Abstract
PURPOSE
Deregulated Myc drives an oncogenic metabolic state, including pseudohypoxic glycolysis, adapted for the constitutive production of biomolecular precursors to feed rapid tumor cell growth. In glioblastoma (GBM), Myc facilitates renewal of the tumor initiating cell reservoir contributing to tumor maintenance. We investigated whether targeting the Myc-driven metabolic state could be a selectively toxic therapeutic strategy for GBM.
METHODS
The glycolytic dependency of Myc-driven GBM was tested using 13C metabolic flux analysis, glucose-limiting culture assays and glycolysis inhibitors, including inhibitors of the NAD+ salvage enzyme nicotinamide phosphoribosyl-transferase (NAMPT), in MYC and MYCN shRNA knockdown and lentivirus overexpression systems and in patient-derived GBM tumorspheres with and without MYC/MYCN amplification. The in vivo efficacy of glycolyic inhibition was tested using NAMPT inhibitors in MYCN amplified patient-derived GBM orthotopic xenograft mouse models.
RESULTS
Enforced Myc overexpression increased glucose flux and expression of glycolytic enzymes in GBM cells. Myc and N-Myc knockdown and Myc overexpression systems demonstrated that Myc activity determined sensitivity and resistance to inhibition of glycolysis. Small molecule inhibitors of glycolysis, particularly NAMPT inhibitors, were selectively toxic to MYC/MYCN amplified patient-derived GBM tumorspheres. NAMPT inhibitors were potently cytotoxic, inducing apoptosis and significantly extended the survival of mice bearing MYCN amplified patient-derived GBM orthotopic xenografts.
CONCLUSION
Myc activation in GBM generates a dependency on glycolysis and an addiction to metabolites required for glycolysis. Glycolytic inhibition via NAMPT inhibition represents a novel metabolically-targeted therapeutic strategy for MYC or MYCN amplified GBM and potentially other cancers genetically driven by Myc.
Keywords: MYC, Glioblastoma, Glycolysis, Metabolism, NAMPT
INTRODUCTION
The MYC gene family (MYC, MYCN and MYCL) consists of potent oncogenes that play critical roles in the pathogenesis of diverse types of human cancers (1, 2). The Myc oncoprotein contributes to malignancy by various mechanisms, including driving cellular proliferation, blocking differentiation, facilitating chromosomal instability, increasing cell migration and inducing angiogenesis (1, 2). In glioblastoma (GBM), Myc enhances the self-renewal capacity of glioma stem-like/tumor neurosphere (tumorsphere) cells and maintains their tumorigenic potential (3–6). High-level amplification of the MYC and MYCN genes are observed in a subset of GBM (7–11). Myc is therefore a compelling therapeutic target in GBM.
Despite extensive efforts, direct inhibition of the Myc transcription factor has remained a challenge. Several indirect strategies that selectively target the pleiotropic Myc-driven downstream effects have recently shown promise, including small molecule inhibitors of BET chromatin adapters (12, 13), Chk1 (14) and CDK7 (15). Another potential Myc-specific target is the Myc-reprogrammed metabolic state, which is evident in GBM (16–18). Deregulated Myc has been shown to increase glycolysis and glutaminolysis to support the increased biosynthetic demand of rapidly proliferating cancer cells, and the altered cell metabolism may render Myc-driven cancers vulnerable to strategic nutrient deprivation (16, 19).
Here, we tested whether inhibition of the Myc-induced glycolytic drive would be a selective strategy for Myc-driven GBM. We confirmed that Myc increases glycolytic flux in GBM cells and found that Myc generates a dependency on glycolysis for survival. Using a panel of patient-derived GBM tumorsphere lines (20, 21) we found glycolytic inhibition to be strikingly selective for lines with highly amplified MYC/MYCN. We identified small molecule inhibitors of the NAD+ salvage enzyme nicotinamide phosphoribosyl-transferase (NAMPT), which mediates cytotoxicity by inhibiting glycolysis (22), as a therapeutic strategy with potential for rapid translation in a genetically defined subset of GBM patients.
MATERIALS AND METHODS
Creation of GBM tumorsphere Lines, Cell culture and Reagents
To create GBM tumorsphere lines, fresh patient GBM surgical specimens were dissociated and cultured in serum-free neural stem cell medium, which enriches for tumor-initiating cells (20, 21). All tumorsphere lines were cryopreserved at passage number 3 or less prior to use for in vitro experiments. For genetic manipulation, lines passaged <10 times were used. All tumor samples were collected with patient consent under protocols approved by the Massachusetts General Hospital (MGH) Institutional Review Board. All tumors were confirmed to be glioblastoma by formal pathological review. U87, H1975, D283 Med, IMR-32, Daoy, and Raji cell lines were obtained from American Type Culture Collection (ATCC) and were cultured using ATCC-formulated EMEM (for IMR-32, D283 Med, and Daoy), ATCC-formulated RPMI-1640 (for H1975 and Raji), or DMEM (for U87). Kelly was obtained from Sigma Aldrich and cultured with EMEM. UACC257 was a gift from David E. Fisher (MGH) and cultured with DMEM. Normal human astrocytes (NHA) were obtained from ScienCell and cultured in DMEM. All standard cell line media were supplemented with 10% fetal bovine serum (FBS) and Penicillin-streptomycin-Amphotericin B. FK866, nicotinamide mononucleotide (NMN), nicotinic acid (NA), nicotinamide adenine dinucleotide (NAD+), and 2-deoxyglucose (2-DG) were purchased from Sigma Aldrich. GMX1778 was purchased from Cayman Chemical.
U87-Myc Cell Line Generation
293T cells were co-transfected with lentivirus vectors containing MYC (pCDH-puro-cMYC, Plasmid #46970, Addgene) or GFP (pCDH-CMV-MCS-EF1-copGFP, CD511B-1, System Bioscience), pCMV-dR8.2 dvpr, and pCMV-VSVG with ScreenFect (Wako). U87 cells were infected with lentivirus with polybrene (8 µg/ml) for 6 hrs, and then selected with puromycin (0.7 µg/ml) for 7 days, then maintained in puromycin (0.2 µg/ml).
Myc shRNA and control shRNA Cell line Generation
To knockdown MYC, MGG4 and H1975 cells were infected with GIPZ Lentiviral Human MYC shRNA (V2LHS_152053, GE Dharmacon) in polybrene (8 µg/ml) for 8 hrs. Two week later, cells were selected with puromycin (0.3µg/ml) for 5 days. To knockdown MYCN, IMR-32 cells were infected with GIPZ Lentiviral Human MYCN shRNA (V2LHS_36751, GE Dharmacon) in polybrene (8 µg/ml) for 8hrs. Three weeks later, cells were selected with puromycin (0.3 µg/ml) for 7 days. GIPZ Non-silencing Lentiviral shRNA Control (RHS4348, GE Dharmacon) was used as control.
Fluorescence in situ Hybridization
Gene amplification status of MYC and MYCN was evaluated by fluorescence in situ hybridization (FISH). BAC clones CTD-3066D1 and RP11-480N14 were used to make the MYC and MYCN probes, respectively, and BAC clones RP11-301H15 (Chr8) and RP11-984I21 (Chr2) were used for centromere controls. The probes were labeled in Cy3-dCTP or FITC-dUTP. Gene-amplified cells were counted in at least 3 different high-power fields, and the proportion of amplification-positive per total cells was calculated. Gene/control probe copy number ratios of >2.0 were considered amplified.
Western Blot Analyses
Cells were lysed in radioimmunoprecipitation buffer (Boston Bioproducts) with a cocktail of protease and phosphatase inhibitors (Roche). 12.5 µg of protein was separated by 4–20% SDS-PAGE and transferred to polyvinylidene difluoride membranes by electroblotting. After blocking with 5% nonfat dry milk in TBST (20 mM Tris [pH, 7.5], 150 mM NaCl, 0.1% Tween20), membranes were incubated at 4C overnight with primary antibodies. After washing and incubation with horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology), blots were washed, and signals were visualized with an ECL kit (Amersham Bioscience). Primary antibodies: c-Myc, N-Myc, Hexokinase II (HKII), PKM2, LDHA, cleaved-PARP, phospho-AMPK, phospho-Raptor, phospho-mTOR, phospho-S6, phospho-4E-BP1, Cleaved PARP, actin (Cell Signaling); NAMPT (Bethyl Laboratories); Naprt1 (Sigma Aldrich); and Vinculin (Thermo Scientific).
Cell Viability and Cytotoxicity Analysis
To assess cell viability, GBM tumorspheres were dissociated into single cells and seeded into 96-well plates at 7,000–8,000 cells/well. After 12–24 hours, chemical inhibitors and/or NMN, NAD+, or NA were serially diluted and added to wells. Cell viability was measured by CellTiter-Glo (Promega) assays and the IC50 values (drug concentrations causing 50% viability of cells) were determined. To determine cytotoxicity, 7,000–8,000 cells were treated with DMSO, 12.5 nM of FK866 or GMX1778, and the number of viable cells that excluded trypan blue was counted using TE2000-U inverted microscope (Nikon) at 48, 72, and 96 hrs after treatment.
Apoptosis Analysis
GBM tumorspheres were dissociated into single cells and 1×105 cells were seeded per well (12-well plates) and 24 hours later treated with NAMPT inhibitors. After 96 hr incubation, cells were collected and stained with propidium iodide (PI) and APC-conjugated Annexin V (Annexin V apoptosis detection kit APC, eBioscience), and analyzed by an Accuri flow cytometer and the BD CSampler software (BD Biosciences). To evaluate caspase-3/7 activities, cells were treated with DMSO or inhibitors (12.5 nM) for 24 hrs and were tested by Caspase-Glo 3/7 Assay (Promega) according to the manufacturer’s recommendations.
NAD+ quantitation
Total NAD (NADt: NADH and NAD+) and NADH were measured using NAD+/NADH Quatification Colorimetric Kit (BioVision Incorporated) following manufacturer’s recommendations. NAD+ was calculated by subtracting NADH from NADt. Data were expressed as pmol/1×106 cells or normalized to DMSO control and presented as % change. To evaluate NAD+ change in tumor tissue, H1975 cells were implanted into the flank of SCID mice. Three weeks after implantation, mice were randomized to GMX1778 (250 mg/kg, n=3) or vehicle (n=3). 48 hrs after treatment, mice were sacrificed, tumor tissues were harvested and NAD+ concentration was measured. Protein concentrations were measured and used for normalization. To evaluate qualitative values of NAD+, the NAD/NADH-Glo™ Assay (Promega) was used as described in the Supplementary Methods.
DHAP quantitation
To measure dihydroxyacetone phosphate (DHAP), Pico probe™ Dihydroxyacetone Phosphate Fluorometric Assay Kit (BioVision Incorporated) was used according to manufacturer’s recommendations. Cells were treated with DMSO, 12.5 nM of FK866 or GMX1778 for 48 hrs, and washed with cold PBS. 3×105 cells were suspended with DHAP Assay Buffer and mixed with PicoProbe, DHAP Enzyme Mix, and DHAP Developer. DHAP signal was measured by Fluorescence (BIOTEK). Data were compared with DMSO treated cells and expressed as % change.
13C Metabolic flux analysis and measurement of glycolytic intermediates
Steady-state labelling of glycolytic intermediates was accomplished by culturing cells (U87-Myc; 2.5×106, U87-GFP 4.5×106 cells/sample) in DMEM-no glucose-medium (Gibco) containing 25mM of D-Glucose (U-13C6, 99%, Cambridge Isotope Laboratories) for 6 hrs. For evaluation of MGG4 with/without NAMPT inhibitor, 4.5×106 cells/sample were treated with DMSO or FK866 (50nM) for 42hrs, washed, and incubated in stem cell medium based on glucose-free Neurobasal medium (Gibco) supplemented with 25mM of D-Glucose (U-13C6, 99%) and DMSO or FK866 (50 nM) for 6 hrs. All treatments were conducted in triplicate. Intracellular metabolites were then extracted and measured by capillary electrophoresis time-of-flight mass spectrometry (CE-TOFMS) by Human Metabolome Technology (Tsuruoka, Japan) as described in the Supplementary Methods (23–26). For the U87 assay, the amounts of each metabolite labeled at specific carbons were measured and C3 or C6 labeled carbons were used for analysis. For the MGG4 assay, the total amounts of each metabolite, both labeled and unlabeled, were measured and used for analysis. Data were expressed as pmol/1×106 cells.
Animal Studies
For GBM orthotopic xenograft survival experiments, 1×105 MGG8 cells were implanted into the right striatum of SCID mice as described (21). After 5 days, animals were randomized to treatments with weekly oral gavage with GMX1778 (250 mg/kg) (n=7) or vehicle [20% Captisol (CyDex) and 5% dextrose in water] (n=7). Mice were monitored daily and body weight was measured 2–3 times a week. Mice were sacrificed when neurologic deficits or general conditions reached the criteria for euthanasia. For H1975 xenograft experiments, 2.5×106 cells were subcutaneously implanted into the right flank of 7–10-week-old female SCID mice. When maximum tumor diameter reached 5 mm, cohorts were randomized to weekly GMX1778 (250 mg/kg) (n=7) or vehicle (n=7) by oral gavage. Tumor diameters were measured 3 times a week using a digital caliper. Calculated volume (mm3) = length (mm) x width (mm)2 x 0.5. All mouse experiments were approved by the Institutional Animal Care and Use Committee at MGH.
Immunohistochemistry
Tumor tissue specimens (n=3 for each group, drug or vehicle) were fixed in 10% neutral-buffered formalin and embedded in paraffin. Hematoxylin and eosin staining was performed using the standard procedures. For Immunohistochemistry, 5µm thick sections were deparaffinized, treated with 0.5% H2O2 in methanol, rehydrated, and heated for 25 minutes for antigen retrieval. Sections were blocked with serum, and incubated with anti-Ki-67 (1:125, Wako) primary antibody at 4C overnight. The next day, sections were washed in PBS, incubated with biotinylated secondary antibody for 60 min at room temperature, and then incubated with ABC solution (PK6200; Vector Laboratories) for 30 min at room temperature. Finally, sections were stained with DAB (Dako) and counterstained with hematoxylin. To detect apoptosis in situ, TUNEL assay was performed using TACS XL (Trevigen) followed by hematoxylin counterstain. For staining quantification, microscopic images of 3 randomly chosen fields / tumor were acquired with SPOT 5.0 software, and more than 1000 cells / tumor were counted with image J 1.48v. Two investigators (K.T. and H.W.) blinded to treatment group quantified signals, and only strongly stained cells were considered positive.
Immunocytochemistry
MGG4, MGG8, and MGG85 cells were treated with DMSO, 12.5 nM of FK866, or GMX1778 for 72 hrs, washed with cold PBS, and mounted on slides by cytospin. Cells were fixed with methanol/acetic acid (3:1) for 20 min, and permeabilized with 0.1% triton X in PBS for 5 min. After blocking with 10% goat serum in PBS, cells were incubated with Ki-67 (1:125) at 4C overnight, and then incubated with fluorophore-conjugated secondary antibody (1:200, Alexa Fluor 546 goat anti-mouse IgG, Life Technologies Corporation) at 4C for 4hr. After washing with PBS, slides were mounted with DAPI (H-1200, Vector Laboratories), and analyzed using a Fluorescent TE2000-U inverted microscope (Nikon). Microscopic images of 3 randomly chosen fields/cell line were acquired with SPOT 5.0 software, and more than 200 cells/cell line were counted with image J 1.48v. Only strongly stained cells were considered positive.
Statistical Analyses
For parametric analyses, 2-tailed student t tests were used, and for analysis of frequencies of nominal data, 2-tailed Fisher exact test was used. Data were expressed as mean ± SE. Survival analysis was performed using the Kaplan-Meier method using the log-rank test to compare treatment arms. P values less than 0.05 were considered statistically significant. All in vitro and in vivo experiments, including cell viability assays, metabolite quantification, Western Blot, and flow cytometry, were performed at least twice to confirm reproducibility. 13C metabolic flux analysis in Myc-transduced U87 GBM cells and MGG4 were also performed twice with at least 3 replicates per condition.
RESULTS
Overactive Myc Drives Aerobic Glycolysis in Glioblastoma Cells
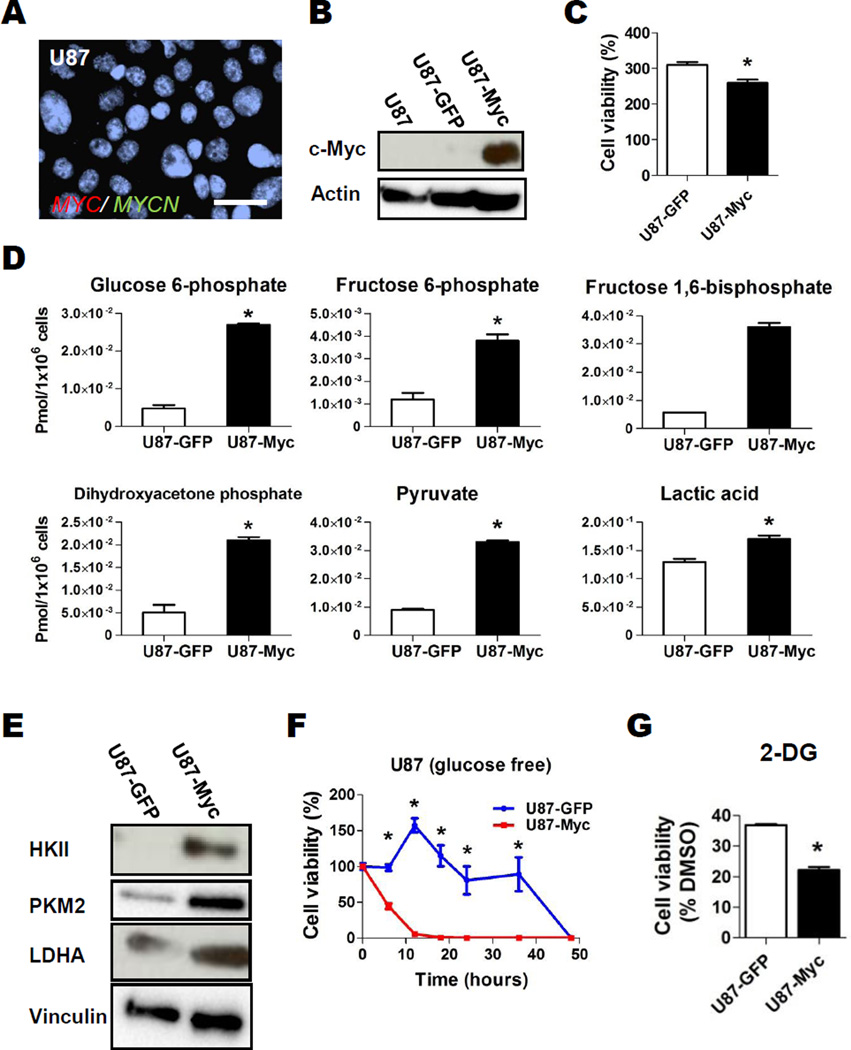
Inappropriate Myc activation drives aerobic glycolysis (“Warburg effect”) in part by inducing expression of key glycolytic pathway regulators (1, 17, 18, 27). To evaluate the effect of overactive Myc on glycolysis in GBM cells, we applied 13C metabolic flux analysis to Myc-transduced U87 GBM cells, which do not have significant Myc or N-Myc expression nor MYC/MYCN gene amplification at baseline (Figs. 1A, 1B). Lentivirus constructs carrying either MYC or GFP driven by a constitutively active cytomegalovirus promoter were used to infect U87, generating U87-Myc and U87-GFP lines. Of note, Myc overexpression induced only a slight reduction in proliferation rate (Fig. 1C). To gauge flux through glycolysis, both U87-Myc and U87-GFP were cultured in normoxia in the presence of [U-13C6] glucose for 6 hours and isotope enrichment in glycolytic metabolites was determined via capillary electrophoresis time-of-flight mass spectrometry (CE-TOFMS). The quantity of 13C labeled glycolytic intermediates glucose 6-phosphate (G6P), fructose 6-phosphate (F6P, C6 labeled), DHAP, pyruvate, and lactic acid (C6 labeled) were significantly increased in U87-Myc compared to U87-GFP, indicating Myc overexpression increased glycolytic flux (Fig. 1D, Table S1). Additionally, Myc overexpression resulted in significantly increased expression of the glycolytic enzymes HKII, PKM2 and LDHA in U87 cells (Fig. 1E).
Figure 1. Overactive Myc Drives Aerobic Glycolysis in Glioblastoma Cells.
A, Representative FISH image with probes for MYC and MYCN in U87 cells. MYC, MYCN and centromere control (Cen) probes in green or red as indicated; bar, 20 µm. B, Western blot analysis of parental and GFP- or MYC-transduced U87 cells. Actin, loading control. C, Cell viability of U87-GFP and U87-Myc at day 3 relative to day 0. Bar, SE; *, P<0.05. D, Effect of Myc overexpression on the levels of major isotope-labeled glycolytic metabolites derived from U-13C6 glucose. Cells were incubated in U-13C6 glucose for 6 hours, and intracellular metabolites were analyzed by Capillary Electrophoresis Time-of-Flight Mass Spectrometry (CE-TOFMS) as described in Materials and Methods. Metabolites labeled at C6 and C3 were the major labeled metabolite species detected and were compared between U87-GFP and U87-Myc. *, P<0.05, Welch’s t-test. E, Western blot analysis of Hexokinase II (HKII), PKM2 and lactate dehydrogenase A (LDHA) expression in GFP- or MYC-transduced U87 cells. Vinculin, loading control. F, U87-GFP (blue) and U87-Myc (red) cells were cultured in normal media for 48 hrs, then cultured in glucose- and FBS-free DMEM. Cell viability was assessed at indicated time points by Cell Titer-Glo; *, P<0.05 for difference between U87-GFP and U87-Myc. G, Relative cell viability of U87-GFP and U87-Myc cells after 2-deoxyglucose (2-DG, glycolysis inhibitor, 1mM, 48hr) treatment compared to DMSO control. Bars, SE; *, P<0.05.
We then tested whether inappropriate Myc activation would induce a dependence on glycolytic metabolism for growth. We assessed the effect of glucose deprivation on growth of cells with or without Myc overexpression. We found that U87-GFP cells survived significantly longer in glucose-free culture conditions than U87-Myc cells, indicating a switch to dependence on glucose for growth was generated when Myc was overexpressed (Fig. 1F). Additionally, we inhibited glycolysis using 2-deoxyglucose (2-DG), a small molecule inhibitor of hexokinase (28), in U87-Myc and U87-GFP. Similar to our findings with glucose deprivation, growth of U87-Myc cells was more sensitive to glycolytic inhibition with 2-DG than U87-GFP (Fig. 1G).
Glycolytic Inhibition is Selectively Toxic to MYC/MYCN amplified GBM Tumorspheres
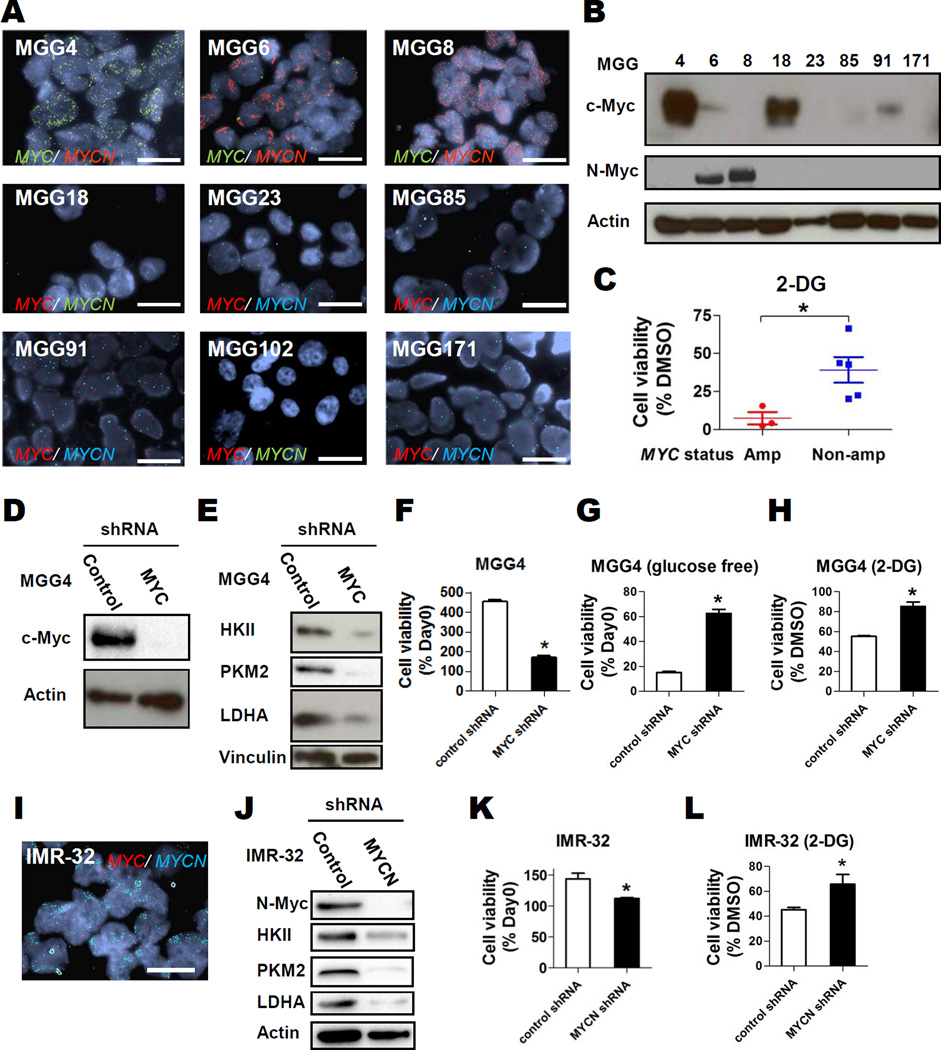
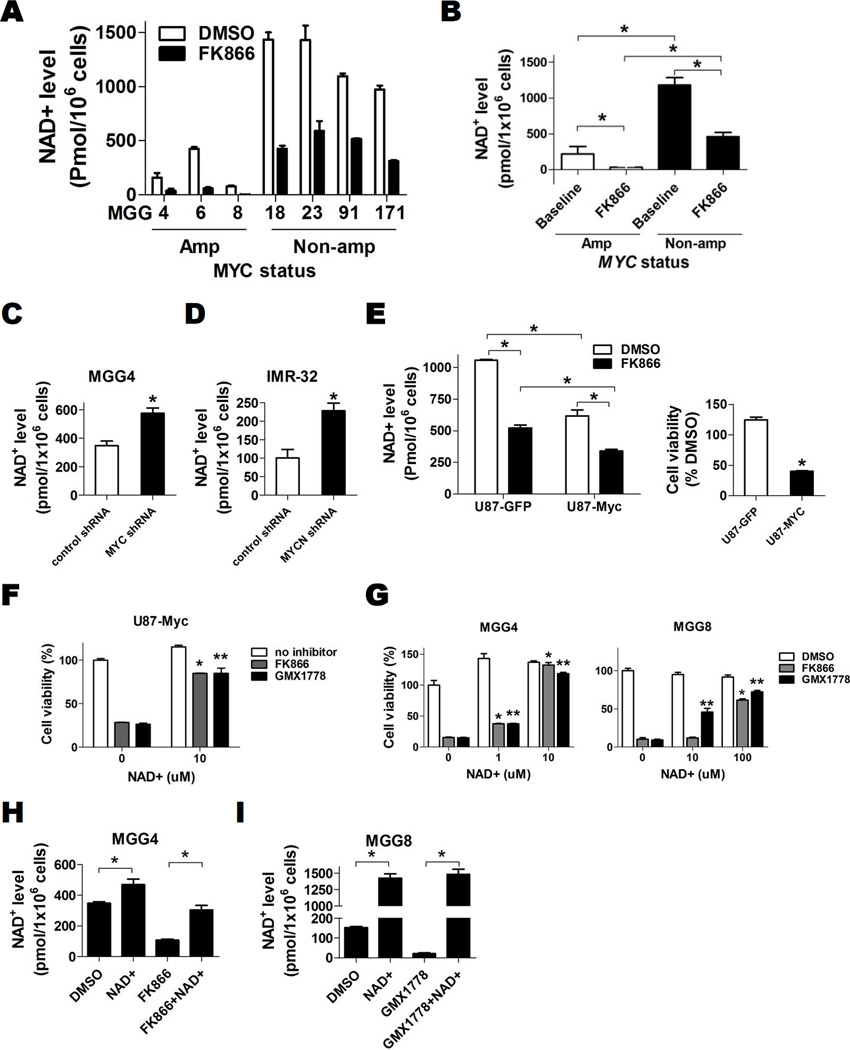
We then tested the effect of glycolytic inhibition in a panel of patient-derived GBM tumorsphere lines, where Myc has been reported to play an important role in self-renewal and maintenance of tumorigenicity (3–6). We tested for MYC and MYCN gene copy number status by fluorescence in situ hybridization and expression of the respective proteins by Western blot. We found that 3 GBM tumorsphere lines in our panel had high-level amplification of MYC (MGG4) or MYCN (MGG 6, MGG8) (Fig. 2A), and that all amplified lines had high-level expression of the corresponding proteins (Fig. 2B). There were two lines that expressed Myc without MYC amplification (MGG18 and MGG91) (Figs. 2A, 2B).
Figure 2. Glycolytic Inhibition is Selectively Toxic to MYC/MYCN amplified cancer cells.
A, Representative Fluorescent in situ hybridization (FISH) images of patient-derived GBM tumorspheres with MYC and MYCN probes in indicated colors; bars, 20 µm. B, Western blot analysis showing expression of c-Myc (Myc) and N-Myc in GBM tumorsphere cells. Actin, loading control. C, Relative cell viability of MYC or MYCN amplified (red) or MYC/MYCN non-amplified GBM tumorspheres (blue) after 48-hr treatment with 2-DG (1 mM) compared to DMSO control. D, Western blot analysis of c-Myc expression in control shRNA- and MYC shRNA-transduced MGG4 cells. Actin, loading control. E, Western blot analysis of, HKII, PKM2, and LDHA expression in control- or MYC-knockdown MGG4 cells. Vinculin, loading control. F and G, Relative cell viability of control shRNA and MYC shRNA transduced MGG4 cells cultured in regular (F) or glucose free (G) neural stem cell medium. Data were obtained at day 4 and normalized to the data at day 0. H, Relative cell viability of control shRNA and MYC shRNA transduced MGG4 cells after 2-DG (1 mM, 48 hrs) treatment compared to DMSO control. I, FISH images of IMR-32 cells with MYC and MYCN probes in indicated colors; bar, 20 µm. J, Western blot analysis of N-Myc, HKII, PKM2, and LDHA expression in control- or MYC-knockdown IMR-32 cells. Actin, loading control. K, Cell viability of control shRNA and MYCN shRNA transduced IMR-32 at day 2 relative to day 0. L, Relative cell viability of control shRNA and MYCN shRNA transduced IMR-32 cells after 2-DG (1 mM, 48 hrs) treatment compared to DMSO control. Bars, SE; *, P<0.05.
We next tested the effect of 2-DG in our panel of GBM tumorspheres, and found that the MYC/MYCN amplified GBM tumorspheres were significantly more sensitive to the deleterious effects of 2-DG (Fig. 2C). We then knocked down Myc expression in MGG4 (MYC amplified) using shRNA and tested the effects on glycolysis (Fig. 2D). In contrast to Myc-overexpression, MYC knockdown inhibited expression of the glycolytic enzymes HKII, PKM2, and LDHA (Fig. 2E). Stable Myc knockdown significantly decreased cell proliferation of MGG4 (Fig. 2F). However, after Myc knockdown we observed significantly enhanced survival of MGG4 in glucose-free culture and with exposure to 2-DG (Figs. 2G, 2H), confirming that Myc activity was a major contributor to the dependence of MGG4 on glycolysis for growth. We attempted to overexpress Myc in two non-MYC/MYCN amplified GBM tumorsphere lines, however stable lines could not be generated after Myc transduction. Although this may have been due to technical limitations, it is known that ecotpic Myc expression can invoke apoptosis in certain cell contexts [reviewed in (2)]
To evaluate the contribution of MYCN to glycolytic dependency in a highly MYCN amplified cell line, we knocked down MYCN in IMR-32 cells (Figs. 2I, 2J). Similar to results observed when knocking down MYC in a highly MYC amplified line, MYCN knockdown mildly decreased IMR-32 cell proliferation (Fig. 2K). MYCN knockdown also inhibited expression of the glycolytic enzymes HKII, PKM2, and LDHA (Fig. 2J). In addition, MYCN knockdown significantly decreased the sensitivity to the glycolysis inhibitor 2-DG (Fig. 2L). These results indicate that MYCN is a driver of cell proliferation in IMR-32 and induces a dependence on glycolysis for cell growth.
Overactive Myc renders GBM cells vulnerable to glycolytic inhibition with NAMPT inhibitors
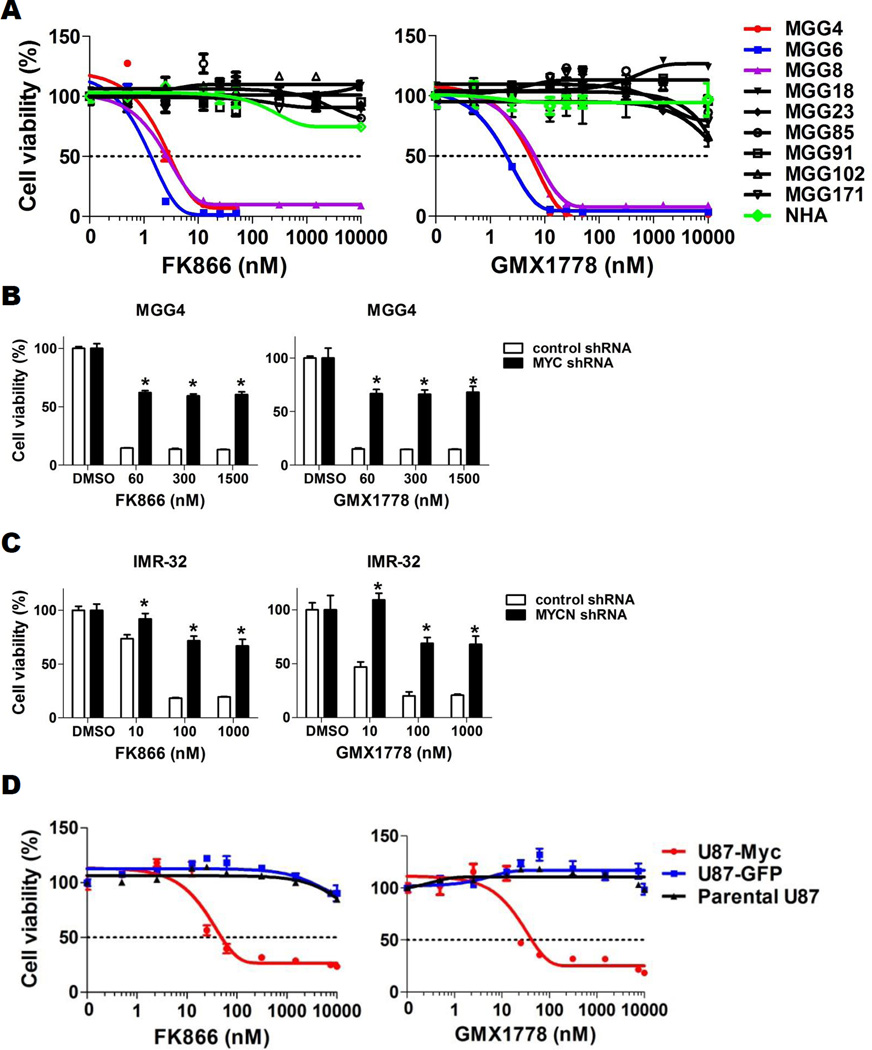
NAMPT is the rate-limiting enzyme of the NAD+ salvage pathway in eukaryotes, and its inhibition mediates cytotoxicity in cancer cells by attenuating glycolysis at the NAD+-requiring glyceraldehyde 3-phosphate dehydrogenase (GAPDH) step, among other metabolic perturbations (22, 29). We therefore tested the effect of small molecule inhibitors of NAMPT in our panel of GBM tumorsphere lines. We tested two chemically distinct, highly specific NAMPT inhibitors, FK866 (30–32) and GMX1778 (33, 34), on the growth of GBM tumorspheres. Both FK866 and GMX1778 significantly decreased NAD+ levels in both MYC/MYCN amplified and MYC/MYCN non-amplified GBM tumorspheres (Fig. S1A). However, reduction of NAD+ with NAMPT inhibitors resulted in strikingly potent cytotoxicity in a MYC/MYCN-dependent manner. In GBM tumorsphere lines tested for MYC/MYCN amplification (n=9), the IC50 values ranged between 1.3–6.4 nM for MYC/MYCN amplified lines (n=3) whereas non-amplified GBM tumorspheres (n=6) and normal human astrocytes (NHA) remained resistant at concentrations up to 10 µM, even up to 5 days (Figs. 3A, S1B, Table S2) (P=0.01, 2-tailed Fisher’s exact test).
Figure 3. Overactive Myc renders GBM cells vulnerable to glycolytic inhibition with NAMPT inhibitors.
A, CellTiter-Glo cell viability assay after 72-hr treatment of GBM tumorsphere lines with FK866 (left panel) and GMX1778 (right panel). Normal human astrocytes (NHA), green. B, CellTiter-Glo cell viability assay after 48-hr treatment of control shRNA (black) and MYC shRNA (red) transduced MGG4 cells with FK866 (left panel) and GMX1778 (right panel). *, P<0.05 for difference between control- and MYC-knockdown MGG4 cells. C, Cell viability assay after 48-hr treatment of control shRNA and MYCN shRNA transduced IMR-32 cells with FK866 (left panel) and GMX1778 (right panel). *, P<0.05 for difference between control- and MYCN-knockdown IMR-32 cells. D, Cell viability assay after FK866 (left panel) and GMX1778 (right panel) treatment of parental and GFP- or MYC-transduced U87 cells (Parental U87, black; U87-GFP, blue; U87-Myc, red) for 72 hrs. Bars, SE.
To test whether sensitivity to NAMPT inhibitors was mediated by Myc or N-Myc overactivity, we tested the effect of FK866 and GMX1778 in our MYC-transduced, MYC knockdown and MYCN knockdown systems. We found that MYC knockdown using shRNA significantly protected MYC amplified MGG4 from the cytotoxic effect of NAMPT inhibition (Fig. 3B), and similarly MYCN knockdown protected highly MYCN amplified IMR-32 from NAMPT inhibitor cytotoxicity (Fig. 3C). Correspondingly, Myc overexpression induced sensitivity to NAMPT inhibitors in U87 cells, which are completely resistant to NAMPT inhibitors at baseline (Fig. 3D).
NAMPT Inhibitors Mediate Cytotoxicity in MYC/MYCN amplified GBM Tumorspheres by Inhibiting Glycolysis
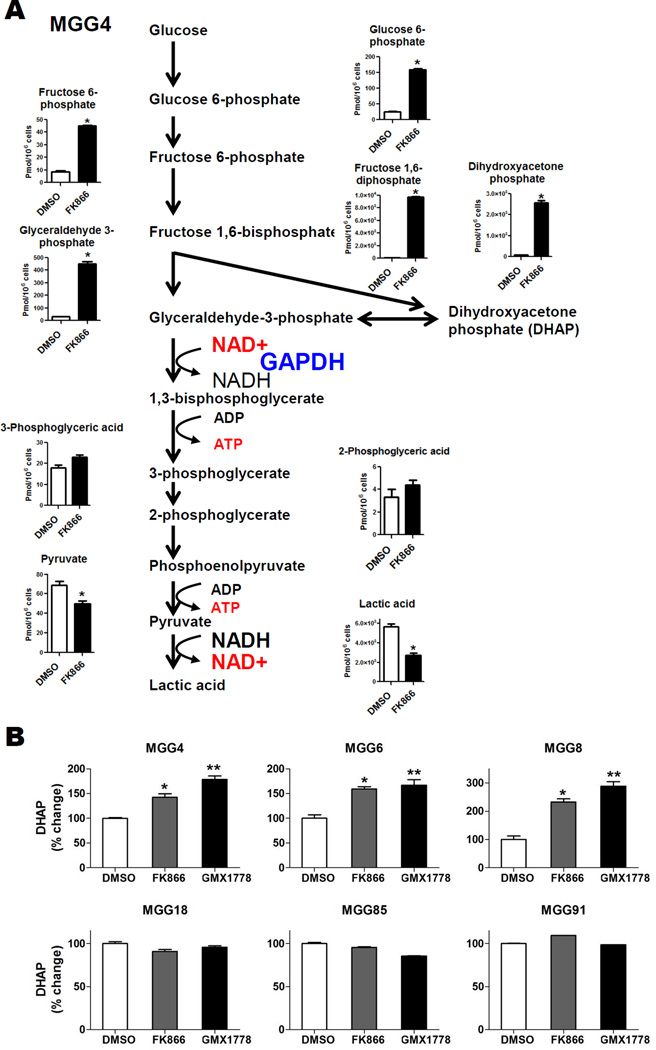
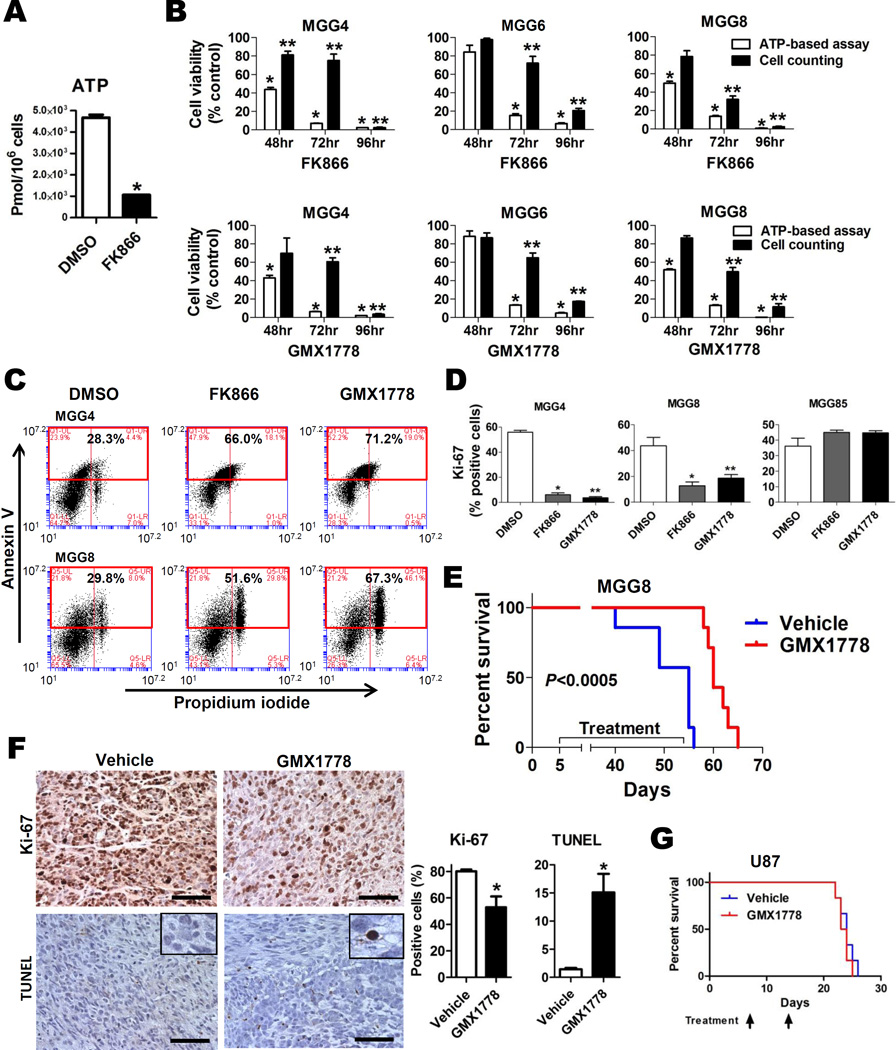
To confirm that NAMPT inhibitors inhibit glycolysis in Myc-driven GBM cells, we measured total levels of glycolytic intermediates in MGG4 (MYC amplified GBM tumorsphere line) with or without NAMPT inhibitor using CE-TOFMS. Incubation with NAMPT inhibitor resulted in marked accumulation of glycolytic intermediates upstream of the GAPDH step [i.e., G6P, F6P, FBP, glyceraldehyde 3-phosphate (G3P) and DHAP] while glycolytic intermediates downstream of GAPDH were not significantly changed (3-phosphoglycerate and 2-phosphoglycerate) or decreased (pyruvate and lactic acid) (Fig. 4A, Table S3). These results indicated NAMPT inhibitor blocked glycolysis at the GAPDH step in MGG4. To determine whether the NAMPT inhibitor-induced glycolysis blockade observed in the MGG4 metabolic flux data was selective for MYC/MYCN amplified GBM cells, we used a second assay to quantify levels of DHAP, which rapidly and reversibly isomerizes with the GAPDH substrate G3P, in our panel of GBM tumorspheres with and without NAMPT inhibition. We found that NAMPT inhibitor exposure significantly increased the levels of DHAP in the MYC/MYCN amplified (and NAMPT inhibitor sensitive) GBM tumorsphere lines, whereas those in non-amplified tumorspheres were unchanged (Fig. 4B). These data confirm that NAMPT inhibitors disrupt glycolysis at the NAD+-dependent GAPDH step in MYC/MYCN amplified GBM tumorspheres.
Figure 4. NAMPT Inhibitors Attenuate Glycolysis in MYC/MYCN amplified GBM Tumorspheres.
A, Capillary Electrophoresis Time-of-Flight Mass Spectrometry (CE-TOFMS) analysis of MGG4 cells demonstrating total levels of glycolytic metabolites upstream and downstream of GAPDH after DMSO (48hr, white bars) or FK866 (50nM, 48hr, black bars) treatment. Data were expressed as pmol/1×106 cells. *, P<0.05, Welch’s t-test. B, Dihydroxyacetone phosphate (DHAP) levels in MYC or MYCN amplified (MGG4, 6, 8, upper panels) and MYC/MYCN non-amplified GBM tumorspheres (MGG18, 85, 91, lower panels) treated with DMSO (white bars, 48 hrs) or FK866 (12.5 nM, 48 hrs, gray bars), or GMX1778 (12.5nM, 48hrs, black bars). *, P<0.05 for difference between DMSO and FK866. **, P<0.05 for difference between DMSO and GMX1778. Bars, SE.
Myc Alters NAD+ Metabolism
Because NAMPT inhibitors deplete intracellular NAD+ to mediate their effects on tumor growth, we further studied the relationship between Myc and NAD+. Intriguingly, we discovered that MYC/MYCN amplified GBM tumorspheres had significantly lower basal intracellular NAD+ levels compared to non-amplified GBM (Figs. 5A, 5B). NAMPT inhibitor exposure significantly reduced the absolute NAD+ levels in both MYC/MYCN amplified and non-amplified lines, however NAD+ levels in amplified lines were nearly completely eliminated while the NAD+ levels in non-amplified lines after NAMPT inhibitor exposure remained higher than the basal levels observed in untreated amplified lines (Figs 5A, 5B). This threshold difference in residual NAD+ levels may have contributed to the profound cytotoxic effect of NAMPT inhibitors observed only in MYC/MYCN amplified lines.
Figure 5. Myc alters NAD+ Metabolism.
A, Intracellular NAD+ levels at baseline (DMSO, 24 hr, white bars) and FK866 treated (12.5 nM, 24 hr, black bars) in GBM tumorspheres lines with MYC or MYCN amplification and those without MYC/MYCN amplification. B, Mean baseline absolute NAD+ levels of GBM tumorshere lines with MYC/MYCN amplification (220.4 ± 105.0 pmol/1×106 cells) and those without MYC/MYCN amplification (1184 ±103.3 pmol/1×106 cells, P=0.0009) and after FK866 treatment (34.0 ±16.8 pmol/1×106 cells with MYC/MYCN amplification and 460.6 ± 59.9 pmol/1×106 cells without MYC/MYCN amplification, P=0.002). C, Absolute intracellular NAD+ levels of control shRNA and MYC shRNA transduced MGG4 cells. D, Absolute intracellular NAD+ levels of control shRNA and MYCN shRNA transduced IMR-32 cells. E, Left panel, Absolute intracellular NAD+ levels at baseline and after FK866 treatment (25nM, 24hr) in U87 cells engineered to overexpress Myc (U87-Myc) or GFP (U87-GFP). Right panel, relative cell viability of U87-GFP and U87-Myc cells after FK866 (25nM, 72hr) treatment compared to DMSO control. F, Effect of exogenous NAD+ on U87-Myc cell viability after DMSO, FK866 (50 nM) or GMX1778 (50 nM) treatment for 72 hrs. *, P<0.05 comparison between DMSO and FK866. **, P<0.05 comparison between DMSO and GMX1778. G, Effect of exogenous NAD+ on cell viability in MYC/MYCN amplified GBM tumorspheres treated with DMSO, FK866 (12.5 nM) or GMX1778 (12.5 nM) for 72 hrs. *, P<0.05 for difference between exogenous NAD+ versus FK866 alone. **, P<0.05 for difference between exogenous NAD+ versus GMX1778 alone. H, NAD+ levels of MGG4 (MYC amplified) after treatment with DMSO or exogenous NAD+ (100uM) with or without FK866 (12.5 nM) for 24 hrs. I, NAD+ levels of MGG8 (MYCN amplified) after treatment with DMSO or exogenous NAD+ (1 mM) with or without GMX1778 (12.5 nM) for 24 hrs. Bars, SE; *, P<0.05.
We then tested the apparent association between Myc activity and NAD+ metabolism using our MYC/MYCN-knockdown and overexpression systems. Knockdown of MYC in MGG4 (MYC amplified) and MYCN in IMR-32 (MYCN amplified) resulted in increased intracellular NAD+ levels (Figs. 5C, 5D), while overexpression of Myc in U87 cells resulted in significant reduction of NAD+ (Fig. 5E). Similar to our observations in GBM tumorspheres (Figs. 5A, 5B), NAMPT inhibitor treatment significantly reduced NAD+ levels in both U87-GFP and U87-Myc cells (Figs. 5E, S2A), but the post-treatment levels in U87-GFP remained comparable to the pre-treatment (basal) levels in U87-Myc (Fig. 5E), again suggesting a threshold NAD+ level for survival was maintained without Myc overactivity.
We then confirmed that the NAMPT inhibitor-mediated effects observed in Myc-driven GBM were dependent on both NAMPT and NAD+. Exogenous supplementation with NAD+ increased intracellular NAD+ levels and rescued U87-Myc and MYC/MYCN amplified cells from NAMPT inhibitor cytotoxicity (Figs. 5F–5I). Furthermore, MYC/MYCN amplified cells were rescued from NAMPT inhibitor cytotoxicity with nicotinamide mononucleotide (NMN), a product of the NAMPT reaction and a direct precursor of NAD+, indicating direct NAMPT inhibition (Fig S2B). In contrast, supplementation with NA, a precursor for an alternate NAD+ salvage pathway rate-limited by nicotinic acid phosphoribosyltransferase (Naprt1), could not rescue cells from the effects of NAMPT inhibition (Fig. S2C).
Because Myc has been reported to induce NAMPT expression (35) and Naprt1 expression is known to modulate sensitivity to NAMPT inhibitors (36, 37), we evaluated the expression of Naprt1 as well as NAMPT in our GBM tumorsphere lines. We found that NAMPT expression was similar in all GBM tumorsphere lines tested (Fig. S3A). However, Naprt1 expression varied and was notably absent in all MYC/MYCN amplified GBM tumorspheres, while in non-amplified lines Naprt1 expression was highly variable and not clearly associated with Myc expression (Fig. S3A). In addition, we found that neither forced overexpression or knockdown of Myc altered the expression of NAMPT or Naprt1 (Figs. S3B, S3C). These results suggest NAMPT and Naprt1 expression per se do not absolutely determine sensitivity to NAMPT inhibitors in GBM tumorspheres. Together with the differences observed in glycolytic intermediates between MYC/MYCN amplified vs. Myc-expressing/non-MYC/MYCN amplified lines after GAPDH inhibition (Fig. 4B), our findings point to MYC/MYCN genetic amplification, rather than expression of NAMPT, Naprt1 or Myc (without gene amplification), as critical for determining sensitivity to glycolytic inhibition.
Glycolytic Inhibition with NAMPT inhibitor Results in Apoptosis and Potent in vivo Activity in Myc-driven GBM
Prior studies have revealed that NAMPT inhibitor-mediated glycolytic blockade causes intracellular depletion of NAD+ as well as ATP, resulting in metabolic perturbation and cytotoxicity (22). We similarly found that NAMPT inhibitors caused ATP depletion and cell death in MYC/MYCN amplified GBM tumorspheres (Figs. 6A–6C). Cytotoxicity was mediated by apoptosis, as indicated by significant increases in Annexin V staining and double staining for Annexin V and propidium iodide (PI) by flow cytometry, and caspase-3/7 activation after treatment with NAMPT inhibitor (Figs. 6C, S4A). We further observed activation of the intracellular energy regulator AMPK, inhibition of mTOR signaling, and induction of PARP cleavage, indicating intracellular metabolic perturbation after NAMPT inhibition in MYC/MYCN amplified cells (Fig. S4B), but not in MGG85 (MYC/MYCN non-amplified) cells (Fig. S4C). To assess whether NAMPT inhibitors affected cell proliferation, we assessed Ki-67 expression in GBM tumorspheres. Whereas the fraction of Ki-67 expressing cells in MGG85 (MYC/MYCN non-amplified) cells was unchanged, NAMPT inhibitors significantly decreased the Ki-67 positive fraction in MYC/MYCN amplified cells (Figs. 6D, S4D).
Figure 6. Glycolytic Inhibition with NAMPT inhibitor Results in Apoptosis and Potent in vivo Activity in Myc-driven GBM.
A, Capillary Electrophoresis Time-of-Flight Mass Spectrometry (CE-TOFMS) analysis quantifying total levels of ATP after DMSO (48 hrs, white bars) or FK866 (50 nM, 48 hrs, black bars) treatment of MGG4. B, Cell viability of GBM tumorsphere lines after treatment with FK866 (upper panels) and GMX1778 (lower panels) measured by ATP-based CellTiter-Glo assay (white bars) and viable cell count (Trypan blue exclusion) assay (black bars). *, P<0.05 for the CellTiter-Glo assay values at 48, 72, and 96 hrs. **, P<0.05 for the Trypan blue exclusion assay values at 48, 72, and 96 hrs. DMSO treated cells were used as control at each time point. C, Flow cytometric analysis of MGG4 (upper panels) and MGG8 (lower panels) cells treated with DMSO (left panel), FK866 (12.5 nM, middle panel) and GMX1778 (12.5 nM, right panel) for 96 hrs, demonstrating increased fraction of cells double positive for Annexin V (Y axis) and propidium iodide (PI; X axis) after FK866 and GMX1778 treatment. D, Ratios of Ki-67 positive cells in MGG4 (MYC amplified, left panel), MGG8 (MYCN amplified, middle panel), and MGG85 (MYC/MYCN non-amplified, right panel) treated with DMSO (white bars), FK866 (12.5nM, gray bars) or GMX1778 (12.5nM, black bars) for 72hrs. *, P<0.05 for difference between DMSO versus FK866. **, P<0.05 for difference between DMSO versus GMX1778. E, Kaplan-Meier survival estimates of SCID mice bearing orthotopic MGG8 xenografts after treatment with weekly oral gavage of GMX1778 (250mg/kg, n=7, red line) or vehicle (n=7, blue line). Data were analyzed by log-rank test (P<0.0005). F, Ki-67 and TUNEL immunohistochemistry of MGG8 orthotopic xenograft tumors 48 hrs after a single dose of vehicle (left panels) or GMX1778 (250 mg/kg) treatment. Representative microscopic pictures with insets depicting staining details and quantification of positivity are shown. Scale bars, 100 µm. Bars, SE; *, P<0.05. G, Kaplan-Meier survival estimates of SCID mice bearing orthotopic U87 xenografts treated with weekly oral gavage of GMX1778 (250mg/kg, n=6, red line) or vehicle (n=6, blue line). Data were analyzed by log-rank test (P=0.72).
To test the effect of glycolytic inhibition on Myc-driven GBM cells in vivo, we investigated NAMPT inhibitors due to their striking selectivity for MYC/MYCN amplified GBM tumorspheres and their feasibility for use in vivo. We tested the effect of GMX1778 on MGG8 (MYCN amplified), which rapidly and diffusely infiltrates mouse brains (21), and U87 (MYC/MYCN non-amplified) orthotopic xenografts. Oral GMX1778 significantly extended the survival of mice bearing MGG8 orthotopic xenografts compared to vehicle dosing (P<0.0005, log-rank test, Fig. 6E). On necropsy, xenografts tumors from animals treated with GMX1778 compared to tumors from vehicle-treated mice had significantly decreased cells positive for Ki-67 staining (80.1±1.4% for vehicle vs. 53.0±8.1% for GMX1778, P=0.03) and increased apoptotic cells as measured by TUNEL staining (1.4±0.2% for vehicle vs.15.1±3.3% for GMX1778, P=0.03, Fig. 6F). In contrast, oral GMX1778 had no effect on the survival of mice bearing U87 orthotopic xenografts (Fig. 6G).
Myc-driven Cancers Are Sensitive to NAMPT Inhibitors
To explore whether our finding that Myc-driven GBM cells are selectively sensitive to glycolytic inhibition is generalizable to other cancer types, we tested the effect of NAMPT inhibitors in several standard cell lines from multiple cancer types known to harbor genetically activated MYC, including H1975 lung (MYC amplified), D283 Med medulloblastoma (MYC amplified), IMR-32 and Kelly neuroblastoma (MYCN amplified), Raji Burkitt’s lymphoma (IGH-MYC translocation) (Figs. 2I, S5). Similar to our findings in MYC/MYCN amplified GBM tumorspheres, NAMPT inhibitors displayed low-nanomolar activity against genetically Myc-driven cell lines (Fig. S6A). Furthermore, consistent with MGG4, shRNA knockdown of Myc in H1975 cells (highly MYC amplified, Fig S5) resulted in increased intracellular NAD+ levels and rescued H1975 from the cytotoxic effects of NAMPT inhibition (Figs. S6B–S6D).
We also tested the effect of GMX1778 on established H1975 (MYC amplified) subcutaneous flank xenograft tumors. Mice bearing H1975 xenografts treated with NAMPT inhibitors had significantly inhibited tumor growth (mean tumor weight ± SE, 101.7± 28.1mg for GMX1778 treatment vs. 612.0 ± 129.8 mg for vehicle treatment, P=0.002, t test) (Figs. S7A–S7C). Tumors from mice treated with GMX1778 had increased TUNEL staining (Fig. S7D) and tumor regressions were observed the day after the weekly dose (Fig. S7A). Notably, detectable NAD+ was nearly eliminated in tumors from GMX1778-treated mice (Fig. S7E). All animals with flank or intracerebral xenografts tolerated oral GMX1778 well, with no significant body weight change or apparent toxicities during treatment (Figs. S8A–S8B).
DISCUSSION
Here, we demonstrate that overactive Myc drives an abnormal metabolic program in GBM cells, increasing glycolytic flux and altering NAD+ metabolism. We found that Myc-driven GBM cells are sensitive to inhibition of the Myc-induced increased glycolytic drive, and identified NAMPT inhibitors, which attenuate glycolysis by depleting NAD+ required for the GAPDH step, as a selective and strikingly potent strategy for MYC/MYCN amplified GBM, Ultimately, our findings support and extend on the work of others that have demonstrated that overactive Myc generates an altered metabolic state in cancer cells that can be exploited by identifying their addiction to specific metabolites (19, 38).
It is now clear that GBM represents a group of molecularly heterogeneous subtypes with distinct clinical phenotypes (9–11) and that therapeutic development strategies must focus on specific biological subtypes in order to derive maximal benefit. Our strategy would target a small proportion of primary GBM, as approximately 4% of primary GBMs harbor high-level MYC or MYCN amplification (7–11). However, most large scale genetic studies have focused on primary (initial) GBM tumors and the few comprehensive studies that analyzed the genetic changes at develop at recurrence included only small numbers of paired tumors (39, 40). The prevalence of MYC/MYCN copy number alterations developing at recurrence in primary GBM is unclear. Interestingly, recent large-scale molecular studies of isocitrate dehydrogenase 1 (IDH1) and IDH2 mutant gliomas have identified a high proportion of MYC amplification (up to 31%) in this subtype of glioma (41–44). Although most MYC amplification events were low-level in these studies, the majority were observed at progression (44) and in the non-1p/19q codeleted subtype (43), suggesting an association with more aggressive clinical behavior in IDH1/2 mutant gliomas (45). Thus, our strategy could apply to a significant proportion of glioma subtypes.
We also identified an intriguing association between overactive Myc and reduced NAD+ levels, suggesting Myc reprograms NAD+ metabolism. Our findings contrast with a prior study that showed Myc induced expression of NAMPT and resulted in increased NAD+ levels (35). In our study, Myc overexpression and knockdown had no effect on NAMPT expression, and increased Myc led to lower NAD+ levels. The reasons for this discrepancy are unclear and may relate to cell context, as all our NAD+ measurements were performed in malignant cells. NAD+ is a fundamental metabolite for numerous critical intracellular pathways and further studies will be needed to elucidate the complex mechanisms of NAD+ regulation, the effects of NAMPT inhibition and the relationship between Myc and NAD+. Intriguingly, we found evidence that a critical NAD+ threshold may exist for cancer cell survival. This preliminary finding warrants further detailed investigation since together with our observation that NAMPT inhibitors nearly completely eliminate detectable NAD+ levels in xenograft tumors, our data suggests intratumoral NAD+ level represents a potential predictive and/or pharmacodynamic biomarker in Myc-driven cancers.
It is likely that other genetic or epigenetic alterations that similarly impact cancer cell metabolism may also sensitize to NAMPT inhibitors, as NAMPT inhibitor activity has been observed in diverse cancer types (34, 46). Whether increased Myc/N-Myc expression without MYC/MYCN genetic alteration sensitizes to glycolytic inhibition remains to be completely determined. We found that the association with glycolytic inhibitor sensitivity in GBM cells was most tightly correlated with gene amplification, particularly with NAMPT inhibitors. It is possible that genetic events that inappropriately activate Myc, such as amplification, translocation, or lentiviral transduction, are more stable or potent means of driving overactive Myc and altering cancer metabolism. In addition, there may be differences in downstream effects between MYC/MYCN genetic alterations and Myc/N-Myc protein expression without genetic alteration. We find evidence of this in our studies as the effect on glycolytic intermediates differed between MYC/MYCN amplified vs. Myc overexpressed (non-amplified) GBM tumorspheres after inhibition of the key glycolytic enzyme GAPDH.
Nevertheless, our findings support the paradigm of glycolytic inhibition as a novel therapeutic strategy for a genetically-defined subgroup within a wide array of cancers. Amplification of the MYC and MYCN genes are among the most frequent amplification events in human cancer (47), and we provide evidence that our findings in GBM may translate to other cancer types, as we observed potent cytotoxicity with NAMPT inhibitors in numerous genetically Myc-driven cancer cells in vitro and in vivo. Additionally, since NAMPT inhibitors have entered clinical development (48), our findings have the potential for rapid translation in a genetically enriched subset of GBM patients.
Supplementary Material
Statement of Translational Relevance.
Glioblastoma (GBM) remains a lethal disease with poor prognosis despite significant recent discoveries in disease pathogenesis, and novel therapeutic strategies are desperately needed. Myc is a potent oncogene that is overactive in a subset of GBM and drives multiple cancer-related phenotypes including altered metabolism to support rapid cell growth and proliferation. Here, we identify a therapeutic strategy that exploits the Myc-induced glycolytic dependency present in a subset of Myc and N-Myc-driven GBM. We show that Myc increases glycolytic flux in GBM cells, and that this metabolic shift generates a vulnerability to glycolytic inhibition. We identify inhibitors of the NAD+ salvage pathway rate-limiting enzyme nicotinamide phosphoribosyl-transferase (NAMPT), which attenuate glycolysis to induce cytotoxicity, as a highly potent and selective therapeutic strategy for MYC/MYCN amplified GBM. As NAMPT inhibitors have entered clinical development, potential exists for rapid clinical translation in a genetically defined subset of GBM.
Acknowledgments
A.S. Chi is supported by a Richard B. Simches Scholars Award as well as the MGH Glioblastoma Research Fund in memory of David M. King, Kirit N. Patel, Sarah E. Rippetoe and Eric W. Schwartz. K. Tateishi is supported by a Society of Nuclear Medicine and Molecular Imaging Wagner-Torizuka Fellowship, the Japan Brain Foundation, and the KANAE foundation for the Promotion of Medical Science. D. Cahill is supported by a Burroughs Wellcome Fund Career Award. A.C., D.C., H.W., are supported by NIH P50CA165962-01A1. We thank Hideyuki Takeuchi, Fares Nigim, and Mara Koerner for technical assistance, Shinichi Esaki and Jianfang Ning for growing GBM tumorsphere lines, and Laura Shelton and Takushi Oga at Human Metabolome Technologies, Inc., for assistance with and analysis of 13C metabolite labeling assays.
Financial Support: This work is supported by U.S. NIH P50CA165962-01A1 (to T.T. Batchelor (PI), D.P. Cahill, H. Wakimoto, and A.S. Chi), K24 CA125440-06 (to T.T. Batchelor), a Burroughs Wellcome Fund Career Award (to D.P. Cahill) and a Society of Nuclear Medicine and Molecular Imaging Wagner-Torizuka Fellowship (to K. Tateishi).
Footnotes
Conflicts of Interest: All authors have no conflicts of interest to report with regard to this manuscript.
REFERENCES
- 1.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 3.Zheng H, Ying H, Yan H, Kimmelman A, Hiller D, Chen A, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Annibali D, Whitfield JR, Favuzzi E, Jauset T, Serrano E, Cuartas I, et al. Myc inhibition is effective against glioma and reveals a role for Myc in proficient mitosis. Nat Commun. 2014;5:4632. doi: 10.1038/ncomms5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerosuo L, Piltti K, Fox H, Angers-Loustau A, Hayry V, Eilers M, et al. Myc increases self-renewal in neural progenitor cells through Miz-1. J Cell Sci. 2008;121:3941–3950. doi: 10.1242/jcs.024802. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Wang H, Li Z, Wu Q, Lathia JD, McLendon RE, et al. c-Myc is required for maintenance of glioma cancer stem cells. PLoS One. 2008;3:e3769. doi: 10.1371/journal.pone.0003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hui AB, Lo KW, Yin XL, Poon WS, Ng HK. Detection of multiple gene amplifications in glioblastoma multiforme using array-based comparative genomic hybridization. Lab Invest. 2001;81:717–723. doi: 10.1038/labinvest.3780280. [DOI] [PubMed] [Google Scholar]
- 8.Hodgson JG, Yeh RF, Ray A, Wang NJ, Smirnov I, Yu M, et al. Comparative analyses of gene copy number and mRNA expression in glioblastoma multiforme tumors and xenografts. Neuro Oncol. 2009;11:477–487. doi: 10.1215/15228517-2008-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The Somatic Genomic Landscape of Glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsons D, Jones S, Zhang X, Lin J, Leary R, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montana MF, et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol. 2011;18:1331–1335. doi: 10.1038/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, Hatheway CM, et al. CDK7 Inhibition Suppresses Super-Enhancer-Linked Oncogenic Transcription in MYCN-Driven Cancer. Cell. 2014;159:1126–1139. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dang CV. Therapeutic targeting of Myc-reprogrammed cancer cell metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:369–374. doi: 10.1101/sqb.2011.76.011296. [DOI] [PubMed] [Google Scholar]
- 17.Masui K, Tanaka K, Akhavan D, Babic I, Gini B, Matsutani T, et al. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 2013;18:726–739. doi: 10.1016/j.cmet.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luan W, Wang Y, Chen X, Shi Y, Wang J, Zhang J, et al. PKM2 promotes glucose metabolism and cell growth in gliomas through a mechanism involving a let-7a/c-Myc/hnRNPA1 feedback loop. Oncotarget. 2015;6:13006–13018. doi: 10.18632/oncotarget.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wakimoto H, Kesari S, Farrell C, Curry WJ, Zaupa C, Aghi M, et al. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res. 2009;69:3472–3481. doi: 10.1158/0008-5472.CAN-08-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wakimoto H, Mohapatra G, Kanai R, Curry WT, Jr, Yip S, Nitta M, et al. Maintenance of primary tumor phenotype and genotype in glioblastoma stem cells. Neuro Oncol. 2012;14:132–144. doi: 10.1093/neuonc/nor195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan B, Young DA, Lu ZH, Wang T, Meier TI, Shepard RL, et al. Pharmacological inhibition of nicotinamide phosphoribosyltransferase (NAMPT), an enzyme essential for NAD+ biosynthesis, in human cancer cells: metabolic basis and potential clinical implications. J Biol Chem. 2013;288:3500–3511. doi: 10.1074/jbc.M112.394510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soga T, Heiger DN. Amino acid analysis by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem. 2000;72:1236–1241. doi: 10.1021/ac990976y. [DOI] [PubMed] [Google Scholar]
- 24.Soga T, Ueno Y, Naraoka H, Ohashi Y, Tomita M, Nishioka T. Simultaneous determination of anionic intermediates for Bacillus subtilis metabolic pathways by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem. 2002;74:2233–2239. doi: 10.1021/ac020064n. [DOI] [PubMed] [Google Scholar]
- 25.Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, Nishioka T. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J Proteome Res. 2003;2:488–494. doi: 10.1021/pr034020m. [DOI] [PubMed] [Google Scholar]
- 26.Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010;6:78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 28.Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tolstikov V, Nikolayev A, Dong S, Zhao G, Kuo MS. Metabolomics analysis of metabolic effects of nicotinamide phosphoribosyltransferase (NAMPT) inhibition on human cancer cells. PLoS One. 2014;9:e114019. doi: 10.1371/journal.pone.0114019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasmann M, Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 2003;63:7436–7442. [PubMed] [Google Scholar]
- 31.Wosikowski K, Mattern K, Schemainda I, Hasmann M, Rattel B, Loser R. WK175, a novel antitumor agent, decreases the intracellular nicotinamide adenine dinucleotide concentration and induces the apoptotic cascade in human leukemia cells. Cancer Res. 2002;62:1057–1062. [PubMed] [Google Scholar]
- 32.Zhang LY, Liu LY, Qie LL, Ling KN, Xu LH, Wang F, et al. Anti-proliferation effect of APO866 on C6 glioblastoma cells by inhibiting nicotinamide phosphoribosyltransferase. Eur J Pharmacol. 2012;674:163–170. doi: 10.1016/j.ejphar.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 33.Hjarnaa PJ, Jonsson E, Latini S, Dhar S, Larsson R, Bramm E, et al. CHS 828, a novel pyridyl cyanoguanidine with potent antitumor activity in vitro and in vivo. Cancer Res. 1999;59:5751–5757. [PubMed] [Google Scholar]
- 34.Watson M, Roulston A, Belec L, Billot X, Marcellus R, Bedard D, et al. The small molecule GMX1778 is a potent inhibitor of NAD+ biosynthesis: strategy for enhanced therapy in nicotinic acid phosphoribosyltransferase 1-deficient tumors. Mol Cell Biol. 2009;29:5872–5888. doi: 10.1128/MCB.00112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menssen A, Hydbring P, Kapelle K, Vervoorts J, Diebold J, Luscher B, et al. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci U S A. 2012;109:E187–E196. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tateishi K, Wakimoto H, Iafrate AJ, Tanaka S, Loebel F, Lelic N, et al. Extreme Vulnerability of IDH1 Mutant Cancers to NAD+ Depletion. Cancer Cell. 2015;28:773–784. doi: 10.1016/j.ccell.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shames DS, Elkins K, Walter K, Holcomb T, Du P, Mohl D, et al. Loss of NAPRT1 expression by tumor-specific promoter methylation provides a novel predictive biomarker for NAMPT inhibitors. Clin Cancer Res. 2013;19:6912–6923. doi: 10.1158/1078-0432.CCR-13-1186. [DOI] [PubMed] [Google Scholar]
- 38.Shim H, Chun YS, Lewis BC, Dang CV. A unique glucose-dependent apoptotic pathway induced by c-Myc. Proc Natl Acad Sci U S A. 1998;95:1511–1516. doi: 10.1073/pnas.95.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J, Lee IH, Cho HJ, Park CK, Jung YS, Kim Y, et al. Spatiotemporal Evolution of the Primary Glioblastoma Genome. Cancer Cell. 2015;28:318–328. doi: 10.1016/j.ccell.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 40.Kim H, Zheng S, Amini SS, Virk SM, Mikkelsen T, Brat DJ, et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015;25:316–327. doi: 10.1101/gr.180612.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47:458–468. doi: 10.1038/ng.3273. [DOI] [PubMed] [Google Scholar]
- 42.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cancer Genome Atlas Research N. Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bai H, Harmanci AS, Erson-Omay EZ, Li J, Coskun S, Simon M, et al. Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat Genet. 2015 doi: 10.1038/ng.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wakimoto H, Tanaka S, Curry WT, Loebel F, Zhao D, Tateishi K, et al. Targetable signaling pathway mutations are associated with malignant phenotype in IDH-mutant gliomas. Clin Cancer Res. 2014;20:2898–2909. doi: 10.1158/1078-0432.CCR-13-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiarugi A, Dolle C, Felici R, Ziegler M. The NAD metabolome--a key determinant of cancer cell biology. Nat Rev Cancer. 2012;12:741–752. doi: 10.1038/nrc3340. [DOI] [PubMed] [Google Scholar]
- 47.Beroukhim R, Mermel C, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galli U, Travelli C, Massarotti A, Fakhfouri G, Rahimian R, Tron GC, et al. Medicinal chemistry of nicotinamide phosphoribosyltransferase (NAMPT) inhibitors. J Med Chem. 2013;56:6279–6296. doi: 10.1021/jm4001049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.