Abstract
Remarkably simple proteins play outsize roles in the execution of developmental complexity within biological systems. Sequence information determines structure and hence function, so how do low complexity sequences fulfill their functions? Recent discoveries are raising the curtain on a new dimension of the sequence-structure paradigm. In it, function derives not from the structures of individual proteins, but instead, from dynamic material properties of entire ensembles of the proteins acting in unison through phase changes. These phases include liquids, one-dimensional crystals, and -- as elaborated herein -- even glasses. The peculiar thermodynamics of glass-like protein assemblies, in particular, illuminate new principles of information flow through and, at times, orthogonal to the central dogma of molecular biology.
Introduction
Proteins are the most sophisticated macromolecules. Each performs a finely tuned function that is defined by the protein’s unique three-dimensional structure, which is in turn specified by its unique amino acid sequence. The sequence-structure paradigm is too simplistic, however, for the approximately one third of eukaryotic proteins with long regions that are devoid of regular structure [1]. Such “intrinsically disordered regions” or IDRs each exist as a cloud of interconverting structures, no one of which alone suffices to carry out protein function. Rather, function manifests over relatively large spatiotemporal scales that include entire ensembles of structures. Disorder even allows certain proteins known as prions to switch between distinct structural states at such low frequency that multiple generations of cells come and go in only one state or the other.
Enzymatic and structural activities of proteins require precise three-dimensional geometries. IDRs rarely perform those tasks directly. Instead, they organize and regulate them. Disorder abounds in eukaryotic signaling pathways and also features prominently in transcriptional and post-transcriptional regulatory machinery. Disordered proteins interact broadly with other proteins, serve as hubs in protein interaction networks, and in the process orchestrate the assembly of massive supramolecular complexes [1,2].
How, with an aversion to structure, do IDRs assemble some of the largest structures in the cell? The answer is deceptively simple. As if for water vapor condensing into dew droplets, the proteins coalesce out of the bulk cellular milieu into their own liquid phases. Unlike structured macromolecular complexes, individual polypeptides remain disordered within the liquid protein droplets, fleeting between self-solvated, energetically comparable intermolecular conformations. Attached globular domains and interacting macromolecules are pulled in along with their disordered partners. The compartmentalization of proteins within the liquid phase facilitates regulatory processes, including signaling, transcription, mRNA processing, and nucleation of cytoskeletal polymers [2,3]. The droplet-dependent localization of these activities enforces cell polarity, symmetry breaking, and cell differentiation. Intriguingly, droplets have been observed to solidify, or “mature”, over time, and this may be a basis for both functional and pathological differentiation of droplet activities [4–8].
As for any fundamental cellular process, misregulation of phase behavior can be catastrophic. Accelerated maturation or accumulation of mRNA granules is now heavily implicated in the pathogenesis of amyotrophic lateral sclerosis (ALS) and related proteopathies [2,4–6,9]. Disease-causing mutations in the low complexity proteins that drive granule condensation accelerate a time-dependent reduction in their fluidity. The mutations cause pathology not by inducing aggregation de novo, but rather, by subtly altering pre-existing phase behaviors. These dynamics mirror those long observed in classic pathological phase transitions in the form of protein aggregation [10]. The IDRs behind the pathogenesis of ALS, Alzheimer’s, Parkinson’s, and prion diseases, among others, progress in vitro from disordered monomers through “molten”, or liquid like, oligomeric states en route to semicrystalline fibrillar aggregates known as amyloids [10].
The progressive reduction in fluidity that characterizes both physiological and pathological phases suggests a simple underlying principle. Extrapolating from an extensive literature in polymer physics and protein folding, I propose that that principle is vitrification -- the transition of a supercritical liquid to a non-equilibrium amorphous solid, or more precisely, a glass. Herein I discuss the biophysical bases for vitrification of low complexity sequences (LCSs), and outline its theoretical relationship to protein function and malfunction.
Sequence complexity and phase behavior
A protein’s function is the indirect manifestation of information contained within its amino acid sequence. The more complex, or less redundant that information, the more precisely a single polypeptide of that sequence can fold into a single conformation among the universe of possible conformations. In the parlance of energy landscapes, information determines topology; the steeply funneled landscape of a well-folded globular protein requires a highly complex sequence. Amino acid compositional biases and repeated sequence elements lower the information content such that the landscape flattens and is no longer dominated by one particular funnel. Low complexity sequences tend therefore not to “fold” in the usual sense; they instead tend to interconvert between energetically comparable conformations [1,11]. The types of conformations within each ensemble are determined by the balance of attractive and repulsive interactions in the sequence: attractive interactions favor collapsed conformations while repulsive interactions favor more extended ones. Attractions dominate for most low complexity sequences due to dipole-dipole interactions in the polypeptide backbone. Hence the “default” LCS ensemble -- that is, in the absence of abundant repulsive interactions -- appears to be a collapsed molten globule [12]. Amino acid side chains enhance or counteract this behavior according to their individual physicochemical properties [1,13].
It is this cohesiveness of LCSs that drives phase behavior. Even transient, low affinity interactions between polypeptides raise the local concentrations of other potentially interacting moieties elsewhere in the sequence, leading the polypeptides to remain associated through avidity effects. But every intermolecular interaction displaces an energetically equivalent intramolecular interaction, rendering the unsatisfied moieties more likely themselves to find intermolecular partners. LCSs therefore tend to assemble cooperatively into polydisperse (i.e. lacking discrete stoichiometry) clusters, or phases, with critical concentrations for nucleation [1,13]. The analogous chain reaction for structured proteins, called “domain swapping”, renders tandem repeat multidomain proteins particularly susceptible to form solid phases -- that is, to aggregate [11,14]. The intrinsic polyvalency of LCSs also explains their well-documented promiscuity: disordered proteins have more binding partners than structured proteins; interact aberrantly with other proteins, leading to dosage-sensitive proteotoxicity [15]; and are enriched in the insoluble fractions of proteomes [16].
In general, the higher the interaction valency of a sequence, the tighter its concentration window for assembly. Intracellular LCS-driven phases including RNA granules, the nuclear pore permeability barrier, Cajal bodies, and nucleoli, among others, exploit repetitive low-affinity sequence elements such as RG, SR, and FG dipeptides [1,17,18]. The strength of the polyvalent interactions determines the dynamics of conformational interconversion and hence the viscosity of the LCS phase. Weakly-interacting tandem repeats also characterize extracellular LCS assemblies such as elastin, resilin, and spider silks. The repeats not only drive their assembly through coacervation, they also endow the extraordinary adhesiveness and plasticity essential to their functions [1,19].
Cells exploit the concentration thresholds for LCS assembly to control when and where phase separation occurs. They do so in part through post-translational modifications to LCS valency or interaction strength, including phosphorylation, glycosylation, and sumoylation [1,2]. Unlike membrane-bound organelles, LCS droplets readily coalesce and dissolve even with small changes in the local subcellular environment. Droplets frequently nucleate through heterotypic interactions with other multivalent molecules. Most of the recently characterized phase forming LCS do so physiologically in complex with other multivalent proteins or nucleic acids, and the inclusion of such ligands in vitro greatly lowers concentration thresholds for liquid or gel coalescence [1,2,13,20].
From LCS droplets are born amyloids and glasses
Although LCS phase separation can be likened to water vapor condensing into dew droplets, there is one essential difference. Liquid water is stable once it has formed, but liquid protein phases are not. The lowest energy state of supersaturated protein solutions is believed to be crystalline. Even though the proteins may first coalesce into liquids, such transitions are inherently metastable and ultimately give way to crystals [21]. That LCS droplets are indeed supersaturated is underscored by abundant recent observations that they solidify, or “mature” over time both in vitro and in vivo [4–9,22–25].
Naturally, because disordered proteins lack a stable three-dimensional structure, they cannot readily crystallize in three dimensions. Their only structural regularity is the linkage of amino acids in the polypeptide backbone. Consequently, crystallization proceeds orthogonally to the backbone. The resulting one-dimensional paracrystals, known as amyloids, take the form of an open-ended polymer of repeating polypeptide subunits. Additional subunits join the polymer only at its ends. The orientation of each polypeptide relative to its neighbors can differ, but most low complexity sequences prefer an in-register parallel orientation, in which the polypeptides align such that each residue forms a continuous stack with the identical residues in other subunits [26,27]. The structure is defined by backbone hydrogen bonding and hence is considered a “default” structure of polypeptides [10].
Because intra- and intermolecular interactions are thermodynamically interchangeable in the context of LCS liquid droplets, individual polypeptides can populate more extended conformations than they would otherwise be able to (as monomers). This enables their backbones to form continuous interactions with those of other polypeptides, which facilitates amyloid nucleation [13,28]. The formation of amyloid from dynamic oligomeric precursors was first reported for a low complexity prion-forming protein [29] and is now extensively documented for a multitude of other amyloid-forming proteins in vitro [10]. Once nucleation has occurred, polymerization can proceed extremely rapidly within a dynamic liquid phase due to the templated conformational conversion of other molecules in the droplet [29,30].
Even though LCS liquids are inherently metastable, different sequences solidify with different kinetics. As exemplified with the following proteins, those differences are paramount to the physiological and pathophysiological activities of the proteins.
Multiple proteins fulfill their physiological functions in an amyloid form. These include extracellular matrix proteins from bacteria and fungi as well as conserved signaling modules in programmed cell death pathways [31–34]. These proteins transition from metastable monomers to amyloids relatively quickly, without significantly populating disordered oligomeric forms [35–37]. The sequences of functional amyloids feature relatively diverse amino acid utilization and imperfect tandem or palindromic repeats. Hence they tend to be less complex than typical globular proteins, yet significantly more complex than most aggregation-prone LCSs. Extrapolating to higher sequence complexity, we find sophisticated signaling machinery composed of globular, rather than disordered, signaling proteins that nevertheless utilize the same principle of nucleated polymerization [31,38]. In these cases, non-amyloid polymers nucleate within dispersed monomeric phases, presumably bypassing metastable liquid phases entirely.
The kinetics can be quite different for very low complexity sequences. Polyglutamine is a minimum complexity sequence common to multiple eukaryotic regulatory proteins. It tends to form liquid droplets that are thought to facilitate dynamic low-affinity interactions with transcriptional and translational machinery [8,39]. The length of the polyglutamine tract directly relates to the dynamics of these phases and of the molecules within them. Excessively long tracts cause aberrantly stable complexes and proteotoxicity responsible for at least ten neurologic diseases [40]. The pathway to polyglutamine aggregates begins with collapsed disordered monomers [40]. Above a critical concentration, the monomers coalesce into disordered oligomers that ultimately foster amyloid nuclei. Paradoxically, however, amyloid nucleation is rate-limited within these oligomers by the same lack of sequence information that enabled their coalescence in the first place (Figure 1a). The polypeptides within the ensemble conformationally interconvert with exceptionally slow, or “glass-like” dynamics [28].
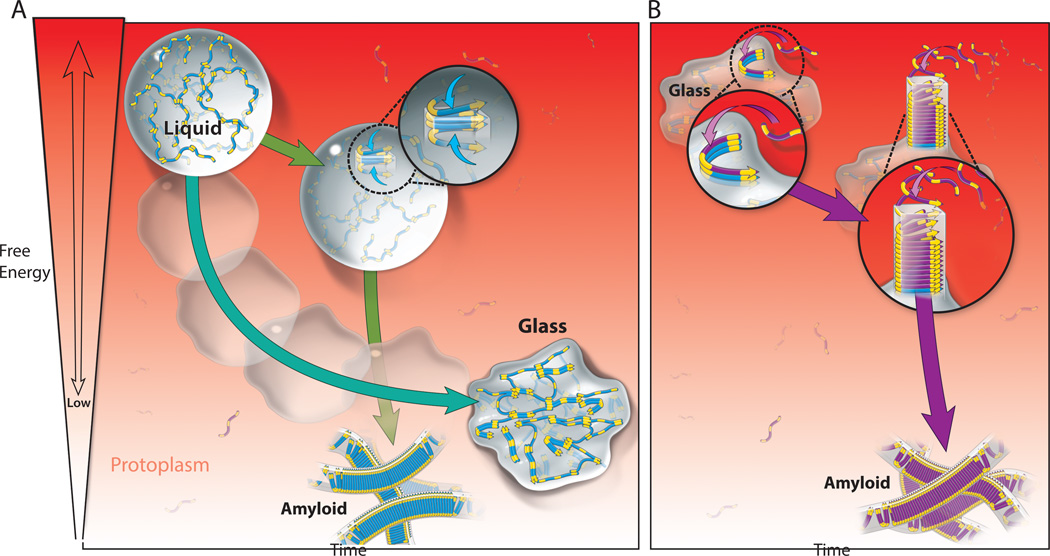
Figure 1. Phase transitions of LCS proteins.
A) Multivalent weak attractions (occurring between yellow moieties in the schematized polypeptides) facilitate the coalescence of polypeptides into a liquid droplet distinct from the bulk protoplasm. The high concentration and dynamics within the liquid can facilitate homogenous nucleation of amyloid, a lower energy solid phase that then templates the polymerization of other polypeptides in the droplet. Alternatively, the polypeptides can incrementally achieve lower energy states (represented by increased structuring and local beta-sheet formation) without nucleating amyloid. This process is called vitrification and results in conversion of the liquid to an amorphous solid, or glass.
B) The glass surface is a high energy template that can interact with other polypeptides in the bulk protoplasm, facilitating their nucleation to amyloid in trans. Nucleation occurring on a pre-existing surface – or heterogenous nucleation – may be the principal pathway for disease-associated amyloid formation de novo.
As the polypeptide chains within a liquid phase acquire increasingly stable arrangements, they become more attractive to other polypeptides while necessarily restricting their own explorations of alternative arrangements. Hence, even though amyloid-like configurations may occur locally, their ability to perpetuate throughout the network will be limited if similarly stable configurations have occurred elsewhere in the droplet that compete for potentially templatable sequence elements. As a consequence available degrees of freedom successively fall out of equilibrium and the liquid becomes increasingly viscous, ultimately becoming a non-crystalline solid state known to condensed matter physicists as a “glass”. Glass formation is not formally a liquid to solid phase transition, but merely a kinetic trap en route to it. Hence, a sufficiently low complexity system, like polyglutamine, may be driven in ratchet-like fashion toward the thermodynamic ground state without ever actually achieving it [11,41]. Consistent with a glass transition, polyglutamine inclusions appear to undergo a protracted intermolecular compaction in vivo [42].
Notwithstanding the physicochemical differences between specific amino acids and the very significant contributions these make to phase behavior, the above examples suggest that sequence complexity itself facilitates amyloid formation within liquid phases. The more information within a sequence, the more restricted its conformational space, and the lower the entropic penalty for accessing a highly ordered amyloid state. Within a multimeric liquid droplet this translates to a reduction in conformational discursions that delay amyloid nucleation and polymerization. Sequence manipulations that add complexity should therefore tend to reduce glassiness and accelerate amyloid formation. Consistent with this expectation, interruption of a pure hyper-elongated glutamine tract (Q85) with any of eleven other amino acids enhanced amyloid formation as determined by the kinetics and/or frangibility of SDS-resistant polydisperse multimers [43]. As explained in Box 1, frangibility is made possible by the one-dimensionality of amyloids, a characteristic that distinguishes them from liquids and glasses and enables their propagation as protein-based elements of inheritance. Insertion of hydrophobic residues similarly enhanced amyloid formation by predominantly polar, uncharged low complexity sequences [44].
Box 1. Prions are brittle one-dimensional phases.
Prions are protein-based transmissible elements that are implicated in a growing number of age-associated diseases including Alzheimer’s, Parkinson’s, and ALS. They also perform key functional roles in antiviral responses, inflammation, and cell fate determination in organisms ranging from fungi to humans [31,66]. The semi-stochastic appearance of some prions may even produce adaptive phenotypic diversity that buffers cell populations against environmental fluctuations [71].
Prions can be described physically as self-templating particles of a nucleated solid phase. The conversion of a polypeptide from one phase to another changes its conformation and/or cellular localization, and hence, its cellular activity.
Any phase transition that is kinetically limited by a sufficiently high nucleation energy will ensure that some cells spend their entire lives without ever acquiring the lower energy phase. Such nucleation barriers produce bimodal distributions of cells, in which some have acquired the lower energy phase and others have not. This is a necessary but insufficient requirement for a protein to form a prion. For the nucleated phase to propagate, or “infect” another cell containing the non-nucleated phase, a piece of it must break off from the original and enter the second cell, wherein it will template the conversion of that cell’s polypeptides to the same phase. In other words, the phase must readily divide into smaller pieces even under the miniscule forces that occur within cells. This requirement likely explains why all biochemically characterized prions are one-dimensional solid phases -- that is, amyloids and other linear polymers. What makes one-dimensional polymers able to act as prions is that they, unlike liquids, crystals, or glasses, can be fragmented by point applications of force. Even a single protein -- most prominently the AAA+ ATPase Hsp104 -- pulling on a single monomer within the amyloid suffices to snap it into two pieces [72]. Most characterized prions require Hsp104 activity; in its absence, the amyloids fail to fragment and hence cannot transmit to other cells. The more brittle the amyloid, the more robustly it propagates as a prion [73].
The wild menagerie: Glass promiscuity drives proteotoxicity
The distinction between amyloids and glasses may seem inconsequential. In fact, it could not be more pertinent to the pathology of protein misfolding diseases. Amyloids historically shouldered the blame for proteotoxicity, but a wealth of evidence now implicates amorphous aggregates and oligomers that may be more akin to glasses [10,28]. The oligomers are generally spherical and have exposed hydrophobic surfaces that aberrantly bind other proteins, leading to their co-aggregation. Amyloids, in contrast, are now viewed as relatively benign end products of less structured, and more toxic, precursors.
What makes glass so destructive? Unlike amyloids, glass is kinetically “frozen” in a relatively high energy state. This means that some regions of the sequence that prefer to be buried may instead remain solvent-exposed and available to interact with other macromolecules. Moreover, because the polypeptides are immobilized, the entropic penalties for engaging in intermolecular interactions are effectively prepaid. In short, protein glasses are “sticky”. As a result, amorphous aggregates of polyglutamine and other LCSs abundantly precipitate disordered proteins on their surfaces [7,45,46]. Remarkably, even small molecule aggregates have the same effect [47–49].
Although glass formation directly counteracts amyloid nucleation within droplet interiors, it may on balance promote amyloid formation by interacting with other molecules in trans (Figure 1b). The sticky surfaces can template newly arriving and still flexible molecules, which then go on to template additional molecules whose conformational dynamics are entirely unencumbered by the glass. Theory and experimental evidence suggest that such “heterogeneous” nucleation, i.e. at surfaces, may far outpace homogeneous nucleation as the principal pathway for intracellular amyloidogenesis [10,50,51]. In vivo observations increasingly support this notion. Amyloid inclusions characteristic of Alzheimer’s, Huntington’s, and prion diseases contain a structurally disordered and presumably catalytic core [52–54]; aggregates of Alzheimer’s-associated Aβ42 rampantly catalyze new aggregates on their surfaces, in living brain tissue [55]; and pathological aggregates in ALS, Alzheimer's, and other dementias colocalize with glass-like LCS structures known as stress granules [56].
Commensurate with its potential to unbalance proteome homeostasis, nature combats glass promiscuity in a variety of ways, both at the protein and cellular levels. Flanking regions and amphipathic character force liquid-like droplets into micellar architectures that reduce surface tension [13]. Protein chaperones bind to and mask promiscuous surfaces [10,55]. A variety of cellular mechanisms sequester the aggregates such that their stickiness neutralizes each other, and the larger size facilitates their asymmetric distribution during cell division [57].
The domesticated menagerie: Glass transition kinetics drive protein function
The inevitability that supersaturated LCSs will, eventually, solidify begs the question: Why are they supersaturated in the first place? Given the enormous energy expenditures of translation and protein homeostasis [57,58], one assumes that evolution has not predestined newly synthesized proteins to aggregate unless doing so confers some advantage.
The physiological dynamics of mRNA granules suggest that cells do, indeed, exploit the perpetual disequilibrium of glass transitions. A dense “hydrogel” (porous glass-like solid) of polyvalent LCSs comprises the selective barrier that governs all nucleocytoplasmic transport in eukaryotes [17]. Nuclear transport receptors compete with low affinity interactions between the LCSs, resulting in a local “melting” that allows them to pass through unimpeded. The gel then resolidifies behind them. Other LCS droplets solidify over time in the cytoplasm, and this may be a basis for functional differentiation of droplet activities [5,13]. It can also preserve the proteins’ functions by precluding the nucleation of relatively irreversible aggregates elsewhere in the cell [59]. LCS droplets also solidify during stress [22,24,25], suggesting that cells under normal growth conditions invest energy to continually postpone otherwise inevitable glass transitions. This situation plausibly constitutes a fail-safe whereby the glass functions to insulate the transcriptome and associated cellular machinery from stress. This idea has ample precedence in the known stress-protective roles of other LCS proteins. For example, plants and even some animals survive extreme desiccation through cytosolic vitrification mediated by disordered LCS proteins [60,61]. Bacterial and yeast cytoplasms also undergo protective glass-like transitions upon starvation [62–64], although in these cases any role for LCSs remains speculative (especially for bacteria, as prokaryotes generally contain far fewer LCSs than eukaryotes). These latter discoveries nevertheless suggest that vitrification may be a universal cell biological principle, within which LCSs are merely an exceptional example.
When a crystalline solid absorbs energy, its component molecules simply vibrate about a global energy minimum. No structural changes occur (short of melting). However, when a glass absorbs energy, some molecules are displaced from their local minima and subsequently relax into other local minima. The physical and biological properties of a glass are changed by the event. At any one time, the exact molecular configuration of a glass depends on its history. In other words, glasses intrinsically respond to, and remember, environmental inputs [11,41]. The most intriguing emerging functions of low complexity sequences suggest that the behavioral complexities of eukaryotes derive, in part, from this phenomenon.
When cycling cells of budding yeast are exposed to mating pheromone, the glutamine-rich LCS, Whi3, immediately begins to coalesce into large, amorphous aggregates. If the cells fail to find a mating partner, those aggregates ultimately de-repress cyclin translation to enable the cells to resume cycling [65]. Remarkably, the Whi3 aggregates then persist in the original cells as a permanent molecular memory that precludes subsequent attempts to mate. The apparent ability of Whi3 multimers to progressively integrate an environmental signal and thereby determine developmental outcome is by no means unique: multiple other LCS regulatory proteins also form stimulus-dependent aggregates that decide cell fate [8,39,66,67]. Most intriguingly, neuronal isoforms of the translation regulatory protein, CPEB, undergo a liquid to solid transition that enciphers long term memory in animals [68]. Consistent with phase behaviors of other LCS proteins of similar sequence and function, CPEB localizes to dynamic mRNA granules in vivo [69,70], and forms transient pre-amyloid oligomers in vitro [23]. Upon neuronal stimulation, however, CPEB acquires a highly stable oligomeric form that localizes and activates the translation of specific mRNAs that stabilize the activated synapse [68]. The structural nature of this oligomer -- specifically, whether it comprises a subunit of a hierarchically-structured amyloid or instead a vitrified droplet -- remains to be elucidated.
Conclusion
Interesting biology arises from the tension between order and chaos. Low complexity sequences are no exception. Unable to form a single structure, LCSs tend instead to populate disordered ensembles that lend themselves to phase separation. Those phases coalesce, fuse, and dissolve when and where needed to regulate the localization and dynamics of cellular processes. But the liquid phase is tenuous -- the chains constantly seek lower energy configurations leading to solidification either as amyloid or glass. This protracted struggle for equilibrium fundamentally relates LCS physical properties with time and environmental inputs, and that struggle plays out in the diverse functions and pathologies of LCS proteins.
Highlights.
Proteins of low sequence complexity are predisposed to form metastable liquid phases.
Liquid phases give way to amyloids or glasses.
Glasses may be exceptionally proteotoxic.
Glasses respond to time and stress, enabling proteins to store information.
Acknowledgments
I am grateful to members of the Halfmann lab for constructive feedback while preparing this manuscript, and to Mark Miller for assistance with scientific illustrations. I apologize to those researchers whose primary works I was unable to cite due to space constraints. This work was supported by an NIH Director’s Early Independence Award (DP5-OD009152) and the Stowers Institute for Medical Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review have been highlighted as:
* of special interest
** of outstanding interest
- 1. Van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. * An exhaustive review on intrinsically disordered proteins, co-authored by many of the major players in this field. It discusses the sequence determinants, functions, biophysics, evolution, and regulation of IDRPs, including their involvement in liquid-liquid phase separation.
- 2.Mitrea DM, Kriwacki RW. Phase separation in biology; functional organization of a higher order. Cell Commun Signal. 2016;14:1. doi: 10.1186/s12964-015-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang H, Wang S, Huang Y, He X, Cui H, Zhu X, Zheng Y. Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell. 2015;163:108–122. doi: 10.1016/j.cell.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, McKnight SL. The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei. Cell. 2015;163:829–839. doi: 10.1016/j.cell.2015.10.040. * One of a series of papers from this group demonstrating hydrogel (porous glass) formation by RNA-binding LCS proteins. Xiang et al. show that protein dynamics within phases anticorrelates with detectable structure, and provide evidence that the LCS phase contains a small amount of fibrillar structure in vivo.
- 5. Lin Y, Protter DSW, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. ** One of multiple recent papers that demonstrate that RNA-binding LCS proteins form liquid droplets through multivalent low affinity interactions, and those droplets can subsequently solidify. Lin et al. document a continuous increase in viscosity over time within individual droplets.
- 6. Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. ** One of multiple recent papers that demonstrate that RNA-binding LCS proteins form liquid droplets through multivalent low affinity interactions, and those droplets can subsequently solidify. As for Lin et al., Patel et al. demonstrates that the droplets become increasingly viscous until they are effectively solid. They also observe amyloids nucleating from the droplets.
- 7. Murakami T, Qamar S, Lin JQ, Schierle GSK, Rees E, Miyashita A, Costa AR, Dodd RB, Chan FTS, Michel CH, et al. ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. ** One of multiple recent papers that demonstrate that RNA-binding LCS proteins form liquid droplets through multivalent low affinity interactions, and those droplets can subsequently solidify. Murakami et al. further link solidification to pathology.
- 8.Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. RNA Controls PolyQ Protein Phase Transitions. Mol Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. * One of multiple recent papers that demonstrate that RNA-binding LCS proteins form liquid droplets through multivalent low affinity interactions, and those droplets can subsequently solidify. Similarly to Patel et al., Molliex et al. observe amyloids nucleating within the droplets.
- 10. Knowles TPJ, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. * A thorough review on current topics in amyloids, the molecular mechanisms of their formation, and the involvement of pre-amyloid oligomers in pathology.
- 11. Ferreiro DU, Komives EA, Wolynes PG. Frustration in biomolecules. Q Rev Biophys. 2014;47:285–363. doi: 10.1017/S0033583514000092. * A comprehensive and highly stimulating review of glass-like dynamics in proteins and its implications for information processing and storage by biological systems.
- 12.Das RK, Ruff KM, Pappu RV. Relating sequence encoded information to form and function of intrinsically disordered proteins. Curr Opin Struct Biol. 2015;32:102–112. doi: 10.1016/j.sbi.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brangwynne CP, Tompa P, Pappu RV. Polymer physics of intracellular phase transitions. Nat Phys. 2015;11:899–904. [Google Scholar]
- 14.Borgia A, Kemplen KR, Borgia MB, Soranno A, Shammas S, Wunderlich B, Nettels D, Best RB, Clarke J, Schuler B. Transient misfolding dominates multidomain protein folding. Nat Commun. 2015;6:8861. doi: 10.1038/ncomms9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138:198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 16.Albu RF, Chan GT, Zhu M, Wong ETC, Taghizadeh F, Hu X, Mehran AE, Johnson JD, Gsponer J, Mayor T. A feature analysis of lower solubility proteins in three eukaryotic systems. J Proteomics. 2015;118:21–38. doi: 10.1016/j.jprot.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 17. Schmidt HB, Görlich D. Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. elife. 2015;4 doi: 10.7554/eLife.04251. ** This paper and its predecessors from the same group provide arguably the most comprehensive validation of functional biological glasses formed by LCS proteins. Schmidt and Gorlich reveal that Nup98 homologs from diverse eukaryotes all readily phase separate in vitro into hydrogels that recapitulate the sieving properties of nuclear pores. They further confirm that gelation is driven foremost by low-affinity multivalent interactions which, in some cases, lead also to local amyloid-like conformations within the gel.
- 18.Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quiroz FG, Chilkoti A. Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat Mater. 2015;14:1164–1171. doi: 10.1038/nmat4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grøfte M, Rask M-BD, Streicher W, Jungmichel S, Nielsen ML, et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose) Nat Commun. 2015;6:8088. doi: 10.1038/ncomms9088. ** Demonstrate that a multivalent ligand (poly(ADP-ribose)) not only functionally nucleates liquid droplet formation by multiple LCSs in vivo, but that such LCS liquids ultimately give way to aggregates.
- 21.Asherie N. Protein crystallization and phase diagrams. Methods. 2004;34:266–272. doi: 10.1016/j.ymeth.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 22.Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hervás R, Li L, Majumdar A, Fernández-Ramírez MDC, Unruh JR, Slaughter BD, Galera-Prat A, Santana E, Suzuki M, Nagai Y, et al. Molecular Basis of Orb2 Amyloidogenesis and Blockade of Memory Consolidation. PLoS Biol. 2016;14:e1002361. doi: 10.1371/journal.pbio.1002361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kroschwald S, Maharana S, Mateju D, Malinovska L, Nüske E, Poser I, Richter D, Alberti S. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. elife. 2015;4:e06807. doi: 10.7554/eLife.06807. * Demonstrate that LCS mRNA granules can solidify in vivo upon inactivation of cellular fluidizing activities.
- 25.Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tycko R, Wickner RB. Molecular structures of amyloid and prion fibrils: consensus versus controversy. Acc Chem Res. 2013;46:1487–1496. doi: 10.1021/ar300282r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 28.Vitalis A, Wang X, Pappu RV. Quantitative characterization of intrinsic disorder in polyglutamine: insights from analysis based on polymer theories. Biophys J. 2007;93:1923–1937. doi: 10.1529/biophysj.107.110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 30.Ogi H, Fukukshima M, Hamada H, Noi K, Hirao M, Yagi H, Goto Y. Ultrafast propagation of β-amyloid fibrils in oligomeric cloud. Sci Rep. 2014;4:6960. doi: 10.1038/srep06960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai X, Chen J, Xu H, Liu S, Jiang Q-X, Halfmann R, Chen ZJ. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daskalov A, Habenstein B, Sabaté R, Berbon M, Martinez D, Chaignepain S, Coulary-Salin B, Hofmann K, Loquet A, Saupe SJ. Identification of a novel cell death-inducing domain reveals that fungal amyloid-controlled programmed cell death is related to necroptosis. Proc Natl Acad Sci U S A. 2016;113:2720–2725. doi: 10.1073/pnas.1522361113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 2012;20:66–73. doi: 10.1016/j.tim.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao Y-S, Damko E, Moquin D, Walz T, McDermott A, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Smith DR, Jones JW, Chapman MR. In vitro polymerization of a functional Escherichia coli amyloid protein. J Biol Chem. 2007;282:3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan W, Stubbs G. Fungal prion HET-s as a model for structural complexity and self-propagation in prions. Proc Natl Acad Sci U S A. 2014;111:5201–5206. doi: 10.1073/pnas.1322933111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Simone A, Kitchen C, Kwan AH, Sunde M, Dobson CM, Frenkel D. Intrinsic disorder modulates protein self-assembly and aggregation. Proc Natl Acad Sci U S A. 2012;109:6951–6956. doi: 10.1073/pnas.1118048109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schröder GF, Fitzgerald KA, Wu H, Egelman EH. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee C, Occhipinti P, Gladfelter AS. PolyQ-dependent RNA-protein assemblies control symmetry breaking. J Cell Biol. 2015;208:533–544. doi: 10.1083/jcb.201407105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wetzel R. Physical chemistry of polyglutamine: intriguing tales of a monotonous sequence. J Mol Biol. 2012;421:466–490. doi: 10.1016/j.jmb.2012.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stillinger FH, Debenedetti PG. Glass Transition Thermodynamics and Kinetics. Annu. Rev. Condens. Matter Phys. 2013;4:263–285. [Google Scholar]
- 42. Bhardwaj V, Panicker MM, Udgaonkar JB. Fluorescence anisotropy uncovers changes in protein packing with inclusion growth in a cellular model of polyglutamine aggregation. Biochemistry. 2014;53:3621–3636. doi: 10.1021/bi500383h. * This paper provides evidence that intracellular polyglutamine inclusions become increasingly compact as they grow, consistent with a gradual acquisition of increasingly low energy states characteristic of glass transitions.
- 43.Alexandrov AI, Polyanskaya AB, Serpionov GV, Ter-Avanesyan MD, Kushnirov VV. The effects of amino acid composition of glutamine-rich domains on amyloid formation and fragmentation. PLoS ONE. 2012;7:e46458. doi: 10.1371/journal.pone.0046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gonzalez Nelson AC, Paul KR, Petri M, Flores N, Rogge RA, Cascarina SM, Ross ED. Increasing prion propensity by hydrophobic insertion. PLoS ONE. 2014;9:e89286. doi: 10.1371/journal.pone.0089286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsumoto G, Kim S, Morimoto RI. Huntingtin and mutant SOD1 form aggregate structures with distinct molecular properties in human cells. J Biol Chem. 2006;281:4477–4485. doi: 10.1074/jbc.M509201200. [DOI] [PubMed] [Google Scholar]
- 46.England JL, Kaganovich D. Polyglutamine shows a urea-like affinity for unfolded cytosolic protein. FEBS Lett. 2011;585:381–384. doi: 10.1016/j.febslet.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 47.Feng BY, Toyama BH, Wille H, Colby DW, Collins SR, May BCH, Prusiner SB, Weissman J, Shoichet BK. Small-molecule aggregates inhibit amyloid polymerization. Nat Chem Biol. 2008;4:197–199. doi: 10.1038/nchembio.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coan KED, Shoichet BK. Stoichiometry and physical chemistry of promiscuous aggregate-based inhibitors. J Am Chem Soc. 2008;130:9606–9612. doi: 10.1021/ja802977h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vácha R, Linse S, Lund M. Surface effects on aggregation kinetics of amyloidogenic peptides. J Am Chem Soc. 2014;136:11776–11782. doi: 10.1021/ja505502e. [DOI] [PubMed] [Google Scholar]
- 51.Galvagnion C, Buell AK, Meisl G, Michaels TCT, Vendruscolo M, Knowles TPJ, Dobson CM. Lipid vesicles trigger α-synuclein aggregation by stimulating primary nucleation. Nat Chem Biol. 2015;11:229–234. doi: 10.1038/nchembio.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hazeki N, Tsukamoto T, Yazawa I, Koyama M, Hattori S, Someki I, Iwatsubo T, Nakamura K, Goto J, Kanazawa I. Ultrastructure of nuclear aggregates formed by expressing an expanded polyglutamine. Biochem Biophys Res Commun. 2002;294:429–440. doi: 10.1016/S0006-291X(02)00498-9. [DOI] [PubMed] [Google Scholar]
- 53.Jin L-W, Claborn KA, Kurimoto M, Geday MA, Maezawa I, Sohraby F, Estrada M, Kaminksy W, Kahr B. Imaging linear birefringence and dichroism in cerebral amyloid pathologies. Proc Natl Acad Sci U S A. 2003;100:15294–15298. doi: 10.1073/pnas.2534647100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nilsson KPR, Aslund A, Berg I, Nyström S, Konradsson P, Herland A, Inganäs O, Stabo-Eeg F, Lindgren M, Westermark GT, et al. Imaging distinct conformational states of amyloid-beta fibrils in Alzheimer’s disease using novel luminescent probes. ACS Chem Biol. 2007;2:553–560. doi: 10.1021/cb700116u. [DOI] [PubMed] [Google Scholar]
- 55.Cohen SIA, Arosio P, Presto J, Kurudenkandy FR, Biverstål H, Dolfe L, Dunning C, Yang X, Frohm B, Vendruscolo M, et al. A molecular chaperone breaks the catalytic cycle that generates toxic Aβ oligomers. Nat Struct Mol Biol. 2015;22:207–213. doi: 10.1038/nsmb.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolozin B. Physiological protein aggregation run amuck: stress granules and the genesis of neurodegenerative disease. Discov Med. 2014;17:47–52. [PMC free article] [PubMed] [Google Scholar]
- 57.Amen T, Kaganovich D. Dynamic droplets: the role of cytoplasmic inclusions in stress, function, and disease. Cell Mol Life Sci. 2015;72:401–415. doi: 10.1007/s00018-014-1740-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shelkovnikova TA, Robinson HK, Connor-Robson N, Buchman VL. Recruitment into stress granules prevents irreversible aggregation of FUS protein mislocalized to the cytoplasm. Cell Cycle. 2013;12:3194–3202. doi: 10.4161/cc.26241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hughes SL, Schart V, Malcolmson J, Hogarth KA, Martynowicz DM, Tralman-Baker E, Patel SN, Graether SP. The importance of size and disorder in the cryoprotective effects of dehydrins. Plant Physiol. 2013;163:1376–1386. doi: 10.1104/pp.113.226803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sakurai M, Furuki T, Akao K-I, Tanaka D, Nakahara Y, Kikawada T, Watanabe M, Okuda T. Vitrification is essential for anhydrobiosis in an African chironomid, Polypedilum vanderplanki. Proc Natl Acad Sci U S A. 2008;105:5093–5098. doi: 10.1073/pnas.0706197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Munder MC, Midtvedt D, Franzmann T, Nüske E, Otto O, Herbig M, Ulbricht E, Müller P, Taubenberger A, Maharana S, et al. A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. elife. 2016;5 doi: 10.7554/eLife.09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joyner RP, Tang JH, Helenius J, Dultz E, Brune C, Holt LJ, Huet S, Müller DJ, Weis K. A glucose-starvation response regulates the diffusion of macromolecules. elife. 2016;5 doi: 10.7554/eLife.09376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Parry BR, Surovtsev IV, Cabeen MT, O’Hern CS, Dufresne ER, Jacobs-Wagner C. The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell. 2014;156:183–194. doi: 10.1016/j.cell.2013.11.028. * This paper first demonstrated that the cytoplasm itself is effectively supersaturated, and vitrifies in the absence of metabolic activity. Munder et al. 2015 and Joyner et al. 2015 subsequently demonstrated the same phenomenon in eukaryotes. These works collectively frame the relatively localized glass-transitions discussed herein as part of a broader cell biological principle.
- 65.Caudron F, Barral Y. A super-assembly of Whi3 encodes memory of deceptive encounters by single cells during yeast courtship. Cell. 2013;155:1244–1257. doi: 10.1016/j.cell.2013.10.046. [DOI] [PubMed] [Google Scholar]
- 66.Holmes DL, Lancaster AK, Lindquist S, Halfmann R. Heritable remodeling of yeast multicellularity by an environmentally responsive prion. Cell. 2013;153:153–165. doi: 10.1016/j.cell.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berchowitz LE, Kabachinski G, Walker MR, Carlile TM, Gilbert WV, Schwartz TU, Amon A. Regulated Formation of an Amyloid-like Translational Repressor Governs Gametogenesis. Cell. 2015;163:406–418. doi: 10.1016/j.cell.2015.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Si K, Kandel ER. The Role of Functional Prion-Like Proteins in the Persistence of Memory. Cold Spring Harb Perspect Biol. 2016;8 doi: 10.1101/cshperspect.a021774. ** A review of the role of CPEB and its homologs in memory, particularly evidence that a prion-like state of CPEB may be responsible for the stimulation of locally restricted translation at specific synapses. This body of work exemplifies the critical role of phase behavior in low complexity protein function. A specific physiological stimulus triggers the permanent solidification of CPEB mRNA granules, which simultaneously switches and compartmentalizes the protein’s activity.
- 69.Si K, Choi Y-B, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 70.Stephan JS, Fioriti L, Lamba N, Colnaghi L, Karl K, Derkatch IL, Kandel ER. The CPEB3 Protein Is a Functional Prion that Interacts with the Actin Cytoskeleton. Cell Rep. 2015;11:1772–1785. doi: 10.1016/j.celrep.2015.04.060. [DOI] [PubMed] [Google Scholar]
- 71.Newby GA, Lindquist S. Blessings in disguise: biological benefits of prion-like mechanisms. Trends Cell Biol. 2013;23:251–259. doi: 10.1016/j.tcb.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 72.Sweeny EA, Shorter J. Mechanistic and Structural Insights into the Prion-Disaggregase Activity of Hsp104. J Mol Biol. 2015 doi: 10.1016/j.jmb.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]