Summary
Overexpression of exogenous fate-specifying transcription factors can directly reprogram differentiated somatic cells to target cell types. Here, we show that similar reprogramming can also be achieved through the direct activation of endogenous genes using engineered CRISPR/Cas9-based transcriptional activators. We use this approach to induce activation of the endogenous Brn2, Ascl1, and Myt1l genes (BAM factors) to convert mouse embryonic fibroblasts to induced neuronal cells. This direct activation of endogenous genes rapidly remodeled the epigenetic state of the target loci and induced sustained endogenous gene expression during reprogramming. Thus, transcriptional activation and epigenetic remodeling of endogenous master transcription factors is sufficient for conversion between cell types. The rapid and sustained activation of endogenous genes in their native chromatin context by this approach may facilitate reprogramming with transient methods that avoid genomic integration and provides a new strategy for overcoming epigenetic barriers to cell fate specification.
eTOC Summary
Black et al. show that reprogramming of fibroblasts to induced neurons via CRISPR/Cas9-based activation of endogenous neurogenic genes leads to rapid epigenetic remodeling at the targeted endogenous loci and sustained gene expression throughout the reprogramming process.
INTRODUCTION
Direct reprogramming of somatic cells has tremendous potential to advance applications in disease modeling, drug discovery, and gene and cell therapies. Common approaches to achieve cellular reprogramming rely on the ectopic expression of transgenes encoding lineage-specific transcription factors (Davis et al., 1987; Takahashi and Yamanaka, 2006; Vierbuchen et al., 2010). To demonstrate stable cellular reprogramming to an autonomous cell phenotype, the expression of exogenous transcription factors should be transient. Thus the establishment of positive feedback networks regulating endogenous genes is necessary to sustain a transgene-independent cellular identity (Vierbuchen and Wernig, 2011). In many cases, the endogenous genes are occluded by cis-acting repressive chromatin marks that are slow to remodel (Vierbuchen and Wernig, 2012). This slow remodeling process typically necessitates prolonged expression of the exogenous factors, limiting the efficacy of transient delivery methods, and poses a major bottleneck to improving the efficiency, speed, and robustness of reprogramming (Hanna et al., 2009).
The type II clustered regularly interspaced short palindromic repeat (CRISPR) system and the CRISPR-associated Cas9 nuclease have recently been repurposed from an adaptive immune system in bacteria and archaea to a gene editing tool (Cong et al., 2013; Jinek et al., 2012; Mali et al., 2013b) and transcriptional regulator (Cheng et al., 2013; Gilbert et al., 2013; Konermann et al., 2013; Maeder et al., 2013b; Mali et al., 2013a; Perez-Pinera et al., 2013; Qi et al., 2013) of endogenous genes in mammalian cells. The ability to program these transcription factors to target any genomic locus of interest through the simple exchange of the 20 nucleotide targeting sequence of the guide RNA (gRNA) enables a simple, robust and highly scalable method for control of complex transcriptional networks (Thakore et al., 2016). Furthermore, dCas9-based transcription factors can target stably silenced genes within compacted chromatin to initiate chromatin remodeling and transcriptional activation (Perez-Pinera et al., 2013; Polstein et al., 2015). Thus this technology may provide a method to deterministically initiate expression of endogenous gene networks of alternate cell lineages.
The CRISPR/Cas9 system and other platforms for programmable transcriptional regulation have been incorporated into methods for cellular reprogramming in a few recent studies. Gao et al. used TALE-based transactivators targeting an enhancer of Oct4 to generate mouse induced pluripotent stem cells. Notably, that study required co-delivery of vectors directly encoding ectopic C-MYC, KLF4, and SOX2 to achieve pluripotency (Gao et al., 2013). More recently, we have demonstrated the direct conversion of primary mouse embryonic fibroblasts (PMEFs) to skeletal myocytes using a dCas9-based transactivator targeting the endogenous Myod1 gene (Chakraborty et al., 2014). Several groups have also applied CRISPR/Cas9-based transcriptional regulation to direct the differentiation of human induced pluripotent and embryonic stem cells (Balboa et al., 2015; Chavez et al., 2015; Wei et al., 2016).
The above examples involve the targeted activation of a single transcription factor to guide reprogramming or differentiation, but many approaches require concurrent expression of multiple factors to efficiently establish a mature phenotype (Takahashi and Yamanaka, 2006; Vierbuchen et al., 2010). There have been no examples demonstrating multiplex endogenous gene activation to induce cellular reprogramming, and the versatility of that approach for direct conversion to other cell phenotypes is not known. Moreover, only the report of TALE transcription factors targeting Oct4 evaluated changes to epigenetic marks at the target loci (Gao et al., 2013), and this group later reported that dCas9-based transcriptional activators were inefficient at endogenous gene activation and reprogramming (Gao et al., 2014). In this study, we tested the hypothesis that targeting and epigenetic reprogramming of the regulatory elements controlling expression of lineage-specific transcription factors are sufficient for direct conversion between cell types by applying dCas9-based transactivators to the activation of endogenous genes that directly convert PMEFs to induced neuronal cells (iNs).
RESULTS
Multiplex Endogenous Gene Activation of Neurogenic Factors in PMEFs
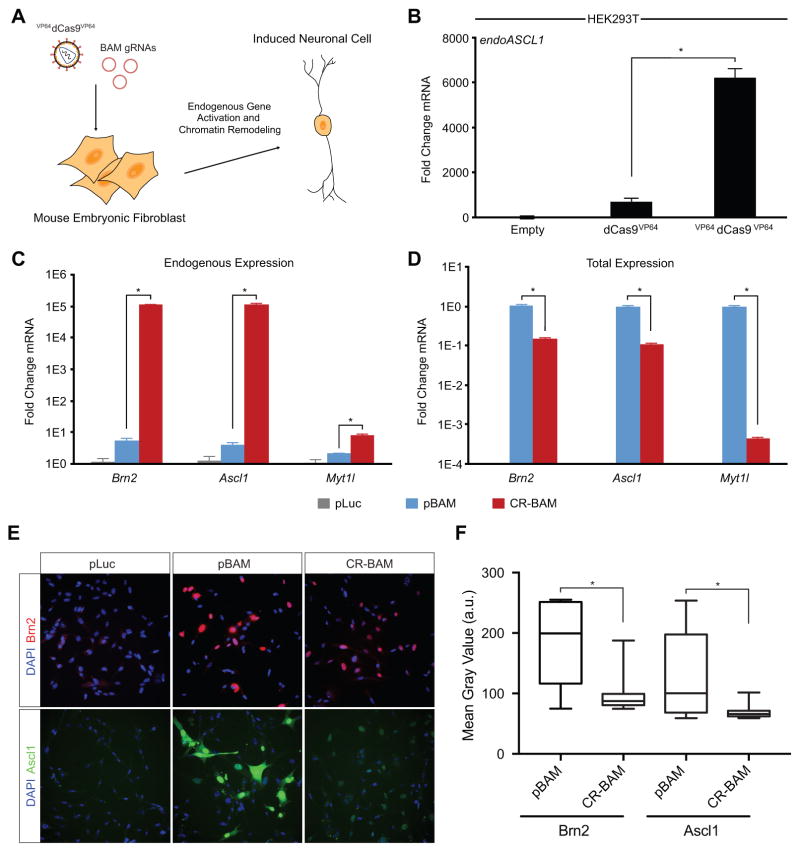
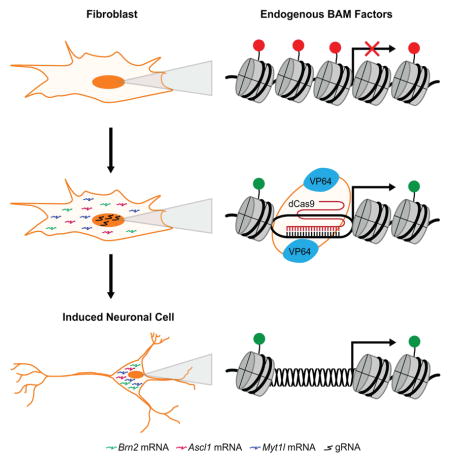
Overexpression of transgenes encoding the transcription factors Brn2, Ascl1, and Myt1l (BAM factors) has been shown to directly convert cultured PMEFs to functional induced neuronal cells (Vierbuchen et al., 2010). We hypothesized that the targeted activation of the endogenous genes encoding these same factors in their native chromatin context via a dCas9-based transactivator could more rapidly and deterministically remodel the chromatin at the target loci and provide an alternate method to achieve the reprogramming of PMEFs to iNs (Figure 1A). To achieve targeted gene activation, we used a transactivator with both N-terminal and C-terminal VP64 transactivation domains (VP64dCas9VP64) (Chakraborty et al., 2014) that generated a ~10-fold improvement in activation of ASCL1 in HEK293T cells at three days post-transfection compared to the first-generation dCas9 transcription factor with a single C-terminal VP64 domain (Maeder et al., 2013b; Perez-Pinera et al., 2013) (Figure 1B). We used VP64dCas9VP64 for the remainder of this study.
Figure 1. Endogenous Gene Activation of Neuronal Transcription Factors in PMEFs.
(A) Reprogramming of PMEFs to neuronal cells via transduction of VP64dCas9VP64 and transfection of gRNA expression plasmids targeting the endogenous BAM factors. (B) Transcriptional activation of ASCL1 in HEK293T cells with dCas9VP64 or VP64dCas9VP64 (*p<0.05). (C) Endogenous expression and (D) total expression of the BAM factors in PMEFs with targeted activation (CR-BAM) or ectopic overexpression (pBAM; *p<0.05). (E) Immunofluorescence staining of Brn2 and Ascl1 in PMEFs demonstrated protein expression through targeted activation of the endogenous loci or expression from ectopic plasmids (scale bar = 50 μm). (F) Automated image analysis of fluorescence intensity revealed significantly more single-cell Brn2 and Ascl1 protein with pBAM transfection compared to CR-BAM (*p<0.05 between distributions of single-cell mean fluorescence; Z-test). All gRNAs used are listed in Table S1. All assays were performed on day three post-transfection. qRT-PCR data are represented as mean ± s.e.m. for n = 3 biological replicates. P-values for qRT-PCR data were determined by global one-way ANOVA with Holm-Bonferroni post hoc tests (α = 0.05). See also Figure S1.
We used lentiviral delivery to constitutively express VP64dCas9VP64 in PMEFs. Initially, we delivered the gRNAs through transient transfection of plasmid DNA in order to assess stable reprogramming of cell phenotype following transient activity of transactivators. The induction of Brn2 and Ascl1 gene expression by VP64dCas9VP64 was attained by delivering four gRNAs targeted to the putative promoter region directly upstream of the transcription start site (TSS). The decision to deliver four gRNAs for each gene was based on the reported synergistic effects of multiple gRNAs on gene activation (Maeder et al., 2013b; Mali et al., 2013a; Perez-Pinera et al., 2013). The optimal gRNAs were selected from a pool of eight gRNAs through elimination screening (Figure S1A). The gRNAs targeting regions proximal to the TSS of the Myt1l locus did not induce detectable levels of activation, but targeting an intronic region directly upstream of the first coding exon of Myt1l was sufficient to activate expression (Figure S1B).
Co-transfection of twelve gRNA expression plasmids (CR-BAM), targeting each of the three endogenous BAM factors with 4 gRNAs, into PMEFs stably expressing VP64dCas9VP64 was sufficient to induce transcriptional upregulation of all three endogenous genes when compared to the transfection of a plasmid encoding firefly luciferase (pLuc, Figure 1C). We also detected Brn2 and Ascl1 protein expression by western blot (Figure S1C), although we could not detect Myt1l protein using commercially available antibodies. In addition to gRNA transfections, we transfected three plasmids encoding the BAM factor transgenes under the control of the EF1α/HTLV promoter (pBAM) into the same cells and observed a modest increase in the mRNA levels of the corresponding endogenous genes (Figure 1C).
To attain successful reprogramming, it is generally considered necessary to express the exogenous factors at high levels (Vierbuchen and Wernig, 2011). Therefore we compared the total mRNA and protein levels of Brn2, Ascl1, and Myt1l produced three days after CR-BAM and pBAM plasmid transfections (Figure 1D–1F). Despite the higher levels of transcriptional activation from the endogenous loci by CR-BAM (Figure 1C), pBAM transfection generated significantly more total mRNA encoding each BAM factor than induction by CR-BAM, as determined by qRT-PCR (Figure 1D). Quantitation of single-cell protein levels from immunofluorescence staining also revealed significantly higher single-cell levels of Brn2 and Ascl1 in cells transfected with pBAM compared to those transfected with CR-BAM (Figures 1E and 1F).
Induction of Neuronal Cells from PMEFs via VP64dCas9VP64-Mediated Gene Activation
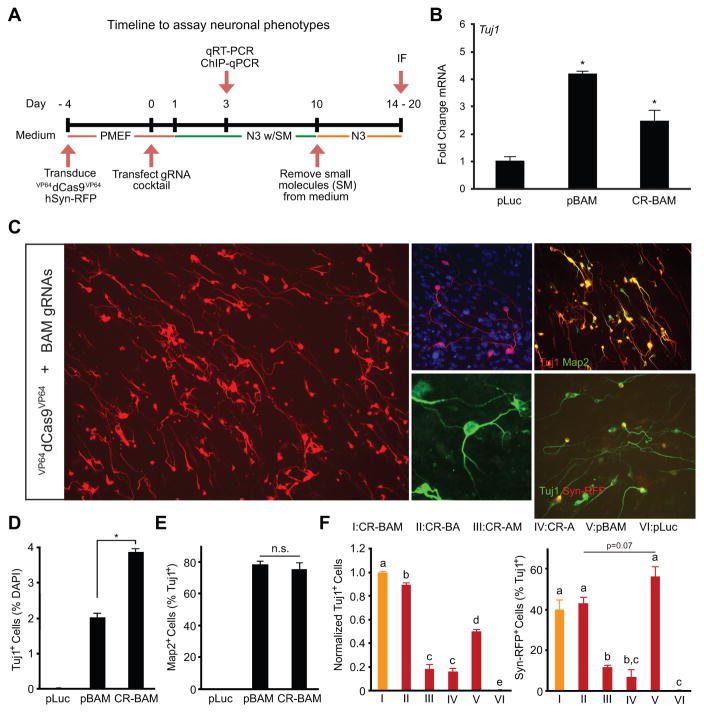
Treated PMEFs were assayed for neuronal phenotypes as detailed schematically in Figure 2A. We observed an increase in mRNA of the early pan-neuronal marker βIII tubulin (Tuj1) three days after transfection with either pBAM or CR-BAM when compared to a pLuc control (Figure 2B). We cultured the cells for two weeks in neurogenic medium and analyzed expression of pan-neuronal markers by immunofluorescence staining. We identified cells with neuronal morphologies that expressed Tuj1 in populations transfected with CR-BAM (Figure 2C). A subset of Tuj1+ cells also expressed the more mature pan-neuronal marker Map2 (Figure 2C). The generation of Tuj1+Map2+ cells with neuronal morphologies following treatment with VP64dCas9VP64 and gRNAs was contingent on the addition of a small molecule cocktail to the medium that has been used previously for neural differentiation of embryonic stem cells and has been shown to improve the efficiency of the direct conversion of human fibroblasts to neurons when used in parallel with ectopic expression of neural transcription factors (Ladewig et al., 2012).
Figure 2. Induction of Neuronal Cells from PMEFs via VP64dCas9VP64-Mediated Gene Activation.
(A) PMEFs were transduced with a lentivirus encoding the VP64dCas9VP64 transactivator and subsequently transfected with gRNAs targeting Brn2, Ascl1, and Myt1l. Neuronal phenotypes were assayed as indicated. (B) Transcriptional activation of Tuj1 was detected in PMEFs at day 3 post-transfection of pBAM or CR-BAM (*p<0.05 relative to transfection of a plasmid encoding firefly luciferase (pLuc)). (C) Immunofluorescence staining revealed numerous Tuj1+ cells with neuronal morphologies co-expressing Map2 at day 14 post-transfection of CR-BAM. The cells with the most elaborate neuronal morphologies activated the synapsin promoter in a Syn-RFP lentiviral reporter (scale bars = (i) 100 μm, (ii–v) 50 μm). (D) Quantitation of Tuj1+ cells as percent nuclei at day 14 post-transfection of either pLuc, pBAM, or CR-BAM (*p<0.05). (E) Quantitation of Map2+ cells as percent Tuj1+ cells at day 14 post-transfection of either pLuc, pBAM, or CR-BAM (n.s., not significant). (F) Quantitation of Tuj1+ and RFP+ cells with transfection of different combinations of gRNAs. Tuj1+ cells are normalized to CR-BAM transfection. Conditions that share the same letter (a-e) are not significantly different. P-values were determined by global one-way ANOVA with Holm-Bonferroni post hoc tests (α = 0.05). See also Figure S2.
We used a lentiviral fluorescent reporter encoding DsRed-Express under the control of the synapsin I promoter (Syn-RFP) as a proxy to define the most functionally mature iNs in the heterogeneous population of reprogrammed cells (Adler et al., 2012). We readily identified RFP+ cells with elaborate arborizations in CR-BAM-transfected PMEFs (Figure 2C). We also identified rare cells with fibroblastic morphologies reactive to the Tuj1 antibody in PMEFs following pLuc transfection (Figure S2A), but these cells were never reactive to the Map2 antibody. Consistent with previous studies, direct overexpression of the ectopic BAM factors via transfection of constitutive expression plasmids generated Tuj1+Map2+ cells with neuronal morphologies (Figure S2B) (Adler et al., 2012; Vierbuchen et al., 2010).
Image analysis revealed that CR-BAM transfection generated a modest, but statistically significant and reproducible, increase in the number of Tuj1+ cells compared to pBAM transfection after 14 days in culture post-transfection (Figure 2D), despite much lower overall expression of the BAM factors (Figure 1D–F). There was no difference in the percentage of Tuj1+ cells that also expressed Map2 (Figure 2E). To evaluate the contribution of each neurogenic factor to the generation of Tuj1+ cells and to the level of neuronal maturation, we transfected gRNAs targeting different combinations of the endogenous factors. Removal of gRNAs targeting the Brn2 locus attenuated iN production ~5-fold when compared to that generated with targeted activation of all three endogenous factors (Figure 2F). We detected a slight reduction in Tuj1+ cell production with the removal of Myt1l gRNAs (Figure 2F). Neuronal maturity was assessed as the percentage of Tuj1+ cells co-positive for the Syn-RFP reporter. Removal of Brn2 gRNAs reduced the percentage of RFP+ cells over 2-fold, but no change was detected with removal of Myt1l gRNAs (Figure 2F). pBAM transfection generated a higher percentage of RFP+ cells than CR-BAM transfection, though it was not statistically significant (Figure 2F).
Induction of Endogenous Gene Expression is Rapid and Sustained
For any reprogramming strategy, activation of the endogenous genes encoding the master fate-specifying transcription factors is an important step to the successful reprogramming and stability of the new cellular phenotype (Vierbuchen and Wernig, 2011). Consequently, we compared the kinetics of endogenous gene expression through late stages of reprogramming with pBAM or CR-BAM transfection. We observed activation of all three endogenous genes as early as one day post-transfection with CR-BAM that remained at high levels through day 18 in culture (Figure S3A). Expression of the BAM factors from the endogenous loci was significantly higher with targeted activation via CR-BAM compared to ectopic overexpression via pBAM transfection throughout the time course of the experiment. Reactivation of the endogenous genes by pBAM transfection was delayed, and a significant and sustained increase over baseline levels was only detected for endogenous Ascl1 and Myt1l (Figure S3A).
We next assessed the kinetics of expression of the downstream pan-neuronal marker Tuj1. Both pBAM and CR-BAM treatment generated a significant increase in Tuj1 expression throughout the time course of the experiment (Figure S3B). At early time points, Tuj1 levels were higher with pBAM treatment than CR-BAM. However, Tuj1 levels with pBAM treatment peaked seven days post-transfection and declined thereafter, whereas expression following CR-BAM treatment remained stable through day 18 in culture (Figure S3B). Importantly, the exogenous BAM factors and gRNAs were significantly depleted by day 18 in culture after transient transfection (Figure S3C), though levels of activation from the endogenous genes remained high in cells treated with CR-BAM (Figure S3A).
Direct Activation via VP64dCas9VP64 Rapidly Remodels Chromatin at Target Loci
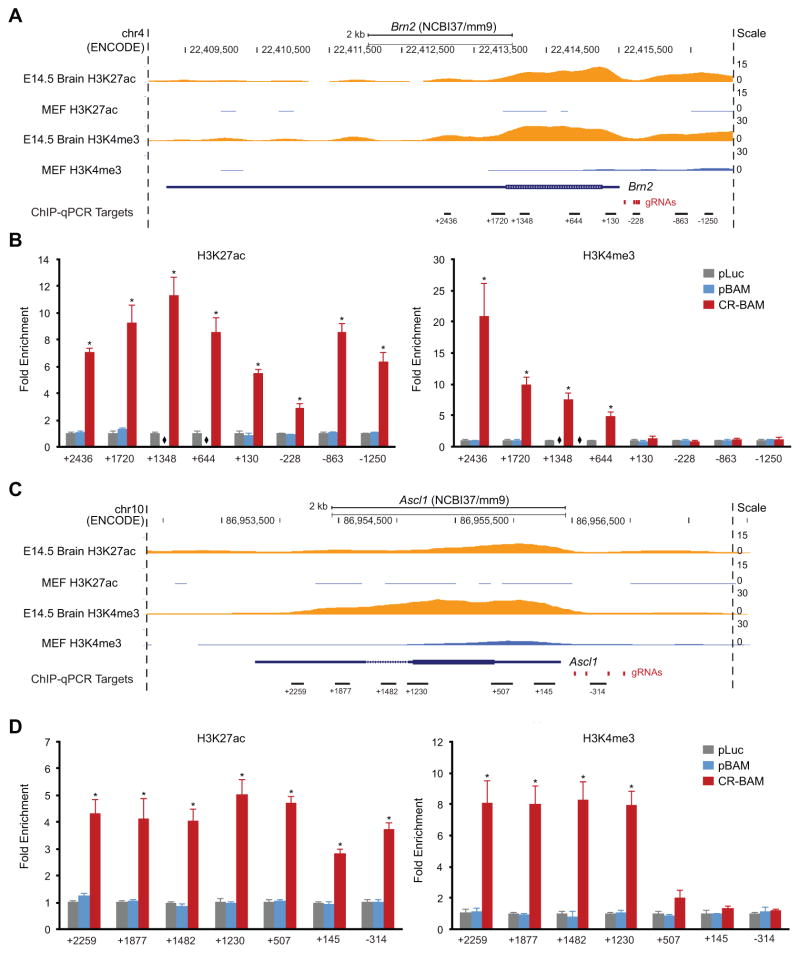
The kinetics of gene activation led us to speculate whether the rapid and sustained elevated levels of endogenous gene expression achieved with CR-BAM corresponded to an altered epigenetic program at the target loci. We used chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) data generated as part of the Encyclopedia of DNA Elements (ENCODE) Project (Consortium, 2012) to identify histone modifications enriched at the transcriptionally active BAM factor loci in mouse embryonic brain tissue, including H3K27ac and H3K4me3 (Figures 3A, 3C, and S4A). We hypothesized that targeting the endogenous BAM factors for activation with VP64dCas9VP64 in PMEFs could recapitulate the chromatin signatures found at these loci in developing brain tissue.
Figure 3. VP64dCas9VP64 Rapidly Remodels Epigenetic Marks at Target Loci.
(A) & (C) Mouse genomic tracks depicting histone H3 modifications H3K27ac and H3K4me3 at the Brn2 and Ascl1 loci in embryonic brain tissue and fibroblasts (data from Mouse ENCODE; GSE31039). Red bars indicate gRNA target sites near the transcription start site, and black bars indicate the location of ChIP-qPCR amplicons along the gene locus. (B) & (D) Targeted activation of endogenous Brn2 and Ascl1 in PMEFs induced significant enrichment of H3K27ac and H3K4me3 at multiple sites along the genomic loci at day 3 post-transfection (*p < 0.05, one-way ANOVA with Holm-Bonferroni post hoc tests, n = 3 biological replicates). Overexpression of the BAM factors via transfection of expression plasmids encoding BAM factor transgenes did not induce a significant change in these chromatin marks. qPCR primers targeting coding regions of the genes are not included for the pBAM transfection condition, as contaminating plasmid DNA biased enrichment values. All fold enrichments are relative to transfection of a plasmid encoding firefly luciferase and normalized to a region of the Gapdh locus. See also Figure S3 and S4.
To investigate the effects of BAM-factor induction on the epigenetic programming at the target loci, we performed ChIP-qPCR in PMEFs transduced with VP64dCas9VP64 and transfected with either pLuc, pBAM, or CR-BAM plasmids (Figures 3 and S4). We used qPCR primers tiled along intragenic and regulatory regions of the Brn2, Ascl1, and Myt1l loci. We detected a significant enrichment in H3K27ac and H3K4me3 at the Brn2 and Ascl1 loci on day three post-transfection of CR-BAM (Figures 3B and 3D). H3K4me3 was enriched along the gene bodies of Brn2 and Ascl1. H3K27ac was enriched along the gene bodies and regions surrounding the putative promoter sequences of both genes. In contrast, targeted activation of Myt1l only induced modest detectable enrichment in H3K27ac at the gRNA target sites directly upstream of the first coding exon (Figure S4B). No significant change in H3K27ac or H3K4me3 was measured within the putative Myt1l promoter. Though overexpression of the BAM factors induced modest levels of expression of the endogenous genes by day three post-transfection (Figures 1C and S3A), we did not detect corresponding enrichment in H3K27ac and H3K4me3 at the endogenous loci (Figures 3B, 3D, and S4B).
Generation of Induced Neuronal Cells with Multiplex gRNA Lentiviral Vectors
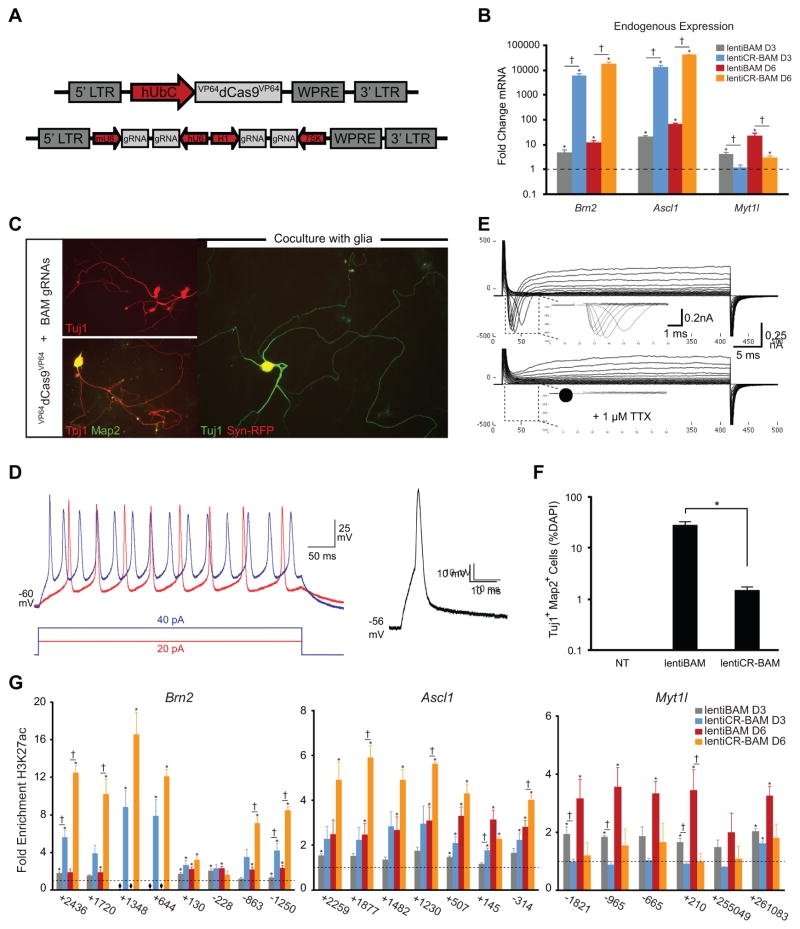
To explore a strategy for stable expression of the CRISPR/Cas9 transcription factors, and to see if the same outcomes observed with transient expression held true with constitutive expression, we used a single lentiviral vector capable of expressing four gRNAs from four independent RNA Polymerase III promoters (Kabadi et al., 2014) (Figure 4A). Co-transduction of lentiviruses encoding VP64dCas9VP64 and a set of four gRNAs targeting each of the three BAM factors (lentiCR-BAM) permitted concurrent activation of the endogenous BAM factors in PMEFs by day six post-transduction (Figure 4B). For comparison, we used lentiviral vectors directly encoding the BAM factors (lentiBAM), and demonstrated activation of the corresponding endogenous genes by day six post-transduction (Figure 4B). Similar to the results we obtained with transient transfection of expression plasmids, targeted activation of the endogenous genes via lentiviral delivery generated significantly more endogenous transcript from the Brn2 and Ascl1 loci than that induced through ectopic expression of the BAM factors. However, unlike the transfection experiments, endogenous Myt1l expression was significantly higher with transduction of lentiBAM compared to lentiCR-BAM (Figure 4B).
Figure 4. Generation of Functionally Mature iNs with Multiplex gRNA Vectors.
(A) Schematic of VP64dCas9VP64 and multiplex gRNA lentiviral constructs used to enable stable integration and constitutive expression. (B) Relative mRNA expression of the endogenous BAM factors following transduction of transgenes encoding the BAM factors (lentiBAM) or VP64dCas9VP64 and gRNAs targeting the endogenous BAM factors (lentiCR-BAM; p<0.05 relative to non-treated PMEFs; †p<0.05 between lentiBAM versus lentiCR-BAM transduction). (C) Immunofluorescence staining of PMEFs following transduction of lentiCR-BAM. Cells were co-positive for Tuj1 and Map2 and exhibited complex neuronal morphologies (scale bar = 50 μm). (D) Action potentials were evoked from VP64dCas9VP64-induced neuronal cells in response to 5 (right) or 500 (left) ms step depolarizing current injection (6/7 cells analyzed) after empiric hyperpolarizing current injection to hold membrane potential at ~ −60 mV. (E) Representative whole-cell currents recorded with or without perfusion of 1 μM tetrodotoxin (TTX). (F) Quantitation of Tuj1+Map2+ cells as percent nuclei (*p<0.05 between lentiBAM versus lentiCR-BAM transduction; NT, non-treated PMEFs). (G) Timecourse of H3K27ac enrichment along the Brn2, Ascl1, and Mytl1 loci (*p<0.05 relative to non-treated PMEFs; †p<0.05 between lentiBAM versus lentiCR-BAM transduction). All p-values calculated by global ANOVA with Holm-Bonferroni post hoc tests (α = 0.05).
Following extended culture for two weeks in neurogenic medium, we readily identified Tuj1+Map2+ cells with complex neuronal morphologies (Figure 4C). All Tuj1+ cells identified also co-expressed Map2. To promote further neuronal maturation and for electrophysiological assessments, PMEFs were replated onto a previously established monolayer of primary rat astrocytes following transduction of VP64dCas9VP64 and gRNAs (Vierbuchen et al., 2010). Synapsin-RFP expression and cell morphology were used to select the most mature neuronal cells for patch-clamp analysis after 21 days in culture. In current clamp mode, single or multiple action potentials were readily elicited in response to depolarizing current injections (6/7 cells analyzed; Figure 4D). The same cells displayed voltage-dependent inward and outward currents. The transient inward currents were abolished in the presence of the voltage-gated Na+ channel blocker tetrodotoxin (TTX; Figure 4E). The average resting membrane potential, action potential (AP) threshold and AP amplitude were −41 ± 3.8 mV, −33 ± 2.6 mV and 49 ± 9.7 mV, respectively (mean ± s.e.m., n = 7 cells).
In contrast to what we observed by transient transfection of the reprogramming factors, constitutive expression of the BAM factor transgenes via lentiviral vectors generated significantly more Tuj1+Map2+ cells than that detected with VP64dCas9VP64 (Figure 4F). We hypothesized that the prolonged and high levels of expression of the BAM factor transgenes enabled by lentiviral delivery permitted further epigenetic and transcriptional reprogramming that improved the efficiency of iN generation when compared to transient transfection methods. Consequently, we revisited the analysis of chromatin remodeling at the endogenous BAM factor loci in the context of lentiviral delivery of the reprograming factors. We found that, as shown with transient transfection, targeted activation of the endogenous genes via lentiCR-BAM transduction led to the rapid deposition of H3K27ac at the Brn2 and Ascl1 loci as early as day 3 post-transduction that persisted at day 6 (Figure 4G). Also, as seen with transient transfection, we did not detect enrichment of H3K27ac at the Myt1l locus with lentiCR-BAM transduction, although we did measure an increase in Myt1l mRNA (Figure 4B and 4G). In contrast to what we observed with transient transfection of the BAM factors, we detected enrichment of H3K27ac along regions of all three endogenous genes with lentiBAM transduction (Figure 4G). Furthermore, we only detected minor enrichment in H3K27ac at all three genes at day 3 post-transduction of lentiBAM, however, both Ascl1 and Myt1l showed a substantial increase in H3K27ac deposition by day 6 post-transduction (Figure 4G).
DISCUSSION
In this study we demonstrate direct cellular reprogramming to induced neuronal cells through targeted activation of endogenous genes. We utilized the CRISPR/Cas9 system as a programmable, locus-specific transcriptional regulator for the multiplex activation of the neurogenic factors Brn2, Ascl1, and Myt1l (BAM factors). We hypothesized that targeted activation of the endogenous genes in PMEFs, as opposed to the forced overexpression of the corresponding transgenes, could more directly access the endogenous loci and rapidly remodel their epigenetic signatures, thus potentially reflecting a more natural mechanism of action and serving as an alternate method to achieve cell lineage conversion.
In PMEFs, the cis-repressive chromatin landscape at neuronal loci may preclude binding of regulatory factors, in turn impeding transcriptional activation. As a result, expression of the BAM factors in PMEFs from exogenous vectors likely relies on stochastic processes for reactivation of the corresponding endogenous genes. Furthermore, transient delivery of the BAM factors, as done in our initial experiments (Figures 1–3), limits the time window within which the endogenous networks and positive feedback loops can be established. We demonstrated that targeting the endogenous genes directly induced the enrichment of histone H3 modifications H3K27ac and H3K4me3 at the Brn2 and Ascl1 loci at three days post-transfection, whereas transgene overexpression via transfection of plasmids encoding the reprogramming factors did not alter these chromatin marks (Figures 3 and S4). Additionally, we observed sustained high levels of expression from the endogenous genes at later stages of reprogramming despite the transient delivery of the gRNA plasmids (Figure S3).
In contrast, we found that stable integration and constitutive expression of the exogenous reprogramming factors via lentiviral delivery led to the eventual deposition of H3K27ac at their endogenous loci with a concomitant improvement in reprogramming capacity (Figure 4F–G). We did not observe a similar improvement with constitutive expression of VP64dCas9VP64 and gRNAs, which is possibly attributable to the lower levels of overall expression of the neuronal transcription factors achieved by transactivation of the endogenous genes compared to ectopic overexpression. Consequently, the direct activation of the endogenous genes via VP64dCas9VP64 may be more amenable to transient delivery approaches that avoid undesired consequences of vector integration into the genome. Such transient methods, including the direct delivery of ribonucleoprotein Cas9-gRNA complexes, may be a more clinically translatable method of generating reprogrammed cells that are genetically unmodified.
Achieving robust and well-defined reprogrammed cell populations is still a central challenge. Reprogrammed cells often fail to acquire completely mature phenotypes and can retain epigenetic remnants of the native cell type (Kim et al., 2010). Moreover, a recent study demonstrated that reprogramming efficiency can be limited by divergence to a competing cell identity (Treutlein et al., 2016). The molecular mechanisms and practical consequences of these limitations are largely unknown. As the toolkit of designer transcription factors expands to precisely modify the epigenome (Hilton et al., 2015; Kearns et al., 2015; Maeder et al., 2013a; Mendenhall et al., 2013; Thakore et al., 2016), these tools may be used to prime specific genomic loci in diverse cell types, promote endogenous transcription factor binding, and directly correct regions of epigenetic remnants that prove to be problematic for a given application. This may lead to improved reprogramming fidelity and extension of the breadth of donor cells amenable to reprogramming.
EXPERIMENTAL PROCEDURES
Cell Culture, transfections and viral transductions
Primary mouse embryonic fibroblasts were maintained in high serum media during transduction and transfection of expression plasmids, and subsequently cultured in neurogenic serum-free medium for the duration of the experiments to promote neuronal survival and maturation. Lentivirus was produced in HEK293T cells using the calcium phosphate precipitation method. All transfections were performed using Lipofectamine 3000 (Invitrogen) in accordance with the manufacturer’s protocol. All expression plasmids used in this study can be found in Table S2.
Immunofluorescence Staining & qRT-PCR
All sequences for qRT-PCR primers can be found in Table S3. Total RNA was isolated using the QIAGEN RNeasy and QIAshredder kits, reverse transcribed using the SuperScript VILO Reverse Transcription Kit (Invitrogen), and analyzed using Perfecta SYBR Green Fastmix (Quanta BioSciences). All qRT-PCR data are presented as fold change in RNA normalized to Gapdh expression. For immunofluorescence staining, cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and incubated with primary and secondary antibodies.
Electrophysiology
A synapsin-RFP lentiviral reporter was used to identify cells in co-culture with primary rat astroglia for patch clamp analysis at indicated time points. Action potentials and inward and outward currents were recorded in whole-cell configuration. Data were analyzed and prepared for publication using MATLAB.
Chromatin Immunoprecipitation qPCR
Chromatin was immunoprecipitated using antibodies against H3K27ac and H3K4me3, and gDNA was purified for qPCR analysis. All sequences for ChIP-qPCR primers can be found in Table S3. qPCR was performed using SYBR Green Fastmix (Quanta BioSciences), and the data are presented as fold change gDNA relative to negative control and normalized to a region of the Gapdh locus.
Mouse ENCODE ChIP-seq data sets
H3K4me3 and H3K27ac ChIP-seq data from C57BL/6 E14.5 whole brain and mouse embryonic fibroblasts (GSE31039) were acquired from the Mouse ENCODE Consortium generated in Bing Ren’s laboratory at the Ludwig Institute for Cancer Research.
Statistical Methods
Statistical analysis was done using GraphPad Prism 7. All data were analyzed with at least three biological replicates and presented as mean ± s.e.m. See figure legends for details on specific statistical tests run and p-values calculated for each experiment.
Supplementary Material
Highlights.
Multiplexed CRISPR/Cas9 activators induce expression of endogenous neurogenic genes
Induced endogenous gene expression directly converts fibroblasts to neuronal cells
Targeted activation of endogenous genes rapidly remodels chromatin at target loci
High expression from the endogenous genes is sustained throughout reprogramming
Acknowledgments
The authors thank Ami Kabadi, Pablo Perez-Pinera, and Syandan Chakraborty for providing plasmid constructs; Christopher Grigsby for insights on PMEF transfections and providing the pUNO-Brn2 plasmid; Rui Dai for technical assistance, and Jeff Stogsdill and Cagla Eroglu for supplying astrocytes and providing insights on co-culture experiments. This work was supported by US National Institutes of Health (NIH) grants to C.A.G. and T.E.R. (R01DA036865 and U01HG007900), a NIH Director’s New Innovator Award (DP2OD008586) and National Science Foundation (NSF) Faculty Early Career Development (CAREER) Award (CBET-1151035) to C.A.G., and NIH grant P30AR066527. J.B.B. was supported by a NIH Biotechnology Training Grant (T32GM008555).
Footnotes
Supplemental Information for this article includes four figures, three tables and Supplemental Experimental Procedures.
AUTHOR CONTRIBUTIONS
J.B.B., A.F.A., K.W.L., and C.A.G. designed experiments. J.B.B., A.F.A., H.W., A.M.D., and H.A.H. performed the experiments. All authors analyzed the data. J.B.B. and C.A.G. wrote the manuscript with contributions by all authors.
CONFLIC OF INTEREST
C.A.G. and J.B.B. have filed patent applications related to genome engineering technologies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adler AF, Grigsby CL, Kulangara K, Wang H, Yasuda R, Leong KW. Nonviral direct conversion of primary mouse embryonic fibroblasts to neuronal cells. Molecular therapy Nucleic acids. 2012;1:e32. doi: 10.1038/mtna.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa D, Weltner J, Eurola S, Trokovic R, Wartiovaara K, Otonkoski T. Conditionally Stabilized dCas9 Activator for Controlling Gene Expression in Human Cell Reprogramming and Differentiation. Stem Cell Reports. 2015;5:448–459. doi: 10.1016/j.stemcr.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Ji H, Kabadi AM, Gersbach CA, Christoforou N, Leong KW. A CRISPR/Cas9-Based System for Reprogramming Cell Lineage Specification. Stem cell reports. 2014;3:940–947. doi: 10.1016/j.stemcr.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, Iyer EPR, Lin S, Kiani S, Guzman CD, Wiegand DJ, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, Rangarajan S, Shivalila CS, Dadon DB, Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, M.E. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biology. 2012:13. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Gao X, Tsang JC, Gaba F, Wu D, Lu L, Liu P. Comparison of TALE designer transcription factors and the CRISPR/dCas9 in regulation of gene expression by targeting enhancers. Nucleic Acids Res. 2014;42:e155. doi: 10.1093/nar/gku836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Yang J, Tsang JC, Ooi J, Wu D, Liu P. Reprogramming to Pluripotency Using Designer TALE Transcription Factors Targeting Enhancers. Stem cell reports. 2013;1:183–197. doi: 10.1016/j.stemcr.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nature biotechnology. 2015 doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014;42:e147. doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015 doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladewig J, Mertens J, Kesavan J, Doerr J, Poppe D, Glaue F, Herms S, Wernet P, Kogler G, Muller FJ, et al. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat Methods. 2012;9:575–578. doi: 10.1038/nmeth.1972. [DOI] [PubMed] [Google Scholar]
- Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, Tsai SQ, Ho QH, Sander JD, Reyon D, Bernstein BE, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nature biotechnology. 2013a;31:1137–1142. doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013b;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature biotechnology. 2013a;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013b;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Williamson KE, Reyon D, Zou JY, Ram O, Joung JK, Bernstein BE. Locus-specific editing of histone modifications at endogenous enhancers. Nature biotechnology. 2013;31:1133–1136. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein LR, Perez-Pinera P, Kocak DD, Vockley CM, Bledsoe P, Song L, Safi A, Crawford GE, Reddy TE, Gersbach CA. Genome-wide specificity of DNA binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome Res. 2015;25:1158–1169. doi: 10.1101/gr.179044.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thakore PI, Black JB, Hilton IB, Gersbach CA. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods. 2016;13:127–137. doi: 10.1038/nmeth.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Lee QY, Camp JG, Mall M, Koh W, Shariati SA, Sim S, Neff NF, Skotheim JM, Wernig M, et al. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature. 2016 doi: 10.1038/nature18323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Wernig M. Direct lineage conversions: unnatural but useful? Nature biotechnology. 2011;29:892–907. doi: 10.1038/nbt.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Wernig M. Molecular roadblocks for cellular reprogramming. Molecular cell. 2012;47:827–838. doi: 10.1016/j.molcel.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Zou Q, Lai S, Zhang Q, Li L, Yan Q, Zhou X, Zhong H, Lai L. Conversion of embryonic stem cells into extraembryonic lineages by CRISPR-mediated activators. Sci Rep. 2016;6:19648. doi: 10.1038/srep19648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.