Abstract
Mesenchymal stem cells (MSCs) are multipotent cells that represent a promising source for regenerative medicine. MSCs are capable of osteogenic, chondrogenic, adipogenic and myogenic differentiation. Efficacy of differentiated MSCs to regenerate cells in the injured tissues requires the ability to maintain the differentiation toward the desired cell fate. Since MSCs represent an attractive source for autologous transplantation, cellular and molecular signaling pathways and micro-environmental changes have been studied in order to understand the role of cytokines, chemokines, and transcription factors on the differentiation of MSCs. The differentiation of MSC into a mesenchymal lineage is genetically manipulated and promoted by specific transcription factors associated with a particular cell lineage. Recent studies have explored the integration of transcription factors, including Runx2, Sox9, PPARγ, MyoD, GATA4, and GATA6 in the differentiation of MSCs. Therefore, the overexpression of a single transcription factor in MSCs may promote trans-differentiation into specific cell lineage, which can be used for treatment of some diseases. In this review, we critically discussed and evaluated the role of transcription factors and related signaling pathways that affect the differentiation of MSCs toward adipocytes, chondrocytes, osteocytes, skeletal muscle cells, cardiomyocytes, and smooth muscle cells.
Keywords: Mesenchymal Stem Cells, Transcription factors, Multilineage Differentiation
Introduction
Mesenchymal stem cells (MSCs) are multipotent stem cells that are capable of self-renewing and differentiating into functional cell types. The ease of isolation, the high migratory capacity, the relatively high expansion rates, and the ability to avoid the allogeneic responses after transplantation (Chen et al., 2004; Fouillard et al., 2007; Le Blanc and Ringden, 2005; Ripa et al., 2007; Sekiya et al., 2002) make them attractive candidates in regenerative medicine. Over the last few years, MSCs have been isolated from various tissues and organs including adipose tissue, bone marrow, placental tissue, umbilical cord blood, the testes, the liver, the pancreas, the spleen, amniotic fluid, menstrual blood, dental pulp, the dermis and the lung (De Coppi et al., 2007; Guan et al., 2006; in ’t Anker et al., 2003; Kruse et al., 2006; Meng et al., 2007; Pierdomenico et al., 2005; Ringe et al., 2008; Sabatini et al., 2005; Sellheyer and Krahl, 2010). They are characterized by their spindle shaped morphology and their ability to differentiate in vitro into adipocytes, chondrocytes and osteocytes. Previous reports suggest that there is no single specific marker to distinguish MSCs from other cells that exhibit similar fibroblastic characteristics. Hence, these cells are immunophenotypically characterized by positive and negative expression of multiple surface antigens. MSCs express surface antigens such as CD44, CD73, CD29, CD90 and CD105 and lack hematopoietic and endothelial markers such as CD11, CD14, CD31, CD34 and CD45 (Haynesworth et al., 1992; Lodie et al., 2002; Suva et al., 2004). In vitro, MSCs usually grow as a monolayer culture in a medium containing 10% fetal bovine serum and L-glutamine.
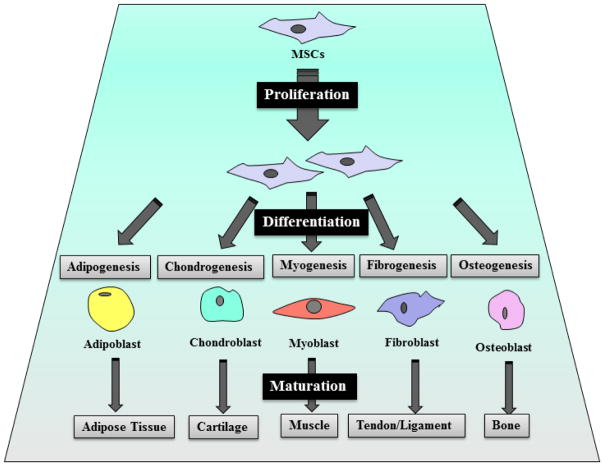
The multi-lineage differentiation of MSCs has been extensively studied in vitro and in vivo since their first discovery. These studies have demonstrated that MSCs have the potential to differentiate into several mesoderm-type lineages, including myogenic, adipogenic, osteogenic and chondrogenic lineages (Figure 1).
Figure 1.
Mesenchymal stem cells differentiate into osteoblast, chondrocyte, myoblast, fibroblast and adipocyte, depending on the stimuli in the differentiation-induction media (in vitro).
During the differentiation of MSCs toward a specific cell type, there are a multitude of stimuli and inhibitors that play important roles in the initial commitment and later stages of differentiation. The differentiation of MSCs into specific mature cell types is controlled by various cytokines, growth factors, extracellular matrix molecules, and transcription factors (TFs). In vitro, the differentiation of MSCs requires certain inducers including some growth factors (Table 1).
Table 1.
Factors in inducing differentiation of MSCs to specific lineage.
| Inducer | Concentration | Effect | Direction of differentiation | Reference |
|---|---|---|---|---|
| Dexamethasone + 3-isobutyl-1-methylxanthine + insulin | 1 μM 0.5 mM 0.01 mg/ml |
Increase fat deposition in the cytoplasm | Adipocytes | (Yu et al., 2012) |
| Ascorbic acid + dexamethasone | 50 μM 100 nM |
Increases the activity of alkaline phosphatase | Osteocytes | (Yu et al., 2012) |
| TGF- β1 + ascorbic acid + dexamethasone | 10 ng/ml 50 μg/ml 100 nM |
Chondrocytes | (Vater et al., 2011) | |
| TGF-β1 | 1, 2, 5 and 10 ng/ml | Stimulate SMC-specific genes expression | Smooth muscle cells | (Vater et al., 2011; Floren et al., 2016) |
| 5-azacytidine | 3 μM 10 μM |
Inhibits DNA methyltransferase | Cardiomyocytes | (Yamada et al., 2007; Rowlands et al., 2008; Wei et al., 2011)) |
| VEGF | 10, 50 ng/ml | Phosphorylate VEGF-RII and increase its Activity | Endothelial | (Pankajakshan et al., 2013; Vater et al., 2011) |
To date, many studies have shown that transcription factors can affect the differentiation of MSCs toward different mature cell types. Through the activation of transcription factors, the effect may occur by upregulating the expression of genes responsible for induction and progression of specific-cell type differentiation. In the following sections, we will discuss the characteristics and function of various transcription factors that affect the differentiation of MSCs (isolated from different sources) toward adipocytes, chondrocytes, osteocytes, skeletal muscle cells, cardiomyocytes, and smooth muscle cells.
Osteogenic differentiation
Differentiation of MSCs toward osteocytes is induced in vitro by incubating a monolayer of MSCs with a differentiation medium containing β-glycerophosphate, dexamethasone, ascorbic acid-2-phosphate and combinations of transforming growth factor-beta (TGF-β), bone morphogenetic proteins (BMPs) and vitamin D3 (Friedenstein et al., 1987; Okamoto et al., 2002). At the molecular level, interactions between hormones and transcription factors control the differentiation of MSCs into osteocytes. The major transcription factors that have key roles in the differentiation of MSCs into osteocytes are CBFA-1/Runx2 and Osterix (Augello and De Bari, 2010).
The osteogenic differentiation is regulated by various transcription factors such as Runt-related transcription factor 2 (Runx2), osterix, and β-catenin. The Runx gene consists of a runt domain (DNA binding domain) and Runx protein forms heterodimers with core binding factor β (Cbfβ)/ polyma enhancer binding protein 2β (Pebp2β) in vitro. Various studies have shown that the Runt-related transcription factor (Runx2) is required for osteogenic differentiation. The Runx2 transcription factor is an essential regulator for bone formation and the osteogenic differentiation of MSCs. It directs MSCs to differentiate into pre-osteoblast and inhibits the adipogenic and chondrogenic differentiation (Komori, 2006). Zhao and his colleague transduced mice with Runx2 in order to examine the osteogenic differentiation of bone morrow-derived MSCs. The data showed that Runx2-induced MSCs were able to form more osteocytes than the control, which consisted of virus- transduced MSCs. These findings indicated that Runx2 plays an essential role in osteogenic differentiation and demonstrated the ability to use Runx2 gene transfer to enhance the differentiation of MSCs into osteocytes (Zhao et al., 2005). The post-mitotic symmetric segregation of Runx2 mRNA to progeny cells has been also found to support the maintenance of osteogenic lineage commitment and osteoblast phenotype (Varela et al., 2015). Homeobox protein Hox-B7 (HOXB7) over-expression affected the mRNA expression of the key transcription factor, Runx2, and promotes osteogenic differentiation. HOXB7 can enhance the osteogenic differentiation by up-regulating Runx2 (Gao et al., 2015). Hoxa2 was also found up-regulated during osteogenic differentiation, while Hoxa9 was down-regulated (Seifert et al., 2015). In addition, overexpression of Hoxa2 induced the osteogenic differentiation, which inhibits Sox9 and chondrogenic differentiation (Seifert et al., 2015).
The expression of Runx2 is regulated by many signaling pathways, including Wnt, BMP, and Notch signaling pathways. BMP binds to BMPR and activates intracellular Smad, which translocate to the nucleus and serve as a transcription factor (Fujii et al., 1999). Therefore, deletion of the BMP ligands disrupts osteogenic differentiation (Bandyopadhyay et al., 2006). Also, BMP9 promotes the activation of Smad1, 5, and 8 and osteogenic differentiation in MSCs (Xu et al., 2012). Smad-Runx2 interaction is required for osteogenic differentiation. Induced mutations in the C-terminal domain of Runx2 disrupt Runx2-Smad transcriptional activities, which suppress osteogenic differentiation (Javed et al., 2008). The TWIST transcription factor also acts as a downstream of Hypoxia-inducible factor-1α (HIF-α), and suppresses expression of Runx2 in MSCs. This suppression results in the regulation of the osteogenic differentiation (Yang et al., 2011b). Overexpression of HIF-1α was found to enhance proliferation, cell survival and expression of pro-angiogenic genes. However, the expression of the osteogenic markers, including BMP-2 and Runx2, were decreased, indicating no effect of HIF-1α overexpression on osteogenic differentiation of MSCs (Lampert et al., 2015).
Osterix belongs to the specificity protein 1 family (Sp1) of transcription factors. It has three zinc finger motifs. Nakashima et al. (2002) reported that the formation of cortical bone and bone trabeculae through either intramembranous or endochondral ossification did not occur in the absence of osterix. In the Runx2/Cbfa1 null mice, osterix was not expressed, which suggested that osterix acts as a downstream of Runx2. It was concluded that Osterix is required in order to direct MSCs to osteoblasts, and hence, necessary for bone formation (Nakashima et al., 2002). Additionally, activation of the Wnt signaling pathway in MSCs induces expression of Osterix and suppresses PPAR-γ (Kang et al., 2007).
β-catenin also plays a crucial role in the differentiation of MSCs into osteoblasts. The absence of β-catenin blocks osteogenic differentiation, and allows MSCs to potentially differentiate into chondrocytes (Day et al., 2005). β-catenin activity is regulated by Wnt signaling pathway. Wnt binds to the Frizzled receptor and LRP5 co-receptor, which leads to accumulation of β-catenin in the cytoplasm. From there, it translocates to the nucleus and interacts with the transcription factor LEF/TCF. The transcription factor β-catenin activates the transcription of downstream gene by binding LEF/TCF (Huelsken and Behrens, 2002). Indeed, the transduction of MSCs with LRP5 and treatment with Wnt3a increase the mineralized bone formation in mice (Qiu et al., 2007).
The transcription factor core binding factor-1 alpha (CBF-1α) plays an important role in osteogenic differentiation of MSCs. In bone marrow-derived MSCs, hypoxia gradually increases the expression of CBF-1α, which enhances the potential of MSCs to differentiate into osteocytes (Huang et al., 2011). CBF-1α is regulated by the Notch signaling pathway, which promotes the formation of Notch intracellular domain (NICD). This event leads to activation of CBF-1α in the nucleus. NICD-overexpression in transgenic mice results in osteosclerotic bone. Conversely, the loss of Notch signaling results in age-related osteoporosis (Engin et al., 2008). TNF-α has also been found to promote osteogenic differentiation of umbilical cord derived-MSCs through NF-kB signaling pathway (Marupanthorn et al., 2015). Several studies have shown that osteogenic differentiation of MSCs can be impaired or stimulated by other transcription factors. The DLX5 transcription factor belongs to the homeoprotein family and was expressed during the formation of bone. In vitro, overexpression of DLX5 prevents the terminal differentiation of MSCs into osteocytes, which is mediated by SOX2 (Muraglia et al., 2008). The Foxc2 transcription factor, which belongs to the winged helix/ forkhead family, stimulates osteogenic differentiation of MSCs, and the Wnt signaling pathway plays an essential role in this process (Kim et al., 2009). Yes-associated protein is a transcriptional co-activator driven by its association with the TEAD family of transcription factors, and it is involved in development, growth, repair, homeostasis, and progression of multiple cancers as a transcriptional regulator (Marupanthorn et al., 2015). It has been found that osteogenic differentiation was enhanced by the activity of YAP, which act as a mechanosensitive transcriptional regulator (Choi et al., 2015).
A range of transcription factors are known to have major regulatory roles in osteogenic differentiation of MSCs including the most widely studied transcription factors CBFA-1/Runx2 and Osterix. There are some other transcription factors that have been also studied and reported to have functional roles in the differentiation of MSCs to osteocytes including HOXB7, CBF-1α, TNF-α, FOXC2, YAP, HOXA2, BMP9 and β-catenin. TWIST and HIF-1α have inhibitory effect on MSC differentiation into osteocytes through their direct or indirect interaction with Runx2.
Chondrogenic differentiation
In vitro, chondrogenic differentiation of MSCs is induced by a medium supplemented with ascorbic acid phosphate, dexamethasone, bovine serum albumin, linoleic acid, sodium pyruvate, transferrin, selenous acid, proline, L-glutamine, and TGF-β1 (Okamoto et al., 2002; Sottile et al., 2002; Suva et al., 2004). During differentiation, the morphology of MSCs changes from a fibroblast-like morphology into a round shape. Transcription factors play an important role in the regulation of the gene expression of collagen type 2, type 9, type 10, type 11, aggrecan and cartilage link protein, which are known as markers for chondrocytes (Bridgewater et al., 1998; Kou and Ikegawa, 2004; Lefebvre et al., 1997; Sekiya et al., 2000; Zhang et al., 2003). However, only a few genetic factors regulating chondrogenesis of MSCs have been identified.
The major transcription factor that have a key role in the differentiation of MSCs into chondrocytes is SRY-related high mobility group-box gene 9 (Sox9) (Augello and De Bari, 2010). Sox9 is an early transcription factor of chondrogenic differentiation and controls the expression of key genes in chondrogenesis. It controls the expression of collagen type 9 by binding to the promoter of this gene and forms trans-activating complexes with other proteins (Bridgewater et al., 1998; Wang et al., 2014). Targeting Sox9 directly by overexpression or inhibition of mRNA-145, has been shown to either decrease or increase the mRNA levels for the chondrogenic marker genes collagen type 2, type 9 type 11, and aggrecan (Yang et al., 2011a). The up-regulation miR-574-3p inhibits Sox9 and chondrogenic differentiation of MSCs (Guerit et al., 2013). Furthermore, a combination of Sox5, Sox6, and Sox9 was transfected into MSCs. The transfection resulted in a significant increase in the chondrogenic differentiation of MSCs (Park et al., 2011). TNF-α has been also found to upregulate Sox9 gene expression (Jagielski et al., 2014). Adenovirus-mediated BMP2 and Sox9 expression in mice embryonic MSCs have shown to effectively enhance chondrogenic differentiation in vitro (Liao et al., 2014).
The role of FOXO3A in chondrogenic differentiation of MSCs was investigated. Cells were transfected with miR-29a, whose direct target is FOXO3A. The overexpression of miR-29a has down-regulated the expression of FOXO3A and chondrocyte-specific markers during MSCs chondrogenic differentiation. The data revealed that down-regulation of miR-29a, and up-regulation of FOXO3A are important in the chondrogenic differentiation of MSCs (Guerit et al., 2014). Hoxa2 was found to be decreased during the chondrogenic differentiation of MSCs, and forced over-expression of Hoxa2 resulted in inhibition of MSCs differentiation toward chondrocytes lineage (Seifert et al., 2015). It was also reported that HOXD9 and HOXD13 were upregulated during chondrogenic differentiation of MSCs, and the inhibition of HOXD10, HOXD11 and HOXD13 inhibits MSCs differentiation into chondrocytes (Seifert et al., 2015).
Zinc-finger protein 145 (ZNF145) is a transcription factor that has been reported to play a role in the differentiation of MSCs into chondrocytes (Liu et al., 2007). Therefore, Liu and his research team examined the role of ZNF145 in chondrogenesis of MSCs. They found that the inhibition of ZNF145 decreased chondrogenic differentiation of MSCs, whereas overexpression of ZNF145 enhanced the expression of Sox9 and chondrogenesis (Liu et al., 2011). Smads have shown to function as regulators of chondrogenic differentiation of MSCs. Activation Smad 2 and 3 are dependent on the effect of TGF-β1 in the early stages of chondrogenesis (Zhang et al., 2015). Furumatsu et al. demonstrated that Smad3 binds the transcription factor Sox9, thereby impairing chondrogenic differentiation (Furumatsu et al., 2005). YAP, which was mentioned earlier in this paper as a regulator of osteogenic differentiation of MSCs, has been also found to have an inhibitory effect on the differentiation of MSCs to chondrocytes (Karystinou et al., 2015). Kondo and his colleagues have demonstrated that STAT3 plays a key role in the commitment of MSCs to chondrogenic lineage through the activation of STAT3 pathway by IL-6 (Kondo et al., 2015). Lui and colleagues (2014) investigated the role of Wnt11 in the chondrogenic differentiation of MSCs. Their data showed that Wnt11 overexpression stimulated the expression of chondrogenic gene regulators. In addition, the overexpression of Wnt11, in synergism with TGF-β, promoted MSCs chondrogenesis (Liu et al., 2014).
There are apparent master regulators of chondrogenic differentiation of MSCs that have been widely studied Including Sox9 and ZNF145. Other transcription factors have been also reported to have functional roles in the differentiation of MSCs to chondrocytes including FOXO3A, HOXD9, HOXD10, HOXD11, HOXD13, STAT3 and Wnt11. However, Smad3, YAP and Hoxa2 have inhibitory effect on MSC differentiation into chondrocytes through their direct or indirect interaction with Sox9. TNF-α was also reported to increase the expression of Sox9 leading to the chondrogenic differentiation of MSCs.
Adipogenic differentiation
Adipogenic differentiation of MSCs is stimulated by the incubation of MSCs in a medium containing 3-isobutyl-1-methyl-xanthine, insulin, indomethacin, triiodothyonine, Asc-2-P, basic FGF, and the glucocorticoid dexamethasone (Suva et al., 2004; Zhang et al., 2009). The differentiation of MSCs into adipocytes results in the accumulation of lipids in intracellular vacuoles (Prawitt et al., 2008). The inhibition or activation of some transcription factors is vital to the cellular commitment of MSCs to adipogenic differentiation.
To date, several transcription factors have been identified to play crucial roles in the differentiation of MSCs into adipocytes. The adipogenic-specific peroxisome proliferation-activated receptor γ (PPARγ) is one of the transcription factors that regulates the expression of genes responsible for adipogenic differentiation (Nuttall and Gimble, 2004; Zhuang et al., 2015). PPARγ has been reported to be up-regulated during the adipogenic differentiation of MSCs, and the inhibition of this transcription factor suppresses adipogenesis (Bionaz et al., 2015; Morganstein et al., 2010; Yu et al., 2012; Zou et al., 2008). PPARγ2 and PPARγ1 isoforms were reported by Wei-Hua Yu and his colleagues to play a vital role in promoting the differentiation of MSCs into adipocytes. Interestingly, the knockdown of C/EBPα inhibited PPARγ2 but not PPARγ1, suggesting that PPARγ1 plays a lesser role in adipogenic differentiation (Yu et al., 2012). The binding of PPARγ to several ligands induces the activation and repression of PPARγ. TAZ was reported to function as a co-repressor of PPARγ, therefore blocking the adipogenic differentiation (Hong et al., 2005). A recent study also showed that up-regulated expression of PPARγ2 alone or combined with CEBPB or PRDM16 promoted the adipogenic differentiation with 90% efficiency. Furthermore, another study demonstrated that myocyte enhancer factor-2 interacting transcriptional repressor plays an important role in suppressing the adipogenic differentiation of MSCs through interaction with PPARγ2 and inhibiting the activity of this transcription factor (Chen et al., 2012).
The early B cell factor EBF-1 is a member of a cascade of transcriptions that play major roles in cellular function and differentiation. Also, EBF-1 plays a crucial role in promoting the differentiation of MSCs into adipocytes and osteocytes (Hesslein et al., 2009). Gene expression analysis also indicated that EBF-1 and PPARγ2 induced two different sets of genes and both are linked to adipogenic differentiation. The author concluded that EBF-1 and PPARγ2 induce the differentiation of MSCs into adipocytes with comparable efficiency (Akerblad et al., 2005).
GATA-2, a member of the GATA family of zinc finger transcription factors, is known to control the proliferation and differentiation of hematopoietic stem cells and various cell lineages. Interestingly, it was reported that GATA-2 maintains the hematopoietic differentiation by regulating adipogenic differentiation. The suppression of this transcription factor enhanced the differentiation of MSCs into adipocytes, whereas the activation of GATA-2 suppressed the adipogenic differentiation (Okitsu et al., 2007). Similarly, the knockdown of the forkhead transcription factor (Foxa1) increases adipogenic differentiation of MSCs and increases the expression of PPARγ, C/EBPα, which are key transcription factors in adipogenesis (Fujimori and Amano, 2011). These findings indicate that GATA-2 and Foxa1 play a suppressive role in the differentiation of MSCs into adipocytes. HOXC8 was also found downregulated during the differentiation of MSCs to adipocytes, and forced over-expression of HOXC8 in MSCs inhibited the adipogenic differentiation (Seifert et al., 2015).
The TWIST family of basic helix-loop-helix transcription factors has been reported to play a regulatory role in adipogenic differentiation. It has been reported that enforced high expression of Twist-1 and Dermo-1 in MSC cultures was associated with an increase in the gene expression of adipocyte-associated markers (Isenmann et al., 2009). These findings indicate that Twist-1 and Dermo-1 play mediatory roles in the differentiation of MSCs into adipocytes (Isenmann et al., 2009). Reduced expression of miR-194 was found with concomitant increases in COUP-TFII expression during adipogenic differentiation of MSCs suggesting that COUP-TFII has a key role in this process (Jeong et al., 2014). Sox2 and Oct4 were also found to have a key regulatory effect on the differentiation of MSCs to adipocytes. The overexpression of these two transcription factors showed higher adipogenic differentiation in comparison to control (Han et al., 2014).
A group of transcription factors have been described to have major regulatory roles in adipogenic differentiation of MSCs including the most widely studied transcription factors PPARγ1, PPARγ2 and EBF-1. There are some other transcription factors that have been also reported to have functional roles in the differentiation of MSCs to adipocytes including CEBPB, PRDM16, Twist-1, Dermo-1, COUP-II, Sox2 and Oct4. However, GATA2, Foxa1 and HOXC8 have inhibitory effect on the differentiation of MSCs into adipocytes.
Myogenic differentiation
Skeletal muscle cells
In recent years, many studies have reported that MSCs possess the ability to differentiate into skeletal muscle cell lineage when treated with the demethylating agent 5-azacytidine (Jackson et al., 2007; Rowlands et al., 2008). Myogenic differentiation has also been promoted by co-culturing MSCs with skeletal myocytes, neonatal fibroblasts, and neonatal cardiomyocytes (Lee et al., 2005; Ramkisoensing et al., 2011). The inhibition or activation of some transcription factors is vital to the cellular commitment of MSCs to skeletal muscle cell differentiation. Myogenic differentiation of MSCs occurs via activation of some specific myogenic transcription factors, including paired box 3 (Pax3), MyoD, Myf-5, and myogenin (Braun and Arnold, 1996; Charytonowicz et al., 2011; Gang et al., 2008). Signals from these transcription factors lead to formation of sclerotome and dermo-myotome.
During the expression of Pax3, cells migrate through the dorsomedial lip of the dermomyotome to form the myotome and promote myogenic differentiation (Charytonowicz et al., 2011). Pax3 and Pax7 are members of the paired box family of transcription factors. They are known as master regulators of myogenic differentiation, as it contributes to early striated muscle development during skeletal muscle development and regeneration. The overexpression of Pax3 in MSCs after transduction promotes the myogenic differentiation and blocks the adipogenic, osteogenic, and chondrogenic differentiation of MSCs (Gang et al., 2008). In another study, MSCs were transfected with Pax3 and Pax7 genes and the resulting data implied that Pax3 and Pax7 transcription factors are required for commitment of MSCs to myocytes (Charytonowicz et al., 2011).
MyoD, Myf-5, and myogenin, which are members of the helix-loop-helix family transcription factors, play a key role in regulating myogenic differentiation. Interestingly, the expression of MyoD and Myf-5 does not occur within the same committed MSCs; therefore, MyoD and Myf-5 transcription factors determine different muscle cell lineages arising from different committed MSCs (Braun and Arnold, 1996). MyoD overexpression was found to inhibit Twist-1 through miR-206 induction, resulting in an increase in muscle cell differentiation (Koutalianos et al., 2015). Overexpression of MyoD1, using a vector to human-induced pluripotent stem cells, promotes these cells to undergo myogenic differentiation (Shoji et al., 2015). The overexpression of TAZ, which is known as a modulator of osteogenic and adipogenic differentiation of MSCs, was found to increase MyoD-mediated myogenic differentiation. This indicates that TAZ plays a regulatory role in the differentiation of MSCs toward myocytes (Jeong et al., 2010). Incubation of MSCs in myogenic medium significantly upregulates the expression of MyoD and myogenin, sugesting the regulatory role of the two transcription factors in the commitment of MSCs to myogenic lineages (Gang et al., 2004). Myogenic differentiation can be induced or inhibited by signaling factors. Insulin-like growth factor-II (IGF-II) induces the myogenic differentiation through the insulin-like growth factor receptor-1, which targets co-regulators of important cofactors for MyoD (Wilson and Rotwein, 2006). Tumor necrosis factor-α (TNF-α) also has a regulatory role in the differentiation of MSCs into myocytes. It down-regulates the expression of the MyoD transcription factor, which is required for myocyte development (Sitcheran et al., 2003). TNF-α inhibits myogenic differentiation through NF-κB activation and reduction of IGF-1 signaling pathway (Zhao et al., 2015). Similarly, Smad3, which belongs to receptor-regulated Smad family, suppresses myogenic differentiation through its association with myogenic transcription factors. TGF-β-activated Smad3 directly suppresses the transcriptional activity of MyoD and myogenin (Liu et al., 2001; Liu et al., 2004).
Several transcription factors have been reported to have key regulatory roles in skeletal muscle cell differentiation of MSCs including Pax3, Pax7, Myf-5 and MyoD. Few other transcription factors have been reported to have functional roles in the differentiation of MSCs to skeletal muscle cells including Myogenin, TAZ and IGF-II. However, TNF-α was found to have inhibitory effects on the differentiation of MSCs through the activation of NF-κB signaling pathway. Smad3 as well was reported to inhibit skeletal muscle differentiation of MSCs.
Cardiomyocytes
Myogenic differentiation has been promoted by co-culturing MSCs with skeletal myocytes, neonatal fibroblasts, and neonatal cardiomyocytes (Lee et al., 2005; Ramkisoensing et al., 2011). MSCs were reported to have the potential to differentiate into cardiomyocytes after two to three weeks of treatment with 5-azacytidine in a medium consisting of low-glucose DMEM supplemented with 10% FBS (Wei et al., 2011). At the molecular level, several transcription factors have been identified as regulators for cardiomyogenic differentiation of MSCs.
GATA4, which belongs to the GATA zinc finger transcription factor family, has been shown to regulate differentiation and growth of different cell types. GATA4-transduced MSCs were shown to have a higher expression of GATA4 than the control MSCs during cardiomyogenic differentiation, suggesting that the overexpression of GATA4 increases the potential of MSCs to differentiate into cardiomyocytes (Li et al., 2010). In the presence of myocytes, MSCs were able to differentiate into cardiomyocytes with a higher expression of GATA4 than the undifferentiated MSCs (Hatzistergos et al., 2010; Xu et al., 2004). Moreover, the overexpression of Nkx2.5 and GATA4 promotes differentiation of MSCs into cardiomyocytes, and Nkx2.5 and GATA4 are required for cardiomyogenic differentiation (Arminan et al., 2010; Yamada et al., 2007). During differentiation of MSCs into cardiomyocytes, the expression of Nkx2.5 and GATA4 was significantly enhanced by VEGF. Anti-VEGF antibodies also suppress the expression of cardiomyocyte markers (Song et al., 2007). The transduction of Wnt11 gene promotes differentiation of MSCs toward cardiomyocytes by up-regulating GATA4 (He et al., 2011). However, the mechanism behind the up-regulation of GATA4 by Wnt11 is still not clear.
Myocardin is a smooth muscle cell and cardiomyogenic transcription factor that contains two or more essential binding sites for serum response factor (SRF). In vitro, overexpression of myocardin gene in MSCs induced the expression of cardiomyogenic genes (van Tuyn et al., 2005). It was reported that forced myocardin expression in MSCs, after transduction of MSCs with human adenovirus vectors expressing myocardin, promotes the expression of several cardiomyogenic markers in vitro. However, myocardin-transduced MSCs did not result in complete cardiomyogenic differentiation suggesting that it only stimulates the expression of early developmental genes (Grauss et al., 2008). Thioredoxin-1 (Trx1), an antioxidant, transcription factor, and growth-factor regulator, was found to significantly enhance the differentiation of MSCs to cardiomyocytes. Genetically modified MSCs with an adenovector expressing Trx1, compared to control MSCs without adeno-vector expressing Trx1, showed significant increase in the proliferation and differentiation of MSCs to cardiomyocytes in vitro and in vivo after transplantation into rat heart tissue (Suresh et al., 2015). Moreover, Ding et al. demonstrated that genetically modified MSCs with an adenovector over-expressing Notch1 intracellular domain (NICD) showed increased differentiation capability of MSCs favoring differentiation into cardiomyocytes (Ding et al., 2015).
GATA4 and NKX2.5 are the most widely studied transcription factors that are known to have major regulatory roles in cardiomyogenic differentiation of MSCs. There are some other transcription factors that have been studied and reported to have functional roles in the differentiation of MSCs to osteocytes including myocardin, Trx1, Wnt11 and Notch1.
Smooth Muscle cells
The most effective inducer of vascular smooth muscle cell differentiation of MSCs is TGF-β. It up-regulates the genes of smooth muscle cell markers alpha smooth muscle actin (α-SMA), smooth muscle myosin heavy chain (SMMHC) and calponin (Deaton et al., 2005; Sinha et al., 2004). It has been also reported that TGF-β1 inhibits the proliferation of MSCs and promotes the avascular smooth muscle differentiation (Ross et al., 2006), and 5-azacytidine and amphotericin B treatments induce MSCs differentiation into myoblasts (Chamberlain et al., 2007). Previous studies have reported that MSCs possess the ability to differentiate into smooth muscle cell lineage (Jackson et al., 2007; Rowlands et al., 2008).
There are a few transcription factors, including myocardin, GATA6, and serum response factor (SRF), that are commonly known to assess smooth muscle cell differentiation (Ross et al., 2006). The treatment of MSCs with the MEK inhibitor up-regulated the expression of smooth muscle cell markers and induced the expression of myocardin, indicating that the inhibition of MEK signaling induces the differentiation of MSCs into smooth muscle cells (Tamama et al., 2008). Additionally, another study demonstrated that sphingosylphosphrylcholine (SPC) induces the differentiation MSCs into smooth muscle cells through a RhoA/Rho kinase-dependent mechanism. SPC was found to up-regulate the expression of the myocardin-related transcription factor (Jeon et al., 2008). It has recently been reported that GATA6, which is up-regulated by sphingosine 1-phosphate (S1P), is a novel player in smooth muscle cell differentiation (Donati et al., 2011). Moreover, TGF-β activates the transcription factors, GATA6 and SRF, during smooth muscle differentiation. This activation enhances the expression of smooth muscle marker genes α-SMA, SM22-α, SMMHC, and calponin in MSCs (Deaton et al., 2005). It was also reported that the expression of specific SMC markers were significantly increased with TGF-β supplementation when MSCs cultured on silk hydrogels compared to MSCs without TGF-β supplementation in media (Floren et al., 2016). PPAR γ was found to have an inhibitory effect on the differentiation of MSCs to myofibroblasts, which have some of the characteristics of smooth muscle cells. Transfection of MSCs with PPARγ-siRNA resulted in increased expression of alpha smooth muscle actin (α-SMA) with TGF-β treatment (Jia et al., 2015). The expression of the laminin isoform LM-521 during the differentiation process, in media containing TGF-β, was demonstrated to enhance the differentiation of MSCs to smooth muscle cells (Seeger et al., 2015). Moreover, olfactomedin 2 (Olfm2) was found to play an important role in TGF-β-induced differentiation of MSCs to smooth muscle cells, which was found to be upregulated during the differentiation. The knockdown of Olfm2 resulted in downregulation of smooth muscle cell markers expression, while the overexpression of Olfm2 increased the expression of the same markers (Shi and Chen, 2015). Olfm2 binds to serum response factor (SRF) and promotes SRF/CArG box interaction leading to increased expression of smooth muscle cell markers (Shi and Chen, 2015).
The major transcription factors that have been reported to have key roles in the differentiation of MSCs into smooth muscle cells are GATA6 and SRF. Forced expression of Olfm2 and treatment with TGF-β were found to increase the expression of smooth muscle cell markers during the differentiation of MSCs. However, PPARγ expression has inhibitory effect on the differentiation of MSCs into smooth muscle cells. There is still ongoing need for more studies to investigate the role of other transcription factors on the differentiation of MSCs to smooth muscle cells as well as their signaling pathways.
Endothelial cell differentiation
MSC therapy can be used for regeneration of the endothelial layer. Identification of cellular regulators that facilitate MSC differentiation to endothelial cells (ECs) is essential to the future of MSC therapy (Pankajakshan et al., 2013). Transcription factors could be transiently manipulated to induce differentiation. To date, there is limited published data that show the relationship between transcription factors and endothelial differentiation of MSCs. In our laboratory, we have been successful in differentiating MSCs into endothelial cells using gene based modifications. Using our protocol, we have identified Sox18 as a transcription factor that can be manipulated to enhance the differentiation of MSCs to ECs (Ikhapoh et al., 2015). We also found that the expression level of Sox18 was up regulated during the differentiation process through VEGFR-II. Additionally, we found that MSCs deficient in Sox18 maintain their undifferentiated phenotype (Ikhapoh et al., 2015). These results provided evidence that the overexpression of Sox18 in MSCs stimulates the translation of EC markers (Ikhapoh et al., 2015). Therefore, Sox18 is a critical regulator of MSC differentiation to ECs which can provide a new clinical application of MSC therapy in cardiovascular disease. The expression of HOXA7 and HOXB3 were also found to be upregulated during the differentiation of MSCs to endothelial cells, while the expression of HOXA3 and HOXB13 were significantly downregulated (Chung et al., 2009). Moreover, it was reported that HOXB5 increases the expression of vascular endothelial growth factor receptor-2 (VEGFR-II), which is a major player in the differentiation of MSCs to endothelial cells (Seifert et al., 2015). Bago et al. (2013) studied the effect of αNotch signaling on the differentiation of MSCs to endothelial cells and the formation of capillary-like structure in vitro and in vivo. Notch1 knock down by shRNA resulted in a lack of capillary-like structures formation. The expression of endothelial cell specific markers was also significantly decreased after Notch1 knock down (Bago et al., 2013). Other biomechanical and biochemical stimuli were found to induce the expression of endothelial cell markers (Ikhapoh et al., 2015b; Kim et al., 2016). Steady shear stress was reported to increase the differentiation of MSCs to endothelial cells. A fabrication of double-layered tubular scaffolds was used to mimic the structural microenvironment of blood vessels with a bioreactor system to apply fluid shear stress. MSCs that were cultured on the inner layer developed endothelial cell phenotype, and the expression of endothelial markers were significantly elevated (Kim et al., 2016). Furthermore, MSCs that were treated with angiotensin type II (ATII) and VEGF-A showed a higher expression of specific endothelial cell markers compared the MSCs that were treated with VEGF-A alone indicating the crucial role of ATII with VEGF-A in the differentiation of MSCs to endothelial cells (Ikhapoh et al., 2015b). However, the mechanism underlying the effect of ATII on the endothelial cell differentiation of MSCs is still not understood.
A group of few transcription factors have been reported to have functional effects on the differentiation of MSCs to endothelial cells including Sox18, HOXA7, HOXB3, HOXB5 and Notch1, while HOXA3 and HOXB13 were found to be downregulated during endothelial cell differentiation of MSCs. Moreover, treatment of MSCs with VEGF-A and ATII has been reported to stimulate the expression of endothelial cell markers during the differentiation of MSCs.
Conclusion
Among cell types, MSCs are a powerful candidate for regenerative medicine, and for the study of cellular differentiation. They represent an attractive cell source for transplantation because they can be isolated from different tissues. In addition to their ability to differentiate into adipocytes, chondrocytes, osteocytes, smooth muscle cells, endothelial cells, and cardiomyocytes, MSCs are capable of differentiating into additional cell lineage such as neurons and hepatocytes (Pacary et al., 2006; Talens-Visconti et al., 2006; Yuan et al., 2012). Each of these types of cells is associated with expression of a distinct set of proteins. However, there are many challenges to fully understand the mechanisms underlying the differentiation of MSCs in various lineages. These challenges include the identification of the signaling and transcription factors as well as the crosstalk between signaling pathways that promote the self-renewal and lineage differentiation in MSCs. The potential of MSCs to differentiate into a particular mesenchymal lineage relies upon up-regulation or suppression of genes specific to a lineage. During the differentiation, up-regulation or suppression of transcription factors occurs via specific signaling pathways or interaction with other transcription factors that act as co-regulators.
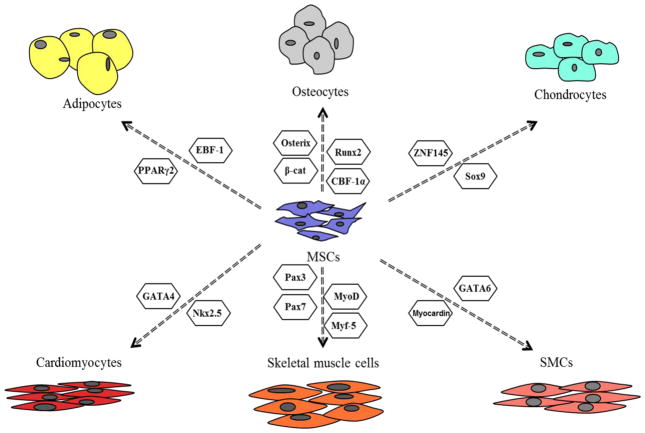
The differentiation of MSC into each of these cell lineages is promoted by specific transcription factors associated with a key cell lineage. In osteogenic differentiation, Runx2 is a key transcription factor that elevates the osteoblast differentiation and inhibits the adipogenic and chondrogenic differentiation. The expression of Runx2 is regulated by many signaling pathways such as the Wnt, BMP, and Notch signaling pathways. Chondrogenic differentiation is driven by Sox9. However, Nkx3.2 is required expression of Sox9 and suppression of osteogenic differentiation. Also, Sox9 can physically interact with and inhibit Runx2 (Augello and De Bari, 2010). This interaction can determine the commitment and fate of MSCs to differentiate into either osteocytes or chondrocytes. Adipogenic differentiation is mainly controlled by PPARγ, which cooperates with other transcription factors to promote the expression of adipogenic markers. The differentiation of MSCs into either adipocytes or osteocytes is also regulated by interactions of different transcription factors including the major players Runx2 and PPARγ, and the induction into one lineage blocks the commitment of MSCs to differentiate to the other (James, 2013). The commitment of MSCs to differentiate into either adipocytes or osteocytes is regulated through different signaling pathways including Wnt, Hedgehog, NELL-1, BMP, and IGF signaling (James, 2013). Finally, members of the helix-loop-helix transcription factor family form heterodimeric complexes with E proteins and promote myogenic differentiation. Their activities are dependent upon the interaction with MEF proteins. Other transcription factors, including GATA6, MyoD, and GATA4, function as drivers for smooth muscle cell, skeletal muscle cells, and cardiomyocytes, respectively (Figure 2).
Figure 2.
Major transcription factors that promote differentiation of mesenchymal stem cells into osteocytes, chondrocytes, adipocytes, smooth muscle cells, skeletal muscle cells, and cardiomyocytes.
There are few studies that have discussed and examined the differentiation of MSCs into fibroblasts, and the role of transcription factors in this process. Indeed, MSCs are capable of differentiating into fibroblasts-like cells in appropriate in vitro conditioned medium containing 100 ng/ml connective tissue growth factor (CTGF) and 50 mg/ml ascorbic acid (Hu et al., 2014; Tong et al., 2011). It has been also demonstrated that CTGF is sufficient to differentiate MSCs into fibroblasts, and the differentiated cells lost their ability to differentiate into other lineages (Lee et al., 2010). CTGF supplement decreases the expression of the MSC markers CD44 and STRO-1, and increases the expression of a fibroblast specific marker, fibroblast specific protein 1 (FSP-1) (Tong et al., 2011). The treated MSCs showed significant increases in the expression of type I collagen and tenascin-C (Lee et al., 2006). The fibroblast-like cells, differentiated from MSCs, were able to produce extracellular matrix proteins (Hu et al., 2014). Although the differentiation of MSCs into fibroblasts has many implications in the tissue engineering including tendons and ligaments repair (Lee et al., 2010), limited studies warrants further investigation. Additional studies are required to identify the signaling factors, master transcription factors, and signaling pathways that mediate fibroblastic differentiation of MSCs.
Several approaches have been tested and employed to introduce transcription factor genes into MSCs for use in regenerative therapy. Transfection methods have shown low efficiency in delivering DNA plasmid into MSCs and high mortality rates. Conversely, viral transduction methods have shown the ability to deliver DNA plasmid with high efficiency and low toxicity. However, the safety concern associated with viral transduction is a controversial topic. To date, numerous studies have shown that the delivery of transcription factors into MSCs increases and maintains the potential of these cells to differentiate into intended cell lineage. Therefore, forced expression of a single transcription factor in MSCs can promote trans-differentiation into several cell lineages, allowing for the treatment of many diseases. Further studies are required in order to avoid any activation of the innate tendency of these cells to differentiate into other (unintended) mesenchymal tissues.
The identification of specific transcription factors, receptors, and signaling molecules during differentiation is important in the understanding of the link between intracellular and extracellular signaling networks. Additional studies are warranted in order to deepen the understanding of the regulation of MSC differentiation by transcription factors at distinct levels during progression of a differentiation pathway. Since transcription factors naturally act as master regulators of cellular processes, they are expected to be excellent candidates for controlling differentiation of MSCs into any cell lineage, and transcription factor-based technologies are likely to be a prominent part of the next generation of MSC-based therapy.
Acknowledgments
The authors thank Dane Marvin for providing writing assistance and proof reading the article. This work was supported by research grants R01 HL116042, R01 HL112597, and R01 HL120659 to DK Agrawal from the National Heart, Lung and Blood Institute, National Institutes of Health, USA. The content of this review article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Sami Almalki is financially supported by Majmaah University in Saudi Arabia to pursue PhD program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akerblad P, Mansson R, Lagergren A, Westerlund S, Basta B, Lind U, Thelin A, Gisler R, Liberg D, Nelander S, Bamberg K, Sigvardsson M. Gene expression analysis suggests that EBF-1 and PPARgamma2 induce adipogenesis of NIH-3T3 cells with similar efficiency and kinetics. Physiol Genomics. 2005;23:206–216. doi: 10.1152/physiolgenomics.00015.2005. [DOI] [PubMed] [Google Scholar]
- Arminan A, Gandia C, Garcia-Verdugo JM, Lledo E, Mullor JL, Montero JA, Sepulveda P. Cardiac transcription factors driven lineage-specification of adult stem cells. Journal of cardiovascular translational research. 2010;3:61–65. doi: 10.1007/s12265-009-9144-3. [DOI] [PubMed] [Google Scholar]
- Augello A, De Bari C. The regulation of differentiation in mesenchymal stem cells. Human gene therapy. 2010;21:1226–1238. doi: 10.1089/hum.2010.173. [DOI] [PubMed] [Google Scholar]
- Bago JR, Alieva M, Soler C, Rubio N, Blanco J. Endothelial differentiation of adipose tissue-derived mesenchymal stromal cells in glioma tumors: implications for cell-based therapy. Molecular therapy : the journal of the American Society of Gene Therapy. 2013;21:1758–1766. doi: 10.1038/mt.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay A, Tsuji K, Cox K, Harfe BD, Rosen V, Tabin CJ. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2:e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bionaz M, Monaco E, Wheeler MB. Transcription Adaptation during In Vitro Adipogenesis and Osteogenesis of Porcine Mesenchymal Stem Cells: Dynamics of Pathways, Biological Processes, Up-Stream Regulators, and Gene Networks. PloS one. 2015;10:e0137644. doi: 10.1371/journal.pone.0137644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Arnold HH. Myf-5 and myoD genes are activated in distinct mesenchymal stem cells and determine different skeletal muscle cell lineages. EMBO J. 1996;15:310–318. [PMC free article] [PubMed] [Google Scholar]
- Bridgewater LC, Lefebvre V, de Crombrugghe B. Chondrocyte-specific enhancer elements in the Col11a2 gene resemble the Col2a1 tissue-specific enhancer. J Biol Chem. 1998;273:14998–15006. doi: 10.1074/jbc.273.24.14998. [DOI] [PubMed] [Google Scholar]
- Chamberlain G, Fox J, et al. Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells. 2007;25(11):2739–2749. doi: 10.1634/stemcells.2007-0197. [DOI] [PubMed] [Google Scholar]
- Charytonowicz E, Matushansky I, Castillo-Martin M, Hricik T, Cordon-Cardo C, Ziman M. Alternate PAX3 and PAX7 C-terminal isoforms in myogenic differentiation and sarcomagenesis. Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2011;13:194–203. doi: 10.1007/s12094-011-0640-y. [DOI] [PubMed] [Google Scholar]
- Chen SL, Fang WW, Ye F, Liu YH, Qian J, Shan SJ, Zhang JJ, Chunhua RZ, Liao LM, Lin S, Sun JP. Effect on left ventricular function of intracoronary transplantation of autologous bone marrow mesenchymal stem cell in patients with acute myocardial infarction. Am J Cardiol. 2004;94:92–95. doi: 10.1016/j.amjcard.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Chen YH, Yeh FL, Yeh SP, Ma HT, Hung SC, Hung MC, Li LY. Myocyte enhancer factor-2 interacting transcriptional repressor (MITR) is a switch that promotes osteogenesis and inhibits adipogenesis of mesenchymal stem cells by inactivating peroxisome proliferator-activated receptor gamma-2. J Biol Chem. 2012;286:10671–10680. doi: 10.1074/jbc.M110.199612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CK, Xu Y, Wang B, Zhu M, Zhang L, Bian L. Substrate Coupling Strength of Integrin-Binding Ligands Modulates Adhesion, Spreading, and Differentiation of Human Mesenchymal Stem Cells. Nano letters. 2015 doi: 10.1021/acs.nanolett.5b02323. [DOI] [PubMed] [Google Scholar]
- Chung N, Jee BK, Chae SW, Jeon YW, Lee KH, Rha HK. HOX gene analysis of endothelial cell differentiation in human bone marrow-derived mesenchymal stem cells. Molecular biology reports. 2009;36:227–235. doi: 10.1007/s11033-007-9171-6. [DOI] [PubMed] [Google Scholar]
- Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- De Coppi P, Bartsch G, Jr, Siddiqui MM, Xu T, Santos CC, Perin L, Mostoslavsky G, Serre AC, Snyder EY, Yoo JJ, Furth ME, Soker S, Atala A. Isolation of amniotic stem cell lines with potential for therapy. Nat Biotechnol. 2007;25:100–106. doi: 10.1038/nbt1274. [DOI] [PubMed] [Google Scholar]
- Deaton RA, Su C, Valencia TG, Grant SR. Transforming growth factor-beta1-induced expression of smooth muscle marker genes involves activation of PKN and p38 MAPK. J Biol Chem. 2005;280:31172–31181. doi: 10.1074/jbc.M504774200. [DOI] [PubMed] [Google Scholar]
- Ding R, Jiang X, Ha Y, Wang Z, Guo J, Jiang H, Zheng S, Shen Z, Jie W. Activation of Notch1 signalling promotes multi-lineage differentiation of c-Kit(POS)/NKX2.5(POS) bone marrow stem cells: implication in stem cell translational medicine. Stem cell research & therapy. 2015;6:91. doi: 10.1186/s13287-015-0085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati C, Marseglia G, Magi A, Serrati S, Cencetti F, Bernacchioni C, Nannetti G, Benelli M, Brunelli S, Torricelli F, Cossu G, Bruni P. Sphingosine 1-phosphate induces differentiation of mesoangioblasts towards smooth muscle. A role for GATA6. PloS one. 2011;6:e20389. doi: 10.1371/journal.pone.0020389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin F, Yao Z, Yang T, Zhou G, Bertin T, Jiang MM, Chen Y, Wang L, Zheng H, Sutton RE, Boyce BF, Lee B. Dimorphic effects of Notch signaling in bone homeostasis. Nat Med. 2008;14:299–305. doi: 10.1038/nm1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floren M, Bonani W, Dharmarajan A, Motta A, Migliaresi C, Tan W. Human mesenchymal stem cells cultured on silk hydrogels with variable stiffness and growth factor differentiate into mature smooth muscle cell phenotype. Acta biomaterialia. 2016;31:156–166. doi: 10.1016/j.actbio.2015.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouillard L, Chapel A, Bories D, Bouchet S, Costa JM, Rouard H, Herve P, Gourmelon P, Thierry D, Lopez M, Gorin NC. Infusion of allogeneic-related HLA mismatched mesenchymal stem cells for the treatment of incomplete engraftment following autologous haematopoietic stem cell transplantation. Leukemia. 2007;21:568–570. doi: 10.1038/sj.leu.2404550. [DOI] [PubMed] [Google Scholar]
- Friedenstein AJ, Chailakhyan RK, Gerasimov UV. Bone marrow osteogenic stem cells: in vitro cultivation and transplantation in diffusion chambers. Cell Tissue Kinet. 1987;20:263–272. doi: 10.1111/j.1365-2184.1987.tb01309.x. [DOI] [PubMed] [Google Scholar]
- Fujii M, Takeda K, Imamura T, Aoki H, Sampath TK, Enomoto S, Kawabata M, Kato M, Ichijo H, Miyazono K. Roles of bone morphogenetic protein type I receptors and Smad proteins in osteoblast and chondroblast differentiation. Mol Biol Cell. 1999;10:3801–3813. doi: 10.1091/mbc.10.11.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori K, Amano F. Forkhead transcription factor Foxa1 is a novel target gene of C/EBPbeta and suppresses the early phase of adipogenesis. Gene. 2011;473:150–156. doi: 10.1016/j.gene.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Furumatsu T, Tsuda M, Yoshida K, Taniguchi N, Ito T, Hashimoto M, Asahara H. Sox9 and p300 cooperatively regulate chromatin-mediated transcription. J Biol Chem. 2005;280:35203–35208. doi: 10.1074/jbc.M502409200. [DOI] [PubMed] [Google Scholar]
- Gang EJ, Bosnakovski D, Simsek T, To K, Perlingeiro RC. Pax3 activation promotes the differentiation of mesenchymal stem cells toward the myogenic lineage. Exp Cell Res. 2008;314:1721–1733. doi: 10.1016/j.yexcr.2008.02.016. [DOI] [PubMed] [Google Scholar]
- Gang EJ, Jeong JA, Hong SH, Hwang SH, Kim SW, Yang IH, Ahn C, Han H, Kim H. Skeletal myogenic differentiation of mesenchymal stem cells isolated from human umbilical cord blood. Stem Cells. 2004;22:617–624. doi: 10.1634/stemcells.22-4-617. [DOI] [PubMed] [Google Scholar]
- Gao RT, Zhan LP, Meng C, Zhang N, Chang SM, Yao R, Li C. Homeobox B7 promotes the osteogenic differentiation potential of mesenchymal stem cells by activating RUNX2 and transcript of BSP. International journal of clinical and experimental medicine. 2015;8:10459–10470. [PMC free article] [PubMed] [Google Scholar]
- Grauss RW, van Tuyn J, Steendijk P, Winter EM, Pijnappels DA, Hogers B, Gittenberger-De Groot AC, van der Geest R, van der Laarse A, de Vries AA, Schalij MJ, Atsma DE. Forced myocardin expression enhances the therapeutic effect of human mesenchymal stem cells after transplantation in ischemic mouse hearts. Stem Cells. 2008;26:1083–1093. doi: 10.1634/stemcells.2007-0523. [DOI] [PubMed] [Google Scholar]
- Guan K, Nayernia K, Maier LS, Wagner S, Dressel R, Lee JH, Nolte J, Wolf F, Li M, Engel W, Hasenfuss G. Pluripotency of spermatogonial stem cells from adult mouse testis. Nature. 2006;440:1199–1203. doi: 10.1038/nature04697. [DOI] [PubMed] [Google Scholar]
- Guerit D, Brondello JM, Chuchana P, Philipot D, Toupet K, Bony C, Jorgensen C, Noel D. FOXO3A regulation by miRNA-29a Controls chondrogenic differentiation of mesenchymal stem cells and cartilage formation. Stem cells and development. 2014;23:1195–1205. doi: 10.1089/scd.2013.0463. [DOI] [PubMed] [Google Scholar]
- Guerit D, Philipot D, Chuchana P, Toupet K, Brondello JM, Mathieu M, Jorgensen C, Noel D. Sox9-regulated miRNA-574-3p inhibits chondrogenic differentiation of mesenchymal stem cells. PloS one. 2013;8:e62582. doi: 10.1371/journal.pone.0062582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SM, Han SH, Coh YR, Jang G, Chan Ra J, Kang SK, Lee HW, Youn HY. Enhanced proliferation and differentiation of Oct4- and Sox2-overexpressing human adipose tissue mesenchymal stem cells. Experimental & molecular medicine. 2014;46:e101. doi: 10.1038/emm.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzistergos KE, Quevedo H, Oskouei BN, Hu Q, Feigenbaum GS, Margitich IS, Mazhari R, Boyle AJ, Zambrano JP, Rodriguez JE, Dulce R, Pattany PM, Valdes D, Revilla C, Heldman AW, McNiece I, Hare JM. Bone marrow mesenchymal stem cells stimulate cardiac stem cell proliferation and differentiation. Circulation research. 2010;107:913–922. doi: 10.1161/CIRCRESAHA.110.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynesworth SE, Baber MA, Caplan AI. Cell surface antigens on human marrow-derived mesenchymal cells are detected by monoclonal antibodies. Bone. 1992;13:69–80. doi: 10.1016/8756-3282(92)90363-2. [DOI] [PubMed] [Google Scholar]
- He Z, Li H, Zuo S, Pasha Z, Wang Y, Yang Y, Jiang W, Ashraf M, Xu M. Transduction of Wnt11 promotes mesenchymal stem cell transdifferentiation into cardiac phenotypes. Stem cells and development. 2011;20:1771–1778. doi: 10.1089/scd.2010.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesslein DG, Fretz JA, Xi Y, Nelson T, Zhou S, Lorenzo JA, Schatz DG, Horowitz MC. Ebf1-dependent control of the osteoblast and adipocyte lineages. Bone. 2009;44:537–546. doi: 10.1016/j.bone.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp PA, Hopkins N, Yaffe MB. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- Hu R, Ling W, Xu W, Han D. Fibroblast-like cells differentiated from adipose-derived mesenchymal stem cells for vocal fold wound healing. PloS one. 2014;9:e92676. doi: 10.1371/journal.pone.0092676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Deng F, Wang L, Xiang XR, Zhou WW, Hu N, Xu L. Hypoxia induces osteogenesis-related activities and expression of core binding factor alpha1 in mesenchymal stem cells. The Tohoku journal of experimental medicine. 2011;224:7–12. doi: 10.1620/tjem.224.7. [DOI] [PubMed] [Google Scholar]
- Huelsken J, Behrens J. The Wnt signalling pathway. J Cell Sci. 2002;115:3977–3978. doi: 10.1242/jcs.00089. [DOI] [PubMed] [Google Scholar]
- Ikhapoh IA, Pelham CJ, Agrawal DK. Sry-type HMG box 18 contributes to the differentiation of bone marrow-derived mesenchymal stem cells to endothelial cells. Differentiation; research in biological diversity. 2015a;89:87–96. doi: 10.1016/j.diff.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikhapoh IA, Pelham CJ, Agrawal DK. Synergistic effect of angiotensin II on vascular endothelial growth factor-A-mediated differentiation of bone marrow-derived mesenchymal stem cells into endothelial cells. Stem cell research & therapy. 2015b;6:4. doi: 10.1186/scrt538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- in ’t Anker PS, Noort WA, Scherjon SA, Kleijburg-van der Keur C, Kruisselbrink AB, van Bezooijen RL, Beekhuizen W, Willemze R, Kanhai HH, Fibbe WE. Mesenchymal stem cells in human second-trimester bone marrow, liver, lung, and spleen exhibit a similar immunophenotype but a heterogeneous multilineage differentiation potential. Haematologica. 2003;88:845–852. [PubMed] [Google Scholar]
- Isenmann S, Arthur A, Zannettino AC, Turner JL, Shi S, Glackin CA, Gronthos S. TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment. Stem Cells. 2009;27:2457–2468. doi: 10.1002/stem.181. [DOI] [PubMed] [Google Scholar]
- Jackson L, Jones DR, Scotting P, Sottile V. Adult mesenchymal stem cells: differentiation potential and therapeutic applications. Journal of postgraduate medicine. 2007;53:121–127. doi: 10.4103/0022-3859.32215. [DOI] [PubMed] [Google Scholar]
- Jagielski M, Wolf J, Marzahn U, Volker A, Lemke M, Meier C, Ertel W, Godkin O, Arens S, Schulze-Tanzil G. The influence of IL-10 and TNFalpha on chondrogenesis of human mesenchymal stromal cells in three-dimensional cultures. International journal of molecular sciences. 2014;15:15821–15844. doi: 10.3390/ijms150915821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AW. Review of Signaling Pathways Governing MSC Osteogenic and Adipogenic Differentiation. Scientifica. 2013;2013:684736. doi: 10.1155/2013/684736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed A, Bae JS, Afzal F, Gutierrez S, Pratap J, Zaidi SK, Lou Y, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Structural coupling of Smad and Runx2 for execution of the BMP2 osteogenic signal. J Biol Chem. 2008;283:8412–8422. doi: 10.1074/jbc.M705578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon ES, Park WS, Lee MJ, Kim YM, Han J, Kim JH. A Rho kinase/myocardin-related transcription factor-A-dependent mechanism underlies the sphingosylphosphorylcholine-induced differentiation of mesenchymal stem cells into contractile smooth muscle cells. Circulation research. 2008;103:635–642. doi: 10.1161/CIRCRESAHA.108.180885. [DOI] [PubMed] [Google Scholar]
- Jeong BC, Kang IH, Hwang YC, Kim SH, Koh JT. MicroRNA-194 reciprocally stimulates osteogenesis and inhibits adipogenesis via regulating COUP-TFII expression. Cell death & disease. 2014;5:e1532. doi: 10.1038/cddis.2014.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Bae S, An SY, Byun MR, Hwang JH, Yaffe MB, Hong JH, Hwang ES. TAZ as a novel enhancer of MyoD-mediated myogenic differentiation. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24:3310–3320. doi: 10.1096/fj.09-151324. [DOI] [PubMed] [Google Scholar]
- Jia S, Liu X, Li W, Xie J, Yang L, Li L. Peroxisome Proliferator-Activated Receptor Gamma Negatively Regulates the Differentiation of Bone Marrow-Derived Mesenchymal Stem Cells Toward Myofibroblasts in Liver Fibrogenesis. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2015;37:2085–2100. doi: 10.1159/000438567. [DOI] [PubMed] [Google Scholar]
- Kang S, Bennett CN, Gerin I, Rapp LA, Hankenson KD, Macdougald OA. Wnt signaling stimulates osteoblastogenesis of mesenchymal precursors by suppressing CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma. J Biol Chem. 2007;282:14515–14524. doi: 10.1074/jbc.M700030200. [DOI] [PubMed] [Google Scholar]
- Karystinou A, Roelofs AJ, Neve A, Cantatore FP, Wackerhage H, De Bari C. Yes-associated protein (YAP) is a negative regulator of chondrogenesis in mesenchymal stem cells. Arthritis research & therapy. 2015;17:147. doi: 10.1186/s13075-015-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Heo SJ, Kang YG, Shin JW, Park SH. Shear stress and circumferential stretch by pulsatile flow direct vascular endothelial lineage commitment of mesenchymal stem cells in engineered blood vessels. Journal of materials science Materials in medicine. 2016;27:60. doi: 10.1007/s10856-016-5670-0. [DOI] [PubMed] [Google Scholar]
- Kim SH, Cho KW, Choi HS, Park SJ, Rhee Y, Jung HS, Lim SK. The forkhead transcription factor Foxc2 stimulates osteoblast differentiation. Biochemical and biophysical research communications. 2009;386:532–536. doi: 10.1016/j.bbrc.2009.06.071. [DOI] [PubMed] [Google Scholar]
- Komori T. Regulation of osteoblast differentiation by transcription factors. Journal of cellular biochemistry. 2006;99:1233–1239. doi: 10.1002/jcb.20958. [DOI] [PubMed] [Google Scholar]
- Kondo M, Yamaoka K, Sakata K, Sonomoto K, Lin L, Nakano K, Tanaka Y. Contribution of the Interleukin-6/STAT-3 Signaling Pathway to Chondrogenic Differentiation of Human Mesenchymal Stem Cells. Arthritis Rheumatol. 2015;67:1250–1260. doi: 10.1002/art.39036. [DOI] [PubMed] [Google Scholar]
- Kou I, Ikegawa S. SOX9-dependent and -independent transcriptional regulation of human cartilage link protein. J Biol Chem. 2004;279:50942–50948. doi: 10.1074/jbc.M406786200. [DOI] [PubMed] [Google Scholar]
- Koutalianos D, Koutsoulidou A, Mastroyiannopoulos NP, Furling D, Phylactou LA. MyoD transcription factor induces myogenesis by inhibiting Twist-1 through miR-206. J Cell Sci. 2015 doi: 10.1242/jcs.172288. [DOI] [PubMed] [Google Scholar]
- Kruse C, Kajahn J, Petschnik AE, Maass A, Klink E, Rapoport DH, Wedel T. Adult pancreatic stem/progenitor cells spontaneously differentiate in vitro into multiple cell lineages and form teratoma-like structures. Annals of anatomy = Anatomischer Anzeiger : official organ of the Anatomische Gesellschaft. 2006;188:503–517. doi: 10.1016/j.aanat.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Lampert FM, Kutscher C, Stark GB, Finkenzeller G. Overexpression of Hif-1alpha in Mesenchymal Stem Cells Affects Cell-Autonomous Angiogenic and Osteogenic Parameters. Journal of cellular biochemistry. 2015 doi: 10.1002/jcb.25361. [DOI] [PubMed] [Google Scholar]
- Le Blanc K, Ringden O. Immunobiology of human mesenchymal stem cells and future use in hematopoietic stem cell transplantation. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2005;11:321–334. doi: 10.1016/j.bbmt.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Lee CH, Moioli EK, Mao JJ. Fibroblastic differentiation of human mesenchymal stem cells using connective tissue growth factor. Conference proceedings : ... Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Annual Conference. 2006;1:775–778. doi: 10.1109/IEMBS.2006.259866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Shah B, Moioli EK, Mao JJ. CTGF directs fibroblast differentiation from human mesenchymal stem/stromal cells and defines connective tissue healing in a rodent injury model. The Journal of clinical investigation. 2010;120:3340–3349. doi: 10.1172/JCI43230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kosinski PA, Kemp DM. Contribution of human bone marrow stem cells to individual skeletal myotubes followed by myogenic gene activation. Exp Cell Res. 2005;307:174–182. doi: 10.1016/j.yexcr.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Molecular and cellular biology. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zuo S, He Z, Yang Y, Pasha Z, Wang Y, Xu M. Paracrine factors released by GATA-4 overexpressed mesenchymal stem cells increase angiogenesis and cell survival. American journal of physiology. Heart and circulatory physiology. 2010;299:H1772–1781. doi: 10.1152/ajpheart.00557.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Zhou N, Lin L, Yi S, Fan T, Zhao C, Hu N, Liang X, Si W, Huang W. Co-expression of BMP2 and Sox9 promotes chondrogenic differentiation of mesenchymal stem cells in vitro. Nan fang yi ke da xue xue bao = Journal of Southern Medical University. 2014;34:317–322. [PubMed] [Google Scholar]
- Liu D, Black BL, Derynck R. TGF-beta inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes & development. 2001;15:2950–2966. doi: 10.1101/gad.925901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Kang JS, Derynck R. TGF-beta-activated Smad3 represses MEF2-dependent transcription in myogenic differentiation. EMBO J. 2004;23:1557–1566. doi: 10.1038/sj.emboj.7600179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Zhang E, Yang M, Lu L. Overexpression of Wnt11 promotes chondrogenic differentiation of bone marrow-derived mesenchymal stem cells in synergism with TGF-beta. Molecular and cellular biochemistry. 2014;390:123–131. doi: 10.1007/s11010-014-1963-0. [DOI] [PubMed] [Google Scholar]
- Liu TM, Guo XM, Tan HS, Hui JH, Lim B, Lee EH. Zinc-finger protein 145, acting as an upstream regulator of SOX9, improves the differentiation potential of human mesenchymal stem cells for cartilage regeneration and repair. Arthritis and rheumatism. 2011;63:2711–2720. doi: 10.1002/art.30430. [DOI] [PubMed] [Google Scholar]
- Liu TM, Martina M, Hutmacher DW, Hui JH, Lee EH, Lim B. Identification of common pathways mediating differentiation of bone marrow- and adipose tissue-derived human mesenchymal stem cells into three mesenchymal lineages. Stem Cells. 2007;25:750–760. doi: 10.1634/stemcells.2006-0394. [DOI] [PubMed] [Google Scholar]
- Lodie TA, Blickarz CE, Devarakonda TJ, He C, Dash AB, Clarke J, Gleneck K, Shihabuddin L, Tubo R. Systematic analysis of reportedly distinct populations of multipotent bone marrow-derived stem cells reveals a lack of distinction. Tissue engineering. 2002;8:739–751. doi: 10.1089/10763270260424105. [DOI] [PubMed] [Google Scholar]
- Marupanthorn K, Tantrawatpan C, Tantikanlayaporn D, Kheolamai P, Manochantr S. The Effects of TNF-alpha on Osteogenic Differentiation of Umbilical Cord Derived Mesenchymal Stem Cells. Journal of the Medical Association of Thailand = Chotmaihet thangphaet. 2015;98(Suppl 3):S34–40. [PubMed] [Google Scholar]
- Meng X, Ichim TE, Zhong J, Rogers A, Yin Z, Jackson J, Wang H, Ge W, Bogin V, Chan KW, Thebaud B, Riordan NH. Endometrial regenerative cells: a novel stem cell population. Journal of translational medicine. 2007;5:57. doi: 10.1186/1479-5876-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morganstein DL, Wu P, Mane MR, Fisk NM, White R, Parker MG. Human fetal mesenchymal stem cells differentiate into brown and white adipocytes: a role for ERRalpha in human UCP1 expression. Cell research. 2010;20:434–444. doi: 10.1038/cr.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraglia A, Perera M, Verardo S, Liu Y, Cancedda R, Quarto R, Corte G. DLX5 overexpression impairs osteogenic differentiation of human bone marrow stromal cells. European journal of cell biology. 2008;87:751–761. doi: 10.1016/j.ejcb.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Nuttall ME, Gimble JM. Controlling the balance between osteoblastogenesis and adipogenesis and the consequent therapeutic implications. Current opinion in pharmacology. 2004;4:290–294. doi: 10.1016/j.coph.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Okamoto T, Aoyama T, Nakayama T, Nakamata T, Hosaka T, Nishijo K, Nakamura T, Kiyono T, Toguchida J. Clonal heterogeneity in differentiation potential of immortalized human mesenchymal stem cells. Biochemical and biophysical research communications. 2002;295:354–361. doi: 10.1016/s0006-291x(02)00661-7. [DOI] [PubMed] [Google Scholar]
- Okitsu Y, Takahashi S, Minegishi N, Kameoka J, Kaku M, Yamamoto M, Sasaki T, Harigae H. Regulation of adipocyte differentiation of bone marrow stromal cells by transcription factor GATA-2. Biochemical and biophysical research communications. 2007;364:383–387. doi: 10.1016/j.bbrc.2007.10.031. [DOI] [PubMed] [Google Scholar]
- Pacary E, Legros H, Valable S, Duchatelle P, Lecocq M, Petit E, Nicole O, Bernaudin M. Synergistic effects of CoCl(2) and ROCK inhibition on mesenchymal stem cell differentiation into neuron-like cells. J Cell Sci. 2006;119:2667–2678. doi: 10.1242/jcs.03004. [DOI] [PubMed] [Google Scholar]
- Pankajakshan D, Kansal V, Agrawal DK. In vitro differentiation of bone marrow derived porcine mesenchymal stem cells to endothelial cells. Journal of tissue engineering and regenerative medicine. 2013;7:911–920. doi: 10.1002/term.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Yang HN, Woo DG, Jeon SY, Do HJ, Lim HY, Kim JH, Park KH. Chondrogenesis of human mesenchymal stem cells mediated by the combination of SOX trio SOX5, 6, and 9 genes complexed with PEI-modified PLGA nanoparticles. Biomaterials. 2011;32:3679–3688. doi: 10.1016/j.biomaterials.2011.01.063. [DOI] [PubMed] [Google Scholar]
- Pierdomenico L, Bonsi L, Calvitti M, Rondelli D, Arpinati M, Chirumbolo G, Becchetti E, Marchionni C, Alviano F, Fossati V, Staffolani N, Franchina M, Grossi A, Bagnara GP. Multipotent mesenchymal stem cells with immunosuppressive activity can be easily isolated from dental pulp. Transplantation. 2005;80:836–842. doi: 10.1097/01.tp.0000173794.72151.88. [DOI] [PubMed] [Google Scholar]
- Prawitt J, Niemeier A, Kassem M, Beisiegel U, Heeren J. Characterization of lipid metabolism in insulin-sensitive adipocytes differentiated from immortalized human mesenchymal stem cells. Exp Cell Res. 2008;314:814–824. doi: 10.1016/j.yexcr.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Qiu W, Andersen TE, Bollerslev J, Mandrup S, Abdallah BM, Kassem M. Patients with high bone mass phenotype exhibit enhanced osteoblast differentiation and inhibition of adipogenesis of human mesenchymal stem cells. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2007;22:1720–1731. doi: 10.1359/jbmr.070721. [DOI] [PubMed] [Google Scholar]
- Ramkisoensing AA, Pijnappels DA, Askar SF, Passier R, Swildens J, Goumans MJ, Schutte CI, de Vries AA, Scherjon S, Mummery CL, Schalij MJ, Atsma DE. Human embryonic and fetal mesenchymal stem cells differentiate toward three different cardiac lineages in contrast to their adult counterparts. PloS one. 2011;6:e24164. doi: 10.1371/journal.pone.0024164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringe J, Leinhase I, Stich S, Loch A, Neumann K, Haisch A, Haupl T, Manz R, Kaps C, Sittinger M. Human mastoid periosteum-derived stem cells: promising candidates for skeletal tissue engineering. Journal of tissue engineering and regenerative medicine. 2008;2:136–146. doi: 10.1002/term.75. [DOI] [PubMed] [Google Scholar]
- Ripa RS, Haack-Sorensen M, Wang Y, Jorgensen E, Mortensen S, Bindslev L, Friis T, Kastrup J. Bone marrow derived mesenchymal cell mobilization by granulocyte-colony stimulating factor after acute myocardial infarction: results from the Stem Cells in Myocardial Infarction (STEMMI) trial. Circulation. 2007;116:I24–30. doi: 10.1161/CIRCULATIONAHA.106.678649. [DOI] [PubMed] [Google Scholar]
- Ross JJ, Hong Z, Willenbring B, Zeng L, Isenberg B, Lee EH, Reyes M, Keirstead SA, Weir EK, Tranquillo RT, Verfaillie CM. Cytokine-induced differentiation of multipotent adult progenitor cells into functional smooth muscle cells. The Journal of clinical investigation. 2006;116:3139–3149. doi: 10.1172/JCI28184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlands AS, George PA, Cooper-White JJ. Directing osteogenic and myogenic differentiation of MSCs: interplay of stiffness and adhesive ligand presentation. American journal of physiology. Cell physiology. 2008;295:C1037–1044. doi: 10.1152/ajpcell.67.2008. [DOI] [PubMed] [Google Scholar]
- Sabatini F, Petecchia L, Tavian M, Jodon de Villeroche V, Rossi GA, Brouty-Boye D. Human bronchial fibroblasts exhibit a mesenchymal stem cell phenotype and multilineage differentiating potentialities. Laboratory investigation; a journal of technical methods and pathology. 2005;85:962–971. doi: 10.1038/labinvest.3700300. [DOI] [PubMed] [Google Scholar]
- Seeger T, Hart M, Patarroyo M, Rolauffs B, Aicher WK, Klein G. Mesenchymal Stromal Cells for Sphincter Regeneration: Role of Laminin Isoforms upon Myogenic Differentiation. PloS one. 2015;10:e0137419. doi: 10.1371/journal.pone.0137419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert A, Werheid DF, Knapp SM, Tobiasch E. Role of Hox genes in stem cell differentiation. World journal of stem cells. 2015;7:583–595. doi: 10.4252/wjsc.v7.i3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya I, Larson BL, Smith JR, Pochampally R, Cui JG, Prockop DJ. Expansion of human adult stem cells from bone marrow stroma: conditions that maximize the yields of early progenitors and evaluate their quality. Stem Cells. 2002;20:530–541. doi: 10.1634/stemcells.20-6-530. [DOI] [PubMed] [Google Scholar]
- Sekiya I, Tsuji K, Koopman P, Watanabe H, Yamada Y, Shinomiya K, Nifuji A, Noda M. SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. J Biol Chem. 2000;275:10738–10744. doi: 10.1074/jbc.275.15.10738. [DOI] [PubMed] [Google Scholar]
- Sellheyer K, Krahl D. Cutaneous mesenchymal stem cells: status of current knowledge, implications for dermatopathology. Journal of cutaneous pathology. 2010;37:624–634. doi: 10.1111/j.1600-0560.2009.01477.x. [DOI] [PubMed] [Google Scholar]
- Shi N, Chen SY. From nerve to blood vessel: a new role of Olfm2 in smooth muscle differentiation from human embryonic stem cell-derived mesenchymal cells. Journal of biomedical research. 2015;29:261–263. doi: 10.7555/JBR.29.20150027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji E, Woltjen K, Sakurai H. Directed Myogenic Differentiation of Human Induced Pluripotent Stem Cells. Methods Mol Biol. 2015 doi: 10.1007/7651_2015_257. [DOI] [PubMed] [Google Scholar]
- Sinha S, Hoofnagle MH, Kingston PA, McCanna ME, Owens GK. Transforming growth factor-beta1 signaling contributes to development of smooth muscle cells from embryonic stem cells. American journal of physiology. Cell physiology. 2004;287:C1560–1568. doi: 10.1152/ajpcell.00221.2004. [DOI] [PubMed] [Google Scholar]
- Sitcheran R, Cogswell PC, Baldwin AS., Jr NF-kappaB mediates inhibition of mesenchymal cell differentiation through a posttranscriptional gene silencing mechanism. Genes & development. 2003;17:2368–2373. doi: 10.1101/gad.1114503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Gehmert S, Sadat S, Pinkernell K, Bai X, Matthias N, Alt E. VEGF is critical for spontaneous differentiation of stem cells into cardiomyocytes. Biochemical and biophysical research communications. 2007;354:999–1003. doi: 10.1016/j.bbrc.2007.01.095. [DOI] [PubMed] [Google Scholar]
- Sottile V, Halleux C, Bassilana F, Keller H, Seuwen K. Stem cell characteristics of human trabecular bone-derived cells. Bone. 2002;30:699–704. doi: 10.1016/s8756-3282(02)00674-9. [DOI] [PubMed] [Google Scholar]
- Suresh SC, Selvaraju V, Thirunavukkarasu M, Goldman JW, Husain A, Alexander Palesty J, Sanchez JA, McFadden DW, Maulik N. Thioredoxin-1 (Trx1) engineered mesenchymal stem cell therapy increased pro-angiogenic factors, reduced fibrosis and improved heart function in the infarcted rat myocardium. International journal of cardiology. 2015;201:517–528. doi: 10.1016/j.ijcard.2015.08.117. [DOI] [PubMed] [Google Scholar]
- Suva D, Garavaglia G, Menetrey J, Chapuis B, Hoffmeyer P, Bernheim L, Kindler V. Non-hematopoietic human bone marrow contains long-lasting, pluripotential mesenchymal stem cells. Journal of cellular physiology. 2004;198:110–118. doi: 10.1002/jcp.10396. [DOI] [PubMed] [Google Scholar]
- Talens-Visconti R, Bonora A, Jover R, Mirabet V, Carbonell F, Castell JV, Gomez-Lechon MJ. Hepatogenic differentiation of human mesenchymal stem cells from adipose tissue in comparison with bone marrow mesenchymal stem cells. World journal of gastroenterology : WJG. 2006;12:5834–5845. doi: 10.3748/wjg.v12.i36.5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamama K, Sen CK, Wells A. Differentiation of bone marrow mesenchymal stem cells into the smooth muscle lineage by blocking ERK/MAPK signaling pathway. Stem cells and development. 2008;17:897–908. doi: 10.1089/scd.2007.0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z, Sant S, Khademhosseini A, Jia X. Controlling the fibroblastic differentiation of mesenchymal stem cells via the combination of fibrous scaffolds and connective tissue growth factor. Tissue engineering Part A. 2011;17:2773–2785. doi: 10.1089/ten.tea.2011.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tuyn J, Knaan-Shanzer S, van de Watering MJ, de Graaf M, van der Laarse A, Schalij MJ, van der Wall EE, de Vries AA, Atsma DE. Activation of cardiac and smooth muscle-specific genes in primary human cells after forced expression of human myocardin. Cardiovascular research. 2005;67:245–255. doi: 10.1016/j.cardiores.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Varela N, Aranguiz A, Lizama C, Sepulveda H, Antonelli M, Thaler R, Moreno RD, Montecino M, Stein GS, van Wijnen AJ, Galindo M. Mitotic Inheritance of mRNA Facilitates Translational Activation of the Osteogenic-Lineage Commitment Factor Runx2 in Progeny of Osteoblastic Cells. Journal of cellular physiology. 2015 doi: 10.1002/jcp.25188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vater C, Kasten P, Stiehler M. Culture media for the differentiation of mesenchymal stromal cells. Acta biomaterialia. 2011;7:463–477. doi: 10.1016/j.actbio.2010.07.037. [DOI] [PubMed] [Google Scholar]
- Wang ZH, Li XL, He XJ, Wu BJ, Xu M, Chang HM, Zhang XH, Xing Z, Jing XH, Kong DM, Kou XH, Yang YY. Delivery of the Sox9 gene promotes chondrogenic differentiation of human umbilical cord blood-derived mesenchymal stem cells in an in vitro model. Brazilian journal of medical and biological research = Revista brasileira de pesquisas medicas e biologicas / Sociedade Brasileira de Biofisica ... [et al ] 2014;47:279–286. doi: 10.1590/1414-431X20133539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Wang T, Liu J, Du Y, Ma A. The subpopulation of mesenchymal stem cells that differentiate toward cardiomyocytes is cardiac progenitor cells. Exp Cell Res. 2011;317:2661–2670. doi: 10.1016/j.yexcr.2011.08.011. [DOI] [PubMed] [Google Scholar]
- Wilson EM, Rotwein P. Control of MyoD function during initiation of muscle differentiation by an autocrine signaling pathway activated by insulin-like growth factor-II. J Biol Chem. 2006;281:29962–29971. doi: 10.1074/jbc.M605445200. [DOI] [PubMed] [Google Scholar]
- Xu DJ, Zhao YZ, Wang J, He JW, Weng YG, Luo JY. Smads, p38 and ERK1/2 are involved in BMP9-induced osteogenic differentiation of C3H10T1/2 mesenchymal stem cells. BMB reports. 2012;45:247–252. doi: 10.5483/bmbrep.2012.45.4.247. [DOI] [PubMed] [Google Scholar]
- Xu M, Wani M, Dai YS, Wang J, Yan M, Ayub A, Ashraf M. Differentiation of bone marrow stromal cells into the cardiac phenotype requires intercellular communication with myocytes. Circulation. 2004;110:2658–2665. doi: 10.1161/01.CIR.0000145609.20435.36. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Sakurada K, Takeda Y, Gojo S, Umezawa A. Single-cell-derived mesenchymal stem cells overexpressing Csx/Nkx2.5 and GATA4 undergo the stochastic cardiomyogenic fate and behave like transient amplifying cells. Exp Cell Res. 2007;313:698–706. doi: 10.1016/j.yexcr.2006.11.012. [DOI] [PubMed] [Google Scholar]
- Yang B, Guo H, Zhang Y, Chen L, Ying D, Dong S. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PloS one. 2011a;6:e21679. doi: 10.1371/journal.pone.0021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DC, Yang MH, Tsai CC, Huang TF, Chen YH, Hung SC. Hypoxia inhibits osteogenesis in human mesenchymal stem cells through direct regulation of RUNX2 by TWIST. PloS one. 2011b;6:e23965. doi: 10.1371/journal.pone.0023965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu WH, Li FG, Chen XY, Li JT, Wu YH, Huang LH, Wang Z, Li P, Wang T, Lahn BT, Xiang AP. PPARgamma suppression inhibits adipogenesis but does not promote osteogenesis of human mesenchymal stem cells. The international journal of biochemistry & cell biology. 2012;44:377–384. doi: 10.1016/j.biocel.2011.11.013. [DOI] [PubMed] [Google Scholar]
- Yuan H, Liu H, Tian R, Li J, Zhao Z. Regulation of mesenchymal stem cell differentiation and insulin secretion by differential expression of Pdx-1. Molecular biology reports. 2012;39:7777–7783. doi: 10.1007/s11033-012-1619-7. [DOI] [PubMed] [Google Scholar]
- Zhang HH, Huang J, Duvel K, Boback B, Wu S, Squillace RM, Wu CL, Manning BD. Insulin stimulates adipogenesis through the Akt-TSC2-mTORC1 pathway. PloS one. 2009;4:e6189. doi: 10.1371/journal.pone.0006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Jimenez SA, Stokes DG. Regulation of human COL9A1 gene expression. Activation of the proximal promoter region by SOX9. J Biol Chem. 2003;278:117–123. doi: 10.1074/jbc.M208049200. [DOI] [PubMed] [Google Scholar]
- Zhang T, Wen F, Wu Y, Goh GS, Ge Z, Tan LP, Hui JH, Yang Z. Crosstalk between TGF-beta/SMAD and integrin signaling pathways in regulating hypertrophy of mesenchymal stem cell chondrogenesis under deferral dynamic compression. Biomaterials. 2015;38:72–85. doi: 10.1016/j.biomaterials.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Yang ST, Wang JJ, Zhou J, Xing SS, Shen CC, Wang XX, Yue YX, Song J, Chen M, Wei YY, Zhou QP, Dai T, Song YH. TNF alpha inhibits myogenic differentiation of C2C12 cells through NF-kappaB activation and impairment of IGF-1 signaling pathway. Biochemical and biophysical research communications. 2015;458:790–795. doi: 10.1016/j.bbrc.2015.02.026. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zhao M, Xiao G, Franceschi RT. Gene transfer of the Runx2 transcription factor enhances osteogenic activity of bone marrow stromal cells in vitro and in vivo. Molecular therapy : the journal of the American Society of Gene Therapy. 2005;12:247–253. doi: 10.1016/j.ymthe.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Zhuang H, Zhang X, Zhu C, Tang X, Yu F, Shang GW, Cai X. Molecular Mechanisms of PPAR-gamma Governing MSC Osteogenic and Adipogenic Differentiation. Current stem cell research & therapy. 2015 doi: 10.2174/1574888x10666150531173309. [DOI] [PubMed] [Google Scholar]
- Zou L, Zou X, Chen L, Li H, Mygind T, Kassem M, Bunger C. Multilineage differentiation of porcine bone marrow stromal cells associated with specific gene expression pattern. Journal of orthopaedic research : official publication of the Orthopaedic Research Society. 2008;26:56–64. doi: 10.1002/jor.20467. [DOI] [PubMed] [Google Scholar]