Abstract
Ewing sarcomas are characterized by the presence of EWS/ETS fusion genes in the absence of other recurrent genetic alterations and mechanisms of tumor heterogeneity that contribute to disease progression remain unclear. Mutations in the Wnt/beta-catenin pathway are rare in Ewing sarcoma but the Wnt pathway modulator LGR5 is often highly expressed, suggesting a potential role for the axis in tumor pathogenesis. We evaluated beta-catenin and LGR5 expression in Ewing sarcoma cell lines and tumors and noted marked intra- and inter-tumor heterogeneity. Tumors with evidence of active Wnt/beta-catenin signaling were associated with increased incidence of tumor relapse and worse overall survival. Paradoxically, RNA sequencing revealed a marked antagonism of EWS/ETS transcriptional activity in Wnt/beta-catenin activated tumor cells. Consistent with this, Wnt/beta-catenin activated cells displayed a phenotype that was reminiscent of Ewing sarcoma cells with partial EWS/ETS loss of function. Specifically, activation of Wnt/beta-catenin induced alterations to the actin cytoskeleton, acquisition of a migratory phenotype and up regulation of EWS/ETS-repressed genes. Notably, activation of Wnt/beta-catenin signaling led to marked induction of tenascin C (TNC), an established promoter of cancer metastasis, and an EWS/ETS-repressed target gene. Loss of TNC function in Ewing sarcoma cells profoundly inhibited their migratory and metastatic potential. Our studies reveal that heterogeneous activation of Wnt/beta-catenin signaling in subpopulations of tumor cells contributes to phenotypic heterogeneity and disease progression in Ewing sarcoma. Significantly, this is mediated, at least in part, by inhibition of EWS/ETS fusion protein function that results in de-repression of metastasis-associated gene programs.
Keywords: Ewing sarcoma, Wnt/beta-catenin, tumor heterogeneity, TNC, metastasis
INTRODUCTION
Ewing sarcomas are aggressive bone and soft tissue tumors with a high propensity for metastasis (1). Tumors are characterized by chromosomal translocations that generate tumor-initiating EWS/ETS fusion oncoproteins. The two most common translocations are t(11;22)q(24;12), which encodes the EWS/FLI1 fusion protein in about 85% of cases, and t(21;22)(q22;q12), which encodes EWS/ERG in 5-10% of cases (1). Ewing sarcomas otherwise display few recurrent genetic mutations (2-4) and little is yet known about the molecular and cellular mechanisms that contribute to metastatic progression (1). Studies of Ewing sarcoma cells and tumors have, to date, identified several potential mediators of tumor relapse and metastatic progression, including activation of chemokine receptors (5, 6), tyrosine kinases (7) and WNT signaling (8, 9), as well as suppression of NOTCH signaling (10). Significantly, a recently described model of EWS/FLI1-induced transformation in primary murine cells also highlighted the Wnt/beta-catenin axis as a key regulator of tumor initiation and progression (11). However, despite these findings, the contribution of Wnt/beta-catenin signaling to Ewing sarcoma pathogenesis remains unclear.
Wnt/beta-catenin signaling is required for both embryonic development and adult tissue homeostasis, and the pathway is often deregulated in epithelial cancers (12). Canonical Wnt ligands associate with frizzled protein and LRP5/6 protein co-receptor complexes at the cell membrane to activate Wnt signaling, resulting in inhibition of beta-catenin degradation by the destruction complex, which includes the AXIN, APC and GSK3β proteins. Stabilized beta-catenin translocates to the nucleus where it can activate transcription by TCF/LEF transcription factors. The transcriptional targets of Wnt/beta-catenin/TCF signaling are context- and cell type-dependent and their modulation can alter numerous cellular functions including proliferation, patterning, migration, and self-renewal (12, 13). In epithelial tumors, particularly colorectal cancer (14), inactivating mutations in genes encoding destruction complex proteins or activating mutations in beta-catenin itself commonly deregulate the Wnt/beta-catenin signaling pathway (14). Recent studies have also highlighted the possible significance of defects in the Wnt/beta-catenin pathway in some sarcomas (15, 16), however recurrent mutations have not been described in Ewing sarcoma (2-4). In addition to activating genetic mutations, Wnt/beta-catenin signaling can also be aberrantly activated by deregulation of ligand-receptor interactions at the cell surface. In particular, high-level expression of R-spondin (RSPO) ligands and/or their receptors LGR4 and LGR5, has recently been shown to potentiate canonical Wnt signaling and promote tumorigenesis by inhibiting RNF43/ZNRF3-mediated turnover of the frizzled/LRP receptor complex (17-19). We recently showed that LGR5 is highly expressed by some Ewing sarcoma tumors and that LGR5+ cells potentiate Wnt/beta-catenin signaling upon RSPO exposure (20). Moreover, our studies demonstrated increased expression of LGR5 in a group of rapidly progressive tumors, lending further support to the hypothesis that the Wnt/beta-catenin pathway might contribute to an aggressive cellular phenotype (20).
In the current work, we evaluated activation of Wnt/beta-catenin in Ewing sarcoma and defined the transcriptional and functional consequences of pathway activation in these tumors. Paradoxically, our findings reveal that activation of Wnt/beta-catenin partially antagonizes EWS/ETS-dependent transcription and that this antagonism promotes phenotypic transition of tumor cells to cell states that promote migration and metastasis.
MATERIALS AND METHODS
Cell lines and lentiviral transductions
Ewing sarcoma cell lines were maintained in RPMI 1640 media (Gibco) supplemented with 10% FBS (Atlas Biologicals) and 2mM L-glutamine (Life Technologies). Ewing sarcoma cell lines were kindly provided by Dr. Timothy Triche (CHLA, Los Angeles, CA; 2004) Dr. Heinrich Kovar (CCRI, St. Anna Kinderkrebsforschung, Vienna, Austria; 2010), and the COG cell bank (cogcell.org; 2012). CHLA25, CHLA32, and STA-ET-8.2 were grown on plates coated with 0.2% gelatin. shA673-1C cells were kindly provided by Dr. Olivier Delattre (Institut Curie, Paris; 2014) and maintained as previously described (21). L-cells (ATCC CRL-2648) and Wnt3a L-cells (ATCC CRL-2647) were obtained from ATCC in 2011 (atcc.org) and were cultured in DMEM (Gibco) with 10% FBS. Cells were verified to be mycoplasma negative and identities confirmed by STR profiling every 6 months. Lentiviral production and transduction was performed as previously described (20), and the following plasmids were used: Addgene #24305 (7TGP), Addgene #24313 (EβP), Sigma TRCN0000230788 (shTNC-3), Sigma TRCN000015400 (shTNC-5), UM-vector core pLentilox-luciferase/GFP (luc-tagged). Transduced cells were selected in puromycin (2 μg/mL).
Reporter assays and cell sorting
Stably transduced 7TGP cells were stimulated with 1:1 RPMI:L-cell/Wnt3a conditioned media (CM) +/− 20ng/mL recombinant RSPO2 (R&D Systems). Fluorescence was measured and quantified on an Accuri C6 cytometer (BD Biosciences) and cells sorted on the basis of GFP expression using the MoFlo Astrios instrument (University of Michigan Flow Cytometry Core). Peak GFP was detected at 48 hr and cells were sorted for gene expression studies at this time.
RNA sequencing and analysis of gene expression
CHLA25-7TGP cells were stimulated with CM and FACS-sorted on the basis of GFP. Three biological replicates per sample were collected and three technical replicates for each of the samples were sequenced on the Illumina HiSeq 2000 (University of Michigan Sequencing Core). Fastq generation was performed using Illumina’s CASAVA-1.8.2 software, analyzed for quality control using FASTQC, and aligned using Sailfish (22). For differential transcript expression analysis, RPKM was calculated and analyzed using Sailfish and the statistical R package, edgeR (23). RNAseq data have been deposited to GEO (GSE75859) and significant genes are listed in Supplemental Table 1. Gene ontology was determined using DAVID Bioinformatics Resources 6.7 (24). Overlapping datasets were identified using the Molecular Signatures Database v4.05 (MolSigDB) and genesets with false discovery rates (FDR) <0.05 (−log FDR >1.3) were considered significant (25). Gene set enrichment analysis (GSEA) was performed using the GseaPreranked function of GSEA v2.1.0 software. To determine if EWS/ETS targets were specifically enriched among Wnt-modulated genes we performed Chi-squared analysis as previously described (26). Wnt targets were validated by quantitative RT-PCR (qRT-PCR) using standard methods (20). Primer sequences are provided as Supplemental Table 2.
Immunostaining and in situ hybridization
Sequential tumor tissue microarray (TMA) sections were probed with CD99 (BioCare Medical), beta-catenin (BD Biosciences), and DAPI, and automated quantitative analysis (AQUA) was performed as previously described (27). RNA in-situ hybridization (ISH) was performed using a commercially available kit and probes according to the manufacturer’s instructions (Advanced Cell Diagnostics Inc., Haywood, Ca). Cells were grown on gelatin-coated coverslips and protein expression evaluated by standard techniques and immunofluorescence microscopy as previously described (20). Detailed protocols and antibody information are included in the Supplemental Methods.
Western blot analysis
Cells were lysed in 4% LDS (for FLI1 detection) or RIPA buffer (for ERG detection), and electrophoresis and transfer was performed using the BioRad Mini-Protean Tetra System according to manufacturer’s instructions. Primary antibodies FLI1 (Abcam EPR4646, 1:1000) or ERG (Abcam #92513, 1:1000) or GAPDH (Cell Signaling, 1:5000) were incubated in 5% milk overnight at 4°C, followed by incubation with secondary antibodies IRDye 800 and 680 (LiCor, 1:5000). Western blots were imaged and densitometry performed using the LiCor imaging system and software (LiCor, Lincoln, NE).
Migration assays
1×105 cells were added in serum-free RPMI to transwells containing 0.8 μm pores with serum-containing media in the bottom chamber. 500 ng/mL of recombinant Wnt3a (R&D Systems) +/− RSPO2 was added to the bottom chamber. After 24hr cells were fixed using a solution of 25% crystal violet and 25% methanol and membranes were imaged and then cells eluted using crystal violet in 10% acetic acid. Colorimetric absorbance (540 nm) was quantified on a BioTek plate reader.
Colony formation assay
Colony formation was assessed by plating a single cell suspension of 1 × 104 cells per well of six well plates in 0.35% noble agar on a layer of 0.5% noble agar. Colonies were stained with a solution of 0.005% crystal violet and counted three weeks later.
In vivo tumor assays
For subcutaneous tumor assays, 2.5×105 stably transduced control (shNS) or TNC knockdown (shTNC) cells were resuspended in PBS, diluted 1:1 in Matrigel, and injected subcutaneously into NOD SCID mice (strain 394, Charles River Laboratories, Wilmington, MA). Each mouse received an injection of shNS cells on the left flank and shTNC cells on the right flank, and tumor formation and growth was measured by calipers every other day. To assess lung engraftment, 1×106 luciferase-labeled cells were injected via tail vein. Where indicated, cells were stimulated in vitro with either L-cell CM or Wnt3a CM+RSPO2 prior to injection. Satisfactory injection of viable cells was confirmed by detection of light emission in the lungs of recipient mice 30 minutes after cell injection (Xenogen IVIS bioluminescence system (Perkin Elmer, Waltham, MA). Imaging was repeated weekly and tumor burden quantified using Living Image software (Perkin Elmer, Waltham, MA). Mice were monitored for up to 7 weeks, at which point mice were euthanized. Tumor formation and location was confirmed by dissection with the help of Pathology Cores for Animal Research within the Unit for Laboratory Animal Medicine at the University of Michigan.
Clinical correlations analysis
Outcomes were interrogated in a recently published, clinically annotated dataset from the Children’s Oncology Group (COG) (GSE 63157 (6)). Cox regression and Kaplan-Meier analyses were conducted for determination of overall survival and event-free survival relative to median and tertile of LEF1 expression. Log-rank tests were used to compare survival distributions.
Statistics
Unless otherwise indicated, data are expressed as mean±standard error of the mean (SEM) from a minimum of three independent experiments. The data were analyzed using GraphPad Prism software by Student’s t-test, Chi-squared, and Fisher’s exact tests, and p-values < 0.05 considered significant.
RESULTS
Wnt/beta-catenin activation is highly heterogeneous in Ewing sarcoma
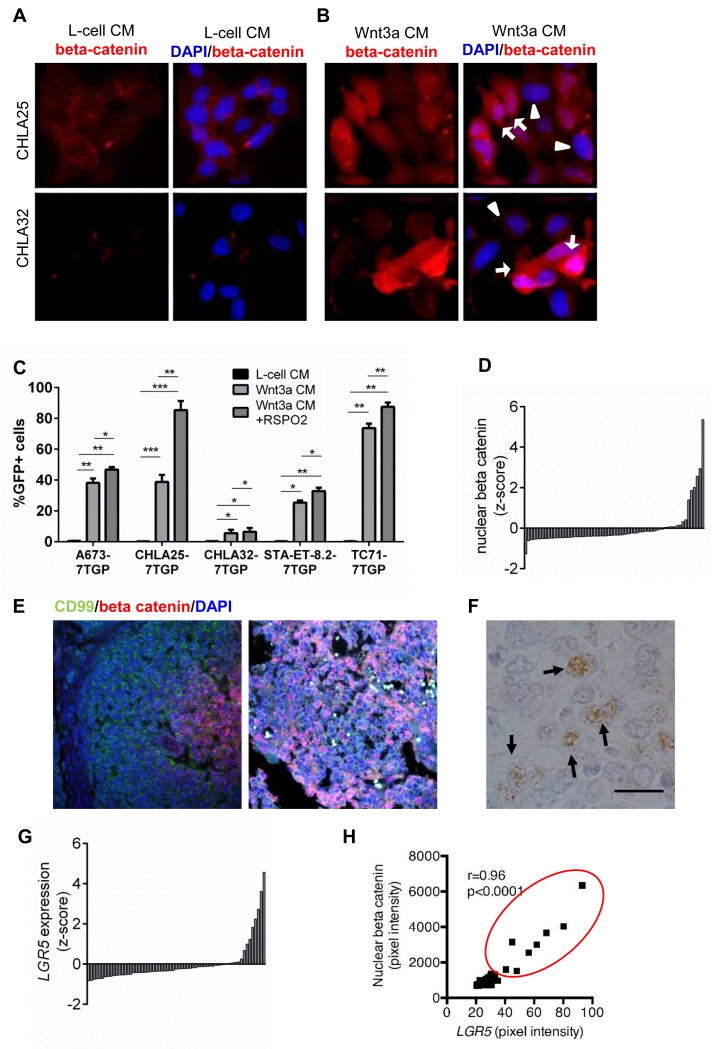
Ewing sarcoma cell lines activate Wnt/beta-catenin signaling in response to exogenous Wnt3a and RSPO2 treatment and our prior studies revealed heterogeneity of response among cell lines (20). To better characterize the basis of the heterogeneous Wnt response we exposed Ewing sarcoma cells to control L-cell conditioned media (CM) or Wnt3a CM and assessed beta-catenin localization in individual cells. While beta-catenin was largely lacking under control conditions (Fig. 1A), stimulation with Wnt3a CM led to an increase in nuclear expression (Fig. 1B). Notably, robust nuclear beta-catenin staining was evident in some cells but little to no beta-catenin was observed in neighboring cells (Fig. 1B). To determine if this heterogeneity resulted in heterogeneous activation of beta-catenin-dependent transcription, we evaluated TCF transcriptional activity in GFP-reporter cells (7TGP). Although all cell lines showed an increase in the proportion of GFP-positive cells following Wnt3a stimulation, the responses were highly variable (Fig. 1C). Addition of RSPO2 potentiated the TCF-dependent transcriptional response but, again, heterogeneity persisted within and between cell lines (Fig. 1C). Thus, individual Ewing sarcoma cells display marked heterogeneity in their responsiveness to exogenous Wnt/beta-catenin activating ligands in vitro.
Figure 1. Heterogeneity of Wnt/beta-catenin activation in Ewing sarcoma.
A) Cells were stained with anti-beta catenin antibody (red) and counterstained with DAPI (blue) after exposure to control (L-cell) conditioned media (CM). Nuclear beta-catenin is absent.
B) After 48hr exposure to Wnt3a, some cells have strong nuclear localization of beta catenin (arrows) while others have weak or no nuclear localization (arrowheads). Scale bars = 20 μm.
C) 7TGP-transduced TCF-reporter cells were stimulated with L-cell, Wnt3a CM, or Wnt3a CM +RSPO2 and GFP+ cells quantified. Heterogeneity of GFP was evident between and within cell lines. Mean ± SEM of 3 independent experiments *p<0.05, **p<0.01 ***p<0.001, ****p<0.0001.
D) AQUA of a Ewing sarcoma TMA detected nuclear beta-catenin expression in a minority of tumors. Z-score: z=(sample – mean)/standard deviation.
E) Left: representative beta-catenin (red) positive tumor. CD99 (green) marks tumor cells (Magnification=200×). Right: 400× image of another positive biopsy shows beta-catenin positive cells adjacent to beta-catenin negative cells.
F) In situ hybridization of LGR5 in a primary Ewing sarcoma. Positive cells are noted by arrows. Scale bar = 20 μm.
G) Quantification of LGR5 expression by SQUISH detected transcript expression in a minority of tumors. Z-score: z=(sample – mean)/standard deviation.
H) Expression of nuclear beta-catenin and LGR5 is highly correlated. r=Pearson’s correlation. Tumors with a z-score > 1 for both beta-catenin and LGR5 are circled.
To determine if Wnt/beta-catenin signaling is similarly heterogeneously activated in vivo, we performed beta-catenin AQUA immunofluorescence staining of a Ewing sarcoma TMA. The majority of over 50 evaluable biopsies showed no evidence of beta-catenin expression; however, elevated cytoplasmic and nuclear beta-catenin expression was detected in eight tumor samples (Fig. 1D). In these tumors expression was limited to discrete subpopulations of tumor cells (Fig. 1E) confirming heterogeneity both within and between individual tumors.
Given that recurrent mutations in Wnt/beta-catenin pathway genes have not been identified in Ewing sarcoma (2-4), and that RSPO2 signaling can augment the response of cells to Wnt3a stimulation (20), we hypothesized that heterogeneity in LGR5 expression may contribute to heterogeneity of beta-catenin activation. To address this we used ISH to measure LGR5 mRNA in tumor biopsies. Non-quantitative ISH identified LGR5+ tumor cells in 8 of 26 evaluable tumors and cell-to-cell heterogeneity was again apparent (Fig. 1F). To determine if LGR5+ tumor cells were more likely to activate beta-catenin we performed fluorescence-based, semi-quantitative ISH on a TMA section adjacent to the section that was used for AQUA studies. This analysis affirmed that only a small proportion of tumors expressed high levels of LGR5 (Fig. 1G) and showed that the tumors with evidence of robust LGR5 expression were the same as those that displayed nuclear beta-catenin staining (Fig. 1H). These data corroborate our in vitro studies (20) and lend support for the hypothesis that Ewing sarcoma cells that express high levels of LGR5 are more responsive to ligand-dependent activation of Wnt/beta-catenin signaling.
Activation of Wnt/beta-catenin signaling is associated with worse clinical outcomes in patients with Ewing sarcoma
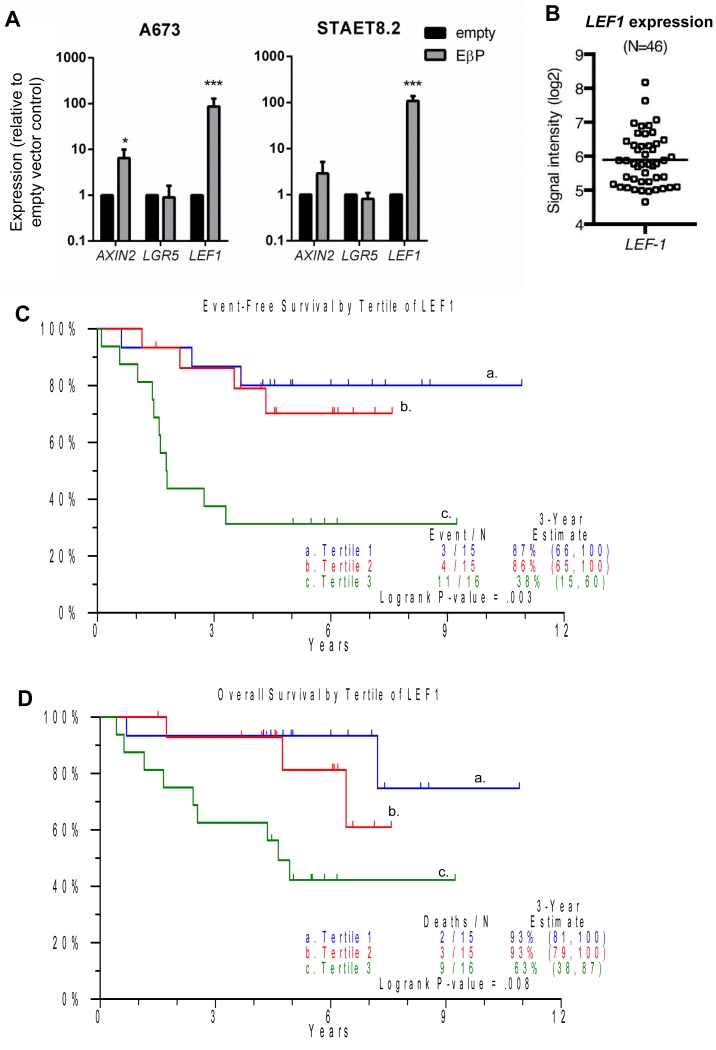
We previously reported that LGR5 mRNA expression was increased in a small cohort of patients with unusually aggressive disease (20). Review of the clinical data for the current TMA samples showed that all tumors with evidence of beta-catenin activation were obtained from patients who later relapsed. To more definitively address whether activation of Wnt/beta-catenin signaling is associated with outcome, we turned to a recently published cohort of patients who were diagnosed with localized Ewing sarcoma and treated on COG therapeutic studies (6). To infer Wnt/beta-catenin activation from the available gene expression data we first needed to identify a biomarker of active beta-catenin signaling. We transduced Ewing sarcoma cells with a constitutively active beta-catenin construct (28) and measured induction of established Wnt/beta-catenin transcriptional targets. Basal expression of LEF1 was low in control cells but more than 100-fold induced by activated beta-catenin (Fig. 2A). In contrast, AXIN2 was more modestly induced and LGR5 was unchanged (Fig. 2A). Therefore, we selected LEF1 as our biomarker and discovered that, consistent with heterogeneity in beta-catenin activation, levels of LEF1 were highly variable in the patient tumors (Fig. 2B). In this clinically homogeneous patient population, high expression of LEF1 was significantly associated with worse event free (Fig. 2C) and overall survival (Fig. 2D).
Figure 2. High LEF1 expression is associated with poor outcomes in Ewing sarcoma.
A) Ewing sarcoma cells were transduced with a constitutively active beta-catenin construct (EβP), and expression of AXIN2, LGR5 and LEF1 was assessed by qRT-PCR.
B) LEF1 gene expression in a cohort of 46 clinically annotated biopsies from patients diagnosed with localized Ewing sarcoma (GSE 63157).
C) Analysis of event free and overall (D) survival shows that patients whose tumors expressed high levels of LEF1 experienced worse clinical outcomes.
Wnt/beta-catenin activation antagonizes EWS/ETS-dependent transcription
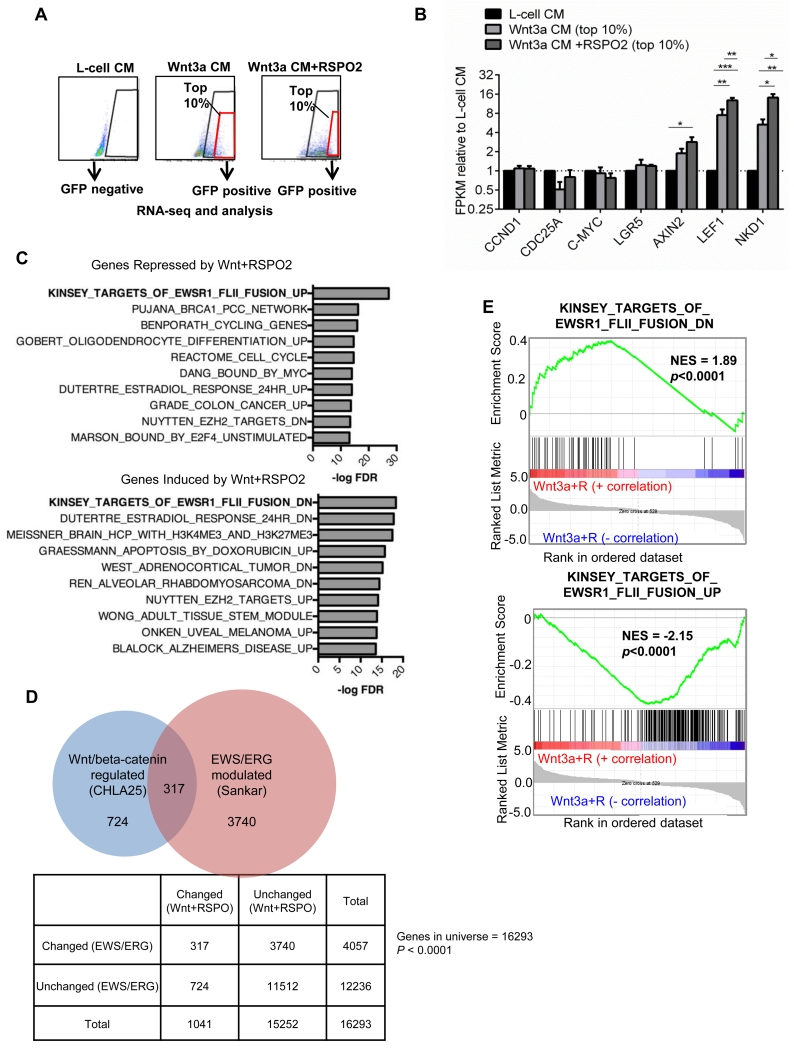
Having established that Wnt/beta-catenin activation is associated with worse clinical outcomes, we next sought to define why this might be. To achieve this we used 7TGP-CHLA25 reporter cells to compare the transcriptional profiles of Wnt/beta-catenin activated cells with non-activated controls (Fig. 3A). RNAseq analysis identified differential expression of over 1,000 transcripts in response to Wnt3a alone or in combination with RSPO2 (Supplementary Fig. S1A, Supplemental Table 1). AXIN2, LEF1, and NKD1 were among the previously established Wnt/beta-catenin target genes that were also induced in Ewing sarcoma (Fig. 3B). Conversely, other established target genes were unaffected, demonstrating the cell-type specificity of this signaling axis (Fig. 3B). This differential Wnt response was validated by independent qRT-PCR analyses of multiple Ewing sarcoma cell lines (Supplementary Fig. S1B & C).
Figure 3. Targets of Wnt/beta-catenin signaling in Ewing sarcoma are cell-type specific and oppositely regulated by EWS/ETS fusions.
A) Sorting strategy to isolate Wnt/beta-catenin-activated and non-activated CHLA25-7TGP reporter cells.
B) Many canonical Wnt/beta-catenin target genes identified in other cell types are not induced by Wnt in Ewing sarcoma. Conversely, established targets AXIN2, LEF1, and NKD1 are induced. Expression plotted as fold change relative to control cells. *p<0.05, **p<0.01, ***p<0.001
C) MSigDB analysis of genes that were significantly and at least 2-fold repressed (top panel) or induced (bottom panel) by Wnt/beta-catenin shows marked overlap with EWS/FLI1 target genes but genesets were inversely regulated.
D) Published EWS/ERG target genes are enriched among Wnt/beta-catenin-modulated genes. Significance was determined using the Chi-square test.
E) GSEA of all Wnt/beta-catenin regulated genes confirms a significant inverse relationship with EWS/FLI1 transcriptional targets. Comparisons of rank-ordered Wnt/beta-catenin-regulated genes to EWS/FLI1- repressed (top panel) and EWS/FLI1-induced (bottom panel) genes are shown. NES indicates the normalized enrichment score.
To gain functional insights into the Wnt/beta-catenin transcriptional response, we compared our list of significant genes to published genesets (25). Strikingly, genes that were repressed two-fold in Wnt/beta-catenin activated cells overlapped most significantly with genes that are induced by EWS/FLI1 (29) (Fig. 3C, top). Conversely, genes that were induced by Wnt/beta-catenin activation overlapped most significantly with genes that are repressed by EWS/FLI1 (Fig. 3C, bottom). This remarkable overlap between EWS/ETS and Wnt/beta-catenin targets in CHLA25 cells was highly significant and not merely a reflection of generalized transcriptional activation as demonstrated by specific enrichment of EWS/ERG target genes in the Wnt/RSPO-responsive geneset (Fig. 3D). To determine if the overlapping genes were indicative of a more generalized inverse relationship between Wnt/beta-catenin and EWS/ERG transcriptional activity we expanded our analysis to include all genes that were significantly modulated by Wnt/beta-catenin irrespective of fold change. GSEA of rank-ordered genes confirmed an inverse correlation between Wnt/beta-catenin and EWS/ETS transcriptional targets (Fig. 3E & Supplementary Fig. S2A). Thus, transcriptional antagonism exists between EWS/ERG and Wnt/beta-catenin in CHLA25 cells, corroborating a prior study of A673 cells which reported an inverse relationship between EWS/FLI1 and TCF/LEF target genes (30).
To evaluate whether transcriptional antagonism between EWS/ETS and Wnt/beta-catenin signaling exists in primary tumors we performed GSEA on gene expression data from the aforementioned COG study. Genes that were positively correlated with LEF1 in these tumors were highly enriched for genes that are normally repressed by EWS/ETS fusions (Supplementary Fig. S2B). These data corroborate a prior study of gene expression in a different tumor dataset wherein a correlation was also discovered between levels of LEF1 and EWS/ETS repressed targets (30). Together these data provide compelling evidence that activation of Wnt/beta-catenin antagonizes the transcriptional function of EWS/ETS fusions in Ewing sarcoma.
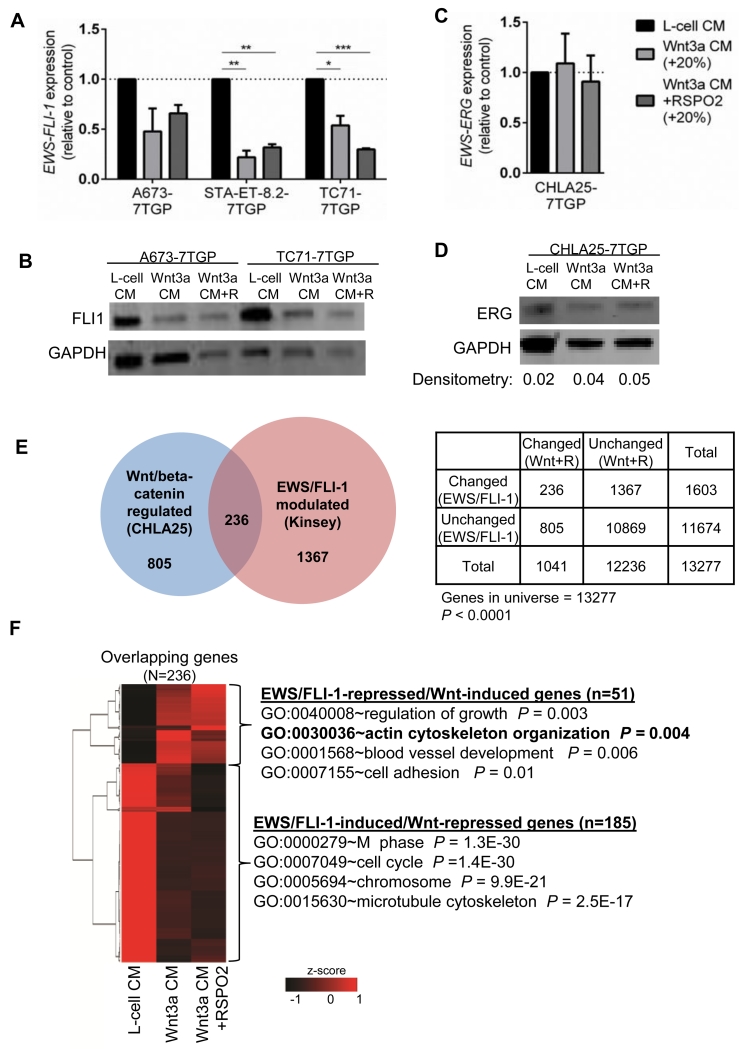
Levels of EWS/FLI1 differ between Wnt/beta-catenin activated and non-activated cells
To gain insight into the potential mechanism of transcriptional antagonism, we assessed whether activation of Wnt/beta-catenin signaling leads to changes in the levels of EWS/ETS fusions. To ensure that we compared expression of the fusions in truly Wnt/beta-catenin active and inactive cells, we measured EWS/ETS levels following FACS-sorting of 7GP cells. Significantly, reduced expression of EWS/FLI1 transcript was reproducibly seen in Wnt/beta-catenin activated cells (Fig. 4A) and a trend to reduced protein expression was also observed (Fig. 4B). In contrast, no significant change in EWS/ERG levels was observed in Wnt-activated CHLA25 cells (Fig. 4C, D). These findings suggest that either Wnt/beta-catenin can inhibit EWS/FLI1 transcription or that heterogeneity in expression of EWS/FLI1 prior to ligand exposure determines Wnt-responsiveness. In support of this latter possibility, Navarro et al. (30) previously showed that cells with reduced expression of EWS/FLI1 were more responsive to Wnt3a, an observation that we have also now independently validated (Supplemental Figure S3A-C).
Figure 4. Overlap between Wnt/beta-catenin and EWS/ETS-modulated genes.
A) qRT-PCR analysis of EWS/FLI1 expression in unstimulated (L-cell/GFP−) and Wnt/beta-catenin-activated cells (Wnt3a CM, Wnt3aCM/RSPO2; top 20% GFP+). *p<0.05, **p<0.01, ***p<0.001
B) Expression of EWS/FLI1 protein was assessed by Western blot in cells as in (A). A trend toward decreased EWS/FLI1 expression is seen in Wnt-active cells.
C) qRT-PCR analysis of EWS/ERG expression in unstimulated (L-cell/GFP−) and Wnt/beta-catenin-activated cells (Wnt3a CM, Wnt3aCM/RSPO2; top 20% GFP+).
D) Expression of EWS/ERG protein was assessed by Western blot in cells as in (C). No change is seen in Wnt-active cells. ERG densitometry values normalized to GAPDH are indicated.
E) Published EWS/FLI1 target genes are enriched among Wnt/beta-catenin-modulated genes. Significance was determined using the Chi-square test.
F) Hierarchical clustering and gene ontologies of 236 overlapping genes from (E).
Activation of Wnt/beta-catenin signaling promotes cell migration and lung engraftment
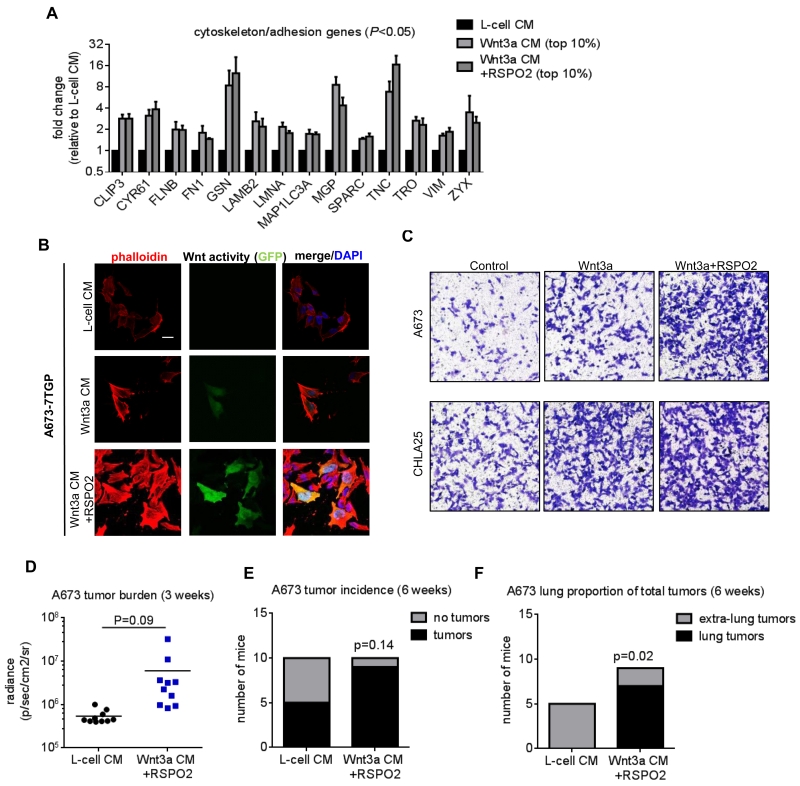
We next sought to understand the functional consequences of Wnt/beta-catenin-dependent antagonism of EWS/ETS activity. To achieve this we focused on the 236 genes that were modulated by Wnt/beta-catenin in CHLA25 cells and EWS/FLI1 in A673 cells (29) (Fig. 4E). Hierarchical clustering of these genes confirmed both the opposing pattern of regulation and the potentiating effect of RSPO2 (Fig. 4E). Gene ontology analysis of the overlapping genes revealed that microtubule genes involved in cell cycle regulation were most prominent among EWS/FLI1-induced/Wnt-repressed genes (Fig. 4E & Supplementary Fig. S3D). However, activation of Wnt/beta-catenin does not lead to significant inhibition of Ewing sarcoma cell proliferation, at least in vitro (20) (Supplementary Fig. S3E). Among EWS/FLI1-repressed/Wnt-induced genes, actin cytoskeleton genes were enriched (Fig. 4E). Thus, activation of Wnt/beta-catenin signaling in Ewing sarcoma results in altered expression of EWS/ETS target genes that are involved in regulation of the microtubule and actin cytoskeletons.
Knockdown of EWS/ETS in Ewing sarcoma cells leads to induction of actin stress fibers, acquisition of a migratory cell phenotype, and enhanced tumor cell adhesion in the lung, phenotypes that are, in part, mediated by de-repression of zyxin (ZYX) (31, 32). Our observation that activation of Wnt/beta-catenin leads to de-repression of numerous cytoskeleton genes (Fig. 5A), including ZYX, led us to test whether activation of Wnt/beta-catenin phenocopied EWS/ETS knockdown. Consistent with EWS/ETS loss-of-function, exposure of tumor cells to activating Wnt-ligands led to the formation of actin stress fibers (Fig. 5B, Supplementary Fig. S4A), an increase in cell spreading (Supplementary Fig. S4B), and enhanced migration (Fig. 5C). Further, podosomes, actin-rich cytoskeletal structures that are essential for cell migration (33, 34), were increased in Wnt/beta-catenin-activated cells (Supplementary Fig. S4C). Similar results were obtained in cells that expressed constitutively active beta-catenin, confirming that the Wnt-stimulated phenotype is, at least in part, mediated by beta-catenin (Supplementary Fig. S4D, E). To test if activated Wnt/beta-catenin signaling promotes lung engraftment of tumors, Wnt-stimulated and non-stimulated cells were injected via tail vein and tumor engraftment monitored by bioluminescence imaging. Stimulation of A673 cells with Wnt3a +RSPO2 prior to injection resulted in a trend towards earlier onset of tumors (Fig. 5D), a higher rate of tumor formation overall (9 of 10 vs. 5 of 10 mice) (Fig. 5E) and a significant increase in the formation of lung tumors (Fig. 5F). However, no statistically significant difference was seen in TC32 cells and all mice that developed tumors showed evidence of lung engraftment, irrespective of ex vivo Wnt-stimulation (Supplementary Fig. S4F-H). Thus, beta-catenin-activation reproducibly activates cytoskeletal changes in Ewing sarcoma cells that are associated with enhanced cell migration in vitro and, in some cases, enhanced lung engraftment.
Figure 5. Activation of Wnt/beta-catenin signaling induces cytoskeleton changes and promotes migration.
A) Relative expression of cytoskeleton and adhesion genes that are repressed by EWS/FLI1 in Ewing sarcoma cells and induced by Wnt/beta-catenin activation.
B) Assessment of F-actin filaments in 7TGP-reporter cells following exposure to Wnt3a +/− RSPO2 shows induction of stress fibers (phalloidin, red) in Wnt/beta-catenin activated (green) cells. Nuclei are counterstained with DAPI (blue). Scale bar = 20 μm.
C) Transwell migration assays of control and recombinant Wnt3a/RSPO2-stimulated cells shows enhanced migration in Wnt/beta-catenin activated cells.
D) Bioluminescence imaging of mice 3 weeks after tail-vein injection of L-cell CM- or Wnt3a CM+RSPO2 treated cells reveals a trend toward higher tumor burden in mice that received Wnt3a CM+RSPO2 cells.
E) At 6 weeks, 5 of 10 mice that received L-cell CM-treated cells had tumors, while 9 of 10 mice that received Wnt3a CM+RSPO2-treated cells had tumors.
F) None of the 5 tumors in L-cell CM-recipient mice were located in the lungs. 7 of 9 tumor-bearing mice that received Wnt-activated cells had lung tumors.
Tenascin C is a Wnt/beta-catenin target gene that promotes cell migration and tumorigenicity
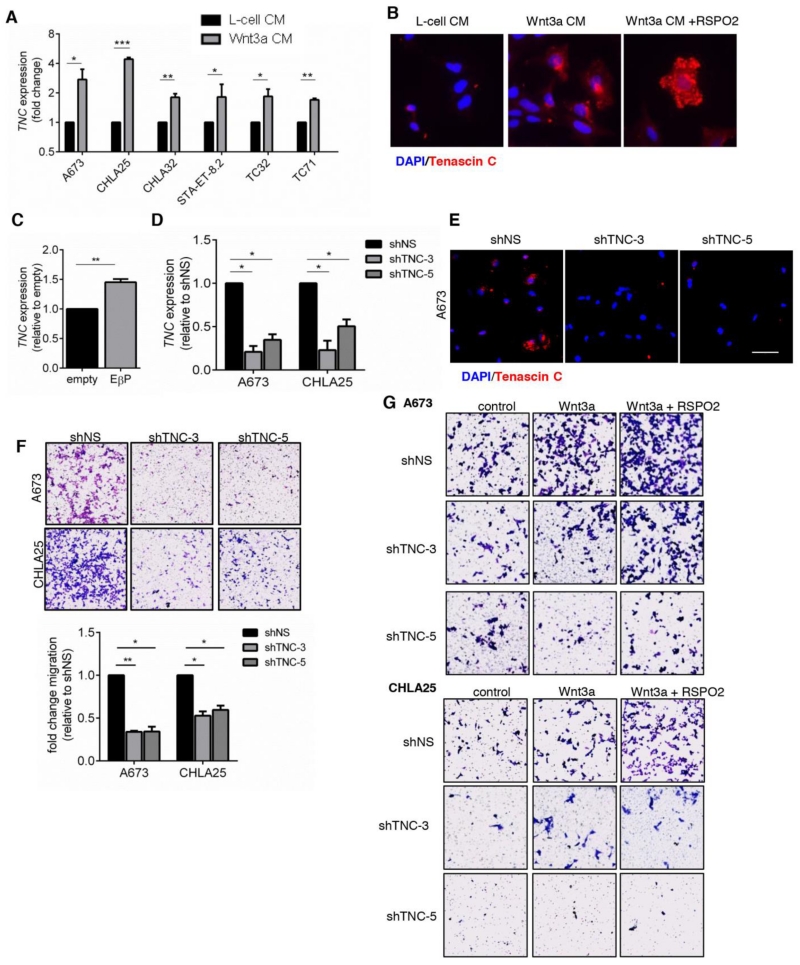
We next interrogated the list of Wnt/beta-catenin-induced/EWS/ETS-repressed genes to identify genes that might contribute to migration and lung engraftment. In addition to ZYX, this analysis identified TNC (see Fig. 5A). TNC encodes tenascin C, a secreted matricellular protein that is essential for successful lung engraftment of breast cancer cells (35). In addition, TNC has also been implicated as a mediator of tumor metastasis in other tumor types and at multiple levels of the metastatic cascade (36). qRT-PCR and immunocytochemistry studies confirmed that TNC is induced by Wnt/beta-catenin in Ewing sarcoma (Fig. 6A-C) and is also subject to EWS/FLI1-dependent repression (Supplementary Fig. S5A).
Figure 6. Tenascin C is a Wnt/beta-catenin target.
A) qRT-PCR of TNC expression in Ewing sarcoma cells following exposure to L-cell or Wnt3a CM. Mean ± SEM of 3 biologic replicates. *p<0.05, **p<0.01, ***p<0.001.
B) Tenascin C protein (red) was assessed by immunocytochemistry in CHLA25 cells following stimulation with L-cell or Wnt3a CM +/− RSPO-2. Cells were counterstained with DAPI (blue).
C) qRT-PCR analysis of TNC in cells stably transduced with constitutively active beta-catenin. Mean ± SEM of 3 biologic replicates. **p<0.01.
D) Knock down of TNC was achieved in Ewing sarcoma cells following stable infection with shTNC vectors (shTNC-3 and shTNC-5). Expression measured by qRT-PCR.
E) Tenascin C protein expression (red) was assessed by immunocytochemistry of A673 cells as in (D). Cells were counterstained with DAPI (blue). Scale bar = 50 μm.
F) Migration of TNC knockdown cells is reduced compared to control shNS cells. Three independent experiments were quantified using crystal violet and expressed as mean ± SEM relative to controls (bottom panel). *p<0.05, **p<0.01.
G) TNC knockdown and control cells were stimulated with PBS (control), recombinant Wnt3a, or recombinant Wnt3a +RSPO2 and allowed to migrate for 24 hr. TNC knockdown partially inhibits Wnt-dependent migration.
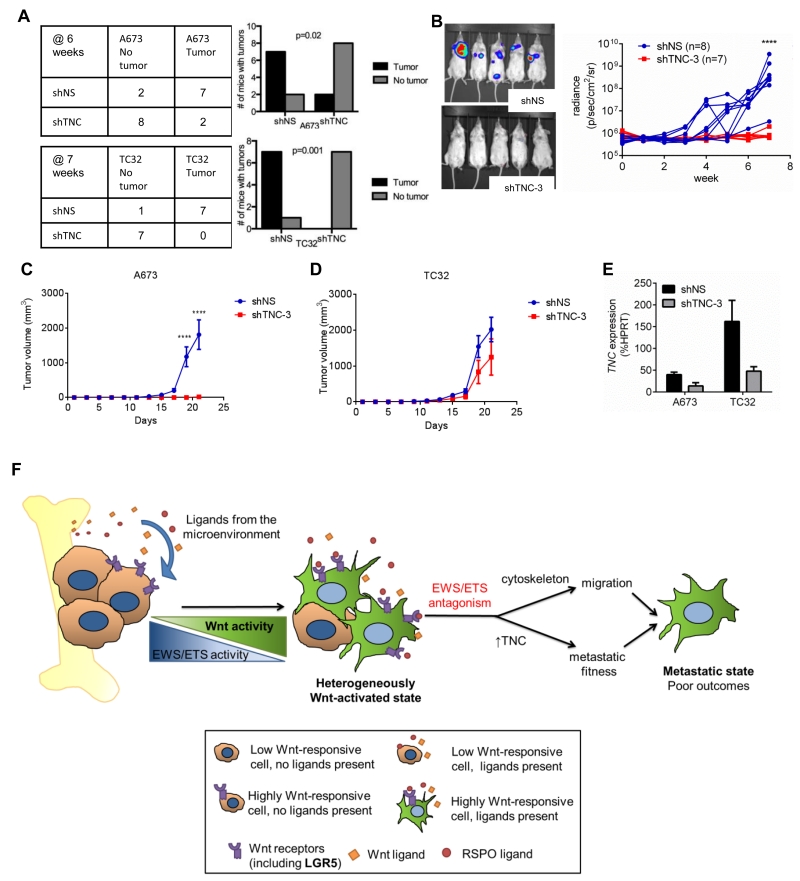
We next assessed the impact of TNC loss-of-function (Fig. 6D, E). Knockdown of TNC had no effect on cell proliferation in vitro but strongly inhibited anchorage independent colony formation (Supplementary Fig. S5B, C). TNC knockdown cells showed diminished migration, both in standard culture conditions (Fig. 6F) and in response to Wnt/RSPO-activation (Fig. 6G), demonstrating that Wnt/beta-catenin-induced migration is, in part, mediated by TNC. With respect to in vivo tumorigenicity, TNC knockdown cells reproducibly showed a reduced capacity to form lung tumors following tail vein injection (Fig. 7A, B). Subcutaneous tumor formation was also abrogated in A673 cells following TNC knockdown (Fig. 7C) but was only mildly impacted in TC32 cells (Fig. 7D). We speculate that the effect of knockdown on subcutaneous engraftment was more pronounced in A673 than TC32 cells due to fact that the absolute levels of TNC expression in A673 cells were lower than in TC32 cells, both at baseline and following knockdown (Fig. 7D). Likewise, we hypothesize that high basal levels of TNC in TC32 cells might make them more amenable to lung engraftment than A673 cells, thus obviating a requirement for additional Wnt/beta-catenin-dependent activation.
Figure 7. Tenascin C promotes lung engraftment of Ewing sarcoma cells.
A) Control and TNC knockdown A673 and TC32 cells were injected via tail vein into NOD-SCID mice and tumor growth monitored by bioluminescence imaging for 6-7 weeks. The efficiency of tumor engraftment in TNC knockdown cells was significantly impaired in both cell lines (Fisher’s exact test).
B) Representative bioluminescence images and growth curves for TC32 mice as in (A).
C) Control and TNC knockdown cells were injected subcutaneously and tumor growth monitored. Tumorigenicity of A673 cells was completely abrogated by TNC-knockdown. ****P<0.0001.
D) Subcutaneous engraftment and growth of TC32 cells was minimally impaired by TNC knockdown.
E) TNC expression determined by qRT-PCR in control and TNC-knockdown cells prior to injection into mice for studies in C & D. Basal expression of TNC was very high in TC32 cells and expression after knockdown was equivalent to that of control A673 cells.
F) The antagonistic effect of Wnt/beta-catenin activation on EWS/ETS-dependent transcription results in transition of cells to a more migratory and metastatic cell state as a consequence of de-repression of cytoskeleton genes and induction of the metastasis promoter TNC. The extent of Wnt/beta-catenin activation in Ewing sarcoma cells is dependent on both the provision of Wnt-activating ligands in the tumor microenvironment and on the density of Wnt-modulating receptors, such as LGR5, that are expressed on the tumor cells themselves.
Together these data demonstrate that TNC plays a key role in promoting Ewing sarcoma cell migration and lung engraftment. Activation of Wnt/beta-catenin signaling in Ewing sarcoma cells leads to up regulated expression of TNC and this contributes to a more tumorigenic cell state. Overall these data support a model in which activation of Wnt/beta-catenin leads to up regulation of metastasis-promoting genes that are normally repressed by EWS/ETS fusions and this, in turn, leads to an increased risk of metastasis and worse clinical outcomes (Fig. 7E).
DISCUSSION
In the current study we investigated the impact of activated Wnt/beta-catenin signaling on Ewing sarcoma. The combined results of these studies demonstrate that activation of canonical Wnt/beta-catenin signaling, by ligands in the tumor microenvironment, results in more clinically aggressive disease. In keeping with the microenvironment being the source of Wnt/beta-catenin activation in these tumors, recurrent somatic mutations in the pathway are rare (2-4, 15) and Wnt and RSPO ligands are highly expressed in developing bone, the predominant site of Ewing tumors in pediatric and young adult patients (37, 38). Mechanistically, our data show that the aggressive clinical phenotype of Wnt/beta-catenin-activated tumors is mediated, at least in part, by a paradoxical antagonism of EWS/ETS-dependent transcription, which results in activation of metastasis-associated genes that are normally subject to EWS/ETS-dependent repression. In particular, we have shown that the extracellular matrix encoding gene, TNC, respectively, is induced by Wnt/beta-catenin activation in Ewing sarcoma and that high-level TNC promotes cell migration, tumorigenicity and lung engraftment.
Tumor cell heterogeneity can arise from genetic as well as from non-mutational mechanisms and non-genetic heterogeneity is emerging as a major contributor to tumor progression and relapse (39-41). As an example, reversible plasticity of tumor cells between proliferative and migratory states, such as occurs in epithelial to mesenchymal transitions, has been linked to metastasis (42, 43) and epigenetic plasticity of chromatin states promotes emergence of drug resistance (44). In concordance with the findings of Endo et al. (45), who described neurite outgrowth in response to Wnt/beta-catenin activation, we also found that Ewing sarcoma cells display cytoskeletal plasticity in response to Wnt stimulation. However, our studies revealed that not all Ewing sarcoma cells respond equally to Wnt stimulation, and that responsiveness can vary widely even between neighboring tumor cells in vitro and in vivo. Numerous factors likely contribute to this heterogeneity of response. Expression of the Wnt-potentiating ligand, LGR5, was found to be heterogeneous on a cell-to-cell basis and we have previously shown that the presence of LGR5 directly impacts on the Wnt response when RSPO ligands are present (20). Variability in the availability of endogenous or exogenous ligands, as well as heterogeneous expression of Wnt-modulating receptors, could all impact on an individual cell’s ability to activate beta-catenin. In addition, other cell-intrinsic factors may be important determinants of the Wnt response. For example, G2/M-phase is, at least in some contexts, most permissive for activation of Wnt signaling (46). Moreover, in colorectal cancer, nuclear beta-catenin is heterogeneous among tumor cells, thus revealing that other factors contribute to pathway activation even when activating mutations are universally present (47). In lung adenocarcinoma, cells isolated from metastatic lesions were more Wnt responsive than their corresponding primary tumors, and aggressive phenotypes were enhanced by exposure to conditioned media from metastatic sites (48). Thus, variability in Wnt responsiveness among tumor cells can be dictated by the differences in the signaling microenvironment, as well as by differences in cell intrinsic factors.
The results of our analyses of gene expression data from both Wnt3a/RSPO2-stimulated cells as well as primary tumor data revealed a striking transcriptional antagonism between Wnt/beta-catenin/LEF1 and EWS/ETS. Our finding that Wnt-stimulation inhibits EWS/ETS is concordant with results from an earlier study in which EWS/FLI1 was shown to inhibit beta-catenin/TCF activity. Thus, the data from Navarro et al. and from our current work show that antagonism between these transcriptional axes is bidirectional. The mechanistic basis of this antagonism remains to be fully elucidated but the studies of Navarro et al. suggest that physical interactions between EWS/FLI1 and LEF1 inhibit transcription of TCF/LEF target genes in response to beta-catenin activation (30). Since LEF1 is strongly induced by Wnt/beta-catenin in Ewing sarcoma cells we speculate that this might lead to sequestration of EWS/ETS proteins by LEF1 and inhibition of EWS/ETS-dependent transcription. However, it should be noted that expression of many established EWS/ETS target genes was unaffected by Wnt/beta-catenin activation, demonstrating that the inverse relationship between Wnt/beta-catenin and EWS/ETS does not extend to all targets of the fusion. Further investigations are required to elucidate the molecular mechanisms that account for differential responses of different EWS/ETS target genes to Wnt/beta-catenin activation. Given that EWS/ETS fusions effect changes to transcription through a variety of epigenetic mechanisms (49-51) it is likely that cooperation between EWS/ETS fusions, TCF/LEF transcription factors, and epigenetic regulators at target gene promoters and enhancers collectively determine the transcriptional output of Wnt/beta-catenin signaling in Ewing sarcoma cells.
In summary, these studies provide compelling biologic and clinical evidence that activation of Wnt/beta-catenin signaling contributes to an aggressive cellular phenotype in Ewing sarcoma tumors. Moreover, they reveal a previously unappreciated degree of inter- and intra-tumoral heterogeneity in Wnt/beta-catenin activation that is mediated by both cell autonomous (e.g. expression of LGR5) and non-cell autonomous (e.g. availability of Wnt and RSPO ligands in the tumor microenvironment) factors. These findings support further investigation of Wnt/beta-catenin pathway-targeted agents as potential adjuvant therapies for metastasis prevention in Ewing sarcoma patients.
Supplementary Material
Acknowledgements
We would like to thank the Lawlor lab members for their helpful discussions and Dr. Patrick Grohar for FLI1 western blot protocol. This research was supported by the SARC Sarcoma SPORE NIH Grant U54 CA168512, an Alex’s Lemonade Stand Foundation Innovation Award and Ruth L. Kirschstein NRSA F30CA183276 (EAP). Additional support was provided the University of Michigan’s Cancer Center Support Grant (P30 CA046592) by the use of the following Cancer Center Core(s): Flow cytometry, cellular imaging, sequencing, and vector.
Footnotes
Conflict of Interest Statement: ---Dr. Rashmi Chugh discloses the following potential conflict of interest: the institution received some research funding to support her salary in order to perform a clinical trial of an unrelated therapeutic study in Ewing sarcoma.
References
- 1.Lawlor ER, Sorensen PH. Twenty Years on: What Do We Really Know about Ewing Sarcoma and What Is the Path Forward? 2015;20(3-4):155–71. doi: 10.1615/critrevoncog.2015013553. 2015-07-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS genetics. 2014 Jul;10(7):e1004475. doi: 10.1371/journal.pgen.1004475. PubMed PMID: 25010205. Pubmed Central PMCID: 4091782. Epub 2014/07/11.eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer discovery. 2014 Nov;4(11):1326–41. doi: 10.1158/2159-8290.CD-13-1037. PubMed PMID: 25186949. Epub 2014/09/05. eng. [DOI] [PubMed] [Google Scholar]
- 4.Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer discovery. 2014 Nov;4(11):1342–53. doi: 10.1158/2159-8290.CD-14-0622. PubMed PMID: 25223734. Pubmed Central PMCID: 4264969. Epub 2014/09/17. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennani-Baiti IM, Cooper A, Lawlor ER, Kauer M, Ban J, Aryee DN, et al. Intercohort gene expression co-analysis reveals chemokine receptors as prognostic indicators in Ewing’s sarcoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010 Jul 15;16(14):3769–78. doi: 10.1158/1078-0432.CCR-10-0558. PubMed PMID: 20525755. Pubmed Central PMCID: 2905506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Volchenboum SL, Andrade J, Huang L, Barkauskas DA, Krailo M, Womer RB, et al. Gene Expression Profiling of Ewing Sarcoma Tumors Reveals the Prognostic Importance of Tumor-Stromal Interactions: A Report from the Children’s Oncology Group. The journal of pathology Clinical research. 2015 Apr;1(2):83–94. doi: 10.1002/cjp2.9. PubMed PMID: 26052443. Pubmed Central PMCID: 4457396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendoza-Naranjo A, El-Naggar A, Wai DH, Mistry P, Lazic N, Ayala FR, et al. ERBB4 confers metastatic capacity in Ewing sarcoma. EMBO molecular medicine. 2013 Jul;5(7):1019–34. doi: 10.1002/emmm.201202343. PubMed PMID: 23681745. Pubmed Central PMCID: 3721475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hauer K, Calzada-Wack J, Steiger K, Grunewald TG, Baumhoer D, Plehm S, et al. DKK2 mediates osteolysis, invasiveness, and metastatic spread in Ewing sarcoma. Cancer research. 2013 Jan 15;73(2):967–77. doi: 10.1158/0008-5472.CAN-12-1492. PubMed PMID: 23204234. [DOI] [PubMed] [Google Scholar]
- 9.Schaefer KL, Eisenacher M, Braun Y, Brachwitz K, Wai DH, Dirksen U, et al. Microarray analysis of Ewing’s sarcoma family of tumours reveals characteristic gene expression signatures associated with metastasis and resistance to chemotherapy. European journal of cancer. 2008 Mar;44(5):699–709. doi: 10.1016/j.ejca.2008.01.020. PubMed PMID: 18294840. [DOI] [PubMed] [Google Scholar]
- 10.Ban J, Aryee DN, Fourtouna A, van der Ent W, Kauer M, Niedan S, et al. Suppression of deacetylase SIRT1 mediates tumor-suppressive NOTCH response and offers a novel treatment option in metastatic Ewing sarcoma. Cancer research. 2014 Nov 15;74(22):6578–88. doi: 10.1158/0008-5472.CAN-14-1736. PubMed PMID: 25281719. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka M, Yamazaki Y, Kanno Y, Igarashi K, Aisaki K, Kanno J, et al. Ewing’s sarcoma precursors are highly enriched in embryonic osteochondrogenic progenitors. The Journal of clinical investigation. 2014 Jul 1;124(7):3061–74. doi: 10.1172/JCI72399. PubMed PMID: 24911143. Pubmed Central PMCID: 4071408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006 Nov 3;127(3):469–80. doi: 10.1016/j.cell.2006.10.018. PubMed PMID: 17081971. [DOI] [PubMed] [Google Scholar]
- 13.Mao CD, Byers SW. Cell-context dependent TCF/LEF expression and function: alternative tales of repression, de-repression and activation potentials. Critical reviews in eukaryotic gene expression. 2011;21(3):207–36. doi: 10.1615/critreveukargeneexpr.v21.i3.10. PubMed PMID: 22111711. Pubmed Central PMCID: 3434703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fearon ER. Molecular genetics of colorectal cancer. Annual review of pathology. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. PubMed PMID: 21090969. Epub 2010/11/26. eng. [DOI] [PubMed] [Google Scholar]
- 15.Vijayakumar S, Liu G, Rus IA, Yao S, Chen Y, Akiri G, et al. High-frequency canonical Wnt activation in multiple sarcoma subtypes drives proliferation through a TCF/beta-catenin target gene, CDC25A. Cancer cell. 2011 May 17;19(5):601–12. doi: 10.1016/j.ccr.2011.03.010. PubMed PMID: 21575861. Pubmed Central PMCID: 3116447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barham W, Frump AL, Sherrill TP, Garcia CB, Saito-Diaz K, VanSaun MN, et al. Targeting the Wnt pathway in synovial sarcoma models. Cancer discovery. 2013 Nov;3(11):1286–301. doi: 10.1158/2159-8290.CD-13-0138. PubMed PMID: 23921231. Pubmed Central PMCID: 3823757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao HX, Xie Y, Zhang Y, Charlat O, Oster E, Avello M, et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012 May 10;485(7397):195–200. doi: 10.1038/nature11019. PubMed PMID: 22575959. [DOI] [PubMed] [Google Scholar]
- 18.Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2011 Jul 12;108(28):11452–7. doi: 10.1073/pnas.1106083108. PubMed PMID: 21693646. Pubmed Central PMCID: 3136304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang X, Hao HX, Growney JD, Woolfenden S, Bottiglio C, Ng N, et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2013 Jul 30;110(31):12649–54. doi: 10.1073/pnas.1307218110. PubMed PMID: 23847203. Pubmed Central PMCID: 3732970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scannell CA, Pedersen EA, Mosher JT, Krook MA, Nicholls LA, Wilky BA, et al. LGR5 is Expressed by Ewing Sarcoma and Potentiates Wnt/beta-Catenin Signaling. Frontiers in oncology. 2013;3:81. doi: 10.3389/fonc.2013.00081. PubMed PMID: 23596566. Pubmed Central PMCID: 3625903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer cell. 2007 May;11(5):421–9. doi: 10.1016/j.ccr.2007.02.027. PubMed PMID: 17482132. [DOI] [PubMed] [Google Scholar]
- 22.Patro R, Mount SM, Kingsford C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nature biotechnology. 2014 May;32(5):462–4. doi: 10.1038/nbt.2862. PubMed PMID: 24752080. Pubmed Central PMCID: 4077321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010 Jan 1;26(1):139–40. doi: 10.1093/bioinformatics/btp616. PubMed PMID: 19910308. Pubmed Central PMCID: 2796818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. PubMed PMID: 19131956. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005 Oct 25;102(43):15545–50. doi: 10.1073/pnas.0506580102. PubMed PMID: 16199517. Pubmed Central PMCID: 1239896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sankar S, Theisen ER, Bearss J, Mulvihill T, Hoffman LM, Sorna V, et al. Reversible LSD1 inhibition interferes with global EWS/ETS transcriptional activity and impedes Ewing sarcoma tumor growth. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014 Sep 1;20(17):4584–97. doi: 10.1158/1078-0432.CCR-14-0072. PubMed PMID: 24963049. Pubmed Central PMCID: 4155010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. The Journal of molecular diagnostics : JMD. 2012 Jan;14(1):22–9. doi: 10.1016/j.jmoldx.2011.08.002. PubMed PMID: 22166544. Pubmed Central PMCID: 3338343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuerer C, Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PloS one. 2010;5(2):e9370. doi: 10.1371/journal.pone.0009370. PubMed PMID: 20186325. Pubmed Central PMCID: 2826402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinsey M, Smith R, Lessnick SL. NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing’s sarcoma. Molecular cancer research : MCR. 2006 Nov;4(11):851–9. doi: 10.1158/1541-7786.MCR-06-0090. PubMed PMID: 17114343. [DOI] [PubMed] [Google Scholar]
- 30.Navarro D, Agra N, Pestana A, Alonso J, Gonzalez-Sancho JM. The EWS/FLI1 oncogenic protein inhibits expression of the Wnt inhibitor DICKKOPF-1 gene and antagonizes beta-catenin/TCF-mediated transcription. Carcinogenesis. 2010 Mar;31(3):394–401. doi: 10.1093/carcin/bgp317. PubMed PMID: 20019092. [DOI] [PubMed] [Google Scholar]
- 31.Chaturvedi A, Hoffman LM, Jensen CC, Lin YC, Grossmann AH, Randall RL, et al. Molecular dissection of the mechanism by which EWS/FLI expression compromises actin cytoskeletal integrity and cell adhesion in Ewing sarcoma. Molecular biology of the cell. 2014 Sep 15;25(18):2695–709. doi: 10.1091/mbc.E14-01-0007. PubMed PMID: 25057021. Pubmed Central PMCID: 4161506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaturvedi A, Hoffman LM, Welm AL, Lessnick SL, Beckerle MC. The EWS/FLI Oncogene Drives Changes in Cellular Morphology, Adhesion, and Migration in Ewing Sarcoma. Genes & cancer. 2012 Feb;3(2):102–16. doi: 10.1177/1947601912457024. PubMed PMID: 23050043. Pubmed Central PMCID: 3463921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Webb BA, Eves R, Mak AS. Cortactin regulates podosome formation: roles of the protein interaction domains. Experimental cell research. 2006 Apr 1;312(6):760–9. doi: 10.1016/j.yexcr.2005.11.032. PubMed PMID: 16434035. [DOI] [PubMed] [Google Scholar]
- 34.Murphy DA, Courtneidge SA. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nature reviews Molecular cell biology. 2011 Jul;12(7):413–26. doi: 10.1038/nrm3141. PubMed PMID: 21697900. Pubmed Central PMCID: 3423958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nature medicine. 2011 Jul;17(7):867–74. doi: 10.1038/nm.2379. PubMed PMID: 21706029. Pubmed Central PMCID: 4020577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowy CM, Oskarsson T. Tenascin C in metastasis: A view from the invasive front. Cell adhesion & migration. 2015;9(1-2):112–24. doi: 10.1080/19336918.2015.1008331. PubMed PMID: 25738825. Pubmed Central PMCID: 4422797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nam JS, Turcotte TJ, Yoon JK. Dynamic expression of R-spondin family genes in mouse development. Gene expression patterns : GEP. 2007 Jan;7(3):306–12. doi: 10.1016/j.modgep.2006.08.006. PubMed PMID: 17035101. [DOI] [PubMed] [Google Scholar]
- 38.Zhong Z, Ethen NJ, Williams BO. WNT signaling in bone development and homeostasis. Wiley interdisciplinary reviews Developmental biology. 2014 Nov-Dec;3(6):489–500. doi: 10.1002/wdev.159. PubMed PMID: 25270716. Pubmed Central PMCID: 4199871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pisco AO, Huang S. Non-genetic cancer cell plasticity and therapy-induced stemness in tumour relapse: ‘What does not kill me strengthens me’. British journal of cancer. 2015 May 26;112(11):1725–32. doi: 10.1038/bjc.2015.146. PubMed PMID: 25965164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013 Sep 19;501(7467):328–37. doi: 10.1038/nature12624. PubMed PMID: 24048065. Pubmed Central PMCID: 4521623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oshimori N, Oristian D, Fuchs E. TGF-beta promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell. 2015 Feb 26;160(5):963–76. doi: 10.1016/j.cell.2015.01.043. PubMed PMID: 25723170. Pubmed Central PMCID: 4509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Developmental cell. 2008 Jun;14(6):818–29. doi: 10.1016/j.devcel.2008.05.009. PubMed PMID: 18539112. [DOI] [PubMed] [Google Scholar]
- 43.Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes & development. 2013 Oct 15;27(20):2192–206. doi: 10.1101/gad.225334.113. PubMed PMID: 24142872. Pubmed Central PMCID: 3814640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010 Apr 2;141(1):69–80. doi: 10.1016/j.cell.2010.02.027. PubMed PMID: 20371346. Pubmed Central PMCID: 2851638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Endo Y, Beauchamp E, Woods D, Taylor WG, Toretsky JA, Uren A, et al. Wnt-3a and Dickkopf-1 stimulate neurite outgrowth in Ewing tumor cells via a Frizzled3- and c-Jun N-terminal kinase-dependent mechanism. Molecular and cellular biology. 2008 Apr;28(7):2368–79. doi: 10.1128/MCB.01780-07. PubMed PMID: 18212053. Pubmed Central PMCID: 2268413. Epub 2008/01/24. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davidson G, Shen J, Huang YL, Su Y, Karaulanov E, Bartscherer K, et al. Cell cycle control of wnt receptor activation. Developmental cell. 2009 Dec;17(6):788–99. doi: 10.1016/j.devcel.2009.11.006. PubMed PMID: 20059949. [DOI] [PubMed] [Google Scholar]
- 47.Vermeulen L, De Sousa EMF, van der Heijden M, Cameron K, de Jong JH, Borovski T, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nature cell biology. 2010 May;12(5):468–76. doi: 10.1038/ncb2048. PubMed PMID: 20418870. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M, et al. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009 Jul 10;138(1):51–62. doi: 10.1016/j.cell.2009.04.030. PubMed PMID: 19576624. Pubmed Central PMCID: 2742946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomazou EM, Sheffield NC, Schmidl C, Schuster M, Schonegger A, Datlinger P, et al. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell reports. 2015 Feb 24;10(7):1082–95. doi: 10.1016/j.celrep.2015.01.042. PubMed PMID: 25704812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suva ML, et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer cell. 2014 Nov 10;26(5):668–81. doi: 10.1016/j.ccell.2014.10.004. PubMed PMID: 25453903. Pubmed Central PMCID: 4492343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sankar S, Bell R, Stephens B, Zhuo R, Sharma S, Bearss DJ, et al. Mechanism and relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma. Oncogene. 2013 Oct 17;32(42):5089–100. doi: 10.1038/onc.2012.525. PubMed PMID: 23178492. Pubmed Central PMCID: 3899696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.