Abstract
As a master regulator of chromatin function, the lysine methyltransferase EZH2 orchestrates transcriptional silencing of developmental gene networks. Overexpression of EZH2 is commonly observed in human epithelial cancers, such as non-small cell lung carcinoma (NSCLC), yet definitive demonstration of malignant transformation by deregulated EZH2 remains elusive. Here, we demonstrate the causal role of EZH2 overexpression in NSCLC with new genetically-engineered mouse models of lung adenocarcinoma. Deregulated EZH2 silences normal developmental pathways leading to epigenetic transformation independent from canonical growth factor pathway activation. As such, tumors feature a transcriptional program distinct from KRAS- and EGFR-mutant mouse lung cancers, but shared with human lung adenocarcinomas exhibiting high EZH2 expression. To target EZH2-dependent cancers, we developed a novel and potent EZH2 inhibitor JQEZ5 that promoted the regression of EZH2-driven tumors in vivo, confirming oncogenic addiction to EZH2 in established tumors and providing the rationale for epigenetic therapy in a subset of lung cancer.
Keywords: EZH2, non-small cell lung cancer, methyltransferase inhibitor, EZH2 inhibitor, JQEZ5, EZH2-driven lung cancer GEMM
Introduction
Lung cancer is the most common and one of the most deadly cancers worldwide (1). Non-small cell lung cancers (NSCLC) are the most prevalent type of lung cancer, comprising a heterogeneous set of diseases (2). The identification of recurrent mutations and amplifications in many targetable oncogenes has significantly improved overall survival of subsets of NSCLC patients. Activating mutations in BRAF, KRAS and the epidermal growth factor receptor (EGFR), as well as fusions involving anaplastic lymphoma kinase (ALK), have been associated with response to kinase inhibition (3–6). Furthermore, with the advent of improved genomic profiling and next-generation sequencing, recurrent mutations and amplifications have been identified in HER2, MET, fibroblast growth factor receptor 1 (FGFR1) and FGFR2, the ROS1 receptor tyrosine kinase, neuregulin 1 (NRG1), neurotrophic tyrosine kinase receptor type 1 (NTRK1) and RET (2). While together these alterations account for many cases of lung adenocarcinoma, a considerable population of NSCLC patients lacks identifiable genetic lesions in therapeutically tractable targets.
Beyond growth factor signaling pathways, chromatin-associated complexes have recently been identified as recurrently altered or transcriptionally deregulated in NSCLC, including TET methylcytosine dioxygenase 2 (TET2), DNA methyltransferase 3A (DNMT3A) and enhancer of zeste homologue 2 (EZH2) (7). Notably, each of these factors influences heterochromatin structure, and has been linked to coordinated regulation of normal developmental transcriptional pathways (8–11). These data establish the hypothesis that disruption of chromatin architecture is a common event in lung cancer pathogenesis, either permissive with or distinct from oncogenic signaling pathways, functioning to deregulate transcriptional programs associated with cellular differentiation.
The dynamic structure of chromatin is influenced by post-translational modifications (PTMs) to DNA and to the unstructured amino-terminal tails of histone proteins within nucleosomal particles. Control of gene expression pathways by DNA-binding transcriptional activators and repressors influences the recruitment of chromatin-associated enzyme complexes that confer covalent PTMs to chromatin. In general, side-chain acetylation of lysine residues on histone tails is associated with active, euchromatin, notably at histone 3 lysine 27 as associated with active cis-regulatory enhancer elements (H3K27ac) (12). Modification of H3K27 exhibits switch-like behavior, as mono-, di- and tri-methylation of H3K27 (H3K27me1, -me2, -me3) is associated with repressive, facultative heterochromatin (13). H3K27 methylation is principally mediated by the polycomb group repressive complex 2 (PRC2), a multi-protein assembly that activates and directs the function of a core catalytic enzyme EZH2 mediating S-adenosyl methionine dependent lysine methylation.
Recurrent alteration of EZH2 is observed in solid and hematologic malignancies, underscoring the unexpected centrality of chromatin structure in the pathogenesis of cancer. Interestingly, both activation (recurrent mutation, overexpression) and inactivation (deletions, inactivating mutations) of EZH2 have been characterized, supporting a tissue-specific role for EZH2 as either an oncogene or tumor suppressor. EZH2 activating mutations have been characterized in B-cell lymphoma (14, 15). More broadly than these focused genetic events, over-expression of EZH2 is found in a wide range of cancers (16, 17). While overexpression is associated with increased global H3K27me3, prompts silencing of tumor suppressors and developmental regulators and often confers a poor prognosis, it can also restrain tumorigenesis in specific epithelial contexts (18–21). Of relevance to lung adenocarcinoma, several recent studies reproducibly demonstrated a correlation between increased EZH2 expression and poor outcome (22–24).
EZH2 has thus emerged as a pressing target for cancer therapeutic development. Strategies have been undertaken to develop disruptors of complex assembly (25), as well as SAM-competitive inhibitors of the canonical SET lysine methyltransferase domain (26–28). Selective EZH2 inhibition using these chemical probes has established EZH2 as a context-specific tumor dependency while providing pharmacologic target validation in B-cell lymphoma (26–28) and defined soft-tissue sarcomas (29, 30). Accordingly, human clinical investigation has been initiated using drug-like EZH2 inhibitors (ClinicalTrial.gov identifier: NCT01897571, NCT02082977, NCT02395601, NCT02601937, NCT02601950).
The evident overexpression of EZH2 in lung adenocarcinoma and the feasibility of clinical investigation motivated the present effort to characterize the effect of transcriptional deregulation of EZH2 on lung cancer pathogenesis. Using genetic and chemical genetic approaches, we demonstrate for the first time an oncogenic role for wild-type EZH2 overexpression in lung cancer and the opportunity for epigenomic therapy in this disease. Specifically, we generated genetically-engineered mouse models (GEMMs) overexpressing wild-type human EZH2 systemically and specifically in lung. We show that EZH2 overexpression promotes the formation of lung tumors that exhibit biochemical and transcriptional features akin to the subset of human tumors that express high levels of EZH2. Analysis of chromatin state in EZH2 overexpressing lung tumors revealed the aberrant spread of H3K27me3 notably at developmental regulator gene loci, many of which are known tumor suppressors in lung cancer. To overcome limitations in potency, availability and in vivo utility of current EZH2 inhibitors, we developed and characterized a novel and open-source EZH2 chemical probe, JQEZ5. In GEMM and human NSCLC models, JQEZ5 exhibits excellent exposure and pharmacodynamic target modulation. Long-term treatment of EZH2-addicted, tumor-bearing mice with JQEZ5 uniformly led to decreases in tumor burden. Together, these studies reveal a role for EZH2 as a NSCLC driver gene and an opportunity for targeted epigenomic therapy.
Results
EZH2 overexpression causes murine lung cancer
To investigate the causal role of EZH2 overexpression in cancer, we ubiquitously enforced EZH2 expression in the mouse using two different strategies to control for temporal specificity. All mice were engineered to carry one copy of a transgene expressing full-length human EZH2 containing a STOP cassette flanked by loxP sites between the CAG promoter and the EZH2 gene (LSL-EZH2) (Supplementary Figure S1A–B). We utilized two different strategies to induce EZH2 overexpression using Cre recombinase (Figure 1A). First, Actin-Cre was used to constitutively overexpress EZH2 in all tissues of the mouse (Actin-Cre:LSL-EZH2). Secondly, Ubiquitin-Cre-ERT2 (UBC:LSL-EZH2) was used to ubiquitously overexpress EZH2 upon treatment with tamoxifen at 6 weeks of age.
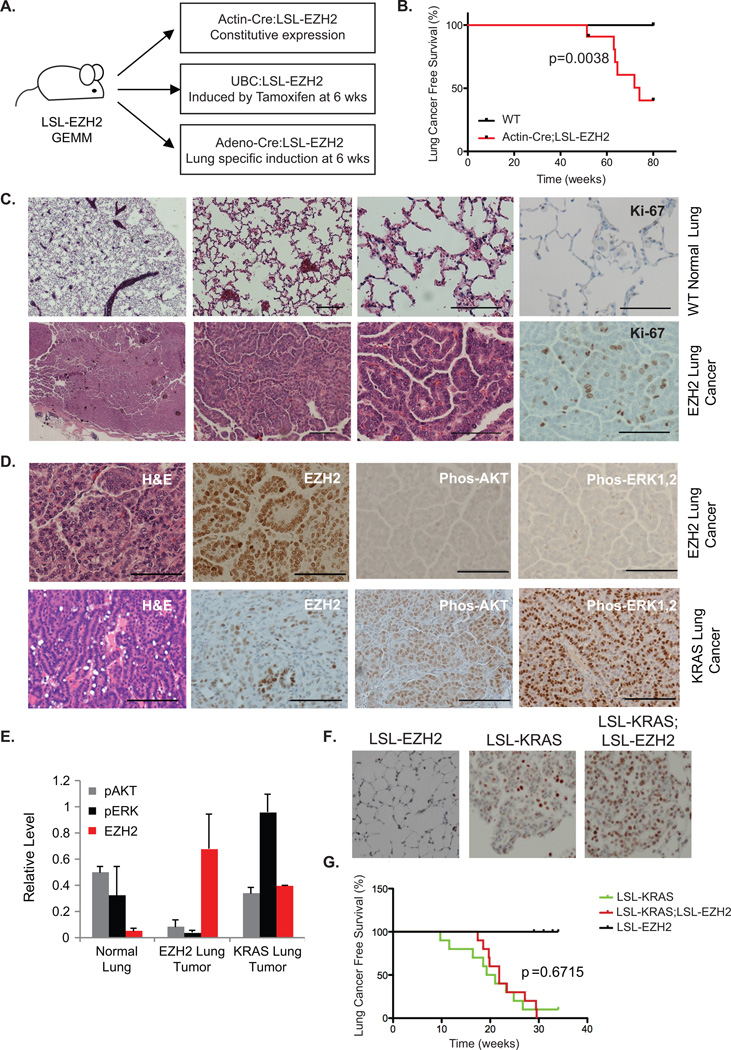
Figure 1. EZH2 Overexpression Induces Murine Lung Cancer.
(A) Schematic depiction of LSL-EZH2 genetically engineered mouse model utilizing 3 different strategies to express Cre recombinase to induce EZH2 overexpression.
(B) Kaplan–Meier lung cancer-free survival summary plot for Actin-Cre:LSL-EZH2 transgenic mice (EZH2) versus wildtype mice (WT).
(C) Histology of wildtype lung (top) and EZH2-induced lung adenocarcinomas (bottom) Sections stained with hematoxylin and eosin (H&E) or immunostained for Ki-67. Scale bar=50µm.
(D) H&E staining or immunostaining for EZH2, p-AKT, and p-ERK1/2 in EZH2-induced mouse lung tumors (top) and KRAS-induced mouse lung tumors (bottom). Scale bar=50µm.
(E) Relative protein expression levels of p-AKT, p-ERK1,2 and EZH2 in normal, EZH2-induced, and KRAS-induced tumor lung tissues measured by Western blot and quantified with ImageJ. n=3 for normal lung and EZH2-induced tumor, n=2 for KRAS-induced tumor.
(F) Indicated mice were sacrificed 6 weeks post Cre-induction for immunostaining for EZH2.
(G) Kaplan–Meier lung cancer-free survival summary plot for the indicated mice.
Actin-Cre:LSL-EZH2 mice overexpressed EZH2 as demonstrated by both immunohistochemistry (IHC) and Western blotting (Supplementary Figure S1C–D). These animals were viable, fertile, developmentally normal, and indistinguishable from their littermates that did not express Cre recombinase through adulthood (data not shown). Thus, overexpression of EZH2 is tolerable during embryonic and developmental growth. As the Actin-Cre:LSL-EZH2 mice entered adulthood, multiple tumor types were observed including lymphoma, histiocytic sarcoma of the liver and lung adenomas/adenocarcinomas. Half of the mice developed lung adenomas/adenocarcinomas without apparent metastases (Supplementary Table S1–2). Likewise, UBC:LSL-EZH2 mice that were administered tamoxifen at 6 weeks of age also developed lung adenocarcinomas with 40% penetrance. As expected, wild-type mice had no evident phenotype and all harvested lungs were normal at 80 weeks, suggesting a causal role of EZH2 overexpression in lung tumorigenesis. To extend these findings, we restricted EZH2 overexpression to lung in a third GEMM using inhaled Adeno-Cre virus to direct Cre expression to the pulmonary epithelium of LSL-EZH2 mice (Figure 1A) (31). Adeno-Cre was administered to animals at 6-weeks of age and 42% of these animals developed lung adenocarcinoma, demonstrating that EZH2 overexpression in lung epithelial cells is sufficient to induce cancer. In sum, our data demonstrate that 45% of EZH2-overexpressing mice develop lung adenocarcinomas with an average survival to 73.6 weeks of age (Figure 1B).
The histology of mouse lung tumors demonstrated features of human grade 1–2 lung adenoma/adenocarcinoma. As compared to normal mouse lung, EZH2-overexpressing lung adenocarcinomas showed high cellularity and less differentiation, all consistent with low- and intermediate-grade adenocarcinoma (Figure 1C). IHC analysis of tumors arising from EZH2 overexpression demonstrated an increase in the proliferative marker, Ki67, as compared to normal lung tissue (Figure 1C). In a comparison to murine lung cancers driven by expression of activated KRAS (KRASG12D), EZH2 expression was markedly higher in the EZH2-driven lung tumors (Figure 1D). Analysis for the expression of pathway markers typically identified in KRAS-driven lung cancer, such as phosphorylated AKT (p-AKT) and phosphorylated ERK (p-ERK), revealed low p-AKT and p-ERK expression in EZH2-induced mouse lung tumors (Figure 1D). Western blot analysis further confirmed that EZH2 mouse lung tumors have significantly less p-AKT and p-ERK than both KRAS-induced mouse lung tumors and normal mouse lung (Figure 1E and Supplementary Figure S1E). Taken together, these data suggest that lung tumors driven by EZH2-mediated epigenomic deregulation are histologically similar but molecularly distinct from lung tumors driven by KRAS-dependent oncogenic signaling.
As KRAS and EZH2 expression are not mutually exclusive (Figure 1D), we wanted to explore possible synergy between KRAS and EZH2 in lung tumorigenesis. KRASG12D (LSL-KRAS) was introduced into the lung epithelium with or without EZH2 overexpression (+/− LSL-EZH2) via inhalation of viral Adeno-Cre. Six weeks post induction, LSL-KRAS and LSL-KRAS;LSL-EZH2 mice had comparable tumors with comparable morphology, even though EZH2 overexpression was only evident in the LSL-KRAS;LSL-EZH2 mice (Figure 1F). Significantly, we did not observe an increase in tumorigenesis or a decrease in survival in the presence of EZH2 overexpression (Figure 1G), suggesting that EZH2 overexpression does not modify tumorigenesis driven by the KRAS oncogene.
EZH2-driven lung cancer as a molecularly distinct entity
To dissect the molecular features of EZH2-induced mouse lung cancer, we first performed RNA sequencing (RNA-seq) to compare the gene expression profiles of EZH2-overexpressing pre-cancerous normal lung tissue and EZH2-overexpressing lung adenocarcinoma tumors from mice (Supplementary Table S3). Using unsupervised hierarchical clustering, gene expression profiles from these samples were compared to expression profiles of EGFR-mutated and KRAS-mutated lung adenocarcinoma mouse tumors (Figure 2A). EZH2-overexpressing tumors segregated as transcriptionally distinct from EZH2-overexpressing normal lung, EGFR- and KRAS-mutated lung tumors. These data suggest that EZH2 modulation of chromatin leads to alteration of transcriptional pathways distinct from canonical lung adenocarcinomas driven by EGFR and KRAS hyperactivating mutations. Additionally, analysis of RNA-Seq data for secondary driver mutations revealed that there are no recurrent somatic alterations in the coding portion of expressed known driver oncogenes or tumor repressors in murine EZH2-overexpressing tumors (data not shown).
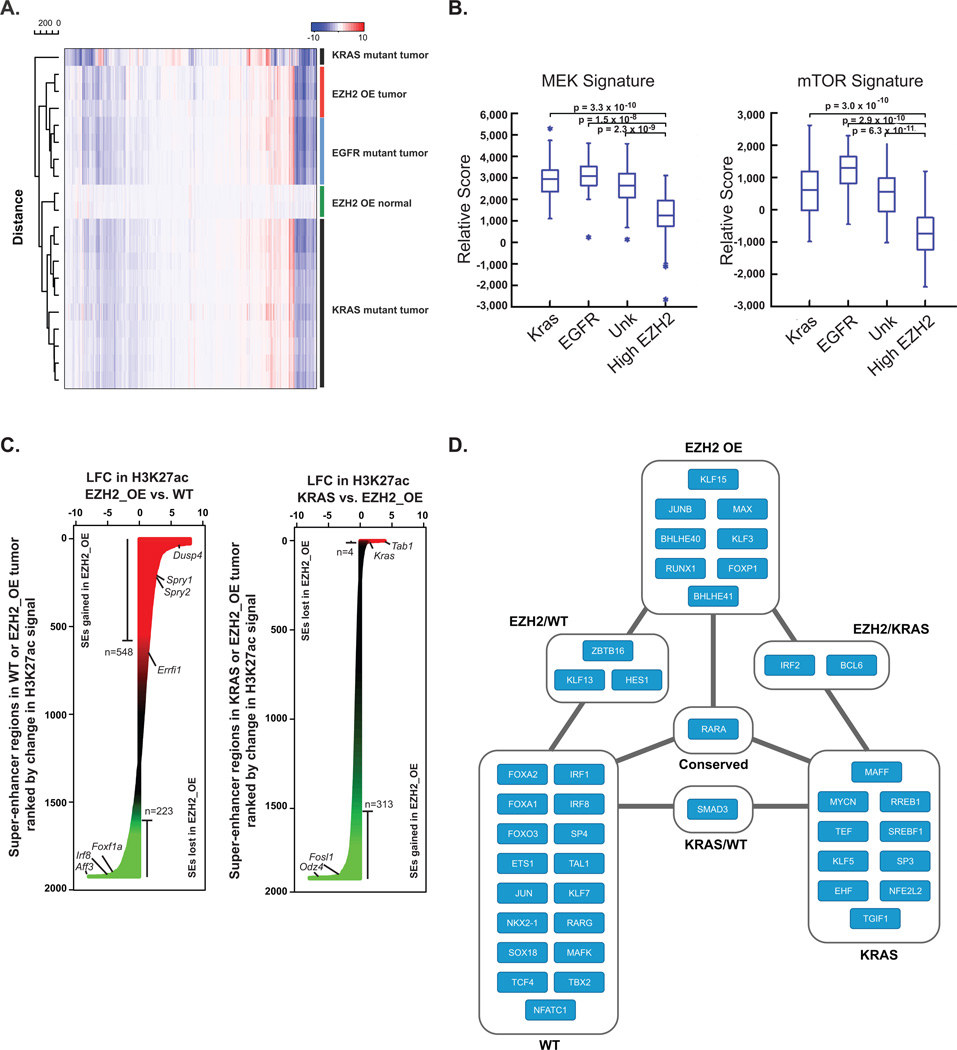
Figure 2. EZH2-Driven Lung Cancer as a Molecularly Distinct Entity.
(A) Heatmap of log2 fold-change (LFC) gene expression in murine EZH2-overexpressing (OE) normal lungs (green), KRAS-mutant lung tumors (black), EGFR-mutant lung tumors (blue), and EZH2-OE lung tumors (red). All genes were selected across all samples for clustering.
(B) Box plot of ssGSEA comparing the enrichment of a MEK (left) and mTOR (right) gene sets in human TCGA lung adenocarcinomas with specific driver mutations (KRAS, EGFR, unknown) or high EZH2 levels.
(C) Waterfall plot showing rank-ordered change in H3K27ac signal at SE-containing regions between mouse WT lung and EZH2_OE (left), and KRAS and EZH2_OE (right). X-axis depicts the LFC in H3K27ac signal. SEs are ranked by LFC in signal with regions gaining the most H3K27ac in tumor at the top.
(D) Core transcriptional regulatory circuitry in murine wildtype (WT) and tumor lung tissues (EZH2- or KRAS-driven) as defined by ChIP-seq for H3K27ac. Nodes are TFs that are associated with an SE. Edges indicate a regulatory interaction between two TFs as defined by an enrichment of TF binding motifs in the respective SE.
Having defined an EZH2-dependent, tumor-specific transcriptional state in murine lung adenocarcinoma, we next assessed whether a comparable subset of human NSCLC exists. Using publicly available data from The Cancer Genome Atlas (TCGA), we identified a cohort of lung adenocarcinoma patients with elevated tumor EZH2 expression (Supplementary Figure S2A). Nearly all lung tumors overexpress EZH2 as compared to normal tissue. EZH2 expression in human lung cancer was found to be broadly distributed over a >50-fold range and high EZH2 levels were not mutually exclusive with KRAS or EGFR mutations. In tumors with the highest EZH2 expression (mean + 2 SD, 28 tumors), 21% harbor canonical “driver mutations” (e.g. EGFR and KRAS) whereas 43% of tumors harbored these mutations in the overall dataset (230 tumors). We further selected highly EZH2-overexpressing tumors (top 20%) with wild-type KRAS and EGFR to emulate the genetics of our murine models. Pathway enrichment was assessed by Gene Set Enrichment Analysis (GSEA) (32, 33). Transcriptional signatures associated with MEK and mTOR activation were repressed in EZH2-overexpressing tumors as compared to tumors with low EZH2 expression regardless of the presence or absence of oncogenic KRAS or EGFR mutations (Figure 2B), corroborating again that EZH2 driven tumors are molecularly distinct from tumors driven by canonical signaling pathways.
Influence of EZH2 overexpression on chromatin structure in murine lung cancer
To understand the dynamic effects of EZH2 overexpression on chromatin structure in the context of malignant transformation, we performed comparative epigenomic analysis of normal and malignant murine lung tissues. We and others have used genome-wide assessment of enhancer-promoter activity, measured by H3K27ac chromatin immunoprecipitation with massively parallel DNA sequencing (ChIP-seq), to comparatively study malignant and inflammatory cell states (34, 35). Focusing epigenomic analysis on regions of massive H3K27ac enrichment, so-called super enhancers (SEs), has afforded inferences into oncogenic signaling (36) and the sub-classification of human tumors (34). Therefore, we first mapped active enhancers across the three tissue types by H3K27ac ChIP-seq, and identified regions of differential hyperacetylation (SEs), as we previously reported (34, 37). Unsupervised hierarchical clustering segregated murine tumor (KRAS- and EZH2-driven tumors) and pulmonary tissue (WT), demonstrates a distinct euchromatin epigenome structure (Supplementary Figure S2B). Differential analysis of highly occupied H3K27ac regions in EZH2-overexpressing tumors and normal lung revealed global redistribution of H3K27ac, with 771 individual loci exhibiting a greater than log2 1.5-fold change in H3K27ac (Figure 2C, left). The same analysis between EZH2-overexpressing and KRAS-driven tumors also revealed a distinct redistribution of H3K27ac, with 317 individual loci exhibiting a greater than log2 1.5-fold change in H3K27ac (Figure 2C, right).
Differences in gene expression and the SE signatures upon EZH2 overexpression versus KRAS-driven transformation in mice may be due to changes in the activated signaling molecules leading to modulation of different transcription factor (TF) networks. To identify TFs responsible for the gene expression changes and SE signatures upon EZH2 versus KRAS transformation, we identified TF binding sites enriched at SEs activated upon transformation. We constructed an SE-defined transcriptional network for normal and tumorigenic cell states by integrating maps of SE-associated master TFs with TF binding site data to reconstruct the core transcriptional regulatory circuitry (38). Our analysis defines three independent core transcriptional regulatory circuitries, which further underscores the mechanistically unique tumorigenic activity of EZH2 versus KRAS (Figure 2D). Thus, we have further established a unique signature of EZH2-overexpressing murine lung tumors, which exhibit divergent core circuitries where deregulated master TFs are enforced by a unique network of super-enhancer associated TFs.
Influence of super enhancers on gene expression in EZH2-overexpressing lung cancer
To determine whether changes in H3K27ac at SEs resulted in reciprocal changes in gene expression at adjacent expressed genes, we analyzed RNA-seq data from WT and EZH2-overexpressing murine tumors and found that the modulation of chromatin impacted gene expression in these tumors (Figure 3A). Unbiased leading edge analysis of genes proximal to regions of lost H3K27ac in murine tumors identified polycomb repressive target gene signatures, implicating PRC2-mediated repression of these regulatory elements (Figure 3B,C).
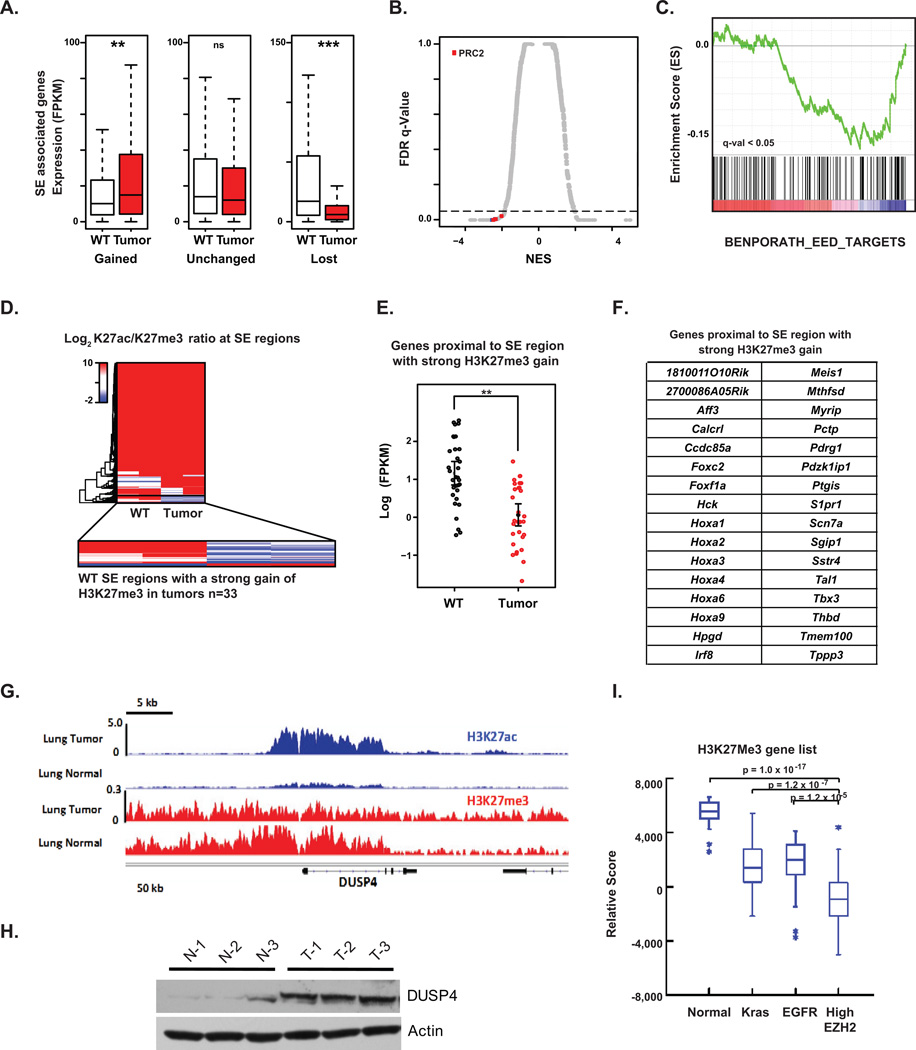
Figure 3. EZH2 Overexpression Establishes a Unique and Conserved Super Enhancer-Associated Transcriptional Landscape.
(A) Box plot of RNA-seq expression in units of FPKM of murine genes associated with SEs that are gained (1812 genes), unchanged (4421 genes), or lost (432 genes) in tumor versus WT lung tissues. Significance was calculated using a two-tailed t test. ** p<2e–4, *** p <2e–6
(B) Scatter plot of normalized enrichment score (NES) versus false discovery rate (FDR) q-value comparing MSigDB curated gene set enrichment in murine tumor versus WT SE-associated genes. X-axis shows NES for evaluated gene sets. Y-axis shows false FDR q-value for each gene set. Gene sets upregulated in tumors have a high positive NES, while downregulated gene sets have a negative NES. Dotted line indicates significance cutoff q-value of 0.05. Red dots indicate PRC2 associated signatures, n = 8.
(C) SE-associated gene set enrichment analysis showing downregulation of EED targets in murine tumor versus WT tissues from RNA-seq analysis.
(D) Heatmap of LFC in H3K27ac over H3K27me3 signals at SE-containing regions. Blue regions indicate SEs with strong gains of H3K27me3 in tumor versus WT, while red regions indicate those with strong losses.
(E) Dot plot of RNA-seq expression in units of log10 FPKM for genes proximal to SE regions with a strong gain of H3K27me3 in murine tumor versus WT. Significance was calculated with a two-tailed t test. **p<1e–5.
(F) 32 mouse genes proximal to SE regions with strong H3K27me3 gain in EZH2-overexpressing tumors.
(G) Gene tracks of ChIP-seq signals in units of rpm/bp for H3K27ac and H3K27me3 at the DUSP4 locus in either murine WT or tumor lung tissues.
(H) Western blot analysis of lysates prepared from murine normal lung (N-1, N-2 and N-3) and lung tumor (T-1, T-2 and T-3) samples.
(I) Box plot of ssGSEA comparing the enrichment of our mouse H3K27me3 gene set in human TCGA lung adenocarcinomas with high EZH2 levels and normal lung tissue. The H3K27me3 gene set is comprised of the 32 mouse genes proximal to SE regions with strong H3K27me3 gain in murine EZH2-overexpressing tumors.
To link EZH2 function to lost SEs, we next performed H3K27me3 ChIP-seq on WT and EZH2-overexpressing mouse samples. Among regions of lost H3K27ac SEs, we identified a distinct subgroup of 33 cis-regulatory regions where loss of H3K27ac was accompanied by strong gain of the polycomb H3K27me3 mark (Figure 3D). Functionally, 32 genes associated with these 33 regulatory regions showed decreased gene expression in murine tumors by RNA-seq (Figure 3E), and were comprised of numerous developmental transcriptional regulators, including Foxf1a, Irf8, Hoxa9, and Meis1, as well as other chromatin factors such as Aff3 (Figure 3F). Repression of Hoxa9, Meis1, Irf8, and Foxf1 have been observed in NSCLC, and their activity has been functionally linked to decreased tumorigenesis, suggesting a tumor suppressive role for these gene regulator (39, 40). Visual inspection of master developmental TFs repressed in tumor samples confirmed an epigenomic switch from large hyperacetylated enhancer elements to broad regions of H3K27 trimethylation, (Supplementary Figure S2C–E).
Among the gained SE-associated regions in murine tumor samples, notably, were genes encoding well-characterized, negative regulators of the MAPK-ERK pathway: Dusp4, Spry1, Spry2 and Errfi1 (Figure 2C, left). Visual inspection corroborated that all four genes featured robust gain in H3K27ac at cis-regulatory elements (Figure 3G–H), and RNA-seq confirmed elevated expression in tumors. These data identify enhancer remodeling attributable to overexpressed EZH2 in the progression to lung adenocarcinoma, specifically silencing normal differentiation genes and activating negative regulators of MAPK-ERK signaling consistent with the signal transduction immunophenotyping of the EZH2-driven GEMMs.
To explore the relevance of these findings to human lung cancer pathophysiology, we next asked whether downregulation of the PRC2 hypermethylated, SE-associated genes identified in murine tumors is observed in EZH2-overexpressing human lung adenocarcinoma. Indeed, strong downregulation of the functional, 32 gene set is observed in human lung cancers with the highest (top 20%) expression of EZH2 and not in normal lung or tumors driven by KRAS or EGFR (Figure 3I). Taken together, integrated epigenomic analysis argues that there is a distinct subset of human lung cancers that are characterized by 1) high levels of EZH2, 2) low activation of Ras effectors, and 3) suppression of a distinct set of EZH2 target genes.
Human NSCLC with high EZH2 expression is sensitive to EZH2 depletion
To examine the oncogenic potential of EZH2 in human cells, we overexpressed EZH2 in an immortalized normal human lung epithelial cell line (hTBE) and monitored oncogenic potential (Supplementary Figure S3A). EZH2-overexpressing hTBE cells (oeEZH2) formed colonies on soft agar and acquired higher transformation capacity over time (serial passages) as compared to control cells (Supplementary Figure S3B–C). Differential SE landscape analysis from hTBE and hTBE-oeEZH2 cells illustrates a significant gain and loss of SEs when EZH2 is overexpressed (Supplementary Figure S3D). These data demonstrate that overexpressing EZH2 modifies the SE landscape as a primary effect of its overexpression in human cells.
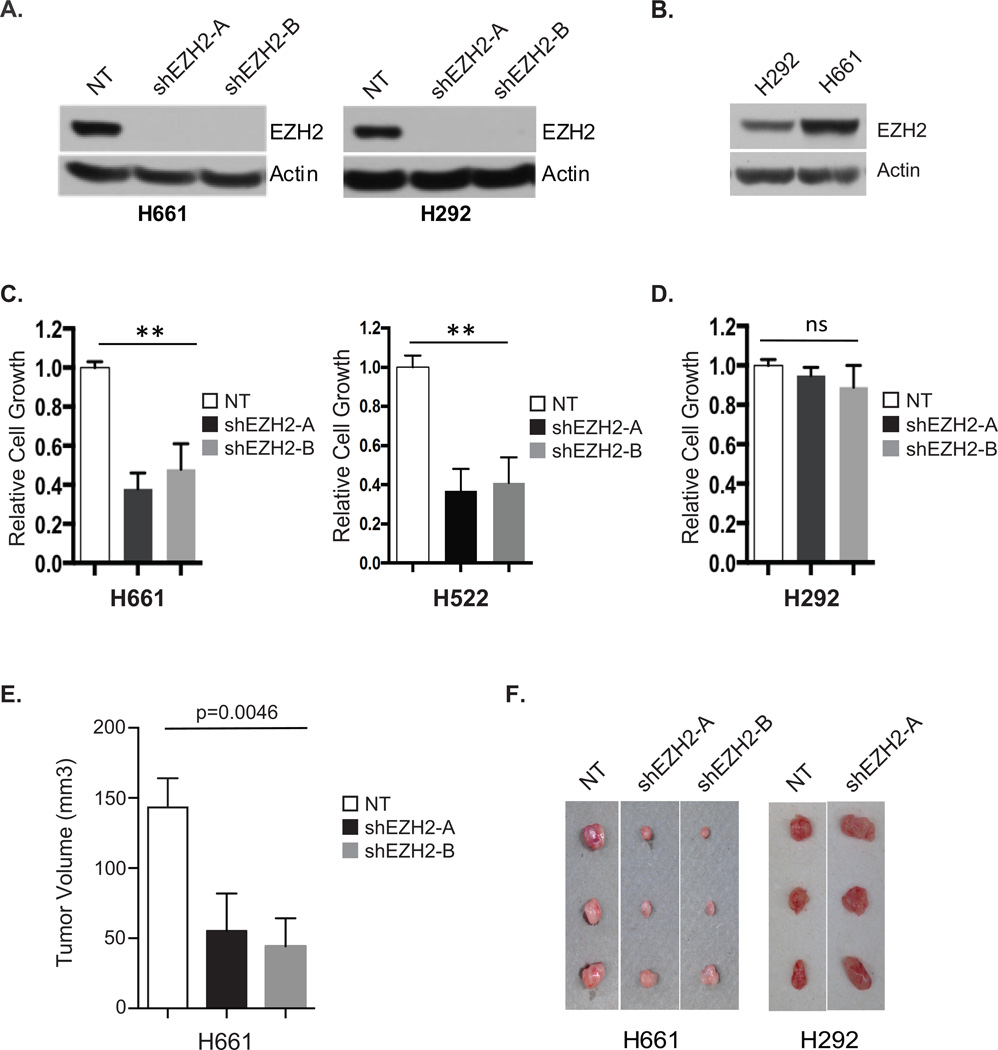
The observation that EZH2 overexpression produces lung cancer in mice and transforms normal human lung epithelia cells suggests that EZH2 may play an essential role in a subset of human NSCLCs. Using two previously verified EZH2 shRNAs, we knocked down EZH2 expression in a panel of rare, human oncogene wild-type NSCLC cell lines (41–43). Both shRNAs showed nearly complete inhibition of EZH2 protein expression as compared to cells with a non-targeting shRNA control (Figure 4A). Cell proliferation assays revealed that H522 and H661 cells, which express high levels of EZH2 with no other known oncogenic mutations, displayed more than 50% growth inhibition in response to EZH2 knockdown compared to control cells (Figure 4B–C, Supplementary Figure S3E). Growth of human NSCLC cell lines expressing slightly lower levels of EZH2, namely H292, H969, H2250, and H2258 cells, were largely unaffected by knockdown of EZH2 (Figure 4B,D, Supplementary Figure S3E–F). Finally, we tested whether EZH2 expression was required for tumor formation in mouse xenograft models of human NSCLC. H661 and H292 cells expressing either control or shEZH2 vectors were injected subcutaneously into mice and tumor formation was monitored bi-weekly. EZH2 knockdown significantly inhibited growth of H661 tumors in vivo while having no effect on H292 tumor formation (Figure 4E, F). Therefore, we identified human lung cancer cell lines that are oncogene wildtype and highly dependent on EZH2 overexpression.
Figure 4. A Subset of Human NSCLC Cells are Dependent on EZH2 Overexpression.
(A) Human NSCLC H661 (left) and H292 (right) cells expressing non-targeting control shRNA (NT) or two different shRNAs targeting EZH2 (shEZH2-A and shEZH2-B) were analyzed for EZH2 expression by Western blotting.
(B) Western blots comparing EZH2 expression levels between human NSCLC cell lines H661 and H292.
(C–D) Relative cell growth of H661, H522 (C) or H292 (D) cells expressing non-targeting control shRNA (NT) or two different shRNAs targeting EZH2 (shEZH2-A and shEZH2-B) was measured by MTS assay. Error bars represent S.E.M., n= 3. ** p < 0.001.
(E–F) Human NSCLC cell lines, H661 and H292, infected with lentivirus containing control (NT) or shEZH2 (shEZH2-A and shEZH2-B) were subcutaneously injected into the flank of nude mice. When the biggest tumor reached approximately 150 mm3, mice were euthanized and tumors were quantified relative to shNT tumor size (E) and documented (F). Mean ± SEM, n=3/treatment.
Dependency of EZH2-overexpressing lung cancer on catalytically active EZH2
To assess the dependency of EZH2-overexpressing lung tumors on sustained EZH2 enzymatic activity, we employed a chemical genetic approach. Recently, several pyridinone-based small molecule inhibitors of EZH2 were reported as chemical probes (e.g. GSK-126, UNC1999 and EPZ-6438; Supplementary Figure S4A) (28, 44, 45). Both of these near structural analogues are potent and selective inhibitors of EZH2, however the broader utility of these probes in biological research in vivo may be limited by low potency (high dose administration), limited bioavailability (twice daily dosing), and uncertain availability (cost of synthesis, pharmaceutical material transfer, undisclosed structure of EPZ-6438 while we conducted these studies). We therefore undertook to develop a novel EZH2 inhibitor as an open-source chemical probe for the scientific community. Lacking the guidance of crystallographic data, structure-activity relationships were deduced empirically from iterative analogue synthesis and biochemical testing.
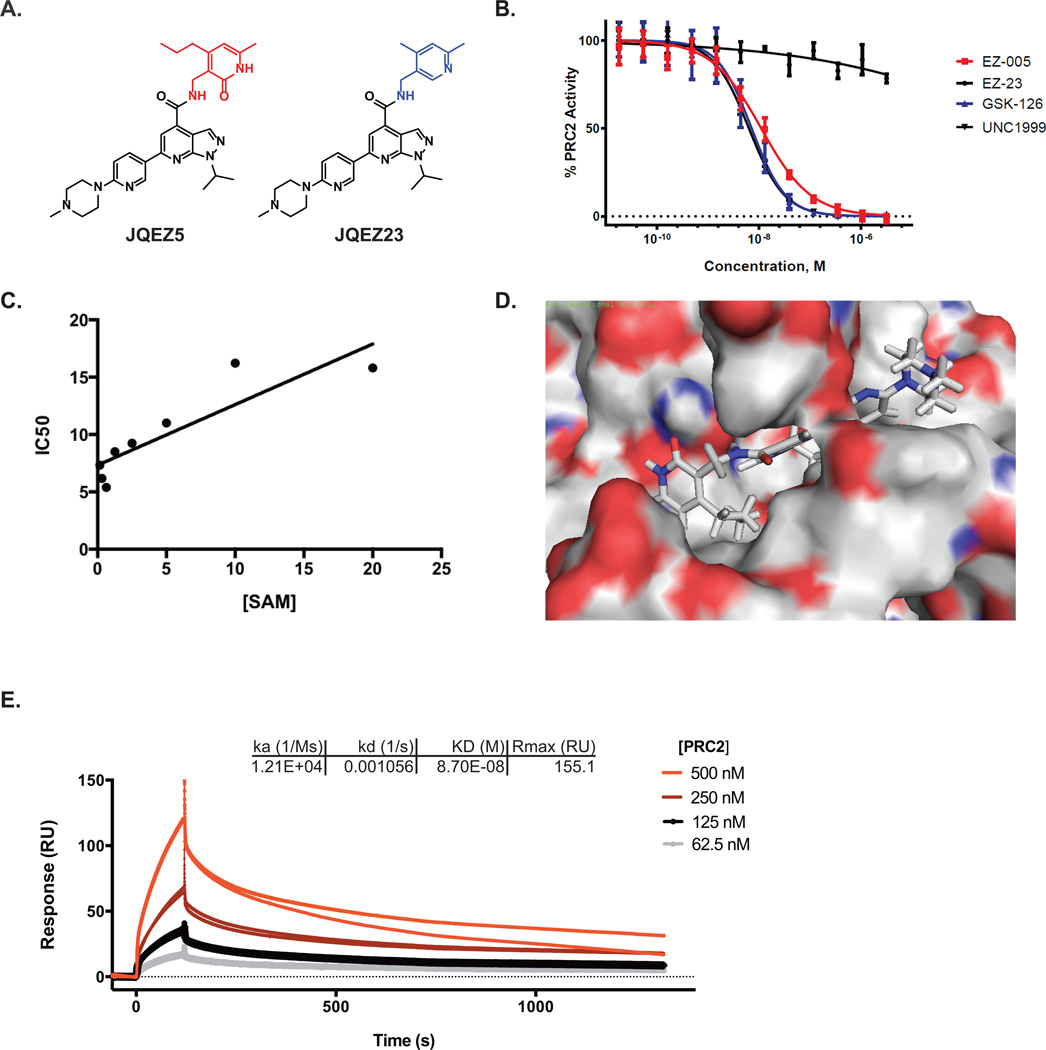
Emerging from follow-up chemistry is JQEZ5, which features a pyrazolo-pyridine core displaying a 6-substituted solubilizing feature and a preserved pyridinone warhead (highlighted in red; Figure 5A). The synthesis of JQEZ5 is nine linear steps, high-yielding and scalable (see Supplementary Methods), to support broad distribution. As a paired control, we developed JQEZ23 with the substitution of the active pyridinone to a predicted inactive pyridinium ring (highlighted in blue; Figure 5A). Both compounds were evaluated in enzymatic assays with a five-component PRC2 complex with radiometric labeled S-adenosyl methionine (SAM). JQEZ5 inhibited enzymatic functionality of PRC2 with a biochemical IC50 of 80nM, similar to GSK-126 and UNC1999, while JQEZ23 had little inhibitory activity towards purified PRC2 (Figure 5B, Supplementary Figure S4B). JQEZ5 exhibited SAM-competitive inhibition of PRC2, as determined by biochemical inhibition assessed in the presence of escalating unlabeled SAM co-factor concentration (Figure 5C, Supplementary Figure S4C). To understand the putative mode of molecular recognition of EZH2 by our inhibitor, we modeled binding of JQEZ5 to EZH2 using a recently reported computational model (Figure 5D) (46). The binding model we established indicates that the D-pyridinone ring of JQEZ5 binds to Asn78 on EZH2, and that the pyrazolo-pyridine ring is deeply buried in the SAM-binding pocket of EZH2. The ligand interaction diagram (LID) of JQEZ5 and EZH2 (Supplementary Figure S4D) also predicts that the piperazine ring on JQEZ5 extends out of the SAM-binding pocket of EZH2 and is thus, amenable to further modification. Based on this prediction, we appended a biotin tag to the piperazine ring of JQEZ5 to create JQEZ6 (Supplementary Figure S4E). Using surface plasma resonance (SPR), we measured the binding affinity of the five-component PRC2 complex for JQEZ6 immobilized on a streptavidin chip surface. The binding affinity measured by SPR (87.0nM) is similar to other biochemical assays that we performed (Figure 5E). Negative control compounds (ex. biotinylated-JQ1) did not exhibit binding to the PRC2 complex (data not shown), further demonstrating the specificity of JQEZ6 for the PRC2 complex. Finally, the specificity of JQEZ5 for EZH2 was confirmed via an orthogonal study of a panel of 22 recombinant, purified lysine methyltransferases (Reaction Biology, Supplementary Figure S4F) (47).
Figure 5. Small Molecule EZH2 Inhibitor Development.
(A) Chemical structures of small molecule EZH2 inhibitor, JQEZ5, and negative control compound, JQEZ23.
(B) Small molecule inhibitory activity of JQEZ5, JQEZ23, GSK-126 and UNC1999 were measured in a five-component PRC2 complex radiometric Scintillation Proximity Assay (SPA) using radiolabeled S-adenosyl methionine (SAM).
(C) The IC50 of JQEZ5 as measured with increasing SAM concentrations to confirm its SAM competitive binding activity.
(D) Computational docking model of JQEZ5 binding to EZH2 using reported model.
(E) Biacore SPR sensorgram from single-cycle kinetics runs with four concentrations of the PRC2 five component complex. A biotinylated derivative of JQEZ5, JQEZ6, was immobilized on streptavidin SPR chip. The affinity (KD) of PRC2 for JQEZ6 was determined to be 87 nM.
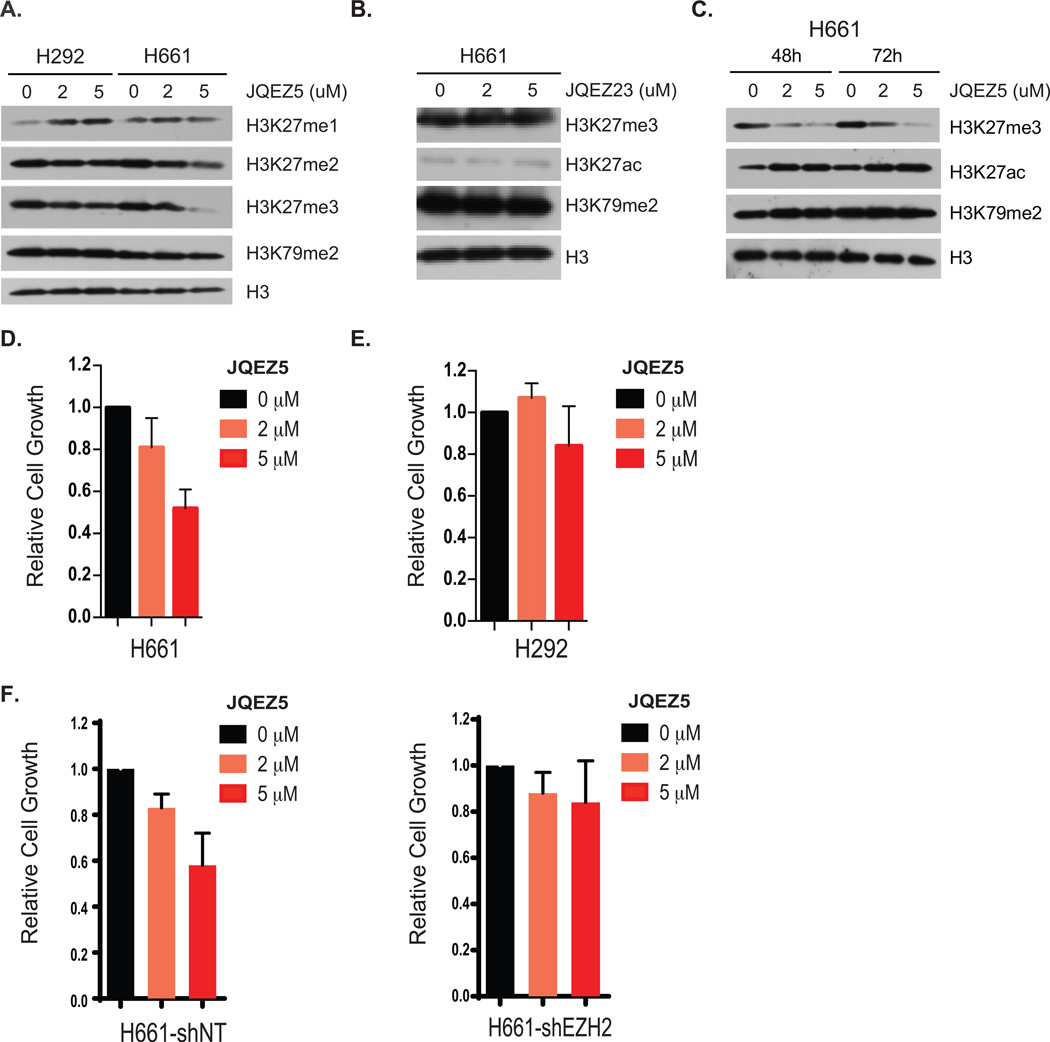
After biochemically validating the specificity and potency of JQEZ5, we treated human oncogene wild-type NSCLC cells in dose-ranging biochemical and cellular studies. H661 cells treated with increasing concentrations of JQEZ5 demonstrated acutely reduced levels of H3K27me3 without affecting H3K27 mono- or di-methylation, as assessed by Western blotting (Figure 6A). Treatment had no affect on H3K27me3 levels in the non-sensitive H292 human NSCLC cell line. Treatment with the negative control compound, JQEZ23, did not have any effect on H3K27 methylation or acetylation status (Figure 6B). H3K27me3 reduction correlated with compound concentration as well as length of treatment (Figure 6C). Similar to our EZH2 shRNA studies, JQEZ5 suppressed the proliferation of EZH2-overexpressing H661 and H522 cells after treatment for 4 days without affecting the proliferation of cell lines that were deemed insensitive to EZH2 knockdown (Figure 6D–E, Supplementary Figure S5A–B). In a direct comparison to alternate SAM-competitive EZH2 inhibitors, JQEZ5 inhibited cell growth more effectively that GSK-126 and GSK-343 in the H522 cell line (Supplementary Figure S5B). Finally, shRNA knockdown of EZH2 in H661 cells rendered cells insensitive to JQEZ5-induced cell growth arrest (Figure 6F). Together, these data demonstrate the limited off-target activity of the chemical probe and support the notion that JQEZ5 is effectively targeting EZH2 in human lung cancer cell lines.
Figure 6. JQEZ5 Inhibits Lung Cancer Growth of EZH2 Overexpressed Human Lung Cancer Cell.
(A) H661 and H292 human lung cancer cells were incubated with increasing concentrations of JQEZ5. Cell lysates were prepared and subjected to SDS-PAGE and analysis by Western blotting with the indicated antibodies.
(B) Western blots of methylation levels in human lung cancer cell line H661, 72h after treatment with increasing concentrations of JQEZ23. H3 is a loading control.
(C) Western blots of methylation levels in H661 human lung cancer cell line after 48 h or 72 h of treatment with increasing concentrations of JQEZ5. H3 is a loading control.
(D) H661 and (E) H292 human lung cancer cells were incubated with increasing concentrations of JQEZ5 and relative cell growth was assessed by MTS assay. Error bars represent SD, n=3.
(F) H661 cells infected with lentivirus containing control (NT) or shEZH2 were incubated with increasing concentrations of JQEZ5 and relative cell growth was assessed by MTS assay. Error bars represent SD, n=3.
JQEZ5 inhibits tumor growth in EZH2-overexpressed lung cancer
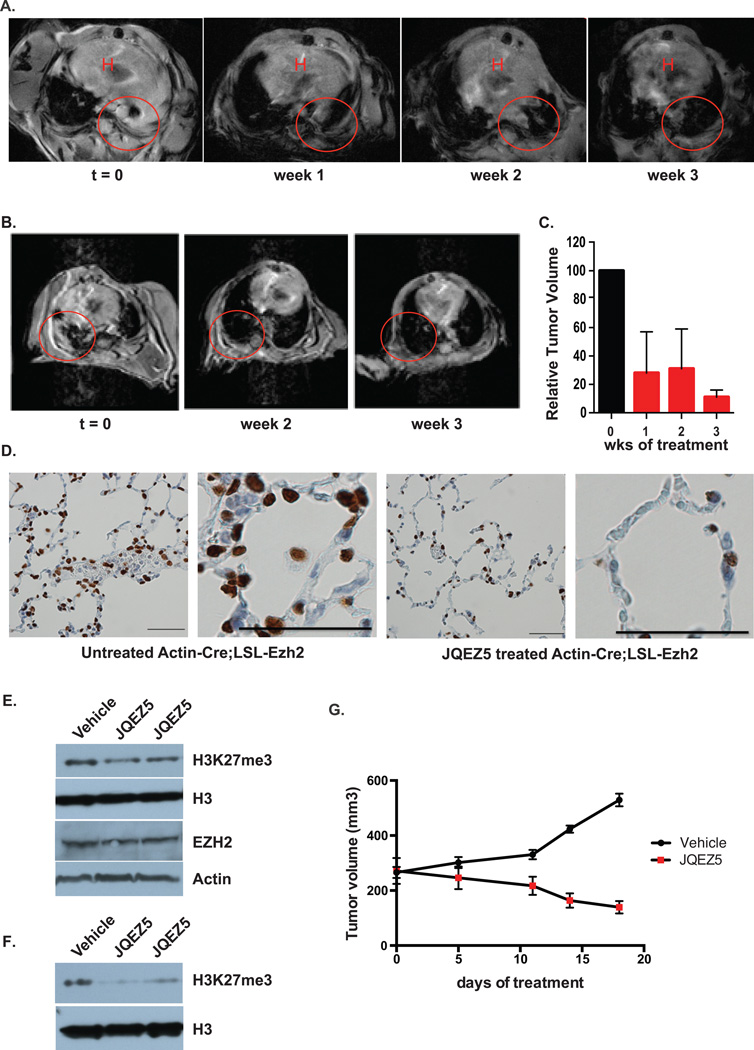
To further explore the translational relevance of this research, we undertook therapeutic trials of JQEZ5 in tumor-bearing GEMMs. JQEZ5 was formulated for intraperitoneal (IP) administration and repeat dosing studies established 75 mg/kg IP daily as a tolerated dose and schedule with no observed toxicities (Supplementary Figure S5C). Pharmacokinetic studies confirmed excellent exposure to JQEZ5 without the need for twice daily dosing (Supplementary Figure S5D). To prepare for therapeutic studies, Actin-Cre:LSL-EZH2 mice and UBC:LSL-EZH2 mice (treated with tamoxifen at 6 weeks of age) were monitored weekly for the onset of symptoms of lung adenocarcinoma (breath distress). At that time (t=0), lung cancers were visualized and confirmed by MRI (Figure 7A, B). Tumor-bearing mice were then treated with JQEZ5 for three weeks (75 mg/kg IP daily) and the tumor volume of the lungs was comparatively visualized by MRI. Animals treated with JQEZ5 exhibited rapid and pronounced tumor regression over the three week treatment course, as demonstrated by two-dimensional MRI and volumetric measurements (Figure 7C). Additionally, H3K27me3 levels were largely reduced with treatment further confirming the on-target effect of JQEZ5 in mice (Figure 7D). To model the therapeutic opportunity of EZH2 inhibition in vivo, we evaluated the tolerability and anti-tumor efficacy of JQEZ5 in a murine xenograft model of human NSCLC cells (H661 cells). Tumor-bearing mice were treated with JQEZ5 administered by intraperitoneal injection (75 mg/kg daily) or vehicle control for 18 days. Administration of JQEZ5 was well tolerated. The level of H3K27me3 was largely reduced with the treatment without effect on EZH2 levels in both the tumor (Figure 7E) and in the lung tissue of treated animals (Figure 7F). More importantly, JQEZ5 treatment attenuated tumor progression as determined by serial volumetric measurement (Figure 7G) to further confirm the therapeutic potential of EZH2 inhibitors in this subset human lung cancer.
Figure 7. JQEZ5 Inhibits Lung Cancer Growth In Vivo.
(A–B) MRI scans of individual tumor-bearing Actin-Cre;LSL-EZH2 mice (t=0) and after 1–3 weeks of treatment with JQEZ5 at 75mpk, daily. Lung tumor is indicated by the red circle. H, Heart.
(C) Quantification of relative tumor volume of mouse lungs based on MRIs using 3D Slicer. Relative tumor volume was compared before and after 3 weeks of JQEZ5 treatment (mean ± SEM, n=2).
(D) Tumor-bearing Actin-Cre;LSL-EZH2 mice were untreated or treated with JQEZ5 at 75mpk for three weeks. Lung sections were prepared and immunostained for H3K27me3. Bar represents 50µm.
(E) Nude mice were injected subcutaneously with 2×106 H661 human NSCLC cells. When tumors reached ~200 mm3, mice were randomized and treated with vehicle or JQEZ5 (75 mg/kg/d, i.p.) for 18 days. Western blots analysis was performed on tumors following either vehicle or JQEZ5 treatment for 18 days.
(F) Western blots analysis of lung tissue from mice after 18 days of treatment with vehicle or JQEZ5 at 75 mpk.
(G) Tumor volume from mouse xenograft model of human lung cancer was measured by caliper (mean ± SEM, n=3/vehicle, n=6/JQEZ5).
Discussion
As the leading cause of death from cancer, lung cancer comprises a profound unmet medical need. A subset of lung cancer patients have benefited from targeted therapies in the past decade, yet the majority of patients will not benefit from these approaches and all metastatic disease remains incurable. There exist a large number of patients who cannot be effectively treated due to the lack of druggable oncogenic drivers (e.g. KRAS). As such, discovering new actionable drivers and tumor dependencies in these remaining tumors is an urgent and important endeavor.
Though not affected by somatic alteration, deregulated overexpression of EZH2 is observed in a subset of human lung cancers (22–24). To date, a causal role in lung tumor development has not been established. In this study, we explored the oncogenic potential of EZH2 deregulation by generating a series of genetically engineered mouse models with conditional EZH2 overexpression. Three GEMMs establish evident oncogenic activity for EZH2 in NSCLC formation. Indeed, 45% of mice engineered to overexpress EZH2 developed lung adenocarcinomas. These tumors lacked recurrent somatic mutations in in known driver oncogenes and tumor suppressors and EZH2 overexpression had no effect on KRAS-mutant lung tumor formation, demonstrating that EZH2 has weak, but apparent oncogenic activity in lung. These data resonate with prior studies that exemplified the transforming activity of mutant EZH2 in B-cell lymphoma (14, 15). A prior study showed that EZH2 overexpression in the myeloid compartment elaborated a myeloproliferative disorder, but to date firm evidence of neoplastic transformation by EZH2 overexpression has been elusive (48). EZH2 expression has even been shown to restrain tumorigenesis in some epithelial cancer models (21). To our knowledge, this work is a first demonstration of cancer resulting from wild-type EZH2 overexpression in vivo. Overexpression of EZH2 is, again, a common feature of numerous solid tumors, and the reagents created in this study will be helpful to further define the role of EZH2 in the pathogenesis of cancer more broadly in different contexts.
Cell biology studies and integrated epigenomic analysis of the resultant EZH2-driven murine tumors emulate the cytosolic and transcriptional signaling of a defined subset of the human disease. Interestingly, lung adenocarcinomas induced by EZH2 overexpression displayed low levels of p-AKT and p-ERK, which are accompanied by elevated expressions of known negative regulators of the MAPK-ERK pathway such as dual-specificity protein phosphatase 4 (DUSP4) and sprouty homologs 1 and 2 (SPRY1 & SPRY2). Consistent with these findings, transcriptional signatures associated with MEK and mTOR activation are repressed in human tumors that expressed high levels of EZH2. Given that many of the known oncogenes in lung cancer activate these pathways, EZH2 appears to promote tumorigenesis through mechanisms that do not involve these canonical pathways, which has important mechanistic and therapeutic implications. Some EGFR and KRAS mutant human cancers also express high levels of EZH2, but our data in mouse models suggest that EZH2 expression does not modify the tumorigenesis of strong oncogenic alterations in KRAS. However, in some settings these pathways may cooperate. Consistent with this concept, DUSP4 has been implicated as a growth suppressor in EGFR-mutant lung adenocarcinoma (49) and as a positive activator of ERK in EGFR-mutant lung cancer cell lines (50). A unifying model for EZH2-mediated malignant transformation based on these findings could be the remodeling of chromatin architecture toward a de-differentiated cell state that facilitates proliferative transformation by additional genetic drivers. By further analyzing the changes on super-enhancer landscape and the TF connection through CRC, we have established the unique signature of EZH2 lung cancers, and distinguished it from other lung cancer types, such as KRAS.
Using functional genetic (shRNA) and chemical genetic approaches, we have demonstrated the dependency of EZH2-overexpressing human and murine lung cancer models on EZH2. Toward pharmacologic target validation, we have created a novel, SAM-competitive inhibitor that establishes an evident therapeutic index for targeting EZH2-overexpressing tumors in vivo. JQEZ5 demonstrates consistent anti-tumor activity at 75 mg/kg versus reported studies of GSK-126 at doses as high as 150 mg/kg (28). We expect the JQEZ5 chemical probe to be useful to the broader research community, in a manner similar to our experience openly distributing probes for BET bromodomains (51). Successful clinical translation of targeted lung cancer therapeutics has been facilitated by genomic or immunohistochemical biomarkers. Here, we show that EZH2 is essential for the growth of human lung cancer cell lines that express high levels of EZH2, while being dispensable for cell lines with lower levels of EZH2. As such, EZH2 expression may be a useful biomarker for patient selection or planned stratification in downstream clinical trials. These findings support prior work in prostate cancer, where EZH2 was found to be indispensable for cell growth in LNCaP-abl cells with higher EZH2 levels, but not in LNCaP cells with lower EZH2 levels (43).
EZH2 inhibition has previously been proposed as a therapeutic strategy in NSCLC in the context of BRG1 or EGFR mutations (41). This study found that EZH2 inhibitors can sensitize NSCLC cells with EGFR or BRG1 mutations to chemotherapy and a combination of EZH2 inhibition with Topo II inhibition was proposed. Here, we establish EZH2 inhibition as an effective single-agent therapy in the defined subset of NSCLC that overexpress EZH2 without other known concurrent oncogenic mutations. Still, we anticipate that the genetic complexity and tumor heterogeneity that is a common feature lung adenocarcinoma will require pairing EZH2 inhibition with other effective therapeutic strategies. Pairing epigenomic EZH2 inhibition with oncogene-directed therapy or immune checkpoint therapies are appealing concepts.
In sum, this work establishes an oncogenic role for EZH2 deregulation in lung adenocarcinoma, creates faithful models of a unique subset of human disease, describes and characterizes a novel chemical probe for studying EZH2 function in both xenograft and GEM models, and provides the rationale for human clinical investigation.
Methods
Cell culture
H661, H292 and H522 human lung carcinoma cell lines were purchased from and authenticated through routine STR analysis and cytogenetic studies by ATCC. H969, H2250 and H2258 cell lines were a kind gift from Dr. Kenneth Huffman at UT Southwestern Medical Center; hTBE cells were a kind gift from Dr. William C. Hahn at Dana-Farber Cancer Institute. hTBE cells were received and last authenticated in 2011; all other cells were received and last authenticated by standard methods in 2014. Human lung carcinoma cell lines were cultured in RPMI-1640/10% FBS/1% penicillin–streptomycin. 293ft cells (Invitrogen) were cultured in Dulbecco's Modified Eagle Medium (DMEM)/10% FBS/1% penicillin–streptomycin. Fresh murine lung tumor nodules were minced and cultured in 100-mm dishes with RPMI-1640/10% FBS/1% penicillin–streptomycin. All cells were cultured at 37°C in a humidified incubator with 5% CO2.
Western blot
Tissues were lysed in 1× lysis buffer (Cell Signaling Technology, #9803) with protease inhibitor cocktail (EMD Biosciences, 539131). Cell lysates were separated using standard western blotting procedures, and membranes were probed with the following antibodies: mouse monoclonal EZH2 (BD Transduction Laboratories, #612666) at 1:1000; total AKT (Cell Signaling Technology, #9272) at 1:2000; total Erk1,2 (Cell Signaling Technology, #4695S) at 1:2000; pAKT antibody (Cell Signaling Technology, #4060S) at 1:1000; pERK1,2 (Cell Signaling Technology, #4376S) at 1:1000) all incubated overnight at 4µC. β-actin-HRP (SIGMA A3854, 1:20000) was used as loading control. Secondary antibody Anti-rabbit IgG, HRP-linked Antibody (Cell Signaling 7074, 1:2000) or Anti-mouse IgG, HRP-linked Antibody (Cell Signaling 7076, 1:2000) was incubated for 1 hour at room temperature. After washing, chemiluminescence was visualized with Western Lightning Plus-ECL (PerkinElmer) and exposure onto KODAK BioMax XAR film. Relative quantifications were performed using ImageJ software.
Chromatin preparation
Mouse lung tissue was pulverized using a Covaris Tissue Smasher (Model #CP02) following the CryoPrep Dry Pulverization Manuel. Lung tissue was smashed 1–2 times on setting 4 in the tissueTUBE (Covaris #520071). Approximately 50mg of pulverized lung tissue was cross-linked with prewarmed 1% formaldehyde (ThermoScientific #28906 diluted in PBS) for 20 minutes at 37°C. The tissue was spun down at 1,000 rpm for 2 min and quenched with 0.125M glycine in PBS + 0.5% BSA for 20 min at room temperature. It was spun down at 1,000 rpm for 2 min and washed with PBS + 2× Protease Inhibitor Cocktail (PIC) (Roche #11873580001) + 5 mM Sodium Butyrate (Millipore #19-137) and then spun down at 1,000 rpm for 2 min. The cross-linked tissue was lysed with 390uL ChIP Lysis Buffer (1% SDS, 10nM EDTA pH8.0, 50mM Tris-HCl pH 8.0, 2× PIC and 5mM Sodium Butyrate) on ice for 1 hr. The lysate was split into 3 microTUBEs (Covaris #520045) and sheared on the Covaris E210 Series with 5% Duty Cycle, 5 Intensity, 200 Cycles per Burst for a total of 27 minutes. The sheared chromatin was spun down at 14,000 rpm for 15 min at 4°C. An aliquot of input was saved while the remaining chromatin was snap frozen and stored at −80°C. Input was brought up to 100ul with TE, 10ug of RNAseA (Roche) added and incubated for 30 minutes at 37°C followed by addition of 100ug of Proteinase K (Roche) and incubation at 65°C overnight. Input was purified with Qiagen PCR Purification Kit (#28104) and quantified.
Library preparation and ChIP
The prepared chromatin was thawed on ice while 10ug of antibodies against either H3K27ac (Abcam #Ab4729) or H3K27me3 (Cell Signaling #CS9733S) was conjugated to a mix of magnetic Protein A and Protein G coupled beads (Invitrogen #100.02D and #100.04D, respectively) in the presence of 0.5% BSA in PBS with rotation at 4°C for 2 hours. Beads were washed 3 times with 0.5%BSA in PBS and either 5ug of chromatin was added to the H3K27ac ChIP or 10ug of chromatin was added to the H3K27me3 ChIP and rotated overnight at 4°C. The beads were washed 2 times with Tris-based RIPA buffer (0.1% SDS, 1% Triton X-100, 10mM Tris-HCl pH 7.4, 1mM EDTA pH 8.0, 0.1% Sodium Deoxycholate), 2 times with 0.3M NaCl RIPA (0.1% SDS, 1% Triton X-100, 10mM Tris-HCl pH 7.4, 1mM EDTA pH 8.0, 0.1% Sodium Deoxycholate, 0.3M NaCl), 2 times with LiCl Buffer (250mM LiCl, 0.5% NP-40, 0.5% Sodium Deoxycholate, 1mM EDTA pH 8.0, 10mM Tris-HCl pH 8.0) and 2 times with TE buffer pH 7.6 (Fisher Scientific cat. no. BP2474-1). The beads were resuspended in 100uL of TE and RNAseA and PK digested/reverse crosslinked and purified as described in the chromatin prep. Chromatin libraries were prepared with 10–20ng of Input or ChIP’d DNA according to the ThruPLEX-FD Prep Kit (Rubicon #R40048) and sequenced by SE75 Next-Seq.
PRC2 methyltransferase assay
Recombinant five-component PRC2 (EZH2/EED/SUZ12/RBBP4/AEBP2) was co-expressed in Sf9 and purified as described (52). PRC2 activity was measured using a radiometric Scintillation Proximity Assay (SPA) performed in 384-well OptiPlates (Perkin Elmer). For IC50 determination, 2.3 nM PRC2 was incubated for 90 min at RT with 1 uM histone H3 (21–44)-lys(biotin) (Anaspec), 1.5 uM SAM (NEB), and 500 nM 3H-SAM in 20 uL reaction buffer (50 mM Tris pH 8.5, 5 mM DTT, and 0.01% Tween-20) containing compound or DMSO. Reactions were quenched with TCA and, following the addition of PVT streptavidin-coated SPA beads (Perkin Elmer; 40 uL of 140 ng diluted in PBS), incubated for 1 hr at RT. CPM values were measured using the TopCount NXT plate reader. Percent activity values were calculated by setting the average background (no-enzyme wells) to 0% and the average DMSO wells to 100% activity. Standard deviations were determined from four replicate measurements for each compound concentration. Data were analyzed and plotted using GraphPad PRISM v6, using the ‘log(inhibitor) vs normalized response –variable slope’ analysis module to calculate IC50.
For determination of JQEZ5 mechanism of action and Ki values, reactions were carried out as described above in the presence of varying concentrations SAM/3H-SAM (at a 1:20 ratio) with a fixed concentration of 1 uM histone H3 peptide. Data were analyzed and plotted using ‘Enzyme Kinetics –inhibition’ and ‘Enzyme Kinetics – substrate versus velocity’ analysis modules in GraphPad PRISM v6.
In vivo studies with JQEZ5
JQEZ5 was dissolved in DMSO and then diluted 1:10 in 10% (2-Hydroxypropyl)-β-cyclodextrin (Sigma-Aldrich). DMSO (vehicle) was dissolved 1:10 in 10% (2-Hydroxypropyl)-β-cyclodextrin. Tumor-bearing GEMMs were monitored for onset of symptoms (breath distress) and then treated with JQEZ5 for three weeks (75 mg/kg IP daily). Tumors were visualized by MRI and tumor volume of the lungs was calculated using 3D Slicer (53). For xenograft experiments, H661 cells were dissociated into single cells, counted and resuspended at 2×106 cells per 250 µl of 1:1 media/matrigel (BD). Eight- to 12-week-old female Foxn1nu/Foxn1nu (Nude) mice (Harlan) were injected subcutaneously with 2 × 106 cells in two to three spots on the flanks. Tumors were allowed to grow to an approximate size of 200 mm3 (~10 weeks) and the mice were randomized for vehicle (n=3) or JQEZ5 administration (n=6, 75 mg/kg/d, i.p.) for 18 days. Tumor growth was measured by caliper measurements and tumor volume was calculated by standard methods. All mice were housed in pathogen-free animal facilities, and all experiments were performed with the approval of the Animal Care and Use Committee at Harvard Medical School and Dana-Farber Cancer Institute.
Sequencing Data
Sequencing data reported in this paper have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE70047.
Synthesis of small molecule inhibitors
GSK-126, GSK-343 and UNC1999 were directly purchased from Sigma-Aldrich, Inc. The structure and purity of these two compounds were further confirmed by NMR and LCMS. The detailed syntheses of compound JQEZ5, JQEZ6 and JQEZ23 are described in Supplementary Methods.
Supplementary Material
Statement of Significance.
EZH2 overexpression induces murine lung cancers that are similar to human NSCLC with high EZH2 expression and low levels of phosphorylated AKT and ERK, implicating biomarkers for EZH2 inhibitor sensitivity. Our novel EZH2 inhibitor, JQEZ5, promotes regression of these tumors, revealing a potential role for anti-EZH2 therapy in lung cancer.
Acknowledgments
Financial Support: G.S.H.-S. was supported by the Deutsche Forschungsgemeinschaft (HE 6897/1-1). K-K.W. and C.M.P are supported by NIH/NCI 1R01CA195740-01. K-K.W. is also supported by NIH/NCI P01CA120964, 5R01CA163896-04, 5R01CA140594-07, 5R01CA122794-10 and 5R01CA166480-04 grants and support from Gross-Loh Family Fund for Lung Cancer Research and Susan Spooner Family Lung Cancer Research Fund at Dana-Farber Cancer Institute. J.E.B. is supported by the Burroughs-Wellcome Fund, the William Lawrence & Blanche Hughes Foundation and a Leukemia & Lymphoma Society SCOR grant.
Footnotes
Conflicts of Interest: P.K.R. is a consultant to Selecta Biosciences Inc, Watertown, MA. J.E.B. is a Scientific Founder of SHAPE Pharmaceuticals, Acetylon Pharmaceuticals, Tensha Therapeutics and C4 Therapeutics and is the inventor on IP licensed to these entities. J.E.B. currently an employee of Novartis Institutes of Biomedical Research.
Author Contributions
H.Z., J.Q., P.S.H., J.E.B. and K.-K.W. designed this study. H.Z. designed and performed biology experiments. J.Q. and J.E.B designed and synthesized JQEZ5, JQEZ6 and JQEZ23. H.Z., C.Y.L., L.L., P.K.R., F.L., J.M.R., A.J.F., H.L., C.M.P. and M.B. performed all sequencing and sequencing data analysis. J.P. and J.Q. performed biochemical assays. A.J.F. and J.Q. performed computational modeling. H.Z., J.Q., J.A.P., J.E.B. and K.-K.W. analyzed results and wrote manuscript with comments from all authors.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14(8):535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. New England Journal of Medicine. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 5.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):U561–U563. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 7.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10(10):697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 9.Chen BF, Chan WY. The de novo DNA methyltransferase DNMT3A in development and cancer. Epigenetics. 2014;9(5):669–677. doi: 10.4161/epi.28324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamidi T, Singh AK, Chen T. Genetic alterations of DNA methylation machinery in human diseases. Epigenomics. 2015;7(2):247–265. doi: 10.2217/epi.14.80. [DOI] [PubMed] [Google Scholar]
- 11.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25(23):2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 13.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A. 2010;107(49):20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22(20):5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647(1–2):21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Gong Y, Huo L, Liu P, Sneige N, Sun X, Ueno NT, et al. Polycomb group protein EZH2 is frequently expressed in inflammatory breast cancer and is predictive of worse clinical outcome. Cancer. 2011;117(24):5476–5484. doi: 10.1002/cncr.26179. [DOI] [PubMed] [Google Scholar]
- 19.Alford S, Toy K, Merajver S, Kleer C. Increased risk for distant metastasis in patients with familial early-stage breast cancer and high EZH2 expression. Breast Cancer Research and Treatment. 2012;132(2):429–437. doi: 10.1007/s10549-011-1591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bachmann I, Halvorsen O, Collett K, Stefansson I, Straume O, Haukaas S, et al. EZH2 Expression Is Associated With High Proliferation Rate and Aggressive Tumor Subgroups in Cutaneous Melanoma and Cancers of the Endometrium, Prostate, and Breast. J Clin Oncol. 2006;24(2):268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 21.Wassef M, Rodilla V, Teissandier A, Zeitouni B, Gruel N, Sadacca B, et al. Impaired PRC2 activity promotes transcriptional instability and favors breast tumorigenesis. Genes Dev. 2015;29(24):2547–2562. doi: 10.1101/gad.269522.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behrens C, Solis LM, Lin H, Yuan P, Tang X, Kadara H, et al. EZH2 Protein Expression Associates with the Early Pathogenesis, Tumor Progression, and Prognosis of Non-Small Cell Lung Carcinoma. Clinical Cancer Research. 2013;19(23):6556–6565. doi: 10.1158/1078-0432.CCR-12-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kikuchi J, Kinoshita I, Shimizu Y, Kikuchi E, Konishi J, Oizumi S, et al. Distinctive expression of the polycomb group proteins Bmi1 polycomb ring finger oncogene and enhancer of zeste homolog 2 in nonsmall cell lung cancers and their clinical and clinicopathologic significance. Cancer. 2010;116(12):3015–3024. doi: 10.1002/cncr.25128. [DOI] [PubMed] [Google Scholar]
- 24.Lv Y, Yuan C, Xiao X, Wang X, Ji X, Yu H, et al. The expression and significance of the enhancer of zeste homolog 2 in lung adenocarcinoma. Oncol Rep. 2012;28(1):147–154. doi: 10.3892/or.2012.1787. [DOI] [PubMed] [Google Scholar]
- 25.Kim W, Bird GH, Neff T, Guo G, Kerenyi MA, Walensky LD, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9(10):643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8(11):890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 27.Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proceedings of the National Academy of Sciences. 2012;109(52):21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 29.Knutson SK, Warholic NM, Wigle TJ, Klaus CR, Allain CJ, Raimondi A, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A. 2013;110(19):7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Wang Y, Qiu J, Li Q, Yuan C, Zhang W, et al. The polycomb group protein EZH2 is a novel therapeutic target in tongue cancer. Oncotarget. 2013;4(12):2532–2549. doi: 10.18632/oncotarget.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4(7):1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24(6):777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown JD, Lin CY, Duan Q, Griffin G, Federation AJ, Paranal RM, et al. NF-kappaB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell. 2014;56(2):219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, et al. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell. 2015;58(2):362–370. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153(2):320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang JA, Lee BB, Kim Y, Hong SH, Kim YH, Han J, et al. HOXA9 inhibits migration of lung cancer cells and its hypermethylation is associated with recurrence in non-small cell lung cancer. Mol Carcinog. 2015;54(Suppl 1):E72–E80. doi: 10.1002/mc.22180. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki M, Ikeda K, Shiraishi K, Eguchi A, Mori T, Yoshimoto K, et al. Aberrant methylation and silencing of expression in non-small cell lung cancer. Oncol Lett. 2014;8(3):1025–1030. doi: 10.3892/ol.2014.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fillmore CM, Xu C, Desai PT, Berry JM, Rowbotham SP, Lin YJ, et al. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature. 2015;520(7546):239–242. doi: 10.1038/nature14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tzatsos A, Paskaleva P, Ferrari F, Deshpande V, Stoykova S, Contino G, et al. KDM2B promotes pancreatic cancer via Polycomb-dependent and -independent transcriptional programs. The Journal of Clinical Investigation. 2013;123(2):727–739. doi: 10.1172/JCI64535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 Oncogenic Activity in Castration-Resistant Prostate Cancer Cells Is Polycomb-Independent. Science. 2012;338(6113):1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konze KD, Ma A, Li F, Barsyte-Lovejoy D, Parton T, Macnevin CJ, et al. An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem Biol. 2013;8(6):1324–1334. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knutson SK, Kawano S, Minoshima Y, Warholic NM, Huang KC, Xiao Y, et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol Cancer Ther. 2014;13(4):842–854. doi: 10.1158/1535-7163.MCT-13-0773. [DOI] [PubMed] [Google Scholar]
- 46.Kalinic M, Zloh M, Eric S. Structural insights into binding of small molecule inhibitors to Enhancer of Zeste Homolog 2. J Comput Aided Mol Des. 2014;28(11):1109–1128. doi: 10.1007/s10822-014-9788-1. [DOI] [PubMed] [Google Scholar]
- 47.Horiuchi KY, Eason MM, Ferry JJ, Planck JL, Walsh CP, Smith RF, et al. Assay development for histone methyltransferases. Assay Drug Dev Technol. 2013;11(4):227–236. doi: 10.1089/adt.2012.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herrera-Merchan A, Arranz L, Ligos JM, de Molina A, Dominguez O, Gonzalez S. Ectopic expression of the histone methyltransferase Ezh2 in haematopoietic stem cells causes myeloproliferative disease. Nat Commun. 2012;3:623. doi: 10.1038/ncomms1623. [DOI] [PubMed] [Google Scholar]
- 49.Chitale D, Gong Y, Taylor BS, Broderick S, Brennan C, Somwar R, et al. An integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant tumors. Oncogene. 2009;28(31):2773–2783. doi: 10.1038/onc.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Britson JS, Barton F, Balko JM, Black EP. Deregulation of DUSP activity in EGFR-mutant lung cancer cell lines contributes to sustained ERK1/2 signaling. Biochem Biophys Res Commun. 2009;390(3):849–854. doi: 10.1016/j.bbrc.2009.10.061. [DOI] [PubMed] [Google Scholar]
- 51.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15(1):57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 53.Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging. 2012;30(9):1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.