content in ‘Jiangtangdao1’ was caused by sbe3-rs, in the present study, we conducted a genetic complementation of the sbe3-rs gene in ‘Jiangtangdao1’ with the wild-type copy of SBEIIb using transgenic technology.
Materials and Methods
Plant materials
This study was conducted using the rice line ‘Jiangtangdao1’, which is a high-RS rice mutant from a double haploid (DH) population, derived from ‘Huaqingdao’ by treating its young panicles with 0.015% N-methylnitrosourea (NMU; Yang et al. 2012). The original parent ‘Huaqingdao’ was used as a wild-type control. The seeds of ‘Jiangtangdao1’ and ‘Huaqingdao’ were harvested at 15 days after flowering (DAF), immediately frozen in liquid nitrogen, and stored at −80°C until use for analysis of gene expression and enzyme activity.
Gene expression analysis by RNA-seq and quantitative RT-PCR
For RNA-seq, total RNA was extracted from seeds of ‘Huaqingdao’ and ‘Jiangtagndao1’ 15 DAF using a Quick RNA isolation kit (Biotech Corporation, Beijing, China), according to the manufacturer’s protocol. RNA-seq analysis was performed on an Illumina High-Seq 2500 platform by a commercial service provider (Shanghai Hanyu Bio-Tech, China). Raw sequences in FASTQ format obtained from the Illumina platform were analyzed using freely available tools (http://hannonlab.cshl.edu/fastx_toolkit/). The sequences were mapped to the rice reference genome sequence using several programs. The expression level of each transcript was expressed as the fragments per transcript kilobase per million fragments mapped value, which was calculated based on the number of mapped reads. Log2 transformation was performed on the reads per kilobases per million reads (RPKM) data to facilitate graphical comparison of the expression of the genes (Fernandez-Aparicio et al. 2013). Quantitative real-time PCR (qRT-PCR) was performed in a QuantStudio™ 6 Flex Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) using 100 ng of cDNA template for amplification. The primers using for the branching enzyme were SBEIIb-F (5′-GACGACTTGCTGTCCTCT-3′) and SBEIIb-R (5′-CAACCTTTATGCTTGCTT-3′). Rice Actin1 cDNA, amplified with the primers ActinF (5′-CATCGTTCTCAGTGGTGG-3′) and ActinR (5′-CATCTGCTGGAATGTGCT-3′), was used as a reference. Real-time PCR amplification was conducted using SYBR Green I PCR Master Mix Kit (TaKaRa). Relative expression levels were calculated using the ΔΔCT method (Livak and Schmittgen 2001).
Determination of starch branching enzyme activities
Starch branching enzyme (SBE) activity was quantified as described in a previous report (Cheng et al. 2001). The panicles were de-hulled manually, and the fresh weight of 20 grains was recorded. The enzymes were extracted and analyzed according to the methods described by Cheng et al. (2001). The optical density of the final product was measured at 540 nm. The experiments were performed on ice in triplicate.
Cloning and sequencing of the SBEIIb gene and its promoter
Total RNA was isolated from seeds (15 DAF) of ‘Huaqingdao’ using an RNA extraction kit (SK8661; Sangon Biotech, Shanghai, China) according to the manufacturer’s instructions. First-strand cDNA synthesis of rice genes was carried out using a cDNA synthesis kit (TaKaRa, Japan). The complete coding sequence of SBEIIb was amplified from these cDNAs using gene-specific primers containing SalI and XbaI restriction sites (upstream primer: 5′-GATCGTCGACTGAGGAGGGTTTAGGTGGAAG-3′; downstream primer: 5′-GTACTCTAGAGCCTCTTGGTGTTCTCATTCC-3′). The amplified cDNA products were cloned into the pMD18-T vector (TaKaRa). The resulting plasmid, pMD18-SBEIIb, containing a 2563-bp cDNA fragment including the 2478-bp SBEIIb coding region, was verified by sequencing. The 2.254-Kb SBEIIb promoter was obtained via polymerase chain reaction (PCR) using the following primers containing HindIII and SalI sites: 5′-GATCAAGCTTCAGCAAGTGACGGTGTTCG-3′ and 5′-GTACGTCGACTTCCACCTAAACCCTCCTCA-3′. Thereafter, the promoter was cloned into a pMD18-T vector (TaKaRa) to create the pMD18-T-SBEP plasmid. The resulting plasmid was sequenced and analyzed using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). The correct promoter was digested with HindIII and SalI and ligated into an expression plasmid, p1300nos (constructed by inserting a Nos terminator in the multiple cloning site of pCAMBIA1300) to create p1300-SBEp-nos. Finally, the plasmid pMD18-SBEIIb was digested with SalI and XbaI and ligated into p1300-SBEp-nos to create the binary vector p1300-SBEp-SBE-nos.
Transformation of the high-RS mutant, ‘Jiangtangdao1’, and screening of transgenic lines
The procedures for rice tissue culture and transformation with Agrobacterium tumefaciens were as described by Yang et al. (2011). The plasmid p1300-SBEp-SBE-nos was introduced into the A. tumefaciens strain EHA105, which was used to transform the rice embryogenic callus cultured from ‘Jiangtangdao1’. The weight of the T1 generation seeds from three T0 lines (No. 1, 19, and 20) and ‘Huaqingdao’ was examined; seed yield-related traits of the transgenic line were evaluated for 10 plants of each line. The host ‘Jiangtangdao1’ and the wild-type ‘Huaqingdao’ plants were used as controls.
DNA extraction and transgene copy number analysis by quantitative real-time PCR
The plant genomic DNA samples used for qPCR were extracted and purified using a genomic DNA extraction kit (Axygen, Union City, CA, USA). Genomic DNA was quantified by determining the absorbance at 260/280 nm and stored at −20°C until further analysis. The oligonucleotide primers were designed using the Primer Premier 5.0 software (Premier Biosoft, Palo Alto, CA, USA). All primers were synthesized by Shanghai Sangon Biotech (China). The primers for the endogenous gene SPS were: SPSF (5′-TTGCGCCTGAACGGATAT-3′) and SPSR (5′-CGGTTGATCTTTTCGGGATG-3′). The primers for target gene HPT were HPTF (5′-AACTCCCCAATGTCAAGC-3′) and HPTR (5′-GTCCTGCGGGTAAATAGC-3′). qPCR was performed in a QuantStudio™ 6 Flex Real-Time PCR system (Applied Biosystems). The reaction mixtures containing 2 μL of DNA and 18 μL of PCR master mixture were transferred to 96-well plates for amplification. The PCR master mixture contained 10 μL of 2X TaKaRa SYBR Premix Ex Taq™ (RR041, TaKaRa, Tokyo, Japan), 0.4 μL of ROX Reference Dye (50X), 0.4 μL of each primer (10 mM), and 6.8 μL of ddH2O. Reactions were performed at 95°C for 1 min (1 cycle), followed by 95°C for 15 s and 60°C for 30 s (40 cycles). Each reaction was performed using three replicates and was repeated three times. The SPS gene, which is a single copy gene in the rice genome, was used as the endogenous reference gene. We used a real-time quantitative PCR method to calculate the HPT copy number according to the methods of Yang et al. (2005).
Measurement of apparent amylose content and RS content
RS content was measured using the Megazyme RS assay kit (Megazyme, Co. Wicklow, Ireland) according to manufacturer’s instructions; this kit has been widely used for RS determination in crops (McCleary et al. 2002). The apparent amylose content (AAC) of starch was measured using a simplified I2/KI assay as described by Yang et al. (2006). Four standard samples, each with a different AAC (1.2%, 11.2%, 16.8% and 26.8%), were procured from the China National Rice Research Institute (CNRRI, China). The experiments were performed in triplicate.
Scanning electron microscopy (SEM)
Brown (unpolished) rice was broken manually and cut into sections approximately 2 mm thin. After coating with Pt ions in an argon atmosphere for 25 min (using an IB-5 Ion coater, Eiko Engineering, Ibaraki, Japan), the cross-sections were adhered to a double-adhesive tape that was fixed to a metallic stud. Starch granules in the sections were visualized using an electron microscope (FEI NovaNano SEM430, FEI Co., Hislsboro, OR, USA).
Determination of the chain length distribution of disbranched α-glucans
The chain length distribution of α-glucans in the transformants, ‘Jiangtangdao1’ and ‘Huaqingdao’, was determined according to the capillary electrophoresis method (O’Shea and Morell 1996). The measurements were conducted using a BioLC system (model DX-500, Dionex, Sunnyvale, CA, USA).
X-ray diffraction analysis of starch
These procedures were performed according to the methods described by Sawada et al. (2009).
Results
Gene expression analyses and detection of branching enzyme activity
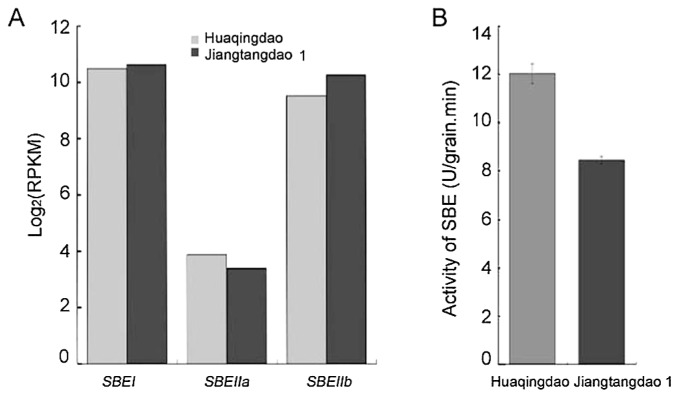
Our previously published work indicated that sbe3-rs might control the biosynthesis of RS and its accumulation to high levels in the ‘Jiangtangdao1’ rice line. To assess the expression of SBE genes (SBEI, SBEIIa, and SBEIIb) in immature rice seeds of the wild type and the mutant, RNA-seq and qPCR analyses were performed. The expression level of each transcript was denoted in terms of the FPKM value. The RPKM values for the three genes are presented in Fig. 1A. The RNA-seq results revealed that the expression levels of the three SBE isoforms in ‘Jiangtangdao1’ were not significantly different from those in ‘Huaqingdao’. SBEIIb transcript levels in ‘Jiangtangdao1’ and control plants were checked by qRT-PCR. The results revealed that the level of SBEIIb in the immature seeds of the wild-type ‘Huaqingdao’ was 1.6 times higher than in ‘Jiangtangdao1’ seeds. There were no significant differences in the SBEIIb expression levels between the mutant and the wild type.
Fig. 1.
Expression analyses and activity detection of starch branching enzyme (SBE). A. Transcriptional levels of three SBE isoforms determined as RPKM values based on RNA-seq analysis and presented as log2. B. Activities of SBE in immature seeds of ‘Jiangtangdao1’ and ‘Huaqingdao’.
The activity of SBEs was measured in the soluble extracts of immature seeds harvested 15 days after flowering (Fig. 1B). The activity of SBEI, SBEIIa, and SBEIIb was evaluated. The specific activity in the mutant ‘Jiangtangdao1’ was approximately 70% of that in the wild-type ‘Huaqingdao’. This result was similar to that of amylose-extender mutants. The activity of the branching enzyme in the rice amylose-extender mutants was approximately 60–90% of that in the wild-type plants (Mizuno et al. 1993). Thus, these results indicated that the high content of RS in ‘Jiangtangdao1’ likely to have resulted from reduced activity of SBEIIb and was not a consequence of the differential expression of the gene.
Generation of transgenic plants with a single copy number of the transgene
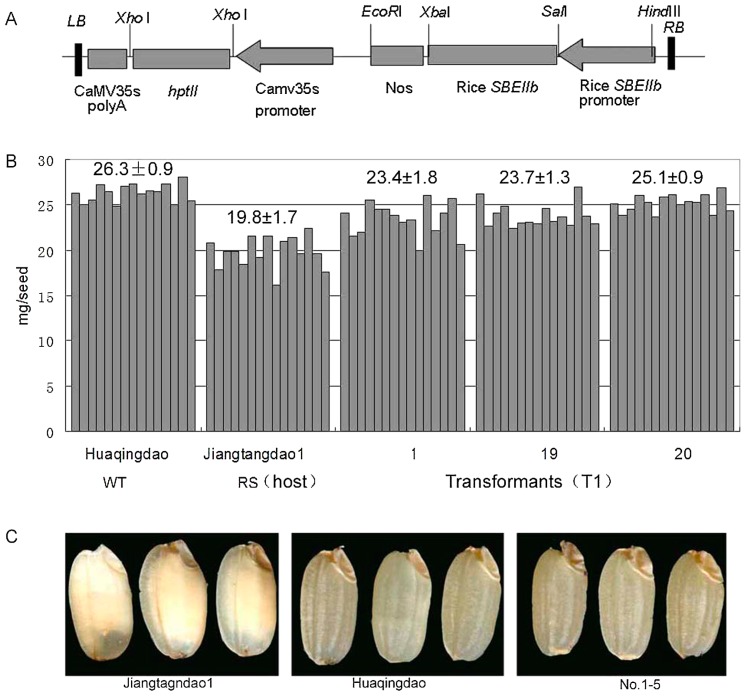
To complement sbe3-rs in ‘Jiangtangdao1’, a native SBEIIb promoter-driven SBEIIb gene expression cassette was constructed (Fig. 2A). The wild-type SBEIIb coding sequence and SBEIIb promoter of ‘Huaqingdao’ were amplified by PCR and inserted into a binary plasmid. Transgenic rice plants were produced by introducing the construct into ‘Jiangtangdao1’. In total, 50 independent T0 transgenic lines were obtained and grown in a glasshouse maintained at 28°C during the day (12 h) and 24°C during the night (12 h). We employed a simple and effective quantitative method to estimate the transgene copy number in the transgenic rice. In this approach, the copy number was determined relative to an endogenous reference gene, SPS, which is a single-copy gene in rice (Ding et al. 2004, Yang et al. 2005). The analysis revealed the presence of a single transgene copy in 21 independent T0 lines. Of these, three independent T0 complemented lines (T0-1, 19, and 20) were chosen for further examination.
Fig. 2.
Transformation of high-RS mutant rice line ‘Jiangtangdao1’ with rice wild-type SBEIIb gene. A. Construct with the OsSBEIIb gene used for transformation of ‘Jiangtagndao1’. The clone contained the promoter region up to 2.3 kb upstream from the transcription start site and the transcribed region for the OsSBEIIb gene. B. Weight of individual mature T1 kernels of the three transformed rice lines No-1, 19, and 20, the host mutant line ‘Jiangtangdao1’ and the wild-type cultivar ‘Huaqingdao’. The numbers above the bars indicate the average values (mg) ± SD (mg) (n = 15). C. Endosperm of ‘Jiangtangdao1’, ‘Huaqingdao’ and T3 seeds of the transformed line No. 1–5, respectively.
OsSBEIIb complemented sbe3-rs phenotypes in Jiangtangdao1
We examined the phenotypic changes in three individual T0 lines. The dry weight of dehulled seeds of ‘Jiangtangdao1’ was significantly lower (19.8 ± 1.7 mg) than that of the wild-type cultivar, ‘Huaqingdao’ (26.3 ± 0.9 mg). The dry weight of mature T1 seeds of the three transgenic lines (T0-1, 19, and 20) was significantly higher than that of the T1 seeds of ‘Jiangtangdao1’ (Fig. 2B). Twelve randomly chosen T1 seeds of T0-1 plants were grown in the greenhouse, and T2 seeds obtained from them were further examined for their phenotypes. The homozygous phenotype was inherited in the seeds of plant No. 1–5 in the following generation; biochemical and morphological analyses were performed using the maturing T3 seeds of these plants. The unpolished grains of the high-RS mutant, ‘Jiangtangdao1’, had an almost opaque endosperm because of increased chalkiness (Fig. 2C). In contrast, the grains of ‘Huaqingdao’ were quite transparent (Fig. 2C). However, all the seeds of the No. 1–5 transgenic plants reverted to the transparent endosperm phenotype, which was similar to the wild type (Fig. 3C).
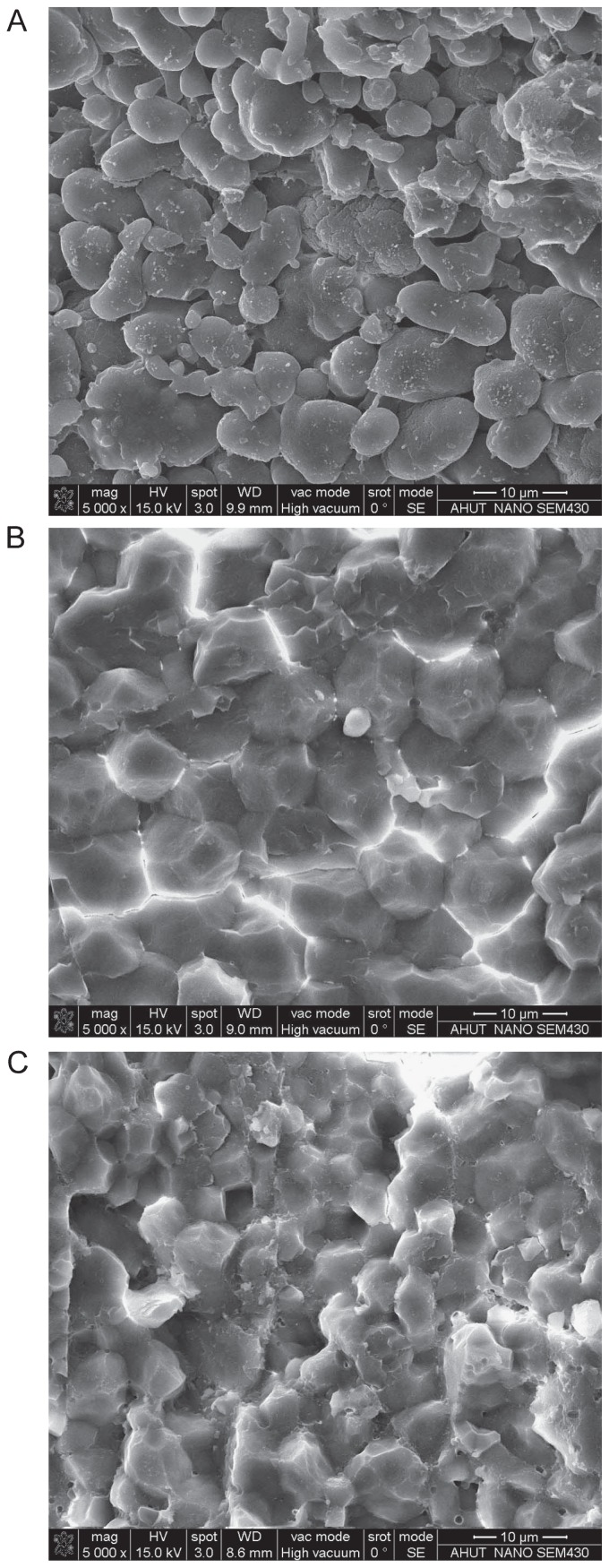
Fig. 3.
Scanning electron micrographs of starch granules in endosperm. A. ‘Jiangtangdao1’. B. ‘Huaqingdao’. C. Transgenic lines No. 1–5.
The content of amylose and RS in rice grains was measured. ‘Jiangtangdao1’ had high RS (11.67 ± 0.43%) and amylose (31.1 ± 0.15%) content, whereas ‘Huaqingdao’ had low RS (0.38 ± 0.12%) and amylose (16.27 ± 0.21%) content. The RS (1.21 ± 0.14%) and amylose contents (19.19 ± 0.19%) in the complemented lines (No. 1–5) were significantly lower than those in ‘Jiangtangdao1’ and almost equal to those in the wild type.
The effects of the SBEIIb gene on several traits related to seed yield were further evaluated in plants of the T3 generation obtained from the complemented lines 1–5. No marked changes were observed in the number of panicles per plant, number of grains per panicle, or rate of seed-set. However, the thousand-seed weight was significantly increased in the transgenic plants (Table 1).
Table 1.
Traits related to seed yield in transformants, host ‘Jiangtangdao1’, and the control ‘Huaqingdao’ (Shanghai, China, 2015)
| Line | Panicle length (cm) | Panicles per plant | Grains per panicle | Seed-setting rate (%) | Thousand-seed weight (g) |
|---|---|---|---|---|---|
| Huaqingdao (CK) | 16.4 ± 1.3 | 11.2 ± 1.2 | 121.6 ± 23.2 | 94.2 ± 2.5 | 24.4 ± 1.1 |
| Jiangtangdao1 | 16.7 ± 1.8 | 12.3 ± 0.9 | 112.1 ± 19.3 | 91.1 ± 2.9 | 19.9 ± 2.1* |
| No. 1–5 | 16.2 ± 1.2 | 10.9 ± 1.1 | 115.1 ± 20.2 | 92.7 ± 2.4 | 22.3 ± 1.8 |
Note: 10 plants from the transformed and the control lines were evaluated and the measurements were taken when the plants were mature.
Significant at the 0.05 probability level.
Recovery of morphological properties of the starch granules in the complemented lines
As the endosperm chalkiness was higher in ‘Jiangtangdao1’, we speculated that the morphological properties of the starch granules in the endosperm could have changed. To test the morphological differences of starch granules among ‘Jiangtangdao1’ plants, the transformants, and the wild-type ‘Huaqingdao’ plants, we used SEM imaging (Fig. 3). The starch granules of the high-RS mutant ‘Jiangtangdao1’ were large, non-angular, and rounded; some were elongated, whereas others were small and polygonal (Fig. 3A). Starch granules in the wild-type ‘Huaqingdao’ mainly consisted of similar-sized polygonal granules with sharp angles and edges. The granules of ‘Jiangtangdao1’ were much more variable in size than the polygonal granules of the wild-type ‘Huaqingdao’. However, the shape of starch granules of OsSBEIIb transformants approached those of the wild-type plants, except for the size of the starch granules, which were slightly smaller than the granules in the wild type (Fig. 3B, 3C). This result demonstrated the chalky properties of sbe3-rs mutant were fully complemented by the introduction of wild-type copy of OsSBEIIb.
Distribution of chain length in amylopectin and X-ray diffraction pattern of the starch granules in the ‘Jiangtangdao1’ transformants
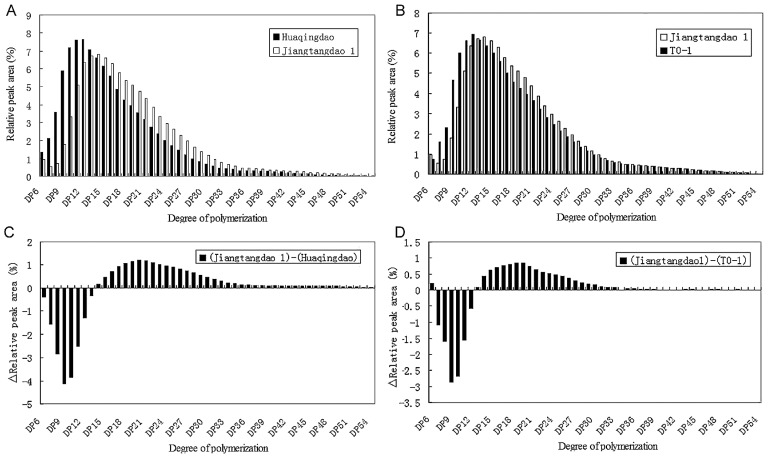
As SBEIIb played a specific role in the formation of branches in the crystalline lamellae of the amylopectin cluster in rice endosperm, it affected the distribution of chain length in amylopectin (Nishi et al. 2001). Fig. 4 shows that in the high-RS-containing ‘Jiangtangdao1’ rice, the proportion of short chains (DP ≤ 13) was apparently lower than that in the wild-type ‘Huaqingdao’. In contrast, the proportion of longer chains in the cluster linking of long chains (DP ≥ 15) was increased considerably (Fig. 4A). The amylopectin chain length distribution profile of OsSBEIIb transformants was very similar to that of the wild type (Fig. 4B).
Fig. 4.
Comparison of chain length distribution of amylopectin in mature endosperm in the transformed rice, its high-RS-containing host ‘Jiangtangdao1’, and the wild-type cultivar ‘Huaqingdao’.
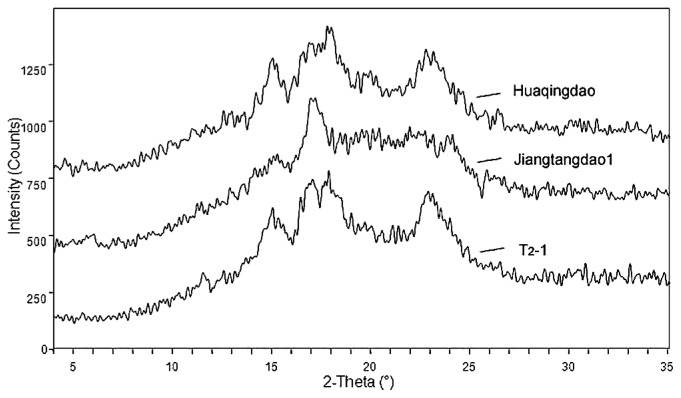
As revealed by X-ray powder diffraction, the starch granule crystals exhibited three different structures designated as A-, B-, and C-type allomorphs. The C-type allomorph was a mixture of both A- and B-type allomorphs. Previous reports have indicated that the starch granules of rice amylose-extender mutants have a B-type X-ray diffraction pattern, whereas those of the wild-type show a typical A-type pattern (Nishi et al. 2001). As shown in Fig. 5, the endosperm starch of ‘Huaqingdao’ had the A-type X-ray diffraction pattern, in contrast to the B-type diffraction pattern observed in the mutant ‘Jiangtangdao1’. The typical B-type diffraction pattern of the starch granules in ‘Jiangtangdao1’ was changed to a typical A-type pattern in the transformants.
Fig. 5.
X-ray diffraction patterns of purified starch granules in the mature endosperm of the rice transformant, its mutant host ‘Jiangtangdao1’, and the wild-type cultivar ‘Huaqingdao’.
All of these results indicated that OsSBEIIb could complement the sbe3-rs in the high-RS-content line, ‘Jiangtangdao1’.
Discussion
In cereals, SBEIIb plays a specific role in the formation of short chains within the amylopectin cluster during starch synthesis (Nishi et al. 2001). It should be noted that in this study, the effect of SBEIIb on the structure of amylopectin was entirely dependent on its activity. Many researchers have reported that down-regulation of SBEII could lead to high amylose content in mutant maize, wheat, barley, and rice (Butardo et al. 2011, Wei et al. 2010, Zhao et al. 2015). The inactivation of SBEIIb in rice is traditionally associated with elevated AAC, increased gelatinization temperature, and a decreased proportion of short amylopectin branches (Butardo et al. 2011, Nishi et al. 2001). In general, the RS content of granular starch is positively correlated with the level of amylose (Sang et al. 2008). High RS always results from high amylose content (Perera et al. 2010). The mutant ‘Jiangtangdao1’ was identified from a double haploid (DH) population derived from ‘Huaqingdao’. The RS content was significantly increased from 0.38 ± 0.12% in the wild-type ‘Huaqingdao’ to 11.67 ± 0.43% in ‘Jiangtangdao1’. Using an F2 population, we previously mapped sbe3-rs to SBEIIb, a recessive allele for high RS in ‘Jiangtangdao1’ (Yang et al. 2012). Previous studies showed that the activity of SBE was highest from 13 DAF to 15 DAF (Cheng et al. 2001, Shu et al. 2014). Therefore, in the present study, gene expression in the mutant ‘Jiangtangdao1’ and the wild-type ‘Huaqingdao’ was analyzed 13–15 DAF by RNA-seq. No differential expression of the three rice SBE isoforms (SBEI, SBEIIa, and SBEIIb) was observed. The nucleotide sequence is the basis of gene expression and gene function, and single-nucleotide polymorphism (SNP) and insertions or deletions in genes related to amylopectin structure among different rice varieties have been reported. In a previous study, a G-A SNP was identified in a high-AAC rice mutant, IR36ae, which resulted in an improper stop codon that could potentially lead to the formation of a truncated SBEIIb (Butardo et al. 2012). Although the structure of starch and the resulting functional properties of ‘Jiangtangdao1’ were consistent with those of IR36ae, a T-C mutation was found in ‘Jiangtangdao1’ (Yang et al. 2012) that did not affect the expression of SBEIIb. This indicated that the mechanism of high RS and AAC accumulation in ‘Jiangtangdao1’ was different from that previously reported for IR36ae.
High-amylose mutants (amylose-extender) of rice are characterized by higher AAC, decreased levels of short chains of amylopectin, higher gelatinization temperature, B-type X-ray diffraction, and a particular morphology of starch granules (Yano et al. 1985). ‘Jiangtangdao1’ was similar to rice amylose-extender mutants with high AAC and B-type X-ray diffraction but for its high RS content. An earlier study demonstrated that branching enzyme activity was significantly decreased in the amylose-extender mutants (Mizuno et al. 1993, Nishi et al. 2001). Our results were consistent with these previously reported findings (Fig. 1B). The level of the specific activity in the mutant ‘Jiangtangdao1’ was approximately 70% of that in the wild-type ‘Huaqingdao’, which indicated that the mutation in SBEIIb (leading to the replacement of Leu with Pro) might have affected the SBE activity, thereby resulting in the high AAC and RS content in ‘Jiangtangdao1’.
We further performed functional complementation of the sbe3-rs gene in ‘Jiangtangdao1’ with wild-type SBEIIb. Wild-type rice SBEIIb was introduced into the high-RS mutant ‘Jiangtangdao1’. The introduction of the gene had no physiological effects on the vegetative tissues of the transgenic plants because SBEIIb was specifically expressed in the endosperm of rice and its expression was driven by its own promoter. When SBEIIb was expressed in the ‘Jiangtangdao1’ mutant, the RS content of ‘Jiangtangdao1’ (11.67%) was reduced to 1.21%, thus indicating that sbe3-rs was an allele of SBEIIb. The ae mutants in cereals are often associated with slight reductions in grain weight; this was also observed in ‘Jiangtangdao1’ (Fig. 2). The dry weight of grain in ‘Jiangtangdao1’ was significantly lower (19.8 ± 1.7 mg) than in the wild-type ‘Huaqingdao’ (26.3 ± 0.9 mg). However, the seed weight in the transformants was similar to that in the wild type. The structure of amylopectin from the mutant Jiangtangdao1 was clearly distinct from that of the amylopectin of the wild-type plants. In ‘Jiangtangdao1’, the proportion of short chains with DP ≤ 13 was markedly lower and the proportion of long chains of DP = 13–30 and DP ≥ 40 was higher than that observed in the wild type (Fig. 4); this was consistent with previous observations (Butardo et al. 2011, Nishi et al. 2001). The fine structure of amylopectin in the transformants of ‘Jiangtangdao1’ exhibited great variations (Figs. 4, 5).
In conclusion, the present study indicated that the variation in SBEIIb enzyme activity probably caused the different phenotypes of ‘Jiangtangdao1’ and that the wild-type rice SBEIIb gene could complement the mutant sbe3-rs gene in Jiangtangdao1. The high RS content in ‘Jiangtagndao1’ was, in fact, caused by a missense mutation that changed Leu599 to Pro599. Further molecular genetic and biochemical analyses of this mutation are required to fully elucidate the exact mechanism of high RS accumulation in Jiangtangdao1.
Acknowledgments
This work was supported by Key Basic Research Project in Shanghai (13JC1408600), Shanghai Natural Science Foundation (13ZR1460800), Shanghai Rising-Star Program (16QB1402500), The Youth Talent Development Plan of Shanghai Municipal Agricultural System, China (Grant No. 201501 and 201510) and International Cooperation Project of China and South Korea (PJ01125703).
Literature Cited
- Bhattacharyya, M.K., Smith, A.M., Ellis, T.H., Hedley, C. and Martin, C. (1990) The wrinkled-seed character of pea described by Mendel is caused by a transposon-like insertion in a gene encoding starch-branching enzyme. Cell 60: 115–122. [DOI] [PubMed] [Google Scholar]
- Bird, A.R., Vuaran, M.S., King, R.A., Noakes, M., Keogh, J., Morell, M.K. and Topping, D.L. (2008) Wholegrain foods made from a novel high-amylose barley variety (Himalaya 292) improve indices of bowel health in human subjects. Br. J. Nutr. 99: 1032–1040. [DOI] [PubMed] [Google Scholar]
- Blauth, S.L., Kim, K.N., Klucinec, J., Shannon, J.C., Thompson, D. and Guiltinan, M. (2002) Identification of Mutator insertional mutants of starch-branching enzyme 1 (sbe1) in Zea mays L. Plant Mol. Biol. 48: 287–297. [DOI] [PubMed] [Google Scholar]
- Boyer, C.D. and Preiss, J. (1978) Mutiple forms of (1→4)-α-d-gulcan, (1→4)-α-d-gulcan-6-glycosyl transferase from developing zea mays L. kernels. Carbohydr. Res. 61: 321–334. [Google Scholar]
- Butardo, V.M., Fitzgerald, M.A., Bird, A.R., Gidley, M.J., Flanagan, B.M., Larroque, O., Resurreccion, A.P., Laidlaw, H.K., Jobling, S.A., Morell, M.K.et al. (2011) Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA- and hairpin RNA-mediated RNA silencing. J. Exp. Bot. 62: 4927–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butardo, V.M.Jr., Daygon, V.D., Colgrave, M.L., Campbell, P.M., Resurreccion, A., Cuevas, R.P., Jobling, S.A., Tetlow, I., Rahman, S., Morell, M.et al. (2012) Biomolecular analyses of starch and starch granule proteins in the high-amylose rice mutant Goami 2. J. Agric. Food Chem. 60: 11576–11585. [DOI] [PubMed] [Google Scholar]
- Cheng, F.M., Jiang, D.A., Wu, P. and Shi, C.H. (2001) The dynamic change of starch synthesis enzymes during the grain filling stage and effects of temperature upon it. Acta Agron. Sin. 27: 201–206. [Google Scholar]
- Ding, J., Jia, J., Yang, L., Wen, H., Zhang, C., Liu, W. and Zhang, D. (2004) Validation of a rice specific gene, sucrose phosphate synthase, used as the endogenous reference gene for qualitative and real-time quantitative PCR detection of transgenes. J. Agric. Food Chem. 52: 3372–3377. [DOI] [PubMed] [Google Scholar]
- Englyst, H., Wiggins, H.S. and Cummings, J.H. (1982) Determination of the non-starch polysaccharides in plant foods by gas-liquid chromatography of constituent sugars as alditol acetates. Analyst 107: 307–318. [DOI] [PubMed] [Google Scholar]
- Fernandez-Aparicio, M., Huang, K., Wafula, E.K., Honaas, L.A., Wickett, N.J., Timko, M.P., Depamphilis, C.W., Yoder, J.I. and Westwood, J.H. (2013) Application of qRT-PCR and RNA-Seq analysis for the identification of housekeeping genes useful for normalization of gene expression values during Striga hermonthica development. Mol. Biol. Rep. 40: 3395–3407. [DOI] [PubMed] [Google Scholar]
- Fujita, N., Toyosawa, Y., Utsumi, Y., Higuchi, T., Hanashiro, I., Ikegami, A., Akuzawa, S., Yoshida, M., Mori, A., Inomata, K.et al. (2009) Characterization of pullulanase (PUL)-deficient mutants of rice (Oryza sativa L.) and the function of PUL on starch biosynthesis in the developing rice endosperm. J. Exp. Bot. 60: 1009–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallström, E., Sestili, F., Lafiandra, D., Björck, I. and Östman, E. (2011) A novel wheat variety with elevated content of amylose increases resistant starch formation and may beneficially influence glycaemia in healthy subjects. Food Nutr. Res. 55 10.3402/fnr.v55i0.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennen-Bierwagen, T.A., Liu, F., Marsh, R.S., Kim, S., Gan, Q., Tetlow, I.J., Emes, M.J., James, M.G. and Myers, A.M. (2008) Starch biosynthetic enzymes from developing maize endosperm associate in multisubunit complexes. Plant Physiol. 146: 1892–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, H., Lio, J., Blanco, M., Campbell, M. and Jane, J.L. (2010) Resistant-starch formation in high-amylose maize starch during Kernel development. J. Agric. Food Chem. 58: 8043–8047. [DOI] [PubMed] [Google Scholar]
- Jobling, S.A., Schwall, G.P., Westcott, R.J., Sidebottom, C.M., Debet, M., Gidley, M.J., Jeffcoat, R. and Safford, R. (1999) A minor form of starch branching enzyme in potato (Solanum tuberosum L.) tubers has a major effect on starch structure: cloning and characterisation of multiple forms of SBE A. Plant J. 18: 163–171. [DOI] [PubMed] [Google Scholar]
- Larsson, C.T., Hofvander, P., Khoshnoodi, J., Ek, B., Rask, L. and Larsson, H. (1996) Three isoforms of starch synthase and two isoforms of branching enzyme are present in potato tuber starch. Plant Sci. 117: 9–16. [Google Scholar]
- Lu, F.H. and Park, Y.J. (2012) An SNP downstream of the OsBEIIb gene is significantly associated with amylose content and viscosity properties in rice (Oryza sativa L.). J. Cereal Sci. 56: 706–712. [DOI] [PubMed] [Google Scholar]
- McCleary, B.V., McNally, M. and Rossiter, P. (2002) Measurement of resistant starch by enzymatic digestion in starch and selected plant materials: collaborative study. J AOAC Int. 85: 1103–1111. [PubMed] [Google Scholar]
- Mizuno, K., Kawasaki, T., Shimada, H., Satoh, H., Kobayashi, E., Okumura, S., Arai, Y. and Baba, T. (1993) Alteration of the structural properties of starch components by the lack of an isoform of starch branching enzyme in rice seeds. J. Biol. Chem. 268: 19084–19091. [PubMed] [Google Scholar]
- Mo, Y.J., Jeung, J.U., Shin, W.C., Kim, K.Y., Ye, C., Redona, E.D. and Kim, B.K. (2014) Effects of allelic variations in starch synthesis-related genes on grain quality traits of Korean nonglutinous rice varieties under different temperature conditions. Breed. Sci. 64: 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell, M.K., Blennow, A., Kosar-Hashemi, B. and Samuel, M.S. (1997) Differential expression and properties of starch branching enzyme isoforms in developing wheat endosperm. Plant Physiol. 113: 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi, A., Nakamura, Y., Tanaka, N. and Satoh, H. (2001) Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol. 127: 459–472. [PMC free article] [PubMed] [Google Scholar]
- O’Shea, M.G. and Morell, M.K. (1996) High resolution slab gel electrophoresis of 8-amino-1,3,6-pyrenetrisulfonic acid (APTS) tagged oligosaccharides using a DNA sequencer. Electrophoresis 17: 681–686. [DOI] [PubMed] [Google Scholar]
- Perera, A., Meda, V. and Tyler, R.T. (2010) Resistant starch: A review of analytical protocols for determining resistant starch and of factors affecting the resistant starch content of foods. Food Res. Int. 43: 1959–1974. [Google Scholar]
- Regina, A., Bird, A., Topping, D., Bowden, S., Freeman, J., Barsby, T., Kosar-Hashemi, B., Li, Z., Rahman, S. and Morell, M. (2006) High-amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proc. Natl. Acad. Sci. USA 103: 3546–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regina, A., Kosar-Hashemi, B., Ling, S., Li, Z., Rahman, S. and Morell, M. (2010) Control of starch branching in barley defined through differential RNAi suppression of starch branching enzyme IIa and IIb. J. Exp. Bot. 61: 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang, Y.H., Bean, S., Seib, P.A., Pedersen, J. and Shi, Y.C. (2008) Structure and functional properties of sorghum starches differing in amylose content. J. Agric. Food Chem. 56: 6680–6685. [DOI] [PubMed] [Google Scholar]
- Satoh, H., Nishi, A., Yamashita, K., Takemoto, Y., Tanaka, Y., Hosaka, Y., Sakurai, A., Fujita, N. and Nakamura, Y. (2003) Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol. 133: 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada, T., Francisco, P.B.Jr, Aihara, S., Utsumi, Y., Yoshida, M., Oyama, Y., Tsuzuki, M., Satoh, H. and Nakamura, Y. (2009) Chlorella starch branching enzyme II (BEII) can complement the function of BEIIb in rice endosperm. Plant Cell Physiol. 50: 1062–1074. [DOI] [PubMed] [Google Scholar]
- Shu, X., Sun, J. and Wu, D. (2014) Effects of grain development on formation of resistant starch in rice. Food Chem. 164: 89–97. [DOI] [PubMed] [Google Scholar]
- Stinard, P.S., Robertson, D.S. and Schnable, P.S. (1993) Genetic isolation, cloning, and analysis of a mutator-induced, dominant antimorph of the maize amylose extender1 locus. Plant Cell 5: 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, C., Sathish, P., Ahlandsberg, S., Deiber, A. and Jansson, C. (1997) Identification of four starch-branching enzymes in barley endosperm: partial purification of forms I, IIa and IIb. New Phytol. 137: 215–222. [DOI] [PubMed] [Google Scholar]
- Tanaka, N., Fujita, N., Nishi, A., Satoh, H., Hosaka, Y., Ugaki, M., Kawasaki, S. and Nakamura, Y. (2004) The structure of starch can be manipulated by changing the expression levels of starch branching enzyme IIb in rice endosperm. Plant Biotechnol. J. 2: 507–516. [DOI] [PubMed] [Google Scholar]
- Topping, D.L. and Clifton, P.M. (2001) Short chain fatty acids and human colonic function-roles of resistant starch and non-starch polysaccharides. Physiol. Rev. 81: 1031–1064. [DOI] [PubMed] [Google Scholar]
- Wattebled, F., Dong, Y., Dumez, S., Delvalle, D., Planchot, V., Berbezy, P., Vyas, D., Colonna, P., Chatterjee, M., Ball, S.et al. (2005) Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol. 138: 184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, C., Qin, F., Zhou, W., Yu, H., Xu, B., Chen, C., Zhu, L., Wang, Y., Gu, M. and Liu, Q. (2010) Granule structure and distribution of allomorphs in C-type high-amylose rice starch granule modified by antisense RNA inhibition of starch branching enzyme. J. Agric. Food Chem. 58: 11946–11954. [DOI] [PubMed] [Google Scholar]
- Wolever, T.M. (2003) Carbohydrate and the regulation of blood glucose and metabolism. Nutr. Rev. 61: S40–48. [DOI] [PubMed] [Google Scholar]
- Wong, J.M., de Souza, R., Kendall, C.W., Emam, A. and Jenkins, D.J. (2006) Colonic health: fermentation and short chain fatty acids. J. Clin. Gastroenterol. 40: 235–243. [DOI] [PubMed] [Google Scholar]
- Xia, H., Yandeau-Nelson, M., Thompson, D.B. and Guiltinan, M.J. (2011) Deficiency of maize starch-branching enzyme I results in altered starch fine structure, decreased digestibility and reduced coleoptile growth during germination. BMC Plant Biol. 11: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.Z., Shu, X.L., Zhang, L.L., Wang, X.Y., Zhao, H.J., Ma, C.X. and Wu, D.X. (2006) Starch properties of mutant rice high in resistant starch. J. Agric. Food Chem. 54: 523–528. [DOI] [PubMed] [Google Scholar]
- Yang, L., Ding, J., Zhang, C., Jia, J., Weng, H., Liu, W. and Zhang, D. (2005) Estimating the copy number of transgenes in transformed rice by real-time quantitative PCR. Plant Cell Rep. 23: 759–763. [DOI] [PubMed] [Google Scholar]
- Yang, R., Tang, Q., Wang, H., Zhang, X., Pan, G., Wang, H. and Tu, J. (2011) Analyses of two rice (Oryza sativa) cyclin-dependent kinase inhibitors and effects of transgenic expression of OsiICK6 on plant growth and development. Ann. Bot. 107: 1087–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, R.F., Sun, C.L., Bai, J.J., Luo, Z.X., Shi, B., Zhang, J.M., Yan, W.G. and Piao, Z.Z. (2012) A putative gene sbe3-rs for resistant starch mutated from SBE3 for starch branching enzyme in rice (Oryza sativa L.). PLoS ONE 7: e43026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, M., Okuno, K., Kawakami, J., Satoh, H. and Omura, T. (1985) High amylose mutants of rice, Oryza sativa L. Theor. Appl. Genet. 69: 253–257. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Li, N., Li, B., Li, Z., Xie, G. and Zhang, J. (2015) Reduced expression of starch branching enzyme IIa and IIb in maize endosperm by RNAi constructs greatly increases the amylose content in kernel with nearly normal morphology. Planta 241: 449–461. [DOI] [PubMed] [Google Scholar]
- Zhu, L., Gu, M., Meng, X., Cheung, S.C., Yu, H., Huang, J., Sun, Y., Shi, Y. and Liu, Q. (2012) High-amylose rice improves indices of animal health in normal and diabetic rats. Plant Biotechnol. J. 10: 353–362. [DOI] [PubMed] [Google Scholar]