Abstract
Inactivation of the WASF3 gene suppresses invasion and metastasis of breast cancer cells. WASF3 function is regulated through a protein complex that includes the NCKAP1 and CYFIP1 proteins. Here we report that silencing NCKAP1 destabilizes the WASF3 complex, resulting in a suppression of the invasive capacity of breast, prostate and colon cancer cells. In an in vivo model of spontaneous metastasis in immunocompromized mice, loss of NCKAP1 also suppresses metastasis. Activation of the WASF protein complex occurs through interaction with RAC1, and inactivation of NCKAP1 prevents the association of RAC1 with the WASF3 complex. Thus, WASF3 depends on NCKAP1 to promote invasion and metastasis. Here we show that stapled peptides targeting the interface between NCKAP1 and CYFIP1 destabilize the WASF3 complex and suppress RAC1 binding, thereby suppressing invasion. Using a complex-disrupting compound identified in this study termed WANT3, our results offer a mechanistic proof of concept to target this interaction as a novel approach to inhibit breast cancer metastasis.
Keywords: WASF3, NCKAP1, stapled peptide, cancer invasion, metastasis, RAC1
Introduction
Tumor progression is associated with an accumulation of genetic changes that define the transition to more aggressive forms (1). Acquisition of the invasion and metastasis phenotypes has been associated with genes that promote metastasis in combination with the loss of metastasis suppressor genes (2–4). Where it has been studied, these two classes of metastasis-related genes operate independently of the genetic defects that lead to uncontrolled proliferation, survival or cell death. The WASF3 gene has been shown to be a promoter of cell invasion in vitro (5–9) and metastasis in vivo (10–12) in different cancer cell types. Inactivation of WASF3 in breast and prostate cancer cells, for example, leads to reduced cell migration and invasion and, in experimental metastasis models in mice (10–11) and zebrafish (12), metastasis is suppressed. Upregulation of WASF3 was part of the gene signature associated with the highly aggressive “claudin-low” subgroup of breast cancers (13). Immunohistochemical (IHC) data showed that WASF3 is upregulated in high-grade breast tumors (10, 14). Similar analyses show the relationship between WASF3 expression levels and poor prognosis in non-small cell lung cancer (15), gastric cancer (16), hepatocellular carcinoma (17) and prostate cancer (11,18). Thus, analysis of WASF3 expression levels in primary cancers, are consonant with the in vitro studies showing WASF3 promotes cancer cell invasion. In addition, the role of WASF3 in metastasis of osteosarcoma has recently been shown as a result of its down regulation by miRNA-217 (19).
The WASF3 protein contains a verprolin-cofilin-acidic (VCA) motif that binds monomeric actin and the Arp2/3 complex at the C-terminus (20,21). In resting cells, the protein is held in an inactive conformation through association with a complex of proteins that bind to the WASF homology domain (WHD) at the N-terminus (22). These proteins include CYFIP1, NCKAP1, ABI1 and BRK1, collectively known as the WASF regulatory complex (WRC). Following stimulation of cells with cytokines or growth factors, for example, RAC1 binds to CYFIP1, initiating WASF3 phosphoactivation and leading to conformational changes in the WASF3 protein. As a result, the VCA domain is exposed, facilitating reorganization of the actin cytoskeleton through actin polymerization. This activation of WASF3 is dependent on its phosphorylation on tyrosine residues (5,6,9,23) and the protein moves to the leading edge of the cell where the lamellipodia that are responsible for cell movement are being formed. In highly aggressive cancer cells, these lamellipodia also facilitate invasion through an artificial matrix in vitro and metastasis in vivo (24). WASF3 has also been shown to have a signaling function where, through suppression of KISS1, NFκB is released from its suppression by IκBα and moves to the nucleus where it activates invasion-promoting genes such as MMP-9 and ZEB1 (7,25). We recently showed a feed forward loop as a result of the IL6 activation of the JAK2/STAT3 where STAT3 acts as a promoter of WASF3 transcription and JAK2 activates WASF3 (9,23). WASF3 upregulation leads to downregulation of E-cadherin and members of the miR-200 family (25), both of which have been shown to suppress epithelial-to mesenchyme transition (EMT). WASF3 can also be upregulated by HIF1 as a result of hypoxia (26) and facilities communication between endoplasmic reticulum and mitochondria through its interaction with the ATAD3A mitochondrial protein and GRP78 (27).
The central role for WASF3 in invasion and metastasis, combined with its selective overexpression in high-grade tumors, suggests that directly targeting its function may serve as a strategy for controlling metastasis. Here we investigated using stapled peptides (SPs) to disrupt WASF3 function and suppress invasion. Stapled peptides are synthetically designed to stabilize and constrain an α-helical structure through N-methylation and macrocyclic ring formation. Stabilization of the secondary structure introduces an entropically favorable, pre-ordered binding state where key interacting residues are spatially poised for target binding. Further, these locked peptides exhibit drug-like properties including enhanced cell permeability, non-immunogenicity, increased binding affinity and resistance to cellular degradation (28, 29). In this report, we demonstrate that NCKAP1 is required for WASF3 function and its regulation of invasion, and that targeting the interaction between two members of the WRC, CYFIP and NCKAP1, using SPs, leads to suppression of invasion. Thus, targeting this complex may serve as a means to inhibit metastasis and the lead compound identified from this study, WANT3, may offer a therapeutic approach for selectively targeting metastasis.
Materials and methods
Cell culture and standard assays
MDA-MB-231, Hs578T and T47D breast cancer cell lines were obtained directly from American Type Culture Collection (Rockville, MD). MDA-MB-231 and T47D and have been verified using SNP-CGH (7,27) for characteristic cytogenetic changes. The ATCC Cell Authentication Testing service confirmed the identity of Hs578T, PC3 and SW620 (August 2015) using STR DNA fingerprinting analysis (30). Standard cell culture, transient transfections, RT-PCR, western blotting, immunoprecipitation (IP), flow cytometry, Biotin-Avidin pulldown, lentiviral transduction, cell proliferation and Transwell invasion assays were carried out as described previously (7–9,30).
DNA constructs, antibodies and other reagents
Lentiviral pCDH-CMV-MCS-EF1-PURO-HA-WASF3 (pCDH-HA-WASF3) was generated as described previously (9). To construct the HA-NCKAP1 overexpression vector, the full-length human NCKAP1 was amplified from the template NCKAP1 cDNA clone (OriGene, Rockville, MD) and inserted into the pCDH-CMV-MCS-EF1-GFP lentiviral vector (System Biosciences, Mountain View, CA) as described previously (25). To stably knock down NCKAP1, pLKO.1 lentiviral vectors harboring shRNA-targeting NCKAP1 were obtained from Open Biosystems (Huntsville, AL). pcDNA3-EGFP-RAC1-T17N (RAC1DN) was a gift from Dr. Gary Bokoch (Addgene plasmid #12982). The RAC1 NSC24766 inhibitor was obtained from Selleckchem (Houston, TX). For western blot and IP assays, the following primary antibodies were used: NCKAP1, WASF1 (Abcam, Cambridge, MA), WASF2, WASF3 (Cell Signaling Technology, Beverly, MA), HA, GST, RAC1, β-Actin (Sigma, St Louis, MO).
Glutathione S-transferase (GST) fusion protein interaction assays
To determine the interaction between NCKAP1 and WASF3, GST-fusion protein pulldown assays were performed as described previously (5,31). A GST-WASF3 (GST-W3) fusion protein was expressed in BL21 bacteria and purified using MagneGST glutathione particles (Promega, Madison, WI). Once the correct size protein was confirmed, using Coomassie Brilliant Blue staining following SDS-PAGE, the immobilized fusion protein was used immediately. Cell lysates from MDA-MB-231 cells that had been transfected with a pCDH-CMV-MCS-EF1-PURO-NCKAP1 construct were incubated in 500 µl of binding buffer (20 mm Tris-HCl, pH 7.5, 140 mm NaCl, 1% Nonidet P-40 and 0.5% BSA) with the GST fusion protein tethered to the glutathione particles for 4 h at 4°C. Precipitates were resolved by SDS-PAGE and analyzed by western blotting.
Protein complementation assays (PCAs)
To identify the interaction between NCKAP1 and WASF3 in live cells, PCAs were performed as described previously (32). In brief, expression vectors encoding NCKAP1 and WASF3 fused to N- and C-terminal fragments of GFP were constructed respectively. The NCKAP1-venus1 (NCKAP1-v1) and WASF3-venus2 (WASF3-v2) constructs were either transiently transfected individually or co-transfected into MDA-MB-231 cells and 12 hours after transfection, GFP was detected by fluorescence microscopy (Carl Zeiss, Jena, Germany).
In vivo tumor growth and metastasis analysis
All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Augusta University. Six-week-old female NSG (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA) and maintained in accordance with IACUC guidelines. The animal experiments were performed using the NSG mouse model as described previously (27). The mice were euthanized on day 56 post-injection and dissected tumors were individually weighed. The lungs were also removed from these mice and the number of nodules on the surface of the lungs was counted. For histological analyses, the lungs were fixed in 10% neutral buffered formalin, embedded in paraffin blocks, sectioned at 5 µm, and subjected to hematoxylin and eosin (H&E) staining.
Peptide Synthesis
Peptides were synthesized on rink amide MBHA resin using 9-fluorenylmethoxycarbonyl (Fmoc) solid phase synthesis in 1-methyl-2-pyrolidinone (NMP). Fmoc protecting groups were removed using 25% (v/v) piperidine in NMP for 20–30 min. For couplings using standard N-α-Fmoc protected amino acids, 10 equivalents were added (0.25 M final concentration) along with 2-(6-chloro-1H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium hexafluorophosphate (HCTU, 0.23 M final concentration) and 8% (v/v) N,N-diisopropyl ethylamine (DIEA) in NMP. (S)-N-Fmoc-2-(4’-pentenyl) alanine couplings were performed using 4 equivalents. The ring-closing metathesis (RCM) reaction was performed prior to addition of N-terminal labeling using 0.4 equivalents bis(tricyclohexylphosphine) benzylidine ruthenium(IV) dichloride (1st generation Grubbs Catalyst, Sigma) in 1,2-dichloroethane (DCE) for two 1-hour reaction periods with agitation.
Prior to N-terminal labeling, β-alanine was added to the N-terminus of all peptides before the addition of 5(6)-carboxyfluorescein. N-terminal fluorescein labeling was performed using 2 equivalents of 5(6)-carboxyfluorescein (Acros) in DMF along with 0.046 M HCTU and 2% (v/v) DIEA. Resin cleavage was performed using a solution containing 95% trifluoroacetic acid, 2.5% water and 2.5% triisopropylsilane (Sigma) for 4 hours at room temperature. Peptides were precipitated in methyl-tert-butyl ether at 4 °C and lyophilized. Peptides were purified by high-performance liquid chromatography (HPLC) and verified by ESI mass spectrometry (ESI-MS). Fluorescein-labeled peptides were quantified by measuring the absorbance of 5(6)-carboxyfluorescein at 495 nm. Absorbance values were measured using a Synergy 2 microplate reader (Bio-Tek). The masses of the purified peptides are as follows: WANT1 = 1849.5 (expected mass = 1849.0), WANT2 = 2014.5 (expected mass = 2015.2), WANT3 = 1848.6 (expected mass = 1849.1), WANT3 scr = 1848.6 (expected mass = 1849.1).
Results
The NCK/NCKAP1 complex interacts with WASF3
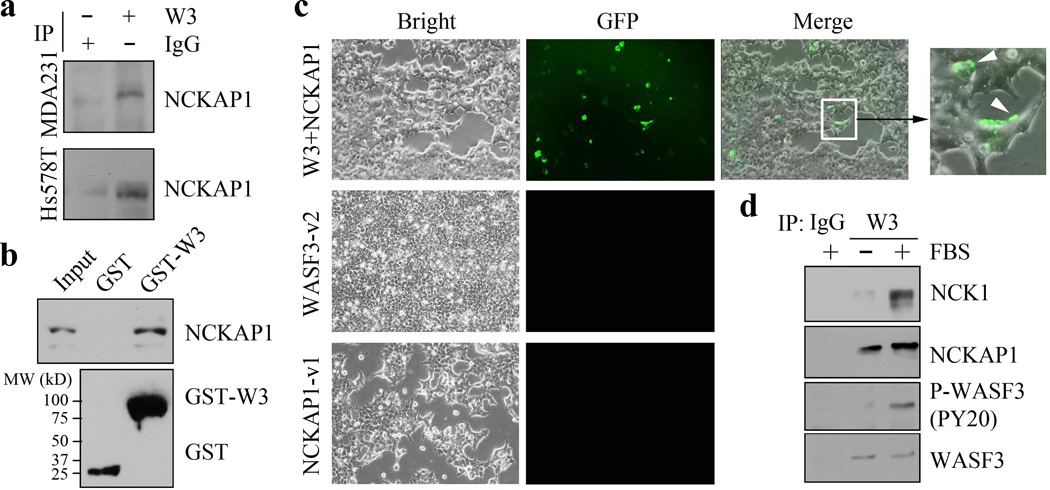
WASF3 is essential for invasion and metastasis in different cancer cell types (10–11). By mass spectrometry, NCK associated protein 1 (NCKAP1) was found to associate with the WASF3 immunocomplex (Supplementary Figure 1). Immunoprecipitation (IP) analysis of the WASF3 immunocomplex from MDA-MB-231 and Hs578T cells confirmed the presence of NCKAP1 (Figure 1a). To further validate the interaction between NCKAP1 and WASF3, a GST-WASF3 fusion protein was immobilized on glutathione-agarose and incubated with lysates from NCKAP1 overexpressing MDA-MB-231 cells. In these pull-down assays, NCKAP1 was recovered using the GST-WASF3 fusion protein, but not GST alone (Figure 1b), indicating that NCKAP1 indeed interacts with WASF3. This interaction was confirmed using protein-fragment complementation (PCA) assays, based on split green fluorescent protein (GFP). Interestingly, the GFP signal was identified at the plasma membrane when NCKAP1-v1 and WASF3-v2 were co-transfected into MDA-MB-231 cells, while no fluorescence was observed when either construct was transfected alone (Figure 1c). IP analysis showed that serum starvation suppressed WASF3 phosphorylation, without affecting the engagement of NCKAP1 in the WASF3 complex (Figure 1d).
Figure 1. NCKAP1 interacts with WASF3.
(a) Following immunoprecipitation (IP) of WASF3 from MDA-MB-231 and Hs578T breast cancer cells, western blot analysis identified NCKAP1 in the IP. The interaction between NCKAP1 and WASF3 was further demonstrated in a GST fusion-protein pulldown assays (b). Lysates from MDA-MB-231 cells were incubated with the GST-tagged WASF3 prepared in BL21 bacterial cells, where the correct size fusion protein was confirmed using anti-GST antibodies (below). The presence of NCKAP1 was then demonstrated in the WASF3-GST (GST-W3) complex using anti-NCKAP1 antibodies. Interaction between NCKAP1 and WASF3 was also demonstrated in vivo following transfection of the NCKAP1-venus1 (NCKAP1-v1) and WASF3-venus2 (WASF3-v2) constructs into MDA-MB-231 cells (c). After 12 hours, GFP was detected by fluorescence microscopy in cells where both constructs were expressed but not in cells where either of the constructs was expressed alone. In the co-transfected cells, a membrane localization of the GFP signal could be seen (arrows).When the WASF3 complex was recovered using immunoprecipitation from MDA-MB-231 cells grown in the presence or absence of FBS (d), NCKAP1 was detected in the complex whether FBS was present or not. The presence of NCK1, however, was only seen in cells treated with FBS, where WASF3 (P-WASF3) was activated.
The NCK1 protein contains multiple SH2/SH3 domains (33) and is the target of several cell surface tyrosine kinase receptors. It was shown that ligand binding activates NCK1 by phosphorylation, and that this event triggers downstream effectors to activate cell motility (34–36). As we have shown, growth factors such as PDGF and cytokines such as IL-6 activate WASF3, thereby leading to increased migration and invasion and WASF3 is recruited to the cell membrane to facilitate actin reorganization at leading edges of cells (6,9,10,23). However, NCK1 was not found in the WASF3 immunocomplex in starved cells as shown by IP analysis (Figure 1d).
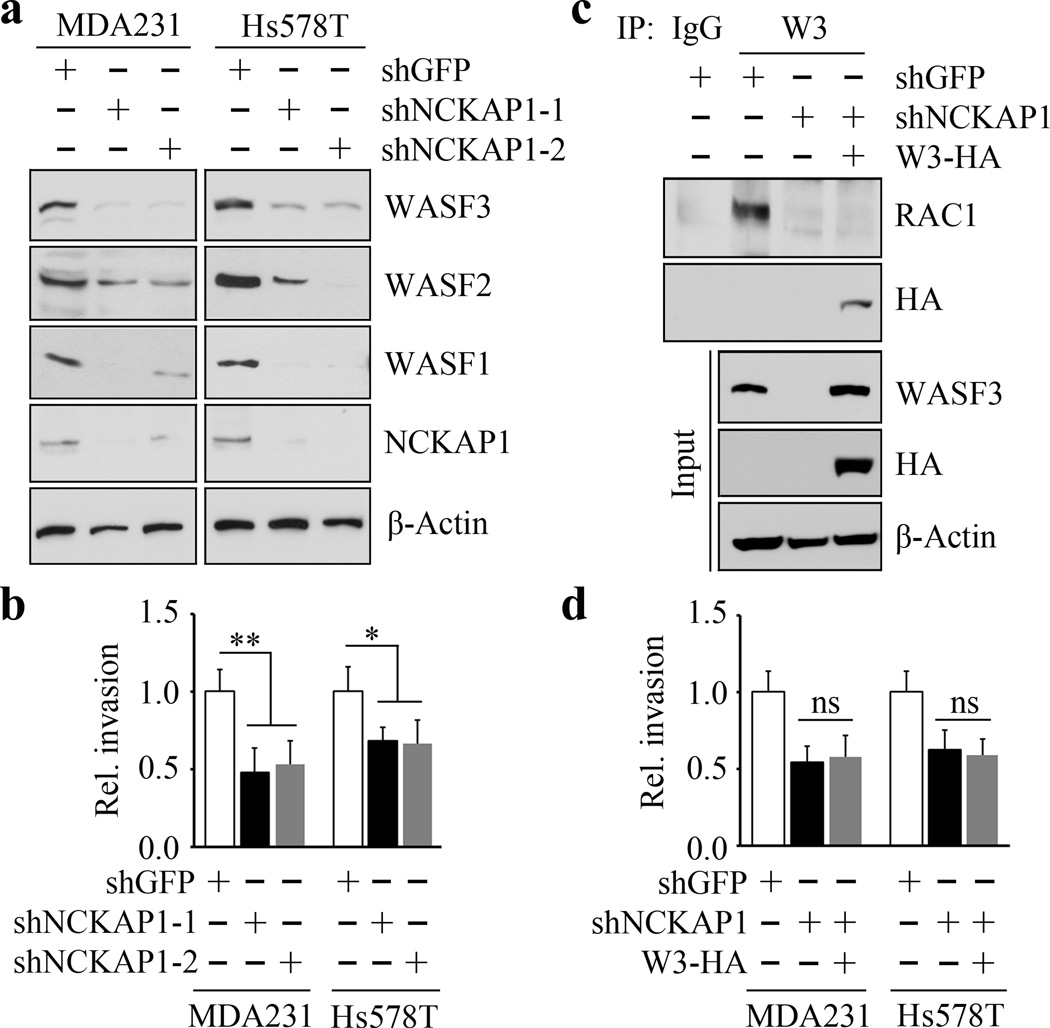
NCKAP1 is required for WASF3 protein stability and invasion potential
To investigate this relationship between NCKAP1 and WASF3, we used two different shRNA constructs to suppress NCKAP1 expression in MDA-MB-231 and Hs578T cells which led to a reduction in WASF3 protein levels (Figure 2a), while not affecting its transcript levels (Supplementary Figure 2), supporting the idea that NCKAP1 protects WASF3 from degradation. Since NCKAP1 is also found in complex with other members of the WASF family (36–38), we analyzed the effect of its knockdown on their protein levels, where levels of both WASF1 and WASF2 were reduced in MDA-MB-231 and Hs578T cells (Figure 2a). Thus, NCKAP1 is also required for the stability of these proteins. Invasion potential, however, is not affected by knockdown of WASF1 and WASF2 in MDA-MB-231 cells (30).
Figure 2. Molecular and cell invasion analysis following NCKAP1 knockdown.
Breast cancer MDA-MB-231 and Hs578T cells in which NCKAP1 had been stably knocked down (shNCKAP1-1 and shNCKAP1-2) show significantly reduced levels of WASF3 (a) compared with cells carrying a control shRNA (shGFP). Similarly, reduced levels of the WASF1 and WASF2 proteins were also seen in the NCKAP1 knockdown cells. When NCKAP1 knockdown cells were analyzed using Transwell invasion assays (b), their invasion potential was suppressed. Immunoprecipitation of HA-tagged WASF3 from MDA-MB-231 cells in which NCKAP1 had been knocked down shows the absence of RAC1 in the WASF3 immunocomplex (c), compared with parental cells expressing the control shRNA (shGFP). When WASF3 was overexpressed in NCKAP1 knockdown MDA-MB-231 and Hs578T cells, there was no recovery of invasion potential (d). *p<0.05, **p<0.01 and ns indicates no statistical significance.
Knockdown of NCKAP1 in both MDA-MB-231 and Hs578T cells leads to a significant reduction in invasion (Figure 2b), but does not impact cell proliferation (Supplementary Figure 3). The Rho-GTPase, RAC1, relays signals to WASF proteins, leading to activation of Arp2/3-mediated actin polymerization (38,39). To determine whether depletion of NCKAP1 suppresses the interaction of RAC1 with the WASF3 complex, we transfected an HA-tagged WASF3 construct into NCKAP1-knockdown MDA-MB-231 cells. IP analysis in these cells using WASF3 antibodies showed that RAC1 was not co-immumoprecipitated with the exogenous WASF3 protein (Figure 2c), in the absence of NCKAP1. Moreover, suppression of invasion in NCKAP1 knockdown cells was not reversed by forced expression of WASF3 (Figure 2d). Thus, NCKAP1 is essential for the RAC1 interaction with the WASF3 complex in order to promote cell invasion.
Knockdown of NCKAP1 suppresses metastasis in breast cancer cells in vivo
To relate the in vitro observation that links NCKAP1 expression with invasion to clinical parameters, we evaluated the correlation between NCKAP1 expression and survival of patients with breast cancer. Using an online gene profiling database we related NCKAP1 expression levels with relapse-free survival data from 3,554 cancer patients based on relative NCKAP1 expression levels as described (40). Univariate survival analysis (Kaplan-Meier method and log-rank test) revealed that high NCKAP1 expression significantly correlates with poor, relapse-free, survival (Figure 3a), which is likely related to its involvement in metastasis.
Figure 3. Metastasis in vivo is suppressed following NCKAP1 knockdown.
Kaplan-Meier plot analyses with the log-rank test, shows that higher NCKAP1 expression was associated with lower relapse-free survival rates compared with low NCKAP1 expression (a). When MDA-MB-231 cells were implanted subcutaneously into six-week-old female NSG mice (b) primary tumor growth was not affected by knockdown of NCKAP1 (shNCKAP1-1 and shNCKAP1-2), compared to control knockdown (shGFP) cells. When the lungs were removed from these mice, however, the number of nodules on the surface of the lungs was significantly reduced in the NCKAP1 knockdown cells (c). Histological analysis of these lungs demonstrated that, while animals receiving the control cells showed extensive tumor infiltration throughout the lung (d) the NCKAP1 knockdown cells showed relatively few, small tumor foci. Images on the right derived from the boxed areas on the left. **p<0.01.
The suppression of invasion seen in vitro following knockdown of WASF3 correlates with suppression of metastasis in vivo in both zebrafish (12) and mouse models (10,11). We have previously used the NSG mouse model for in vivo metastasis studies where, unlike SCID mouse models, primary tumor formation and metastasis occurs coincidentally within 2 months (27). MDA-MB-231 cells with NCKAP1 knockdown were injected into the mammary fat pads of NSG mice and tumor development and metastasis was monitored over 8 weeks. There was no significant difference in primary tumor size (Figure 3b) between the mice injected with either the NCKAP1 knockdown or knockdown control cells. When pulmonary metastasis was examined at the conclusion of the experiment, mice injected with the control cells showed multiple surface tumors. In contrast, those animals injected with NCKAP1 knockdown cells showed a significantly reduced number of metastases (Figure 3c). Histological analysis further demonstrated that, while the mice injected with the control cells show multiple large tumors throughout the lungs, there are relatively few, small metastases in the mice injected with the NCKAP1 knockdown cells (Figure 3d). Thus, loss of NCKAP1 expression inhibits in vivo metastasis, confirming that disrupting its interaction with the WASF3 complex may be a means of suppressing this aggressive stage of cancer.
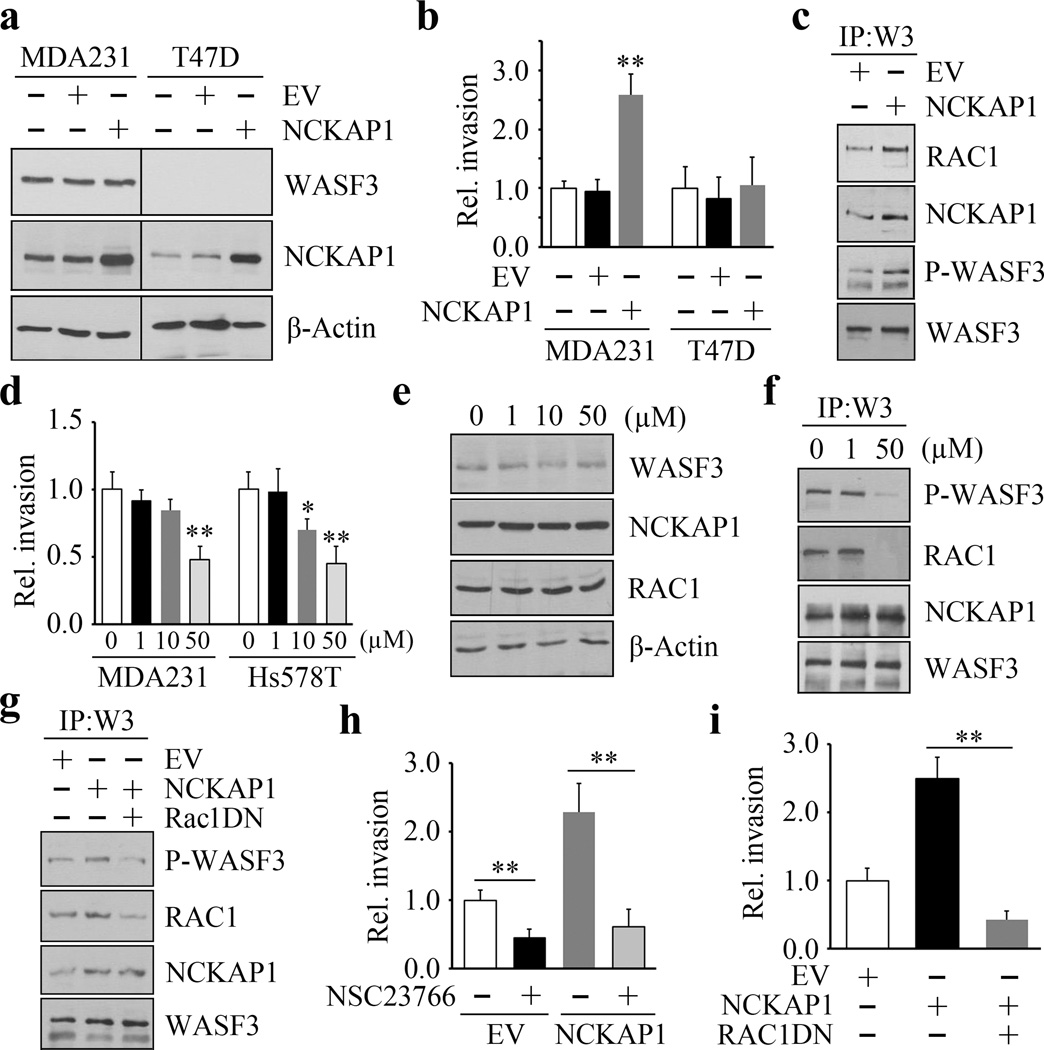
RAC1 binding to the WASF3 complex is required for the NCKAP1-WASF3 invasive signaling axis in breast cancer cells
Knockdown of NCKAP1 does not affect WASF3 transcription levels, but leads to a destabilization of the WASF3 protein (see above). To further investigate whether NCKAP1 is functionally associated with WASF3, we overexpressed NCKAP1 in invasive MDA-MB-231 and non-invasive T47D cells, and analyzed cell invasion potential. Overexpression of NCKAP1 did not increase WASF3 protein levels in either cell line (Figure 4a) but significantly increased the invasion potential in MDA-MB-231 cells while making no difference in T47D cells which lack WASF3 expression (Figure 4b). Thus, WASF3 is essential for NCKAP1-mediated invasion in breast cancer cells. IP analysis of WASF3 from MDA-MB-231 cells demonstrated increased levels of NCKAP1 and RAC1 in the WASF3 immunocomplex, concomitant with increased WASF3 activation levels (Figure 4c). These observations suggest that overexpressing NCKAP1 leads to increased engagement of RAC1 with the WASF3 complex. Treating MDA-MB-231 and Hs578T cells with NSC23766, an inhibitor of RAC1 (41), led to a dose-dependent reduction in invasion in both cell lines (Figure 4d), without affecting cell proliferation (Supplementary Figure 4) or affecting the protein levels of either WASF3, NCKAP1 or RAC1 (Figure 4e). IP analysis shows that NSC23766 does not disrupt engagement of NCKAP1 with the WASF3 complex, although high dose NSC23766 (50 µM) significantly inhibited both WASF3 phosphorylation and RAC1 binding to the WASF3 complex (Figure 4f). This is likely due to the reduced levels of RAC1 expression. To further determine the role of RAC1 on the NCKAP1-WASF3 complex, we transfected a T17N dominant-negative RAC1 construct (RAC1DN) into MDA-MB-231 cells overexpressing NCKAP1. Similar to NSC23677 treatment, expression of RAC1DN disrupted the WASF3 interaction with RAC1 and subsequently impaired WASF3 phosphoactivation (Figure 4g). Moreover, both NSC23677 treatment (Figure 4h) and overexpression of RAC1DN (Figure 4i) led to a significant reduction of invasion potential in cells expressing NCKAP1, suggesting that inhibition of RAC1 activation attenuates NCKAP1-mediated cell invasion. Taken together, these data demonstrate that RAC1 binding to the WASF3 complex is critical for promoting invasion in breast cancer cells.
Figure 4. RAC1 binding to the WASF3 complex is required for NCKAP1-mediated invasion of breast cancer cells.
NCKAP1 overexpression in MDA-MB-231 cells does not affect WASF3 levels and, in T47D cells which do not express WASF3, overexpression of NCKAP1 does not increase WASF3 levels (a). Transwell assays demonstrate that overexpressing NCKAP1 in MDA-MB-231 cells significantly increases invasion potential, although T47D cells are unaffected (b). IP of WASF3 (W3) from MDA-MB-231 cells shows increased RAC1 levels in the WASF3 complex and increased WASF3 phosphorylation when NCKAP1 is overexpressed (c). Treatment of MDA-MB-231 and Hs578T breast cancer cells with the NSC23766 RAC1 inhibitor, leads to a dose-dependent reduction in invasion potential (d) but does not affect protein levels of either WASF3, NCKAP1 or RAC1 (e). IP of WASF3 (W3) from MDA-MB-231 cells treated with NSC23766 shows that, at high (50 uM) concentration, activation of WASF3 is suppressed and RAC1 engagement in the complex is virtually eliminated (f). When a dominant-negative RAC1 (RAC1DN) is introduced into MDA-MB-231 cells overexpressing NCKAP1, levels of phosphoactivated WASF3 are significantly reduced in concert with reduced RAC1 levels (g). In Transwell assays, NSC23766 leads to a significant reduction in invasion in both MDA-MB-231 parental cells containing the empty vector (EV) and cells overexpressing NCKAP1 (h). Similarly, the RAC1 dominant-negative construct (RAC1DN) significantly suppresses invasion in MDA-MB-231 cells overexpressing NCKAP1 (i). *p<0.05 and **p<0.01.
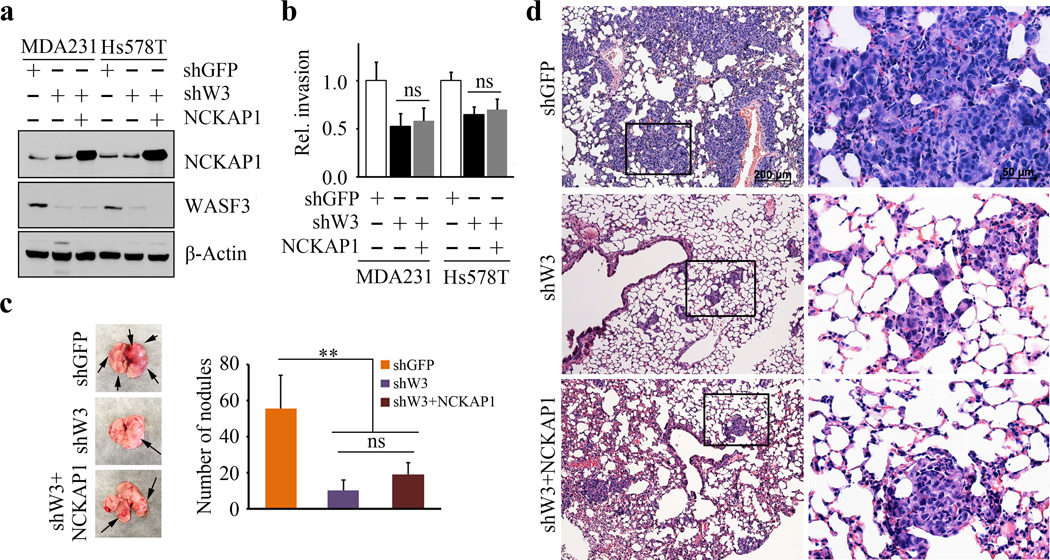
NCKAP1 engagement with the WASF3 complex is required for metastasis of breast cancer cells
Knockdown of WASF3 in MDA-MB-231 and Hs578T cells (Figure 5a) led to a significant reduction in invasion potential (Figure 5b), which is consistent with our previous reports (5–12,25–27). However, when NCKAP1 is overexpressed in WASF3 knockdown cells, there was no significant increase in invasion potential compared with the parental cells (Figures 5a and 5b). When in vivo metastasis assays were performed using NSG mice, increased numbers of tumor nodules were seen on the surface of the lungs in the knockdown control cells compared with either the WASF3 knockdown cells or the WASF3 knockdown cells overexpressing NCKAP1 (Figure 5c). In addition, there is no significant change in the number of tumor nodules on the surface of the lungs when NCKAP1 was overexpressed in WASF3 knockdown cells (Figure 5c). Histological examination of the lungs of these mice shows large tumor foci in the knockdown control cells, compared with the small tumor foci seen in WASF3 knockdown cells (Figure 5d). Thus, there was no increased metastasis potential to the lung in the WASF3 knockdown MDA-MB-231 cells expressing NCKAP1, compared with the WASF3 knockdown cells (Figure 5d). This is consistent with the in vitro invasion assays. Collectively, these data demonstrate that the NCKAP1-WASF3 complex is essential for cell invasion and metastasis in breast cancer cells.
Figure 5. Invasion and metastasis analysis after NCKAP1 overexpression in WASF3 knockdown cells.
When NCKAP1 was overexpressed in WASF3 knockdown MDA-MB-231 and Hs578T cells (a), cell invasion was not significantly affected (b). Following subcutaneous implantation of MDA-MB-231 cells overexpressing NCKAP1 into NSG mice, the number of nodules on the surface of the lungs after 8 weeks in these animals was not significantly different compared with the WASF3 (shW3) knockdown cells (c). Histological analyses showed the same distribution of tumors in the lungs of these mice carrying the NCKAP1 overexpressing cells as seen for the WASF3 knockdown cells (d). **p<0.01 and ns indicates no statistical significance.
Suppression of invasion by targeting the CYFIP1-NCKAP1 interaction using SPs
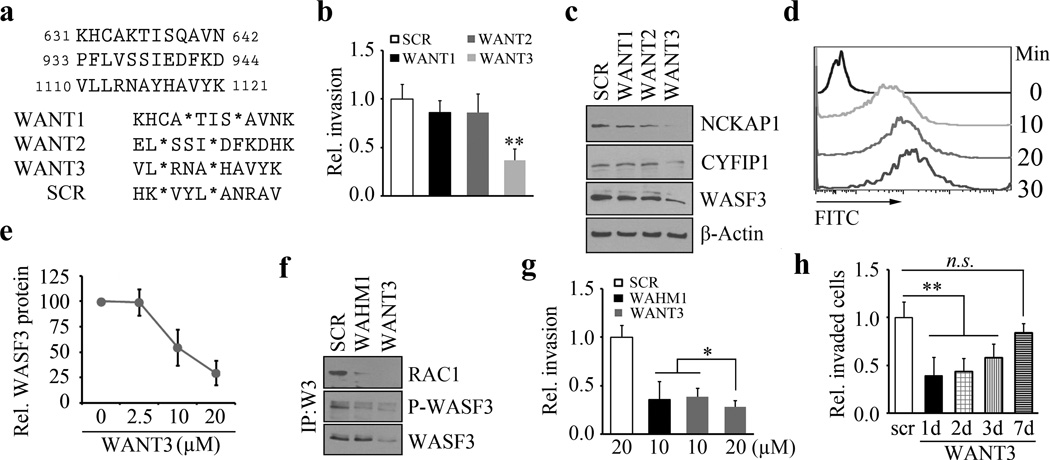
Since the NCKAP1-WASF3 interaction is required for invasion and metastasis, we explored whether targeting this complex could suppress invasion. Knockdown of NCKAP1 in highly invasive cancer cells leads to suppression of invasion (Figure 2b), which is accompanied by decreased WASF3 levels. Reduced WASF3 levels are also seen after knockdown of CYFIP1 (30), which associates with WASF3 as a dimer with NCKAP1 (42–43). Loss of either member of this trimeric complex leads to loss of WASF3 protein levels (Supplementary Figure 5) and suppresses invasion. These observations suggest that disrupting the engagement of NCKAP1 with the WASF3 complex could lead to disruption of the WRC and loss of invasion. Currently, no inhibitors target WASF3 function directly, but targeting the WRC may provide a means to suppress invasion. We have previously reported, for example, that targeting a direct protein-protein interaction between CYFIP1 and WASF3 using stapled peptides (SPs) can effectively suppress invasion in WASF3 overexpressing cancer cells (30). It is possible, therefore, that disrupting the WRC by targeting NCKAP1 might have a similar consequence. Analysis of the crystal structure of the WRC complex, however, showed no direct contact between NCKAP1 and WASF3 (42,43), even though loss of NCKAP1 leads to reduced WASF3 stability (Figure 2a). SPs can be designed to mimic α-helical surfaces between proteins to serve as disruptors of protein-protein interactions. In order to disrupt the WRC complex, several α-helices were identified at the interface between CYFIP1 and NCKAP1 (Supplementary Figure 6) that could potentially serve as disruptors of the WRC and hence suppress invasion. We therefore targeted three regions derived from NCKAP1 that had α-helical interactions with CYFIP1. These PPIs encompassed amino acids 631–642, 933–944 and 1110–1121. Stapled peptides, designated WASF3-NCKAP1 Targets (WANT), were designed against the NCKAP1 surface (Figure 6a) at these sites. Highly invasive MDA-MB-231 and HS578T breast cancer cells were treated with each of the three WANT peptides independently at a final concentration of 10 µM. When MDA-MB-231 cells were then challenged to invade in Transwell chamber assays, no significant effect on invasion potential was observed in the presence of WANT1 and WANT2 (Figure 6b), albeit with only a single peptide designed against these regions. The lack of activity may be due to the fact that these particular compounds were not able to sufficiently disrupt the targeted protein-protein interface. In contrast, treatment with WANT3 resulted in a highly significant suppression of invasion (Figure 6b) without affecting cell proliferation (Supplementary Figure 7a). To investigate the mechanism of WANT3 action, we analyzed WASF3 stability using western blotting. Compared with treatment with a scrambled peptide control, WANT3 resulted in a significant reduction of WASF3 levels (Figure 6c) and this coincided with loss of NCKAP1. Flow cytometry analysis demonstrated that, in the two different breast cancer cell lines, uptake of these peptides was rapid, achieving maximal levels after only 10–20 minutes (Figure 6d). Significantly, in vitro cell toxicity at 10 µM was minimal (Supplementary Figure 7) and the ability of WANT3 to suppress WASF3 levels was dose dependent (Supplementary Figure 8 and Figure 6e), achieving a maximal effect at 20 µM. Thus, the WANT3 stapled peptide serves as a successful tool to suppress invasion.
Figure 6. Targeting the NCKAP1-WASF3 complex using stapled peptides leads to loss of invasion in breast cancer cells.
Sequence of amino acid regions 631–642, 933–944 and 1110–1121 in NCKAP1 (a) used to design stapled peptides (above). The three stapled peptides WANT1, 2 and 3 were designed to target interaction surfaces between CYFIP1 and NCKAP1 where (*) represent the position of the non-natural amino acids (below). The scrambled peptide was used as a negative control. Transwell invasion assays show that only WANT3 significantly suppresses MDA-MB-231 cell invasion (b) and suppresses both WASF3 and NCKAP1 protein levels (c). A time course of WANT3-FITC uptake using flow cytometry over the first 30 minute of exposure (d) shows progressive fluorescein labeling in breast cancer MDA-MB-231 cells. WANT3 suppresses WASF3 protein levels in a dose-dependent manner (e). WANT3 suppresses phosphoactivation of WASF3 more significantly than the WASF3-CYFIP1 peptide mimic WAHM1 (f). Using a high dose of WANT3 (20 µM) leads to a more remarkable reduction in MDA-MD-231 cell invasion compared with low dose treatment (g). WANT3 peptides were preincubated in serum-containing medium at 37°C for 1–7 days. When this medium was then used in invasion assays, significant suppression of invasion in MDA-MB-231 cells was still observed for up to three days (h).
Since targeting the WASF3-CYFIP1 interface also led to disruption of the WRC and suppression of invasion (30), we investigated the relative ability of targeting the WASF3-CYFIP1 (using the WHAM1 peptide) or CYFIP1-NCKAP1 (using the WANT3 peptide) interaction to disrupt the WRC. Following treatment of MDA-MB-231cells with WHAM1 or WANT3, it was clear that targeting the CYFIP1-NCKAP1 interface was more efficient in decreasing WASF3 levels compared to targeting the WASF3-CYFIP1 interface (Figure 6f). WASF3 is activated through its interaction with RAC1 binding to CYFIP1 (44) and so, as expected, since targeting the CYFIP1-NCKAP1 interaction leads to destabilization of the WRC, the engagement of RAC1 in the absence of NCKAP1 is suppressed. The engagement of RAC1 with the WASF3 complex was more significant following treatment with WANT3 than WHAM1 (Figure 6f). Despite this variance in the level of destabilization of the WRC, Transwell invasion assays demonstrated that both WAHM1 and WANT3 peptides produced a comparable ability to suppress invasion (Figure 6g). Further, WANT3 could successfully suppress invasion in a concentration-dependent manner (Figure 6g).
WASF3 also promotes metastasis in prostate and colon cancer (11, 27). To extend these studies to other cancer cell types, we treated highly invasive PC3 prostate and SW620 colon cancer cells with WANT3 where again invasion was suppressed (Supplementary Figure 9). Thus targeting the WASF3 complex may have wider efficacy in suppressing metastasis in different cancer subtypes.
The observation that targeting the WRC with SPs leads to suppression of invasion, provides pre-clinical evidence that this target may serve as a means to suppress invasion and potentially metastasis. To evaluate the stability of these SPs, we incubated WANT3 in serum containing medium for varying lengths of time (1–7 days) at 37°C and then added this medium to MDA-MB-231 cells to evaluate the ability of the preincubated peptide to suppress invasion. As shown in Figure 6h, WANT3 that had been preincubated for up to three days was still able to significantly suppress invasion. Even after 7 days, although not significant, there was a residual effect on suppression of invasion, demonstrating that this peptide has an appreciable half-life in enriched medium. Going forward, WANT3 may serve as a viable candidate for targeted inhibition of the WRC by disrupting the trimeric protein complex and thereby suppress the ability of cancer cells to invade and metastasize.
Discussion
Reorganization of the actin cytoskeleton to facilitate cell invasion and metastasis is a complex regulatory process involving many interacting pathways (44–46). One of the key initiating events is the activation of RAC1, which is known to signal actin cytoskeleton reorganization following stimulation with growth factor receptors (34, 47). This facilitates recruitment of WASF family members to the membrane to promote invasion. Here, we demonstrate that RAC1 cannot be recruited to the WASF3 complex in the absence of NCKAP1. High level expression of NCKAP1 is associated with poorer survival in breast cancer patients. This may be due to the increased stability of WASF3 since, when NCKAP1 levels are increased in MDA-MB-231 cells, invasion potential also increases and this is associated with both increased RAC1 levels in the WRC complex and increased WASF3 activation. The cascade leading to increased invasion, however, is dependent on WASF3 expression, since non-invasive cells do not respond to increased NCKAP1 expression.
WASF3 is part of a three-member family which share similar structural motifs that define their function in actin cytoskeletal reorganization (20,48). Knockdown of WASF3 leads to suppression of invasion and metastasis in breast and prostate cancer cells. WASF1 and WASF2 are redundant in this function since they clearly cannot compensate for the suppression of invasion even with sustained expression. We have also shown recently that knockdown of WASF1 and WASF2 in these same breast cancer cells does not lead to suppression of invasion or metastasis (30). While WASF3 regulates lamellipodia formation (5), which is essential for the development of the invasion and metastasis phenotypes, WASF1 appears to regulate dorsal ruffle formation (49) while WASF2 regulates filopodia production (50). It is possible, therefore, that while controlling similar actin dynamics, the specificity of WASF3 in influencing metastasis depends on the mechanisms of its regulation and possibly the proteins it binds to. WASF3, for example, is under the regulation of the STAT3 transcription factor (9, 23) and is activated following cytokine and growth factor stimulation, but WASF1 and WASF2 do not have consensus STAT binding sites in their promoters and do not respond to IL-6 stimulation (9). Knockdown of NCKAP1, however, also leads to destabilization of WASF1 and WASF2 protein complexes but no specific resultant cell phenotypes were evident.
Part of the mechanism proposed for WASF protein function is through recruitment to membrane locations following growth factor stimulation resulting from actin cytoskeleton reorganization through interactions with NCK1 (51, 52). Consistent with this idea, NCK1 is not present in the WASF3 immunocomplex in the absence of serum and WASF3 remains inactive, although addition of serum growth factors activates WASF3 and a sub pool of protein interacts with NCK1. In contrast, NCKAP1 is associated with WASF3 in both its inactive and active forms, consistent with the idea that its presence is required for protein stability. It is tempting to speculate that NCK1 is an important protein for the recruitment of WASF3 to tyrosine kinase receptor complexes through an interaction with NCKAP1 upon extracellular stimulation. Thus, NCK1-NCKAP1-RAC1 signaling may be critical for WASF3 activation leading to the significant consequence of cell invasion.
The central role of WASF3 in regulating invasion and metastasis (5–12,23,25–27,48), together with its overexpression in high-grade and metastatic tumors (13,14), provides an ideal target to suppress metastasis. Here we show that SPs targeting the large interaction interface between two key proteins that maintain the integrity of WASF3, leads to loss of WASF3 and subsequently suppression of invasion, suggesting this complex may serve as a viable target to suppress metastasis. While only one of the three designed peptides demonstrated efficacy, this may simply be due to their ability or inability to effectively disrupt the protein-protein interface. The interface is composed of multiple helical contact points and each will have differing energetic contributions to the stabilized interface. It is possible that the active compound, WANT3, may additionally have unanticipated off-target effects that may be contributing to the observed phenotypes. This will be explored to fully elucidate the biological mechanism of WANT3. Importantly, since there are currently no inhibitors that directly target WASF3, and the interface lacks pockets required for small molecule targeting, the development of the WANT3 peptide described here, and the WHAM peptides targeting the WASF3-CYFIP1 interaction (30), provide a potential approach to suppress WASF3 function and suppress metastasis. The emerging field of SPs as therapeutic agents is gaining traction through clinical trials currently underway targeting the MDM2/MDMX-p53 protein interaction (53,54) for cancer patients with tumors expressing wild type p53. Although there are many conventional, unmodified peptides currently in clinical trials, because of their limited ability to penetrate the cell, most still target extracellular proteins. SPs on the other hand, are constrained in a highly stable helical conformation, and address many of the limitations of standard peptides because (1) their active transport into cells (2) their pharmaceutical stability (3) low immunogenicity and (4) their binding affinity for the target (28). Indeed, we show here that the NCKAP1 peptide mimic, WANT3, retains its ability to suppress invasion after incubation in serum for up to three days. While the NCKAP1 mimic appears to be more effective in destabilizing the WASF3 complex, CYFIP1 mimics that targeting the WASF3-CYFIP1 interaction (30), suppress WASF3 phosphoactivation. Nonetheless, since we have shown that phosphoactivation is required for the ability of WASF3 to regulate invasion, both peptide mimics suppress invasion equally effectively. This approach validates the WASF3 complex as a viable target for suppression of metastasis and identifies WANT3, as a potential therapeutic lead compound to selectively inhibit invasion and metastasis.
Supplementary Material
Acknowledgments
Financial support: This work was supported in part by grants from the National Institutes of Health, CA120510 (JKC) and CA188439 (EJK); and Department of Defense, W81XWH-14-1-0412 (YT).
Footnotes
Disclosure of Potential Conflicts of Interest The authors declare no competing financial interests.
REFERENCES
- 1.Hanahan D, Weinberg RA. Hallmarks of Cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS. Angiogenesis inhibitors: motivators of metastasis? Nat Med. 2003;9:822–823. doi: 10.1038/nm0703-822. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen DX, Massagué J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8:341–352. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- 4.Hurst DR1, Welch DR. Metastasis suppressor genes at the interface between the environment and tumor cell growth. Int. Rev. Cell Mol Biol. 2011;286:107–180. doi: 10.1016/B978-0-12-385859-7.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sossey-Alaoui K, Li X, Ranalli TA, Cowell JK. WAVE3-mediated cell migration and lamellipodia formation are regulated downstream of phosphatidylinositol 3-kinase. J Biol Chem. 2005;280:21748–21755. doi: 10.1074/jbc.M500503200. [DOI] [PubMed] [Google Scholar]
- 6.Sossey-Alaoui K, Li X, Cowell JK. c-Abl-mediated phosphorylation of WAVE3 is required for lamellipodia formation and cell migration. J Biol Chem. 2007;282:26257–26565. doi: 10.1074/jbc.M701484200. [DOI] [PubMed] [Google Scholar]
- 7.Teng Y, Liu M, Cowell JK. Functional interrelationship between the WASF3 and KISS1 metastasis-associated genes in breast cancer cells. Int J Cancer. 2011;129:2825–2835. doi: 10.1002/ijc.25964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teng Y, Ngoka L, Mei Y, Lesoon L, Cowell JK. HSP90 and HSP70 are essential for stabilization and activation of the WASF3 metastasis promoting protein. J Biol Chem. 2012;287:10051–10059. doi: 10.1074/jbc.M111.335000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teng Y, Ghoshal P, Ngoka L, Mei Y, Cowell JK. Critical role of the WASF3 gene in JAK2/STAT3 regulation of cancer cell invasion. Carcinogenesis. 2013;34:1994–1999. doi: 10.1093/carcin/bgt167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sossey-Alaoui K, Safina A, Li X, Vaughan MM, Hicks DG, Bakin AV, Cowell JK. Down-regulation of WAVE3, a metastasis promoter gene, inhibits invasion and metastasis of breast cancer cells. Am J Pathol. 2007;170:2112–2121. doi: 10.2353/ajpath.2007.060975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teng Y, Ren MQ, Cheney R, Sharma S, Cowell JK. Inactivation of the WASF3 gene in prostate cancer cells leads to suppression of tumorigenicity and metastases. Br J Cancer. 2010;103:1066–1075. doi: 10.1038/sj.bjc.6605850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teng Y, Xie X, Walker S, White DT, Mumm JS, Cowell JK. Evaluating human cancer cell metastasis in zebrafish. BMC Cancer. 2013;13:453. doi: 10.1186/1471-2407-13-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulkarni S, Augoff K, Rivera L, McCue B, Khoury T, Groman A, et al. Increased expression levels of WAVE3 are associated with the progression and metastasis of triple negative breast cancer. PLoS One. 2012;7:e42895. doi: 10.1371/journal.pone.0042895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, Wang GC, Chen XJ, Xue ZR. Expression of WASF3 in patients with non-small cell lung cancer: Correlation with clinicopathological features and prognosis. Oncol Lett. 2014;8:1169–1174. doi: 10.3892/ol.2014.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yue Z, Feng W, Xiangke L, Liuxing W, Qingxia F, Jianbo G. WAVE3 promotes epithelial-mesenchymal transition of gastric cancer through upregulation of Snail. Cancer Gene Ther. 2014;21:499–506. doi: 10.1038/cgt.2014.52. [DOI] [PubMed] [Google Scholar]
- 17.Ji Y, Li B, Zhu Z, Guo X, He W, Fan Z, et al. Overexpression of WAVE3 promotes tumor invasiveness and confers an unfavorable prognosis in human hepatocellular carcinoma. Biomed Pharmacother. 2015;69:409–415. doi: 10.1016/j.biopha.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Fernando HS, Sanders AJ, Kynaston HG, Jiang WG. WAVE3 is associated with invasiveness in prostate cancer cells. Urol Oncol. 2010;28:320–327. doi: 10.1016/j.urolonc.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 19.Shen L, Wang P, Yang J, Li X. MicroRNA-217 regulates WASF3 expression and suppresses tumor growth and metastasis in osteosarcoma. PLoS One. 2014;9:e109138. doi: 10.1371/journal.pone.0109138. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Sossey-Alaoui K, Head K, Nowak N, Cowell JK. Characterization of the genomic organization and expression profile of the human and mouse WAVE gene family. Mamm Genome. 2003;14:314–322. doi: 10.1007/s00335-002-2247-7. [DOI] [PubMed] [Google Scholar]
- 21.Takenawa T, Miki H. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J Cell Sci. 2001;114:1801–1809. doi: 10.1242/jcs.114.10.1801. [DOI] [PubMed] [Google Scholar]
- 22.Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- 23.Teng Y, Ross JL, Cowell JK. The involvement of JAK-STAT3 in cell motility, invasion, and metastasis. JAKSTAT. 2014;3:e28086. doi: 10.4161/jkst.28086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogden A, Rida PC, Aneja R. Heading off with the herd: how cancer cells might maneuver supernumerary centrosomes for directional migration. Cancer Metastasis Rev. 2013;32:269–287. doi: 10.1007/s10555-012-9413-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teng Y, Mei Y, Hawthorn L, Cowell JK. WASF3 regulates miR-200 inactivation by ZEB1 through suppression of KISS1 leading to increased invasiveness in breast cancer cells. Oncogene. 2014;33:203–211. doi: 10.1038/onc.2012.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghoshal P, Teng Y, Lesoon LA, Cowell JK. HIF1A induces expression of the WASF3 metastasis-associated gene under hypoxic conditions. Int J Cancer. 2012;131:E905–E915. doi: 10.1002/ijc.27631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng Y, Ren X, Li H, Shull A, Kim J, Cowell JK. Mitochondrial ATAD3A combines with GRP78 to regulate the WASF3 metastasis-promoting protein. Oncogene. 2016;35:333–343. doi: 10.1038/onc.2015.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verdine GL, Hilinski GJ. Stapled peptides for intracellular drug targets. Methods Enzymol. 2012;503:3–33. doi: 10.1016/B978-0-12-396962-0.00001-X. [DOI] [PubMed] [Google Scholar]
- 29.Walensky LD, Bird GH. Hydrocarbon-stapled peptides: principles, practice, and progress. J Med Chem. 2014;57:6275–6288. doi: 10.1021/jm4011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teng Y, Bahassan A, Dong D, Hanold LE, Ren X, Kennedy EJ, et al. Targeting the WASF3-CYFIP1 complex using stapled peptides suppresses cancer cell invasion. Cancer Res. 2016;76:965–973. doi: 10.1158/0008-5472.CAN-15-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong C, Nichols CD, Guo J, Huang W, Lambert NA, Wu G. A triple arg motif mediates α(2B)-adrenergic receptor interaction with Sec24C/D and export. Traffic. 2012;13:857–868. doi: 10.1111/j.1600-0854.2012.01351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Wang G, Dupré DJ, Feng Y, Robitaille M, Lazartigues E, et al. Rab1 GTPase and dimerization in the cell surface expression of angiotensin II type 2 receptor. J Pharmacol Exp Ther. 2009;330:109–117. doi: 10.1124/jpet.109.153460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W, Hu P, Skolnik EY, Ullrich A, Schlessinger J. The SH2 and SH3 domain-containing Nck protein is oncogenic and a common target for phosphorylation by different surface receptors. Mol Cell Biol. 1992;12:5824–5833. doi: 10.1128/mcb.12.12.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rivera GM, Antoku S, Gelkop S, Shin NY, Hanks SK, Pawson T, et al. Requirement of Nck adaptors for actin dynamics and cell migration stimulated by platelet-derived growth factor B. Proc Natl Acad Sci U S A. 2006;103:9536–9541. doi: 10.1073/pnas.0603786103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lebensohn AM, Kirschner MW. Activation of the WAVE complex by coincident signals controls actin assembly. Mol Cell. 2009;36:512–524. doi: 10.1016/j.molcel.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang G, Chen X, Qiu F, Zhu F, Lei W, Nie J. A novel interaction between the SH2 domain of signaling adaptor protein Nck-1 and the upstream regulator of the Rho family GTPase Rac1 engulfment and cell motility 1 (ELMO1) promotes Rac1 activation and cell motility. J Biol Chem. 2014;289:23112–23122. doi: 10.1074/jbc.M114.549550. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Nakao S, Platek A, Hirano S, Takeichi M. Contact-dependent promotion of cell migration by the OL-protocadherin-Nap1 interaction. J Cell Biol. 2008;182:395–410. doi: 10.1083/jcb.200802069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le J, Mallery EL, Zhang C, Brankle S, Szymanski DB. Arabidopsis BRICK1/HSPC300 is an essential WAVE-complex subunit that selectively stabilizes the Arp2/3 activator SCAR2. Curr Biol. 2006;16:895–901. doi: 10.1016/j.cub.2006.03.061. [DOI] [PubMed] [Google Scholar]
- 39.Steffen A, Rottner K, Ehinger J, Innocenti M, Scita G, Wehland J, et al. Sra-1 and Nap1 link Rac1 to actin assembly driving lamellipodia formation. EMBO J. 2004;23:749–759. doi: 10.1038/sj.emboj.7600084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 41.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac1 GTPase-specific small molecule inhibitor. Proc Natl Acad Sci U S A. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, et al. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell. 2014;156:195–207. doi: 10.1016/j.cell.2013.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, et al. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468:533–538. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev Cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 45.Yilmaz M, Christofori GEMT. the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 46.Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol Cell Biol. 2014;15:577–590. doi: 10.1038/nrm3861. [DOI] [PubMed] [Google Scholar]
- 47.Akin O, Zipursky SL. The shape of things to come. Cell. 2014;156:13–14. doi: 10.1016/j.cell.2013.12.037. [DOI] [PubMed] [Google Scholar]
- 48.Teng Y, Pi W, Wang Y, Cowell JK. WASF3 provides the conduit to facilitate invasion and metastasis in breast cancer cells through HER2/HER3 signaling. Oncogene. 2016 doi: 10.1038/onc.2015.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goh WI, Lim KB, Sudhaharan T, Sem KP, Bu W, Chou AM, et al. mDia1 and WAVE2 proteins interact directly with IRSp53 in filopodia and are involved in filopodium formation. J Biol Chem. 2012;287:4702–4714. doi: 10.1074/jbc.M111.305102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suetsugu S, Yamazaki D, Kurisu S, Takenawa T. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev Cell. 2003;5:595–609. doi: 10.1016/s1534-5807(03)00297-1. [DOI] [PubMed] [Google Scholar]
- 51.Dart AE, Donnelly SK, Holden DW, Way M, Caron E. Nck and Cdc42 co-operate to recruit N-WASP to promote FcγR-mediated phagocytosis. J Cell Sci. 2012;125:2825–2830. doi: 10.1242/jcs.106583. [DOI] [PubMed] [Google Scholar]
- 52.Pils S, Kopp K, Peterson L, Delgado Tascón J, Nyffenegger-Jann NJ, et al. The adaptor molecule Nck localizes the WAVE complex to promote actin polymerization during CEACAM3-mediated phagocytosis of bacteria. PLoS One. 2012;7:e32808. doi: 10.1371/journal.pone.0032808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang YS, Graves B, Guerlavais V, Tovar C, Packman K, To KH, et al. Stapled α-helical peptide drug development: a potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc Natl Acad Sci U S A. 2013;110:E3445–E3454. doi: 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chu Q, Moellering RE, Hilinski GJ, Kim YW, Grossmann TN, Yeh JT, et al. Towards understanding cell penetration by stapled peptides. Med Chem Commun. 2015;6:111–119. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.