Summary
Pluripotency is increasingly recognized as a spectrum of cell states, defined by their growth conditions. Although naïve and primed pluripotency states have been characterized molecularly, our understanding about events regulating state acquisition is wanting. Here, we performed comparative RNA sequencing of mouse embryonic stem cells (ESCs) and defined a pluripotent cell fate (PCF) gene signature associated with acquisition of naive and primed pluripotency. We identify Zfp281 as a key transcriptional regulator for primed pluripotency that also functions as a barrier towards achieving naive pluripotency in both mouse and human ESCs. Mechanistically, Zfp281 interacts with Tet1, but not Tet2, and its direct transcriptional target miR-302/367 negatively regulate Tet2 expression to establish and maintain primed pluripotency. Conversely, ectopic Tet2 alone, but not Tet1, efficiently reprograms primed cells towards naive pluripotency. Our study reveals a molecular circuitry in which opposing functions of Tet1 and Tet2 control acquisition of alternative pluripotent states.
Keywords: naive, primed, ZNF281, miR-302/367 cluster, ESCs, EpiSCs
Graphical abstract
Introduction
Cell fate decisions involve coordinated gene regulation at transcriptional and post-transcriptional levels, but the precise mechanisms underlying these complex processes are still poorly defined. Notably, manipulation of key signaling pathways is sufficient to force specific cell types to undergo global transcriptional changes in order to acquire new identities, including pluripotency (Chou et al., 2008). A great interest has thus emerged to understand how these alternative pluripotent identities are regulated (Weinberger et al., 2016; Wu and Izpisua Belmonte, 2015). In mouse embryonic stem cells (ESCs), standard culture conditions containing serum and leukemia inhibitory factor (LIF) (hereafter “SL”) support the self-renewal and maintenance of a heterogeneous or metastable pluripotent state (Marks et al., 2012). This in vitro cell identity can be reprogrammed into two inter-convertible and defined pluripotent states by activating distinct signaling pathways. Specifically, serum-free medium containing MEK and GSK3β kinase inhibitors and LIF (hereafter “2iL”) supports naive pluripotency that mimics the naive inner cell mass (ICM) of the blastocyst (Nichols and Smith, 2009). Alternatively, a more committed primed pluripotent state resembling post-implantation epiblast cells can be induced in serum-free medium containing the cytokines fibroblast growth factor 2 (Fgf2) and Activin A (hereafter “FA”) (Guo et al., 2009; Han et al., 2010).
The functional conservation of pluripotency hallmarks, i.e. unlimited self-renewal and differentiation into all somatic cell types, among these three pluripotent states (naive, metastable, and primed) suggests that common regulatory networks may participate in the pluripotent cell fate (hereafter “PCF”) determination. Considerable progress has been made in identifying transcriptional and epigenetic regulators required for maintenance of gene expression patterns associated with pluripotency identity, mainly under SL and 2iL conditions (Hackett and Surani, 2014). However, our knowledge regarding the molecular players responsible for rewiring the epigenome for proper transcriptional and epigenetic control of naive and primed cell fate acquisition is still limited.
Manipulation of specific signaling cues to directly induce SL ESCs towards naive (2iL) or primed (FA) pluripotent states provides an opportunity to dissect the molecular mechanisms underlying PCF determination. Here, by using this experimental system, we identified the expression pattern of gene sets associated with naive and primed cell identities, referred to as PCF gene signature hereafter, which was found to be evolutionarily conserved in mammals. Using this PCF gene signature as the discovery tool, we performed RNAi screen to search for transcriptional and epigenetic regulators that control the two pluripotent state transitions. We report here our findings of opposing functions of DNA dioxygenases Tet1 and Tet2 in regulating primed and naïve pluripotency, respectively, and of Zfp281 as a key pluripotency factor that coordinates Tet1 and Tet2 functions, transcriptionally and post-transcriptionally, in activation of primed and repression of naïve genes for primed pluripotency.
Results
Identification of a PCF Gene Signature for Alternative Pluripotent States
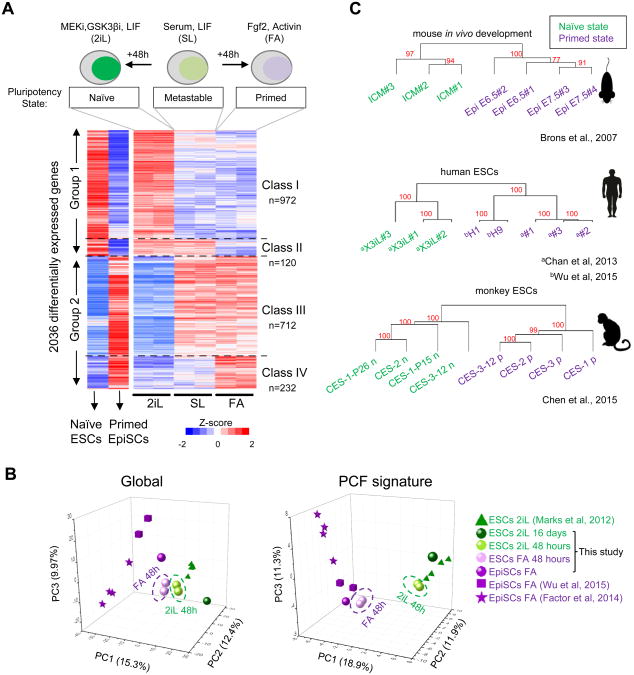
To deconstruct the complex and dynamic processes involved in global gene expression changes for alternative pluripotent states, we reasoned that identification of distinct gene expression patterns in response to 2iL and FA signaling pathways could provide insight into the regulatory networks governing naive and primed PCF acquisition. We first monitored the kinetics of direct conversion of SL ESCs to naive and primed pluripotent states with a green fluorescent protein (GFP) reporter under the control of the Oct4 distal enhancer (Oct4-ΔPEGFP) (Figure S1A), whose activity is active in the 2iL and SL, but extinguished in the primed epiblast (Bao et al., 2009). Flow cytometry analysis showed stable expression of GFP in ESCs grown in SL throughout multiple passages (Figure S1A). However, these ESCs displayed markedly different reporter activity under 2iL or FA conditions over extended passages (Figures S1A and S1B). In particular, we found that 48-hour culture was sufficient to increase or decrease Oct4-ΔPE-GFP activity by 50% under 2iL and FA, respectively, representing an ideal time frame to examine changes in gene expression associated with alternative PCF acquisition. We then performed RNA sequencing (RNA-seq) to characterize transcriptome changes of ESCs grown in SL upon transfer to 2iL and FA conditions for 48 h. To identify genes that are most closely associated with naive and primed states, we compared these transcriptomes to those of ESCs cultured for 16 days in 2iL, and to EpiSCs in FA culture conditions. Differential expression analysis revealed a set of 2,036 genes with dynamic expression patterns comprising two major groups (Figure 1A, and see Supplemental Experimental Procedures): i) group 1 genes are upregulated or actively expressed during metastable to naive transition, encompassing two classes (I and II), and ii) group 2 genes are actively expressed or upregulated during metastable to primed transition, encompassing two additional classes (III and IV) (Table S1). While classes I and IV are induced in response to 2iL and FA, respectively, classes II and III are silenced in response to FA and 2iL, respectively. PCA analysis of transcriptomes of our cultured cells and those of established ESCs and EpiSCs under 2iL and FA conditions revealed a better separation of naive and primed pluripotent cells based on our PCF signature genes than global transcripts (Figure 1B). Gene ontology (GO) analysis of these four classes of genes revealed a significant presence of GO terms linked to developmental processes (Figure S1C). While naive state active gene classes (I and II) are enriched for GO terms associated with metabolic processes, primed state active gene classes (III and IV) are enriched for post-implantation developmental terms, such as morphogenesis and gastrulation (Figure S1C). These pluripotent state associated GO terms are consistent with previously reported findings in naive 2iL ESCs (Marks et al., 2012) and compatible with the transition into a developmentally primed state.
Figure 1. Identification of a Pluripotent Cell Fate (PCF) gene signature for alternative pluripotency. See also Figure S1.
(A) Raw normalized expression of 2,036 genes differentially expressed in SL ESCs and during transitions under 2iL or FA for 48 hours. ESCs under 2iL (naive) for 16 days and previously established EpiSCs (primed) are shown for reference. The scale represents Z-score.
(B) Principle component analysis (PCA) analysis of transcriptome comparison between this study and published datasets. PCA was performed for the RNA-seq expression (cRPKM) data of all genes (global) or the PCF signature genes. The expression data matrix was imported by Cluster 3.0 software. Mean center and normalize genes were performed, then PCA was applied to arrays and results were virtualized by the Origin software.
(C) Unbiased clustering analysis of 2,036 PCF genes across previously published transcriptomes for mouse in vivo development, human and Cynomolgus monkey naive and primed pluripotent cells. The approximately unbiased (AU) p-value, which is computed by multiscale bootstrap resampling, is used to estimate robustness of each cluster. A cluster with AU larger than 95% indicates that this cluster significantly exists.
We next tested whether this PCF gene signature was consistent across multiple genetic backgrounds. Unbiased hierarchical clustering of the 2,036 differentially expressed genes using our own as well as published RNA-seq data sets from mouse ESC lines of various genetic backgrounds cultured for several passages in 2iL as well as EpiSC lines cultured in FA (Factor et al., 2014; Marks et al., 2012; Wu et al., 2015) revealed that naive cells clustered together in a group distinct from primed cells (Figure S1D). Notably, similar results were also obtained by analyzing available transcriptomes of in vivo mouse naive ICM and post-implantation primed epiblast (Brons et al., 2007) (Figure 1C, top panel). Cross-species comparison of the expression of human and monkey orthologs of 2,036 mouse genes (Chan et al., 2013; Chen et al., 2015; Wu et al., 2015) demonstrated that the identified PCF gene signature could also distinguish naive from primed primate cells (Figure 1C, middle and bottom panels).
Together, our studies reveal a novel and dynamic PCF gene expression signature associated with naive and primed pluripotency transitions that is evolutionarily conserved across different species and genetic backgrounds. This PCF gene signature provides a tool for interrogating molecular regulators that orchestrate pluripotent state transitions.
Zfp281 Contributes to PCF Signature Gene Regulation
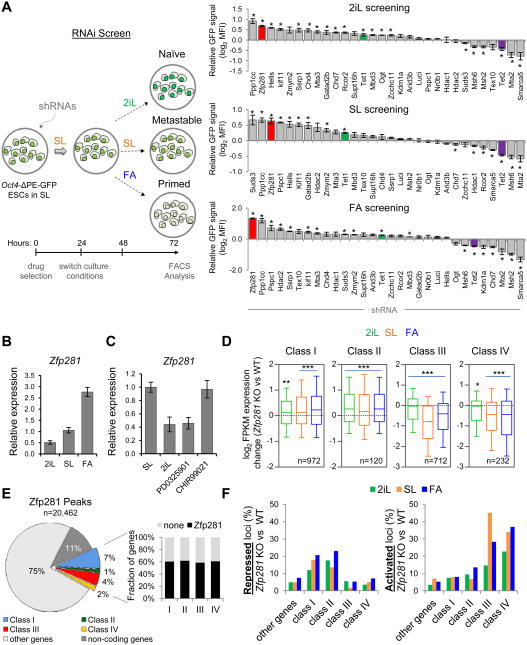
To search for master regulators that control this newly defined PCF gene signature, we performed chromatin enrichment analysis using the Network2Canvas computational tool for a large number of transcription factors (TFs) and chromatin regulators in ESCs (see Supplemental Experimental Procedures). We observed the separation of enrichment patterns according to the pluripotent states. That is, naive classes I and II (Group 1) and primed classes III and IV (Group 2) are more similar within the group than across groups (Figure S2A). Notably, we observed that the core pluripotency factors Oct4, Sox2, and Nanog (OSN), were significantly enriched throughout the four gene classes (Figure S2A), consistent with their well-established dual roles in promoting naive pluripotency and suppressing differentiation program (Orkin, 2005). We also identified specific Polycomb enrichment (Suz12, Mtf2) in class III and IV genes, but not in class I and II genes. This is consistent with the deposition of H3K27me3 marks at developmental regulators in EpiSCs (Gafni et al., 2013), and is also in line with reported H3K27me3 depletion (Marks et al., 2012) and dispensability of Polycomb repression under naive 2i culture (Galonska et al., 2015). To investigate how other transcriptional cofactors and epigenetic regulators within the OSN pluripotency network (Huang and Wang, 2014) may contribute to the regulation of both naive and primed PCF transitions, we conducted RNA interference (RNAi) screens focusing on our previously reported high-confidence OSN-interacting partners (Costa et al., 2013; Ding et al., 2015; Ding et al., 2012; Wang et al., 2006) (Figure S2B). These include TFs, epigenetic regulators, and RNA processing factors whose roles in the pluripotent state transitions have not been explored before (Figure 2A, right panel). We tested whether the naive reporter (Oct4-ΔPE-GFP) in ESCs might be affected by depletion of these OSN partners during pluripotent state transitions (Figure 2A, left panel). We found that loss of several factors including the TF Zfp281 noticeably increased GFP reporter activity in all conditions tested, while the loss of other factors reduced GFP reporter activity (Figure 2A, right panel). Interestingly, we detected opposite effects of the ten-eleven translocation (TET) family methylcytosine hydroxylases Tet1 and Tet2, with loss of Tet1 increasing, and loss of Tet2 decreasing, Oct4-ΔPE-GFP activity (Figure 2A, right panel).
Figure 2. RNAi Screen Identifies Zfp281 as a Critical Regulator for Transcriptional Control of PCF Signature Genes. See also Figure S2.
(A) A schematic overview of RNAi screen strategy (left panel) and results (right panel). Oct4-ΔPE-GFP reporter ESCs were infected with two independent shRNAs against each candidate, and an average of mean fluorescence intensity in the cell population is measured. Error bars indicate average ± SEM (n=2). Student's unpaired t test (*P<0.05) relative to shRNA against luciferase (shLuci) was performed.
(B) Relative expression of Zfp281 analyzed by qPCR during pluripotent state transitions after 48 hours medium switch as indicated.
(C) Relative expression of Zfp281 by qPCR in SL ESCs under treatment of the MEK and GSKβ inhibitors. Note that Zfp281 is downregulated by the MEK inhibitor (PD0325901) but not the GSKb inhibitor (CHIR99021).
(D) Expression changes of PCF gene classes in SL and under 48-hr transition to 2iL and FA in Zfp281 KO relative to WT cells (mRNA-seq, 2 biological replicates per condition). Whiskers extend to the 10-90 percentile range. P values between KO and WT ESCs were calculated using one-side Wilcoxon signed rank test (***P<0.0001, **P<0.001, *P<0.01). n indicates number of genes analyzed per gene class.
(E) Distribution of total Zfp281 binding peaks (left panel), and percentages of PCF gene class occupancy by Zfp281 (right panel).
(F) Transcriptional effects of Zfp281 in target loci. Percentages of repressed and activated loci were calculated by comparative gene expression changes between Zfp281 KO cells and WT cells (> 2 fold differences).
To gain insight into the molecular control of the PCF gene signature, we focused on Zfp281, a zinc finger protein expressed in mouse ESCs, whose depletion is compatible with pluripotency in SL condition (Fidalgo et al., 2011). Interestingly, Zfp281 has also been identified in an siRNA screen as a factor required for exiting naive pluripotency (Betschinger et al., 2013) and in another study as a specific partner of Oct4 in the primed EpiLC state (Buecker et al., 2014). Network analyses also suggest an important role of Zfp281 in regulating the pluripotency network (MacArthur et al., 2012). These observations together suggest that Zfp281 might be an important player in regulating naive to primed transition. While loss of Zfp281 caused a significant enhancement of Oct4-ΔPE-GFP activity (Figure 2A), ectopic expression of Zfp281 confirmed the repressive role of Zfp281 on Oct4-ΔPE-GFP activity (Figure S2C), further suggesting that Zfp281 could be an important transcriptional regulator that promotes primed pluripotency. In agreement with this, we observed a stepwise upregulation of Zfp281 during naive to primed transition (Figure 2B), and found that Zfp281 is positively regulated by FGFERK signaling pathway (Figures 2C and S2D-G). RNA-seq analysis comparing Zfp281 knockout (KO) ESCs to wild-type (WT) ESCs indicated significant changes in global gene expression among all four gene classes during pluripotent state transitions (Figure 2D). Notably, we found that naive active genes (classes I and II) were significantly increased and primed active genes (classes III and IV) were significantly reduced upon Zfp281 depletion (Figure 2D).
We next examined global genomic targets of Zfp281 in SL ESCs by chromatin immunoprecipitation followed by sequencing (ChIP-seq) to further investigate how Zfp281 regulates its transcriptional targets within the PCF gene signature. We indentified 20,462 genome-wide peaks and found that, although all 4 classes of genes represent only 14% of Zfp281 binding targets, there is a pronounced binding of Zfp281 to all four gene classes (∼60% of all targets within each class of genes) (Figure 2E). Using unbiased hierarchical clustering and published ChIP-seq datasets of histone marks, we detected a striking correlation between Zfp281 and epigenetic marks associated with transcriptional regulation (Figure S2H). The presence of both activating and repressive histone marks on PCF signature gene loci suggests that Zfp281 may mediate both transcriptional activation and repression. Consistent with this, analysis of gene expression changes in Zfp281 KO relative to WT cells and Zfp281 occupancy confirmed a predominantly repressive and activating function of Zfp281 at naive (classes I and II) and primed (classes III and IV) genes, respectively, across the three pluripotent states (Figures 2F and S2I-K).
Taken together, our data indicate that Zfp281 is a critical regulator for the PCF signature gene expression with dual functions in transcriptional activation and repression of primed and naïve genes, respectively.
Zfp281 is Required for Establishment and Maintenance of Primed Pluripotency
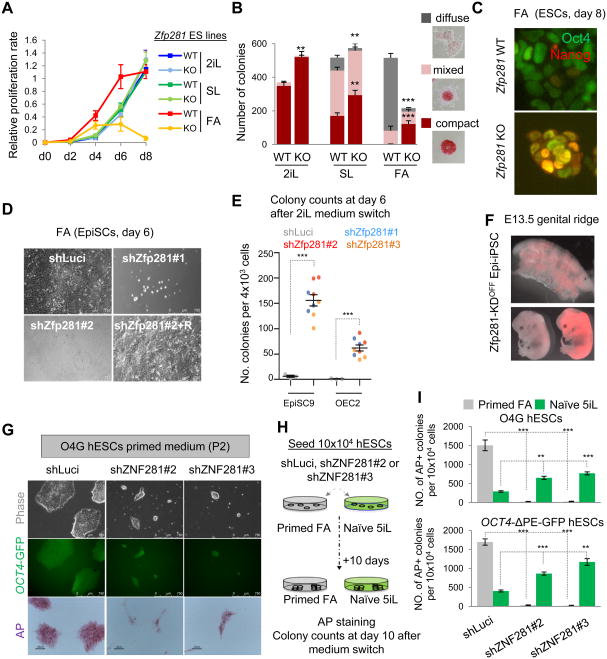
To directly test the function of Zfp281 in pluripotent state transition, we cultured Zfp281 WT and KO ESCs in defined media and found that Zfp281 KO ESCs could not be maintained in the FA culture condition for more than three passages, whereas no significant proliferation differences were found in 2iL or SL conditions (Figure 3A). We next examined whether self-renewal properties of each specific pluripotent state were affected by the absence of Zfp281. Colony formation assays indicated that Zfp281 KO ESCs yielded significantly more naive compact dome-shaped colony numbers under all culture conditions (Figure 3B). Remarkably, the typical primed diffuse-shaped colonies as well as expression of lineage primed markers were severely compromised by Zfp281 depletion during primed transition (Figure S3A), demonstrating that Zfp281 KO promotes naive pluripotency, as evident by increased Nanog expression in the primed culture condition (Figure 3C). Conversely, we found that ectopic Zfp281 expression was sufficient to promote primed features including flattened colony morphology and downregulation of naive and upregulation of primed pluripotency markers in SL ESCs and during SL to FA transition (Figure S3B). Furthermore, we found that EpiSCs could not be stably maintained in the primed state upon Zfp281 knockdown, due to compromised proliferation, elongated G1 phase, and increased cell death (Figures 3D and S3C-F). Within 6 days upon Zfp281 depletion, EpiSCs upregulate naive markers (Nanog, Prdm14, Esrrb, and Nr0b1) and downregulate primed makers (Fgf5, Otx2, Brachyury, and Eomes) (Figures S3G-H). These loss-of-function defects can be rescued by transgenic Zfp281 re-expression (“+R”; Figures 3D and S3C-H). Together, our data clearly demonstrate that Zfp281 functions as a key factor that is critical for both induction and maintenance of primed pluripotency. While EpiSCs undergo massive cell death during the transition to the naive state under 2iL as previously reported (Guo et al., 2009), we found that, when cultured in 2iL immediately following Zfp281 knockdown, EpiSCs showed significant enhancement of reprogramming efficiency towards the naive state (Figure 3E). We confirmed bona fide naive pluripotency of the resulting Epi-iPSCs by their contribution to the germ lineage as well as chimeric embryos after blastocyst injection (Figure 3F). These data suggest that Zfp281 functions as a major barrier for primed to naive reversion.
Figure 3. Zfp281 is Critical for Primed Pluripotency. See also Figure S3.
(A) Proliferation analysis of WT and Zfp281 KO ESCs cultured in SL, 2iL or FA. Error bars indicate average ± SEM (n=3).
(B) Colony formation assay for WT and Zfp281 KO ESCs. Colonies were stained for alkaline phosphatase (AP) activity at day 6 of culture in specified conditions, and positive colonies were scored into three categories (compact, mixed and diffuse as indicated). Error bars indicate average ± SEM (n=6). Student's unpaired t test (***P<0.001, **P<0.01) relative to WT ESCs was performed.
(C) Immunostaining of Nanog and Oct4 at day 8 under FA. Scale bars represent 100 μm.
(D) Representative bright-field images at day 6 of Zfp281 KD (shZfp281) EpiSCs and rescued (+R) line compared with control luciferase knockdown (shLuci) EpiSCs.
(E) Zfp281 KD facilitates reprogramming of EpiSCs towards naive-like cells (Epi-iPSCs). Two different EpiSC lines were transfected with three independent shRNAs against Zfp281, and Epi-iPSC colony numbers were quantified at day 6 in 2iL. Error bars indicate average ± SEM (n=3). Student's unpaired t test (***P<0.001) relative to shLuci was performed.
(F) Contribution of Epi-iPSCs to the germline (top) and chimeric E13.5 mouse embryos (bottom). Epi-iPSCs were generated by reprogramming EpiSCs with pTRIPZ-shZfp281. EpiiPSCs were labeled with a Tomato reporter transgene and maintained without Dox (OFF).
(G) ZNF281 dependency of conventional hESCs. Representative colony morphology under bright-field (top) and fluorescent (for OCT4-GFP; middle) microscope as well as AP-stained colonies (bottom) are shown. ZNF281-depleted and control shLuci O4G hESCs were passaged twice in primed culture conditions before microscopy.
(H) A strategy to test effects of ZNF281 depletion on converting primed hESCs to a naive state using the published 5iL condition.
(I) Quantification of AP-positive colony numbers of hESCs at day 10 of primed human ES medium (FA) or naive culture conditions (5iL) upon control or ZNF281 KD. Error bars indicate average ± SEM (n=3). Student's unpaired t test (***P<0.001, **P<0.01) relative to shLuci hESCs.
Conventional human ESCs (hESCs) are also considered to be at a primed pluripotent state. As the PCF gene signature is conserved in human (Figure 1C), we also tested whether the human ortholog ZNF281 plays similar roles in maintaining the primed pluripotency of hESCs and whether it also poses as a barrier in converting primed hESCs to a naive state. Using hESCs harboring an OCT4-2A-GFP reporter (O4G) (Hockemeyer et al., 2011), we found that ZNF281 knockdown compromised self-renewal of these hESCs under conventional culture conditions as shown by smaller GFP+ and AP+ colonies (Figure 3G). Nonetheless, these small colonies became less flattened and maintained (e.g., OCT4 and PRDM14) or even upregulated (e.g., KLF2/4/5, Dppa3/5, TCFP2L1 and TET2) naïve pluripotency marker gene expression (Figures S3I-J). When switching the culture conditions from conventional medium to the defined naive medium containing five small molecule inhibitors and LIF (5iL) (Theunissen et al., 2014), we observed a dramatic increase in the number of naive AP+ (Figures 3H-I) and GFP+ (Figure S3K) hESCs using two independent GFP reporter lines upon ZNF281 depletion. Together, these results establish Zfp281/ZNF281 as a key primed pluripotency regulator in both mouse and human ESCs with its depletion promoting naive pluripotency.
Zfp281 Functions in Epigenomic Reconfiguration during Pluripotent State Transitions
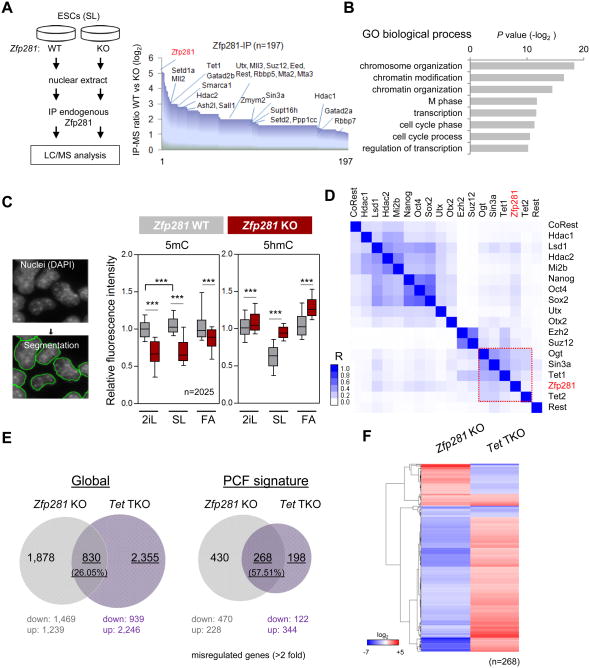
To explore the molecular mechanisms by which Zfp281 regulates PCF signature gene expression during pluripotent state transitions, we performed affinity purification of Zfp281 protein complexes followed by mass spectrometry (AP-MS) in SL ESCs and identified 196 high confidence Zfp281-interacting partners (Figure 4A and Table S4). GO analysis of Zfp281 interactome revealed a significant association of Zfp281 with chromatin regulatory functions due to the presence of many histone and DNA modifying enzymes including Tet1, HDAC-containing NuRD and Sin3a complexes, MLL complex, and Polycomb complex proteins (Figures 4A-B). In accord with this, global alterations of certain histone modifications (e.g., increased H3K27ac under SL and FA) with corresponding histone code reader/writer changes (e.g., reduced chromatin bound HDAC2 under SL and FA) upon Zfp281 depletion were observed (Figures S4A-B). As DNA hypomethylation is a prominent feature of naive pluripotency (Marks et al., 2012) and developmental progression from naive to primed cell fate is intrinsically associated with DNA methylation (Clark, 2015), we decided to address how Zfp281 may contribute to this important epigenomic reconfiguration in pluripotent state transitions by assaying global alterations of 5mC and 5hmC levels in Zfp281 WT and KO ESCs. Quantitative immunofluorescence analysis of single cells and dot-blot analysis revealed a significant reduction of 5mC and increase of 5hmC upon Zfp281 depletion under the three pluripotent states (Figures 4C and S4C-D).
Figure 4. Characterization of Epigenetic Reconfiguration Dynamics in Zfp281 KO ESCs. See also Figure S4.
(A) Affinity purification and mass spectrometry (AP-MS) identification of Zfp281-interacting partners. Depiction of Zfp281 antibody-based AP-MS procedure (left panel), and candidate partners (n=197) ordered by fold change in peptide numbers between Zfp281 WT and KO ESCs (right panel). Chromatin modifiers are shown.
(B) Gene Ontology (GO) analysis for biological processes associated with Zfp281-interacting partners identified by AP-MS.
(C) Global decrease of 5mC is accompanied by an increase of 5hmC upon Zfp281 depletion. Single cell quantitative immunofluorescence was performed in Zfp281 WT and KO cells cultured in SL and after 48 hours of medium switch to 2iL or FA. Signals were normalized against DAPI nuclei staining. Whiskers extend to the 10-90 percentile range. P values between KO and WT ESCs were calculated using one-side Wilcoxon signed rank test (***P<0.0001). “n” indicates number of cells analyzed per condition.
(D) Hierarchical clustering of Zfp281 and chromatin associated factors in genome-wide occupancy. The scale represents a Pearson correlation coefficient (R), and the red rectangle indicates a group of factors with a high correlation.
(E) Pairwise comparisons of the number of genes mis-regulated (>2 fold alteration compared to WT cells) in Zfp281 KO and Tet triple knockout (Tet-TKO) ESCs under SL culture.
(F) Relative expression of 268 mis-regulated genes in Zfp281 KO cells compared with Tet-TKO cells. The scale represents fold changes compared to WT cells.
Analysis of global target binding of Zfp281 and many pluripotency TFs and epigenetic regulators revealed a strong correlation in genome-wide occupancy between Zfp281 and Tet1/2 (Figure 4D), prompting us to test whether active DNA demethylation via Tet-mediated 5hmC may contribute to the transcriptional regulatory functions of Zfp281 during pluripotent state transitions. We therefore compared the global transcriptional profiles of Zfp281 KO and Tet1/2/3 triple-knockout (Tet-TKO) ESCs (Dawlaty et al., 2014). While we observed that global deregulation (> 2-fold expression change) of 830 shared genes in both Zfp281 KO and Tet-TKO cells represents only 26.05% of the total misregulated genes in the absence of Tet proteins, a significantly higher proportion (57.51%) of misregulated genes in Tet-TKO cells was shared with Zfp281 KO cells when only the PCF signature genes were compared (Figure 4E), highlighting a potential functional relationship between Zfp281 and Tet proteins in regulating PCF signature genes. Interestingly, the majority of 268 identified shared PCF genes showed opposite expression changes in Zfp281 KO versus Tet-TKO ESCs (Figure 4F). The significant upregulation of 5hmC in Zfp281 KO ESCs (Figure 4C) suggests potentially activated Tet functions upon Zfp281 depletion as the most parsimonious explanation. Together with the physical association between Zfp281 and Tet1, but not Tet2 (Figure 4A), our data indicate a complex interplay among Tet1, Tet2 and Zfp281 in transcriptional and epigenetic control of PCF signature gene expression during pluripotent state transitions.
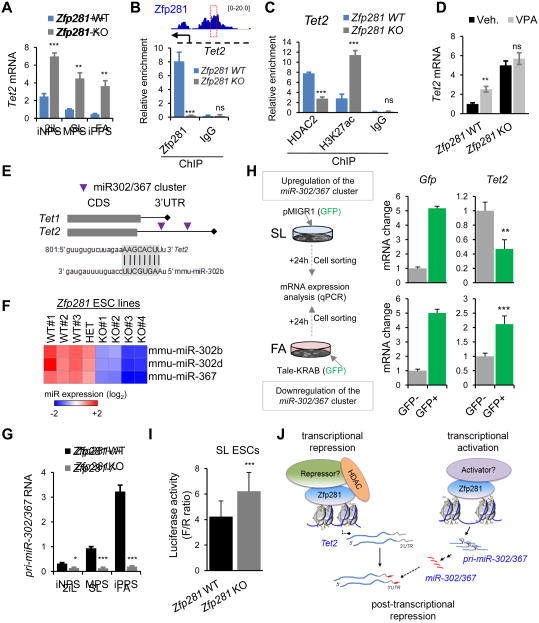
Tet2 is Regulated at the Transcriptional and Post-transcriptional Levels by Zfp281
To explore which Tet protein might be activated upon Zfp281 depletion, we examined our RNAseq data and found that Tet2, but not Tet1, is significantly upregulated in Zfp281 KO ESCs (Figure S5A), which was validated by qPCR (Figure 5A). We also found three ChIP-seq peaks of Zfp281 binding to the regulatory regions of the Tet2 locus bearing enhancer marks (Figure S5B). Because histone deacetylase (HDAC) complexes are present in the Zfp281 interactome (Figure 4A), and because depletion of Zfp281 also increases global H3K27ac levels (Figure S4A), we asked whether Zfp281 might modulate histone acetylation levels at the Tet2 enhancer. Indeed, we found that loss of Zfp281 reduced binding of HDAC2 with a concomitant increase in H3K27ac enrichment at the Tet2 enhancer (Figures 5B-C), indicating that Zfp281 may regulate Tet2 enhancer activity through modulating histone acetylation. In line with this, we found that Tet2 was upregulated upon knockdown of HDAC proteins, especially HDAC2 (Figure S5C). In addition, chemical inhibition of HDACs with valproic acid (VPA) increased Tet2 expression in WT ESCs, but was unable to promote further upregulation of Tet2 in Zfp281 KO ESCs (Figure 5D), suggestive of Zfp281-dependent action of HDACs on Tet2 enhancer. Interestingly, we detected a lower level of Tet2 in WT ESCs treated with VPA than that in Zfp281 KO ESCs regardless of VPA treatment, suggesting that Zfp281 must also regulate Tet2 beyond the transcriptional level.
Figure 5. Transcriptional and Post-transcriptional Regulation of Tet2 by Zfp281. See also Figure S5.
(A) Relative expression of Tet2 by qPCR showing upregulation of Tet2 in Zfp281 KO relative to WT ESCs cultured during 48-hour pluripotent state transitions.
(B) Enrichment of Zfp281 at the regulatory locus of Tet2 by ChIP-qPCR validating Tet2 as a direct transcriptional target of Zfp281.
(C) ChIP-qPCR analysis of relative enrichment of HDAC2 and H3K27ac at the Zfp281 binding Tet2 loci as indicated in (B).
(D) Chemical inhibition of HDAC activity increases Tet2 expression in Zfp281 WT, but not KO cells. Expression analysis was determined by qPCR in Zfp281 KO and WT ESCs treated with VPA (0.5 mM) for 24 hours.
(E) Schematic depiction of seed sequences for miR-302/367 cluster on Tet1 and Tet2 genes. Seed match sequences of miR-302b within Tet2 3′-UTR were shown.
(F) Relative expression of mature miR-302b, miR-302d, and miR-367 detected by miRCURY™ LNA Array in Zfp281 WT, HET (heterozygous) and KO ESCs. The color scale bar represents fold change.
(G) Relative expression of pri-miR-302/367 cluster measured by qPCR in Zfp281 KO and WT ESCs at SL and 48 hours at 2iL or FA.
(H) Depiction of miR-302/367 cluster gain- and loss-of-function assays (left), and qPCR analysis of Tet2 expression levels (right).
(I) Zfp281 depletion enhances firefly luciferase activity. Zfp281 KO and WT ESCs were transiently transfected with the luciferase reporter tethering to the 3′-UTR of Tet2. Renilla reporter was included for normalization (F/R ratio).
(J) A model illustrating the transcriptional activation and repression roles of Zfp281 in regulating Tet2 expression.
All error bars in this figure indicate average ± SEM (n=3). P values were determined by Student's unpaired t test (ns, not significant; ***P<0.0001, **P<0.001).
To investigate whether and how Zfp281 may participate in the regulation of Tet2 levels in a post-transcriptional manner, we first examined the presence of Zfp281 in non-coding gene regions (Figure 2E) and found the enrichment of a significant fraction of miRNAs (Figure S5D). We therefore performed global mature miRNA expression profiling in Zfp281 WT and KO ESCs and identified candidate miRNAs that are regulated by Zfp281 (Figure S5E). We hypothesized that miRNAs that are positively regulated by Zfp281 would be the candidates to directly target Tet2 mRNA for repression. Among them, we focused our interest on the miR-302/367 cluster (Figure S5F) because i) only Tet2, but not Tet1, is a predicted target of miR-302/367 (Figure 5E), ii) the presence of this mature miRNA family is greatly diminished in the absence of Zfp281 (Figure 5F), and iii) the expression of this pluripotency miRNA family shows the opposite trend to that of Tet2 expression during pluripotent state transitions (compare Figure 5A with Figure 5G). Q-PCR analysis confirmed that Zfp281 is required for transcriptional activation of primiRNA-302/367 (Figures 5G and S5G). We next tested whether this miRNA cluster participates in the post-transcriptional regulation of Tet2 in ESCs, and found that enforced expression of this miRNA cluster in SL ESCs downregulated Tet2, whereas its inhibition in FA cells led to a significant upregulation of Tet2 (Figure 5H). We further confirmed this potential post-transcriptional regulation of Tet2 expression by Zfp281-controlled miR-302/367 using a luciferase reporter tethering to the Tet2 3′-UTR, which contains the miR-302/367 seed sequences. Our data show increased luciferase activity in Zfp281 KO relative to WT ESCs (Figure 5I) and an inverse expression pattern between luciferase activity/Tet2 and Zfp281/primiR-302/367 under the three pluripotent states (Figure S5H).
Collectively, our data establish Zfp281 as a major regulator of Tet2 expression in pluripotency control by direct transcriptional repression through histone deacetylation of the Tet2 enhancer, and indirect post-transcriptional repression through activation of miR-302/367 (Figure 5J).
Distinct Functions of Tet1 and Tet2 in Zfp281-Mediated Transcriptional Control of Naive to Primed Transition
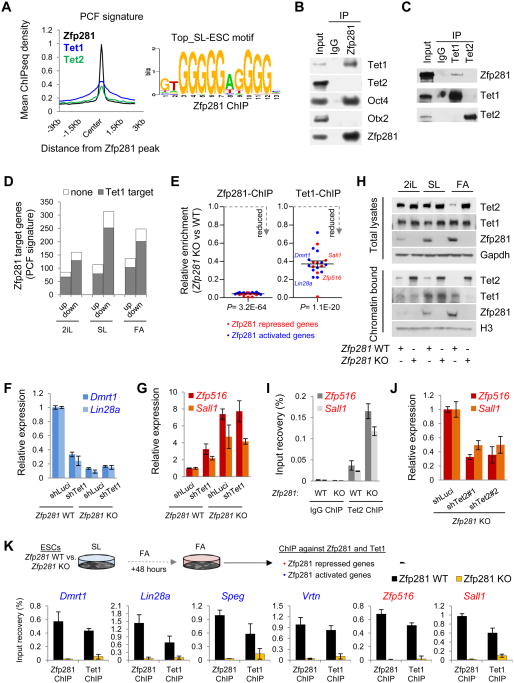
To understand how Tet1 and Tet2 contribute to transcriptional regulatory functions of Zfp281, we compared our Zfp281 ChIP-seq data (Figure 2E) with genomic occupancy of Tet1 (Wu et al., 2011) and Tet2 (Chen et al., 2013) under the same SL conditions. We found that Zfp281 has a significant overlap with Tet1 and Tet2 in controlling specific PCF signature gene occupancy (Figures 6A and S6A). Remarkably, 81.8% of the signature genes (505 out of 617) that are bound by Zfp281 and misregulated in Zfp281 KO ESCs are also directly bound by at least one Tet protein (predominantly Tet1) (Figure S6B), strongly suggesting that Tet proteins are required for proper transcriptional regulation of Zfp281 target genes. Moreover, expression analysis of these 505 misregulated genes indicated that they were subjected to both transcriptional activation and repression in all pluripotent states (Figure S6C), consistent with the dual functions of Tet1 in transcriptional regulation in mouse ESCs (Wu et al., 2011).
Figure 6. Distinct Functions of Tet1 and Tet2 in Modulating Zfp281-Mediated Transcriptional Control. See also Figure S6.
(A) Average ChIP-seq read density of Zfp281, Tet1, and Tet2 near the Zfp281 peak center on PCF genes (left) and the top Zfp281 binding motif in SL ESCs (right).
(B) Zfp281 interacts with Tet1 and Oct4, but not Tet2 or Otx2 in SL ESCs. IP was performed using anti-Zfp281 antibody followed by western blotting with indicated antibodies.
(C) Zfp281 interacts with Tet1, but not Tet2, in SL ESCs. IP was performed using anti-Tet1 and anti-Tet2 antibodies followed by western blotting with indicated antibodies.
(D) Comparison of Zfp281-regulated PCF gene targets with Tet1 targets showing the majority of Zfp281-regulated PCF target genes that are mis-regulated in Zfp281 KO ESCs are also Tet1 targets during pluripotent state transitions.
(E) Validation of Zfp281-dependent Tet1 binding to the target genes by ChIP-qPCR. Error bars indicate average ± SEM (n=20). P values were determined by Student's unpaired t test.
(F) Zfp281-dependent transcriptional activation function of Tet1. Error bars indicate average ± SEM (n=3).
(G) Zfp281-dependent transcriptional repression function of Tet1. Error bars indicate average ± SEM (n=3).
(H) Western blot analysis of total and chromatin-bound Zfp281, Tet1, and Tet2 levels in Zfp281 WT and KO cells cultured in SL, 2iL, or FA for 48 hours.
(I) Analysis of Tet2 enrichment at the Zfp281 repressed loci (Zfp516 and Sall1) in the presence (WT) and absence (KO) of Zfp281 in SL ESCs. Error bars indicate average ± SEM (n=3).
(J) Knockdown of Tet2 partially rescues Zfp516 and Sall1 upregulation in Zfp281 KO ESCs under SL culture condition. Error bars indicate average ± SEM (n=3).
(K) Zfp281-dependent binding of Tet1 to the candidate target genes under FA condition by ChIP-qPCR.
The identification of Tet1, but not Tet2, as a partner of Zfp281 in SL ESCs (Figure 4A), and the marked reduction of Tet2 expression from naive to primed transition (Figure S6D), prompted us to consider whether Tet1 might be the critical Tet family member for modulating Zfp281 transcriptional regulatory functions during the transition. We confirmed the physical interaction between Zfp281 and Tet1, but not Tet2, in SL ESCs (Figures 6B-C). We also observed the majority of Zfp281 direct targets within the PCF signature that are misregulated upon Zfp281 depletion being also Tet1 targets during the transitions (Figure 6D). ChIP-qPCR experiments showed that loss of Zfp281 reduced Tet1 binding to several common targets that are either repressed (e.g. Zfp516 and Sall1) or activated (e.g. Dmrt1 and Lin28a) by Zfp281 in ESCs during pluripotent state transitions (Figures 6E and S6C). More importantly, we detected reduced 5hmC levels at targets that are downregulated in the absence of Zfp281 (e.g., Lin28a) during the pluripotent state transitions (Figure S6E). These data suggest that Zfp281 may recruit Tet1 to a subset of shared target genes to promote transcriptional activation through 5hmC deposition during the transitions. This Zfp281-dependent transcriptional activation function of Tet1 was confirmed by downregulation of Dmrt1 and Lin28a upon Tet1 knockdown in WT but not Zfp281 KO ESCs (Figure 6F). In contrast, we detected increased 5hmC levels at several shared target genes (e.g., Zfp516 and Sall1) (Figure S6E) that showed significant upregulation in the absence of Zfp281 (Figures 6G and S6C). This result is quite surprising as chromatin bound Tet1 is greatly diminished upon Zfp281 depletion in FA cells despite the presence of similar amounts of total Tet1 proteins (Figure 6H), which prompted us to examine the effect of Zfp281 KO on total and chromatin-bound protein levels of Tet2. In agreement with the repressive role of Zfp281 on Tet2 expression (Figure 5), we found that loss of Zfp281 increased the protein levels of both total and chromatin-bound Tet2 during transitions, whereas the levels of chromatin-bound Tet1 were only affected, i.e. reduced, in FA culture (Figure 6H). These results suggest that Tet2 may be responsible for the elevated 5hmC levels observed in the absence of Zfp281, leading to transcriptional activation. We confirmed that Zfp281 KO increased Tet2 binding to the regulatory loci of upregulated genes Zfp516 and Sall1 (Figure 6I). Conversely, we observed decreased expression of both genes after Tet2 knockdown in Zfp281 KO ESCs (Figure 6J). Thus, we established a role for Tet2 in contributing to transcriptional activation of genes that are upregulated upon Zfp281 depletion. Interestingly, a Zfp281-dependent repressive role of Tet1 was also observed for Zfp516 and Sall1 expression as shown by their upregulation upon Tet1 knockdown in WT but not Zfp281 KO cells (Figure 6G). We further confirmed the recruitment of Tet1 to the chromatin by Zfp281, but not the other way around, for transcriptional activation and repression by Zfp281 and Tet1 (Figures S6F-G). Such an effect is less obvious under 2iL due to the already low Zfp281 expression in naive cells but becomes more pronounced in SL and particularly in FA condition (Figure 6K) where Zfp281 is highly abundant and plays a major role as the key primed pluripotency factor.
Together our results reveal a Zfp281-coordinated distinct functions of Tet1 and Tet2 in transcriptional control of the PCF gene signature during naive and primed transition.
Tet1 and Tet2 Play Opposing Roles in Regulating Primed and Naive Pluripotent States
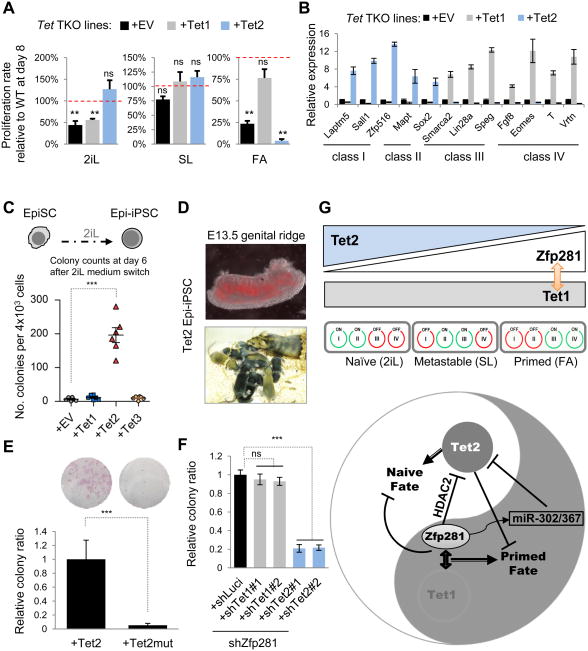
In support of their distinct functions in PCF gene regulation during pluripotent state transitions described above, we found that ectopic expression of Tet2, but not Tet1, was sufficient to promote naive features (i.e., dome-shaped colony formation and increased clonogenicity) in primed culture conditions (Figures S7A-B). Additionally, we found that the ability of Tet2 to promote naive pluripotency is dependent on its catalytic activity, as ectopic expression of a catalytic activity deficient Tet2 mutant (Tet2Mut) abolished the ability of this enzyme to promote naive pluripotency (Figures S7A-B). To conclusively establish a pluripotent state-specific function of Tet1 and Tet2, we employed Tet-TKO ESCs, which self-renew normally in SL culture, and introduced individual Tet1 or Tet2 (and Tet2Mut) for transgenic rescue (Figure S7C) followed by the pluripotent state transition test. We found that Tet-TKO cells are defective in transitioning to either naive or primed pluripotent state (Figure 7A, black bars), supporting that Tet functions are important in regulating alternative pluripotent states. Our finding that Tet1 is critical for primed and Tet2 for naive pluripotency was substantiated by efficient transition to primed pluripotency and inhibition of naive pluripotency upon Tet1 rescue (Figure 7A, gray bars), and by efficient transition to naive pluripotency but inhibition of primed pluripotency upon Tet2 rescue (Figure 7A, blue bars). Such a critical role of Tet2 in promoting naive while inhibiting primed pluripotency is dependent on its catalytic activity (Figure S7D) and is manifested with rescued cells under FA for 6 days still expressing highly abundant Nanog (Figure S7E). Accordingly, we found that Tet2 promoted the upregulation of naive active class I/II genes and the repression of primed active class III/IV genes, whereas opposite effects on the same genes were detected upon the rescue with Tet1 (Figure 7B).
Figure 7. Opposing Functions of Tet1 and Tet2 in Primed and Naive Pluripotent State Transitions. See also Figure S7.
(A) Pluripotent state transition test of Tet-TKO ESCs rescued with empty vector (EV), Tet1, or Tet2 and cultured in SL, 2iL, and FA for 8 days. Proliferation rates of rescued Tet-TKO cells relative to WT cells (100%) were presented. Error bars indicate average ± SEM (n=3). Student's unpaired t test was performed (ns, not significant; **P<0.01).
(B) Q-PCR analysis for naïve (class I/II) and primed (class III/IV) marker expression in indicated transgenic lines under SL culture conditions. Error bars indicate average ± SEM (n=3).
(C) Ectopic expression of Tet2 promotes EpiSC reprogramming to a naive state. Quantification of the Epi-iPSC colony numbers was shown at day 6 of 2iL treatment.
(D) Contribution of Tet2 Epi-iPSCs (labeled with a Tomato reporter transgene) to the germline at E13.5 (top) and live-born chimeras (bottom) after blastocyst injection.
(E) Ectopic expression of a catalytic activity deficient Tet2 mutant (Tet2Mut) fails to reprogram EpiSCs. Representative images of alkaline phosphatase (AP) staining are also shown.
(F) Knockdown of Tet2, but not Tet1, compromises efficient EpiSC reprogramming mediated by Zfp281 depletion. EpiSC reprogramming was performed under the combinatorial KDs as indicated. All error bars in the reprogramming assays indicate average ± SEM (n=3). Student's unpaired t test was performed (ns, not significant; ***P<0.001).
(G) A model for Zfp281 functions in coordinating opposing functions of Tet1 and Tet2 for primed and naive pluripotency. (Top) The relative expression of Tet1/2, Zfp281, and PCF gene classes during pluripotent state transitions. (Bottom) In the primed pluripotent state, Zfp281 recruits Tet1 and HDAC-containing complexes to shared loci, leading to transcriptional activation of primed and repression of naive genes. In contrast, the downregulation of Zfp281 during naive pluripotent transition releases the Tet2 transcriptional and post-transcriptional (via primed specific miR-302/367) repression, leading to Tet2 upregulation and naive gene activation.
To further investigate the opposing functions of Tet1/2 proteins in regulating primed and naive pluripotent states, we examined how individual Tet proteins might affect the reprogramming of primed EpiSCs to a naive state. Remarkably, we found that Tet2 alone, but not Tet1 or Tet3, was able to facilitate the reprogramming of EpiSCs to naive Epi-iPSCs under 2iL condition (Figure 7C), supporting the naive specific function of Tet2. Germline contribution and live-born chimeras after blastocyst injection confirmed the pluripotency status of these Epi-iPSCs (Figure 7D). In contrast, Tet2Mut failed to promote EpiSC reprogramming to a naive state (Figure 7E). Given that endogenous Tet2 was not detectable in EpiSCs (Figure S7F) and upregulated at both protein and RNA levels upon Zfp281 knockdown (Figures S7G-H), and that we detected increased 5hmC levels at naive loci in Zfp281-knockdown EpiSCs (Figure S7I), we tested whether knockdown of Tet2 could abrogate the reprogramming effect of Zfp281 depletion. In agreement with our observations, downregulation of Tet2, but not Tet1, led to a significant reduction in the 5hmC levels at naive pluripotency gene loci (Figure S7I) and compromised reprogramming efficiency of Zfp281 KD EpiSCs (Figure 7F). Taken together, our data establish primed and naïve pluripotent state specific functions of Tet1 and Tet2, respectively, in regulating pluripotency.
Discussion
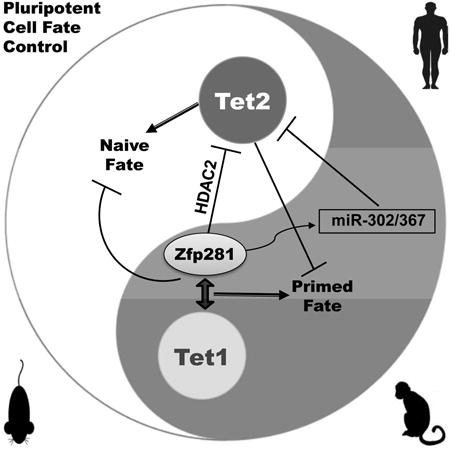
We have established a unique PCF gene signature that is evolutionarily conserved in distinguishing naive from primed pluripotent state. Investigation into the upstream regulators of the PCF gene expression signature underlying genetically identical and homogeneous naive and primed pluripotent cell populations led us to the discovery of Zfp281 as a key transcriptional regulator for primed pluripotency as well as opposing functions of the DNA hydroxylases Tet1 and Tet2 in regulating primed and naive pluripotent states, respectively. Our study demonstrates an important contribution of Tet2 to the epigenetic reprogramming of primed pluripotency to a naive state, whereas Tet1 and Zfp281 act together in promoting the naive to primed transition and safeguarding the primed pluripotency by inhibiting the naive cell fate (Figure 7G). Like Tet1 (Wu et al., 2011), Zfp281 exerts dual functions in transcriptional regulation of primed pluripotency, which is exemplified by activation of the primed specific gene miR-302/367 and repression of the naive specific gene Tet2 (Figure 7G).
Although global analyses of DNA methylation and relative Tet1 and Tet2 expression in 2iL and SL ESCs revealed dynamic changes of both 5hmC and Tet1/2 transcriptional levels during the time course of 2iL and SL conversion (Ficz et al., 2013; Habibi et al., 2013; Hackett et al., 2013; Leitch et al., 2013), our study for the first time elucidates how Tet1 can play a critical role in primed pluripotency and how Tet2 is regulated, i.e., repressed, at both transcriptional and post-transcriptional levels in primed cells to safeguard the primed pluripotency. It was proposed that demethylation involves oxidation, replicative loss, and repression of de novo methyltransferases Dnmt3a/b by Nanog and/or Prdm14 in 2iL naïve ESCs (Ficz et al., 2013; Habibi et al., 2013; Hackett et al., 2013; Leitch et al., 2013). Our study advocates regulatory roles of the key pluripotency factor Zfp281 in suppressing naïve and promoting primed pluripotency by activation of a subset of primed genes via recruitment of Tet1 and its DNA demethylation activity and repression of naive genes, exemplified by Tet2, through a dual HDAC2 and miR-302/367 repression mechanism (Figure 7G). Such a model was further supported by a close physical association of Zfp281 with epigenetic regulators HDAC2, Tet1, and Sin3a in EpiSCs (Figure S7J). Future studies using ChIP-seq to interrogate global binding profiles of both Tet1 and Zfp281 under FA conditions or in EpiSCs will be instrumental to further define how Zfp281 coordinates Tet1 functions in chromatin and target gene regulation, globally or locally, during the establishment and maintenance of primed pluripotency. Since Zfp281 in addition to Otx2 was also reported to be associated with primed specific function of Oct4 in EpiLCs (Buecker et al., 2014), whether it can function together with or independently of Otx2 in affecting Oct4 binding and reshaping core pluripotency factor binding profiles during naïve to primed state transition is worthy of future investigation. In this regard, an Otx2-independent function of Zfp281 is most likely due to the lack of a physical association between Zfp281 and Otx2 (Figure 6B).
Distinct roles of Tet1 and Tet2 in pluripotency and reprogramming have been suggested in the literature, although a detailed molecular understanding is lacking. While mapping 5hmC global distribution in Tet1- versus Tet2-depleted mouse ESCs, Huang et al. reported distinct roles for Tet1 and Tet2 in regulating promoter, exon, and polyadenylation site usage (Huang et al., 2014). While performing the fusion reprogramming, Piccolo et al. found that Tet1 was critically required for the imprint erasure during EGC and somatic cell fusion, whereas Tet2 was only required for the efficient reprogramming capacity of EGCs by upregulating pluripotency-associated genes after fusion (Piccolo et al., 2013). In contrast, imprinting is stably maintained in mouse ESC and somatic cell fusion, and a rapid accumulation of 5hmC was observed at pluripotency loci (e.g., Oct4) with a primary involvement of Tet2 function as observed in EGC and somatic cell fusion (Piccolo et al., 2013). These fusion reprogramming data lend further support on our findings that Tet2 promotes naive pluripotency, although it remains to be seen whether and how distinct functions of Tet1/2 may play a role in imprinting control during pluripotent state transition. In this regard, it is noteworthy that potential issues were recently reported (Gkountela et al., 2015; Pastor et al., 2016) to associate with the two well-recognized naive hESCs reverted from their conventional primed counterparts (Takashima et al., 2014; Theunissen et al., 2014), including the hotspots of “unbridled DNA demethylation”, a nonblastocyst DNA methylation pattern, and a near total loss of imprinting. Therefore, understanding the molecular determinants of primed and naive pluripotency encompassing ZNF281 and TET1/2 proteins and their mechanistic action in hESCs warrants further investigation.
Experimental Procedures
Additional experimental procedures are provided in the Supplemental Experimental Procedures.
In vitro Metastable to Naive and Primed Pluripotent Cell Fate Conversions
For conversion of mouse metastable pluripotent ESCs (SL) into naive or primed pluripotent cells, 3×104 cells/cm2 were seeded on 0.1% gelatin-coated plates using 2iL culture conditions or fibronectin-coated plates using FA culture conditions. Converted cells were passaged with Accutase (Innovative Cell Technologies, Inc.) at a 1:6 ratio every 2 days. Details of cell culture are given in the Supplemental Experimental Procedures.
Ectopic Expression and Endogenous Repression of miR-302/367
The miR-302/367 cluster was overexpressed by retroviral infection of pMIGR1-miR-302/367 construct and endogenous miR-302/367 was repressed by lentiviral infection with the vector pLenti-EF12-KRAB-302s-TALE1. Infected cells were FACS-sorted according to the expression of GFP, and RNA was extracted and processed for expression analysis using qPCR.
Luciferase Assay
Zfp281 KO and WT ESCs were transfected with psiCheck2 empty vector or containing Tet2 3′-UTR, as previously reported (Cheng et al., 2013). Twenty-four hours after transfection, the cells were cultured in 2iL, SL or FA medium for another 24 h. Luciferase and renilla activity were determined using Dual-Glo Luciferase Assay kit (#E2920, Promega) according to manufacturer's instructions.
ChIP Coupled with Sequencing (ChIP-seq)
ChIP was performed as previously described (Lee et al., 2006) by using Zfp281 antibody (ab101318, Abcam) in SL ESCs. See detailed analysis in the Extended Experimental Procedures. High throughput sequencing HiSeq2500 (Illumina) was performed in Mount Sinai Sequencing Core Facility.
RNA-seq and Analysis
Two biological replicates of RNA-seq for Zfp281 KO and WT ESCs cultured 48 hours in SL, 2iL, or FA conditions, and WT ESCs under long culture in 2iL (16 days) and stable EpiSCs in FA medium, respectively, were performed. See detailed analysis in the Extended Experimental Procedures.
Supplementary Material
Highlights.
Identification of gene expression signatures for alternative pluripotent states
Requirement of Zfp281 for the establishment and maintenance of primed pluripotency
Opposite functions of Tet1 and Tet2 for primed and naive pluripotency
Transcriptional and post-transcriptional repression of Tet2 for primed pluripotency
Acknowledgments
We thank Dr. Jun Lu (Yale) for the Tet2 3′-UTR luciferase reporter construct, Dr. Rudolf Jaenisch (MIT) for the OCT4-2A-GFP and OCT4-DPE-GFP reporter human ESCs, and Tet triple knockout ESCs. This research was funded by grants from the National Institutes of Health (NIH) to J.W. (1R01-GM095942), the Empire State Stem Cell Fund through New York State Department of Health (NYSTEM) to J.W. (C028103, C028121). J.W. is a recipient of Irma T. Hirschl and Weill-Caulier Trusts Career Scientist Award. M.F. has a “Ramon y Cajal” contract (RYC-2014-16779) from the Ministerio de Economia y Competitividad of Spain. A.S. is an awardee of the Traineeship of NIDCR-Interdisciplinary Training in Systems and Developmental Biology and Birth Defects (T32HD075735).
Footnotes
Author Contributions: M.F. conceived, designed and conducted the studies and wrote the manuscript; D.G., C.S-P., J.V., A.S., J.D., W.-S. W., C.C. provided reagents and performed experiments; X.H. provided bioinformatics support; J.W. conceived the project, designed the experiments, prepared and approved the manuscript.
Accession Numbers: Zfp281 ChIP-seq and RNA-seq data in mouse ESCs have been deposited with the Gene Expression Omnibus under the accession ID (GSE81045).
Supplemental Information: Supplemental Information includes Extended Experimental Procedures, seven figures and six tables that can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K, Surani MA. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461:1292–1295. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J, Nichols J, Dietmann S, Corrin PD, Paddison PJ, Smith A. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell. 2013;153:335–347. doi: 10.1016/j.cell.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Buecker C, Srinivasan R, Wu Z, Calo E, Acampora D, Faial T, Simeone A, Tan M, Swigut T, Wysocka J. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YS, Goke J, Ng JH, Lu X, Gonzales KA, Tan CP, Tng WQ, Hong ZZ, Lim YS, Ng HH. Induction of a Human Pluripotent State with Distinct Regulatory Circuitry that Resembles Preimplantation Epiblast. Cell Stem Cell. 2013;13:663–675. doi: 10.1016/j.stem.2013.11.015. [DOI] [PubMed] [Google Scholar]
- Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone OGlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Niu YY, Li YJ, Ai ZY, Kang Y, Shi H, Xiang Z, Yang ZH, Tan T, Si W, et al. Generation of Cynomolgus Monkey Chimeric Fetuses using Embryonic Stem Cells. Cell Stem Cell. 2015;17:116–124. doi: 10.1016/j.stem.2015.06.004. [DOI] [PubMed] [Google Scholar]
- Cheng J, Guo S, Chen S, Mastriano SJ, Liu C, D'Alessio AC, Hysolli E, Guo Y, Yao H, Megyola CM, et al. An Extensive Network of TET2-Targeting MicroRNAs Regulates Malignant Hematopoiesis. Cell reports. 2013 doi: 10.1016/j.celrep.2013.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YF, Chen HH, Eijpe M, Yabuuchi A, Chenoweth JG, Tesar P, Lu J, McKay RD, Geijsen N. The growth factor environment defines distinct pluripotent ground states in novel blastocyst-derived stem cells. Cell. 2008;135:449–461. doi: 10.1016/j.cell.2008.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AT. DNA methylation remodeling in vitro and in vivo. Curr Opin Genet Dev. 2015;34:82–87. doi: 10.1016/j.gde.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Barrasa MI, Raddatz G, Gao Q, Powell BE, Cheng AW, Faull KF, Lyko F, et al. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell. 2014;29:102–111. doi: 10.1016/j.devcel.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Huang X, Shao N, Zhou H, Lee DF, Faiola F, Fidalgo M, Guallar D, Saunders A, Shliaha PV, et al. Tex10 Coordinates Epigenetic Control of Super-Enhancer Activity in Pluripotency and Reprogramming. Cell Stem Cell. 2015;16:653–668. doi: 10.1016/j.stem.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Xu H, Faiola F, Ma'ayan A, Wang J. Oct4 links multiple epigenetic pathways to the pluripotency network. Cell Res. 2012;22:155–167. doi: 10.1038/cr.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Factor DC, Corradin O, Zentner GE, Saiakhova A, Song L, Chenoweth JG, McKay RD, Crawford GE, Scacheri PC, Tesar PJ. Epigenomic Comparison Reveals Activation of “Seed” Enhancers during Transition from Naive to Primed Pluripotency. Cell Stem Cell. 2014;14:854–863. doi: 10.1016/j.stem.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Hore Timothy A, Santos F, Lee Heather J, Dean W, Arand J, Krueger F, Oxley D, Paul YL, Walter J, et al. FGF Signaling Inhibition in ESCs Drives Rapid Genome-wide Demethylation to the Epigenetic Ground State of Pluripotency. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo M, Shekar PC, Ang YS, Fujiwara Y, Orkin SH, Wang J. Zfp281 functions as a transcriptional repressor for pluripotency of mouse embryonic stem cells. Stem Cells. 2011;29:1705–1716. doi: 10.1002/stem.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013 doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Galonska C, Ziller MJ, Karnik R, Meissner A. Ground State Conditions Induce Rapid Reorganization of Core Pluripotency Factor Binding before Global Epigenetic Reprogramming. Cell Stem Cell. 2015 doi: 10.1016/j.stem.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S, Zhang KX, Shafiq TA, Liao WW, Hargan-Calvopina J, Chen PY, Clark AT. DNA Demethylation Dynamics in the Human Prenatal Germline. Cell. 2015;161:1425–1436. doi: 10.1016/j.cell.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi E, Brinkman AB, Arand J, Kroeze LI, Kerstens HH, Matarese F, Lepikhov K, Gut M, Brun-Heath I, Hubner NC, et al. Whole-Genome Bisulfite Sequencing of Two Distinct Interconvertible DNA Methylomes of Mouse Embryonic Stem Cells. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Hackett JamieA, Dietmann S, Murakami K, Down ThomasA, Leitch HarryG, Surani MA. Synergistic Mechanisms of DNA Demethylation during Transition to Ground-State Pluripotency. Stem Cell Reports. 2013;1:518–531. doi: 10.1016/j.stemcr.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Surani MA. Regulatory Principles of Pluripotency: From the Ground State Up. Cell Stem Cell. 2014;15:416–430. doi: 10.1016/j.stem.2014.09.015. [DOI] [PubMed] [Google Scholar]
- Han DW, Tapia N, Joo JY, Greber B, Arauzo-Bravo MJ, Bernemann C, Ko K, Wu G, Stehling M, Do JT, et al. Epiblast stem cell subpopulations represent mouse embryos of distinct pregastrulation stages. Cell. 2010;143:617–627. doi: 10.1016/j.cell.2010.10.015. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nature biotechnology. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wang J. The extended pluripotency protein interactome and its links to reprogramming. Curr Opin Genet Dev. 2014;28:16–24. doi: 10.1016/j.gde.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Chavez L, Chang X, Wang X, Pastor WA, Kang J, Zepeda-Martinez JA, Pape UJ, Jacobsen SE, Peters B, et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2014;111:1361–1366. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch H, McEwen K, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich J, Smith A, et al. Naive pluripotency is associated with global DNA hypomethylation. Nature structural & molecular biology. 2013;20:311–316. doi: 10.1038/nsmb.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur BD, Sevilla A, Lenz M, Muller FJ, Schuldt BM, Schuppert AA, Ridden SJ, Stumpf PS, Fidalgo M, Ma'ayan A, et al. Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity. Nat Cell Biol. 2012;14:1139–1147. doi: 10.1038/ncb2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Orkin SH. Chipping away at the embryonic stem cell network. Cell. 2005;122:828–830. doi: 10.1016/j.cell.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Chen D, Liu W, Kim R, Sahakyan A, Lukianchikov A, Plath K, Jacobsen SE, Clark AT. Naive Human Pluripotent Cells Feature a Methylation Landscape Devoid of Blastocyst or Germline Memory. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo FM, Bagci H, Brown KE, Landeira D, Soza-Ried J, Feytout A, Mooijman D, Hajkova P, Leitch HG, Tada T, et al. Different Roles for Tet1 and Tet2 Proteins in Reprogramming-Mediated Erasure of Imprints Induced by EGC Fusion. Molecular cell. 2013 doi: 10.1016/j.molcel.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, et al. Resetting Transcription Factor Control Circuitry toward Ground-State Pluripotency in Human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, et al. Systematic Identification of Culture Conditions for Induction and Maintenance of Naive Human Pluripotency. Cell Stem Cell. 2014 doi: 10.1016/j.stem.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Weinberger L, Ayyash M, Novershtern N, Hanna JH. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol. 2016 doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Eve Sun Y, Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Izpisua Belmonte JC. Dynamic Pluripotent Stem Cell States and Their Applications. Cell Stem Cell. 2015;17:509–525. doi: 10.1016/j.stem.2015.10.009. [DOI] [PubMed] [Google Scholar]
- Wu J, Okamura D, Li M, Suzuki K, Luo C, Ma L, He Y, Li Z, Benner C, Tamura I, et al. An alternative pluripotent state confers interspecies chimaeric competency. Nature. 2015;521:316–321. doi: 10.1038/nature14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.