SUMMARY
DEAD-box RNA helicases play central roles in RNP biogenesis. We reported earlier that LAF-1, a DEAD-box RNA helicase in C. elegans, dynamically interacts with RNA and that the interaction likely contributes to the fluidity of RNP droplets. Here, we investigate the molecular basis of the interaction of RNA with LAF-1 and its human homolog, DDX3X. We show that both LAF-1 and DDX3X, at low concentrations, are monomers that induce tight compaction of single stranded RNA. At high concentrations, the proteins are multimeric and dynamically interact with RNA in an RNA length-dependent manner. The dynamic LAF-1-RNA interaction stimulates RNA annealing activity. ATP adversely affects the RNA remodeling ability of LAF-1 by suppressing the affinity, dynamics, and annealing activity of LAF-1, suggesting that ATP may promote disassembly of the RNP complex. Based on our results, we postulate a plausible molecular mechanism underlying the dynamic equilibrium of the LAF-1 RNP complex.
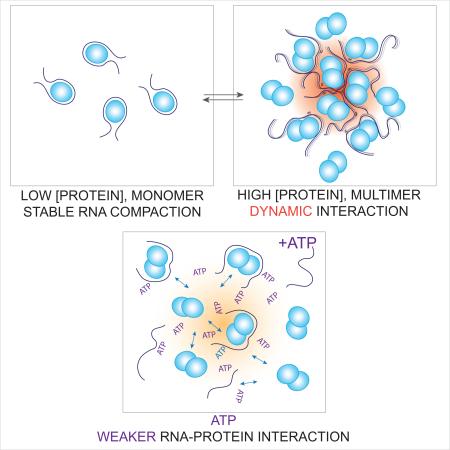
Graphical abstract
(A) At low protein concentration, the protein-RNA interaction is static as the protein wraps RNA tightly. (B) As the local protein concentration increases, protein-RNA interaction becomes dynamic and this promotes association between protein-RNA complexes and RNA annealing. (C) In the presence of ATP, protein-RNA and interaction between protein-RNA complexes becomes weaker, favoring dissociation mode. Protein concentration and ATP concentration can contribute to the dynamic nature of RNP assembly and disassembly.
INTRODUCTION
LAF-1 and DDX3X are orthologous DEAD-box RNA helicases found in C. elegans and humans, respectively. They belong to the DDX3 subfamily (Fig 1A) and are core protein components of ribonucleoprotein (RNP) bodies known as P granules (Updike and Strome, 2010). The helicases in DDX3 subfamily including LAF-1 (C. elegans), Ded1p (yeast) and Belle (Drosophila) play a role in RNP assembly and remodeling (Beckham et al., 2008; Shih et al., 2012; Yarunin et al., 2011). Our previous study demonstrated that dynamic LAF-1-RNA interaction may be involved in the phase separation of RNP granules in vitro (Elbaum-Garfinkle et al., 2015). Both LAF-1 and DDX3X consist of a DEAD-box helicase core and long stretches of flexible amino acids chains in their N-terminal domain (NTD) and C-terminal domain (CTD). Motif analysis defines characteristic amino acid sequences at both NTD and CTD as intrinsically disordered regions (IDR) (Goujon et al., 2010; Sigrist et al., 2013) (Fig 1B, C). The IDRs on RNA binding proteins have been shown to guide targeting of particular proteins, TIA and FUS to stress granules (Gilks et al., 2004; Kato et al., 2012) and promote P body formation in yeast (Decker et al., 2007; Reijns et al., 2008). Owing to the known propensity of IDRs to self-assemble, they have been proposed to play a major role in stimulating RNP granule assembly (Buchan, 2014), which is believed to occur spontaneously when the constituent components reach a critical concentration (Brangwynne et al., 2011). Interestingly, the RNP granules display a liquid-like behavior (Brangwynne et al., 2009; Feric and Brangwynne, 2013; Wippich et al., 2013). Numerous recent studies demonstrated liquid-like phase separated granules formed with RNA binding proteins harboring intrinsically disordered or low complexity domains which are implicated in neurodegenerative diseases. (Burke et al., 2015; Lin et al., 2015; Molliex et al., 2015; Patel et al., 2015; Zhang et al., 2015). Some of these proteins including the ones with patients’ mutations exhibited liquid to solid transition over time, resembling a pathological fibrilization observed in patients with neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS) and frontotemperal dementia (FTD). A recent proteomics study reported that stress granules undergo highly dynamic exchange at the surface, likely by forming an outer shell which surrounds a more stable core. This study demonstrated the role of ATP in both assembly and dynamics of the RNP granules (Jain et al., 2016). Despite the emerging view on RNP formation and dynamics, molecular details unclear. Here, we chose two key proteins found in RNP granules, LAF-1 and DDX3X as model systems to dissect RNA-protein, protein-protein and RNP-RNP interactions and thus elucidate the underlying molecular events that may contribute to RNP granule formation and dynamics.
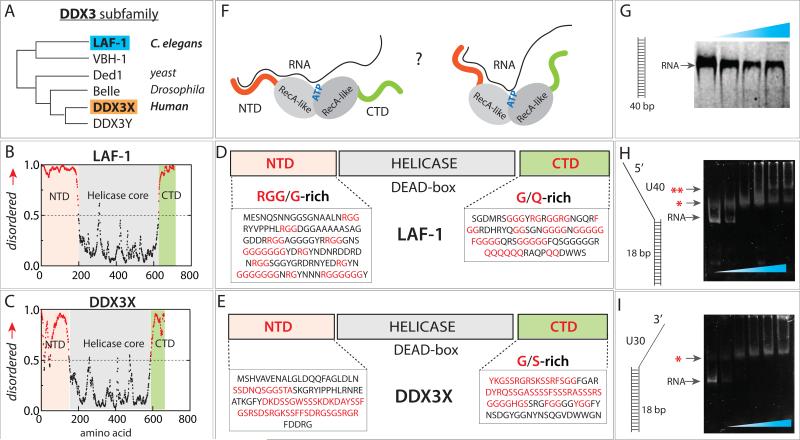
FIGURE 1. Domain composition of LAF-1 and DDX3X.
(A) DDX3 subfamily phylogeny. (B, C) Analysis of intrinsically disordered domain of LAF-1 and DDX3X. (D, E) Composition both proteins containing intrinsically disordered N- and C-terminal domains and central helicase. (F) Unknown protein-RNA interface between LAF-1 and RNA. (G) EMSA showing no LAF-1 binding to 40bp dsRNA. (H, I) EMSA demonstrating LAF-1 binding to partially duplexed, partially single strand RNA with 5’ tail (H) and 3’ tail (I). Single and double red star denote one and two unites of LAF-1 bound band, respectively. See also Figure S1.
The helicase core domain in the center of both LAF-1 and DDX3 features a Q-motif containing the DEAD box peptide sequence, and contains the ATP and RNA-binding sites. Based on previous structural studies (Sengoku et al., 2006), we expect that the helicase core that consists of two RecA-like domains will interact with single strand RNA. However, the functional significance of the IDRs at both N and C-termini of LAF-1 and DDX3 is still poorly understood. The NTD of LAF-1 has unusually high number of RGG/RG boxes that are known to bind single stranded RNA (ssRNA) (Mattaj, 1993). In addition, the NTD of both proteins consist of multiple consecutive sequence patches that are predicted to be of low complexity and high disorder(Dyson and Wright, 2005) (Fig 1D, E). The CTD of LAF-1 is composed of many consecutive glycines, FGG boxes, and consecutive glutamines, which is the signature of a prion domain(Taylor et al., 2002) (Fig 1D). The FGG box is a hydrophilic-hydrophobic interaction motif, mainly found in nuclear membrane interacting proteins (Suntharalingam and Wente, 2003). Glutamine-rich low complexity regions are often found in RNA interacting proteins, especially in the context of processing bodies or stress granules (Buchan et al., 2008; Gilks et al., 2004). DDX3X also entails a prion-like domain at its CTD, signified by the presence of FGG box and YGG box, known to be involved in protein-protein assembly (Fig 1E). Despite the presence of several signature sequences, and their known function in RNP assembly, the mechanistic details of how the NTD and CTD of LAF-1 and DDX3 may contribute to RNA binding, and how such RNA binding may modulate protein conformation (Fig 1F), is unknown. Furthermore, in light of our previous study that implicates these domains in forming a hub of RNA and proteins during RNP assembly(Elbaum-Garfinkle et al., 2015), questions arise as to how the protein-RNA interaction mode changes as a function of RNA length, protein concentration and ATP concentration.
To address these questions, here we probed the molecular interaction between LAF-1 and RNA. Using single molecule fluorescence detection and biochemical measurements, we mapped the changes in protein-RNA binding mode as a function of protein concentration, stoichiometric distribution, RNA length and ATP concentration. At low concentration, LAF-1 displayed a stable interaction with ssRNA where the monomer protein appears to wrap the RNA strand tightly. In contrast, at high concentration of LAF-1, on long ssRNA substrate (≥ 40nt) multiple units of LAF-1 occupied RNA and induced dynamic conformational change within the ssRNA. The same concentration dependent pattern in RNA binding mode was also displayed by DDX3X, raising a possibility of a conserved mechanism shared in this subfamily of RNA helicases. Our mutational analysis indicates that the N-terminal RGG rich domain lowers affinity of LAF-1 toward RNA and is directly responsible for inducing the dynamics in RNA, in agreement with our previous finding (Elbaum-Garfinkle et al., 2015). Interestingly, such dynamics lead to accelerated RNA annealing activity, likely reflecting an improved interaction between protein-RNA complexes. In the presence of ATP, the protein-RNA affinity, dynamics and RNA annealing activity by LAF-1 were all diminished, reflecting a role of ATP in reducing protein-RNA and protein-protein interactions. Taken together, we propose a model where the RNP formation, disassembly and remodeling is dynamically modulated by parameters such as local protein concentration, length of RNA and ATP concentration.
RESULTS
ssRNA compaction by LAF-1
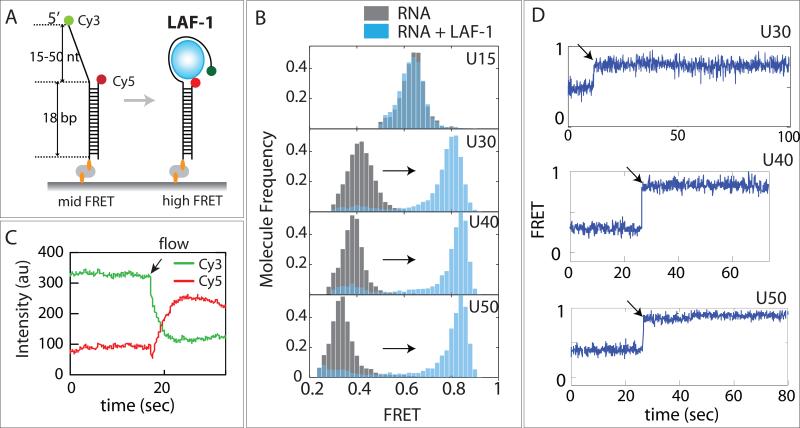
We tested LAF-1 binding to double strand (ds) and ssRNA substrates by electrophoretic gel mobility shift assay (EMSA). LAF-1 displayed binding to RNA containing ssRNA at either 3’ or 5’ end, but not to purely dsRNA (Fig. 1G-I). We further confirmed the substrate specificity by single molecule protein induced fluorescence enhancement assay (Hwang et al., 2011; Hwang and Myong, 2014) which is consistent with the EMSA data. (Fig. S1) This substrate specificity is in agreement with the previous structural study of DDX3X (Epling et al., 2015). Based on this finding, we prepared a partially duplexed RNA labeled with Cy3 (donor, green) and Cy5 (acceptor, red) situated at either end of the ssRNA (See Table S1 for RNA sequence), to probe the conformational change of ssRNA induced by LAF-1 binding by single molecule FRET (Roy et al., 2008). To immobilize this RNA substrate to the single molecule surface by biotin-NutrAvidin linkage, we constructed biotinylated strands with 15, 30, 40 and 50 nucleotide of poly Uracil tail (Fig 2A). We used poly Uracil ssRNA rather than mixed bases to avoid formation of unintended secondary structure within ssRNA. We collected FRET values from more than 5000 molecules and built FRET histogram for each length of ssRNA. Due to the flexibility of single strand nucleic acid (Murphy et al., 2004a), the resulting FRET values of U15, U30, U40 and U50 ranged between 0.6 (for U15) and 0.3 (for U50) (Fig 2B, gray bars). When LAF-1 protein (20nM) was applied to this set of ssRNA, we detected a distinct FRET increase for U30, U40 and U50, but not for U15 (Fig 2B, light blue bars). LAF-1 binding to poly-U substrates occurred immediately after the protein addition as shown in ensemble averaged donor and acceptor intensities (Fig 2C) and in representative single molecule FRET traces (Fig 2D). This result indicates the following. First, U15 is not long enough for LAF-1 association, but U30-50 ssRNA can accommodate LAF-1 binding. Second, the discrete FRET shift to higher level observed in U30-50 suggests that the end to end distance of RNA is diminished in all three substrates. Third, the high FRET peaks remained high even after three times of buffer wash (Fig S2), indicating that the association of LAF-1 to ssRNA is highly stable. In a typical FRET experiment, any protein binding to this type of substrate is expected to occupy space in ssRNA or ssDNA and lower FRET (Hwang et al., 2012; Hwang et al., 2014; Qiu et al., 2013). We tested this effect by subjecting an RNA helicase, Sen1 to the U30-FRET RNA construct. The helicase binding to U30 resulted in FRET decrease, suggesting that the protein binding increased the dye to dye distance by stretching the two ends of the ssRNA (Fig. S3). The unusual FRET increase seen here suggests that the protein induces a tight compaction of the flexible ssRNA such that the two dyes come to close vicinity. The similar high FRET level obtained for U30, 40 and 50 suggests a length-independent compaction generated by LAF-1.
FIGURE 2. LAF-1 induces ssRNA compaction.
(A) FRET DNA constructs containing 5’ U15-50 tail and Cy3/Cy5 dyes across ssRNA. (B) FRET histograms before (gray) and after (light blue) LAF-1 addition. (C) Averaged Cy3 and Cy5 intensities obtained from smFRET experiment. Arrow indicates when LAF-1 was added (D) Representative single molecule FRET traces for U30, 40 and 50. Rapid FRET increase occurs immediately after addition of LAF-1 (20nM). See also Figure S2 and 3.
Multimer of LAF-1 induces dynamic interaction with ssRNA
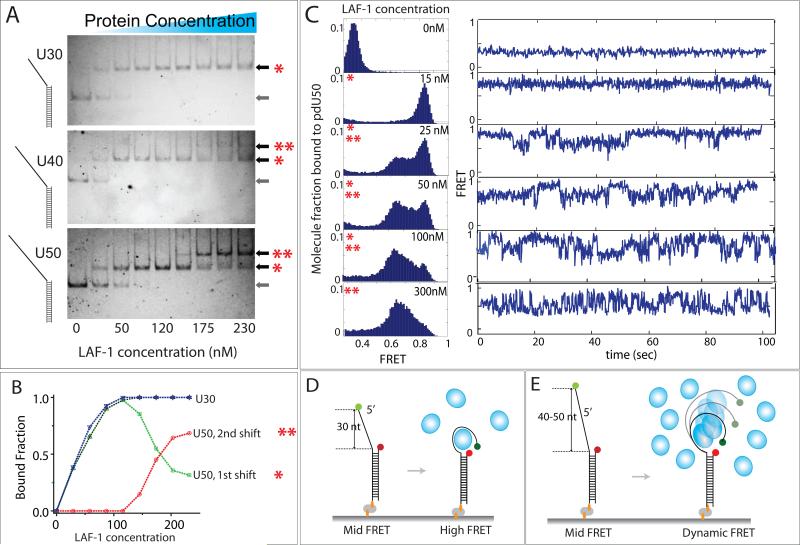
We reported earlier that LAF-1 induces dynamics on ssRNA selectively at high concentrations where LAF-1 can self-organize into viscous liquid like droplet in vitro (Elbaum-Garfinkle et al., 2015). Based on this observation, we hypothesized that LAF-1 may form into multimers/oligomers in this concentration range. To test this effect, we applied varying concentrations (1-230 nM) of LAF-1 to U30, 40 and 50 RNA substrates and performed EMSA analysis. For U30, only a single band shift appeared throughout all concentrations whereas U40 and U50 displayed two shifts denoted by red asterisks at higher LAF-1 concentrations (Fig 3A). This result clearly shows that shorter length (≤ 30 nt) cannot, but longer length ssRNA (≥ 40 nt) can accommodate multimers of LAF-1 (Fig 3A). Quantification of first and second shift obtained for U50 shows that the transition from first to second shift occurs approximately at 100nM LAF-1 concentration (Fig 3B). We then tested similar concentration range of LAF-1 in smFRET experiment with U30, 40 & 50. When 15nM LAF-1 was added to U50 RNA, the low FRET peak shifted completely to a high FRET, again reflecting tight compaction of ssRNA also observed in Figure 2. When the protein concentrations were raised to 25, 50 and 100nM, we observed an appearance of a broad mid FRET peak and diminishing level of the high FRET peak. In agreement, as the protein concentration increased, single molecule traces displayed a transition from static high FRET to dynamic FRET fluctuation. At 300nM LAF-1, for the U50 RNA, dynamic FRET dominates (Fig 3C). On the contrary, at high LAF-1 concentration, U30 RNA showed mostly static, non-dynamic high FRET traces.
FIGURE 3. Mutimer LAF-1 induces dynamic interaction with ssRNA.
(A) EMSA image of U30, 40 and 50 binding to varying concentrations of LAF-1. One and two asterix denotes monomer and multimer status, respectively while the arrow without any asterix indicates RNA-only. (B) Quantitation of the EMSA image for U30 and 50 shown in (A). (C)FRET histograms and representative smFRET traces for U50 at varying LAF-1 concentrations (D, E) Schematic depiction of LAF-1 interaction at low (D) vs. high (D) LAF-1 concentrations. See also Figure S4 and 5.
Based on these results, we interpret that the first band shift in EMSA corresponds to a monomeric LAF-1 which wraps RNA tightly at low concentration (< 15nM) (Fig 3D) and the second shift corresponds to multimers of LAF-1 that induce dynamic interaction with ssRNA at high concentrations (Fig 3E). We note that FRET fluctuation is not likely due to successive protein binding because the FRET range in the fluctuation (0.45-0.85) is above that of RNA-only FRET level of 0.35 (Elbaum-Garfinkle et al., 2015). Further examination of smFRET traces obtained for U40 and U50 revealed a small fraction of molecules (10-15%) exhibiting two-step FRET increase followed by FRET fluctuation, suggesting that successive binding of two units of LAF-1 may be responsible for the dynamic FRET fluctuation (Fig S4A,B). In addition, fluorescently labeled N-terminal domain, which also multimerized (Figure 5E) and exhibited dynamic FRET (Figure 5D) displayed two-step photobleaching, also reflecting two LAF-1-NTD molecules bound to RNA (Fig. S4C-E).The emerging picture is that the two protein units stay on the RNA and continuously induces dynamic wrapping and unwrapping of RNA. LAF-1 gives rise to static high FRET and dynamic fluctuating FRET, depending on whether it is present as a monomeric or dimeric species, respectively. In the intermediate concentration ranges (25-100nM), we observed mixed behavior, corresponding to mixture of monomer and dimer LAF-1 binding to RNA. Thus, the two parameters controlling the static versus dynamic protein-RNA interaction interface is LAF-1 concentration and the length of ssRNA. The tight protein-RNA interface at monomer state changes to dynamic interaction mode when two units of protein binds RNA together (Fig 3D, E).
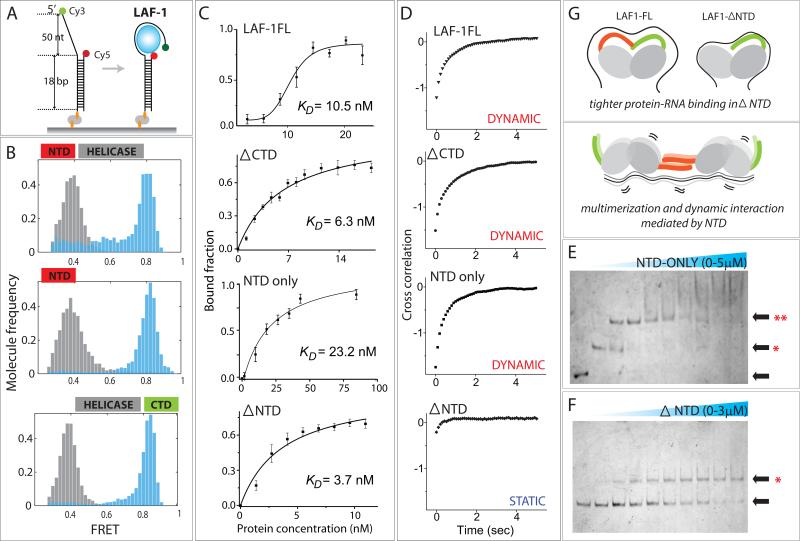
FIGURE 5. Role of RGG-rich NTD of LAF-1.
(A) Schematic of smFRET experiment for all truncation variants of LAF-1. (B) FRET histogram before (gray) and after (light blue) LAF-1 addition. (C) Binding isotherm for LAF-1FL and mutants. All data are represented as mean +/− SEM. (D) Cross correlation of FRET data for all proteins tested. The strong cross correlation in LAF-1FL, ΔCTD and NTD-only results from analysis of FRET fluctuations induced by these proteins. The lack of cross correlation in ΔNTD mutant indicates that NTD is required for the dynamic LAF-1-RNA interaction probed by FRET fluctuation. (E, F) EMSA of NTD-only (E) and ΔNTD (F). Single and double asterix denote monomer and multimer status, respectively. (G) Schematic depiction of plausible role of NTD in reducing affinity to RNA while inducing mutimer formation and stimulating dynamic interaction with ssRNA.
DDX3X displays concentration dependent dynamics on RNA
Based on its high similarity to LAF-1, we prepared DDX3X to test if it exhibits similar binding behavior to ssRNA. As before, the U50 RNA FRET construct produced FRET peak at 0.35. Upon addition of DDX3X (720 nM), the low FRET peak shifted to high FRET (~0.8), suggesting a similar tight compaction of ssRNA as observed for LAF-1 (Fig 4A, top). In fact, the high FRET value is similar to what was seen for LAF-1 bound to U50 (Fig 3C, 15nM), suggesting a similar mechanism involved in protein-RNA interaction. As the protein concentration of DDX3X is increased, a broad mid FRET peak emerges, again in a similar manner as seen in LAF-1 (Fig 4A bottom). In agreement, the single molecule traces display a steady high FRET in low DDX3X concentration and dynamic FRET fluctuation in high concentration (Fig 4B). In summary, DDX3X induces tight compaction or wrapping of ssRNA at low concentration (Fig 4C) and imparts dynamic fluctuations in ssRNA at high concentration (Fig 4D), in a similar manner exhibited by LAF-1. This result signifies that both LAF-1 and DDX3X undergo the same inherent change in the way they interact with RNA, in a concentration dependent manner.
FIGURE 4. DDX3X displays similar concentration dependent, biphasic behavior as LAF-1.
(A) FRET histograms of U50 taken at low to high DDX3X concentrations. (B) smFRET traces obtained for the conditions in (A). (C, D) Schematic depiction of DDX3X interaction with ssRNA at low (C) and high (D) DDX3X concentration.
N-terminal RGG domain of LAF-1 lowers affinity to RNA and induces dynamics on RNA
We previously reported about the role of N-terminal RGG-rich domains of LAF-1 in inducing dynamics on RNA and promoting droplet formation (Elbaum-Garfinkle et al., 2015). Here, we used smFRET and biochemical assays to probe how the intrinsically disordered N- and C-terminal domains of LAF-1 may contribute to RNA binding affinity and dynamics (Fig 5A). We compared three truncation mutants of LAF-1, C-terminal deletion (ΔCTD), N-terminal domain (NTD-only) and N-terminal deletion (ΔNTD) (Fig 5B). On U50, all three LAF-1 protein variants showed FRET shift to 0.8, suggesting a similar mode of RNA binding and compaction in all cases. Using the fraction of high FRET molecules as binding criteria, we generated the binding isotherm curve for the LAF-1FL and the three truncation mutants of LAF-1 (Fig 5C). Interestingly, all three truncation mutants displayed tighter binding to RNA than the LAF-1FL, reflecting that both the NTD and CTD negatively regulate RNA binding in the context of full length LAF-1. When applied in high concentration (2μM), the LAF-1FL, ΔCTD and NTD-only all exhibited dynamic FRET fluctuation whereas the ΔNTD remained bound in a static manner (Elbaum-Garfinkle et al., 2015). In agreement, the single molecule FRET data analyzed by cross correlation displayed a clear presence of FRET fluctuation in all but in ΔNTD mutant (Fig 5D). We asked if the loss of dynamic interaction with RNA in ΔNTD can be due to its inability to mutlimerize. To test this effect, we performed EMSA analysis with NTD-only and ΔNTD on U50. Indeed, the result showed that while NTD-only mutant multimerized, giving rise to a clear second band shift at low concentration (Fig 5E), the ΔNTD mutant exhibited a single shift all throughout the concentration range tested (1-3μM) (Fig 5F). Based on these findings, we propose a plausible model which defines a role of NTD in modulating protein-RNA interaction. NTD serves to reduce the protein-RNA affinity, enables multimerization of LAF-1 on a long ssRNA and induces dynamics on RNA (Fig 5G).
Dynamic interaction leads to accelerated RNA annealing
Next, we asked how the LAF-1 multimer induced dynamics play a role in RNA remodeling. As in other DEAD box helicases (Bizebard et al., 2004; Jarmoskaite and Russell, 2011; Linder and Jankowsky, 2011; Rogers et al., 1999; Tijerina et al., 2006), LAF-1 does not lead to active unwinding tested by gel electrophoresis and single molecule fluorescence assays (Fig S5). In light of the correlation between the dynamic interaction of LAF-1 and RNA and the formation of droplets (Elbaum-Garfinkle et al., 2015), we sought to test if the RNA-protein dynamics can promote efficient RNA annealing by bringing RNP complexes in close proximity. In our single molecule platform, we immobilized a partially duplexed RNA labeled with FRET dyes at either end of ssRNA, which exhibits high FRET (~0.75) due to the flexibility of ssRNA (Murphy et al., 2004b). We note that the FRET value is substantially higher than expected from 25 poly-Uracil substrate (U25), likely due to the mixed base composition (with A, U, G, C) that led to an increased intramolecular interactions within the ssRNA. The same RNA used in unwinding study revealed the same high FRET (Koh et al., 2014). We applied LAF-1 and ssRNA (10nM) which bears a complementary sequence to the FRET RNA construct such that annealing will be detected as a decrease in FRET signal (Fig 6A). When the pre-incubated mixture of LAF-1 and 10nM complementary RNA is applied to FRET-RNA immobilized surface, the free LAF-1 is expected to interact with the RNA on surface while the complementary RNA is already in complex with LAF-1. Therefore, we monitor the annealing induced by interaction amongst LAF-1-RNA complexes. We tested conditions of no protein (RNA-only), LAF-1FL, ΔNTD, NTD-only and ΔCTD LAF-1 at binding isotherm saturation point of 40-60nM. FRET histograms collected from over 100,000 molecules were plotted over time for LAF-1FL, ΔNTD and ΔCTD (Fig 6B-D). Fastest annealing was achieved by the ΔCTD mutant followed by NTD-only and WT. To obtain the annealing rate, the same FRET histograms were dissected into finer segments from which the fraction of annealed RNA was extracted and plotted (Fig 6E). The calculated annealing rates show that ΔCTD induces the most efficient annealing with a rate approximately 15 times higher than the RNA-only condition, revealing the CTD of LAF-1 as a negative regulator of RNA annealing activity, possibly by inhibiting interaction between LAF-1-RNA complexes. In contrast, ΔNTD yielded negligible difference in annealing rate while NTD-only induced a moderate improvement. Wild type LAF-1 showed a concentration dependent annealing. When concentration is low, it barely promoted RNA annealing, but in high protein concentration, it accelerated RNA annealing (Fig S6, Fig 6F). Interestingly, the conditions that promoted dynamic protein-RNA interaction led to enhanced annealing rate whereas the two conditions which induce tight compaction of RNA as a monomer (Fig 2, 4) resulted in no improvement (Fig 6F). This result strongly suggests that the dynamic protein-RNA interaction, mediated by the NTD of LAF-1 is directly responsible for enhanced interaction between LAF-1-RNA complexes, which leads to rapid annealing between two complementary RNA strands (Fig 6G).
FIGURE 6. Dynamic LAF-1-RNA interaction stimulates RNA annealing.
(A) Schematic diagram of annealing assay in which high FRET is expected to transition to low FRET upon RNA annealing. (B, C, D) FRET histograms obtained for annealing reactions of LAF-1FL (B), ΔNTD (C) and ΔCTD (D). The pink shadow reflects annealed fraction of molecules. (E) Annealing reaction of LAF-1 LAF-1FL and mutants captured in real time. (F) Annealing rates calculated from (E). (G) Schematic depiction of NTD-driven dynamics that induce increased interaction between LAF-1-RNA complexes, leading to enhanced RNA annealing. See also Figure S6.
ATP diminishes protein-RNA interaction and dynamics
All the assays performed and discussed above were done in the absence of ATP. We next asked if or how ATP might play a role in modulating the interaction between LAF-1 and RNA. First, we examined RNA binding affinity by performing the experiment shown in Figure 2 in the presence of ATP (1mM). Briefly, the fraction of LAF-1 bound U30 ssRNA molecules that produced high FRET were plotted as a function of LAF-1 concentration. As shown, the binding affinity was reduced by ATP, evidenced by a higher Kd (Fig 7A). The binding rate of LAF-1 to RNA was calculated by tabulating the LAF-1 bound fraction (high FRET) over time. The difference in half time deduced from the binding kinetic curve reveals that the binding is slower in presence of ATP (Fig 7B). This effect is likely due to ATP hydrolysis, not ATP binding since ATPγS does not change the binding rate of LAF-1 (Fig. S7A). To test if LAF-1-RNA dynamics may also be influenced by ATP, we applied LAF-1 at a concentration of 1 μM to monitor LAF-1 induced dynamics on ssRNA in varying ATP concentration. As we increased the ATP concentration, we observed lower frequency of the FRET signal fluctuation (Fig 7C). To quantify this effect, we collected FRET peak-to-peak dwell times from over 200 molecules (Fig 7C, double arrow in red) and plotted the average rate of FRET fluctuation as a function of ATP concentration (Fig 7D). The anti-correlated relationship between the FRET fluctuation frequency and ATP concentration indicates that ATP acts to reduce the dynamic interaction between LAF-1 and ssRNA.
FIGURE 7. ATP effect.
(A) Binding isotherm of LAF-1 with and without ATP. (B) LAF-1 binding rate to U50 with and without ATP. (C) smFRET traces taken at varying ATP concentrations. (D) Rate of FRET fluctuations measured at different ATP concentrations. (E) RNA annealing rate with and without ATP. (F) Summary of ATP effect. See also Figure S7.
We then sought to test if the decreased dynamics influences RNA annealing rate. The annealing experiment performed in the presence and absence of ATP was plotted in the same way shown in Figure 6E. We applied 1 μM concentration of LAF-1 so that the annealing outcome would not be controlled by differential binding affinity between the two conditions. In presence of ATP, the annealing was significantly reduced both in rate and in the total amount of annealed product (Fig 7E). We note that the result is not affected by limited ATP based on the low rate of ATP hydrolysis by LAF-1. (Fig. S7B). This result is consistent with the observation that the dynamic protein-RNA interaction promotes RNA annealing. In summary, ATP serves to reduce LAF-1's affinity to RNA, decrease its binding rate, lessen dynamics on RNA and diminish RNA annealing activity (Fig 7F).
DISCUSSION
Concentration dependent binding mode
Many studies have reported on protein concentration dependent unwinding by helicases. For example, super family (SF)1 helicases Rep, PcrA, UvrD in e. coli and Srs2 from yeast translocate on single stranded DNA in low concentration while they unwind partially duplexed DNA in elevated concentration range (Ali et al., 1999; Myong et al., 2005; Park et al., 2010; Qiu et al., 2013). Such results are consistent with the prediction that more than one protein unit is required for efficient unwinding. DEAD box proteins are classified as a non-processive helicase because they often display weak unwinding activity limited to short length of base pairs of RNA (Linder and Jankowsky, 2011; Sengoku et al., 2006). Here, we examined LAF-1 and DDX3X, both of which are RNA DEAD-box helicases critical for assembly of RNP granules. Unlike the SF1 helicases which unwind DNA in a concentration dependent manner, here we report that LAF-1 and DDX3X exhibit different RNA binding mode dependent upon the protein concentration. At low concentration (10-20 nM), monomer unit of protein wraps ssRNA tightly and stably, as evidenced by a stable high FRET signal which persist even after a buffer wash. The same high FRET value observed in U30, U40 and U50 indicates that the RNA compaction occurs in such way that the end of RNA strand come in close proximity to the ssRNA/dsRNA junction in all RNA substrates. Although we cannot infer the conformational state of RNA, we expect that the multivalent interaction between the protein and RNA enables a tight compaction which may lead to a complete sequestration of RNA. Previous structure of DDX3X revealed an unusual helical element that extends highly positively charged sequence in a loop which contributed to increased protein-RNA contact (Hogbom et al., 2007). Such structural feature, together with the N- and C-terminal IDR domains may be responsible for the extensive RNA compaction we observe here. In contrast, a high concentration of LAF-1 allows multimer binding on a long ssRNA substrate and induces continuous and repetitive conformational dynamics on ssRNA, represented by long lived FRET fluctuations. In light of their function in assembly of RNP complex, the compaction of RNA seen at low protein concentration may represent an inactive or dormant state in which the protein sequesters RNA into its reservoir rather than interacting with neighboring molecules. In contrast, the dynamic interaction obtained at high protein concentration on long ssRNA substrate may reflect an activated or charged state in which the dynamic protein-RNA complex actively associates with neighboring protein-RNA components. In this capacity, the dynamic state may reflect a nucleating condition primed for assembly into RNP droplet. Although functionally distinct, such nucleation may be similar to the nucleation event found in Rad51 and RecA (Joo et al., 2006; Qiu et al., 2013) in which 4-5 monomer cluster represents the active state which can lead to rapid filament extension suited for homology searching.
Contribution of N- and C-terminal domains
Both N- and C-termini of LAF1 consist of IDRs that are composed of elements such as RGG/RG, G/Q and G/S rich chains of amino acids. These low complexity domains are expected to contribute to RNA binding and increased propensity to self-aggregate. Although the NTD of LAF-1 contains RGG rich segments which enables RNA binding, the NTD is not solely responsible for RNA binding affinity of LAF-1 as ΔNTD also exhibits RNA binding. Unexpectedly, the ΔNTD displays higher RNA binding affinity than the full-length protein, suggesting that NTD reduces RNA binding affinity in the context of the full length protein. Our results reveal that the NTD of LAF-1 is directly responsible for the concentration dependent static and dynamic binding modes exhibited by the full length protein. We propose that NTD of LAF-1 acts as a switch turned off as a monomer which allows capturing of RNA in a tightly packed format, and turned on by forming a multimer which imparts a slippery and fluidic protein-RNA interface and induces dynamic mobility on RNA. The lack of multimerization in ΔNTD strongly suggests that the NTD is directly involved in protein-protein interaction mediated by a long RNA substrate (Fig 5G). Taken together, our result is consistent with a model in which the differing conformation of NTD between monomer and multimer dictates its interaction mode with RNA as depicted in Figure 5G. While the deletion of C-terminus (ΔCTD) did not induce a drastic change i.e the binding affinity and the concentration dependent static vs. dynamic RNA binding, it displayed a significant reduction in RNA annealing activity. We note that the CTD-only could not be purified due to heavy precipitation which likely arose from self-aggregation.
Insights about RNA remodeling function
The core proteins in RNP are expected to exhibit multifaceted ability to interact with RNA, protein and RNA-protein (RNP) complexes. First, they bind RNA with high affinity in order to capture RNA for storage or transport functions. However, such binding cannot be too tight because RNA molecules are required to be dislodged and released as needed for translation, degradation and transport. Second, the RNP proteins interact with neighboring RNP components to form an organized meshwork to be assembled into granules. Again, the interaction should be strong enough to hold up the constituent and to support further growth of granules, yet not entirely locked into a rigid structure since the RNP components undergo fluidic exchange with other granules in cells (Jain et al., 2016). Third, they remodel RNA for different purposes. Such activity includes unwinding of duplexed RNA, unfolding of secondary structured regions, annealing of complementary strands, packaging or condensing RNA for storage or transport and sequestering RNA from degrading enzymes. We demonstrate that LAF-1 does not exhibit unwinding activity, but promotes RNA annealing in conditions that induce dynamic RNA-protein interaction (Fig 6F). Interestingly, the same conditions led to phase separation in our previous study (Elbaum-Garfinkle et al., 2015). Based on these findings, the increased annealing activity we observe here may be a proxy for droplet assembly i.e a signal informing that the local concentration of protein-RNA complexes has exceeded the threshold required for initiating nucleation for droplet assembly.
Our earlier work demonstrated that LAF-1 forms into liquid-like droplet at physiologically relevant concentration of protein (1μM) and salt (125mM NaCl) in the absence of RNA. In the presence of RNA, the droplet exhibits higher dynamics and fluidity as displayed by microrheology and FRAP experiments (Elbaum-Garfinkle et al., 2015). Based on these observations, while RNA is not required for the droplet formation, it acts like a fluidizer that may contribute to the dynamic nature of RNP granules in cells. In light of the recent findings of in vitro protein-only droplets developing into fiber-like state over time (Molliex et al., 2015; Patel et al., 2015), RNA may play a critical role in generating and maintaining the liquid like property of granules. In addition, the extremely high annealing rate achieved in ΔCTD reveals the role of CTD in downregulating LAF-1's annealase activity. It can be due to a conformational change of CTD that results in decreased interaction between protein-RNA complexes.
Role of ATP and dynamic equilibrium in RNP complex
In many helicases, ATP binding and hydrolysis lead to catalytic activities such as unwinding, translocation and protein displacement. Unlike these well-known effects, ATP served to reduce LAF-1's affinity to RNA, diminish the RNA-LAF-1 dynamics and dampen RNA annealing. If we interpret the high protein-RNA affinity, increased dynamics and enhanced annealing as the condition that favors nucleation and assembly of RNP complex, the ATP induced effects demonstrated here suggest a condition that promotes disassembly of RNP complex. In this regard, the ATP concentration, in addition to protein concentration, may be an important parameter that controls formation and disassembly of LAF-1 RNP granules. Recent study reported a structure of DDX3X bearing an ATP-binding loop (ABL) which is critical for RNA stimulated ATPase activity. The NMR chemical shift suggested that the ABL interacts dynamically with ATP (Epling et al., 2015). The weaker protein-RNA interaction in the presence of ATP may arise from such dynamic fluctuation of the ABL-like domain. In agreement, earlier DDX3X structure was crystalized bound to AMP despite the provided conditions of ATPƴS and ADP (Hogbom et al., 2007), suggesting that the ATP bound form is less structured and more flexible. Another structure of a closely related RNA helicase, Vasa with ATP and RNA exhibited a sharply bent RNA that avoids clash with the N-terminal domain (Sengoku et al., 2006). This conformation was interpreted to be suited to unwind duplexed RNA by Vasa. In light of our findings, such arrangement may explain the weakened interaction between the ATP bound LAF-1 and RNA. The emerging picture based on our results is depicted in the graphical abstract.
Our results reveal three parameters that can contribute to tuning the RNP assembly and dynamics: (i) the local protein concentration, (ii) length of ssRNA and (iii) ATP. At low protein concentration, individual LAF-1 unit may interact with a single RNA molecule by tightly wrapping it around perhaps to sequester it from other RNA binding proteins or degrading enzymes. As the local protein concentration increases, more than one LAF-1 occupies single RNA and induce dynamic interaction interface, which in turn triggers enhanced interaction between protein-RNA complexes and improved RNA annealing activity. Such interaction may be sufficient to drive nucleation of protein-RNA complexes toward formation of RNP droplets. In contrast, ATP acts to reduce affinity and dynamics in LAF-1-RNA interactions, thereby lowers the propensity to assemble RNP complex. The effect induced by ATP, which appears to favor disassembly of RNP, can represent a critical means to separate and disperse the RNP components to other parts of cytoplasm for delivery or exchange purposes. Our result strongly suggests that ATP acts as a dispersing agent that disaggregate protein-RNA complex formation. Such role played by ATP can be critical in a crowded cellular environment in which the extremely high protein concentration can easily induce aggregations and pathological inclusions bodies (Lin et al., 2015; Molliex et al., 2015; Patel et al., 2015). The molecular probing of LAF-1 to RNA interaction reported here presents a plausible mechanism by which RNP complex dynamics can be tuned by several key parameters.
EXPERIMENTAL PROCEDURES
Materials
Vectors pET28a and pUC19 were procured from Novagen and Fisher Scientific, respectively, and E. coli strain BL21(DE3) from Stratagene. Restriction endonucleases, T4 ligase, Taq polymerase, Phusion polymerase, and calf intestinal alkaline phosphatase were purchased from New England Biolabs, while Kapa taq polymerase was purchased from Kapa BioSciences. LB powder mix (Luria and Miller Broth) and glycerol were from Fischer Scientific and IPTG from Roche. LB agar plates with antibiotics were obtained from UIUC cell media center. Custom DNA oligonucleotides were purchased from Integrated DNA Technologies, Coralville, IA. Materials for DNA purification via agarose gel electrophoresis including a QIAquick Gel Extraction kit and a Qiaprep Spin Miniprep kit was purchased from QIAGEN, Valencia, CA.
Molecular cloning
Standard molecular biology protocols were followed for molecular cloning ((2011)). Plasmid DNA was prepared using miniprep kit (QIAGEN), and vectors were digested by appropriate restriction endonucleases (New England Biolabs). Digested inserts and linearized vectors were purified by Qiagen PCR cleaning kit. All molecular sub-cloning steps were performed using E. coli strain DH5-alpha and NEB high competent cells. The amino acid sequence of LAF1 protein (shown below) was codon-optimized. A codon optimized, synthesized DNA sequence was inserted to contain 6-histidin at the N-terminus. Truncated mutants of LAF1 were generated via PCR with LAF-1 encoding DNA. Phusion high fidelity polymerase-based PCR reactions (20 uL) were carried out as per the following program: 2 min at 95 °C, followed by 24 cycles of 15 seconds each at 95 °C; 15 sec at appropriate annealing temperature (gradient range); and 2 mins at 72 °C.. The PCR amplicon encoding for LAF1 was ligated. Positive colonies were selected the next day, amplified and DNA was sequenced at UIUC sequencing center.
Protein Expression
The pET28a expression plasmid encoding full length or deletion mutants of LAF-1 were transformed into E. coli expression strain BL21(DE3)pLysS. For protein expression, 10 mL Luria Broth (LB) culture medium supplemented with 100 ug/L Kanamycin was inoculated with a single colony and incubated overnight (16 hr) in Erlenmeyer flask at 250rpm, 37 °C. Following overnight growth, the culture was diluted 100-fold into 1000 mL LB culture supplemented with 100 ug/L Kanamycin. Absorbance was monitored at a wavelength of 600 nm, and upon reaching an optical density (OD600) of 0.3, protein expression was induced by addition of 1.0 mM isopropyl-D-thiogalactopyranoside (IPTG). After incubating cultures at 18°C and 200rpm for overnight protein expression, cell pellets were harvested by centrifugation (6,000 g, 15 min, 4°C), followed by snap freezing in dry ice and ethanol. All the amino acid sequences for LAF-1 was published previously (Elbaum-Garfinkle et al., 2015).
Protein Purification
LAF1 and DDX3X were purified using affinity columns followed by a size exclusion column as done previously (Elbaum-Garfinkle et al., 2015).
RNA sample preparation
All RNA oligonucleotides substrates were purchased from IDT labeled with either Cy3 or Cy5 dyes. The complementary RNA was modified with Cy3 (GE Healthcare) at its 5’ end. RNA substrates were assembled via annealing, by mixing Cy3 labeled strand and Cy5 labeled complementary strand RNA at a molar ratio of 1:1 in T100 (10 mM Tris–HCl pH 7.5, 100 mM NaCl). The annealing mix was incubated at 65°C for 5 min followed by slow cooling to room temperature over 3 hours.
Single molecule imaging buffers
For single molecule imaging, 1.0 mg/ml glucose oxidase, 0.2% glucose, 2 mM 6-hydroxy-2,5,7,8- tetramethylchromane-2-carboxylic (Trolox), and 0.01 mg/ml catalase were added to the buffer (125 mM NaCl, 50mM Tris–HCl pH 7.5). All FRET and single molecule imaging measurements were carried out in the same buffer (50 mM Tris–HCl pH 7.5 and 125 mM NaCl) at room temperature (22 ± 1°C).
Single molecule fluorescence data acquisition
Single molecule fluorescence experiments utilized quartz slides (Finkenew nbeiner) coated with polyethylene glycol (PEG), prepared as described previously (Roy et al., 2008).
Electrophoretic Mobility Shift Assay (EMSA)
Protein- RNA interaction was assayed by EMSA as described (Hellman L. M et al., Nat.Protocols, 2007, 3, 1849). Briefly, the same Cy3 or Cy5-labeled RNA used for single molecule FRET with Cy3 or Cy5 labeling was utilized. Fluorophore labeled RNA was diluted to 100nM and used for EMSA. For EMSA, protein and RNA were mixed at the appropriate ratio and incubated at room temperature for 20 min. After incubation, the reaction mixture was mixed with DNA loading dye and subjected to gel electrophoresis (6% PAA gel). The gel was imaged using Typhoon fluorescence scanner (GE healthcare).
smFRET data analysis
All data analysis was carried out by scripts written in Matlab and an additional analysis software was coded in C++ and Matlab. The software for analyzing single-molecule FRET data is available for download from https://physics.illinois.edu/cplc/software/ and http://vbfret.sourceforge.net/.
Binding isotherm calculation
Binding isotherm was calculated from different FRET status in different protein concentrations. The fraction of FRET values were counted with the custom Matlab code. Also the number of RNA was counted to normalize the fraction numbers. The resulting data collection was plotted with Origin 8.0 software using Hill equation.
Supplementary Material
HIGHLIGHTS.
LAF-1 and DDX3X induce tight compaction of single strand RNA at low concentration.
At high concentration, LAF-1 forms a dimer and dynamically interact with RNA.
Dynamic RNA-protein interaction stimulates RNA annealing activity.
ATP modulates RNA remodeling activity by reducing affinity, dynamics and annealing.
ACKNOWLEDGEMENTS
The authors would like to thank Shana Elbaum, Clifford Brangwynne at Princeton University and Krzysztof Szczepaniak, Christian Eckmann at Martin-Luther-University, Halle-Wittenberg, Germany for helpful discussions. This work was supported by the Human Frontier Science Program (RGP0007/2012), American Cancer Society RSG-12-066-01-DMC, NIH 1DP2GM105453, National Science Foundation and Physics Frontiers Center Program (0822613) through the Center for the Physics of Living Cells to Y.K and S. M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Y. K performed all the experiments included in the manuscript. S.M guided the project and wrote the manuscript.
REFERENCES
- Current Protocols in Molecular Biology (Wiley) 2011.
- Ali JA, Maluf NK, Lohman TM. An oligomeric form of E. coli UvrD is required for optimal helicase activity. J Mol Biol. 1999;293:815–834. doi: 10.1006/jmbi.1999.3185. [DOI] [PubMed] [Google Scholar]
- Beckham C, Hilliker A, Cziko AM, Noueiry A, Ramaswami M, Parker R. The DEAD-box RNA helicase Ded1p affects and accumulates in Saccharomyces cerevisiae P-bodies. Mol Biol Cell. 2008;19:984–993. doi: 10.1091/mbc.E07-09-0954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizebard T, Ferlenghi I, Iost I, Dreyfus M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–7866. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 2014;11:1019–1030. doi: 10.4161/15476286.2014.972208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke KA, Janke AM, Rhine CL, Fawzi NL. Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nature Reviews Molecular Cell Biology. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci U S A. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epling LB, Grace CR, Lowe BR, Partridge JF, Enemark EJ. Cancer-associated mutants of RNA helicase DDX3X are defective in RNA-stimulated ATP hydrolysis. J Mol Biol. 2015;427:1779–1796. doi: 10.1016/j.jmb.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Brangwynne CP. A nuclear F-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nat Cell Biol. 2013;15:1253–1259. doi: 10.1038/ncb2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic acids research. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogbom M, Collins R, van den Berg S, Jenvert RM, Karlberg T, Kotenyova T, Flores A, Karlsson Hedestam GB, Schiavone LH. Crystal structure of conserved domains 1 and 2 of the human DEAD-box helicase DDX3X in complex with the mononucleotide AMP. J Mol Biol. 2007;372:150–159. doi: 10.1016/j.jmb.2007.06.050. [DOI] [PubMed] [Google Scholar]
- Hwang H, Buncher N, Opresko PL, Myong S. POT1-TPP1 Regulates Telomeric Overhang Structural Dynamics. Structure. 2012 doi: 10.1016/j.str.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci U S A. 2011;108:7414–7418. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H, Kreig A, Calvert J, Lormand J, Kwon Y, Daley JM, Sung P, Opresko PL, Myong S. Telomeric overhang length determines structural dynamics and accessibility to telomerase and ALT-associated proteins. Structure. 2014;22:842–853. doi: 10.1016/j.str.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H, Myong S. Protein induced fluorescence enhancement (PIFE) for probing protein-nucleic acid interactions. Chemical Society reviews. 2014;43:1221–1229. doi: 10.1039/c3cs60201j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 2016 doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmoskaite I, Russell R. DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip Rev RNA. 2011;2:135–152. doi: 10.1002/wrna.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh HR, Xing L, Kleiman L, Myong S. Repetitive RNA unwinding by RNA helicase A facilitates RNA annealing. Nucleic Acids Res. 2014;42:8556–8564. doi: 10.1093/nar/gku523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- Mattaj IW. RNA recognition: a family matter? Cell. 1993;73:837–840. doi: 10.1016/0092-8674(93)90265-r. [DOI] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M, Rasnik I, Cheng W, Lohman TM, Ha T. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophysical Journal. 2004a;86:2530–2537. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys J. 2004b;86:2530–2537. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, Lohman TM, Ha T. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Antony E, Doganay S, Koh HR, Lohman TM, Myong S. Srs2 prevents Rad51 filament formation by repetitive motion on DNA. Nature communications. 2013;4:2281. doi: 10.1038/ncomms3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. Journal of cell science. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GW, Jr., Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nature methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- Shih JW, Wang WT, Tsai TY, Kuo CY, Li HK, Wu Lee YH. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem. 2012;J441:119–129. doi: 10.1042/BJ20110739. [DOI] [PubMed] [Google Scholar]
- Sigrist CJ, De Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Bougueleret L, Xenarios I. New and continuing developments at PROSITE. Nucleic acids research. 2013;41:D344–D347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam M, Wente SR. Peering through the pore: nuclear pore complex structure, assembly, and function. Dev Cell. 2003;4:775–789. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- Tijerina P, Bhaskaran H, Russell R. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci U S A. 2006;103:16698–16703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike D, Strome S. P granule assembly and function in Caenorhabditis elegans germ cells. J Androl. 2010;31:53–60. doi: 10.2164/jandrol.109.008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- Yarunin A, Harris RE, Ashe MP, Ashe HL. Patterning of the Drosophila oocyte by a sequential translation repression program involving the d4EHP and Belle translational repressors. RNA Biol. 2011;8:904–912. doi: 10.4161/rna.8.5.16325. [DOI] [PubMed] [Google Scholar]
- Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. RNA Controls PolyQ Protein Phase Transitions. Mol Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.