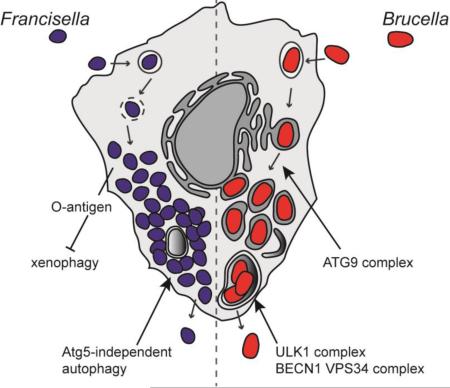
Abstract
Autophagy is a lysosomal recycling process conserved in eukaryotes, which maintains cellular homeostasis during stress and starvation conditions by degrading and recycling proteins, lipids and carbohydrates, ultimately increasing nutrient availability. An additional function of autophagy, termed xenophagy, is to detect, capture and destroy invading microorganisms, such as viruses, bacteria and protozoa, providing autophagy with a role in innate immunity. Many intracellular pathogens have however developed mechanisms to avoid xenophagy, and have evolved strategies to take advantage of select autophagic processes to undergo their intracellular lifecycle. This review article will discuss the molecular mechanisms used by the intracellular bacterial pathogens Francisella spp. and Brucella spp. to manipulate components of the autophagic pathway, promoting cytosolic growth in the case of Francisella spp., and facilitating cellular egress and cell-to-cell spread in the case of Brucella spp. These examples highlight how successful, highly infectious bacterial pathogens avoid or subvert host autophagy mechanisms normally employed to maintain eukaryotic homeostasis.
Keywords: infection, cellular egress, nutrient acquisition, O-antigen, bacterial secretion system, xenophagy
Graphical abstract
Introduction on Autophagy
Autophagy is an essential and conserved eukaryotic process for intracellular breakdown and disposal of damaged organelles and protein aggregates that are too large to be degraded by the proteasome. Three mechanistically distinct types of autophagy occur in eukaryotic cells, (1) microautophagy, (2) chaperone-mediated autophagy and (3) macroautophagy [1,2]. Macroautophagy, commonly referred to as autophagy, is a membrane trafficking pathway that captures material for delivery to the lysosomal degradative compartment. Autophagy is initiated by the capture of cytosolic components, specifically damaged organelles, protein aggregates and intracellular microbes into double-membrane-bound compartments called autophagosomes. These autophagosomes then mature into degradative autolysosomes via fusion with lysosomes along the late endocytic pathway [2]. The content of the autolysosome is then degraded, to either regenerate nutrients or destroy invading pathogens. Lysosomal permeases and transporters export amino acids and other by-products of degradation into the cytosol, where they can be recycled and used as building blocks of macromolecules [2,3]. Autolysosomes are then recycled through lysosome reformation [4]. Autophagy is integral to human health and occurs at a basal level in eukaryotic cells to maintain cellular homeostasis, and imbalances in autophagy regulation have been associated with many diseases such as cancer, neurodegeneration, aging and microbial infection [5,6].
A Conserved Autophagic Machinery and Activation
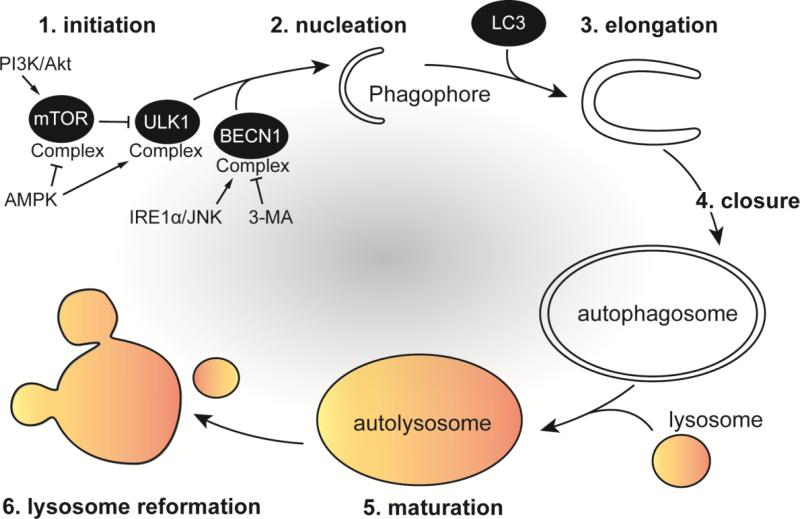
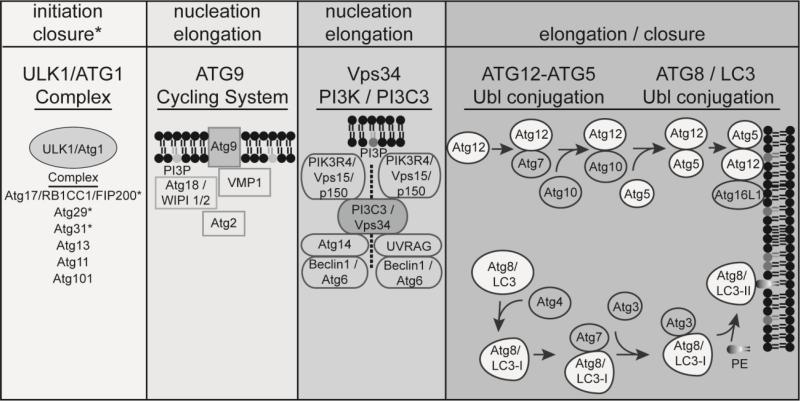
The canonical autophagy process can be divided in six sequential steps: (1) initiation, (2) phagophore formation/nucleation, (3) elongation, (4) closure, (5) maturation and (6) recycling/lysosomal reformation (Figure 1). More than 30 autophagy-related genes (Atg) have been identified in yeast with homologs in humans that encode proteins, which coordinate autophagosome formation and maturation (Figure 2) [1]. In addition, small GTPases involved in the endocytic and secretory pathways are also engaged in autophagosome formation and maturation [7]. Phospholipids such as phosphatidylinositol and phosphatidylethanolamine are also integral components of autophagy [8]. The core machinery of autophagy can be classified into several functional units including: the Atg1/ULK1 complex, the transmembrane protein Atg9 cycling system, the Vps34 phosphatidylinositol 3-kinase (PI3P / PtdIns3K) complex and two ubiquitin-like conjugating systems, Atg12-Atg5 and Atg8/LC3 (Figure 2). Upon autophagy initiation, a crescent-shaped, double-membrane called the phagophore (or isolation membrane) is formed. The phagophore assembly site (PAS) can be derived from membranes from different organelles [9]. One characterized example of a PAS is the omegasome, a structure formed by extension of the endoplasmic reticulum (ER) that contains the protein DFCP1 [10]. Subsequently, Atg9, Atg18 (WIPI1/2) and VMP1 are recruited to the omegasome [11]. Atg9 is a transmembrane protein that promotes lipid recruitment to the expanding phagophore [12]. Phagophore formation is controlled by the Unc51-like kinase 1 (ULK1) complex [13,14], which is composed of Atg1 (ULK1), Atg11, Atg13, Atg17, Atg29 and Atg31 in yeast and ULK1, ATG13, RB1CC1/FIP200 and ATG101 in mammalian cells [15,16]. Following amino acid starvation, the ULK1 complex activates BECN1/Beclin1, which triggers production of phosphatidylinositol 3-phosphate (PI3P). Vps34 phosphorylates the phospholipid, phosphatidylinositol (PI) to form PI3P, which is central for membrane trafficking processes and is the major lipid signal controlling autophagic vesicle formation [17]. PI3P is essential for phagophore elongation by bringing additional Atg proteins to the phagophore that recruit membranes to the elongating phagophore [18]. BECN1 is part of a core complex that contains Atg14, p150 and Vps34, a class III phosphoinositide 3-kinase (PI3K). A pharmacological inhibitor of autophagy called 3-methyladenine (3-MA) binds Vps34, thereby preventing autophagy nucleation and elongation [19]. The phagophore ultimately elongates and expands to engulf and sequester autophagic cargo using a ubiquitin-like conjugation system of proteins, including Atg5-Atg12 and Atg16L1, which mediates the maturation and closure of the phagophore [20]. Another ubiquitin-like conjugation system involves Atg8 in yeasts. Mammals express three subfamilies of Atg8 proteins: (1) microtubule-associated protein 1A/1B-light chain 3 (MAP-LC3 or LC3), (2) γ-aminobutyric acid receptor-associated protein (GABARAP) and (3) Golgi-associated ATPase enhancer of 16 kDa (GATE-16) [21]. Atg8/LC3 proteins are involved in recruiting cargo into autophagosomes and function in autophagosome biogenesis. The cysteine protease Atg4 cleaves Atg8/LC3 to generate LC3-I, which is subsequently conjugated to phosphatidylethanolamine (PE) on the autophagosomal membranes via Atg3 and Atg7, forming the lipidated, membrane-associated LC3-II form. LC3-II is involved in the final sealing steps that allow completion of an autophagosome [22], and is considered a gold standard marker of autophagic compartments [8]. The newly formed autophagosome then fuses with the lysosome to form an acidified autophagolysosome. Autophagosomes can either nonspecifically engulf cytosolic matter, or selectively sequester unwanted material including pathogens using autophagy receptors, such as p62/SQSTM1, NDP52, NBR1 and optineurin, which bind to polyubiquitin-tagged proteins and to autophagosome-associated LC3-II [23].
Figure 1. Sequential steps of canonical autophagy.
In the presence of amino acids, growth factors and energy, the mTOR complex represses autophagy by inhibiting the kinase activity of ULK1. The PI3K/Akt pathway inhibits autophagy, while AMPK activates autophagy by controlling mTOR activation under nutrient-limiting conditions. Upon autophagy induction (1), the ULK1 complex activates the BECN1/VPS34 complex to initiate (2) phagophore formation and nucleation. BECN1 can be activated directly by the IRE1α/JNK pathway or inhibited by the pharmacological drug 3-methyladenine (3-MA). Phagophore elongation (3) proceeds to engulf and sequester autophagic cargo and the phagophore membrane acquires LC3. Ubiquitin-like conjugation systems mediate the closure of the autophagosome (4). Maturation of the autophagosome (5) occurs via fusion with late endocytic/lysosomal compartments, forming the autolysosome where material is degraded. Autolysosomes are then recycled in a process that allows for lysosome reformation (6).
Figure 2. Autophagy complexes that coordinate autophagosome formation.
The ULK1 complex includes ULK1/Atg1, Atg17/RB1CC1/FIP200, Atg29, Atg31, Atg13, Atg11 and Atg101. Atg13 and Atg17/RB1CC1/FIP200 are substrates of ULK1 kinase activity. In addition to autophagy initiation, Atg17, Atg29 and Atg31 are also involved in phagophore closure. The transmembrane protein Atg9 cycling system mediates membrane recruitment during nucleation and elongation and involves interactions between the phosphoinositide PI3P, Atg9, Atg18/WIPI, VMP1 and Atg2 on the phagophore membrane. The Vps34 phosphatidylinositol 3-kinase (PI3P/PtdIns3K/PI3C3) complex includes, either PI3P, Vps15/PIK3R4/p150, BECN1/Atg6 and Atg14L or UVRAG. The Vps34 complex is required for the nucleation of autophagosomal membranes. Two ubiquitin-like conjugating systems, Atg12-Atg5-Atg16L1 and Atg8/LC3, are required for maturation and closure of the autophagosomal membrane. The cysteine protease Atg4 cleaves LC3 to generate LC3-I; Atg3 and Atg7 subsequently conjugate LC3-I to phosphatidylethanolamine (PE), forming the membrane-associated LC3-II. All membranes depicted represent the phagophore membrane.
Autophagy is initiated by a complex and diverse set of stimuli. A number of triggers increase autophagy over basal levels, including cellular stressors such as nutrient starvation, hypoxia, growth factor withdrawal, ER stress and infection [24-26]. A classical trigger of autophagy is nutrient deprivation. When intracellular amino acid concentrations are low, autophagy is induced. A central regulator of autophagy is the kinase mammalian target of rapamycin (mTOR) (Figure 1), which inhibits autophagy in the presence of growth factors and abundant nutrients. When starvation conditions arise, cellular AMP levels rise and ATP levels drop, which in turn activates the energy sensing protein AMP-activating protein kinase (AMPK). AMPK can induce autophagy by phosphorylating TSC2 and RPTOR to inactivate mTOR, and AMPK can directly activate autophagy by phosphorylating ULK1 during nutrient deprivation [13]. mTOR is also a downstream target of the phosphatidylinositol 3-kinase and Akt (PI3K/Akt) pathway. The PI3K/Akt pathway is activated by insulin and growth factors to promote cell growth, differentiation and survival [27]. Activation of the PI3K/Akt pathway inhibits autophagy via increased activation of mTOR [28]. Another critical regulator of autophagy is the class III PI3K, Vps34 (also known as PIK3C3) [29], which acts independently of the class I PI3K/Akt pathway. The IRE1α-JNK pathway is yet another regulator of autophagy and is activated during ER stress [30]. Eukaryotic cells deal with ER stress via the unfolded protein response, involving the induction of molecular chaperones, translation attenuation, ER-associated degradation, and the activation of autophagy, which are important for cell survival [30]. Cell homeostasis depends on the balance between the activation and inhibition of autophagy, and the signaling pathways that converge to control this dynamic process are complex and expanding.
Non-Canonical Autophagy
Canonical autophagy requires the sequential activity of defined molecular complexes, whose roles and importance have been defined [9]. However, recent evidence has revealed that formation of functional autophagosomes can bypass certain steps, eliminating the use of particular molecular autophagy complexes [9]. LC3 conversion still occurs in ulk1/(atg1) and ulk2 knockout mouse embryonic fibroblasts (MEFs) [31], and at a reduced level in rb1cc1/fip200(atg17) knockout MEFs [32], suggesting that the ULK1 complex is not essential for activation of the LC3 conjugation machinery and that the autophagy cascade can be initiated in an ULK1-independent manner [13]. Atg5- and Atg7-independent autophagy has also been recently described, which is not associated with LC3 processing and appears to specifically involve formation of autophagosomes from late endosomes and the trans-Golgi network [33]. The discovery of alternative, non-canonical autophagy pathways argues that autophagic processes are more diverse than initially anticipated. Depending on the stimuli and cellular context, only a subset of autophagy-related functional complexes may be activated at one time to degrade and recycle material.
Xenophagic capture of invading bacteria
Xenophagy is an autophagic process that specifically targets bacterial, viral and protozoan parasites for destruction [34]. Key aspects of xenophagy rely on sensing invading microorganisms and directing them towards autophagic degradation [35]. Mammalian cells express a variety of cell surface and cytosolic pattern recognition receptors (PRR), such as Toll-like receptors (TLRs) or cytosolic nucleotide-binding oligomerization domains, (NOD)-like receptors (NLRs) to detect invading pathogens [36]. These PRRs recognize specific bacterial pathogen-associated molecular patterns (PAMPs), including lipopolysaccharides (LPS), peptidoglycan, flagella and bacterial DNA [37], and likely play important roles in initiating xenophagy. Bacterial PAMPs also activate inflammasome components, which coordinate autophagy and pyroptosis to control infection [38,39]. Galectin 8 is a cytosolic lectin that acts as a versatile danger receptor that restricts bacterial proliferation by binding NDP52 and activating antibacterial autophagy [40]. Upon their detection, intracellular microbes undergo surface polyubiquitination, which is in turn recognized by various adaptor proteins or autophagy receptors, such as p62/SQSTM1, NDP52 and optineurin, which mediate selective autophagy of ubiquitinated pathogens by binding LC3 and recruiting the autophagic machinery [41-43]. Despite the importance of pathogen tagging via ubiquitination in the xenophagic process, the actual microbial targets of ubiquitination are unknown, and LRSAM1 is the only E3 ubiquitin ligase known so far to specifically target bacteria and contribute to xenophagy [44]. Given the antimicrobial function of xenophagy, it is not surprising that many intracellular bacterial pathogens have evolved to prevent or interrupt this cytosolic innate immune defense mechanism, by either preventing their detection or delivery to degradative lysosomes [45].
The Intracellular Niches of Francisella spp. and Brucella spp
Bacteria of the Francisella and Brucella genera include zoonotic pathogens of global importance, which cause tularemia and brucellosis, respectively. Francisella spp. and Brucella spp. are Gram-negative facultative intracellular bacteria that have evolved elaborate mechanisms to infect various eukaryotic cell types and grow within specific intracellular niches. The genus Francisella includes several species, such as F. tularensis, F. novicida, F. noatunensis, and F. philomiragia, which differ in virulence in humans and animals. Human tularemia is caused predominantly by F. tularensis [46]. Two subspecies of F. tularensis, subsp. tularensis and subsp. holarctica, display distinct geographical distribution and disease severity [47]. F. tularensis subspecies tularensis is the most virulent subspecies and causes a lethal disease in humans after exposure to as few as 10 inhaled bacteria [48]. Bacteria of the genus Brucella infect a variety of mammals, among which Brucella abortus, Brucella melitensis and Brucella suis can cause disease in humans [49]. Brucellosis is the most common bacterial zoonotic disease worldwide [50], with over a half-million reported human cases annually. Brucella spp. have a particular tropism for the reproductive system in animals, often leading to abortion and sterility [51]. In both humans and animals, brucellosis can become chronic with persistent relapses [52].
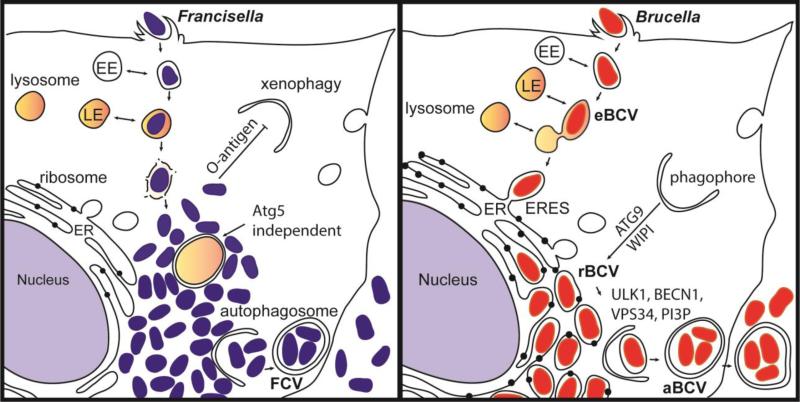
Francisella species grow efficiently in the cytosol of a plethora of cell types and in a diverse array of organisms [53]. Following internalization, Francisella spp. initially resides in a vacuole that sequentially acquires markers of early and late endosomes [54] (Figure 3). Francisella tularensis escape its original phagosome before lysosomal fusion to reach the cytosol where it undergoes rapid replication, a hallmark of its intracellular life cycle [55]. The Francisella pathogenicity island (FPI), a cluster of genes that share homology with type VI secretion systems [56], is essential for intracellular growth and escape from the vacuole [56,57]. The FPI-encoded apparatus is thought to deliver effector proteins into host cells that may modulate host functions to allow Francisella spp. to evade lysosomal fusion and escape from the vacuole, yet direct evidence of bacterial effectors playing a role in these critical steps of the intracellular lifecycle is lacking.
Figure 3. Intracellular niches of Francisella spp. and Brucella spp.
(A) Following internalization, Francisella resides in a vacuole that sequentially acquires markers of early endosomes (EE) and late endosomes (LE). Francisella escapes the original vacuole before lysosomal fusion to reach the cytosol. Once in the cytosol, the F. tularensis capsular and lipopolysaccharide O-antigen is an essential component of xenophagy avoidance. Francisella replicates rapidly in the cytosol of host cells and is found adjacent to autophagosomes. F. tularensis acquires amino acids generated via autophagy in an Atg5 independent manner. After extensive replication in murine macrophages, some bacteria are found in Francisella-containing vacuoles (FCVs). (B) Brucella exploits functions of the host secretory pathway to grow within membrane bound compartments called Brucella-containing vacuoles (BCV) throughout its intracellular life cycle. Brucella first traffics along the endocytic pathway in an endosomal BCV (eBCV), which acquires early endosomal (EE) markers then late endosomal (LE) markers, and partially fuses with lysosomes. Brucella then accesses the secretory pathway via interactions with endoplasmic reticulum (ER) exit sites (ERES) to access the ER. B. abortus requires the autophagy proteins ATG9 and WIPI, for biogenesis of the replicative BCV (rBCV) derived from ER membranes. After extensive replication, some rBCV are engulfed by autophagosome-like structures to become autophagic BCVs (aBCVs), which are involved in bacterial release and cell-to-cell spread. B. abortus selectively subverts several autophagy initiation complexes containing ULK1, BECN1, VPS34 and PI3P to generate an autophagic vacuole important for egress and cell-to-cell spread.
Unlike Francisella spp., Brucella spp. remain within a membrane bound compartment, the Brucella-containing vacuole (BCV), and exploit functions of the host secretory pathway to convert its original, endosomal BCV (eBCV) into a replicative BCV (rBCV) that is derived form the host ER (Figure 3). One virulence factor necessary for Brucella spp. to generate the rBCV, grow intracellularly and establish infection is the VirB type IV secretion system (T4SS) [58,59], a membrane-spanning apparatus that delivers effector proteins into the host cell [60-63]. Protein effectors are thought to modulate host functions to promote rBCV biogenesis. Upon internalization, Brucella spp. traffics along the endocytic pathway in BCVs that acquire markers of early then late endosomes such as the lysosomal glycoprotein LAMP-1 [64-67]. The BCV partially fuses with lysosomes, consistent with the role of the lysosomal small GTPase Rab7 in BCV maturation [67], but subsequently excludes endosomal markers from the BCV membrane, via a process initiated when the BCV intersects with the secretory pathway at endoplasmic reticulum exit sites (ERES) [68]. The small GTPase Sar1 is required for the association between the BCVs and ERES [68], consistent with its role in ERES integrity and functions. Rab2, a small GTPase that controls Golgi to ER transport, is also required for rBCV biogenesis [69]. The Brucella VirB type IV secretion system (T4SS) is required for sustained interactions between BCVs and ERES, which promotes accretion of ER-derived membranes, culminating in the biogenesis of the ER-derived, replication-permissive rBCV [68].
Hence, both Francisella spp. and Brucella spp. have evolved mechanisms to reach or generate a specific intracellular niche, the cytosol in the case of Francisella spp., and the ER-derived rBCV in the case of Brucella spp. Each specific environment provides Francisella spp. and Brucella spp. with replication-permissive conditions that support their intracellular proliferation (Figure 3).
Francisella avoidance of xenophagy
Cytosolic replication places Francisella spp. in an ideal location for autophagic recognition and capture. Yet, these bacteria undergo unrestricted replication, suggesting they can avoid xenophagy. Consistently, the highly virulent strain F. tularensis subsp. tularensis SchuS4 avoids autophagic recognition and capture, ascytosolic SchuS4 does not become ubiquitinated and less than 5% is recognized by the autophagy receptors p62/SQSTM1 and NBR1, suggesting SchuS4 prevents ubiquitin tagging and impairs recruitment of the autophagic machinery [70]. However, another study showed early (1 hr pi) and transient recruitment of p62/SQSTM1 and LC3 on intracellular Francisella tularensis subsp. holarctica live vaccine strain (LVS) [71], an attenuated strain of Francisella [72]. The inability of LVS to prevent early recruitment of autophagic machinery may relate to the strain's attenuation. While wild type SchuS4 bacteria are not targeted by autophagy, various replication-deficient mutants of SchuS4 that die in the cytosol are eventually tagged with ubiquitin and captured into autophagosomes in an Atg5-, LC3- and p62/SQSTM1-dependent manner [70], further suggesting that F. tularensis actively avoids xenophagic recognition. Additionally, autophagy-related genes including BECN1, ATG5, ATG12, ATG16L1, ATG7 and ATG4a are down regulated at 24 hours post infection in human monocytes with the virulent strain F. tularensis SchuS4 and the less virulent strain F. novicida [73,74]. A recent study proposed a mechanism of xenophagic avoidance by F. tularensis [75]. A library of F. tularensis subsp. tularensis transposon mutant in GFP-LC3-expressing murine macrophages was screened by fluorescence microscopy and 11 mutants were identified that are rapidly captured by autophagy (by 6 hours post infection) [75]. These mutants all grouped within four genetic loci involved in lipopolysaccharide and capsular O-antigen biosynthesis, indicating that F. tularensis surface O-antigen prevents xenophagic recognition and clearance [75]. These findings suggest that F. tularensis polysaccharidic O-antigen could act as a shield on cytosolic bacteria, preventing their detection and tagging of PAMPs that would otherwise activate the xenophagic cascade.
Mechanisms of Autophagy Exploitation by Francisella
F. tularensis replicates up to 1000 fold in the cytosol of infected cells within 24 hours [76-78]. To achieve such rapid intracellular growth, these bacteria must efficiently scavenge energy and nutrients from the host cell, in addition to evading cytosolic surveillance systems such as xenophagy. Steele et al. recently found that F. tularensis infection induces autophagy through an Atg5-independent pathway that provides amino acids and bulk carbon to the bacterium [76]. F. tularensis infection of human monocytes also leads to a decrease in expression of the PI3K/Akt pathway, which inhibits pro-inflammatory gene transcription [73], in addition to activating autophagy. Supplementation of non-essential amino acids or pyruvate during infection rescued F. tularensis growth during autophagy inhibition [76], demonstrating that amino acids scavenged from autophagy are used for protein synthesis and are metabolized as a major carbon source [76]. Consistently, F. tularensis is commonly found adjacent to autophagosomes during their replication in mouse embryonic fibroblasts (MEFs) and J774A.1 mouse macrophage-like cells [76]. Hence, Francisella spp. can avoid xenophagic detection early during infection, and use autophagy to acquire amino acids during replication. Additionally, bacteria were observed late during infection in large autophagic Francisella-containing vacuoles (FCVs) in murine BMMs, following extensive cytosolic replication [79]. FCV formation was blocked by chloramphenicol, suggesting bacterial proteins are responsible for this process [79]. Yet, FCV formation is specific to mouse cells and does not occur in human macrophages [80,81], so it remains unclear whether the FCV plays a role in the intracellular cycle of Francisella spp. Future studies addressing which bacterial factors modulate autophagy will be important in further defining how Francisella takes advantage of the autophagic process for nutritional purposes.
Mechanisms of Autophagy Exploitation by Brucella
Pizarro et al. originally reported structural features of BCVs [82] that were consistent with autophagosomes in HeLa epithelial cells infected with B. abortus for 24 hours. This led to the proposal that autophagy was required for the biogenesis of the replicative BCVs early during infection [65,82]. However the marker monodansylcadaverine (MDC) used in these early studies to identify autophagic vacuoles lacks specificity, as it also accumulates in lysosomes and does not colocalize with the autophagic marker LC3 [83]. Additional studies also found that MDC does not accumulate in BCVs during the initial 24 hours of infection with B. abortus in either bone marrow derived macrophages (BMMs) or J774A.1 macrophage-like cells [66,84]. Based on the use of LC3-II recruitment to membranes, a more specific marker for autophagosome formation [8], subsequent studies found that BCVs do not associate with LC3-II [85]. B. abortus or B. melitensis infections do not increase LC3-II protein levels [86], arguing against autophagy induction during infection. Despite these early inconsistencies, additional studies have recently provided a potential link between autophagy and Brucella infection, via activation of one arm of the unfolded protein response (UPR) [87-90]. The UPR aims to restore ER homeostasis upon ER stress through tight control of transcription of genes involved in promoting ER functions, including lipid synthesis, ER-associated degradation (ERAD) and protein chaperones [91]. UPR pathways are controlled by three ER-associated receptors that sense ER stress, (1) inositol-requiring enzyme-1 (IRE-1α), (2) protein kinase RNA-like ER kinase (PERK) and (3) activating transcription factor 6 (ATF6) [92]. Autophagy is also activated upon ER stress to restore homeostasis and promote cell survival [30], and accumulating evidence suggests that UPR-dependent autophagy can also be beneficial for some intracellular pathogens during infection [93]. Smith et al. recently proposed that the UPR is required for B. melitensis replication, since it was induced upon B. melitensis infection and treatment of macrophages with tauroursodeoxycholic acid (TUDCA), a pharmacologic chaperone that counteracts the UPR by assisting protein folding, decreased B. melitensis growth [88]. However, B. suis replication was significantly inhibited at 24 hours post infection in goat trophoblasts in which ER stress was induced using tunicamycin, but restored with the addition of a pharmacologic chaperone 4 phenyl butyric acid (4-PBA) or expression of the chaperone GRP78, which both alleviate ER stress-induced apoptosis [94]. These conflicting results may reflect cell type differences but will need to be reconciled to unequivocally establish the effect of the UPR on Brucella replication. It is also unclear whether Brucella infection triggers a complete or partial UPR response. B. abortus and B. suis only activate IRE1α [87,94], which has been linked to host cell sensing of T4SS secretion [87], while B. melitensis induces ER stress by triggering all three arms of the UPR (IRE1α, PERK and ATF6-dependent pathways) via the secreted effector TcpB [88]. A number of additional effectors including BspC, BspG, BspH and BspK also trigger ER stress when expressed in epithelial cells, suggesting that the mechanisms of UPR induction by B. abortus are complex and involve a growing list of bacterial and host proteins [63]. There is however consensual support that IRE1α is activated by Brucella infection in macrophages, trophoblasts and epithelial cells [87-89,94]. IRE1α is a transmembrane kinase that regulates the UPR, which plays an important role in cell survival after ER stress, and also promotes ER-associated autophagosome biogenesis [30]. B. abortus replication is attenuated in both IRE1α-depleted Drosophila melanogaster S2 cells and IRE1α knockout (KO) murine embryonic fibroblasts (MEFs) [90]. Taguchi et al. (2015) detected IRE1α activation as early as 4 hours post-infection [89], suggesting a role of this UPR component early during the Brucella intracellular cycle. IRE1α activation occurs via the host protein Yip1a, which binds to and phosphorylates IRE1α, triggering XBP-1 dependent transcription [89]. Yip1a activation of IRE1α was associated with an up-regulation of the GTPase Sar1 and subunits of the COPII coat complex [89], which are essential components of ERES and early secretory trafficking, and required for rBCV biogenesis [68]. While it is unclear how up-regulation of COPII components and the GTPase Sar1 benefit Brucella, it may enhance ERES functions and consequently increase ER-derived vesicle budding to promote ER membrane acquisition for rBCV biogenesis. In support of this hypothesis, Yip1A activation of IRE1α triggers formation of large vacuoles, which is dependent on the autophagy associated proteins ATG9 and WIPI [89]. Depletion of either ATG9 or WIPI impairs rBCV biogenesis, suggesting that B. abortus hijacks these components of the autophagic cascade to promote rBCV biogenesis [89].
In addition to the subversion of autophagy components for rBCV biogenesis, B. abortus selectively subverts other components of the autophagic cascade for egress and cell-to-cell spread during late stages of infection [85]. After extensive replication in the ER, some rBCVs are engulfed by autophagosome-like structures and converted into multimembrane vacuoles called aBCVs, for autophagic BCVs [85]. Formation of these newly discovered aBCVs does not however require canonical autophagy, as the elongation complexes including ATG5, ATG16L1, ATG4B, ATG7 and LC3B are not recruited during, or required for, aBCV formation [85]. Biogenesis of the aBCV however requires BECN1, ATG14L and ULK1, three autophagy-associated proteins involved in autophagy initiation. In addition, PI3P was detected on the aBCV [85], consistent with the role of a class III PI3K activity associated with the BECN1-VPS34 autophagy initiation complex, and treatment of infected cells with 3-MA, an inhibitor of class III PI3K, decreased aBCV formation [85]. Additionally, miRNA expression profiles of the autophagy genes ampk, ulk3 and vps34 are altered in late stages of Brucella-infected macrophages [95], suggesting that the bacterium may alter expression of autophagy-related genes it subverts for aBCV formation. However, additional studies are required to substantiate possible links between aBCV formation and these observations. These recent findings support the concept that Brucella subverts a subset of molecular machineries associated with autophagy at a late stage of infection to promote spread to neighboring cells, and may typify pathogenic mechanisms employed by other microorganisms to modulate specific stages of their intracellular cycle. Altogether, these studies reveal a picture of subversion of discrete autophagy-associated membrane trafficking processes by Brucella, whereby the bacterium likely uses a given subset of autophagy associated proteins to acquire ER derived membranes for rBCV biogenesis, and coopts another subset of autophagic proteins to spread form cell-to-cell, in processes that functionally differ from canonical autophagy. Further understanding of these steps of the Brucella intracellular cycle requires the identification and characterization of the bacterial factors that modulate autophagy-associated processes.
Concluding Remarks and Perspectives
Autophagy research has expanded into innate immunity, driven by contemporary recognition of its significance during bacterial infection [45]. Brucella and Francisella are two examples of bacterial pathogens that have evolved unique mechanisms to avoid xenophagy and also exploit selective autophagic machineries to promote their intracellular cycles. The O-antigen polysaccharide of cytosolic Francisella could act as a shield, preventing xenophagic detection [75]. Atg5-independent autophagy is essential for both Francisella nutrient acquisition during replication [76] and for Brucella vacuole acquisition for aBCV biogenesis and egress [85], although it is unclear whether the same non-canonical processes are invoked on both cases. Additionally, Brucella hijacks the autophagy components ATG9 and WIPI to generate the replication-permissive rBCV [89]. A number of intracellular pathogens have been shown to hijack a select set of autophagic components [76,86,96-98]. The next decade of research needs to uncover the microbial effectors responsible for hijacking autophagic components and their mechanisms of action need to be characterized. Limited studies on Coxiella burnetii, Helicobacter pylori and Anaplasma phagocytophilum have begun to uncover such bacterial effectors [93,99-101]. The C. burnetii T4SS effector, Cig2, recruits LC3 to the parasitophoruous vacuole (PV) to aid in maintenance of the PV membrane [101], while A. phagocytophilum T4SS effector Ats-1 binds BECN1 and recruits Atg14L to promote autophagosome nucleation at the bacterial vacuole membrane [99]. Brucella requires BECN1 for aBCV formation [85], so future directions should examine whether the bacterium expresses effectors that directly target this autophagic protein.
Discovering what host factors are involved in phagophore elongation in the absence of the ubiquitin-like conjugation systems during Brucella and Francisella infection will provide important insight into non-canonical autophagic processes. The Atg17/31/29 complex involved in autophagy initiation and found at the phagophore assembly site, is also involved in autophagosome closure [102]. It will be interesting to determine if this complex is required during Francisella and Brucella infections for autophagosome closure in lieu of the Atg5 and LC3 ubiquitin-like conjugation systems. Understanding how pathogens hijack specific non-canonical autophagy machineries may uncover targets for disease prevention, while preserving functions of the canonical autophagy pathway for maintenance of eukaryotic cellular homeostasis. It will therefore be important to expand our knowledge on how autophagosome formation occurs when specific autophagy complexes are bypassed, and whether bacterial effectors drive these processes. Both Brucella and Francisella are useful tools to understand pathogenic manipulation of autophagy, and future research should be directed at resolving the many mechanisms by which intracellular pathogens exploit autophagy.
Highlights.
- Background Contents
- ○ Xenophagy is a host process of pathogen clearance, which is hijacked by Francisella spp. and Brucella spp. for replication purposes and cell-to-cell spread, respectively.
- Conceptual Advances
- ○ F. tularensis capsular and lipopolysaccharide O-antigen is an essential component of xenophagy avoidance.
- ○ F. tularensis acquires amino acids generated via autophagy to grow unrestricted in the host cell cytosol.
- ○ B. abortus requires the autophagy proteins ATG9 and WIPI, for biogenesis of the replicative vacuole.
- ○ B. abortus selectively subverts several autophagy initiation complexes to generate an autophagic vacuole important for egress and cell-to-cell spread.
- Perspectives
- ○ The molecular mechanisms by which Francisella spp. and Brucella spp modulate their interactions with autophagic machinery pathway need to be elucidated.
Acknowledgments
We apologize to colleagues whose work could not be cited due to space limitation. We are grateful to Erin Smith and Jessica Klein for providing critical editing of the manuscript and the figures. This work was supported by the NIH/NIAID grant AI112649, the USDA/NIFA Postdoctoral Fellowship 2016-67012-25179 and the Paul G. Allen School for Global Animal Health.
Abbreviations
- 3-MA
3-methyladenine
- AMPK
AMP-activating protein kinase
- ATF6
activating transcription factor 6
- ATG
autophagy-related genes
- BCV
Brucella-containing vacuole
- BECN1
Beclin1
- BMM
Bone marrow-derived macrophages
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum-associated degradation
- ERES
endoplasmic reticulum exit sites
- FPI
Francisella pathogenicity island
- GABARAP
γ-aminobutyric acid receptor-associated protein
- GATE-16
Golgi-associated ATPase enhancer of 16 kDa
- LC3
microtubule-associated protein 1A/1B-light chain 3
- LPS
Lipopolysaccharide
- LVS
Francisella tularensis subspecies holarctica live vaccine strain
- MEF
mouse embryonic fibroblast cells
- mTOR
kinase mammalian target of rapamycin
- PAMP
Pathogen-associated molecular patterns
- PAS
phagophore assembly site
- PE
phosphatidylethanolamine
- PERK
protein kinase RNA-like ER kinase
- PI3K
phosphoinositide 3-kinase
- PI3P
phosphatidylinositol 3-phosphate
- PI
phosphatidylinositol
- pi
post infection
- PtdIns3K
phosphatidylinositol 3-kinase
- T4SS
Type IV Secretion System
- TLR
Toll-like receptor
- UPR
unfolded protein response
- UBL
ubiquitin-like protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Research. 2013;24:24–41. doi: 10.1038/cr.2013.168. doi:10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. doi:10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 3.Yang Z, Huang J, Geng J, Nair U, Klionsky DJ. Atg22 Recycles Amino Acids to Link the Degradative and Recycling Functions of Autophagy. Mol. Biol. Cell. 2006;17:5094–5104. doi: 10.1091/mbc.E06-06-0479. doi:10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. doi:10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nature Cell Biology. 2010;12:814–822. doi: 10.1038/ncb0910-814. doi:10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. doi:10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chua CEL, Bin Qi Gan BL. Tang, Involvement of members of the Rab family and related small GTPases in autophagosome formation and maturation. Cell. Mol. Life Sci. 2011;68:3349–3358. doi: 10.1007/s00018-011-0748-9. doi:10.1007/s00018-011-0748-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. doi:10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nature Reviews Molecular Cell Biology. 2012;13:7–12. doi: 10.1038/nrm3249. Published Online: 01 August 2001; | Doi:10.1038/35085062 doi:10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
- 10.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. doi:10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvo-Garrido J, King JS, Muñoz Braceras S, Escalante R. Vmp1 Regulates PtdIns3P Signaling During Autophagosome Formation in Dictyostelium discoideum. Traffic. 2014;15:1235–1246. doi: 10.1111/tra.12210. doi:10.1111/tra.12210. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. doi:10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong P-M, Puente C, Ganley IG, Jiang X. The ULK1 complex. Autophagy. 2013;9:124–137. doi: 10.4161/auto.23323. doi:10.4161/auto.23323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan EYW, Kir S, Tooze SA. siRNA Screening of the Kinome Identifies ULK1 as a Multidomain Modulator of Autophagy. J. Biol. Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. doi:10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 15.Mizushima N, Yoshimori T, Ohsumi Y. The Role of Atg Proteins in Autophagosome Formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. doi:10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 16.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–776. doi: 10.4161/auto.6.6.12709. doi:10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dall'Armi C, Devereaux KA, Di Paolo G. The Role of Lipids in the Control of Autophagy. Current Biology. 2013;23:R33–R45. doi: 10.1016/j.cub.2012.10.041. doi:10.1016/j.cub.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nature Cell Biology. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. doi:10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 19.Wu Y-T, Tan H-L, Shui G, Bauvy C, Huang Q, Wenk MR, et al. Dual Role of 3-Methyladenine in Modulation of Autophagy via Different Temporal Patterns of Inhibition on Class I and III Phosphoinositide 3-Kinase. J. Biol. Chem. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. doi:10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuma A. Formation of the 350-kDa Apg12-Apg5middle dotApg16 Multimeric Complex, Mediated by Apg16 Oligomerization, Is Essential for Autophagy in Yeast. J. Biol. Chem. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. doi:10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- 21.Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 2011 doi: 10.1186/gb-2011-12-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sou YS, Waguri S, Iwata JI, Ueno T, Fujimura T, Hara T, et al. The Atg8 Conjugation System Is Indispensable for Proper Development of Autophagic Isolation Membranes in Mice. Mol. Biol. Cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. doi:10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lippai M, Low P. The Role of the Selective Adaptor p62 and Ubiquitin-Like Proteins in Autophagy. BioMed Research International. 2014;2014:1–11. doi: 10.1155/2014/832704. doi:10.1155/2014/832704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gozuacik D, Kimchi A. Autophagy and Cell Death. Elsevier; 2007. pp. 217–245. doi:10.1016/S0070-2153(06)78006-1. [DOI] [PubMed] [Google Scholar]
- 25.Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12:1509–1518. doi: 10.1038/sj.cdd.4401751. doi:10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- 26.Stephan JS, Herman PK. The Regulation of Autophagy in Eukaryotic Cells: Do All Roads Pass through Atg1? Autophagy. 2006;2:146–148. doi: 10.4161/auto.2.2.2485. doi:10.4161/auto.2.2.2485. [DOI] [PubMed] [Google Scholar]
- 27.Heras-Sandoval D, Pérez-Rojas JM, Hernández-Damián J, Pedraza-Chaverri J. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cellular Signalling. 2014;26:2694–2701. doi: 10.1016/j.cellsig.2014.08.019. doi:10.1016/j.cellsig.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 28.Bhaskar PT, Hay N. The Two TORCs and Akt. Developmental Cell. 2007;12:487–502. doi: 10.1016/j.devcel.2007.03.020. doi:10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 29.Jaber N, Dou Z, Chen J-S, Catanzaro J, Jiang Y-P, Ballou LM, et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proceedings of the National Academy of Sciences. 2012;109:2003–2008. doi: 10.1073/pnas.1112848109. doi:10.1073/pnas.1112848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogata M, Hino S-I, Saito A, Morikawa K, Kondo S, Kanemoto S, et al. Autophagy Is Activated for Cell Survival after Endoplasmic Reticulum Stress. Mol. Cell. Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. doi:10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proceedings of the National Academy of Sciences. 2011;108:11121–11126. doi: 10.1073/pnas.1107969108. doi:10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hara T, Takamura A, Kishi C, Iemura S-I, Natsume T, Guan J-L, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. doi:10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. doi:10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 34.Bauckman KA, Owusu-Boaitey N, Mysorekar IU. Selective autophagy: Xenophagy. Methods. 2015;75:120–127. doi: 10.1016/j.ymeth.2014.12.005. doi:10.1016/j.ymeth.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knodler LA, Celli J. Eating the strangers within: host control of intracellular bacteria via xenophagy. Cellular Microbiology. 2011;13:1319–1327. doi: 10.1111/j.1462-5822.2011.01632.x. doi:10.1111/j.1462-5822.2011.01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y, Jagannath C, Liu X-D, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like Receptor 4 Is a Sensor for Autophagy Associated with Innate Immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. doi:10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. International Immunology. 2009;21:317–337. doi: 10.1093/intimm/dxp017. doi:10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joshi AD, Swanson MS. Secrets of a Successful Pathogen: Legionella Resistance to Progression Along the Autophagic Pathway. Frontiers in Microbiology. 2011;2 doi: 10.3389/fmicb.2011.00138. doi:10.3389/fmicb.2011.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dubuisson J-F, Swanson MS. Mouse infection by Legionella, a Model to Analyze Autophagy. Autophagy. 2014;2:179–182. doi: 10.4161/auto.2831. doi:10.4161/auto.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thurston TLM, Wandel MP, von Muhlinen N, Foeglein Á, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. doi:10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perrin AJ, Jiang X, Birmingham CL, So NSY, Brumell JH. Recognition of Bacteria in the Cytosol of Mammalian Cells by the Ubiquitin System. Current Biology. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. doi:10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 42.Thurston TLM, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nature Immunology. 2009;10:1215–1221. doi: 10.1038/ni.1800. doi:10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 43.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, et al. Phosphorylation of the Autophagy Receptor Optineurin Restricts Salmonella Growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. doi:10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huett A, Heath RJ, Begun J, Sassi SO, Baxt LA, Vyas JM, et al. The LRR and RING Domain Protein LRSAM1 Is an E3 Ligase Crucial for Ubiquitin-Dependent Autophagy of Intracellular Salmonella Typhimurium. Cell Host & Microbe. 2012;12:778–790. doi: 10.1016/j.chom.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winchell CG, Steele S, Kawula T, Voth DE. Dining in: intracellular bacterial pathogen interplay with autophagy. Current Opinion in Microbiology. 2016;29:9–14. doi: 10.1016/j.mib.2015.09.004. doi:10.1016/j.mib.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Champion MD, Zeng Q, Nix EB, Nano FE, Keim P, Kodira CD, et al. Comparative Genomic Characterization of Francisella tularensis Strains Belonging to Low and High Virulence Subspecies. PLoS Pathog. 2009;5:e1000459. doi: 10.1371/journal.ppat.1000459. doi:10.1371/journal.ppat.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larsson P, Oyston PCF, Chain P, Chu MC, Duffield M, Fuxelius H-H, et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nature Genetics. 2005;37:153–159. doi: 10.1038/ng1499. doi:10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- 48.SASLAW S, EIGELSBACH HT, PRIOR JA, WILSON HE, CARHART S. Tularemia Vaccine Study: II. Respiratory Challenge. Arch Intern Med. 1961;107:702–714. doi: 10.1001/archinte.1961.03620050068007. doi:10.1001/archinte.1961.03620050068007. [DOI] [PubMed] [Google Scholar]
- 49.Chain PSG, Comerci DJ, Tolmasky ME, Larimer FW, Malfatti SA, Vergez LM, et al. Whole-Genome Analyses of Speciation Events in Pathogenic Brucellae. Infect. Immun. 2005;73:8353–8361. doi: 10.1128/IAI.73.12.8353-8361.2005. doi:10.1128/IAI.73.12.8353-8361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pappas G. The peculiar ways of Brucella survival: Looking through the keyhole. Virulence. 2010 doi: 10.4161/viru.1.6.13200. [DOI] [PubMed] [Google Scholar]
- 51.Bargen K, Gorvel J-P, Salcedo SP. Internal affairs: investigating the Brucella intracellular lifestyle. FEMS Microbiology Reviews. 2012;36:533–562. doi: 10.1111/j.1574-6976.2012.00334.x. doi:10.1111/j.1574-6976.2012.00334.x. [DOI] [PubMed] [Google Scholar]
- 52.Franco MP, Mulder M, Smits HL. Persistence and relapse in brucellosis and need for improved treatment. Trans R Soc Trop Med Hyg. 2007;101:854–855. doi: 10.1016/j.trstmh.2007.05.016. doi:10.1016/j.trstmh.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 53.Hall JD, Woolard MD, Gunn BM, Craven RR, Taft-Benz S, Frelinger JA, et al. Infected-Host-Cell Repertoire and Cellular Response in the Lung following Inhalation of Francisella tularensis Schu S4, LVS, or U112. Infect. Immun. 2008;76:5843–5852. doi: 10.1128/IAI.01176-08. doi:10.1128/IAI.01176-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clemens DL, Lee BY, Horwitz MA. Virulent and Avirulent Strains of Francisella tularensis Prevent Acidification and Maturation of Their Phagosomes and Escape into the Cytoplasm in Human Macrophages. Infect. Immun. 2004;72:3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004. doi:10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Celli J, Zahrt TC. Mechanisms of Francisella tularensis Intracellular Pathogenesis. Cold Spring Harb Perspect Med. 2013;3:a010314–a010314. doi: 10.1101/cshperspect.a010314. doi:10.1101/cshperspect.a010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nano FE, Zhang N, Cowley SC, Klose KE, Cheung KKM, Roberts MJ, et al. A Francisella tularensis Pathogenicity Island Required for Intramacrophage Growth. J. Bacteriol. 2004;186:6430–6436. doi: 10.1128/JB.186.19.6430-6436.2004. doi:10.1128/JB.186.19.6430-6436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barker JR, Chong A, Wehrly TD, Yu JJ, Rodriguez SA, Liu J, et al. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Molecular Microbiology. 2009;74:1459–1470. doi: 10.1111/j.1365-2958.2009.06947.x. doi:10.1111/j.1365-2958.2009.06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sieira R, Comerci DJ, Sánchez DO, Ugalde RA. A Homologue of an Operon Required for DNA Transfer in Agrobacterium Is Required in Brucella abortusfor Virulence and Intracellular Multiplication. J. Bacteriol. 2000;182:4849–4855. doi: 10.1128/jb.182.17.4849-4855.2000. doi:10.1128/JB.182.17.4849-4855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli ML, Bourg G, Foulongne V, et al. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Molecular Microbiology. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. doi:10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 60.De Jong MF, Sun YH, Den Hartigh AB, Van Dijl JM, Tsolis RM. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Molecular Microbiology. 2008;70:1378–1396. doi: 10.1111/j.1365-2958.2008.06487.x. doi:10.1111/j.1365-2958.2008.06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marchesini MI, Herrmann CK, Salcedo SP, Gorvel J-P, Comerci DJ. In search of Brucella abortus type IV secretion substrates: screening and identification of four proteins translocated into host cells through VirB system. Cellular Microbiology. 2011;13:1261–1274. doi: 10.1111/j.1462-5822.2011.01618.x. doi:10.1111/j.1462-5822.2011.01618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Barsy M, Jamet A, Filopon D, Nicolas C, Laloux G, Rual JF, et al. Identification of a Brucella spp. secreted effector specifically interacting with human small GTPase Rab2. Cellular Microbiology. 2011;13:1044–1058. doi: 10.1111/j.1462-5822.2011.01601.x. doi:10.1111/j.1462-5822.2011.01601.x. [DOI] [PubMed] [Google Scholar]
- 63.Myeni S, Child R, Ng TW, Kupko JJ, III, Wehrly TD, Porcella SF, et al. Brucella Modulates Secretory Trafficking via Multiple Type IV Secretion Effector Proteins. PLoS Pathog. 2013;9:e1003556. doi: 10.1371/journal.ppat.1003556. doi:10.1371/journal.ppat.1003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Comerci DJ, Martínez Lorenzo MJ, Sieira R, Gorvel J-P, Ugalde RA. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cellular Microbiology. 2001;3:159–168. doi: 10.1046/j.1462-5822.2001.00102.x. doi:10.1046/j.1462-5822.2001.00102.x. [DOI] [PubMed] [Google Scholar]
- 65.Pizarro-Cerda J, Méresse S, Parton RG, van der Goot G, Sola-Landa A, Lopez-Goñi I, et al. Brucella abortus Transits through the Autophagic Pathway and Replicates in the Endoplasmic Reticulum of Nonprofessional Phagocytes. Infect. Immun. 1998;66:5711–5724. doi: 10.1128/iai.66.12.5711-5724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Celli J, de Chastellier C, Franchini D-M, Pizarro-Cerda J, Moreno E, Gorvel J-P. Brucella Evades Macrophage Killing via VirB-dependent Sustained Interactions with the Endoplasmic Reticulum. J Exp Med. 2003;198:545–556. doi: 10.1084/jem.20030088. doi:10.1084/jem.20030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Starr T, Ng TW, Wehrly TD, Knodler LA, Celli J. Brucella Intracellular Replication Requires Trafficking Through the Late Endosomal/Lysosomal Compartment. Traffic. 2008;9:678–694. doi: 10.1111/j.1600-0854.2008.00718.x. doi:10.1111/j.1600-0854.2008.00718.x. [DOI] [PubMed] [Google Scholar]
- 68.Celli J, Salcedo SP, Gorvel J-P. Brucella coopts the small GTPase Sar1 for intracellular replication. Pnas. 2005;102:1673–1678. doi: 10.1073/pnas.0406873102. doi:10.1073/pnas.0406873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fugier E, Salcedo SP, de Chastellier C, Pophillat M, Muller A, Arce-Gorvel V, et al. The Glyceraldehyde-3-Phosphate Dehydrogenase and the Small GTPase Rab 2 Are Crucial for Brucella Replication. PLoS Pathog. 2009;5:e1000487. doi: 10.1371/journal.ppat.1000487. doi:10.1371/journal.ppat.1000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chong A, Wehrly TD, Child R, Hansen B, Hwang S, Virgin HW, et al. Cytosolic clearance of replication-deficient mutants reveals Francisella tularensisinteractions with the autophagic pathway. Autophagy. 2014;8:1342–1356. doi: 10.4161/auto.20808. doi:10.4161/auto.20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Härtlova A, Link M, Balounova J, Benesova M, Resch U, Straskova A, et al. Quantitative Proteomics Analysis of Macrophage-Derived Lipid Rafts Reveals Induction of Autophagy Pathway at the Early Time of Francisella tularensis LVS Infection. Journal of Proteome Research. 2014;13:796–804. doi: 10.1021/pr4008656. doi:10.1021/pr4008656. [DOI] [PubMed] [Google Scholar]
- 72.Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, et al. Tularemia as a Biological Weapon: Medical and Public Health Management. Jama. 2001;285:2763–2773. doi: 10.1001/jama.285.21.2763. doi:10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- 73.Butchar JP, Cremer TJ, Clay CD, Gavrilin MA, Wewers MD, Marsh CB, et al. Microarray Analysis of Human Monocytes Infected with Francisella tularensis Identifies New Targets of Host Response Subversion. PLoS ONE. 2008;3:e2924. doi: 10.1371/journal.pone.0002924. doi:10.1371/journal.pone.0002924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cremer TJ, Amer AO, Tridandapani S, Butchar JP. Francisella tularensis regulates autophagy-related host cell signaling pathways. Autophagy. 2009;5:125–128. doi: 10.4161/auto.5.1.7305. doi:10.4161/auto.5.1.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Case E, Chong A, Wehrly TD, Hansen B. The Francisella O-antigen mediates survival in the macrophage cytosol via autophagy avoidance. Cellular. 2014;16:862–877. doi: 10.1111/cmi.12246. doi:10.1111/cmi.12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Steele S, Brunton J, Ziehr B, Taft-Benz S, Moorman N, Kawula T. Francisella tularensis Harvests Nutrients Derived via ATG5-Independent Autophagy to Support Intracellular Growth. PLoS Pathog. 2013;9:e1003562. doi: 10.1371/journal.ppat.1003562. doi:10.1371/journal.ppat.1003562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oyston PCF, Sjöstedt A, Titball RW. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat Rev Micro. 2004;2:967–978. doi: 10.1038/nrmicro1045. doi:10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- 78.Anthony LD, Burke RD, Nano FE. Growth of Francisella spp. in rodent macrophages. Infect. Immun. 1991;59:3291–3296. doi: 10.1128/iai.59.9.3291-3296.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Pnas. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. doi:10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Akimana C, Al-Khodor S, Kwaik YA. Host Factors Required for Modulation of Phagosome Biogenesis and Proliferation of Francisella tularensis within the Cytosol. PLoS ONE. 2010;5:e11025. doi: 10.1371/journal.pone.0011025. doi:10.1371/journal.pone.0011025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Edwards JA, Rockx-Brouwer D, Nair V, Celli J. Restricted cytosolic growth of Francisella tularensis subsp. tularensis by IFN-γ activation of macrophages. Microbiology. 2010;156:327–339. doi: 10.1099/mic.0.031716-0. doi:10.1099/mic.0.031716-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pizarro-Cerda J, Moreno E, Sanguedolce V, Mege J-L, Gorvel J-P. Virulent Brucella abortus Prevents Lysosome Fusion and Is Distributed within Autophagosome-Like Compartments. Infect. Immun. 1998;66:2387–2392. doi: 10.1128/iai.66.5.2387-2392.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mizushima N. Methods for monitoring autophagy. The International Journal of Biochemistry & Cell Biology. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. doi:10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 84.Arenas GN, Staskevich AS, Aballay A, Mayorga LS. Intracellular Trafficking of Brucella abortus in J774 Macrophages. Infect. Immun. 2000;68:4255–4263. doi: 10.1128/iai.68.7.4255-4263.2000. doi:10.1128/iai.68.7.4255-4263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Starr T, Child R, Wehrly TD, Hansen B, Hwang S, López-Otín C, et al. Selective Subversion of Autophagy Complexes Facilitates Completion of the Brucella Intracellular Cycle. Cell Host & Microbe. 2012;11:33–45. doi: 10.1016/j.chom.2011.12.002. doi:10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hamer I, Goffin E, De Bolle X, Letesson JJ, Jadot M. Replication of Brucella abortus and Brucella melitensis in fibroblasts does not require Atg5-dependent macroautophagy. BMC Microbiology. 2014;14:223. doi: 10.1186/s12866-014-0223-5. doi:10.1186/s12866-014-0223-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.De Jong MF, Starr T, Winter MG, Den Hartigh AB, Child R, Knodler LA, et al. Sensing of Bacterial Type IV Secretion via the Unfolded Protein Response. mBio. 2013;4:e00418–12–e00418–12. doi: 10.1128/mBio.00418-12. doi:10.1128/mBio.00418-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smith JA, Khan M, Magnani DD, Harms JS, Durward M, Radhakrishnan GK, et al. Brucella Induces an Unfolded Protein Response via TcpB That Supports Intracellular Replication in Macrophages. PLoS Pathog. 2013;9:e1003785. doi: 10.1371/journal.ppat.1003785. doi:10.1371/journal.ppat.1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taguchi Y, Imaoka K, Kataoka M, Uda A, Nakatsu D, Horii-Okazaki S, et al. Yip1A, a Novel Host Factor for the Activation of the IRE1 Pathway of the Unfolded Protein Response during Brucella Infection. PLoS Pathog. 2015;11:e1004747. doi: 10.1371/journal.ppat.1004747. doi:10.1371/journal.ppat.1004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qin Q-M, Pei J, Ancona V, Shaw BD, Ficht TA, de Figueiredo P. RNAi Screen of Endoplasmic Reticulum–Associated Host Factors Reveals a Role for IRE1α in Supporting Brucella Replication. PLoS Pathog. 2008;4:e1000110. doi: 10.1371/journal.ppat.1000110. doi:10.1371/journal.ppat.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. doi:10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 92.Todd DJ, Lee A-H, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nature Reviews Immunology. 2008;8:663–674. doi: 10.1038/nri2359. doi:10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 93.Halder P, Datta C, Kumar R, Sharma AK, Basu J, Kundu M. The secreted antigen, HP0175, of Helicobacter pylori links the unfolded protein response (UPR) to autophagy in gastric epithelial cells. Cellular Microbiology. 2015;17:714–729. doi: 10.1111/cmi.12396. doi:10.1111/cmi.12396. [DOI] [PubMed] [Google Scholar]
- 94.Wang X, Lin P, Li Y, Xiang C, Yin Y, Chen Z, et al. Brucella suis Vaccine Strain 2 Induces Endoplasmic Reticulum Stress that Affects Intracellular Replication in Goat Trophoblast Cells In vitro. Frontiers in Cellular and Infection Microbiology. 2016;6:226. doi: 10.3389/fcimb.2016.00019. doi:10.3389/fcimb.2016.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zheng K, Chen D-S, Wu Y-Q, Xu X-J, Zhang H, Chen C-F, et al. MicroRNA Expression Profile in RAW264.7 cells in Response to Brucella melitensisInfection. International Journal of Biological Sciences. 2012;8:1013–1022. doi: 10.7150/ijbs.3836. doi:10.7150/ijbs.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McDonough JA, Newton HJ, Klum S, Swiss R, Agaisse H, Roy CR. Host Pathways Important for Coxiella burnetii Infection Revealed by Genome-Wide RNA Interference Screening. mBio. 2013;4:e00606–12–e00606–12. doi: 10.1128/mBio.00606-12. doi:10.1128/mBio.00606-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liao C-C, Ho M-Y, Liang S-M, Liang C-M. Recombinant protein rVP1 upregulates BECN1-independent autophagy, MAPK1/3 phosphorylation and MMP9 activity via WIPI1/WIPI2 to promote macrophage migration. Autophagy. 2012;9:5–19. doi: 10.4161/auto.22379. doi:10.4161/auto.22379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Deuretzbacher A, Czymmeck N, Reimer R, Trülzsch K, Gaus K, Hohenberg H, et al. β1 Integrin-Dependent Engulfment of Yersinia enterocolitica by Macrophages Is Coupled to the Activation of Autophagy and Suppressed by Type III Protein Secretion. J Immunol. 2009;183:5847–5860. doi: 10.4049/jimmunol.0804242. doi:10.4049/jimmunol.0804242. [DOI] [PubMed] [Google Scholar]
- 99.Niu H, Rikihisa Y. Ats-1. Autophagy. 2013;9:787–788. doi: 10.4161/auto.23693. doi:10.4161/auto.23693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Niu H, Xiong Q, Yamamoto A, Hayashi-Nishino M, Rikihisa Y. Autophagosomes induced by a bacterial Beclin 1 binding protein facilitate obligatory intracellular infection. Proceedings of the National Academy of Sciences. 2012;109:20800–20807. doi: 10.1073/pnas.1218674109. doi:10.1073/pnas.1218674109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Newton HJ, Kohler LJ, McDonough JA, Temoche-Diaz M, Crabill E, Hartland EL, et al. A Screen of Coxiella burnetii Mutants Reveals Important Roles for Dot/Icm Effectors and Host Autophagy in Vacuole Biogenesis. PLoS Pathog. 2014;10:e1004286. doi: 10.1371/journal.ppat.1004286. doi:10.1371/journal.ppat.1004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu X, Mao K, Yu AYH, Omairi-Nasser A, Austin J, II, Glick BS, et al. The Atg17-Atg31-Atg29 Complex Coordinates with Atg11 to Recruit the Vam7 SNARE and Mediate Autophagosome-Vacuole Fusion. Current Biology. 2016;26:150–160. doi: 10.1016/j.cub.2015.11.054. doi:10.1016/j.cub.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]