Abstract
Manduca sexta, known as the tobacco hornworm or Carolina sphinx moth, is a lepidopteran insect that is used extensively as a model system for research in insect biochemistry, physiology, neurobiology, development, and immunity. One important benefit of this species as an experimental model is its extremely large size, reaching more than 10 g in the larval stage. M. sexta larvae feed on solanaceous plants and thus must tolerate a substantial challenge from plant allelochemicals, including nicotine. We report the sequence and annotation of the M. sexta genome, and a survey of gene expression in various tissues and developmental stages. The Msex_1.0 genome assembly resulted in a total genome size of 419.4 Mbp. Repetitive sequences accounted for 25.8% of the assembled genome. The official gene set is comprised of 15,451 protein-coding genes, of which 2498 were manually curated. Extensive RNA-seq data from many tissues and developmental stages were used to improve gene models and for insights into gene expression patterns. Genome wide synteny analysis indicated a high level of macrosynteny in the Lepidoptera. Annotation and analyses were carried out for gene families involved in a wide spectrum of biological processes, including apoptosis, vacuole sorting, growth and development, structures of exoskeleton, egg shells, and muscle, vision, chemosensation, ion channels, signal transduction, neuropeptide signaling, neurotransmitter synthesis and transport, nicotine tolerance, lipid metabolism, and immunity. This genome sequence, annotation, and analysis provide an important new resource from a well-studied model insect species and will facilitate further biochemical and mechanistic experimental studies of many biological systems in insects.
Keywords: Lepidoptera, Insect, Tobacco hornworm, Synteny, Moth, Insect biochemistry, Innate immunity
1. Introduction
Insects in the order Lepidoptera, moths and butterflies, include more than 150,000 species with enormous diversity. They include some of the most striking and beautiful of insect species, as well as many of the world’s most serious agricultural pests (Powell, 2003). Lepidopteran insects have been the subjects of extensive experimental studies in genetics, molecular biology, and biochemistry of a wide array of physiological processes, and they include model systems and species that have unique ecological or economic importance (Goldsmith and Marek, 2010). Investigation of lepidopteran biology is beginning to benefit from advances in genomic sequencing, with published draft genomes available for the commercial silkworm, Bombyx mori (International Silkworm Genome, Consortium, 2008; Mita et al., 2004; Xia et al., 2004), the first lepidopteran genome sequenced, and several additional species including butterflies Danaus plexippus (Zhan et al., 2011), Heliconius melpomene (Dasmahapatra et al., 2012), Melitaea cinxia (Ahola et al., 2014), Papilio glaucus (Cong et al., 2015), and moths Plutella xylostella (You et al., 2013) and Spodoptera frugiperda (Kakumani et al., 2014). We report here a draft sequence for the genome of Manduca sexta, known as the tobacco hornworm or the Carolina sphinx moth, the first genome from the family Sphingidae. M. sexta is in the same superfamily, Bombycoidea, as B. mori but their biology differs dramatically. While B. mori has been domesticated for silk production and feeds exclusively on mulberry leaves, M. sexta is a wild species that feeds on solanaceous plants as larvae, including the crops tobacco and tomato.
M. sexta has been used extensively as a classic biochemical and physiological model for laboratory research on a wide array of topics over the last 40 years. It is an important model species for investigations of development and metamorphosis (Gilbert et al., 2002; Hiruma and Riddiford, 2010; Nijhout et al., 2014; Truman et al., 2006; Truman and Riddiford, 2007), neurobiology and olfaction (Heinbockel et al., 2013; Martin et al., 2011), lipid metabolism (Canavoso et al., 2001), immunity (Kanost and Nardi, 2010), parasitoid- and pathogen-host interactions (Amaya et al., 2005; Chevignon et al., 2015), mechanisms of Bacillus thuringiensis Cry toxins (Soberon et al., 2010), insect-plant interactions (Schuman et al., 2015), midgut physiology (Wieczorek et al., 2003) and many other aspects of insect biochemistry, physiology, and behavior. Annotation and expression analysis of gene families in the M. sexta genome described here provide new insight into a diversity of important topics in insect biology.
2. Methods
2.1. DNA sequencing and assembly
A M. sexta colony started from eggs obtained from Carolina Biological Supply and maintained at Kansas State University for more than 15 years was the source of genomic DNA for the sequencing project. We carried out four generations of single-pair sibling inbreeding to reduce heterozygosity. We isolated DNA from a single male pupa by proteinase K and RNAse A treatment of homogenized tissues, followed by phenol-chloroform extraction, and ethanol precipitation (Bradfield and Wyatt, 1983). We deposited two male adult and two female adult siblings of the individual M. sexta selected for genome sequencing with the Kansas State University Museum of Entomological and Prairie Arthropod Research (voucher number 212).
We sequenced the genome using 454 sequencing technology, using three whole genome shotgun libraries to produce the assembled sequence. These libraries included a 454 Titanium fragment library, and two 454 mate pair libraries with 3 kbp and 8 kbp insert sizes produced from the single male M. sexta pupa described above. Methods for library construction and 454 sequencing were as described in (Chen et al., 2014). We assembled about 48.3 million reads, representing approximately 80.7x coverage of the M. sexta genome (Table S1). In addition, a library of approximately 7000 BAC sequences (app. 164 kbp inserts) was also used to aid assembly.
The Msex_1.0 release is an assembly of whole genome shotgun reads (WGS) generated with the 454 Newbler assembler (2.3-PreRelease-10/19/2009). Additionally, we grouped reads from each Newbler scaffold, along with any missing mate-pairs, and reassembled using Phrap to close gaps within Newbler scaffolds (Table S2). The N50 of the contigs was 40.4 kbp and the N50 of the scaffolds was 664.0 kbp. The total length of all contigs was 399.7 Mbp. When the gaps between contigs in scaffolds were included, the total span of the assembly was 419.4 Mbp. The M. sexta raw sequence, and assembled genome sequence data are available at the NCBI under bioproject PRJNA81037, Assembly ID GCA_000262585.1, and AIXA00000000.1.
2.2. Tissue RNA preparation
For analysis of M. sexta transcripts, we isolated RNA from a variety of tissues at various developmental stages and times. We obtained insects used for these analyses from a colony maintained at the Boyce Thompson Institute, which was initiated from eggs obtained from Carolina Biological Supply (Burlington, NC). Larvae were reared at 60% relative humidity, under a photoperiod/tem-perature cycle of 16 h light and 25 °C: 8 h dark and 23 °C, and fed an artificial wheat germ based diet (Davidowitz et al., 2003). RNA samples were prepared from a variety of tissues at developmental stages ranging from eggs through adult moths. More specifically, samples included eggs, intact 1st 2nd and 3rd instar larvae, heads from larval and adult stages, midgut from a variety of larval, pupal, and adult stages, muscle from 4th and 5th instar larvae, fatbody from 4th and 5th instar larvae as well as pupae and adults, Malpighian tubules from larvae and adults, and testes and ovaries from pupae and adults. Table S8 provides a listing of individual samples used for RNA-seq. Because RNA-seq data from the 52 tissue samples were not replicated, differences observed between samples should be viewed only as indicative of possible trends, and requiring further confirmation for precise quantitative evaluations. Tissues were homogenized in ice-cold TRIzol reagent (Invitrogen) using a Dounce homogenizer at 4 °C and total RNA was extracted following the manufacturer’s instructions.
2.3. RNA-seq and strand-specific RNA-seq library construction and sequencing
PolyA mRNA samples, prepared as described above, were used for RNA-seq library construction using either standard protocols or for strand-specific RNA-seq construction following a modified protocol (Chen et al., 2013; Zhong et al., 2011). Briefly, for strand-specific RNA-seq, polyadenylated RNA was isolated from 30 µg total RNA using Dynabeads® Oligo (dT)25 (Invitrogen) following the manufacturer’s instructions, then simultaneously eluted and fragmented in 2X SuperScript III buffer in the presence of 500 ng hexamer and 100 ng oligo dT(10)VN (5′ p-TTTTTTTTTTVN 3′, IDT). First-strand cDNA synthesis was carried out using SuperScriptIII (Invitrogen). Second-strand cDNA was synthesized using RNase H (NEB) and DNA polymerase I (NEB) with a dUTP mix (final concentration of 1 mM for each nucleotide). After end-repair and dA-tailing, the DNA fragments were ligated with the TruSeq adapter. The sample was then treated with uracil DNA glycosylase (New England Biolabs) to remove the dUTP-containing strand and then PCR amplified with TruSeq indexed PCR primers. Sequencing was performed on the Illumina HiSeq2000 platform at Weill Cornell Medical College. Individual libraries were barcoded and combined in lanes with the goal of generating ≥10 M reads per sample.
2.4. Transcriptome assemblies of RNA-seq data
Several transcriptome assemblies were generated and combined to generate a consensus transcriptome assembly. An initial transcriptome assembly was generated using TopHat and Cufflinks (Trapnell et al., 2009, 2010) and named “Official Gene Set (OGS) June 2012 transcripts.” Separately, Trinity (Grabherr et al., 2011) was used to generate a second transcriptome assembly. For the second assembly, paired end reads (100 bp) of 33 libraries were trimmed to 80 bp using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit (2010)). The forward and reverse reads were assembled separately using Trinity (Grabherr et al., 2011) to generate two separate assemblies. Nineteen single-end libraries with reads of 50 bp were also assembled. The transcripts of these three Trinity assemblies were combined, and the longest of highly similar transcripts (95% identity) was selected using CD-HIT-EST (Li and Godzik, 2006). The combined Trinity transcriptome assembly was named “2014 Trinity RNA-seq assembly.” A third transcriptome assembly was generated using Oases (Schulz et al., 2012). For Oases assemblies, the 52 libraries were divided into eight groups, and transcripts from each group were assembled with Velvet (Zerbino and Birney, 2008) and Oases with kmer length of 27. These outputs were combined under the same conditions (>95% identity, CD-HIT-EST) to form the assembly named “2014 Oases RNA-seq assembly.”
A final assembly that combined the above information through manual annotation and PASA2 processing is described below as “2014 OGS2 transcripts.” In parallel, a new method was developed to automatically crosscheck and select the best protein-coding gene models from the outputs of MAKER (see below), Cufflinks, Trinity, and Oases to constitute a new assembly known as “2014 MCOT 1.0 transcripts” (Cao and Jiang, 2015).
2.5. Gene annotation
M. sexta gene models were annotated using a combination of automated and manual methods. As a first step, the MAKER annotation pipeline version 2.25 (Cantarel et al., 2008) was trained with CEGMA gene set, M. sexta ESTs from NCBI, and protein homology from Lepidoptera, and insect genome projects available from NCBI as of May 2011, including louse, mosquito, fruit fly, wasp, pea aphid, silkworm, red flour beetle, and honey bee. MAKER output was used to train Augustus and SNAP in three iterations. The resulting output, Official Gene Set (OGS) 1.0, was manually curated by a community of experts using WebApollo (Lee et al., 2013), resulting in 2498 curated genes.
We combined the de novo (i.e., genome-free) transcript assemblies (Trinity and Oases) and genome-guided transcriptome assembly (Cufflinks), and provided this combined dataset to PASA2 (Haas et al., 2003) for a genome-guided transcriptome assembly. As part of the PASA2 process, open reading frames were predicted using TransDecoder (Haas et al., 2013). Therefore, full length transcripts larger than transcripts missing the start or stop codons were weighted more highly. The PASA2 output was then post-processed with the JAMg pipeline (http://jamg.sourceforge.net) to produce a high quality, full length subset (termed PASA_gold) that aligned to the genome. These data were limited to what was supported only by cDNA evidence and therefore in the next step, automated gene predictions and manually curated genes were added to derive a single combined dataset (Official Gene Set, OGS2.0). In order to produce a consensus using EvidenceModeler (Haas et al., 2008), a combined dataset was produced using: the PASA_gold alignments, the first phase of manual curations, the original cufflinks alignments, and the automated, snap_masked, and augustus_masked predictions performed by MAKER. These were weighted based on an arbitrary weight (100, 1000, 7, 4, 1 and 2 respectively) that reflected confidence in the accuracy of the gene models. This final gene set was then re-processed with the PASA2 transcript database in order to add untranslated regions and alternatively spliced transcripts based on the accumulated cDNA evidence. M. sexta OGS 2.0 is available at ftp://ftp.bioinformatics.ksu.edu/pub/Manduca/OGS2/ and with a BLAST site linked to JBrowse at http://agripestbase.org/manduca/ and at the i5k Workspace https://i5k.nal.usda.gov/Manduca_sexta.
3. Results and discussion
3.1. Sequencing, assembly, and annotation
We sequenced DNA from a single male pupa, using 454 sequencing technology. We used a male for sequencing to avoid complications in assembly from the highly repetitive W chromosome present only in females (Sahara et al., 2012). Our Msex_1.0 genome assembly (see Methods for details) had a final size of 419.4 Mbp consistent with a prior measurement of 422 ± 12 Mbp for the M. sexta genome (Hanrahan and Johnston, 2011). The Msex 1.0 assembly has excellent contiguity with contig and scaffold N50s of 40.4 kbp and 664.0 kbp respectively (Table S1). Assessing assembly completeness with Benchmarking Universal Single-Copy Orthologs (BUSCOs) (Simao et al., 2015) recovered 95% of 2675 arthropod BUSCOs, which is slightly more than for B. mori (93%) (Table S2).
We used the MAKER pipeline to produce a preliminary set of gene predictions on the Msex 1.0 assembly, followed by manual curation by a community of experts using WebApollo. This process produced an official gene set (OGS 2.0) using a PASA2 pipeline, with the MAKER and manual annotations as well as Trinity de novo and Cufflinks transcript assemblies as input. OGS 2.0 contains 15,451 protein-coding genes, of which 2498 were manually curated (Table S3). Assessing the completeness of the OGS 2.0 annotation recovered 92% of arthropod BUSCOs, which, like for the assembly, is slightly more than for B. mori (90%) (Table S2). In addition, 91 genes encoding microRNAs were manually curated, for a total of 15,542 genes in OGS 2.0 (available at ftp://ftp.bioinformatics.ksu.edu/pub/Manduca/OGS2/ and https://i5k.nal.usda.gov/Manduca_sexta). Sixteen genes were identified as probable lateral gene transfers from bacterial origins (Table S4).
3.2. Genome structure and analysis
3.2.1. Novel repeats, transposable elements, and repeat masking
Using the automated pipeline RepeatModeler (repeatmasker. org) to scan the genome for repetitive elements, we identified 668 unique, complex repeats, 125 of which were based on structural motifs and sequence similarity to previously described repeat sequences from other species. Among classifiable repeats, we found representatives of 36 repeat families belonging to 5 superfamilies (Table 1), and combined this de novo library of repeats with known repeats from arthropods obtained from RepBase (Jurka et al., 2005) to identify repetitive regions in the M. sexta genome assembly using RepeatMasker software (repeatmasker.org). This process identified ~108 Mbp of repetitive sequence, corresponding to 25.8% of the genome (Table 2). This value was substantially less than the repeat masking statistics reported for Bombyx mori (~35%) (Osanai-Futahashi et al., 2008), although similar to the butterflies H. melpomene (~25%; (Dasmahapatra et al., 2012)) and M. cinxia (28%; (Ahola et al., 2014)), but notably greater than reported for the monarch butterfly, D. plexippus (~13%; (Zhan et al., 2011)). Among classified repeats, retrotransposon elements were more than twice as abundant as DNA-based elements, with LINE and SINE elements being the most abundant superfamilies found in the M. sexta genome. However, the overwhelming majority of masked regions corresponded to complex repetitive sequences yet to be characterized, and the proportion of sequence identified as repetitive within each scaffold showed substantial variation (Fig. S1). For large scaffolds (>10 kbp), the mean proportion masked (identified as repetitive) was 27%, similar to the total for the genome. Yet values ranged from 2% to 76% among all scaffolds, with a standard deviation of 11%.
Table 1.
Summary of transposable elements found in the M. sexta genome.
| Class | Superfamily | Number of subfamilies |
|---|---|---|
| RNA retroelements | 83 | |
| LINE | 67 | |
| LTR | 4 | |
| SINE | 12 | |
| DNA transposons | 42 | |
| Helitron | 6 | |
| TIR | 36 | |
| Unknown | 543 |
Table 2.
Summary of RepeatMasker analysis of M. sexta genome assembly using de novo libraries in conjunction with known arthropod repeats.
| Number of elements | Length occupied (bp) | Percentage of genome | |
|---|---|---|---|
| Retroelements | 148,328 | 25,144,563 | 5.99 |
| SINEs: | 88,700 | 14,539,679 | 3.47 |
| Penelope | 1845 | 331,599 | 0.08 |
| LINEs: | 56,627 | 9,551,917 | 2.28 |
| CRE/SLACS | 21 | 2248 | <0.01 |
| L2/CR1/Rex | 28,706 | 4,773,062 | 1.14 |
| R1/LOA/Jockey | 7172 | 1,460,871 | 0.35 |
| R2/R4/NeSL | 1347 | 358,703 | 0.09 |
| RTE/Bov-B | 16,085 | 2,469,529 | 0.59 |
| L1/CIN4 | 15 | 857 | <0.01 |
| LTR elements: | 3001 | 1,052,967 | 0.25 |
| BEL/Pao | 906 | 215,267 | 0.05 |
| Ty1/Copia | 404 | 213,225 | 0.05 |
| Gypsy/DIRS1 | 1493 | 589,251 | 0.14 |
| Retroviral | 112 | 18,515 | <0.01 |
| DNA transposons | 64,549 | 10,692,867 | 2.55 |
| hobo-Activator | 3743 | 645,599 | 0.15 |
| Tc1-IS630-Pogo | 38,267 | 6,882,672 | 1.64 |
| En-Spm | 243 | 35,611 | 0.01 |
| PiggyBac | 45 | 13,186 | <0.01 |
| Tourist/Harbinger | 2904 | 496,414 | 0.12 |
| Other | 31 | 2851 | <0.01 |
| Unclassified | 434,165 | 68,763,336 | 16.39 |
| Total interspersed repeats | 104,600,766 | 24.94 | |
| Small RNA | 35,719 | 5,108,851 | 1.22 |
| Satellites | 7 | 792 | <0.01 |
| Simple repeats | 69,846 | 3,047,373 | 0.73 |
| Low complexity | 11,854 | 545,828 | 0.13 |
3.2.2. Orthology and molecular species phylogeny
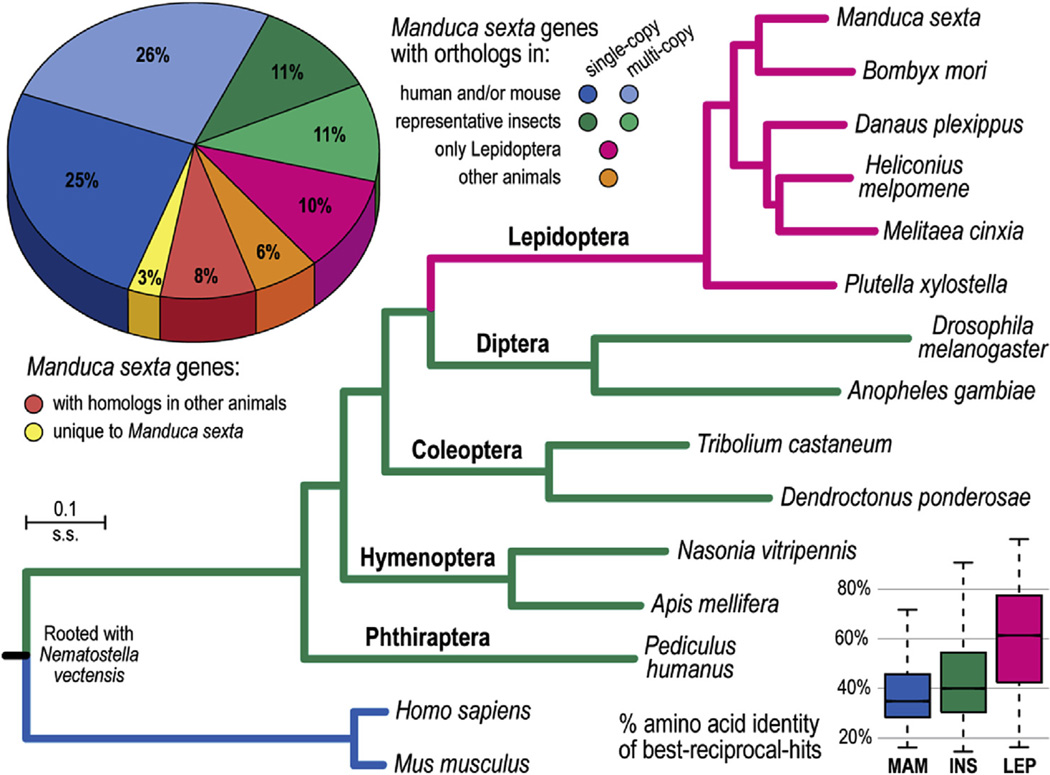
We traced the evolutionary histories of the 15,451 M. sexta OGS 2.0 protein-coding genes using orthology delineation with genes from 172 other animal species at OrthoDB (Kriventseva et al., 2015). Approximately half of the predicted M. sexta genes have identifiable orthologs in representative mammals, a further 22% are shared among all representative insects, and 10% have orthologs only in other lepidopterans. Approximately 3% of M. sexta genes appear to be unique (Fig. 1). The best-reciprocal-hit protein sequence alignments used to identify orthologs of M. sexta genes showed median percent amino acid identities of 60% with the other lepidopterans. This figure dropped to 40% for the representative non-lepidopteran insects and 35% for human and mouse (Fig. 1). To estimate the molecular species phylogeny, we used aligned protein sequences of a subset of orthologs that we identified as single-copy genes (Fig. 1). This subset of single-copy genes was derived through analysis of each of the six lepidopteran species, the seven representative insect species, human, mouse, and the outgroup starlet sea anemone, Nematostella vectensis. Our results clearly resolved the relationships among the six lepidopterans, including the two Bombycoid moth species, the three butterfly species, and the out-group diamondback moth species (Xia et al., 2004; Zhan et al., 2011; Ahola et al., 2014; Dasmahapatra et al., 2012; You et al., 2013). It also revealed that among the examined insect orders, the Lepidoptera, like the Diptera, exhibited relatively rapid molecular evolutionary divergences (i.e. more substitutions per site since the last common insect ancestor). Within the Lepidoptera, the molecular divergence from the outgroup diamondback moth to the Bombycoid moths or the butterflies was similar to that between wasps (Nasonia vitripennis) and bees (Apis mellifera). In contrast, molecular divergence between the Bombycoid moths and the butterflies was about 3.75 times greater than that between human and mouse, highlighting the ancient divergence of these moth and butterfly lineages. Tracing the evolutionary histories of M. sexta genes through comparisons with other animals identified widelyconserved genes for phylogenomic analyses, as well as genes with orthologs only in other Lepidoptera or apparently unique to M. sexta, for which future experiments will be needed to investigate their possible roles in M. sexta biology.
Fig. 1.
The Manduca sexta gene repertoire and molecular species phylogeny. Approximately half of the 15,451 M. sexta genes have identifiable orthologs in the representative genomes of mammals, human and mouse (pie chart, blue), suggesting that these are ancient genes likely to have been present in the metazoan ancestor. A further 32% of M. sexta genes exhibit orthology to genes from the other seven representative insect species (green, 22% or 3427 genes), or only to genes from the other five lepidopteran species (pink, 10% or 1588 genes). Of the remaining genes, some have orthologs (orange) or homologs (red) in other animal species (other metazoan species from OrthoDB), leaving 417 M. sexta genes (app. 3%) without any recognizable homologs (yellow, e-value cutoff 1e-3). Employing aligned protein sequences of universal single-copy orthologs to estimate the molecular species phylogeny rooted with the starlet sea anemone, Nematostella vectensis, shows that the Lepidoptera and Diptera exhibit the fastest rates of molecular divergence. All nodes have 100% bootstrap support. The boxplots show the distributions of percent amino acid identities between M. sexta proteins and their best-reciprocal-hits from mammal species (MAM; median 34.9%), insect species (INS; median 40.1%), and lepidopteran species (LEP; median 60.2%). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2.3. Genome-wide synteny across Lepidoptera and beyond
Previous comparative studies employing linkage maps with limited sets of genomic markers to examine the conservation of gene content and gene order (synteny) suggested very high levels of broad-scale synteny (macrosynteny) conservation among lepidopterans. This conserved macrosynteny allows chromosomal correspondences to be identified, e.g., for comparisons between B. mori and M. sexta (Sahara et al., 2007; Yasukochi et al., 2009), H. melpomene (Pringle et al., 2007; Yasukochi et al., 2006), Bicyclus anynana (Beldade et al., 2009), and Biston betularia (Van’t Hof et al., 2013). Fine-scale genomic conservation (microsynteny) at major color pattern loci (Papa et al., 2008) and regions encoding wing development genes (Conceição et al., 2011) confirmed this well-maintained synteny, but with several inversions and transpositions that disrupted gene co-linearity. Comparing two noctuid moths with the B. mori genome, there are high levels of macrosynteny but also numerous local rearrangements, leading to the suggestion that lepidopteran holocentric chromosomes resist large-scale rearrangements yet generate unusually high levels of localized shuffling (d’Alençon et al., 2010). Comparisons of sequences from additional macrolepidopteran genomes including D. plexippus, H.melpomene, M. cinxia, and B. mori showed strong co-linearity across most chromosomes except for the Z chromosome (Zhan et al., 2011), together with a limited number of fusions (Dasmahapatra et al., 2012), confirming the exceptional stability of the ancestral lepidopteran karyotype (n = 31). Comparisons with P. xylostella were also able to establish confident correspondences, showing that conserved macrosynteny extends beyond the Macrolepidoptera (Baxter et al., 2011; You et al., 2013). However, these genome-wide data have not yet addressed the potential paradox that highly-conserved macrosynteny is apparently accompanied by numerous small-scale rearrangements that lead to the breakdown of microsynteny (d’Alençon et al., 2010; Dasmahapatra et al., 2012). With the addition of M. sexta to the list of fully sequenced lepidopteran genomes, we take the opportunity to further evaluate the evolution of synteny in moths and butterflies in relation to other insects (Figs. S2–S5 and Tables S5–S6).
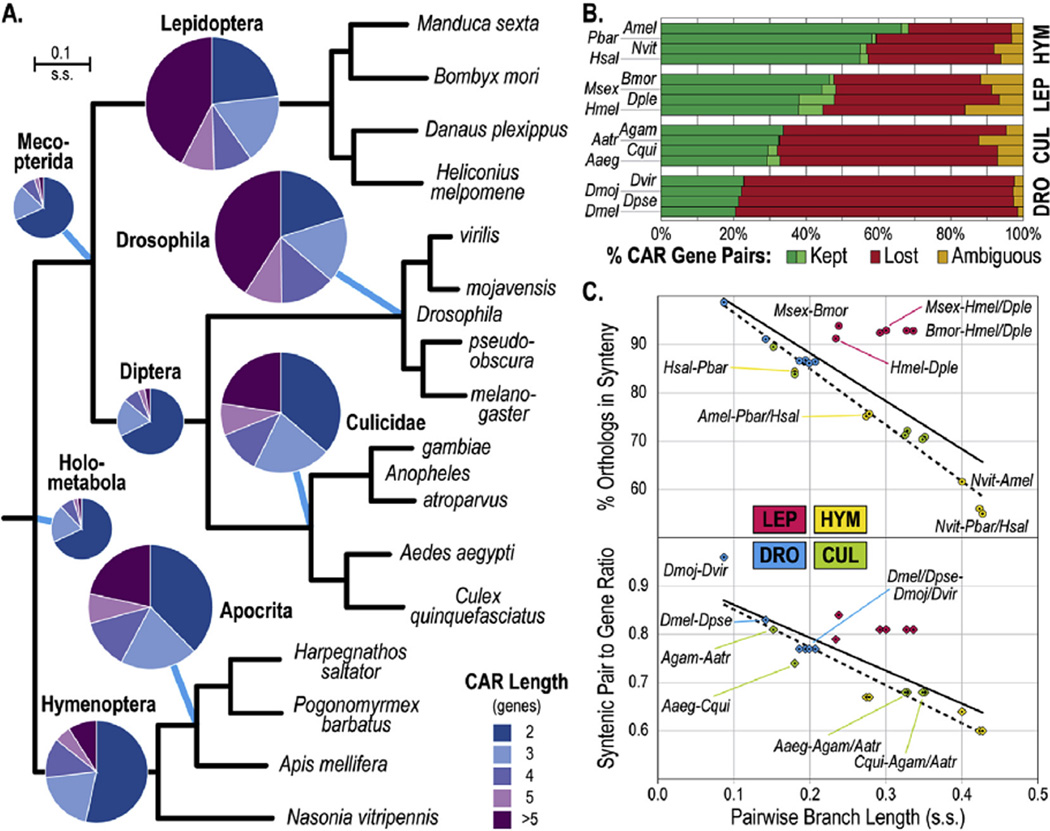
The identification of M. sexta genes with widely-conserved orthologs across the Insecta provided the opportunity to perform genome-wide synteny analyses both within and across four clades from three major insect orders. Employing 7988 single-copy M. sexta – B. mori orthologs with chromosomal assignments in B. mori, ~87% of M. sexta genes spanning ~83% of the genome assembly were mapped to their corresponding chromosomes (Fig. S2). Assigning M. sexta scaffolds to chromosomes in this manner suggested several genomic rearrangements relative to B. mori, many of which correspond to translocations previously detected using cytogenetic techniques (Yasukochi et al., 2009). Contiguous ancestral regions (CARs) were built with the ancestral genomes (ANGES) analysis software (Jones et al., 2012) using 5113 orthologous gene anchors from the outgroup species, the body louse Pediculus humanus, and 16 representative holometabolous species (Table S5). Holometabola, Mecopterida, and Diptera CARs encompassed only 43%–47% of all anchors, the majority of which were found in CARs made up of only two genes, highlighting the large amount of genome shuffling that has occurred over these long evolutionary timescales (Fig. 2A and Table S6). In contrast, many more anchors were captured by the generally much longer CARs of the ancestors of the more closely-related sets of Hymenoptera (72%), Diptera [Culicidae (84%), Drosophila (93%)], and Lepidoptera (93%). Despite moths and butterflies having diverged many millions of years earlier than the fruit flies, the lepidopteran and drosophilid ancestors exhibited similarly high proportions of captured anchors and a majority of long CARs (>5 genes), suggesting less frequent gene rearrangements in lepidopteran genomes. This is supported by examining 1329 neighboring gene pairs from the 873 Holometabola CARs to determine whether they have been rearranged or maintained as neighbors or inferred neighbors (Fig. S3), in the genomes of the 16 extant species (Fig. 2B). Although the Hymenoptera most closely resembled the likely ancestral gene arrangements (~60% maintained), many more ancestrally neighboring gene pairs have been maintained in the Lepidoptera (~47%) than in the Culicidae (~33%) and in the Drosophila (~22%). This was confirmed with the Lepidoptera-Diptera (Mecopterida) ancestor, where the Lepidoptera maintained ~58%of ancestrally neighboring gene pairs, compared with only ~41% for the Culicidae, and ~26% for the Drosophila (Fig. S4).
Fig. 2.
Remarkably conserved synteny among the Lepidoptera. A. Predicted contiguous ancestral regions (CARs) for common ancestors at the major nodes of the insect species phylogeny. Pie charts show distributions of CAR lengths (number of orthologous anchor genes) with diameters proportional to the total number of anchor genes in the CARs. The molecular species phylogeny was built from single-copy orthologs from four representative species from four major groups – Lepidoptera, Drosophila, Culicidae, and Hymenoptera – and rooted with the body louse, Pediculus humanus. B. Examining the fates of the 1329 Holometabola CAR gene neighbor pairs in the sixteen extant species classifies them as kept (dark green, maintained neighbors), likely kept (light green, inferred maintained neighbors), lost (red, no longer neighbors), or ambiguous (orange, missing orthologs). C. Evolutionary distances between species pairs, in terms of branch lengths from the phylogeny in panel A, are plotted against the percentage of orthologous gene anchors maintained in synteny (top) and the syntenic pair to gene ratio (bottom, number of neighboring gene pairs/number of genes maintained in synteny), with linear regressions of all species pairs (solid lines) and all non-lepidopteran species pairs (dashed lines). s.s., substitutions per site; HYM, Hymenoptera; LEP, Lepidoptera; DRO, Drosophila; CUL, Culicidae; Aaeg, Aedes aegypti; Aatr, Anopheles atroparvus; Agam, Anopheles gambiae; Amel, Apis mellifera; Bmor, Bombyx mori; Cqui, Culex quinquefasciatus; Dmel, Drosophila melanogaster; Dmoj, Drosophila mojavensis; Dple, Danaus plexippus; Dpse, Drosophila pseudoobscura; Dvir, Drosophila virilis; Hmel, Heliconius melpomene; Hsal, Harpegnathos saltator; Msex, Manduca sexta; Nvit, Nasonia vitripennis; Pbar, Pogonomyrmex barbatus. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The patterns observed from inferred ancestral genome contents were further explored using pairwise species comparison approaches, similar to the quantifications of synteny and sequence conservation among 12 insects (Zdobnov and Bork, 2007). Comparing pairwise molecular evolutionary divergences from the species phylogeny (Fig. 2A) with synteny quantifications between pairs of species from each of the four clades showed an expected decrease in synteny conservation with increasing evolutionary distances (Fig. 2C). However, these analyses revealed that the Lepidoptera exhibit much higher levels of synteny conservation than would be expected given their levels of molecular evolutionary divergence. This was true for two different measures of the extent of synteny conservation: (i) the proportion of orthologous anchor genes maintained as neighbors, and (ii) synteny block lengths measured as the ratio of the number of pairs of maintained neighbors to the total number of anchor genes maintained as neighbors. These measures employed the same set of anchors as the ANGES-CAR analyses and orthology-inferred neighbors to minimize the under-estimation of conserved synteny due to assembly fragmentation (Table S7 and Fig. S5). Pairwise quantifications therefore support the observations from reconstructed ancestrally-contiguous regions that showed unusually high levels of synteny conservation among the Lepidoptera.
The extent to which this exceptional conservation of synteny results from the properties of holocentric chromosomes remains an intriguing mystery. Holocentric chromosomes appear to have arisen independently at least 13 times in a wide variety of species, including plants, nematodes, and insects (Melters et al., 2012). Like other insects with this distinctive centromere structure, Lepidoptera lack an essential CenH3 histone variant required for kinetochore assembly in species with monocentric chromosomes, and are missing other key inner kinetochore components (Drinnenberg et al., 2014). The idea that holocentricity contributes to increased chromosome diversification and speciation is widely held (Bureš and Zedek, 2014; Jankowska et al., 2015) and is based in part on a high diversity of chromosome numbers in closely-related species, including moths (Nagaraju and Jolly, 1986) and butterflies (Kandul et al., 2007; Vershinina et al., 2015). Yet, the data presented here, together with many published karyotypes (Robinson, 1971), support a well-conserved chromosome number of n = 31 throughout the taxon, along with great stability in microsynteny even after major karyotypic evolution via chromosomal fusions such as in Heliconius butterflies with n = 21, which appears to have been stable for as long as 6 million years (Davey et al., 2016). The low cytogenetic resolution of lepidopteran mitotic and meiotic chromosomes and paucity of genetic maps preclude a broad survey of fusions, translocations or macro- and micro-inversions in lepidopteran species with different chromosome numbers. Nevertheless, we anticipate many whole genome sequences of moths and butterflies will be forthcoming and will provide the information necessary for more definitive answers about the evolutionary stability of lepidopteran chromosomes.
3.3. Gene expression through development
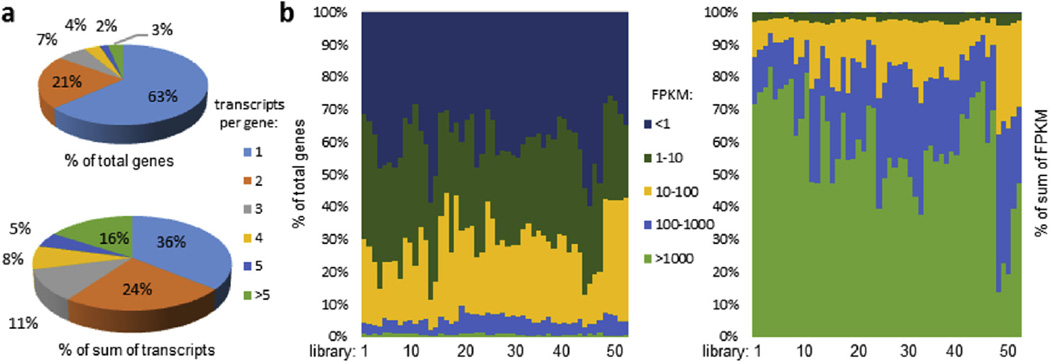
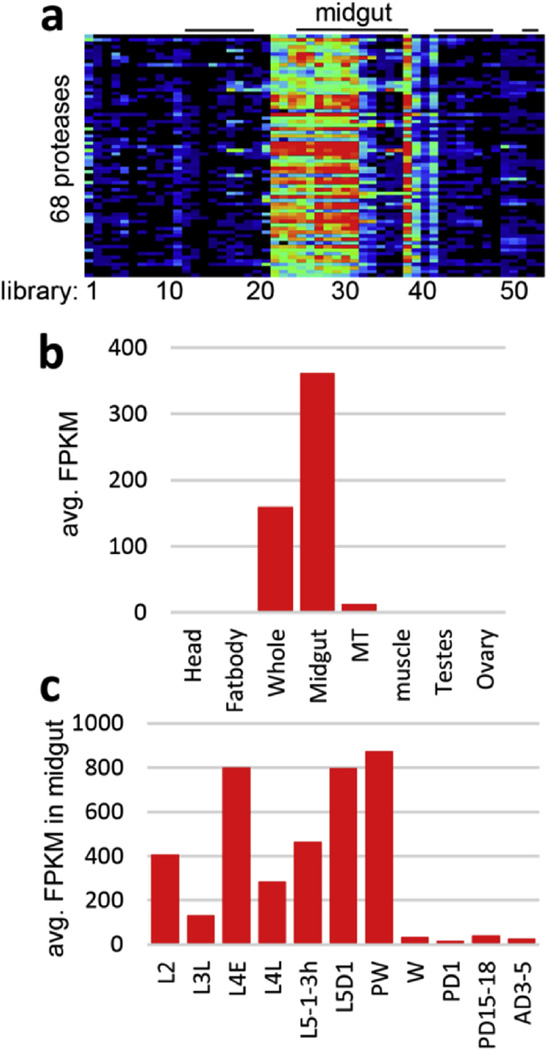
To provide an overview of M. sexta gene expression, we performed a broad RNA-seq survey of gene expression across tissues and developmental stages and times (Table S8). Reads from 52 RNA-seq libraries were mapped to the genome to identify differentially regulated genes and alternative splicing in the tissue samples. Expression patterns (ftp://ftp.bioinformatics.ksu.edu/pub/Manduca/OGS2/OSU_files/) are available for 27,531 transcripts. For most M. sexta genes (63%) we identified a single transcript. However, a small percentage of the genes (3%) had more than five alternative transcripts per gene, and these represented 16% of the total number of M. sexta transcripts (Fig. 3). In general for each library, about 40%, 30%, and 25% of the transcripts had average FPKM values of <1, 1–10, and 10–100, respectively. Highly transcribed genes (FPKM >100) contribute about 85% of total FPKM in the RNA-seq libraries. The tissue- and development-specific expression patterns can be examined in the context of development and gene function. For example, we identified a unique expression pattern for 68 of the total 193 serine protease-like genes, whose mRNAs were substantially more abundant in midgut, as compared with other tissues (Fig. 4). The average FPKM values for this group of serine proteases were higher in the early portion of each larval stage, correlating with the growth-molting cycle. As feeding ceases in the pre-wandering stage, the high level of these transcripts was reduced and almost completely shut down at the onset of wandering stage, remaining low in pupae and adults. Thus, we conclude that these enzymes participate in food digestion and their expression is tightly regulated. This example illustrates the utility of this extensive survey of gene expression and should provide a wealth of information to support future functional studies in this model species.
Fig. 3.
Overview of gene transcripts and their relative levels in the 52 cDNA libraries. (a) Distribution of genes and their transcripts based on splicing variants per gene; (b) Percentages of OGS2.0 genes (left) and sums of their FPKM values (right) in the five FPKM categories. The 52 libraries are in the same order as described in He et al. (2015).
Fig. 4.
Gene expression of 68 gut serine proteases and their close homologs in various tissue samples. (a) The mRNA levels, as represented by log2(FPKM +1) values, are shown in the gradient heat map from blue (0) to red (≥10) (Cao et al., 2015c); (b) Average FPKM values in whole body, Malpighian tubules (MT), and other tissues; (c) stage-dependent transcription in midgut tissues from 2nd instar larvae (L2), late 3rd instar (L3L), early (L4E) and late (L4L) 4th instar, 1–3 h, day 1, pre-wandering (PW) and wandering (W) 5th instar larvae, day 1 and days 15–18 pupae, and days 3–5 adults. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Intracellular pathways, cell biology, and development
M. sexta has provided important insights into many areas of insect science, ranging from flight dynamics to physiological and molecular mechanisms of development. With the availability of the M. sexta genome, we examined a number of representative gene groups from cellular and developmental pathways to structural components of the exoskeleton and egg. The comparative analysis of such representative gene groups will permit an enhanced overview of M. sexta and a more detailed view of similarities and differences with other insect groups and other metazoans. Our analyses showed that while a number of the representative gene family members were highly conserved across the Insecta, there were also important examples of gene expansions or loss that may be related to the unique biology of the lepidopteran group more generally, or M. sexta specifically. As representative groups, we examined gene families associated with intracellular pathways (cathepsin proteins, apoptosis and vacuolar protein sorting), regulatory and developmental processes (growth factors and Hox genes), and structural proteins and associated mechanisms (cuticle proteins, chitin-related proteins, myofilament proteins, and chorion proteins).
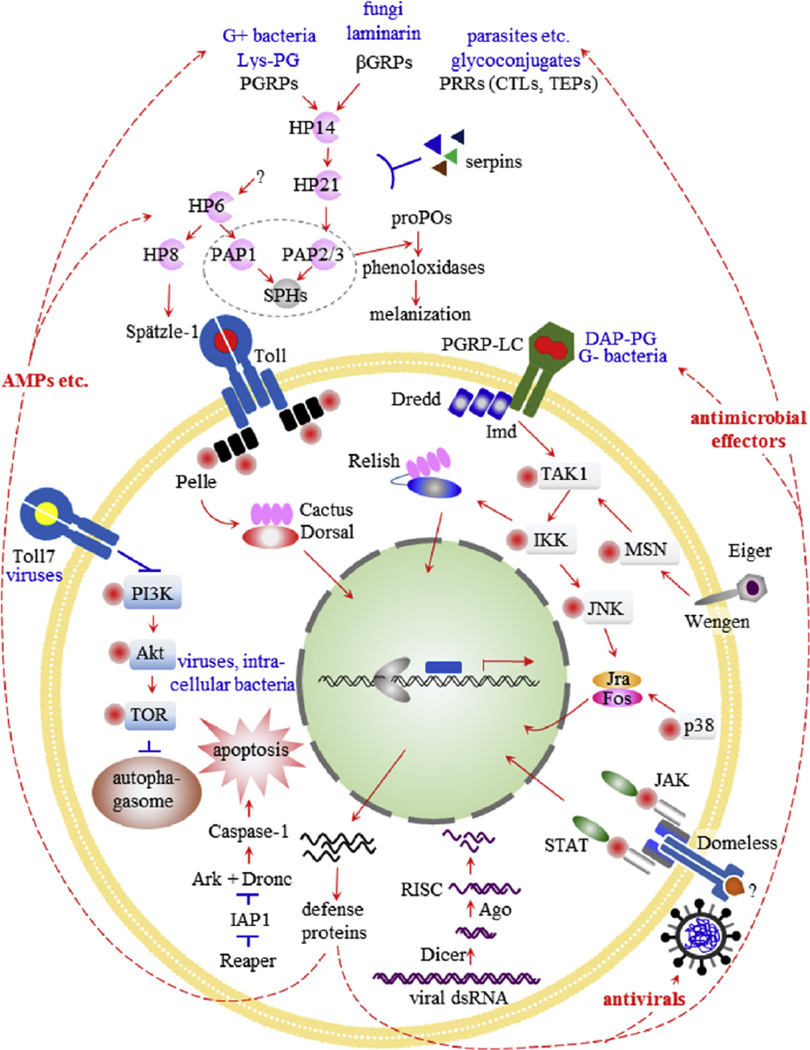
3.4.1. Apoptosis genes
The term “programmed cell death” (PCD) was originally coined to describe the loss of the intersegmental muscles of Lepidoptera (Lockshin and Williams, 1965). The best-characterized mechanism of PCD, apoptosis, is a phylogenetically-conserved process that facilitates an enormous variety of organismal processes that range from defense against pathogens, to developmental reorganization and efficient repurposing of macromolecules. We identified and annotated a total of 23 apoptosis-related genes in the M. sexta genome. These included genes encoding six caspases, five BIR-containing proteins, three p53-related proteins, and single homologs encoding reaper, buffy, Bcl-2, Ark/Apaf-1, ICAD, cytochrome c, AIF, FADD, Htra2, and Dnr1. Caspases are cysteine proteases that are central mediators of apoptosis and inflammation. All are encoded as inactive zymogens with an N-terminal pro-domain. Initiator/ apical caspases cleave the pro-domain and activate downstream effector/executioner caspases, which, in turn cleave a wide range of essential cellular regulators (Bao and Shi, 2007; Courtiade et al., 2011; Fuentes-Prior and Salvesen, 2004). The six M. sexta caspases compare to seven in Drosophila melanogaster. Phylogenetic analyses of selected insect initiator and effector caspases are shown in Fig. S6. The initiator caspases dronc and dredd are conserved one-to-one in the M. sexta, B. mori, and D. plexippus genomes, and conservation of these genes has been observed in all insect genomes sequenced to date (Courtiade et al., 2011). In contrast to the initiator caspases, other caspase types did not exhibit one-to-one orthology. For the damm/dream clade, the M. sexta genome contains two genes that are more similar to each other than any other caspase in the clade (Fig. S6). In addition, these genes reside close together in the genome. Dipteran caspases in the damm/ dream clade show similar phylogeny and gene synteny in their respective genomes (Fig. S6). In comparison, B. mori has one gene in the damm/dream clade, and D. plexippus has no identified gene representative (Fig. S6), in agreement with previous findings (Courtiade et al., 2011). Taken together, these results suggest that damm/dream caspases have undergone a rapid species-specific tandem gene duplication event in most insect species analyzed. Regarding the other caspases, the M. sexta genome contains only two effector caspases, one in the main effector clade and one in a lepidopteran-specific clade, which is similar to B. mori and D. plexippus genomes (Fig. S6). Notably, the D. melanogaster effector caspase decay was not present in M. sexta, B. mori, or D. plexippus (Fig. S6). In summary, the M. sexta genome contains homologs for all non-caspase apoptosis-related genes analyzed, while caspases exhibited one-to-one orthology for initiator caspases but not for effector caspases.
3.4.2. Vacuolar protein sorting (VPS) genes
As much as 30% of the coding capacity of higher eukaryotic genomes is devoted to secreted or membrane bound proteins, requiring membrane or vesicular transport systems (Dancourt and Barlowe, 2010; Kanapin et al., 2003). The vacuolar protein sorting (VPS) proteins perform this task with functions including protein recognition and recruitment to specific vesicular compartments, vesicle formation, vesicle transport, tethering of vesicles to a target membrane, and fusion with a target membrane. Subsets of VPS proteins form multiple functional complexes which include: endosomal sorting complex required for transport (ESCRT), Vps-C, Retromer, GARP, and PI3K sub-complex (Li and Blissard, 2015). The ESCRT machinery of eukaryotic cells is involved primarily in endosomal sorting and trafficking of cargo proteins to multivesicular bodies for protein degradation or autophagy. However, ESCRT proteins are also involved in cytokinesis, and components of the ESCRT machinery are frequently hijacked by viruses to provide the cellular machinery for budding and pinching-off of virus particles exiting from infected cells (Chen and Lamb, 2008; Peel et al., 2011). Of the 17 yeast and 29 human ESCRT pathway proteins, we found 19 genes in M. sexta (Table S9). Analysis of 11 other sequenced insect genomes showed that the same 19 genes are conserved in most cases, including representatives from the Diptera (3 species), Hymenoptera (3 species), Hemiptera (1 species), Phthiraptera (1 species), Coleoptera (1 species), and Lepidoptera (3 species). Eight ESCRT complex genes identified in the human genome were not found in yeast or insect genomes. We also identified 33 M. sexta gene orthologs of proteins that make up the Vps-C, Retromer, GARP, and PI3K complexes and other characterized VPS complexes or genes. Table S9 compares amino acid sequence identities of VPS complex proteins among the 11 representative insect species, between insects, yeast, and humans, and between M. sexta, B. mori and D. melanogaster. Amino acid sequence identities ranged from approximately 30–98% among insect species, with few exceptions. That these protein trafficking components displayed such high sequence conservation is consistent with their central roles in many critical biological processes. Details of specific amino acid sequence and domain conservation and phylogeny of the VPS proteins are provided in a companion publication (Li and Blissard, 2015). In comparison with the yeast genome, VPS gene families are expanded in metazoans. While VPS genes are expanded into various isoforms in insect and human genomes (Schuh and Audhya, 2014), the expansions are less extensive in insects than in the human genome (Table S9). Nevertheless, VPS gene expansions and specific isoforms appear to be highly uniform across the 6 orders of Insecta examined.
3.4.3. C1A peptidases
Proteases perform a wide range of roles in the biology of any organism, from intracellular proteases that activate proenzymes and digest endocytosed proteins, to extracellular proteases that activate immune cascades and digest proteins in the gut and the cuticle. Here we focused on an analysis of C1A peptidases including cathepsins (Rawlings and Salvesen, 2013). In mammals, C1A peptidases are essentially lysosomal enzymes, responsible for primarily intracellular protein degradation. In pathological conditions, C1A cysteine proteases (CPs) can be released from the cell and become involved in remodeling or damaging the extracellular matrix, leading to conditions such as tumor metastasis (Fonovic and Turk, 2014; Tan et al., 2013). In insects, cathepsins are suggested to be involved in tissue degradation during molting and metamorphosis (Hegedus et al., 2002; Homma et al., 1994; Lee et al., 2009; Liu et al., 2006; Zhai and Zhao, 2012), digestion of dietary proteins, or defense against plant toxins or protease inhibitors (Koo et al., 2008; Shindo and Van der Hoorn, 2008; Sojka et al., 2008). 26/29 kDa cathepsins may also have a role in insect immunity (Saito et al., 1992; Serbielle et al., 2009).
Annotation of the M. sexta genome resulted in the identification of 16 new C1A CPs in addition to the 5 cathepsins already described (Miyaji et al., 2007, 2010; Serbielle et al., 2009). Among the C1A CPs, 8 correspond to 26/29 kDa-like cathepsins, 7 encode cathepsin B or B-like proteins, 2 encode cathepsin L proteins, and 2 correspond to a cathepsin F-like protein and the multicystatin procathepsin F. And, for the first time, 2 cathepsin O-like genes were identified. 26/ 29 kDa cathepsins represent the largest C1A CPs gene family in M. sexta. Three gene pairs (cathepsin 26/29 kDa-like 2 and 5; 1 and 3; 7 and 8) are closely linked in the genome and display 80% similarity at the nucleotide level, indicating they likely represent recent gene duplication events. These genes could, however, still be diverging in function since they showed different expression profiles under certain conditions. Globally, these cathepsins were expressed mainly at larval stages in brain, fat body and abdominal muscles. However, we found higher expression levels in late instars and pupae, suggestive of a role in tissue remodeling (Fig. S7). These cathepsins also exhibited interesting expression patterns in fat body and hemocytes of immune-challenged larvae (Zhong et al., 2011). Most 26/29 kDa proteases were down-regulated in these conditions, consistent with results observed on caterpillars parasitized with Cotesia congregata (Chevignon et al., 2015; Serbielle et al., 2009). Only cathepsin 26/29-like 3 was induced after immune challenge in hemocytes.
In the cathepsin B gene family, 5 genes encode cathepsin B-like proteins that lack the occluding loop, and Cathepsin B-like 5 lacks the cysteine residue of the catalytic dyad, suggesting this protein may not be a functional protease. Cathepsin B1 expression in fat body of late larval instars and pupa could be suggestive of a role in molting and metamorphosis. Cathepsin B-like 5 is expressed in the midgut, but whether this protein could act to degrade dietary proteins remains to be tested. A remarkable and high expression level was observed for cathepsin B-like 3 in adult Malpighian tubules. A protein showing similarity to cathepsin B-like proteins has been implicated in renal tubulogenesis in mammals, and it would be very interesting to determine whether cathepsins could also be involved in tubulogenesis in insects (Ikeda et al., 2000; Kanwar et al., 1999). Two cathepsin L genes have been identified in the M. sexta genome. Cathepsin L2 protein sequence lacks the characteristic ERFNIN and RNYD cathepsin-L like motifs normally present in the propeptide. Cathepsin L1 is expressed in most tissues and at different developmental stages, with peaks of expression in pupal fat body and head and midgut of 5th instar larvae, again suggestive of a role in metamorphosis. Cathepsin F, in a manner similar to Cathepsin B-like 3, shows high and specific expression in adult Malpighian tubules.
We conclude that cathepsins identified in the M. sexta genome are likely to be implicated in tissue remodeling during insect development, but could also be involved in organ morphogenesis in adults such as Malpighian tubule tubulogenesis. The role of these proteins in innate immunity is still elusive, but transcriptional regulation of these genes under immune challenge invite further research in this direction.
3.4.4. Growth factor genes
Growth factors are signaling molecules that bind to a receptor on the surface of a target cell, and regulate various cellular processes such as proliferation, growth, and differentiation. Receptor engagement initiates an intracellular signaling cascade. Growth factors can be divided into families, and family member growth factors affect a specific cell type. Together, growth factors are a complex and functionally diverse group of proteins. Growth factor and growth factor-related genes found in the genome of D. melanogaster were identified and annotated in M. sexta (Table S10). A total of 36 growth factor genes were identified. All genes had homologs in B. mori, but five had incomplete sequences for the B. mori homologs. We annotated M. sexta orthologs of D. melanogaster development cell fate proteins defining the Notch signaling pathway (Lai, 2004), a highly conserved cell signaling pathway in multicellular organisms, including Notch ligands delta and serrate homologs (Diaz-Benjumea and Cohen,1995). The Notch binding and antagonizing protein uninflatable (uif) (Jiang et al., 2009) and the transforming growth factor (TGF)-beta superfamily member glass bottom boat (gbb) and its receptor (Ballard et al., 2010; Khalsa et al., 1998) were annotated. Among other genes involved in early development, we found orthologs of decapentaplegic (dpp), which is involved in dorsalventral polarity of organisms (de Celis, 1997), and its receptors punt and thickveins (tkv) (O’Connor et al., 2006). For genes involved in neural development, we identified an ortholog of slit (Kidd et al., 1999), which is a midline repellent expressed in midline glia that binds to the axon guidance Roundabout (Robo) receptor. Among genes with functions associated with insect hemolymph, we identified orthologs of the adenosine deaminase growth factor (adgf), which functions as an adenosine deaminase (Dolezelova et al., 2005); musashi (msi), which encodes Drosophila eye development RNA-binding protein (Nakamura et al., 1994); and a homolog of fat, a factor controlling cell proliferation (Mahoney et al., 1991) and belonging to the cadherin gene superfamily. Two of three adgf homologs were closely clustered in one scaffold (scaffold00232) and another gene copy was found in a separate scaffold (scaffold01562). Similarly, D. melanogaster and B. mori have four clustered copies of adgf. We also identified the fibroblast growth factor (fgf) homolog to the D. melanogaster branchless (bnl; scaffold00044), which encodes the ligand for the breathless receptor (Sutherland et al., 1996). The M. sexta bnl homolog is significantly smaller than the D. melanogaster bnl (770 amino acids in D. melanogaster versus 266 amino acids in M. sexta) but consistent with the size of predicted human FGF polypeptides. However, we were unable to identify pyramus (pyr) and thisbe (ths), which encode fgf ligands of the heartless receptor. Fibroblast growth factors are present in all metazoans, and are also found in an insect virus. The mean predicted protein identity of M. sexta growth factor polypeptides was greater in comparison to B. mori (75.9%) than to D. melanogaster (53.9%) orthologs.
3.4.5. Hox cluster genes
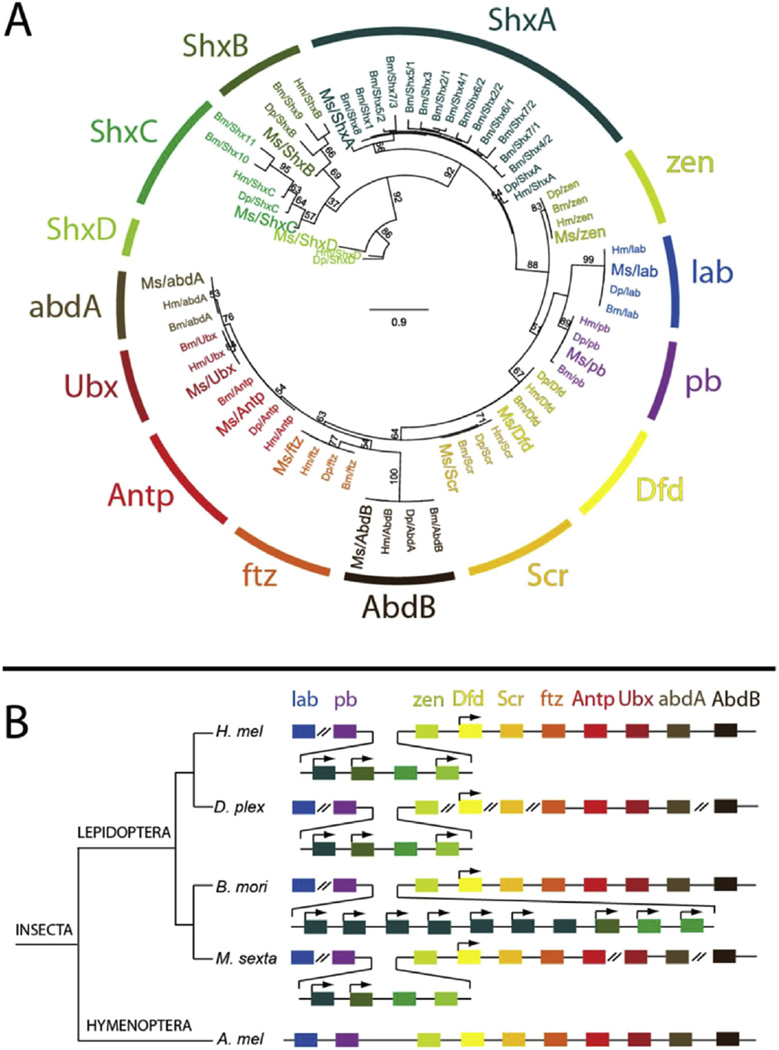
The Hox cluster contains homeodomain transcription factor encoding genes that direct body plan organization by determining segment identity along the anterior-posterior axis. The Hox genes are normally conserved in an organized cluster reflecting the spatial order and developmental timing of affected regions. We identified and annotated the M. sexta Hox cluster using expressed sequences and predicted gene models as a basis for comparison with genome sequences of H. melpomene, D. plexippus and B. mori. We recovered the Hox cluster in 4 scaffolds, with labial (lab) in scaffold00266, proboscipedia (pb) to fushi tarazu (ftz) in scaffold 00164, Ultrabithorax (Ubx) and abdominalA (abdA) in scaffold00058, and AbdominalB (AbdB) in scaffold00007. In addition to identifying all of the canonical Hox genes, M. sexta was found to have four Shx (Special homeobox) genes between pb and zerknullt (zen) (Fig. 5B). Phylogenetic analysis indicated that M. sexta, like H. melpomene and D. plexippus, had orthologs of ShxA, ShxB, ShxC and ShxD (Fig. 5A). B. mori was previously found to have an expansion of ShxA, two ShxC genes (Bm/Shx9 and Bm/Shx10 in Fig. 5A), one ShxB (Bm/Shx9), and no ShxD (Dasmahapatra et al., 2012). The data from M. sexta suggests that these features are likely to be derived in B. mori, and that the ancestor of B. mori and M. sexta had a single copy of ShxA-D. Relative to the butterflies, M. sexta ShxD is reversed in orientation (Fig. 5A). The differences in gene number, type and orientation between M. sexta and the other Lepidoptera that have been studied suggest on-going Hox cluster evolution in this large group of insects.
Fig. 5.
Manduca Hox gene cluster. (A) A phylogenetic tree was constructed using translations of the Hox and Shx gene homeodomains from Manduca sexta (Ms), Heliconius melpomene (Hm), Bombyx mori (Bm) and Danaus plexippus (Dp). Homeodomains were extracted from genomic annotations and aligned using ClustalW. A maximum likelihood tree was generated with 100 bootstrap replicates using PhyML with a LG + G model with parameters sampled from the data. Manduca orthologs are highlighted. (B) A single Manduca scaffold contained the majority of the Hox cluster, including four Shx genes. The orientation of ShxD is reversed relative to the other Lepidoptera. An Apis mellifera (bee) Hox cluster is displayed as a representative ancestral insect cluster.
3.5. Structural molecules
3.5.1. Cuticular protein genes
M. sexta has been an experimental subject for important studies on cuticular morphogenesis (Wolfgang and Riddiford, 1986), identification and hormonal regulation of cuticular protein genes (Riddiford et al., 1986; Rebers and Riddiford, 1988; Horodyski and Riddiford, 1989), and cuticular protein cross-linking during sclerotization (Okot-Kotber et al., 1996; Suderman et al., 2006, 2010). With the annotation of the M. sexta genome, a more complete picture of cuticle synthesis can be developed. Several families of cuticular proteins (CP) have been described based on the presence of conserved sequence motifs (Willis, 2010). These include the Rebers and Riddiford family (CPR, divided into three subgroups: RR-1, RR-2, and RR-3), cuticular proteins with a forty-four amino acid motif (CPF), CPF-like proteins (CPFL) that lack the forty-four amino acid motif, Tweedle proteins (TWDL), and cuticular proteins analogous to peritrophins (CPAPs) (Table 3). The largest family is CPR with gene numbers ranging from as few as 32 in A. mellifera to as many as 156 in Anopheles gambiae. Astonishingly, 207 CPR genes have now been identified in M. sexta (79 RR-1,124 RR-2, 4 RR-3), indicating extensive gene duplication, thus making M. sexta the insect with the greatest number of CPR genes described to date (Dittmer et al., 2015). In comparison, B. mori has 148 CPR genes (56 RR-1, 93 RR-2, 4 RR-3) (Futahashi et al., 2008). Comparison of the M. sexta and B. mori CPR genes (Dittmer et al., 2015) indicated that the greatest difference was evident with the RR-2 genes, for which only 51 orthologous pairs could be established. However, five orthologous groups could be identified, in which 3–13 M. sexta RR-2 genes (42 in total) were clearly related to 3–6 B. mori RR-2 genes (23 in total), although within these groups the M. sexta genes were more closely related to each other than any one of them was to a B. mori gene. This suggests that each group arose from a common ancestral gene that later underwent duplication after speciation. Within the RR-1 group, putative orthologs were found in M. sexta for 52 of the 56 B. mori genes. An additional 16 M. sexta RR-1 genes represent an expansion of just four B. mori genes (CPR2, CPR13, CPR41, CPR46). Only 11 of the M. sexta RR-1 genes did not have an identifiable ortholog in B. mori. In contrast to the CPR genes, M. sexta has similar numbers of CPF, CPFL, TWDL, and CPAP genes in comparison to other insects, with one-to-one orthology identifiable for many of them (Table 3) (Dittmer et al., 2015). A further finding from this annotation is the identification of five additional CPAP1 genes, expanding on the 10 genes originally identified (Jasrapuria et al., 2010).
Table 3.
Cuticular Protein (CP) gene numbers for selected species.
| Species | CPR | CPF & CPFL | TWDL | CPAP |
|---|---|---|---|---|
| An. gambiae | 156 | 11 | 12 | 21 |
| Ap. mellifera | 32 | 3 | 2 | 20 |
| B. mori | 148 | 5 | 4 | 24 |
| D. melanogaster | 101 | 3 | 27 | 22 |
| M. sexta | 207 | 7 | 4 | 25 |
| T. castaneum | 104 | 8 | 3 | 22 |
Abbreviations are CPR (RR family cuticular proteins), CPF (cuticle proteins with 44 amino acid domain), CPFL (CPF-like), TWDL (Tweedle), CPAP (cuticular proteins analogous to peritrophin). Gene numbers for M. sexta and the CPAP family are from (Dittmer et al., 2015). Gene numbers for An. gambiae, Ap. mellifera, and D. melanogaster are from (Willis, 2010) and references within. Gene numbers for B. mori are from (Futahashi et al., 2008). Gene numbers for T. castaneum are from (Dittmer et al., 2012).
The 52 RNAseq libraries created to aid the gene annotation were unfortunately not well suited for examining CP gene expression. Epidermal tissue alone was not specifically collected, but was present in libraries prepared from eggs, whole larvae (first, second, third instar), and heads. Not surprisingly, these libraries showed the highest level of CP gene expression both in the number of CP genes expressed as well as FPKM values. Unexpectedly, CP gene transcripts were also found in libraries prepared from midgut, Malpighian tubules, fat body, testes, and ovaries, likely indicating contamination from trachea or epidermis during the dissection process. It is generally expected that RR-1 proteins are more abundant in soft cuticle, and RR-2 proteins more abundant in hard cuticle, but both groups of proteins can be found in soft or hard cuticle (Willis, 2010). Sixty-one of the 79 RR-1 genes were present in scaffolds as 5 clusters of 5, 11, 14, 14, and 17 genes. However, coordinated RR-1 gene expression was restricted to 5 small groups of 3–4 genes each, with high levels of expression in libraries prepared from larval heads (various stages and both pre- and post-molt), 1st through 3rd instar whole larvae (collected 1 day post-molt), and from abdominal muscle just prior to or just after the 4th to 5th instar molt. Thus, the RR-1 genes displayed a wide variety of expression patterns regardless of their chromosomal location. A library in which RR-2 genes were highly expressed was from heads of 4th instar larvae after head capsule slippage, consistent with an expected role for RR-2 proteins in formation of hard, sclerotized cuticle. Forty-one percent of the CP transcripts from heads of adult day 1 came from just two genes, CPH30 and CPH31; the corresponding proteins contain an 18 amino acid motif identified in several insect CPs (Willis, 2010).
An intriguing finding of this analysis was the low to moderate expression levels in nearly all of the libraries for a group of 8 genes, including five from the CPAP group (CPAP1-C, 1-H,1-M, and CPAP3-D2 and 3-Cb) and two from the RR-3 group (CPR146 and 149). The nearly ubiquitous expression pattern of these genes suggests that they may be important for general cuticle synthesis or synthesis of tracheal cuticle. Another group of 6 genes (the RR-2 genes CPR68 -70 and TWDL2–4) were near exclusively expressed in 7 of the 16 libraries prepared from pre-molt tissues (head, fat body, midgut, and abdominal muscle from 4th instar larvae after head capsule slippage, as well as 3 day old eggs, midgut from 3rd instar larvae after head capsule slippage, and fat body of 15e18 day old pupae); this is similar to B. mori where the TWDL genes were shown to be coordinately expressed, with the highest expression occurring at the larval stage during the molt (Liang et al., 2010). A more comprehensive description of the expression analysis can be found in Dittmer et al. (2015).
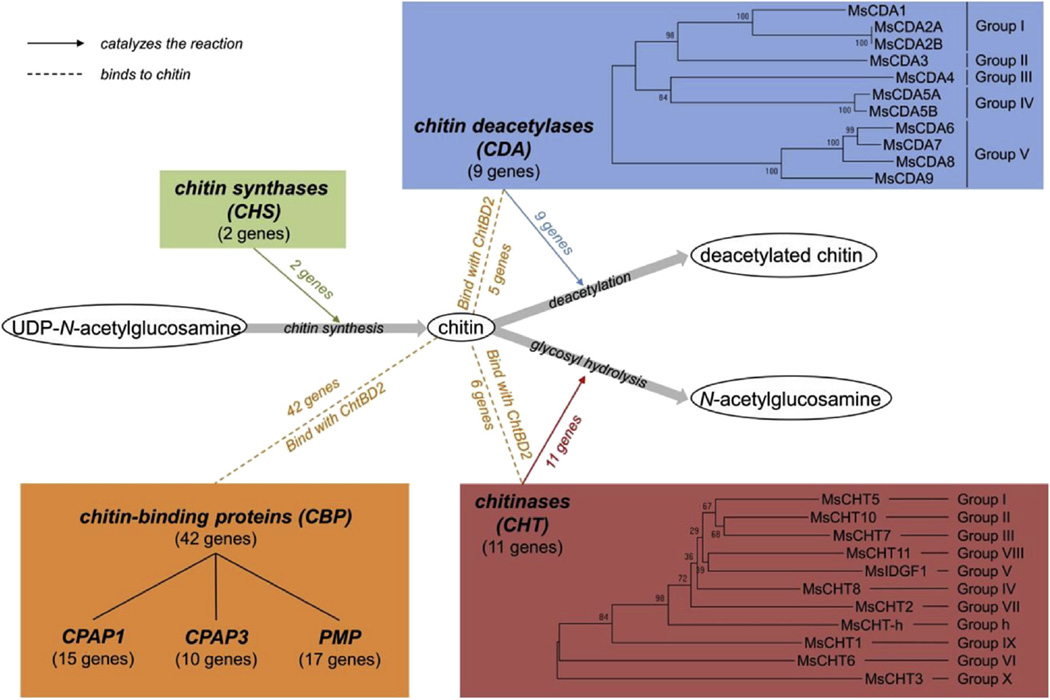
3.5.2. Chitin metabolism-related genes
Chitin, a polymer of N-acetylglucosamine, is one of the most abundant biopolymers on earth, second only to cellulose. Chitin is a major structural component of the arthropod exoskeleton, and in insects it is also a major component of the peritrophic matrix lining the digestive tract. The M. sexta genome contains a large number of genes encoding enzymes for chitin metabolism, including chitin synthases (CHS), chitin deacetylases (CDA), chitinases (CHT), and chitin-binding proteins (CBP) that interact with chitin to modulate its related functions (Fig. 6). The M. sexta genome has only two chitin synthase genes (CHS1 and CHS2), which have been described previously (Hogenkamp et al., 2005; Zhu et al., 2002), consistent with other arthropods (Merzendorfer, 2011). We identified 9 chitin deacetylase genes in the M. sexta genome (MsCDA 1 to 9), with a total of 11 different transcripts coding for the extracellular chitin modifying enzymes. Each CDA gene belongs to one of the 5 phylogenetic groups described to date for the insect CDA family (Dixit et al., 2008). The individual MsCDAs contain a typical carbohydrate esterase 4 domain: CE4-1 (cd10974; in MsCDA from groups I to III) or CE4-2 (cd10975; in MsCDA from groups IV and V), with different degrees of sequence variation. We found 11 chitinase genes in the M. sexta genome, all of which contain the conserved glycosyl hydrolase domain GH18 (smart00636). Nine of the 11 M. sexta chitinases belong to the eight phylogenetic groups of CHTs described to date (Arakane and Muthukrishnan, 2010), whereas two chitinases exhibited protein domain organization and sequence signatures different from the currently known CHTs, therefore creating two new CHT groups (groups IX and X) (Tetreau et al., 2015a). Group IX is a more ancestral chitinase family. It is one of the first chitinase groups with a ChtBD2 domain that appeared in arthropods and a closely related representative has also been found in Echinodermata. Conversely, group X is a recent group with representatives found only in the lepidopteran, coleopteran, hymenopteran and dipteran genomes currently available (Tetreau et al., 2015a). We also confirm the presence of a Lepidoptera specific chitinase-h (group h) in M. sexta, which supports the hypothesis of a recent horizontal transfer from bacteria to Lepidoptera (Tetreau et al., 2015a).
Fig. 6.
Summary of the families of genes coding for chitin metabolism enzymes and chitin binding proteins (CBPs) in the M. sexta genome. Dotted lines indicate the binding of CBPs to chitin by their chitin binding domains (CBD – orange), and lines with an arrow indicate that chitin metabolism enzymes and their functions in chitin synthesis (chitin synthases, CHS – green), deacetylation (chitin deacetylases, CDA – blue) and degradation (chitinases, CHT – red). For CDA and CHT, a phylogenetic tree has been constructed using the neighbor-joining method, with 1000 replications of bootstrap analyses, implemented in MEGA 6.06 (Tamura et al., 2011). CPAP: cuticular proteins analogous to peritrophin; PMP: peritrophic matrix protein. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Chitin is always associated with proteins in nature (Jasrapuria et al., 2010). We identified 53 genes in the M. sexta genome that we classified as coding for chitin-binding proteins (CBPs) based on the presence of at least one chitin-binding domain (ChtBD2, pfam01607) (Tetreau et al., 2015b). Among the CBPs, 11 are chitin metabolism enzymes (5 CDAs and 6 CHTs) that contain a CBD and 42 are structural chitin-binding proteins. Among the structural CBPs, 21 proteins had only one CBD. Fifteen of them were cuticular proteins analogous to peritrophins with 1 ChtBD2 (CPAP1s) and 10 were CPAP3s (CPAPs with 3 copies of ChtBD2). Seventeen of the CBPs were peritrophic matrix proteins (or peritrophic membrane proteins) (PMPs), which contained 1 to 13 ChtBD2 domains and were most abundant in the midgut. We performed a comprehensive analysis of chitin-binding domain evolution in insects (Tetreau et al., 2015b) and found that CPAP1s formed a clearly distinct cluster in the phylogenetic tree and that the three ChtBD2 domains in CPAP3s appeared before the diversification of the CPAP3 family. Similarly, ChtBD2 domains appeared to be associated with the CHT and CDA catalytic domains in CHTs and CDAs before the evolutionary radiation that led to the high diversity of chitin metabolism enzymes observed in M. sexta and in the other Lepidoptera. Finally, the number of PMPs and the organization of ChtBD2 domains in PMPs appear to be species-specific. Therefore, ChtBD2 domains evolved in each species in a species-specific manner, probably driven by environment and feeding patterns.
3.5.3. Myofilament protein genes
An important characteristic of many insects is flight, which is facilitated by large and highly specialized flight muscles in the adult thorax. The physiology of insect thoracic flight muscles may be either synchronous or asynchronous. Synchronous flight muscles contract in a direct one-to-one response to a motor neuron impulse (action potential). In contrast, asynchronous flight muscles (typically found in insects with high wing stroke frequencies) may contract several or many times in response to a single nerve impulse. Insects with asynchronous muscles, such as D. melanogaster, express different myofibrillar protein isoforms relative to those found in the body wall muscles. These isoforms are generated from either several members of a gene family or by alternative splicing of a unique gene. Studies in D. melanogaster have examined the importance of these protein variants in regard to the assembly and stability of the flight muscle sarcomeres, as well as flight performance. This accumulated information from D. melanogaster leads to the question of (1) whether a similar set of protein variants would be found in asynchronous flight muscles but not in synchronous flight muscles of other insects or (2) whether the protein variants described in D. melanogaster flight muscles represent the difference between flight and non-flight muscles irrespective of whether the flight muscles are asynchronous or not. We tested these alternate hypotheses by analyzing the flight muscle proteome of M. sexta, an insect with synchronous flight muscles. The current analysis covers genes encoding the main myofilament proteins, actin, myosin, the troponin complex and tropomyosin, as well as the large proteins of the elastic filament. Except for actin and troponin C, which are represented by gene families, all the other annotated proteins are encoded by single copy genes with multiple isoforms generated by complex alternative splicing options. The myosin and troponin C variants identified in the M. sexta flight muscles are very similar to the ones identified in the D. melanogaster flight muscle system, lending support to the second hypothesis. On the other hand, troponin I and the elastic proteins projectin and Sallimus show unique isoforms (Ayme-Southgate et al., 2015). Fig. S8 shows the arrangement of the M. sexta unique myosin heavy chain gene with exons and alternative splices illustrated. Thus, in M. sexta, the flight muscle proteome may represent a protein composition indicative of muscles capable of generating the power output needed for flight, but not adapted to the asynchronous activation seen in other insects such as Hymenoptera and Diptera.
3.5.4. Chorion protein genes
Chorion proteins of lepidopteran insects assemble to form natural protective amyloids that are the major components of eggshells. As such, these proteins allow gas exchange between the oocyte or the developing embryo and the environment and protect it from viral, bacterial or fungal infections and environmental adversity (Hamodrakas, 1992). Chorion protein sequences have a tripartite structure, which consists of a conserved central region and two flanking arms. The central region contains glycine tandem repeats every 6 amino acid residues (Iconomidou and Hamodrakas, 2008). Based on the central regions, chorion proteins are classified into two main classes (A and B), while the degree of amino acid enrichment for proline, glycine, or cysteine residues in the amino-and carboxy-terminal arm sequences is associated with the stage of choriogenesis (early, middle, or late, respectively) for each chorion protein (Rodakis et al., 1982). The genes and genomic structure of most lepidopteran chorion genes are uniform. Each gene contains two exons. The first exon and the first 9 bp of the second exon encode a signal peptide for secretion. The genes encoding chorion proteins are arranged in divergent non-overlapping pairs (DNOPs). The DNOP of genes share a common <400 bp promoter region and the two genes are transcribed in opposite orientations. In B. mori DNOP genes are clustered into a single genetic locus (Kafatos et al., 1995; Chen et al., 2015a,b). With rare exceptions, each DNOP consists of a gene for a class A chorion protein and a gene for a class B chorion protein. As DNOP genes share the same cis-regulatory elements, they are co-expressed (Lecanidou and Papantonis, 2010a, b) and their corresponding proteins share similar amino acid enrichment patterns in their arm sequences (Lecanidou et al., 1986). These proteins interact during chorion formation.
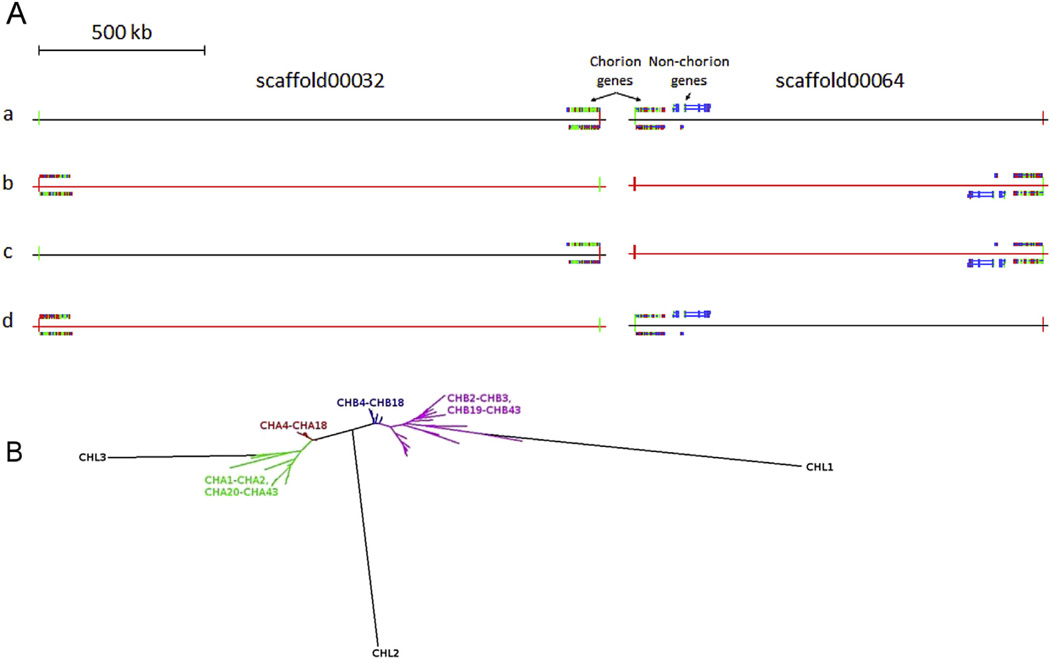
In total, 79 genes and 2 pseudogenes that code for chorion proteins were identified. Of these, 35 genes code for class A chorion proteins, 41 genes code for class B proteins, 3 genes encode chorion-like proteins and 2 genes are pseudogenes. 42 and 34 of the chorion protein genes were early and middle chorion protein genes, respectively, while no late (high-cysteine) chorion protein genes were found. As tandemly repeated domains are difficult to assemble completely, the chorion locus in M. sexta is split into three scaffolds with gaps: The sequenced part of the chorion protein gene cluster(s) comprises 99 kb of the 3′ end of scaffold00032 (1604186–1702971), 88 kb of the 5′ end of scaffold00064 (1–87512) and 2 kb of the entire scaffold04803 (1–1730). As the chorion genetic locus of the M. sexta genome is fragmented, we can only speculate on its reconstitution, taking into consideration syntenic data from other lepidopteran species. Currently, the only fully sequenced lepidopteran chorion locus is a 717 kb region in B. mori, containing 127 chorion protein genes. This locus is split into two distinct chorion gene clusters which are separated by a 197 kb region where 4 non-chorion genes are located. Syntenic analysis shows that orthologs of these non-chorion genes are neighbors of chorion genes in 4 ditrysian families (Chen et al., 2015a,b). Another (possibly only partially) sequenced lepidopteran chorion locus is a region which may exceed 1170 kb, in H. melpomene. A single scaffold (HE671164) also contains two distinct chorion gene clusters which are located in its 5′ and 3′ ends. The two chorion gene clusters are separated by a 326 kb region where 3 non-chorion genes which are orthologous to the ones of B. mori, are also located. These orthologs are also found adjacent to the chorion gene cluster in scaffold00064 in M. sexta. There are 4 main possible configurations for the M. sexta chorion genetic locus (Fig. 7A). Configuration a assumes a single cluster of chorion genes which does not exceed 190 kb. This is supported by the high degree of sequence similarity between the genes of the 3′ and 5′ ends of scaffold00032 and scaffold00064, respectively. On the other hand, this configuration, as well as configuration d, leaves the non-chorion orthologs outside the chorion locus. Configurations b, c and d assume the existence of two distinct gene clusters. The non-chorion genes are flanked by the chorion gene clusters only in configurations b and c. The total size of the chorion locus and the size of the genomic region between the two chorion clusters exceed 2946 kb and 2760 kb or 1342 kb and 1155 kb, according to configuration b or c, respectively. Thus, the most plausible configuration is c, as the sizes of the chorion locus and of the genomic region between the two clusters are closer to those of B. mori and H. melpomene.
Fig. 7.
Location, structure, and phytogeny of the chorion gene cluster and chorion genes of M. sexta. A. Possible configurations of the chorion locus. Represented are the four different relative orientations between chorion gene containing scaffolds, scaffold00032 and scaffold00064. The two annotated contiguous chorion gene clusters and the non-chorion orthologs adjacent to the scaffold00064 cluster are shown. The reverse complementary strands of the scaffolds are represented in red. scaffold04803 may be adjacent to scaf-fold00032 or to scaffold00064. B. Phylogenetic tree of all chorion protein sequences, based on maximum likelihood. Each class is divided into two subclasses which are clustered together in the genome (early A in green, middle A in red, middle B in blue, and early B in magenta). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To identify the transcription start sites of the genes, the 4 scaffolds that contained chorion protein genes were searched with a lepidopteran chorion protein gene promoter Hidden Markov Model, which was built using a multiple sequence alignment of the promoters of lepidopteran chorion protein genes downloaded from the Eukaryotic Promoter Database (Dreos et al., 2013). Gene pairs were numbered according to the order of appearance in the genome, and gene nomenclature was based on their class and the number of the gene pair to which they belonged (i.e., CHA7 refers to “class A chorion protein found in the 7th gene pair of the cluster”). A phylogenetic analysis revealed that each chorion protein class is divided into two main subclasses, which correspond to early and middle protein genes (Fig. 7B); 35 middle protein genes are flanked in the genome by 3 and 40 early protein genes.
3.6. Neurobiology
3.6.1. Vision
A total of 80 genes involved in eye development or phototransduction were annotated and named according to their D. melanogaster homologs (Table S11). Comparison with putative orthologs in other insect species (B. mori, D. plexippus, A. gambiae, A. mellifera, and Tribolium castaneum) verified that one-to-one orthologs for several eye-related genes are lacking in the M. sexta genome, likely due to Drosophila-specific gene duplications (Bao and Friedrich, 2009). Using RNA-seq data, we identified previously undescribed lepidopteran-specific gene duplications in gene families involved in photoreceptor differentiation pathways (corkscrew [csw], embryonic lethal/abnormal vision [elav]) (Fig. S9A.B) and chromophore binding (prolonged depolarization afterpotential is not apparent [PINTA]) in D. melanogaster. One copy of csw in the M. sexta genome contains 11 exons and the other is intronless, suggesting duplication in the genome via insertion of a retrogene from mature mRNA. The elav gene family consists of RNA binding proteins that are restricted to neurons and regulate post-transcriptional processing. These genes are required for embryogenesis and proper neuronal differentiation (Colombrita et al., 2013). Two elav gene duplication events were identified. They appear to be lepidopteran-specific, and all four M. sexta genes in this family were intronless. PINTA, which binds the visual chromophore in Drosophila eyes, appears to be missing in Lepidoptera; however, the M. sexta genome encodes 42 other CRAL-TRIO domain-containing proteins (GenBank Accession Nos. KT943537-KT943566) (Smith and Briscoe, 2015), far more than other insect genomes examined (D. melanogaster, n = 12; A. gambiae, n = 14; T castaneum, n = 18). Many of the genes in this family have duplicated within Lepidoptera. Lastly, we identified five opsins and two opsin-like genes (Fig. S9C), including two moth-specific long-wavelength opsin genes, both of which have been retained in the B. mori genome while another, a cerebral opsin found in B. mori (Shimizu et al., 2001) has been lost in M. sexta. The two opsin-like genes contain features similar to a Limulus polyphemus peropsin-like protein and an Ischnura asiatica RGR-like protein; however, unlike the Limulus protein and other opsins, both M. sexta proteins lack a conserved lysine in the seventh transmembrane domain to which the chromophore is covalently linked. Searches of moth and butterfly transcriptomes (Macias-Munoz et al., 2016; Smith et al., 2014) yielded transcripts for all seven opsin or opsin-like gene family members in adult lepidopteran heads.
3.6.2. Chemosensation
All major gene families involved in chemosensation have been identified in the M. sexta genome (see also companion papers (Vogt et al., 2015; Koenig et al., 2015)). Three receptor families known to participate in chemosensory detection are found in insects: odorant receptors (OR), ionotropic receptors (IR), and gustatory receptors (GR). OR contribute to the detection of volatile chemical cues, GR primarily detect contact chemical cues and CO2, and IR contribute to both olfaction and gustation. The GRs are also likely ancestral to the Polyneoptera-specific ORs (Benton, 2015; Missbach et al., 2014; Penalva-Arana et al., 2009; Robertson and Wanner, 2006). Ionotropic receptors (IRs) are likely derived from ionotropic glutamate receptors and occur across the Protostomia (Benton et al., 2009; Croset et al., 2010). In D. melanogaster a subgroup of IRs is expressed in neurons associated with coeloconic sensilla on the antenna and mediate responses to volatile chemical cues (Benton et al., 2009; Silbering et al., 2011).
In total 71 OR, 45 GR and 21 IR genes were identified in the genome of M. sexta, similar to that of other lepidopteran species. Phylogenetic analysis with GRs from B. mori, D. plexippus, H. melpomene and D. melanogaster revealed an M. sexta-specific expansion of GRs putatively associated with bitter tastants. Of note is the finding that though the present IR types mostly exhibit one-to-one relationships to those identified in B. mori, IR75.p underwent duplication in M. sexta. Furthermore, the two M. sexta orthologs of the B. mori IR pseudogenes IR68a and IR75a appear to be functional in M. sexta. In the chemosensory periphery, two apparently unrelated classes of small, soluble proteins are involved in perireceptor events, the odorant binding proteins (OBP) and chemosensory proteins (CSP) (reviewed in Sanchez-Gracia et al. (2009)). OBPs belong to a large gene family that is partially characterized by sequence similarity, structural motifs and a specific number and spatial pattern of cysteines. While OBPs are known throughout the Neoptera, the first OBPs identified were the lepidopteran general odorant binding proteins (GOBPs) and pheromone binding proteins (PBPs). Although five GOBP/PBP genes were previously reported from M. sexta (Gyorgyi et al., 1988; Robertson et al., 1999; Vogt et al., 1991, 2002), our analysis of the draft M. sexta genome identified six genes in this family. Comparative structural and spatial analysis of this gene complex with three other lepidopteran genomes (B. mori, D. plexippus, H. melpomene) suggested a history of gene gain and loss, strongly associated with long distance sex attraction in moths, and loss of a PBP in butterflies (Vogt et al., 2015).
We identified 19 complete CSP family genes in the M. sexta genome. In addition, we also found three more full-length CSP domains, annotated as part of multi-domain proteins that also include other non-CSP domains. Phylogenetic analysis shows that 15 CSP genes have one-to-one orthologous relationships to B. mori CSP family members, and the remaining genes show complex relations indicating gene duplications or losses (Fig. S10). Remarkably, most of the CSP genes (15 out of 19) are located on the same scaffold, forming a ~160 kbp cluster.
Sensory neuron membrane proteins (SNMPs) associate with chemosensory sensilla, and belong to a moderate sized gene family identified by its similarity to human CD36 transmembrane proteins. Insects have around 15 CD36 homologs, which segregate into 3 clades, one of which includes the SNMPs (Nichols and Vogt, 2008; Vogt et al., 2009). Fifteen CD36 homologs were observed in the M. sexta genome, including 3 members of an SNMP clade (two were previously identified (Rogers et al., 2001)). The two additional CD36 clades are comprised of orthologs of D. melanogaster NinaD (a carotene transporter) and Crq (a protein involved in the recognition of apoptotic cells), and D. melanogaster emp (a protein of unclear function but representative of a group with strongly conserved orthologous relationships in multiple species). Two M. sexta genes belong to the NinaD/Crq, and 10 to the Drosophila emp clade. Comparing M. sexta and D. melanogaster, M. sexta has significantly fewer of the NinaD/Crq-genes (2 vs. 6), but more emp-like genes (10 vs 6). The significance of these contractions/expansions remains to be elucidated. A more detailed analysis of the odorant binding protein gene family and the GOBP/PBP complex genes among moths and butterflies can be found in the related companion publication (Vogt et al., 2015).
3.6.3. TRP channels
Transient receptor potential channels (TRPs) are ion channels involved in a large variety of neurological functions, including mechano-, chemo- and thermo-sensation (Matsuura et al., 2009). We found a total of 19 TRP genes in the M. sexta genome, six more than in D. melanogaster (Fig. S11), including several duplications in the TrpA subgroup, which is associated with thermosensation. Three copies of TrpA5, a gene previously identified only in A. mellifera and T. castaneum, are present in M. sexta. The TrpA5 homologs are clustered together on the same scaffold, suggesting recent duplication. The TrpC gene encodes a protein involved in phototransduction, and in contrast to most other insect species, which have only one TrpC gene, M. sexta has 4 genes belonging to the TrpC group. Phylogenetic analysis suggested that 4 copies may be the ancestral state for insects.
3.6.4. Ion channels
3.6.4.1. GABA and glycine receptors
Ionotropic glycine and γ-aminobutyric acid (GABA) receptors are ligand gated chloride channels that mediate rapid, usually inhibitory, synaptic transmission. Both GABA and glycine receptors are pentameric and belong to the Cysloop superfamify of neurotransmitter receptors. We identified nine different receptors in M. sexta: five GABA receptors and four glycine receptors. GABA receptors are the targets of the cyclodiene and phenylpyrazole insecticides. Lepidoptera, including B. mori, P. xylostella and Heliothis virescens have an expansion of the RDL (resistant to dieldrin) genes in this gene family. The M. sexta genome shows a similar expansion, with 3 RDL genes plus two other GABA receptors that are found in all other insects (Yu et al., 2010). The RDL expansion has occurred independently in each lepidopteran species examined previously, with different genes being duplicated in each case. However, we found that the gene duplication pathway in M. sexta was similar to B. mori. Since both M. sexta and B. mori belong to the Bombycoidea this might be a general characteristic of the superfamify in comparison to other Lepidoptera. Expression levels of GABA and glycine receptors, measured by RNA-seq, were generally low, as is often found with ion channels, although higher levels of expression were found in tissues rich with neurons such as the head (brain). RDL3 (Msex2.14174) appeared to be an exception to this pattern. In addition to displaying relatively higher expression in the brain, RDL3 was expressed in the fat body, Malpighian tubules, testes and ovaries of pupae and adults.
3.6.4.2. Nicotinic acetylcholine receptors
Nicotinic acetylcholine receptors (nAChRs) are Cys-loop ligand gated ion channels that mediate fast synaptic transmission in insect neurons. They are the target of the neonicotinoid class of insecticides and similar genes are found in species as diverse as bacteria and mammals. The M. sexta genome contains 12 nAChR genes (Table S12), the same number as B. mori and similar to other insects (Jones and Sattelle, 2010). The complement of genes is similar, with 9 a subunits (α1–9) and three β subunits (β1–3). Expression of nAChRs as measured by RNA-seq, was generally low as is often found with ion channels, but higher levels of expression were observed in tissue rich with neurons such as the brain. Some exceptions to this pattern are the α9 and the β protein subunits, which seem to be expressed in a variety of tissues including fat body, eggs and muscle.
3.6.5. Cyclic nucleotide signaling
Cyclic nucleotides serve as secondary messengers in many signal transduction pathways, and cyclic nucleotide signaling involves members of several distinct protein families. We identified members of all major gene families involved in the cyclic nucleotide signaling process. Noteworthy is the presence of five receptor guanylyl cyclase (rGCs) genes in the M. sexta genome, in addition to the previously cloned rGC (MsGC-II) (Morton and Nighorn, 2003). One of the newly identified rGCs is predicted to be the eclosion hormone (EH) receptor based on sequence similarity to the medffly EH receptor. Another guanylyl cyclase (MsGC-I) (Nighorn et al., 2001) related to rGCs was previously found in M. sexta. Using the genome data, we confirmed an unusual feature: that MsGC-I does not have an extracellular ligand binding domain. We also identified a second GC (Msex007716) that apparently lacks this extracellular domain. In addition to the soluble guanylyl cyclases previously reported (Nighorn et al., 1998, 1999), we also identified a fourth subunit (Msex002928) that is likely to form an oxygen-sensitive cyclase with Ms-GCβ3.
Cyclic nucleotide-dependent protein kinases are the major intracellular receptors of cyclic nucleotides. Cyclic AMP-dependent protein kinases (PKAs) are formed from two regulatory subunits and two catalytic subunits. The M. sexta genome contains 4 genes that code for catalytic domains of PKA and two genes that code for conventional regulatory subunits. An atypical regulatory subunit which is encoded by the swiss cheese gene (also known as the neuropathy target esterase) was recently identified in D. melanogaster (Bettencourt da Cruz et al., 2008). An ortholog of swiss cheese is also present in M. sexta. In summary, most if not all of the genes known to be involved in cyclic nucleotide signaling identified in other insects appear to be represented in the M. sexta genome.
3.6.6. Neuropeptides
M. sexta has been a model system in pioneering studies of insect endocrinology, used to uncover neural and hormonal controllers of insect development, molting and metamorphosis (King et al., 1974; Nijhout and Williams, 1974; Truman et al., 1974; Zitnan et al., 1996). The genome sequence provided further insights into the neuroendocrine system of this model species, in addition to confirmation of the repertoire of genes previously studied. Neuropeptides are small signaling molecules involved in a broad range of neuronal functions. The M. sexta genome sequence revealed a total of 85 genes encoding neuropeptides and endocrine peptides (Fig. S13), of which 62 have not been previously reported. There are a number of cases showing recent expansions in this group of genes, although there is a general pattern of one-to-one orthology with corresponding neuropeptide genes of B. mori. One such expansion is illustrated by genes encoding insulin-like peptides (ILP), of which 26 were found in the M. sexta genome. Of these, 22 are clustered within 44 kbp, and are found in a pattern suggesting multiple duplications of a block of paired genes (Fig. S13A). A phylogeny of the ilp genes (Fig. S13B) supports the hypothesis that the gene expansion in M. sexta was likely a recent event, independent from that of the expansion in Bombyx which carries 44 ilp genes (or bombyxins). The detection and relative transcription levels of ilp genes indicated that ancestral ilp genes (i.e., ilp-A, B, D, E, F, X, and Y) are expressed in the larval head, whereas recently duplicated ilp genes (i.e., ilp-P2, Q, R, S, and T) are expressed in the adult head (Fig. S13C). Many ilp genes appear to be highly expressed in the adult midgut and in the pre-wandering larval Malpighian tubules (Fig. S13C). Other increased gene copy numbers occurring as clusters are: the adopokinetic hormone (AKH) cluster with three genes, the allatostatin C cluster with two genes, the CCHamide cluster with three genes, and the Trissin cluster with four genes (Fig. S13D). The trissin cluster also contained the crustacean cardioactive peptide (ccap) gene nested between trissin genes. Of note is the observation that there are independent expansions of the ilp gene in two closely related species in the same superfamily Bombycoidae, and moderate levels of copy number increase in each of the neuropeptide genes AKH, CCHamide, and trissin in M. sexta, without gene-loss compared to those in B. mori. The functional importance of the unique gene expansions in M. sexta is still unknown. The large number of newly identified neuropeptides in the genome of this model insect offers exciting possibilities for unveiling new neuropeptide functions.
3.6.7. Neurotransmitters
Biogenic amine and small molecule neurotransmitters, such as dopamine, serotonin, octopamine, tyramine, and acetylcholine, are major classes of invertebrate neurotransmitters that have fundamental roles in nervous system function. We have identified and annotated 13 genes for biogenic amine and small molecule neurotransmitter synthesis and transport in the M. sexta genome. These include genes encoding seven enzymes (choline acetyltransferase, acetylcholinesterase, dopa decarboxylase, tryptophan hydroxylase, tyrosine hydroxylase, tyrosine decarboxylase, and tyramine beta-hydroxylase) and six transporters (vesicular acetylcholine transferase, choline transporter, vesicular monoamine transporter, serotonin transporter, dopamine transporter, and octopamine transporter). All genes had a homolog in B. mori and most were also present in D. melanogaster. Putative M. sexta proteins and their homologs show high sequence identity or similarity, especially within domains required for substrate and cofactor binding. RNA-seq data indicate that expression patterns for many of these enzyme and transporters across developmental stages and tissue types are complex, and in some cases unexpected, suggesting potentially novel roles for these gene products.
3.6.8. Neurohormonal signaling
In insects, neuropeptides, protein hormones, and biogenic amines regulate basic physiological processes such as reproduction, development, behavior, and carbohydrate homeostasis. Most of these neurohormones act via specific G protein-coupled receptors (GPCRs), which are located in the cell membranes of the target cells. Insects have about 40 neuropeptide genes and 70 GPCR genes, although not all neuropeptide-GPCR couples occur in every insect species or family. By comparing a large number of insect genomes, we found a “core” set of GPCRs and their ligands that occur in every insect, and a “variable” set” in which some members may be present or absent in specific insect species (Nygaard et al., 2011). We hypothesize that the “core set” regulates housekeeping processes, while the “variable set” regulates processes specific to insect groups and relates to their specific habitats or life styles (Nygaard et al., 2011).
We found that both the core and variable sets of M. sexta GPCRs and their ligands were identical to those in B. mori, where the neuropeptide GPCRs have been annotated in some detail (Fan et al., 2010). This finding suggests that the overall neuroendocrinological landscapes of the two species are very similar or identical, despite the fact that they belong to two different moth families. M. sexta has 22 biogenic amine GPCRs, a number similar to that found in other insects (Table S13C). In addition, M. sexta also has 52 neuropeptide and protein hormone GPCRs (Table S13, A, B; Fig. S14, Figs. S15, S16, S17). It is remarkable that for several neuropeptides (adipokinetic hormone/corazonin-related peptide, CCAP, natalisin and RYamide) two receptors exist in both M. sexta and Bombyx (adipokinetic hormone/corazonin-related peptide), while all other insects with a sequenced genome appear to have only one receptor or none (Table S13, A). The situation for short neuropeptide F receptors is also unusual, because M. sexta (and B. mori) have 3 short neuropeptide F receptors, while other insects examined have only one or two (Table S13A,; Fig. S14; Fig. S15 B) (Hauser et al., 2008). The reasons for these remarkable receptor gene duplications in M. sexta and B. mori are currently not well understood. We could not find receptors for inotocin (also called arginine/vasopressin-like peptide) or proctolin. In conclusion, the M. sexta genome shows both neuropeptide gene duplications and receptor gene duplications. However, gene duplications were not always congruent: there were neuropeptide gene duplications without receptor gene duplication, as well as the converse. We propose that such neuropeptide and GPCR gene expansions reflect an increased need for these neuropeptide signaling pathways in moths compared to other insects.
3.6.9. Nicotine tolerance
M. sexta is a specialist on Solanaceae, and larvae have an extraordinarily high tolerance of nicotine, which is abundant in many of its hosts. The medial lethal dose by injection of nicotine for M. sexta larvae is 1.5 g/kg body weight, compared with 0.0003 g/kg for mice (Wink and Theile, 2002). Nicotine binds to subunits of the nicotinic acetylcholine receptor (nAchR) and interferes with cholinergic synapses in the nervous system. It has long been wondered whether M. sexta possesses unique amino acid substitutions in nAchR that confer intrinsic insensitivity to nicotine, analogous to substitutions in the Na+,K+-ATPase that render the monarch butterfly resistant to cardiac glycosides in its milkweed host (Holzinger et al., 1992). In previous studies of conserved portions of two nAChR subunits, no obvious substitutions were identified (Eastham et al., 1998; Wink and Theile, 2002). The availability of the M. sexta genome now permits an exhaustive examination of nAchR receptor subunits. M. sexta possesses genes for nine α-subunits (α1-α9) and three β-subunits (β1-β3), similar to B. mori and D. plexippus. The receptor is a pentamer of α and β subunits with six conserved loops (A–F) making up the acetylcholine (ACh) binding site. α-subunits are defined by the presence of two adjacent cysteine residues in Loop C. ACh binds at the interface of two subunits, at a site defined by loops A, B, and C of an α-subunit, and loops D, E, and F of the adjacent subunit, whether α or β. Alignments comparing M. sexta with the nicotine-susceptible B. mori and D. plexippus show high conservation of subunits α1-α8 and β1 (Fig. S18). Similar to the other Lepidoptera and to Drosophila (Grauso et al., 2002), subunits α4, a6, and α8 exhibit alternative splicing with use of alternative exons. M. sexta has no unique amino acid substitutions in Loops A-F of the conserved subunits, and changes due to alternative splicing or RNA editing are also found in B. mori. Sequences in α9, β2, and β3 are highly divergent across the three sequenced lepidopteran species (Fig. S18). Loop substitutions in M. sexta are usually shared in B. mori. Although the physiological roles of these more divergent subunits are unknown, there are no evident unique amino acid substitutions in the M. sexta sequences in regions that might modulate sensitivity to nicotine. This is consistent with studies showing that membrane preparations from M. sexta adult and larval brains bound to nicotine with the same affinity as preparations from nicotine-sensitive insects (Eastham et al., 1998).
In prior studies (Wink and Theile, 2002), injection of nicotine caused convulsions and other symptoms of intoxication, after which the larvae recover, suggesting a rapidly inducible detoxification mechanism. When provided with artificial diet supplemented with nicotine, larvae consumed it at first slowly, then more rapidly, as the rate of aldrin epoxidation and metabolism of nicotine by midgut microsomal preparations increased (Snyder and Glendinning, 1996). Pretreatment with the P450 inhibitor piperonyl butoxide decreased the metabolism of nicotine and reduced the consumption rate of the nicotine diet, implicating P450s in detoxification. In a study comparing wild-type Nicotiana attenuata (a tobacco species) with plants in which nicotine production was eliminated by genetic transformation (Govind et al., 2010), two P450 genes (CYP6B46 and CYP304F1) were expressed at a higher level in midguts of larvae feeding on nicotine-producing plants. These two P450s were also induced in the midgut by feeding on other non-nicotine producing host plants, relative to nicotine-free artificial diet, with CYP6B46 expressed most in the larval antenna and maxilla (Koenig et al., 2015). The genomically adjacent gene, CYP6B45, does not respond transcriptionally to nicotine.
The metabolic consequences of P450 expression are still controversial, with some studies detecting the oxidation products cotinine, cotinine-N-oxide, or nicotine 1-N-oxide in the feces (Snyder et al., 1994; Wink and Theile, 2002). Others report the absence of these compounds and instead assert that the rapid disappearance from the hemolymph is due to excretion of unmodified nicotine in the feces (Kakumani et al., 2014). The Malpighian tubules of larval (but not adult) M. sexta efficiently excrete nicotine (Maddrell and Gardiner, 1976). Properties of the transport system suggest the activity of P-glycoprotein (Gaertner et al., 1998), i.e., subfamily B of the ABC superfamily of proteins, integral membrane proteins that function in the translocation of a broad spectrum of substrates across lipid membranes. The M. sexta genome harbors 54 ABC genes, which group into eight subfamilies (A through H), including 12 subfamily B genes and 7 subfamily C genes, both of which have been shown to export xenobiotics in mammals. P-glycoprotein immunostaining at the blood-brain-barrier suggests that ABC proteins may also actively transport nicotine away from the nervous system (Murray et al., 1994), which may account for the insensitivity of intact and even partially desheathed nerve cords to nicotine (Morris and Harrison, 1984).
N. attenuata plants transiently or stably expressing double-stranded RNA constructs targeting M. sexta genes can induce plant-mediated RNAi causing down-regulation of transcripts in the insects feeding on such plants (Kumar et al., 2012). Targeting of CYP6B46 illuminated an unusual role of this P450 in nicotine metabolism (Kumar et al., 2014). Although plant-mediated RNAi reduced CYP6B46 transcript levels by up to 95% in midguts, there was no effect on larval growth, mortality, or food intake, nor were oxidation products found in bodies or frass of silenced or non-silenced larvae. Instead, CYP6B46 suppression reduced hemolymph nicotine levels and increased the amount excreted in the feces. The higher amount of hemolymph nicotine in non-suppressed larvae promoted more release of volatile nicotine out of the spiracles, which deterred predation by wolf spiders. This resulted in higher predation rates on CYP6B46-suppressed larvae by wolf spiders, but not Geocoris bugs or Myrmeleon antlions. The authors hypothesized that CYP6B46 increases the transfer of dietary nicotine to the hemolymph, by converting it to a short-lived metabolite that is re-converted to nicotine as it enters the hemolymph (Kakumani et al., 2014).
3.6.10. Detoxification-associated genes
Expansions within gene families associated with detoxification and host use.
The cytochrome P450s (P450s), carboxyl/cholinesterases (CCEs), and glutathione S-transferases (GSTs) are widely regarded as the major insect gene/enzyme families involved in xenobiotic detoxification (Despres et al., 2007; Rane et al., 2016), although several members of each family also have other functions, and the UDP-glucuronosyltransferases (UGTs) and ABC transporters (ABCs) also can play a role in detoxification (Ahn et al., 2012; Dermauw and Van Leeuwen, 2014; Merzendorfer, 2014). Comparisons of a few species with sequenced genomes across diverse orders have suggested that the sizes of these three families are correlated with the breadth of the species’ food sources (Claudianos et al., 2006; Oakeshott et al., 2010). We can test this hypothesis by comparing the compositions of the three gene families in M. sexta (which has a wide host range across the Solanaceae) with the corresponding data from published annotations for these three enzyme families for two other lepidopterans: the silkworm B. mori (which is a specialist feeder on mulberry (International Silkworm Genome, Consortium, 2008)) and diamondback moth P. xylostella (which has a wide host range across the Brassicaceae (You et al., 2013)). Table S14 shows that the total number of genes in the three families across the three species (181, 228, and 168 genes for B. mori, M. sexta, and P. xylostella, respectively) shows more limited variation than observed in some of the inter-order comparisons on which the hypothesis was based, and which showed aggregate differences in gene numbers of 2–4 fold across orders (Claudianos et al., 2006; Lee et al., 2010; Oakeshott et al., 2010; Sadd et al., 2015).
However, close examination of the three gene families (and in particular the clades within the families for which functional data for various species implicate directly in detoxification and resistance) (Despres et al., 2007; Rane et al., 2016), showed some relatively large differences between the three species. For the P450s, M. sexta not only has 25% more genes in total but also shows a significant expansion in certain clans (Table S15). Interestingly, there is a significant expansion of nuclear-encoded mitochondrial P450s, specifically in the CYP333B subfamily. However, the largest total expansion is found in clan 3, and consists of smaller expansions in numerous different sub-families, including the CYP6B, CYP6AE, and CYP9A subfamilies (Fig. S19), which are associated with plant allelochemical detoxification (Feyereisen, 2012; Schuler, 2011). The M. sexta expansion of clan 4 P450s is largely composed of CYP4 family genes, and not CYP340, as seen for the other two species. Among the GSTs, the overall numbers of genes remain similar between species (Table S16), but M. sexta shows a significant expansion of the Sigma class on scaffold JH668345.1. Although overall numbers in the Delta and Epsilon classes, which are associated with detoxification and resistance (Enayati et al., 2005; Shi et al., 2012) are comparable, there is considerable variation among the three species in terms of the specific clade that is amplified (see Fig. S20). The overall number of CCEs is noticeably greater for M. sexta than for the other two species, and this is particularly due to the recent rapid expansion of the function groups containing its clade 001 and 016 esterases (Table S17). The clade 001 esterases, which several studies have implicated in detoxification in Lepidoptera (as is also the case for the homologous clade A esterases of Diptera (Oakeshott et al., 2005; Teese et al., 2010), number 27 in M. sexta, but only 8 in B. mori and 7 in P. xylostella. Clade 001 CCE genes in M. sexta represent a single large phylogenetic group (Fig. S21), but only 9 of these genes are found in a genomic cluster with a physical location on scaffold JH668441.1 that shows microsyntenic correspondence to that of the Bombyx clade 001 CCE genes. The remainder are scattered in smaller clusters across 6 other scaffolds, some small (Table S18), possibly due to assembly problems resulting from the very close sequence similarity among the CCE genes (indicated by the very short edge lengths in Fig. S21).
Thus, while annotations of these gene families in other Lepidoptera with diverse feeding habits will be needed before firmer conclusions can be drawn, the hypothesis as simply stated does not appear to apply among the three lepidopterans for which appropriate annotation data are currently available. However, the observation of rapid expansions in the M. sexta lineage of various esterase and P450 gene families implicated in detoxification now warrants further attention in relation to host use and the detoxification of plant defence chemistries.
UDP-glycosyltransferases (UGTs) catalyze the conjugation of a range of diverse small hydrophobic compounds with sugars to produce water-soluble glycosides, playing an important role in the detoxification of xenobiotics and in the regulation of endobiotics. In insects, UGTs play an important role in the detoxification sequestration of a variety of plant allelochemicals and insecticides. Enzyme activities of the M. sexta UGTs are detected mostly in the fat body, midgut and other tissues (Ahmad and Hopkins, 1993b). Endogenous compounds, like ecdysteroid hormones (Svoboda and Weirich, 1995) and cuticle tanning precursors (Ahmad et al., 1996; Hopkins and Kramer, 1992) as well as plant phenolics (Ahmad and Hopkins, 1993a) are glycosylated by M. sexta UGT enzymes. A M. sexta UGT gene is expressed in the antennae of male M. sexta, suggesting a role in odorant degradation (Robertson et al., 1999).
The M. sexta genome contains 44 putative UGT genes including two pseudogenes (Table S19). This is similar to the number found in the genomes of other lepidopteran insects (B. mori with 45 genes and H. melpomene with 52 genes), and in a beetle genome (T. castaneum with 43 genes) (Table S20). More than half of the UGT genes are concentrated in three scaffolds: scaffold00641 (9 genes), scaf-fold00311 (8 genes), and scaffold00405 (6 genes). Recent gene or domain duplications may have increased the gene number in these regions (Fig. S22). The largest UGT33 family (16 M. sexta UGT genes) together with a closely related UGT340 family (6 M. sexta UGTs genes) accounts for 50% of the UGT genes (Fig. S22). An ancestor of these two large families might have resulted from gene duplication and divergence from a common ancestor of UGT34, which is relatively conserved in sequence similarity and genomic position among different lepidopteran species. The second largest family, UGT40, is composed of 9 M. sexta UGTs including a pseudogene (UGT40J1p) clustered with UGT41 and UGT48. It is noteworthy that there is no UGT43 ortholog identified in the M. sexta genome, but UGT42, UGT43, and UGT44 are found grouped in tandem (i.e., BmChr18) in all other known lepidopteran genomes, suggesting that gene duplications occurred in an ancestral lepidopteran species, and the duplicated genes in this location have diverged. Another observation of interest is that UGT45 is found in M. sexta but not in B. mori, Helicoverpa armigera, nor Spodoptera spp. Nonetheless, the UGT45 ortholog is conserved in other lepidopteran genomes including H. melpomene, D. plexipus, and P. xylostela, suggesting the UGT45 might have been lost before the Noctuidae and Bombycidae cluster was branched from Sphingidae in the evolution of Lepidoptera.
The UGT genes vary in their intron-exon structures. Most UGT genes (30 genes, 75%) have four exons, while others (9 genes, 20%) have an eight-exon structure (Table S19). The genes comprised of four exons have a long first exon, which encodes the N-terminal substrate binding domain, while the following three short exons encode a more conserved C-terminal sugar-binding domain. In the UGT genes with eight exons, however, the N-terminal region of the protein is encoded by five separate exons instead of a single long exon. In the UGT33C subfamily, the long first exons (corresponding to the substrate binding domain) are present as multiple alternative exons. Transcript assemblies indicate that in Scaffold00641, 6 UGT33C genes are transcribed from 6 alternative exon1s, with common exons 2, 3 and 4 (Fig. S23). This UGT gene structure seems to be a unique feature of M. sexta and is not detected in the other five lepidopteran genomes examined. Evolution of UGT genes with multiple, alternative substrate-binding domains might have increased the range of substrates, and thereby provided the herbivore with higher adaptability to potentially noxious compounds from its host plants.
3.7. Lipid metabolism
Many biochemical and physiological studies that contributed to our current understanding of the metabolism and transport of lipids in insects were carried out in lepidopterans, and among them, M. sexta has been the most important model. The transport of fatty acids (FA) in the form of diacylglycerol (DG) is a salient feature of most insects. The production and secretion of large quantities of DG by the fat body and midgut and the resulting large concentration of sn-1,2-diacylglycerol in hemolymph (Lok and van der Horst, 1980) represent a clear difference in the metabolism of lipids and the mechanisms of FA mobilization and transport between insects and vertebrates. The main insect lipoprotein, lipophorin, is the carrier of the secreted DG (Beenakkers et al., 1985). In contrast to the apolipoprotein B (apoB) containing lipoproteins of vertebrates, lipophorin particles can pick up and deliver DG molecules without compromising the integrity of the apolipoproteins (Downer and Chino, 1985; van der Horst et al., 2002). Liver and intestine of vertebrates release triglycerides (TG) into circulation, but this process requires the concomitant synthesis of apoB and the intracellular assembly of lipoproteins. The simple comparison of insect genes or insect vs vertebrate genes is not likely to provide explanations for the differences in FA transport between these groups. However, the availability of the genomic information constitutes a potent tool for advancing the biochemical studies that will eventually yield answers to these and related questions. Studies in M. sexta have contributed greatly to the understanding of lipid synthesis and mobilization in fat body and midgut (Arrese and Soulages, 2010).
3.8. Production of diacylglycerols (DG)
The massive production of DG that takes place in fat body and midgut of most insects is achieved through the expression and activity of several proteins: lipases, lipid droplet proteins, synthetic enzymes associated with glycerides, lipid carriers, and others. Fig. 8 illustrates possible pathways for DG synthesis and shows the predicted M. sexta genes coding for the enzymes involved in individual reactions (Table 4). Lipases, which hydrolyze stored triacylglycerol (TG), are important players in the production of DG. Triglyceride lipase (TGL), a major fat body TG-lipase (ACR61720.1), that is perhaps unique to insects, was discovered in M. sexta, and purified and characterized (Arrese and Wells, 1994). However, at least two additional lipases, both highly conserved in the animal kingdom, are involved in the hydrolysis of fat body TG: adipose triglyceride lipase (ATGL; Msex2.12864; Msex2.13342) and hormone sensitive lipase (HSL, Msex2.01196). Studies in Drosophila have shown the importance of ATGL (Gronke et al., 2007) and also suggest a role for HSL (Bi et al., 2012). Still, the roles of these three lipases in the mobilization of lipid in fat body and midgut are not fully understood. The concerted study of the three lipases, ATGL, TGL and HSL, and the role of the lipid droplet proteins, PLIN1 (Msex2.00753) and PLIN2 (Msex2.00753), previously named Lsd1 and Lsd2, should lead to advances in the biochemistry of lipid mobilization. PLIN proteins are lipid droplet resident proteins that play a major role in the regulation of storage of TG and DG production and secretion (Arrese et al., 2008a; Bi et al., 2012; Gronke et al., 2003; Patel et al., 2005; Teixeira et al., 2003). Studies in M. sexta demonstrated that AKH induces PKA-mediated phosphorylation of PLIN1 which activates the hydrolysis of TG (Arrese et al., 2008b; Patel et al., 2005). The genome data predicted the expression of several possible isoforms of PLIN1 as a result of alternative splicing. The cDNAs encoding two PLIN1 isoforms have been cloned (KF835603.1 and KF835604.1).
Fig. 8.
Pathways for the synthesis and secretion of diacylglycerol (DG) in fat body. Fatty acids (FA) entering the fat body, produced de novo, or released from triacylglycerol (TG) by the action of lipases (TGL, ATGL and HSL) on the lipid droplets are reused in part to form DG for export. The acyl-CoA formed by ACS could enter the synthesis of DG through the PA-pathway or through the MG-pathway, which would use monoacylglycerol (MG) produced by the hydrolysis of stored TG. Export of DG to circulating lipophorin is expected to involve the lipid transfer protein (LTP) and other membrane proteins, such as the lipophorin receptor. A similar scheme could be envisaged for the synthesis and export of DG from midgut tissue.
Table 4.
Manduca sexta genes associated with the metabolism and transport of fatty acids.
| Predicted protein | Abbreviation | ID | |
|---|---|---|---|
| Fatty acid synthesis | |||
| 1 | Acetyl CoA carboxylase | ACC | Msex2.07659 |
| 2 | Fatty Acid Synthase | FAS | Msex2.11718 |
| 3 | Fatty Acid Synthase | Msex2.03583 | |
| 4 | Acyl-CoA delta-9 desaturase | SCD | Msex010912 |
| Glyceride synthesis | |||
| 5 | Acyl-CoA Synthetase | ACS | Msex2.00253 |
| 6 | Glycerol-3-Phosphate DH (Mit) | GPDHmit | Msex2.05434 |
| 7 | Glycerol-3-Phosphate DH (Cyt) | GPDHcyt | Msex2.00684 |
| 8 | Glycerol-3-Phosphate Acyltransferase (Mit) | GPAT | Msex2.05807 |
| 9 | Dihydroxyacetone-Phosphate Acyltransferase | DHAPAT | Msex2.09853 |
| 10 | Lysophosphatidic Acid Acyltransferase | LPAAT | Msex2.08511 |
| 11 | Phosphatidic Acid Phosphatase | PAP, Lipin | Msex2.06506 |
| 12 | Monoacylglycerol Acyltransferase | MGAT | Msex2.07183 KF800700.1/KF800699.1 |
| 13 | Diacylglycerol Acyltransferase | DGAT | Msex2.08486 |
| TG Hydrolysis and storage | |||
| 14 | Adipose Triglyceride Lipase | ATGL | Msex2.12864; Msex2.13342 gb:AEJ33048.1 |
| 15 | Hormone Sensitive Lipase | HSL | Msex2.01196 |
| 16 | Triglyceride Lipase | TGL | gb:ACR61720.1 |
| 17 | Monoglyceride Lipase | MGL | Msex2.12997 |
| 18 | Lipid Storage Droplet Protein 1 | Lsd1, PLIN1 | Msex2.00753 gb:EU809925.1 |
| 19 | Lipid Storage Droplet Protein 2 | Lsd2, PLIN2 | Msex2.00759 gb: JF809664.1 |
| Glyceride and fatty acid transport | |||
| 20 | Apolipophorin-I and II | ApoLp-I and ApoLp-II | Msex2.09436 gb:U57651.1 |
| 21 | Apolipophorin-III | ApoLp-III | Msex2.09903 |
| 22 | Lipid Transfer Particle (subunit I and II) | LTP-I&II | Msex2.09991 |
| 23 | Lipid Transfer Particle (subunit III) | LTP-III | Msex2.04122 |
| 24 | Microsomal Triacylglycerol Transfer Protein | MTP | Msex2.05145 |
| 25 | Lipophorin Receptor | Lp-R | Msex2.07918 |
| 26 | Fatty Acid Binding Protein | FABP | Msex2.10635 |
The ability of insects to produce large amounts of DG should also be partly related to the pathways of DG and TG synthesis. Two major pathways are involved in the synthesis of DG in eukaryotes: 1) The stereospecific acylation of monoacylglycerol (MG) to DG by acyl-CoA monoacylglycerol acyltransferases (MGATs; EC 2.3.1.22), known as the MG-pathway, and 2) the phosphatidic acid (PA) pathway, which involves de novo synthesis of DG via acylation of glycerol-3 phosphate and subsequent hydrolysis of the PA produced (Weiss and Kennedy, 1956). These pathways are present in both midgut and fat body tissues of insects (Arrese et al., 1996; Canavoso and Wells, 2000; Hoffman and Downer, 1979; Peled and Tietz, 1974; Tietz et al., 1975), and we identified the key players from the M. sexta genome sequence (Fig. 8 and Table 4).
2-monoglyceride (2-MG) is the main product of the hydrolysis of lipid-droplet TG in vitro (Arrese and Wells, 1994), suggesting the MG-pathway in M. sexta could be the main route for the synthesis and secretion of fat body DG. MGAT activity has been observed in fat body of insects (Arrese et al., 1996; Hoffman and Downer, 1979; Peled and Tietz, 1974; Tietz et al., 1975), including M. sexta. The putative Msex-MGAT gene (Msex2.07183) was identified and two cDNAs corresponding to two isoforms produced by alternative splicing were cloned (KF800699 and KF800700) (Soulages et al., 2015). The gene and protein are highly conserved among lepidopterans and between insects and vertebrates. The net rate of synthesis of DG in fat body and midgut and its subsequent secretion into hemolymph are also dependent on the rate of DG acylation by acyl-CoA diacylglycerol acyltransferases (DGAT, EC 2.3.1.20). DGAT activity is present in both fat body and midgut tissues (Arrese et al., 1996; Buszczak et al., 2002; Canavoso et al., 2004; Canavoso and Wells, 2000). Two distinct DGAT genes, DGAT1 and DGAT2, are found in vertebrates. Both proteins have DGAT activity but have different structural and catalytic properties as well as physiological roles (Cases et al., 2001; Cheng et al., 2008). A search for M. sexta DGATs led to the identification of a single DGAT (Msex2.08486) and cloning of a DGAT cDNA (KF800701). The predicted gene and protein are conserved in insects and vertebrates, and is highly similar to the well-characterized human and mouse DGAT1. Genetic studies in Drosophila have shown that DGAT1 (Midway) plays a central role in lipid metabolism and reproduction (Buszczak et al., 2002). Interestingly, a DGAT2 gene candidate was not found in any of the insects surveyed, suggesting the possibility that insects lack DGAT2 (Soulages et al., 2015). Whether this partially explains why insects are able to produce and export DG remains to be examined experimentally.
3.8.1. Secretion of DG
Another intriguing feature of most insects is their ability to secrete large quantities of DG. The midgut secretes DG without synthesizing lipophorin, whereas the fat body can secrete DG with or without the concomitant assembly and synthesis of a lipoprotein. Although the mechanisms of DG secretion have not been completely elucidated, we know the identity of some of the genes that play a direct role. The apolipophorin (apoLp) gene (Msex2.09436) encodes the structural apolipoproteins of lipophorin, the main acceptor and transporter of DG. The first cDNA encoding apoLp was cloned from M. sexta (Sundermeyer et al., 1996). This gene encodes a single protein that undergoes a post-translational cleavage into apoLp-I and apoLp-II (Sundermeyer et al., 1996) by a furin protease (Msex2.04615). This cleavage could play a role in DG transport by providing a flexible lipoprotein structure that allows the incorporation or removal of large amounts of DG in the absence of lipoprotein synthesis or degradation. This unique ability of lipophorin is also related to the gene encoding the exchangeable apoLp, apoLp-III (Msex2.09903), which reversibly binds or dissociates from lipophorin following changes in DG content (Soulages et al., 1996). Reversible loading and unloading of lipophorin-DG requires a lipid transfer factor that facilitates the exchange of lipids between plasma membranes and the lipoprotein particles. This protein, lipid transfer particle (LTP), was discovered in M. sexta hemolymph in 1986 (Ryan et al., 1986). LTP is a highly active and unique lipid transfer protein that is found in the hemolymph of insects, and that catalyzes the bidirectional lipid transfer/exchange between lipoproteins and tissues (Canavoso and Wells, 2001; Liu and Ryan, 1991; Van Heusden and Law, 1989), and between lipoproteins (Ryan et al., 1988; Tsuchida et al., 1997). The B. mori LTP gene was recently identified and characterized (Yokoyama et al., 2013). BmLTP has three subunits. Two of them, apoLTP-I and II, are encoded by a single gene (4121 amino acids), whereas the third subunit is encoded by a different gene (Yokoyama et al., 2013). We confirmed that Msex2.09991 encodes the two major subunits of LTP, LTP-I & -II. We propose that the third LTP subunit, not yet confirmed, is encoded by Msex2.04122.
The mechanisms of lipophorin assembly and/or transport of DG from the intracellular sites to the plasma membrane are not known. The gene encoding the microsomal triglyceride transfer protein (MTP or MTTP) could be involved in these processes. MTP is a heterodimer that has a large lipid transfer subunit, apoMTP (Msex2.05145), and a protein disulfide isomerase subunit (PDI, Msex2.02333). MTP is needed for lipid loading during the assembly of apolipoprotein-B (apoB) containing lipoproteins in vertebrates (Sellers et al., 2003) and lipophorin in insects (Smolenaars et al., 2007a). Interestingly, the genes encoding ApoLp, LTP and MTP, are members of the large lipid transfer protein (LLTP) superfamily (Avarre et al., 2007; Smolenaars et al., 2007b). This family of proteins, which includes apoB, MTP and vitellogenin, could have originated from a vitellogenin precursor or an ancient MTP precursor (Sellers et al., 2005). A phylogenetic tree including some of these proteins is shown in Fig. S24. The tree emphasizes the comparison of LTP protein sequences, which seem to be unique to insects. In addition to the LTP sequences previously reported from B. mori and D. melanogaster (Yokoyama et al., 2013), the tree includes the predicted LTP sequences from eight more insect species. As previously inferred for BmLTP (Yokoyama et al., 2013), LTPs seem to share a common ancestor with apoLp-I&II and even human apoB. The process of secretion of DG may also require the specific interaction of lipophorin with a lipophorin receptor (Lp-R). The Lp-R was first purified from M. sexta fat body (Tsuchida and Wells, 1990) and subsequently cloned and studied in other insects (Cheon et al., 2001; Dantuma et al., 1999; Gopalapillai et al., 2006; Lee et al., 2003). Four isoforms of the Lp-R produced by alternative splicing are described in B. mori, and similar alternate splicing is predicted for the M. sexta Lp-R gene (Msex2.07918).
3.9. A comprehensive description of the M. sexta immunity toolkit
M. sexta has been used extensively as a model to study the innate humoral and cellular immune responses of insects (Jiang et al., 2010; Kanost and Nardi, 2010), in part because it is possible to obtain large quantities of hemolymph from larvae for studies of plasma proteins and hemocytes. Biochemical analysis of larval plasma proteins has led to identification of families of pattern recognition receptors, serine proteases, serpins, prophenoloxidases (proPOs), and antimicrobial peptides that function in responses to infection. Previous quantitative transcriptome analyses combined with homology searching revealed over 250 M. sexta genes associated with immunity, many of which are differentially regulated in response to an immune challenge (Gunaratna and Jiang, 2013; Zhang et al., 2011). The genome-wide annotation of putative immunity genes described here provides a more complete arthropod immunity protein toolkit (Table S21) for defending against attack by pathogens and parasites.
A first step in innate immune responses is recognition of infection, often by binding of host proteins to conserved molecular patterns (such as cell wall polysaccharides) on the surfaces of microorganisms. In the M. sexta genome, we identified 156 putative pattern recognition receptors, including 14 peptidoglycan recognition proteins, 5 β-1,3-glucanase-related proteins, 6 EGF/Nimdomain proteins, 4 galectins, 3 thioester-containing proteins, 4 fibrinogen-related proteins, 5 Ig-domain proteins, 34 C-type lectin-domain proteins, and 76 leucine-rich repeat proteins (Cao et al., 2015a; Zhang et al., 2015). Together, these proteins are likely to act as an efficient surveillance system to detect pathogens and trigger protective responses (Fig. 9).
Fig. 9.
An overview of the immune system in M. sexta. Pathogens and their surface molecules (in blue font) are recognized by pattern recognition receptors in the plasma or on the immune cells. A serine protease/serine protease homolog system (shown as pink pacmans) is activated by sequential proteolytic cleavage to generate active phenoloxidases and Spätzle-1. Serpins (colored triangles) modulate melanization and cytokine effects by inhibiting immune SPs. The putative intracellular pathways (Toll, Imd, MAPK-JNK-p38, JAK-STAT) are activated by cytokines (e.g., Spätzle-1) and microbial compounds (e.g., DAP-PG) through receptors, adaptors, kinases (red spheres), and transcription factors (colored ovals), which transactivate the expression of immunity-related genes (e.g., AMPs). Newly synthesized proteins either replenish the defense molecules used up in the initial reaction or serve as effectors to kill the survived pathogens. Autophagy, apoptosis, and RNA interference are involved in insect antiviral responses. The stimulatory and inhibitory steps are depicted as red arrows and blue bars, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Recognition of pathogens can stimulate activation of serine protease (SP) cascades, which rapidly amplify an initial signal and result in proteolytic activation of enzymes or cytokines. Protease cascades in M. sexta hemolymph have been extensively investigated through reconstitution of pathways using purified natural and recombinant proteins and by ex vivo analysis of proteins added to hemolymph plasma (An et al., 2009; Gorman et al., 2007; Jiang, 2008; Kanost et al., 2004; Wang and Jiang, 2007, 2008; Wang et al., 2014). In the current studies, we found that serine proteases and catalytically inactive pseudoproteases, termed serine protease homologs (SPH), are encoded by 193 genes in M. sexta. These proteins may function in digestion, development, defense, and other physiological processes. There are 107 SPs and 18 SPHs which are not primarily expressed in midgut and, therefore, are unlikely to function in digestion of food; many of these are likely to participate as immune factors in hemolymph (Cao et al., 2015b). Fifty-two of these proteins have a complex domain structure, with up to ten putative regulatory modules in addition to a catalytic or protease-like domain. Among these, 42 SPs and SPHs contain one or more amino terminal clip domains, a domain structure present in many extracellular arthropod serine proteases that participate in protease cascade pathways. In contrast, B. mori, another lepidopteran model for immunity research, has only 24 members of the clip protease gene family (Table S17). In this regard, M. sexta is more like D. melanogaster and A. gambiae, which have 37 and 41 CLIP family genes, respectively. Perhaps the silkworm genome lost part of its complement of CLIP genes in the course of domestication. Pathways of clip domain proteases and SPHs (acting as cofactors with poorly understood molecular function) that activate proPO and the Toll ligand spaetzle have been identified in M. sexta hemolymph, involving 6 clip SPs and 2 SPHs (Fig. 9) (Kanost and Nardi, 2010; Wang et al., 2014). Sequence alignment and phylogenetic analysis revealed evolutionary relationships among the clip-domain proteins (Fig. 10C), which form four clades. CLIPBs include the three prophenoloxidase-activating proteases and HP8, which activates pro-spaetzle, each as the final protease in a cascade. On the other hand, the CLIPC group included HP21 and HP6, which are penultimate proteases in cascade pathways, consistent with a hypothesis that CLIPC proteases often function to activate CLIPB proteases (An et al., 2009). The CLIPA group included the SPHs, with catalytic serine replaced, most often with glycine. Only a few of these have been studied experimentally. The functions for CLIPD proteases remain unknown and should be the focus of future research efforts. Analysis of mRNA levels for the 125 SP/SPH genes in 52 tissues at different stages (Cao et al., 2015b) revealed patterns of expression that may yield clues to biological roles, as demonstrated in Fig. 10D for SP50 and PAP3. This collection of information on the non-digestive SPs/SPHs is anticipated to facilitate the elucidation of previously unknown SP pathways that mediate immune functions and perhaps other physiological processes in M. sexta and other Metazoa.
Fig. 10.
Domain architecture, structural model, phylogenetic relationships, and expression profiles for some of the nondigestive serine proteases and serine protease homologs (SPs/ SPHs) in M. sexta. a) SP50, representing the 52 multidomain SPs/SPHs, has the domains organized in the same way as those in its ortholog Drosophila Nudel; PAP3, one of the 42 clipdomain SPs/SPHs, activates proPOs in the presence of SPH1 and SPH2. b) 3D model of the clip domain-1 in PAP3 is highly similar to the known structure of PAP2 clip domain-1 (Huang et al., 2007). α helix, red; β strand, yellow; coil, green; Cys, pink. c) Phylogenetic analysis of the entire clip-domain SP/SPH sequences in groups A (black, SPH, group-3 clip domain), B (red, SP, group-2 clip domain), C (green, SP, group-1a clip domain), and D (blue, SP, group-1b or-1c clip domain). d) PAP3 and SP50 mRNA levels in M. sexta tissues from various life stages. X-axis, RNA-seq library number; Y-axes, FPKM values of SP50 (black dotted line) and PAP3 (red solid line). PAP3 transcripts are abundant in fat body of wandering larvae and early pupae; SP50 mRNA levels are high in fat body and ovary of late pupae and adults. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Serpins are a family of proteins that function mostly as serine protease inhibitors, which regulate the clip domain proteases that function in M. sexta immune cascades (Kanost and Nardi, 2010). In total we found 32 serpin genes encoding 49 proteins via alternative mRNA splicing. However, the functions of only seven serpins and the specific proteases they inhibit have been investigated. Some serpin genes were upregulated upon microbial infection and some displayed tissue- and stage-specific expression. Of the 16 serpin genes expressed in the larval fat body or hemocytes, eleven showed 2–20-fold increases in mRNA level after immune challenge.
Experimental analysis of intracellular signal transduction for immune responses is limited in M. sexta. We searched the genome for putative immune pathway members using D. melanogaster sequences known to have such functions and identified 184 genes encoding potential members of immune signal transduction pathways. The observed 1:1 orthology in most of these proteins with their Drosophila homologs suggests that similar processes exist in M. sexta, including Toll, Imd, MAPK-JNK-p38 and JAK-STAT pathways, RNA interference, autophagy, and apoptosis (Cao et al., 2015a) (Fig. 9), although the number of Toll-like genes is variable in different species (Table S21). These pathways are responsible for a broad range of cellular reactions including defense against viruses, bacteria, fungi and parasites. An important consequence of the pathway activation is the nuclear translocation of transcription factors that up-regulate the expression of immunityrelated genes, especially in stimulating synthesis of antimicrobial peptides, which are secreted into the hemolymph and provide protection by killing pathogens (Fig. 9). We identified 86 genes for putative antimicrobial peptides in the M. sexta genome, including expansions of multigene families for attacins (11 genes), cecropins (15 genes), defensins (6 genes), and antifungal diapausins (11 genes) (He et al., 2015). In comparison with B. mori, the number of attacins and defensins (only two of each in B. mori) and Gallerimycins (0 detected in B. mori) are prominently higher in M. sexta. Overall, the number of putative immune genes is greater in M. sexta (272) than in the B. mori genome (219) (Table S21). M. sexta is a rich source for the study of genes for antimicrobial peptides that protect insects from infection.
In summary, there are 583 M. sexta genes encoding over 600 proteins that have putative functions in various phases of antimicrobial immune responses, such as pathogen recognition, extra-and intra-cellular signal transduction and modulation, and pathogen destruction (Fig. 9). Thus, the analysis of immunity-related genes provides a detailed picture of the components or toolkit of a highly complex physiological system, critical for survival. With the players/components identified, much work remains to experimentally validate the proposed physiological roles for most of the proteins encoded by these genes.
4. Conclusions
M. sexta is and has been a powerful and important model system for studies of many areas of insect biology for many decades. Through the reported draft genome sequence, representative transcriptome data, and the detailed studies of M. sexta genes and gene families that are associated with selected genomic, biochemical, and physiological systems, we make a major step on the long journey toward understanding both the specific details and the larger context of the complex biology of this fascinating animal and of the Lepidoptera and the Arthropoda.
Supplementary Material
Acknowledgments
We thank Sandy Youngeberg, Janice Beal, and Marjolein Schat for insect rearing. We also thank Julie Poulain and Corinne Da Silva from the Genoscope (Centre National de Séquencage, Evry, France) for raw transcript sequence data from fat body and hemocyte libraries, which were used to refine annotation of certain M. sexta genes.
Funding
Primary funding for DNA sequencing and assembly and RNA-seq was from grants from NIH(GM041247) to M.R Kanost and from DARPA to G.W. Blissard. Funding to support annotation of specific gene families or analysis of genome features is listed in Supplemental Table S22 (Author Contacts, Topic Areas, and Funding).
List of abbreviations
- 2-MG
2-monoglyceride
- ABCs
ABC transporters
- abdA
abdominalA
- AbdB
abdominalB
- ACh
acetylcholine
- adgf
adenosine deaminase growth factor
- AKH
adopokinetic hormone
- ANGES
ancestral genomes
- apoB
apolipoprotein-B
- apoLp
apolipophorin
- ATGL
adipose triglyceride lipase
- BAC
Bacterial artificial chromosome
- bnl
branchless
- BUSCOs
Benchmarking Universal Single-Copy Orthologs
- CARs
contiguous ancestral regions
- CBP
chitin-binding proteins
- ccap
crustacean cardioactive peptide
- CCEs
carboxyl/cholinesterases
- CDA
chitin deacetylases
- CE
carbohydrate esterase
- CHS
chitin synthases
- CHT
chitinases
- ChtBD
chitin-binding domain
- CPAPs
cuticular proteins analogous to peritrophins
- CPFL
CPF-like proteins
- CPR
Rebers and Riddiford family of Cuticular Proteins
- CP
cuticular proteins
- CPs
cysteine proteases
- CSP
chemosensory proteins
- csw
corkscrew
- DG
diacylglycerol
- DG
diacylglycerols
- DGAT
diacylglycerol acyltransferases
- DNOPs
divergent non-overlapping pairs
- dpp
decapentaplegic
- EGF
epidermal growth factor
- EH
eclosion hormone
- elav
embryonic lethal/abnormal vision
- ESCRT
endosomal sorting complex required for transport
- FA
fatty acids
- fgf
fibroblast growth factor
- ftz
fushi tarazu
- FPKM
fragments per kilobase of transcript per million mapped reads
- GABA
γ-aminobutyric acid
- gbb
glass bottom boat
- GH
glycosyl hydrolase
- GOBPs
general odorant binding proteins
- GPCRs
G protein-coupled receptors
- GR
gustatory receptors
- GSTs
glutathione S-transferases
- Hox
Homeobox
- HSL
hormone sensitive lipase
- ILP
insulin-like peptides
- IR
ionotropic receptors
- lab
labial
- LLTP
large lipid transfer protein
- Lp-R
lipophorin receptor
- LTP
lipid transfer particle
- MG
monoacylglycerol
- MGATs
monoacylglycerol acyltransferases
- msi
musashi
- MTP or MTTP
microsomal triglyceride transfer protein
- nAchR
nicotinic acetylcholine receptor
- OBP
odorant binding proteins
- OGS
official gene set
- OR
odorant receptors
- PA
phosphatidic acid
- pb
proboscipedia
- PBPs
pheromone binding proteins
- PCD
programmed cell death
- PDI
protein disulfide isomerase subunit
- PINTA
prolonged depolarization afterpotential is not apparent
- PKAs
protein kinases
- PMPs
peritrophic matrix proteins
- proPOs
prophenoloxidases
- pyr
pyramus
- RDL
resistant to dieldrin
- rGCs
receptor guanylyl cyclase
- Robo
roundabout
- Shx
special homeobox
- SNMPs
sensory neuron membrane proteins
- SP
serine protease
- SPH
serine protease homologs
- TG
triglycerides
- TGF
transforming growth factor
- TGL
triglyceride lipase
- ths
thisbe
- tkv
thickveins
- TPI
trypsin protein inhibitors
- TRPs
transient receptor potential channels
- TWDL
tweedle proteins
- ubx
ultrabithorax
- UGTs
UDP-glucuronosyltransferases
- uif
uninflatable
- VPS
vacuolar protein sorting
- WGS
whole genome shotgun reads
- zen
zerknullt
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ibmb.2016.07.005.
References
- Ahmad SA, Hopkins TL. Beta-glucosylation of plant phenolics by phenol beta-glucosyltransferase in larval tissues of the tobacco hornworm, Manduca sexta (L) Insect Bioch. Mol. Biol. 1993a;23:581–589. [Google Scholar]
- Ahmad SA, Hopkins TL. Phenol beta-glucosyltransferases in 6 species of insects – properties and tissue Localization. Comp. Biochem. Phys. B. 1993b;104:515–519. [Google Scholar]
- Ahmad SA, Hopkins TL, Kramer KJ. Tyrosine beta-glucosyltransferase in the tobacco hornworm, Manduca sexta (L): properties, tissue localization, and developmental profile. Insect Biochem. Mol. Biol. 1996;26:49–57. [Google Scholar]
- Ahn SJ, Vogel H, Heckel DG. Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochem. Mol. Biol. 2012;42:133–147. doi: 10.1016/j.ibmb.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Ahola V, Lehtonen R, Somervuo P, Salmela L, Koskinen P, Rastas P, Valimaki N, Paulin L, Kvist J, Wahlberg N, Tanskanen J, Hornett EA, Ferguson LC, Luo S, Cao Z, de Jong MA, Duplouy A, Smolander OP, Vogel H, McCoy RC, Qian K, Chong WS, Zhang Q, Ahmad E, Haukka JK, Joshi A, Salojarvi J, Wheat CW, Grosse-Wilde E, Hughes D, Katainen R, Pitkanen E, Ylinen J, Waterhouse RM, Turunen M, Vaharautio A, Ojanen SP, Schulman AH, Taipale M, Lawson D, Ukkonen E, Makinen V, Goldsmith MR, Holm L, Auvinen P, Frilander MJ, Hanski I. The Glanville fritillary genome retains an ancient karyotype and reveals selective chromosomal fusions in Lepidoptera. Nat. Commun. 2014;5:4737. doi: 10.1038/ncomms5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaya KE, Asgari S, Jung R, Hongskula M, Beckage NE. Parasitization of Manduca sexta larvae by the parasitoid wasp Cotesia congregata induces an impaired host immune response. J. Insect Physiol. 2005;51:505–512. doi: 10.1016/j.jinsphys.2004.11.019. [DOI] [PubMed] [Google Scholar]
- An CJ, Ishibashi J, Ragan EJ, Jiang HB, Kanost MR. Functions of Manduca sexta hemolymph proteinases HP6 and HP8 in two innate immune pathways. J. Biol. Chem. 2009;284:19716–19726. doi: 10.1074/jbc.M109.007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakane Y, Muthukrishnan S. Insect chitinase and chitinase-like proteins. Cell. Mol. Life Sci. 2010;67:201–216. doi: 10.1007/s00018-009-0161-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, Mirza S, Rivera L, Howard AD, Chetty PS, Soulages JL. Expression of lipid storage droplet protein-1 may define the role of AKH as a lipid mobilizing hormone in Manduca sexta. Insect Biochem. Mol. Biol. 2008a;38:993–1000. doi: 10.1016/j.ibmb.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, Rivera L, Harnada M, Mirza S, Hartson SD, Weintraub S, Soulages JL. Function and structure of lipid storage droplet protein 1 studied in lipoprotein complexes. Arch. Biochem. Biophys. 2008b;473:42–47. doi: 10.1016/j.abb.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, Rojas-Rivas BI, Wells MA. Synthesis of sn-1,2-diacylglycerols by monoacylglycerol acyltransferase from Manduca sexta fat body. Arch. Insect Biochem. Physiol. 1996;31:325–335. doi: 10.1002/(SICI)1520-6327(1996)31:3<325::AID-ARCH7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Arrese EL, Soulages JL. Insect fat body: energy, metabolism, and regulation. Annu. Rev. Entomol. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, Wells MA. Purification and properties of a phosphorylatable triacylglycerol lipase from the fat body of an insect, Manduca sexta. J. Lipid Res. 1994;35:1652–1660. [PubMed] [Google Scholar]
- Avarre JC, Lubzens E, Babin PJ. Apolipocrustacein, formerly vitellogenin, is the major egg yolk precursor protein in decapod crustaceans and is homologous to insect apolipophorin II/I and vertebrate apolipoprotein B. BMC Evol. Biol. 2007;7:3. doi: 10.1186/1471-2148-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayme-Southgate A, Feldman S, Fulmer D. Myofilament proteins in the synchronous flight muscles of Manduca sexta show both similarities and differences to Drosophila melanogaster. Insect Biochem. Mol. Biol. 2015;62:174–182. doi: 10.1016/j.ibmb.2015.02.008. [DOI] [PubMed] [Google Scholar]
- Ballard SL, Jarolimova J, Wharton KA. Gbb/BMP signaling is required to maintain energy homeostasis in Drosophila. Dev. Biol. 2010;337:375–385. doi: 10.1016/j.ydbio.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Q, Shi Y. Apoptosome: a platform for the activation of initiator caspases. Cell Death Differ. 2007;14:56–65. doi: 10.1038/sj.cdd.4402028. [DOI] [PubMed] [Google Scholar]
- Bao R, Friedrich M. Molecular evolution of the Drosophila retinome: exceptional gene gain in the higher Diptera. Mol. Biol. Evol. 2009;26:1273–1287. doi: 10.1093/molbev/msp039. [DOI] [PubMed] [Google Scholar]
- Baxter SW, Davey JW, Johnston JS, Shelton AM, Heckel DG, Jiggins CD, Blaxter ML. Linkage mapping and comparative genomics using next-generation RAD sequencing of a non-model organism. PLoS One. 2011;6:el9315. doi: 10.1371/journal.pone.0019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenakkers AM, Van der Horst DJ, Van Marrewijk WJ. Insect lipids and lipoproteins, and their role in physiological processes. Prog. Lipid Res. 1985;24:19–67. doi: 10.1016/0163-7827(85)90007-4. [DOI] [PubMed] [Google Scholar]
- Beldade P, Saenko SV, Pul N, Long AD. A gene-based linkage map for Bicyclus anynana butterflies allows for a comprehensive analysis of synteny with the lepidopteran reference genome. PLoS Genet. 2009;5:el000366. doi: 10.1371/journal.pgen.1000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R. Multigene family evolution: perspectives from insect chemoreceptors. Trends Ecol. Evol. 2015;30:590–600. doi: 10.1016/j.tree.2015.07.009. [DOI] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt da Cruz A, Wentzell J, Kretzschmar D. Swiss Cheese, a protein involved in progressive neurodegeneration, acts as a noncanonical regulatory subunit for PKA-C3. J. Neurosci. 2008;28:10885–10892. doi: 10.1523/JNEUROSCI.3015-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J, Xiang Y, Chen H, Liu Z, Gronke S, Kuhnlein RP, Huang X. Opposite and redundant roles of the two Drosophila perilipins in lipid mobilization. J.Cell Sci. 2012;125:3568–3577. doi: 10.1242/jcs.101329. [DOI] [PubMed] [Google Scholar]
- Bradfield JY, Wyatt GR. X-Linkage of a vitellogenin gene in Locusta migratoria. Chromosoma. 1983;88:190–193. [Google Scholar]
- Bureš P, Zedek E. Holokinetic drive: centromere drive in chromosomes without centromeres. Evolution. 2014;68:2412–2420. doi: 10.1111/evo.12437. [DOI] [PubMed] [Google Scholar]
- Buszczak M, Lu X, Segraves WA, Chang TY, Cooley L. Mutations in the midway gene disrupt a Drosophila acyl coenzyme A: diacylglycerol acyltransferase. Genetics. 2002;160:1511–1518. doi: 10.1093/genetics/160.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canavoso LE, Frede S, Rubiolo ER. Metabolic pathways for dietary lipids in the midgut of hematophagous Panstrongylus megistus (Hemiptera: Reduviidae) Insect Biochem. Mol. Biol. 2004;34:845–854. doi: 10.1016/j.ibmb.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Canavoso LE, Jouni ZE, Karnas KJ, Pennington JE, Wells MA. Fat metabolism in insects. Annu. Rev. Nutr. 2001;21:23–46. doi: 10.1146/annurev.nutr.21.1.23. [DOI] [PubMed] [Google Scholar]
- Canavoso LE, Wells MA. Metabolic pathways for diacylglycerol biosynthesis and release in the midgut of larval Manduca sexta. Insect Biochem. Mol. Biol. 2000;30:1173–1180. doi: 10.1016/s0965-1748(00)00094-1. [DOI] [PubMed] [Google Scholar]
- Canavoso LE, Wells MA. Role of lipid transfer particle in delivery of diacylglycerol from midgut to lipophorin in larval Manduca sexta. Insect Biochem. Mol. Biol. 2001;31:783–790. doi: 10.1016/s0965-1748(00)00183-1. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, Holt C, Sanchez Alvarado A, Yandell M. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Ress. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, He Y, Hu Y, Wang Y, Chen YR, Bryant B, Clem RJ, Schwartz LM, Blissard G, Jiang H. The immune signaling pathways of Manduca sexta. Insect Biochem. Mol. Biol. 2015a;62:64–74. doi: 10.1016/j.ibmb.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, He Y, Hu Y, Zhang X, Wang Y, Zou Z, Chen Y, Blissard GW, Kanost MR, Jiang H. Sequence conservation, phylogenetic relationships, and expression profiles of nondigestive serine proteases and serine protease homologs in Manduca sexta. Insect Biochem. Mol. Biol. 2015b;62:51–63. doi: 10.1016/j.ibmb.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jiang H. Integrated modeling of protein-coding genes in the Manduca sexta genome using RNA-Seq data from the biochemical model insect. Insect Biochem. Mol. Biol. 2015;62:2–10. doi: 10.1016/j.ibmb.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao XL, He Y, Hu YX, Zhang XF, Wang Y, Zou Z, Chen YR, Blissard GW, Kanost MR, Jiang HB. Sequence conservation, phylogenetic relationships, and expression profiles of nondigestive serine proteases and serine protease homologs in Manduca sexta. Insect Biochem. Mol. Biol. 2015c;62:51–63. doi: 10.1016/j.ibmb.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases S, Stone SJ, Zhou P, Yen E, Tow B, Lardizabal KD, Voelker T, Farese RV., Jr Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J. Biol. Chem. 2001;276:38870–38876. doi: 10.1074/jbc.M106219200. [DOI] [PubMed] [Google Scholar]
- Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virol. 2008;372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YR, Zhong S, Fei Z, Hashimoto Y, Xiang JZ, Zhang S, Blissard GW. The transcriptome of the baculovirus Autographa californica multiple nucleopolyhedrovirus in Trichoplusia ni cells. J. Virol. 2013;87:6391–6405. doi: 10.1128/JVI.00194-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Nohata J, Guo H, Li S, Liu J, Guo Y, Yamamoto K, Kadono-Okuda K, Liu C, Arunkumar KP, Nagaraju J, Zhang Y, Liu S, Labropoulou V, Swevers L, Tsitoura P, Iatrou K, Gopinathan KP, Goldsmith MR, Xia Q, Mita K. A comprehensive analysis of the chorion locus in silkmoth. Sci. Rep. 2015a;5:16424. doi: 10.1038/srep16424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Nohata J, Guo H, Li S, Liu J, Guo Y, Yamamoto K, Kadono-Okuda K, Liu C, Arunkumar KP, Nagaraju J, Zhang Y, Liu S, Labropoulou V, Swevers L, Tsitoura P, Iatrou K, Gopinathan KP, Goldsmith MR, Xia Q, Mita K. Construction, complete sequence, and annotation of a BAC contig covering the silkworm chorion locus. Sci. Data. 2015b;2:150062. doi: 10.1038/sdata.2015.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZX, Sturgill D, Qu J, Jiang H, Park S, Boley N, Suzuki AM, Fletcher AR, Plachetzki DC, FitzGerald PC, Artieri CG, Atallah J, Barmina O, Brown JB, Blankenburg KP, Clough E, Dasgupta A, Gubbala S, Han Y, Jayaseelan JC, Kalra D, Kim YA, Kovar CL, Lee SL, Li M, Malley JD, Malone JH, Mathew T, Mattiuzzo NR, Munidasa M, Muzny DM, Ongeri E, Perales L, Przytycka TM, Pu LL, Robinson G, Thornton RL, Saada N, Scherer SE, Smith HE, Vinson C, Warner CB, Worley KC, Wu YQ, Zou X, Cherbas P, Kellis M, Eisen MB, Piano F, Kionte K, Fitch DH, Sternberg PW, Cutter AD, Duff MO, Hoskins RA, Graveley BR, Gibbs RA, Bickel PJ, Kopp A, Carninci P, Celniker SE, Oliver B, Richards S. Comparative validation of the D. melanogaster modENCODE transcriptome annotation. Genome Res. 2014;24:1209–1223. doi: 10.1101/gr.159384.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D, Iqbal J, Devenny J, Chu CH, Chen L, Dong J, Seethala R, Keim WJ, Azzara AV, Lawrence RM, Pelleymounter MA, Hussain MM. Acyla-tion of acylglycerols by acyl coenzyme A:diacylglycerol acyltransferase 1 (DGAT1). Functional importance of DGAT1 in the intestinal fat absorption. J. Biol. Chem. 2008;283:29802–29811. doi: 10.1074/jbc.M800494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon HM, Seo SJ, Sun J, Sappington TW, Raikhel AS. Molecular characterization of the VLDL receptor homolog mediating binding of lipophorin in oocyte of the mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 2001;31:753–760. doi: 10.1016/s0965-1748(01)00068-6. [DOI] [PubMed] [Google Scholar]
- Chevignon G, Cambier S, Da Silva C, Poulain J, Drezen JM, Huguet E, Moreau SJ. Transcriptomic response of Manduca sexta immune tissues to parasitization by the bracovirus associated wasp Cotesia congregata. Insect Biochem. Mol. Biol. 2015;62:86–99. doi: 10.1016/j.ibmb.2014.12.008. [DOI] [PubMed] [Google Scholar]
- Claudianos C, Ranson H, Johnson RM, Biswas S, Schuler MA, Berenbaum MR, Feyereisen R, Oakeshott JG. A deficit of detoxification enzymes: pesticide sensitivity and environmental response in the honeybee. Insect Mol. Biol. 2006;15:615–636. doi: 10.1111/j.1365-2583.2006.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombrita C, Silani V, Ratti A. ELAV proteins along evolution: back to the nucleus? Mol. Cell. Neurosci. 2013;56:447–455. doi: 10.1016/j.mcn.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Conceição IC, Long AD, Gruber JD, Beldade P. Genomic sequence around butterfly wing development genes: annotation and comparative analysis. PLoS One. 2011;6:e23778. doi: 10.1371/journal.pone.0023778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong C, Borek D, Otwinowski Z, Grishin NV. Tiger swallotail genome reveals mechanisms for speciation and caterpillar chemical defense. Cell Rep. 2015;10:910–919. doi: 10.1016/j.celrep.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtiade J, Pauchet Y, Vogel H, Heckel DG. A comprehensive characterization of the caspase gene family in insects from the order Lepidoptera. BMC Genomics. 2011;12:357. doi: 10.1186/1471-2164-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, Benton R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6:el001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Alençon E, Sezutsu H, Legeai F, Permal E, Bernard-Samain S, Gimenez S, Gagneur C, Cousserans F, Shimomura M, Brun-Barale A, Flutre T, Couloux A, East P, Gordon K, Mita K, Quesneville H, Fournier P, Feyereisen R. Extensive synteny conservation of holocentric chromosomes in Lepidoptera despite high rates of local genome rearrangements. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7680–7685. doi: 10.1073/pnas.0910413107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancourt J, Barlowe C. Protein sorting receptors in the early secretory pathway. Annu. Rev. Biochem. 2010;79:777–802. doi: 10.1146/annurev-biochem-061608-091319. [DOI] [PubMed] [Google Scholar]
- Dantuma NP, Potters M, De Winther MP, Tensen CP, Kooiman FP, Bogerd J, Van der Horst DJ. An insect homolog of the vertebrate very low density lipoprotein receptor mediates endocytosis of lipophorins. J. Lipid Res. 1999;40:973–978. [PubMed] [Google Scholar]
- Dasmahapatra KK, Walters JR, Briscoe AD, Davey JW, Whibley A, Nadeau NJ, Zimin AV, Hughes DST, Ferguson LC, Martin SH, Salazar C, Lewis JJ, Adler S, Ahn SJ, Baker DA, Baxter SW, Chamberlain NL, Chauhan R, Counterman BA, Dalmay T, Gilbert LE, Gordon K, Heckel DG, Hines HM, Hoff KJ, Holland PWH, Jacquin-Joly E, Jiggins FM, Jones RT, Kapan DD, Kersey P, Lamas G, Lawson D, Mapleson D, Maroja LS, Martin A, Moxon S, Palmer WJ, Papa R, Papanicolaou A, Pauchet Y, Ray DA, Rosser N, Salzberg SL, Supple MA, Surridge A, Tenger-Trolander A, Vogel H, Wilkinson PA, Wilson D, Yorke JA, Yuan FR, Balmuth AL, Eland C, Gharbi K, Thomson M, Gibbs RA, Han Y, Jayaseelan JC, Kovar C, Mathew T, Muzny DM, Ongeri F, Pu LL, Qu JX, Thornton RL, Worley KC, Wu YQ, Linares M, Blaxter ML, Ffrench-Constant RH, Joron M, Kronforst MR, Mullen SP, Reed RD, Scherer SE, Richards S, Mallet J, McMillan WO, Jiggins CD, Consortium HG. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JW, Chouteau M, Barker SL, Maroja L, Baxter SW, Simpson F, Joron M, Mallet J, Dasmahapatra KK, Jiggins CD. Major Improvements to the Heliconius melpomene genome assembly used to confirm 10 chromosome fusion events in 6 million years of butterfly evolution. G3. 2016;15:695–708. doi: 10.1534/g3.115.023655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidowitz G, D’Amico LL, Nijhout HF. Critical weight in the development of insect body size. Evol. Dev. 2003;5:188–197. doi: 10.1046/j.1525-142x.2003.03026.x. [DOI] [PubMed] [Google Scholar]
- de Celis JF. Expression and function of decapentaplegic and thick veins during the differentiation of the veins in the Drosophila wing. Development. 1997;124:1007–1018. doi: 10.1242/dev.124.5.1007. [DOI] [PubMed] [Google Scholar]
- Dermauw W, Van Leeuwen T. The ABC gene family in arthropods: comparative genomics and role in insecticide transport and resistance. Insect Biochem. Mol. Biol. 2014;45:89–110. doi: 10.1016/j.ibmb.2013.11.001. [DOI] [PubMed] [Google Scholar]
- Despres L, David JP, Gallet C. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol. Evol. 2007;22:298–307. doi: 10.1016/j.tree.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Diaz-Benjumea FJ, Cohen SM. Serrate signals through Notch to establish a Wingless-dependent organizer at the dorsal/ventral compartment boundary of the Drosophila wing. Development. 1995;121:4215–4225. doi: 10.1242/dev.121.12.4215. [DOI] [PubMed] [Google Scholar]
- Dittmer NT, Hiromasa Y, Tomich JM, Lu N, Beeman RW, Kramer KJ, Kanost MR. Proteomic and transcriptomic analyses of rigid and membranous cuticles and epidermis from the elytra and hindwings of the red flour beetle, Tribolium castaneum. J. Proteom. Res. 2012;11:269–278. doi: 10.1021/pr2009803. [DOI] [PubMed] [Google Scholar]
- Dittmer NT, Tetreau G, Cao X, Jiang H, Wang P, Kanost MR. Annotation and expression analysis of cuticular proteins from the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol. 2015;62:100–113. doi: 10.1016/j.ibmb.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Arakane Y, Specht CA, Richard C, Kramer KJ, Beeman RW, Muthukrishnan S. Domain organization and phylogenetic analysis of proteins from the chitin deacetylase gene family of Tribolium castaneum and three other species of insects. Insect Biochem. Mol. Biol. 2008;38:440–451. doi: 10.1016/j.ibmb.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Dolezelova E, Zurovec M, Dolezal T, Simek P, Bryant PJ. The emerging role of adenosine deaminases in insects. Insect Biochem. Mol. Biol. 2005;35:381–389. doi: 10.1016/j.ibmb.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Downer RGH, Chino H. Turnover of protein and diacylglycerol components of lipophorin in insect haemolymph. Insect Biochem. 1985;15:627–630. [Google Scholar]
- Dreos R, Ambrosini G, Cavin Perier R, Bucher P. EPD and EPDnew, high-quality promoter resources in the next-generation sequencing era. Nucleic Acids Res. 2013;41:D157–D164. doi: 10.1093/nar/gks1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg IA, deYoung D, Henikoff S, Malik HS. Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. Elife. 2014:3. doi: 10.7554/eLife.03676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastham HM, Lind RJ, Eastlake JL, Clarke BS, Towner P, Reynolds SE, Wolstenholme AJ, Wonnacott S. Characterization of a nicotinic acetylcholine receptor from the insect Manduca sexta. Eur. J. Neurosci. 1998;10:879–889. doi: 10.1046/j.1460-9568.1998.00095.x. [DOI] [PubMed] [Google Scholar]
- Enayati AA, Ranson H, Hemingway J. Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 2005;14:3–8. doi: 10.1111/j.1365-2583.2004.00529.x. [DOI] [PubMed] [Google Scholar]
- Fan Y, Sun P, Wang Y, He X, Deng X, Chen X, Zhang G, Chen X, Zhou N. The G protein-coupled receptors in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2010;40:581–591. doi: 10.1016/j.ibmb.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Feyereisen R. Insect CYP genes and P450 enzymes. In: Gilbert LI, editor. Insect Molecular Biology and Biochemistry. New York: Elsevier; 2012. pp. 236–316. [Google Scholar]
- Fonovic M, Turk B. Cysteine cathepsins and extracellular matrix degradation. Biochim. Biophys. Acta. 2014;1840:2560–2570. doi: 10.1016/j.bbagen.2014.03.017. [DOI] [PubMed] [Google Scholar]
- Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem. J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futahashi R, Okamoto S, Kawasaki H, Zhong YS, Iwanaga M, Mita K, Fujiwara H. Genome-wide identification of cuticular protein genes in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2008;38:1138–1146. doi: 10.1016/j.ibmb.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Gaertner LS, Murray CL, Morris CE. Transepithelial transport of nicotine and vinblastine in isolated malpighian tubules of the tobacco hornworm (Manduca sexta) suggests a P-glycoprotein-like mechanism. J. Exp. Biol. 1998;201:2637–2645. doi: 10.1242/jeb.201.18.2637. [DOI] [PubMed] [Google Scholar]
- Gilbert LI, Rybczynski R, Warren JT. Control and biochemical nature of the ecdysteroidogenic pathway. Annu. Rev. Entomol. 2002;47:883–916. doi: 10.1146/annurev.ento.47.091201.145302. [DOI] [PubMed] [Google Scholar]
- Goldsmith MR, Marek F, editors. Molecular Biology and Genetics of the Lepidoptera. Boca Raton, FL: CRC Press; 2010. [Google Scholar]
- Gopalapillai R, Kadono-Okuda K, Tsuchida K, Yamamoto K, Nohata J, Ajimura M, Mita K. Lipophorin receptor of Bombyx mori: cDNA cloning, genomic structure, alternative splicing, and isolation of a new isoform. J. Lipid Res. 2006;47:1005–1013. doi: 10.1194/jlr.M500462-JLR200. [DOI] [PubMed] [Google Scholar]
- Gorman MJ, Wang Y, Jiang HB, Kanost MR. Manduca sexta hemolymph proteinase 21 activates prophenoloxidase-activating proteinase 3 in an insect innate immune response proteinase cascade. J. Biol. Chem. 2007;282:11742–11749. doi: 10.1074/jbc.M611243200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind G, Mittapalli O, Griebel T, Allmann S, Boecker S, Baldwin IT. Unbiased Transcriptional comparisons of generalist and specialist herbivores feeding on progressively defenseless Nicotiana attenuata plants. PLoS One. 2010;5:e8735. doi: 10.1371/journal.pone.0008735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotech. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grauso M, Reenan RA, Culetto E, Sattelle DB. Novel putative nicotinic acetylcholine receptor subunit genes, D alpha 5, D alpha 6 and D alpha 7 in Drosophila melanogaster identify a new and highly conserved target of adenosine deaminase acting on RNA-mediated A-to-I pre-mRNA editing. Genetics. 2002;160:1519–1533. doi: 10.1093/genetics/160.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronke S, Beller M, Fellert S, Ramakrishnan H, Jackie H, Kuhnlein RP. Control of fat storage by a Drosophila PAT domain protein. Curr. Biol. 2003;13:603–606. doi: 10.1016/s0960-9822(03)00175-1. [DOI] [PubMed] [Google Scholar]
- Gronke S, Muller G, Hirsch J, Fellert S, Andreou A, Haase T, Jackie H, Kuhnlein RP. Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biol. 2007;5:el37. doi: 10.1371/journal.pbio.0050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaratna RT, Jiang HB. A comprehensive analysis of the Manduca sexta immunotranscriptome. Dev. Comp. Immunol. 2013;39:388–398. doi: 10.1016/j.dci.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorgyi TK, Roby-Shemkovitz AJ, Lerner MR. Characterization and cDNA cloning of the pheromone-binding protein from the tobacco hornworm, Manduca sexta: a tissue-specific developmentally regulated protein. Proc. Nat. Acad. Sci. U. S. A. 1988;85:9851–9855. doi: 10.1073/pnas.85.24.9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, Salzberg SL, White O. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi E, Weeks N, Westerman R, William T, Dewey CN, Henschel R, Leduc RD, Friedman N, Regev A. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamodrakas SJ. Molecular architecture of helicoidal proteinaceous eggshells. Results Probl. Cell Differ. 1992;19:115–186. doi: 10.1007/978-3-540-47207-0_6. [DOI] [PubMed] [Google Scholar]
- Hanrahan SJ, Johnston JS. New genome size estimates of 134 species of arthropods. Chromosome Res. 2011;19:809–823. doi: 10.1007/s10577-011-9231-6. [DOI] [PubMed] [Google Scholar]
- Hauser E, Cazzamali G, Williamson M, Park Y, Li B, Tanaka Y, Predel R, Neupert S, Schachtner J, Verleyen P, Grimmelikhuijzen CJ. A genome-wide inventory of neurohormone GPCRs in the red flour beetle Tribolium castaneum. Front. Neuroendocrinol. 2008;29:142–165. doi: 10.1016/j.yfrne.2007.10.003. [DOI] [PubMed] [Google Scholar]
- He Y, Cao XL, Li K, Hu YX, Chen YR, Blissard G, Kanost MR, Jiang HB. A genome-wide analysis of antimicrobial effector genes and their transcription patterns in Manduca sexta. Insect Biochem. Mol. Biol. 2015;62:23–37. doi: 10.1016/j.ibmb.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus D, O’Grady M, Chamankhah M, Baldwin D, Gleddie S, Braun L, Erlandson M. Changes in cysteine protease activity and localization during midgut metamorphosis in the crucifer root maggot (Delia radicum). Insect Biochem. Mol. Biol. 2002;32:1585–1596. doi: 10.1016/s0965-1748(02)00099-1. [DOI] [PubMed] [Google Scholar]
- Heinbockel T, Shields VDC, Reisenman CE. Glomerular interactions in olfactory processing channels of the antennal lobes. J. Comp. Physiol. A. 2013;199:929–946. doi: 10.1007/s00359-013-0842-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruma K, Riddiford LM. Developmental expression of mRNAs for epidermal and fat body proteins and hormonally regulated transcription factors in the tobacco hornworm, Manduca sexta. J. Insect Physiol. 2010;56:1390–1395. doi: 10.1016/j.jinsphys.2010.03.029. [DOI] [PubMed] [Google Scholar]
- Hoffman AGD, Downer RGH. Synthesis of diacylglycerols by mono-acylglycerol acyltransferase from crop, midgut and fat body tissues of the american cockroach, Periplaneta americana L. Insect Biochem. 1979;9:129–134. [Google Scholar]
- Hogenkamp DG, Arakane Y, Zimoch L, Merzendorfer H, Kramer KJ, Beeman RW, Kanost MR, Specht CA, Muthukrishnan S. Chitin syn-thase genes in Manduca sexta: characterization of a gut-specific transcript and differential tissue expression of alternately spliced mRNAs during development. Insect Biochem. Mol. Biol. 2005;35:529–540. doi: 10.1016/j.ibmb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Holzinger E, Frick C, Wink M. Molecular basis for the insensitivity of the monarch (Danaus plexippus) to cardiac glycosides. FEBS Lett. 1992;314:477–480. doi: 10.1016/0014-5793(92)81530-y. [DOI] [PubMed] [Google Scholar]
- Homma K, Kurata S, Natori S. Purification, characterization, and cDNA cloning of procathepsin L from the culture medium of NIH-Sape-4, an embryonic cell line of Sarcophaga peregrina (flesh fly), and its involvement in the differentiation of imaginal discs. J. Biol. Chem. 1994;269:15258–15264. [PubMed] [Google Scholar]
- Hopkins TL, Kramer KJ. Insect cuticle sclerotization. Annu. Rev. Entomol. 1992;37:273–302. [Google Scholar]
- Horodyski FM, Riddiford LM. Expression and hormonal control of a new larval cuticular multigene family at the onset of metamorphosis of the tobacco hornworm. Dev. Biol. 1989;132:292–303. doi: 10.1016/0012-1606(89)90226-1. [DOI] [PubMed] [Google Scholar]
- Huang RD, Lu ZQ, Dai HE, Velde DV, Prakash O, Jiang HB. The solution structure of clip domains from Manduca sexta prophenoloxidase activating proteinase-2. Biochem. 2007;46:11431–11439. doi: 10.1021/bi7010724. [DOI] [PubMed] [Google Scholar]
- Iconomidou VA, Hamodrakas SJ. Natural protective amyloids. Curr. Prot. Pept. Sci. 2008;9:291–309. doi: 10.2174/138920308784534041. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Takemura T, Hino S, Yoshioka K. Molecular cloning, expression, and chromosomal localization of a human tubulointerstitial nephritis antigen. Biochem. Biophys. Res. Commun. 2000;268:225–230. doi: 10.1006/bbrc.2000.2103. [DOI] [PubMed] [Google Scholar]
- International Silkworm Genome, Consortium. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2008;38:1036–1045. doi: 10.1016/j.ibmb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Jankowska M, Fuchs J, Klocke E, Fojtova M, Polanska P, Fajkus J, Schubert V, Houben A. Holokinetic centromeres and efficient telomere healing enable rapid karyotype evolution. Chromosoma. 2015;124:519–528. doi: 10.1007/s00412-015-0524-y. [DOI] [PubMed] [Google Scholar]
- Jasrapuria S, Arakane Y, Osman G, Kramer KJ, Beeman RW, Muthukrishnan S. Genes encoding proteins with peritrophin A-type chitin-binding domains in Tribolium castaneum are grouped into three distinct families based on phytogeny, expression and function. Insect Biochem. Mol. Biol. 2010;40:214–227. doi: 10.1016/j.ibmb.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Jiang H, Vilcinskas A, Kanost MR. Immunity in Lepidopteran insects. Adv. Exp. Med. Biol. 2010;708:181–204. doi: 10.1007/978-1-4419-8059-5_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HB. The biochemical basis of antimicrobial responses in Manduca sexta. Insect Sci. 2008;15:53–66. [Google Scholar]
- Jiang R, Kim EH, Gong JH, Kwon HM, Kim CH, Ryu KH, Park JW, Kurokawa K, Zhang J, Gubb D, Lee BL. Three pairs of protease-serpin complexes cooperatively regulate the insect innate immune responses. J. Biol. Chem. 2009;284:35652–35658. doi: 10.1074/jbc.M109.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AK, Sattelle DB. Diversity of insect nicotinic acetylcholine receptor subunits. Adv. Exp. Med. Biol. 2010;683:25–43. doi: 10.1007/978-1-4419-6445-8_3. [DOI] [PubMed] [Google Scholar]
- Jones BR, Rajaraman A, Tannier E, Chauve C. ANGES: reconstructing ANcestral GEnomeS maps. Bioinformatics. 2012;28:2388–2390. doi: 10.1093/bioinformatics/bts457. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kafatos FC, Tzertzinis G, Spoerel NA, Nguyen HT. Chorion genes: an overview of their structure, function, and transcriptional regulation. In: Goldsmith MA, Wilkins AS, editors. Molecular Model Systems in the Lepidoptera. Cambridge, UK: Cambridge University Press; 1995. pp. 181–215. [Google Scholar]
- Kakumani PK, Malhotra P, Mukherjee SK, Bhatnagar RK. A draft genome assembly of the army worm, Spodoptera frugiperda. Genomics. 2014;104:134–143. doi: 10.1016/j.ygeno.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Kanapin A, Batalov S, Davis MJ, Gough J, Grimmond S, Kawaji H, Magrane M, Matsuda H, Schonbach C, Teasdale RD, Yuan Z, Group RG, Members GSL. Mouse proteome analysis. Genome Res. 2003;13:1335–1344. doi: 10.1101/gr.978703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandul NP, Lukhtanov VA, Pierce NE. Karyotypic diversity and speciation in Agrodiaetus butterflies. Evolution. 2007;61:546–559. doi: 10.1111/j.1558-5646.2007.00046.x. [DOI] [PubMed] [Google Scholar]
- Kanost MR, Jiang HB, Yu XQ. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 2004;198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- Kanost MR, Nardi JB. Innate immune responses of Manduca sexta. In: Goldsmith M, Marec F, editors. Molecular Biology and Genetics of Lepidoptera. Boca Raton, FL: CRC Press; 2010. pp. 271–291. [Google Scholar]
- Kanwar YS, Kumar A, Yang Q, Tian Y, Wada J, Kashihara N, Wallner EI. Tubulointerstitial nephritis antigen: an extracellular matrix protein that selectively regulates tubulogenesis vs. glomerulogenesis during mammalian renal development. Proc. Nat. Acad. Sci. U. S. A. 1999;96:11323–11328. doi: 10.1073/pnas.96.20.11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalsa O, Yoon JW, Torres-Schumann S, Wharton KA. TGF-beta/BMP superfamily members, Gbb-60A and Dpp, cooperate to provide pattern information and establish cell identity in the Drosophila wing. Development. 1998;125:2723–2734. doi: 10.1242/dev.125.14.2723. [DOI] [PubMed] [Google Scholar]
- Kidd T, Bland KS, Goodman CS. Slit is the midline repellent for the robo receptor in Drosophila. Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- King DS, Bollenbacher WE, Borst DW, Vedeckis WV, O’Connor JD, Ittycheriah PI, Gilbert LI. The Secretion of alpha-ecdysone by the pro-thoracic glands of Manduca sexta in vitro. Proc. Natl. Acad. Sci. U. S. A. 1974;71:793–796. doi: 10.1073/pnas.71.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig C, Bretschneider A, Heckel DG, Grosse-Wilde E, Hansson BS, Vogel H. The plastic response of Manduca sexta to host and non-host plants. Insect Biochem. Mol. Biol. 2015;63:72–85. doi: 10.1016/j.ibmb.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Koo YD, Ahn JE, Salzman RA, Moon J, Chi YH, Yun DJ, Lee SY, Koiwa H, Zhu-Salzman K. Functional expression of an insect cathepsin B-like counter-defence protein. Insect Mol. Biol. 2008;17:235–245. doi: 10.1111/j.1365-2583.2008.00799.x. [DOI] [PubMed] [Google Scholar]
- Kriventseva EV, Tegenfeldt F, Petty TJ, Waterhouse RM, Simao FA, Pozdnyakov IA, Ioannidis P, Zdobnov EM. OrthoDB v8: update of the hierarchical catalog of orthologs and the underlying free software. Nucl. Acids Res. 2015;43:D250–D256. doi: 10.1093/nar/gku1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Pandit SS, Baldwin IT. Tobacco rattle virus vector: a rapid and transient means of silencing Manduca sexta genes by plant mediated RNA interference. PLoS One. 2012;7:e31347. doi: 10.1371/journal.pone.0031347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Pandit SS, Steppuhn A, Baldwin IT. Natural history-driven, plant-mediated RNAi-based study reveals CYP6B46’s role in a nicotine-mediated antipredator herbivore defense. Proc. Nat. Acad. Sci. U. S. A. 2014;111:1245–1252. doi: 10.1073/pnas.1314848111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- Lecanidou R, Papantonis A. Modeling bidirectional transcription using silkmoth chorion gene promoters. Organogenesis. 2010a;6:54–58. doi: 10.4161/org.6.1.10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecanidou R, Papantonis A. Silkmoth chorion gene regulation revisited: promoter architecture as a key player. Insect Mol. Biol. 2010b;19:141–151. doi: 10.1111/j.1365-2583.2009.00969.x. [DOI] [PubMed] [Google Scholar]
- Lecanidou R, Rodakis GC, Eickbush TH, Kafatos EC. Evolution of the silk moth chorion gene superfamily: gene families CA and CB. Proc. Nat. Acad. Sci. U. S. A. 1986;83:6514–6518. doi: 10.1073/pnas.83.17.6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Han JH, Lee SM, Hwang JS, Kang SW, Lee BH, Kim HR. Wax moth, Galleria mellonella fat body receptor for high-density lipophorin (HDLp) Arch. Insect Biochem. Physiol. 2003;54:14–24. doi: 10.1002/arch.10095. [DOI] [PubMed] [Google Scholar]
- Lee E, Helt GA, Reese JT, Munoz-Torres MC, Childers CP, Buels RM, Stein L, Holmes IH, Elsik CG, Lewis SE. Web Apollo: a web-based genomic annotation editing platform. Genome Biol. 2013;14:R93. doi: 10.1186/gb-2013-14-8-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Kim BY, Choo YM, Yoon HJ, Kang PD, Woo SD, Sohn HD, Roh JY, Gui ZZ, Je YH, Jin BR. Expression profile of cathepsin B in the fat body of Bombyx mori during metamorphosis. Comp. Biochem. Physiol. B. 2009;154:188–194. doi: 10.1016/j.cbpb.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Lee SH, Kang JS, Min JS, Yoon KS, Strycharz JP, Johnson R, Mittapalli O, Margam VM, Sun W, Li HM, Xie J, Wu J, Kirkness EF, Berenbaum MR, Pittendrigh BR, Clark JM. Decreased detoxification genes and genome size make the human body louse an efficient model to study xenobiotic metabolism. Insect Mol. Biol. 2010;19:599–615. doi: 10.1111/j.1365-2583.2010.01024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cdhit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Li Z, Blissard G. The vacuolar protein sorting genes in insects: a comparative genome view. Insect Biochem. Mol. Biol. 2015;62:211–225. doi: 10.1016/j.ibmb.2014.11.007. [DOI] [PubMed] [Google Scholar]
- Liang J, Zhang L, Xiang Z, He N. Expression profile of cuticular genes of silkworm, Bombyx mori. BMC Genomics. 2010;11:173. doi: 10.1186/1471-2164-11-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Ryan RO. Role of lipid transfer particle in transformation of lipophorin in insect oocytes. Biochim. Biophys. Acta. 1991;1085:112–118. doi: 10.1016/0005-2760(91)90238-d. [DOI] [PubMed] [Google Scholar]
- Liu J, Shi GP, Zhang WQ, Zhang GR, Xu WH. Cathepsin L function in insect moulting: molecular cloning and functional analysis in cotton bollworm, Helicoverpa armigera. Insect Mol. Biol. 2006;15:823–834. doi: 10.1111/j.1365-2583.2006.00686.x. [DOI] [PubMed] [Google Scholar]
- Lockshin RA, Williams CM. Programmed cell death–I. Cytology of degeneration in the intersegmental muscles of the pernyi silkmoth. J. Insect Physiol. 1965;11:123–133. doi: 10.1016/0022-1910(65)90099-5. [DOI] [PubMed] [Google Scholar]
- Lok CM, van der Horst DJ. Chiral 1,2-diacylglycerols in the haemolymph of the locust, Locusta migratoria. Biochim.. Biophys. Acta. 1980;618:80–87. doi: 10.1016/0005-2760(80)90055-7. [DOI] [PubMed] [Google Scholar]
- Macias-Munoz A, Smith G, Monteiro A, Briscoe AD. Transcriptome-wide differential gene expression in Bicyclus anynana butterflies: female vision-related genes are more plastic. Mol. Biol. Evol. 2016;33:79–92. doi: 10.1093/molbev/msv197. [DOI] [PubMed] [Google Scholar]
- Maddrell SHP, Gardiner BOC. Excretion of alkaloids by Malpighian tubules of insects. J. Exp. Biol. 1976;64:267–281. doi: 10.1242/jeb.64.2.267. [DOI] [PubMed] [Google Scholar]
- Mahoney PA, Weber U, Onofrechuk P, Biessmann HJBR, Goodman CS. The fat tumor suppressor gene in Drosophila encodes a novel member of the cadherin gene superfamily. Cell. 1991;67:853–868. doi: 10.1016/0092-8674(91)90359-7. [DOI] [PubMed] [Google Scholar]
- Martin JP, Beyerlein A, Dacks AM, Reisenman CE, Riffell JA, Lei H, Hildebrand JG. The neurobiology of insect olfaction: sensory processing in a comparative context. Prog. Neurobiol. 2011;95:427–447. doi: 10.1016/j.pneurobio.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Matsuura H, Sokabe T, Kohno K, Tominaga M, Kadowaki T. Evolutionary conservation and changes in insect TRP channels. BMC Evol. Biol. 2009;9:228. doi: 10.1186/1471-2148-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters DP, Paliulis LV, Korf IF, Chan SW. Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 2012;20:579–593. doi: 10.1007/s10577-012-9292-1. [DOI] [PubMed] [Google Scholar]
- Merzendorfer H. The cellular basis of chitin synthesis in fungi and insects: common principles and differences. Eur. J. Cell Biol. 2011;90:759–769. doi: 10.1016/j.ejcb.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Merzendorfer H. ABC Transporters and their role in protecting insects from pesticides and their metabolites. Adv. Insect Physiol. 2014;46:1–72. [Google Scholar]
- Missbach C, Dweck HK, Vogel H, Vilcinskas A, Stensmyr MC, Hansson BS, Grosse-Wilde E. Evolution of insect olfactory receptors. eLife. 2014;3:e02115. doi: 10.7554/eLife.02115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita K, Kasahara M, Sasaki S, Nagayasu Y, Yamada T, Kanamori H, Namiki N, Kitagawa M, Yamashita H, Yasukochi Y, Kadono-Okuda K, Yamamoto K, Ajimura M, Ravikumar G, Shimomura M, Nagamura Y, Shin-I T, Abe H, Shimada T, Morishita S, Sasaki T. The genome sequence of silkworm, Bombyx mori. DNA Res. 2004;11:27–35. doi: 10.1093/dnares/11.1.27. [DOI] [PubMed] [Google Scholar]
- Miyaji T, Kouzuma Y, Yaguchi J, Matsumoto R, Kanost MR, Kramer KJ, Yonekura M. Purification of a cysteine protease inhibitor from larval hemolymph of the tobacco hornworm (Manduca sexta) and functional expression of the recombinant protein. Insect Biochem. Mol. Biol. 2007;37:960–968. doi: 10.1016/j.ibmb.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Miyaji T, Murayama S, Kouzuma Y, Kimura N, Kanost MR, Kramer KJ, Yonekura M. Molecular cloning of a multidomain cysteine protease and protease inhibitor precursor gene from the tobacco hornworm (Manduca sexta) and functional expression of the cathepsin F-like cysteine protease domain. Insect Biochem. Mol. Biol. 2010;40:835–846. doi: 10.1016/j.ibmb.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Morris CE, Harrison JB. Central nervous system features of a nicotine-resistant Insect, the tobacco hornworm Manduca sexta. Tissue Cell. 1984;16:601–612. doi: 10.1016/0040-8166(84)90034-x. [DOI] [PubMed] [Google Scholar]
- Morton DB, Nighorn A. MsGC-II, a receptor guanylyl cyclase isolated from the CNS of Manduca sexta that is inhibited by calcium. J. Neurochem. 2003;84:363–372. doi: 10.1046/j.1471-4159.2003.01528.x. [DOI] [PubMed] [Google Scholar]
- Murray CL, Quaglia M, Arnason JT, Morris CE. A putative nicotine pump at the metabolic blood-brain-barrier of the tobacco hornworm. J. Neurobiol. 1994;25:23–34. doi: 10.1002/neu.480250103. [DOI] [PubMed] [Google Scholar]
- Nagaraju J, Jolly MS. Interspecific hybrids of Antheraea roylei and A. pernyi -a cytogenetic reassessment. Theor. Appl. Genet. 1986;72:269–273. doi: 10.1007/BF00267003. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Okano H, Blendy JA, Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:67–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- Nichols Z, Vogt RG. The SNMP/CD36 gene family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem. Mol. Biol. 2008;38:398–415. doi: 10.1016/j.ibmb.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Nighorn A, Byrnes KA, Morton DB. Identification and characterization of a novel beta subunit of soluble guanylyl cyclase that is active in the absence of a second subunit and is relatively insensitive to nitric oxide. J. Biol. Chem. 1999;274:2525–2531. doi: 10.1074/jbc.274.4.2525. [DOI] [PubMed] [Google Scholar]
- Nighorn A, Gibson NJ, Rivers DM, Hildebrand JG, Morton DB. The nitric oxide-cGMP pathway may mediate communication between sensory afferents and projection neurons in the antennal lobe of Manduca sexta. J. Neurosci. 1998;18:7244–7255. doi: 10.1523/JNEUROSCI.18-18-07244.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nighorn A, Simpson PJ, Morton DB. The novel guanylyl cyclase MsGC-I is strongly expressed in higher-order neuropils in the brain of Manduca sexta. J. Exp. Biol. 2001;204:305–314. doi: 10.1242/jeb.204.2.305. [DOI] [PubMed] [Google Scholar]
- Nijhout HF, Riddiford LM, Mirth C, Shingleton AW, Suzuki Y, Callier V. The developmental control of size in insects. Dev. Biol. 2014;3:113–134. doi: 10.1002/wdev.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhout HF, Williams CM. Control of moulting and metamorphosis in the tobacco hornworm, Manduca sexta (L): cessation of juvenile hormone secretion as a trigger for pupation. J. Exp. Biol. 1974;61:493–501. doi: 10.1242/jeb.61.2.493. [DOI] [PubMed] [Google Scholar]
- Nygaard S, Zhang G, Schiott M, Li C, Wurm Y, Hu H, Zhou J, Ji L, Oju F, Rasmussen M, Pan H, Hauser F, Krogh A, Grimmelikhuijzen CJ, Wang J, Boomsma JJ. The genome of the leaf-cutting ant Acromyrmex echinatior suggests key adaptations to advanced social life and fungus farming. Genome Res. 2011;21:1339–1348. doi: 10.1101/gr.121392.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor MB, Umulis D, Othmer HG, Blair SS. Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing. Development. 2006;133:183–193. doi: 10.1242/dev.02214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakeshott JG, Claudianos C, Campbell PM, Newcomb RD, Russell RJ. Biochemical genetics and genomics of insect esterases. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. New York: Elsevier; 2005. pp. 309–380. [Google Scholar]
- Oakeshott JG, Johnson RM, Berenbaum MR, Ranson H, Cristino AS, Claudianos C. Metabolic enzymes associated with xenobiotic and che-mosensory responses in Nasonia vitripennis. Insect Mol. Biol. 2010;1(19 Suppl. 1):147–163. doi: 10.1111/j.1365-2583.2009.00961.x. [DOI] [PubMed] [Google Scholar]
- Okot-Kotber BM, Morgan TD, Hopkins TL, Kramer KJ. Catecholamine-containing proteins from the pharate pupal cuticle of the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol. 1996;26:475–484. doi: 10.1016/0965-1748(96)00003-3. [DOI] [PubMed] [Google Scholar]
- Osanai-Futahashi M, Suetsugu Y, Mita K, Fujiwara H. Genome-wide screening and characterization of transposable elements and their distribution analysis in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2008;38:1046–1057. doi: 10.1016/j.ibmb.2008.05.012. [DOI] [PubMed] [Google Scholar]
- Papa R, Morrison CM, Walters JR, Counterman BA, Chen R, Haider G, Ferguson L, Chamberlain N, ffrench-Constant R, Kapan DD, Jiggins CD, Reed RD, McMillan WO. Highly conserved gene order and numerous novel repetitive elements in genomic regions linked to wing pattern variation in Heliconius butterflies. BMC Genomics. 2008;9:345. doi: 10.1186/1471-2164-9-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RT, Soulages JL, Hariharasundaram B, Arrese EL. Activation of the lipid droplet controls the rate of lipolysis of triglycerides in the insect fat body J. Biol. Chem. 2005;280:22624–22631. doi: 10.1074/jbc.M413128200. [DOI] [PubMed] [Google Scholar]
- Peel S, Macheboeuf P, Martinelli N, Weissenhorn W. Divergent pathways lead to ESCRT-III-catalyzed membrane fission. Trends Biochem. Sci. 2011;36:199–210. doi: 10.1016/j.tibs.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Peled Y, Tietz A. Acylation of monoglycerides by locust fat-body micro-somes. FEBS Lett. 1974;41:65–68. doi: 10.1016/0014-5793(74)80955-5. [DOI] [PubMed] [Google Scholar]
- Penalva-Arana DC, Lynch M, Robertson HM. The chemoreceptor genes of the waterflea Daphnia pulex: many Grs but no Ors. BMC Evol. Biol. 2009;9:79. doi: 10.1186/1471-2148-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JA. Lepidoptera. In: Resh VH, Carde RT, editors. Encyclopedia of Insects. New York: Academic Press; 2003. pp. 631–664. [Google Scholar]
- Pringle EG, Baxter SW, Webster CL, Papanicolaou A, Lee SF, Jiggins CD. Synteny and chromosome evolution in the lepidoptera: evidence from mapping in Heliconius melpomene. Genetics. 2007;177:417–126. doi: 10.1534/genetics.107.073122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane RVWTK, Pearce SL, Jermin LS, Goron KHJ, Richards S, Oakeshott JG. Are feeding preferences and insecticide resistance associated with the size of detoxifying enzyme families in insect herbivores? Curr. Opin. Insect Sci. 2016;13:70–76. doi: 10.1016/j.cois.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Salvesen G. Handbook of Proteolytic Enzymes. third. Elsevier Ltd; 2013. [Google Scholar]
- Rebers JE, Riddiford LM. Structure and expression of a Manduca sexta larval cuticle gene homologous to Drosophila cuticle genes. J. Mol. Biol. 1988;203:411–423. doi: 10.1016/0022-2836(88)90009-5. [DOI] [PubMed] [Google Scholar]
- Riddiford LM, Baeckmann A, Hice RH, Rebers J. Developmental expression of three genes for larval cuticular proteins of the tobacco hornworm, Manduca sexta. Dev. Biol. 1986;118:82–94. doi: 10.1016/0012-1606(86)90075-8. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Martos R, Sears CR, Todres EZ, Walden KK, Nardi JB. Diversity of odourant binding proteins revealed by an expressed sequence tag project on male Manduca sexta moth antennae. Insect Mol. Biol. 1999;8:501–518. doi: 10.1046/j.1365-2583.1999.00146.x. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Wanner KW. The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res. 2006;16:1395–1403. doi: 10.1101/gr.5057506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson R. Lepidoptera Genetics. Oxford, UK: Pergamon Press; 1971. [Google Scholar]
- Rodakis GC, Moschonas NK, Kafatos FC. Evolution of a multigene family of chorion proteins in silkmoths. Mol. Cell. Biol. 1982;2:554–563. doi: 10.1128/mcb.2.5.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers ME, Krieger J, Vogt RG. Antennal SNMPs (sensor neuron membrane proteins) of lepidoptera define a unique family of invertebrate CD36-like proteins. J. Neurobiol. 2001;49:47–61. doi: 10.1002/neu.1065. [DOI] [PubMed] [Google Scholar]
- Ryan RO, Senthilathipan KR, Wells MA, Law JH. Facilitated diacylglycerol exchange between insect hemolymph lipophorins. Properties of Manduca sexta lipid transfer particle. J. Biol. Chem. 1988;263:14140–14145. [PubMed] [Google Scholar]
- Ryan RO, Wells MA, Law JH. Lipid transfer protein from Manduca sexta hemolymph. Biochem. Biophys. Res. Commun. 1986;136:260–265. doi: 10.1016/0006-291x(86)90903-4. [DOI] [PubMed] [Google Scholar]
- Sadd BM, Barribeau SM, Bloch G, de Graaf DC, Dearden P, Elsik CG, Gadau J, Grimmelikhuijzen CJ, Hasselmann M, Lozier JD, Robertson HM, Smagghe G, Stolle E, Van Vaerenbergh M, Waterhouse RM, Bornberg-Bauer E, Klasberg S, Bennett AK, Camara F, Guigo R, Hoff K, Mariotti M, Munoz-Torres M, Murphy T, Santesmasses D, Amdam GV, Beckers M, Beye M, Biewer M, Bitondi MM, Blaxter ML, Bourke AF, Brown MJ, Buechel SD, Cameron R, Cappelle K, Carolan JC, Christiaens O, Ciborowski KL, Clarke DF, Colgan TJ, Collins DH, Cridge AG, Dalmay T, Dreier S, du Plessis L, Duncan E, Erler S, Evans J, Falcon T, Flores K, Freitas FC, Fuchikawa T, Gempe T, Hartfelder K, Hauser F, Helbing S, Humann FC, Irvine F, Jermiin LS, Johnson CE, Johnson RM, Jones AK, Kadowaki T, Kidner JH, Koch V, Kohler A, Kraus FB, Lattorff HM, Leask M, Lockett GA, Mallon EB, Antonio DS, Marxer M, Meeus I, Moritz RF, Nair A, Napflin K, Nissen I, Niu J, Nunes FM, Oakeshott JG, Osborne A, Otte M, Pinheiro DG, Rossie N, Rueppell O, Santos CG, Schmid-Hempel R, Schmitt BD, Schulte C, Simoes ZL, Soares MP, Swevers L, Winnebeck EC, Wolschin F, Yu N, Zdobnov EM, Aqrawi PK, Blankenburg KP, Coyle M, Francisco L, Hernandez AG, Holder M, Hudson ME, Jackson L, Jayaseelan J, Joshi V, Kovar C, Lee SL, Mata R, Mathew T, Newsham IF, Ngo R, Okwuonu G, Pham C, Pu LL, Saada N, Santibanez J, Simmons D, Thornton R, Venkat A, Walden KK, Wu YQ, Debyser G, Devreese B, Asher C, Blommaert J, Chipman AD, Chittka L, Fouks B, Liu J, O’Neill MP, Sumner S, Puiu D, Qu J, Salzberg SL, Scherer SE, Muzny DM, Richards S, Robinson GE, Gibbs RA, Schmid-Hempel P, Worley KC. The genomes of two key bumblebee species with primitive eusotial organization. Genome Biol. 2015;16:76. doi: 10.1186/s13059-015-0623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara K, Yoshido A, Marec F, Fukova I, Zhang HB, Wu CC, Goldsmith MR, Yasukochi Y. Conserved synteny of genes between chromosome 15 of Bombyx mori and a chromosome of Manduca sexta shown by five-color BAC-FISH. Genome. 2007;50:1061–1065. doi: 10.1139/g07-082. [DOI] [PubMed] [Google Scholar]
- Sahara K, Yoshido A, Traut W. Sex chromosome evolution in moths and butterflies. Chromosome Res. 2012;20:83–94. doi: 10.1007/s10577-011-9262-z. [DOI] [PubMed] [Google Scholar]
- Saito H, Kurata S, Natori S. Purification and characterization of a hemocyte proteinase of Sarcophaga, possibly participating in elimination of foreign substances. Eur. J. Biochem. 1992;209:939–944. doi: 10.1111/j.1432-1033.1992.tb17366.x. [DOI] [PubMed] [Google Scholar]
- Sanchez-Gracia A, Vieira FG, Rozas J. Molecular evolution of the major chemosensory gene families in insects. Heredity. 2009;103:208–216. doi: 10.1038/hdy.2009.55. [DOI] [PubMed] [Google Scholar]
- Schuh AL, Audhya A. The ESCRT machinery: from the plasma membrane to endosomes and back again. Crit. Rev. Biochem. Mol. Biol. 2014;49:242–261. doi: 10.3109/10409238.2014.881777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler MA. P450s in plant-insect interactions. Biochim. Biophys. Acta. 2011;1814:36–45. doi: 10.1016/j.bbapap.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuman MC, Allmann S, Baldwin IT. Plant defense phenotypes determine the consequences of volatile emission for individuals and neighbors. eLife. 2015;4:43. doi: 10.7554/eLife.04490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers JA, Hou L, Athar H, Hussain MM, Shelness GS. A Drosophila microsomal triglyceride transfer protein homolog promotes the sssembly and secretion of human apolipoprotein B: implications for human and insect lipid transport and metabolism. J. Biol. Chem. 2003;278:20367–20373. doi: 10.1074/jbc.M300271200. [DOI] [PubMed] [Google Scholar]
- Sellers JA, Hou L, Schoenberg DR, Batistuzzo de Medeiros SR, Wahli W, Shelness GS. Microsomal triglyceride transfer protein promotes the secretion of Xenopus laevis vitellogenin Al. J. Biol. Chem. 2005;280:13902–13905. doi: 10.1074/jbc.M500769200. [DOI] [PubMed] [Google Scholar]
- Serbielle C, Moreau S, Veillard F, Voldoire E, Bezier A, Mannucci MA, Volkoff AN, Drezen JM, Lalmanach G, Huguet E. Identification of parasite-responsive cysteine proteases in Manduca sexta. Biol. Chem. 2009;390:493–502. doi: 10.1515/BC.2009.061. [DOI] [PubMed] [Google Scholar]
- Shi HX, Pei LH, Gu SS, Zhu SC, Wang YY, Zhang Y, Li B. Glutathione S-transferase (GST) genes in the red flour beetle, Tribolium castaneum, and comparative analysis with five additional insects. Genomics. 2012;100:327–335. doi: 10.1016/j.ygeno.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Shimizu I, Yamakawa Y, Shimazaki Y, Iwasa T. Molecular cloning of Bombyx cerebral opsin (Boceropsin) and cellular localization of its expression in the silkworm brain. Biochem. Biophys. Res. Commun. 2001;287:27–34. doi: 10.1006/bbrc.2001.5540. [DOI] [PubMed] [Google Scholar]
- Shindo T, Van der Hoorn RA. Papain-like cysteine proteases: key players at molecular battlefields employed by both plants and their invaders. Mol. Plant Pathol. 2008;9:119–125. doi: 10.1111/j.1364-3703.2007.00439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbering AF, Rytz R, Grosjean Y, Abuin L, Ramdya P, Jefferis GS, Benton R. Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J. Neurosci. 2011;31:13357–13375. doi: 10.1523/JNEUROSCI.2360-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Smith G, Briscoe AD. Molecular evolution and expression of the CRALJ_TRIO protein family in insects. Insect Biochem. Mol. Biol. 2015;62:168–173. doi: 10.1016/j.ibmb.2015.02.003. [DOI] [PubMed] [Google Scholar]
- Smith G, Chen YR, Blissard GW, Briscoe AD. Complete dosage compensation and sex-biased gene expression in the moth Manduca sexta. Genome Biol. Evol. 2014;6:526–537. doi: 10.1093/gbe/evu035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolenaars MM, de Morree A, Kerver J, Van der Horst DJ, Rodenburg KW. Insect lipoprotein biogenesis depends on an amphipathic beta cluster in apolipophorin II/I and is stimulated by microsomal triglyceride transfer protein. J. Lipid Res. 2007a;48:1955–1965. doi: 10.1194/jlr.M600434-JLR200. [DOI] [PubMed] [Google Scholar]
- Smolenaars MM, Madsen O, Rodenburg KW, Van der Horst DJ. Molecular diversity and evolution of the large lipid transfer protein superfamily J. Lipid Res. 2007b;48:489–502. doi: 10.1194/jlr.R600028-JLR200. [DOI] [PubMed] [Google Scholar]
- Snyder MJ, Glendinning JI. Causal connection between detoxification enzyme activity and consumption of a toxic plant compound. J. Comp. Physiol. A. 1996;179:255–261. doi: 10.1007/BF00222792. [DOI] [PubMed] [Google Scholar]
- Snyder MJ, Walding JK, Feyereisen R. Metabolic fate of the allelochemical nicotine in the tobacco hornworm Manduca sexta. Insect Biochem. Mol. Biol. 1994;24:837–846. [Google Scholar]
- Soberon M, Pardo L, Munoz-Garay C, Sanchez J, Gomez I, Porta H, Bravo A. Pore formation by Cry toxins. In: Anderluh G, Lakey J, editors. Proteins: Membrane Binding and Pore Formation. 2010. pp. 127–142. [DOI] [PubMed] [Google Scholar]
- Sojka D, Franta Z, Horn M, Hajdusek O, Caffrey CR, Mares M, Kopacek P. Profiling of proteolytic enzymes in the gut of the tick Ixodes ricinus reveals an evolutionarily conserved network of aspartic and cysteine peptidases. Parasites Vectors. 2008;1:7. doi: 10.1186/1756-3305-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulages JL, van Antwerpen R, Wells MA. Role of diacylglycerol and apolipophorin-III in regulation of physiochemical properties of the lipophorin surface: metabolic implications. Biochem. 1996;35:5191–5198. doi: 10.1021/bi952794d. [DOI] [PubMed] [Google Scholar]
- Soulages JL, Wu Z, Firdaus SJ, Mahalingam R, Arrese EL. Mono-acylglycerol and diacylglycerol acyltransferases and the synthesis of neutral glycerides in Manduca sexta. Insect Biochem. Mol. Biol. 2015;62:194–210. doi: 10.1016/j.ibmb.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suderman RJ, Dittmer NT, Kanost MR, Kramer KJ. Model reactions for insect cuticle sclerotization: cross-linking of recombinant cuticular proteins upon their laccase-catalyzed oxidative conjugation with catechols. Insect Biochem. Mol. Biol. 2006;36:353–365. doi: 10.1016/j.ibmb.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Suderman RJ, Dittmer NT, Kramer KJ, Kanost MR. Model reactions for insect cuticle sclerotization: participation of amino groups in the cross-linking of Manduca sexta cuticle protein MsCP36. Insect Biochem. Mol. Biol. 2010;40:252–258. doi: 10.1016/j.ibmb.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Sundermeyer K, Hendricks JK, Prasad SV, Wells MA. The precursor protein of the structural apolipoproteins of lipophorin: cDNA and deduced amino acid sequence. Insect Biochem. Mol. Biol. 1996;26:735–738. doi: 10.1016/s0965-1748(96)00060-4. [DOI] [PubMed] [Google Scholar]
- Sutherland D, Samakovlis C, Krasnow MA. Branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell. 1996;87:1091–1101. doi: 10.1016/s0092-8674(00)81803-6. [DOI] [PubMed] [Google Scholar]
- Svoboda JA, Weirich GF. Sterol metabolism in the tobacco hornworm, Manduca sexta–a review. Lipids. 1995;30:263–267. doi: 10.1007/BF02537831. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GJ, Peng ZK, Lu JP, Tang FQ. Cathepsins mediate tumor metastasis. World J. Biol. Chem. 2013;4:91–101. doi: 10.4331/wjbc.v4.i4.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teese MG, Campbell PM, Scott C, Gordon KHJ, Southon A, Hoyan D, Robin C, Russell RJ, Oakeshott JG. Gene identification and proteomic analysis of the esterases of the cotton bollworm, Helicoverpa armigera. Insect Biochem. Mol. Biol. 2010;40:1–16. doi: 10.1016/j.ibmb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Teixeira L, Rabouille C, Rorth P, Ephrussi A, Vanzo NF. Drosophila Per-ilipin/ADRP homologue Lsd2 regulates lipid metabolism. Mech. Dev. 2003;120:1071–1081. doi: 10.1016/s0925-4773(03)00158-8. [DOI] [PubMed] [Google Scholar]
- Tetreau G, Cao X, Chen YR, Muthukrishnan S, Jiang H, Blissard GW, Kanost MR, Wang P. Overview of chitin metabolism enzymes in Manduca sexta: identification, domain organization, phylogenetic analysis and gene expression. Insect Biochem. Mol. Biol. 2015a;62:114–126. doi: 10.1016/j.ibmb.2015.01.006. [DOI] [PubMed] [Google Scholar]
- Tetreau G, Dittmer NT, Cao X, Agrawal S, Chen YR, Muthukrishnan S, Haobo J, Blissard GW, Kanost MR, Wang P. Analysis of chitin-binding proteins from Manduca sexta provides new insights into evolution of peritrophin A-type chitin-binding domains in insects. Insect Biochem. Mol. Biol. 2015b;62:127–141. doi: 10.1016/j.ibmb.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietz A, Weintraub H, Peled Y. Utilization of 2-acyl-sn-glycerol by locust fat body microsomes. Specificity of the acyltransferase system. Biochim. Biophys. Acta. 1975;388:165–170. doi: 10.1016/0005-2760(75)90120-4. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman JW, Hiruma K, Allee JP, Macwhinnie SG, Champlin DT, Riddiford LM. Juvenile hormone is required to couple imaginal disc formation with nutrition in insects. Science. 2006;312:1385–1388. doi: 10.1126/science.1123652. [DOI] [PubMed] [Google Scholar]
- Truman JW, Riddiford LM. The morphostatic actions of juvenile hormone. Insect Biochem. Mol. Biol. 2007;37:761–770. doi: 10.1016/j.ibmb.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Truman JW, Riddiford LM, Safranek L. Temporal patterns of response to ecdysone and juvenile hormone in the epidermis of the tobacco hornworm, Manduca sexta. Dev. Biol. 1974;39:247–262. doi: 10.1016/0012-1606(74)90238-3. [DOI] [PubMed] [Google Scholar]
- Tsuchida K, Soulages JL, Moribayashi A, Suzuki K, Maekawa H, Wells MA. Purification and properties of a lipid transfer particle from Bombyx mori: comparison to the lipid transfer particle from Manduca sexta. Biochim. Biophys. Acta. 1997;1337:57–65. doi: 10.1016/s0167-4838(96)00149-5. [DOI] [PubMed] [Google Scholar]
- Tsuchida K, Wells MA. Isolation and charaderization of a lipoprotein receptor from the fat body of an insect, Manduca sexta. J. Biol. Chem. 1990;265:5761–5767. [PubMed] [Google Scholar]
- Van’t Hof AE, Nguyen P, Dalikova M, Edmonds N, Marec E, Saccheri IJ. Linkage map of the peppered moth, Biston betularia (Lepidoptera, Geo-metridae): a model of industrial melanism. Heredity. 2013;110:283–295. doi: 10.1038/hdy.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst DJ, van Hoof D, van Marrewijk WJ, Rodenburg KW. Alternative lipid mobilization: the insect shuttle system. Mol. Cell. Biochem. 2002;239:113–119. [PubMed] [Google Scholar]
- Van Heusden MC, Law JH. An insect lipid transfer particle promotes lipid loading from fat body to lipoprotein. J. Biol. Chem. 1989;264:17287–17292. [PubMed] [Google Scholar]
- Vershinina AO, Anokhin BA, Lukhtanov VA. Ribosomal DNA clusters and telomeric (TTAGG)n repeats in blue butterflies (Lepidoptera, Lycaenidae) with low and high chromosome numbers. Comp. Cytogenet. 2015;9:161–171. doi: 10.3897/CompCytogen.v9i2.4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt RG, Grosse-Wilde E, Zhou JJ. The Lepidoptera odorant binding protein gene family: gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect Biochem. Mol. Biol. 2015;62:142–153. doi: 10.1016/j.ibmb.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Vogt RG, Miller NE, Litvack R, Fandino RA, Sparks J, Staples J, Friedman R, Dickens JC. The insect SNMP gene family. Insed Biochem. Mol. Biol. 2009;39:448–456. doi: 10.1016/j.ibmb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Vogt RG, Rogers ME, Franco MD, Sun M. A comparative study of odorant binding protein genes: differential expression of the PBP1-GOBP2 gene cluster in Manduca sexta (Lepidoptera) and the organization of OBP genes in Drosophila melanogaster (Diptera) J. Exp. Biol. 2002;205:719–744. doi: 10.1242/jeb.205.6.719. [DOI] [PubMed] [Google Scholar]
- Vogt RG, Rybczynski R, Lerner MR. Molecular doning and sequencing of general odorant-binding proteins GOBP1 and GOBP2 from the tobacco hawk moth Manduca sexta: comparisons with other insect OBPs and their signal peptides. J. Neurosci. 1991;11:2972–2984. doi: 10.1523/JNEUROSCI.11-10-02972.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiang H. Reconstitution of a branch of the Manduca sexta proph-enoloxidase activation cascade in vitro: snake-like hemolymph proteinase 21 (HP21) cleaved by HP14 adivates prophenol oxidase-activating proteinase-2 precursor. Insect Biochem. Mol. Biol. 2007;37:1015–1025. doi: 10.1016/j.ibmb.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiang HB. A positive feedback mechanism in the Manduca sexta prophenoloxidase activation system. Insect Biochem. Mol. Biol. 2008;38:763–769. doi: 10.1016/j.ibmb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lu ZQ, Jiang HB. Manduca sexta proprophenoloxidase activating proteinase-3 (PAP3) stimulates melanization by activating proPAP3, proSPHs, and proPOs. Insect Biochem. Mol. Biol. 2014;50:82–91. doi: 10.1016/j.ibmb.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss SB, Kennedy EP. The enzymatic synthesis of triglycerides. J. Am. Chem. Soc. 1956;78:3550. [PubMed] [Google Scholar]
- Wieczorek H, Huss M, Merzendorfer H, Reineke S, Vitavska O, Zeiske W. The insect plasma membrane H+ V-ATPase: intra-, inter-, and supramolecular aspects. J. Bioenerg. Biomembr. 2003;35:359–366. doi: 10.1023/a:1025733016473. [DOI] [PubMed] [Google Scholar]
- Willis JH. Structural cuticular proteins from arthropods: annotation, nomenclature, and sequence characteristics in the genomics era. Insect Biochem. Mol. Biol. 2010;40:189–204. doi: 10.1016/j.ibmb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wink M, Theile V. Alkaloid tolerance in Manduca sexta and phylogenetically related sphingids (Lepidoptera: Sphingidae) Chemoecology. 2002;12:29–46. [Google Scholar]
- Wolfgang WJ, Riddiford LM. Larval cuticular morphogenesis in the tobacco hornworm, Manduca sexta, and its hormonal regulation. Dev. Biol. 1986;113:305–316. doi: 10.1016/0012-1606(86)90166-1. [DOI] [PubMed] [Google Scholar]
- Xia Q, Zhou Z, Lu C, Cheng D, Dai E, Li B, Zhao P, Zha X, Cheng T, Chai C, Pan G, Xu J, Liu C, Lin Y, Qjan J, Hou Y, Wu Z, Li G, Pan M, Li C, Shen Y, Lan X, Yuan L, Li T, Xu H, Yang G, Wan Y, Zhu Y, Yu M, Shen W, Wu D, Xiang Z, Yu J, Wang J, Li R, Shi J, Li H, Su J, Wang X, Zhang Z, Wu Q, Li J, Zhang Q, Wei N, Sun H, Dong L, Liu D, Zhao S, Zhao X, Meng Q, Lan E, Huang X, Li Y, Fang L, Li D, Sun Y, Yang Z, Huang Y, Xi Y, Qi Q, He D, Huang H, Zhang X, Wang Z, Li W, Cao Y, Yu Y, Yu H, Ye J, Chen H, Zhou Y, Liu B, Ji H, Li S, Ni P, Zhang J, Zhang Y, Zheng H, Mao B, Wang W, Ye C, Wong G, Yang H, Group BA. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- Yasukochi Y, Ashakumary LA, Baba K, Yoshido A, Sahara K. A second-generation integrated map of the silkworm reveals synteny and conserved gene order between lepidopteran insects. Genetics. 2006;173:1319–1328. doi: 10.1534/genetics.106.055541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukochi Y, Tanaka-Okuyama M, Shibata E, Yoshido A, Marec E, Wu C, Zhang H, Goldsmith MR, Sahara K. Extensive conserved synteny of genes between the karyotypes of Manduca sexta and Bombyx mori revealed by BAC-FISH mapping. PLoS One. 2009;4:e7465. doi: 10.1371/journal.pone.0007465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama H, Yokoyama T, Yuasa M, Fujimoto H, Sakudoh T, Honda N, Fugo H, Tsuchida K. Lipid transfer particle from the silkworm, Bombyx mori, is a novel member of the apoB/large lipid transfer protein family. J. Lipid Res. 2013;54:2379–2390. doi: 10.1194/jlr.M037093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You M, Yue Z, He W, Yang X, Yang G, Xie M, Zhan D, Baxter SW, Vasseur L, Gurr GM, Douglas CJ, Bai J, Wang P, Cui K, Huang S, Li X, Zhou Q, Wu Z, Chen Q, Liu C, Wang B, Li X, Xu X, Lu C, Hu M, Davey JW, Smith SM, Chen M, Xia X, Tang W, Ke E, Zheng D, Hu Y, Song E, You Y, Ma X, Peng L, Zheng Y, Liang Y, Chen Y, Yu L, Zhang Y, Liu Y, Li G, Fang L, Li J, Zhou X, Luo Y, Gou C, Wang J, Wang J, Yang H, Wang J. A heterozygous moth genome provides insights into herbivory and detoxification. Nat. Genet. 2013;45:220–225. doi: 10.1038/ng.2524. [DOI] [PubMed] [Google Scholar]
- Yu LL, Cui YJ, Lang GJ, Zhang MY, Zhang CX. The ionotropic gamma aminobutyric acid receptor gene family of the silkworm, Bombyx mori. Genome. 2010;53:688–697. doi: 10.1139/g10-056. [DOI] [PubMed] [Google Scholar]
- Zdobnov EM, Bork P. Quantification of insect genome divergence. Trends Genet. 2007;23:16–20. doi: 10.1016/j.tig.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai X, Zhao XF. Participation of haemocytes in fat body degradation via cathepsin L expression. Insect Mol. Biol. 2012;21:521–534. doi: 10.1111/j.1365-2583.2012.01157.x. [DOI] [PubMed] [Google Scholar]
- Zhan S, Merlin C, Boore JL, Reppert SM. The monarch butterfly genome yields insights into long-distance migration. Cell. 2011;147:1171–1185. doi: 10.1016/j.cell.2011.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Gunaratna RT, Zhang X, Najar E, Wang Y, Roe B, Jiang H. Pyrosequencing-based expression profiling and identification of differentially regulated genes from Manduca sexta, a lepidopteran model insect. Insect Biochem. Mol. Biol. 2011;41:733–746. doi: 10.1016/j.ibmb.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, He Y, Cao X, Gunaratna RT, Chen YR, Blissard G, Kanost MR, Jiang H. Phylogenetic analysis and expression profiling of the pattern recognition receptors: insights into molecular recognition of invading pathogens in Manduca sexta. Insect Biochem. Mol. Biol. 2015;62:38–50. doi: 10.1016/j.ibmb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Joung JG, Zheng Y, Chen YR, Liu B, Shao Y, Xiang JZ, Fei Z, Giovannoni JJ. High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harb. Protoc. 2011;2011:940–949. doi: 10.1101/pdb.prot5652. [DOI] [PubMed] [Google Scholar]
- Zhu YC, Specht CA, Dittmer NT, Muthukrishnan S, Kanost MR, Kramer KJ. Sequence of a cDNA and expression of the gene encoding a putative epidermal chitin synthase of Manduca sexta. Insect Biochem. Mol. Biol. 2002;32:1497–1506. doi: 10.1016/s0965-1748(02)00070-x. [DOI] [PubMed] [Google Scholar]
- Zitnan D, Kingan TG, Hermesman JL, Adams ME. Identification of ecdysis-triggering hormone from an epitracheal endocrine system. Science. 1996;271:88–91. doi: 10.1126/science.271.5245.88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.