Abstract
Identifying genetic variants with pleiotropic associations can uncover common pathways influencing multiple cancers. We took a two-staged approach to conduct genome-wide association studies for lung, ovary, breast, prostate and colorectal cancer from the GAME-ON/GECCO Network (61,851 cases, 61,820 controls) to identify pleiotropic loci. Findings were replicated in independent association studies (55,789 cases, 330,490 controls). We identified a novel pleiotropic association at 1q22 involving breast and lung squamous cell carcinoma, with eQTL analysis showing an association with ADAM15/THBS3 gene expression in lung. We also identified a known breast cancer locus CASP8/ALS2CR12 associated with prostate cancer, a known cancer locus at CDKN2B-AS1 with different variants associated with lung adenocarcinoma and prostate cancer and confirmed the associations of a breast BRCA2 locus with lung and serous ovarian cancer. This is the largest study to date examining pleiotropy across multiple cancer-associated loci, identifying common mechanisms of cancer development and progression.
Introduction
Genome wide association studies (GWAS) have identified hundreds of genetic variants that are associated with risk of specific cancers (1). It has been observed that some chromosomal regions show pleiotropic associations, where the same genetic loci are associated with different cancers. One of the first identified pleiotropic loci is at 8q24, where genetic variants are associated with breast, prostate, colorectal and ovarian cancer risk, with some of the variants only associated with one cancer, while others are associated with multiple cancers (2). Similarly, genetic variants at the TERT-CLPTM1L region at 5p15.33 are associated with risk of lung, bladder, prostate, and other cancers (3).
The identification of pleiotropic loci is an important step in elucidating cancer etiology by understanding common pathways that influence carcinogenesis across different tumors, and in improving knowledge of cancer susceptibility. Furthermore, analyzing genomic data across multiple cancer sites might identify novel susceptibility loci, as variants that do not meet the stringent criteria for GWAS significance for any one cancer site, might show significant associations when multiple cancers are analyzed together (4).
Our investigation used data from the Genetic Associations and Mechanisms in Oncology (GAME-ON) Network and the Genetic and Epidemiology of Colorectal Cancer Consortium (GECCO) (5). The GAME-ON Network was launched by the National Cancer Institute (NCI) to capitalize on the extensive investment in GWAS, with the overarching goal to integrate post-GWAS research and to facilitate analyses that address research questions that are common across multiple cancer sites. The GAME-ON Network is focused on tumors that currently represent a major public health burden and has assembled extensive genomic data from consortia investigating the cancer sites which constituted the basis of our cross-cancer analysis. We use these data and independent replication studies to investigate pleiotropic associations across lung, breast, colorectal, ovary and prostate cancer using GWAS results for 61,851 cases and 61,820 controls, the largest investigation of pleiotropic associations to date.
Materials and Methods
Data and contributing consortia
This study used summary level data to perform cross-cancer GWAS analysis of lung, colorectal, prostate, breast and ovarian cancers using a subset-based meta-analytical approach. Forty-six studies from North America and Europe organized into cancer site specific consortia within the GAME-ON Network (http://epi.grants.cancer.gov/gameon/) or GECCO, participated. Table 1 provides details for contributing consortia and studies. Analyses also included the following subtypes: adenocarcinoma and squamous cell carcinoma of the lung; the aggressive form of prostate cancer; estrogen receptor negative breast cancer, and serous and endometrioid cancers of the ovary (Table 1). All studies frequency matched cases and controls on at least age and gender. All subjects were of European descent.
Table 1.
Contributing consortia and characteristics of data sets.
| GAME-ON/GECCO Analysis | Replication Stage (European ancestry) | Generalizability (Other ancestry) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||||
| Cancer Site (Consortium) | No. studies | Cases | Controls* | Variants† | Genotyping Platform | Imputation threshold‡ | Study | Cases | Controls* | Study | Cases | Controls* |
|
|
|
|
||||||||||
| Lung (TRICL)10 | 6 | 12160 | 16838 | 8492272 | Illumina | R2>0.3 | deCODE10,13 | 3865 | 196658 | Nanjing18 | 2331 | 3077 |
| Harvard14,15 | 984 | 970 | ||||||||||
| Adenocarcinoma | 3718 | 15871 | 8472145 | deCODE | 1434 | 198663 | Nanjing | 1304 | 3077 | |||
| Harvard | 597 | 970 | Japan17 | 1575 | 3363 | |||||||
| Squamous cell carcinoma | 3422 | 16015 | 8478230 | deCODE | 784 | 171059 | Nanjing | 822 | 3077 | |||
| Harvard | 216 | 970 | ||||||||||
| Colorectal (CORECT)11 | 6 | 5100 | 4831 | 7229595 | Affymetrix Axiom | None | deCODE | 3546 | 236404 | |||
| Colorectal (GECCO)5 | 13 | 10314 | 12857 | 9193926 | Illumina, Affymetrix | R2>0.3 | ||||||
| Prostate (ELLIPSE) | 14160 | 12724 | 9084781 | Illumina, Affymetrix | R2>0.3 | deCODE | 4858 | 83103 | LAPC/MEC20 | 1034 | 1046 | |
| 6 | iCOGS16 | 20219 | 20440 | AAPC23 | 4853 | 4678 | ||||||
| JAPC/MEC20 | 980 | 1005 | ||||||||||
| Prostate Aggressive | 6 | 4450 | 12724 | 9003304 | ||||||||
| Breast (DRIVE)7,9 | 15748 | 18084 | 9331393 | Illumina, Affymetrix | R2>0.3 | Shanghai19 | 2867 | 2285 | ||||
| 11 | deCODE | 5318 | 280808 | LABC/MEC, SF21 | 1497 | 3213 | ||||||
| iCOGS7 | 46785 | 42892 | ||||||||||
| AABC22 | 3015 | 2743 | ||||||||||
| Breast ER6 | 8 | 4939 | 13128 | 9250406 | ||||||||
| Ovary (FOCI)8 | 3 | 4369 | 9123 | 9911464 | Illumina | R2>0.3 | deCODE | 716 | 111373 | |||
| iCOGS8 | 16283 | 23491 | ||||||||||
| Ovary - Serous | 3 | 2556 | 9123 | 9911279 | iCOGS | 10316 | 23491 | |||||
| Ovary - Endometrial | 3 | 715 | 9123 | 9910229 | iCOGS | 2338 | 23491 | |||||
|
| ||||||||||||
| Total* | 45 | 61851 | 61820 | 9916564 | 55789 | 330490 | 18152 | 21410 | ||||
The number of unique individuals after accounting for cancer subtypes and overlapping controls. Breast iCOGS included only 1q22 variants, so total for replicates without breast iCOGS is shown.
Subset meta-analyses were performed for a specific variant if at least 3 sites (i.e., three of lung, colorectal, prostate, breast or ovary) contributed data.
Imputation performed using the 1000 genome reference panel. Exclusion threshold shown.
Genotyping and imputation
Genotyping was performed on Affymetrix or Illumina platforms (Table 1). Standard marker exclusion criteria were applied in each cancer consortium (5–11), and we have previously reported the summary of QC details in Hung et al. (12). Genotype imputation was conducted for each cancer site using IMPUTE, BEAGLE, MACH and Minimac (imputation threshold of R2 > 0.3) using the 1000 genome reference panel.
Statistical Analysis
Logistic regression analysis using a log additive model was performed previously to test variant associations with cancer risk for the forty-five studies (5–11), providing per-allele odds ratios (ORs) adjusted for age, principal components and gender where applicable. Study-specific results were then combined for each cancer site using a fixed effects model.
A subset-based meta-analysis approach developed by Bhattacharjee et al. (ASSET) was used to investigate pleiotropic effects across cancer sites (4). The method generalizes the standard fixed effect meta-analysis by examining the association between a genetic variant and multiple subsets of cancers, allowing opposing direction of effects and null associations. Associations are summarized with an overall two-sided P value. A multiple-testing adjustment procedure (based on statistical theory for tail-probability approximation) maintains appropriate type I error rates. Analysis was performed when at least three cancer sites had data.
Accounting for subsets of studies with no effects and/or effects in opposing directions (i.e., both increased and reduced risk subsets) is an advantage of the subset-based meta-analysis approach. However, when a large majority of underlying associations are in one direction, subset meta-analysis can have lower power than standard fixed effects analysis. We therefore explored results using standard fixed effects meta-analysis (when data was present for all cancer sites), again using ASSET.
Subjects appearing in several studies with different cancer sub-types (e.g., overlapping controls for lung adenocarcinoma and lung SqCC) and across cancer types (e.g., overlapping controls from the Welcome Trust Case Control Consortium (WTCCC) for UK ovary and UK breast GWAS) were accounted for in the covariance matrix when estimating standard errors for subset-based and standard meta-analyses. In order to find robust pleiotropic associations we first identified variants that showed evidence for pleiotropy and then sought to validate the individual variant cancer site associations in replication data sets. To prioritize variants for replication, we selected those with P < 5 × 10−7 based on the two sided subset-analysis test, the positive and negative associations that contributed to the two-sided subset-analysis test signal, and the fixed effect meta-analysis. We excluded variants where associations were obviously driven by a single cancer site. We also selected variants associated with at least two cancers (including subtypes) at P < 5 × 10−3. Among variants selected, we further prioritized for validation those showing the strongest pleiotropic association in a region (based on subset or standard cross cancer meta-analysis), as well as those representing association peaks in site-specific analyses. We then sought to validate these specific variant-cancer site associations using independent study populations of European descent with all five cancers from deCODE (10,13 ), lung cancer from Harvard (14,15), breast (region 1q22 only) from iCOGS, ovarian from OCAC/iCOGS and prostate from PRACTICAL/iCOGS (7,8,16). For cross-ethnicity generalizability, we examined results for the selected variants in different race/ethnicities using data from Japan (lung (17)), Nanjing (lung (18)), Shanghai (breast (19)), MEC, African American Breast Cancer GWAS Consortium (AABC), African Ancestry Prostate Cancer GWAS Consortium (AAPC), and San Francisco (breast for Latinas) (20–23). An outline of analysis steps is presented in supplemental figure 1.
Further investigation of pleiotropic effects
We further investigated pleiotropy between cancer sites using conditional Q-Q plots to examine enrichment of association signals in one cancer when conditioning on significance of P values of a second cancer. Enrichment is reflected in a leftward deflection of the Q-Q plot with decreasing P value categories of the conditioning cancer (24), indicating higher pleiotropy between cancer sites than expected by chance.
Functional Significance
eQTL analysis
We obtained non-tumor lung eQTL data of 1,111 patients from three studies: Laval University (n = 409, GSE23352), The University of British Columbia (n = 339) and the University of Groningen (n = 363). Gene expression profiles were obtained using an Affymetrix array (GEO platform GPL10379). Genotyping was performed using the Illumina Human1M-Duo genotyping BeadChip. Analyses were adjusted for age, sex and smoking status. eQTL data are deposited in the Gene Expression Omnibus (GEO) database with accession numbers GSE23352, GSE23529 and GSE23545 for the three studies respectively. Further details of this study are published elsewhere (25). We investigated validated lung cancer associated variants from our study. A statistically significant eQTL for a variant was declared if 2 of 3 studies showed a significant P value after Bonferroni correction for multiple comparisons. We also evaluated eQTL data for variants in LD (R2 > 0.7) with our validated variants. We also obtained TCGA eQTL data for 402 high-grade serous ovarian cases and 145 prostate tumor samples again investigating variants with validated associations and those in high LD (R2 > 0.7) with them. Gene expression values for high-grade serous ovarian cases were assessed by P value. Gene expression values for prostate cancer were adjusted for somatic copy number and CpG methylation as previously described (26). Significant associations were defined as those having both P value and false discovery rate (Benjamini-Hochberg method) of less than 0.05.
Function/prediction
We used FuncPred from SNPinfo (http://snpinfo.niehs.nih.gov/) to assist with variant function prediction. The software evaluates a variant’s potential function in splicing regulation, TFBS prediction, miRNA binding site prediction and regulatory potential using in-house algorithms and tools developed elsewhere. In addition, we used RegulomeDB to further assess regulatory potential for variants of interest (http://www.regulomedb.org/). This tool includes high-throughput, experimental data sets from ENCODE and other sources, as well as computational predictions and manual annotations to identify putative regulatory potential and functional variants. We further examined ENCODE data using the UCSC genome browser (https://genome.ucsc.edu/) (see Supplementary Materials: Bioinformatics Tools).
Results
After applying quality control filters 9,916,564 variants were analyzed for pleiotropic associations using 61,851 cases and 61,820 controls of European ancestry across five common cancer sites in the GAME-ON Network and GECCO (GAME-ON/GECCO) (Table 1). Figure S2 displays an overview of the association test results for each of the five main cancer sites investigated.
One hundred and ninety variants in 33 regions (13 to 22 for each cancer site) were prioritized for follow-up for replication in Caucasian populations (55,789 cases and 330,490 controls) and generalizability (18,152 cancer cases and 21,410 controls) after the initial analysis (see Table 1 and Supplementary Table S1). An additional 46,785 cases and 42,892 controls from iCOGS Breast cancer were used for validation of the novel pleiotropic locus at 1q22 (see below). We replicated associations at four of the 33 regions selected for follow-up in Europeans. Variants with replicated pleiotropic associations were not associated with cancer risk in other ethnic populations and therefore these data are not presented. The main findings in each region are described below (see supplementary Table S1 and S2 for summary).
Novel region: 1q22 for lung squamous cell carcinoma and breast cancer
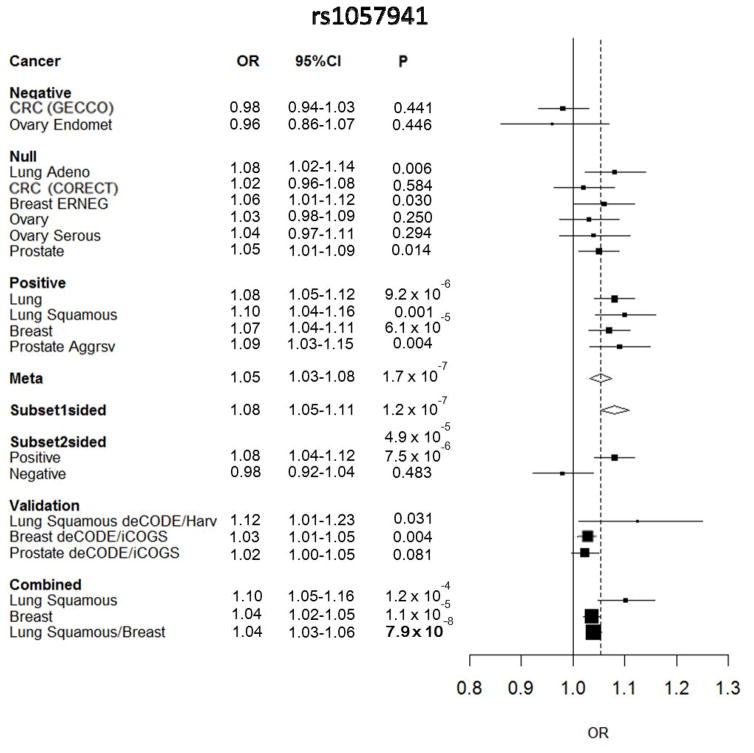
Standard meta-analysis of GAME-ON/GECCO discovery data identified an association between rs1057941 located at 1q22 and overall risk of cancer (P = 1.74 × 10−7, Fig. 1). Overall lung, lung squamous cell carcinoma (lung SqCC) and breast cancer were strongly associated with this variant in GAME-ON/GECCO data (Lung: OR = 1.08, 95% CI 1.05–1.12, P = 9.2 × 10−6; lung SqCC: OR = 1.10, 95% CI 1.04–1.16, P = 0.001; Breast: OR = 1.07, 95% CI 1.04–1.11, P = 6.08 × 10−5). The association for lung SqCC was replicated in deCODE and Harvard studies combined, with OR of 1.12 (95% CI 1.01–1.23, P = 0.03). The association with breast cancer was replicated in deCODE and iCOGS combined (OR = 1.03, 95%CI 1.01–1.05, P = 0.004). The overall P value for association from both stages for lung SqCC and breast cancer approached genome-wide significance (P = 7.9 × 10−8) (Fig. 1).
Figure 1.
Result for rs1057941. Forest plot showing per allele ORs for risk allele A (of A/G). Standard fixed effects meta-analysis (also indicated by dashed line), and subset meta-analysis results (two-sided, one-sided and positive and negative subset associations) are shown. Abbreviations - CRC: colorectal, ERNEG: estrogen receptor negative, Aggrsv: aggressive, Adeno: Adenocarcinoma.
The aggressive form of prostate cancer was also selected by ASSET as part of the subset of cancers associated with rs1057941. We have no replication data for this subtype and did not replicate the association with overall prostate cancer (deCODE and PRACTICAL/iCOGS combined: OR = 1.02, 95% CI 1.00–1.05, P = 0.08).
Regional plots constructed from GAME-ON/GECCO results show a distinct peak in P values represented by rs1057941 in a 40kb region of LD at 1q22 that includes KRTCAP2, GBA, MTX1, MUC1, TRIM46, THBS3, ADAM15 and ASH1L for the cross-cancer analysis (Fig. 2A), while for individual sites MUC1 variant rs4072037 was most significant for lung SqCC (P = 3.21 × 10−4) and the TRIM46 variant, rs3814316, for breast cancer (P = 3.06 × 10−6). These 3 variants are in high to complete LD with each other with pair-wise D′ of 0.86 (rs1057941–rs3814316) or 1.00 (rs1057941–rs4072037, and rs3814316–rs4072037).
Figure 2.
Results for rs1057941 (A) Regional plot showing P values from overall meta-analysis at region 1q22 using GAME-ON/GECCO discovery set data. The top breast cancer hit (rs3814316) and top squamous cell carcinoma hit (rs4072037) are also highlighted. (B) Boxplots of gene expression levels in normal lung tissue for ADAM15 and THBS3 by study (Laval, UBC, Groningen).
eQTL analyses indicated that rs4072037 acted as a normal lung tissue eQTL for two genes in this region, ADAM15 and THBS3, with two studies showing significant associations after adjustment for multiple comparisons and the third showing a nominally significant association consistent in direction with the other two (ADAM15: Laval P = 2.39 × 10−7, University of British Columbia P = 4.09 × 10−5, University of Groningen P = 0.08; THBS3: Laval P = 1.71 × 10−5, University of British Columbia P = 4.15 × 10−6, University of Groningen P = 0.004) (Fig 2B). ADAM15 is of primary interest as it was shown to be overexpressed in lung and breast cancer (27). The risk allele for rs4072037, A, was consistently associated with increased ADAM15 gene expression in all three studies. In the UBC discovery set rs4072037 was the strongest eQTL among 33 variants.
Previously known cancer loci with pleiotropic associations
2q33.1 (known breast and melanoma locus) and prostate cancer
We identified a pleiotropic association between rs13016963 located in 2q33.1 and prostate (OR = 1.08, 95% CI 1.04–1.13, P = 3.05 × 10−5) and breast cancer (OR = 0.93, 95% CI 0.90–0.96, P = 5.75 × 10−5) (Fig. 3A) in GAME-ON/GECCO. This variant is in intron 5 of ALS2CR12, adjacent to CASP8 (Fig. 3B), and was associated with melanoma in a previous GWAS (28). The region was previously identified as harboring variants associated with breast cancer risk (29–31) and the association found with breast cancer can be explained through LD (R2 = 0.74) with one of these (rs1830298, P value for breast cancer association = 1.02 × 10−7). The association with prostate cancer has not been previously reported and we replicated it in deCODE and iCOGS (OR = 1.05, 95% CI 1.03–1.08, P = 7.6 × 10−5) (Fig. 3A). The combined GAME-ON/GECCO, deCODE and iCOGS P value was 1.9 × 10−8.
Figure 3.
Results for rs13016963: (A) Forest plot showing per allele ORs for risk allele G (of A/G). Standard fixed effects meta-analysis (also indicated by dashed line), and subset meta-analysis results (two-sided, one-sided and positive and negative subset associations) are shown. (B) Regional plot showing P values for GAME-ON prostate cancer GWAS at region 2q33.1 using GAME-ON/GECCO discovery set data. Peak is at ALS2CR12. (C) Boxplot of BZW1 gene expression levels in prostate tumor and normal tissue for rs1035142/rs13016963. rs1035142 is presented as a surrogate for rs13016963, with which it shows perfect LD in eQTL data. Abbreviations - CRC: colorectal, ERNEG: estrogen receptor negative, Aggrsv: aggressive, Adeno: Adenocarcinoma.
In Figure 3C, we show eQTL analysis results for rs1035142, which was in perfect LD with rs13016963 in the eQTL data set and was associated with BZW1 (P = 0.001, FDR = 0.04). This variant had the strongest association signal among those investigated. We do note that there were variants with more significant associations with BZW1 expression that fell outside of the LD range of R2≥ 0.70 with rs13016963 (e.g., rs13113, P = 4.9 × 10−6, R2 = 0.62). This suggests there are other potential cis-eQTL candidates for BZW1 in the region.
9p21.3 (known lung SqCC and breast locus) and lung adenocarcinoma
Similar to 8q24, the 9p21.3 region displays a complex nature of associations with multiple cancer sites. It was previously known to be associated with lung SqCC and breast cancer. In our analysis based on GAME-ON/GECCO discovery data, we observed suggestive evidence of association between the variant rs62560775 at CDKN2B-AS1 and lung adenocarcinoma (OR = 1.19, 95% CI =1.08–1.31, P = 2.77 × 10−4), as well as breast cancer (OR = 1.11, 95% CI 1.05–1.17, P = 5.30 × 10−4). Although this region was previously reported as a lung SqCC susceptibility locus, this is the first time that we observed an association with lung adenocarcinoma. The association was replicated based on deCODE and Harvard data (OR = 1.16, 95% CI 1.03–1.30, P = 0.01). The combined (both stages) P value was 1.0 × 10−5 (Fig. 4A). The association of this variant with lung adenocarcinoma was the most significant in the region (Fig. 4B). Our lung eQTL investigation of this variant showed no significant association with gene expression in this region.
Figure 4.
Results for rs62560775: (A) Forest plot showing per allele ORs for risk allele G (of A/G). Standard fixed effects meta-analysis (also indicated by dashed line), and subset meta-analysis results (two-sided, one-sided and positive and negative subset associations) are shown. (B) Regional plot showing P values for GAME-ON lung adenocarcinoma GWAS at region 9p21.3, using GAME-ON/GECCO discovery set data. Peak is at CDKN2B-AS1. Results for rs1011970: (C) Forest plot showing per allele ORs for risk allele T (of T/G). Standard fixed effects meta-analysis (also indicated by dashed line), and subset meta-analysis results (two-sided, one-sided and positive and negative subset associations) are shown. (D) Regional plot showing P values for GAME-ON prostate cancer GWAS at region 9p21.3, using GAME-ON/GECCO discovery set data. Peak is at CDKN2B-AS1. Abbreviations - CRC: colorectal, ERNEG: estrogen receptor negative, Aggrsv: aggressive, Adeno: Adenocarcinoma.
Another variant in the same region, rs1011970, was shown to be associated with prostate cancer (OR = 1.10, 95% CI = 1.05–1.15, P = 7.3 × 10−5). This variant was found to be associated with breast cancer in a previous GWAS (32). The association with prostate cancer was replicated in deCODE and iCOGS combined (P = 0.001). The combined P value based on both stages was 9.5 × 10−7 (Fig 4C). The regional plots shows that this variant had the second most significant association with prostate cancer in the region. (The peak association for prostate cancer is represented by rs72652411, a variant not associated with any cancer other than prostate (Fig. 4D)).
13q13.1 (known breast and lung locus) confirmed for serous ovarian cancer
We observed a significant pleiotropic associations for rs11571833 (ASSET two-sided P value: = 6.14 × 10−10). This variant (Fig. S3) is potentially functional and genome-wide significant associations between this variant and breast and lung cancer (driven by SqCC) were previously reported, with the latter study using a subset of the lung cancer data use here (7,10). More recently, a study focusing specifically on rs11571833 reported an association with serous ovarian cancer using the iCOGS data which formed our replication data set (P = 3 × 10−5). Our results from the GAME-ON data set confirm the association with serous ovarian cancer (GAME-ON OR = 1.76, 95% CI 1.30–2.39; P = 2.49 × 10−4; with the rare allele associated with increased risk)) and reached genome-wide significance when we combined the two data sets (P = 3.95 × 10−8) (Fig S3). We did not replicate the association with breast cancer; however this might be due to a lack of power to detect an association for this rare variant in our sample.
Other evidence for pleiotropic associations
As demonstrated by the leftward deflection of Q-Q plots with decreasing P value category, there is evidence of pleiotropic associations for breast and ovarian cancer (supplementary Fig. S4A), breast and prostate cancer (supplementary Fig. S4B) and prostate and colorectal cancer (supplementary Fig. S4C). There was no evidence of pleiotropic associations for prostate and ovary cancer (Fig. S4D), or lung cancer with any of the other four cancer sites.
Discussion
Using data from the GAME-ON Network and GECCO we conducted a cross-cancer GWAS analysis investigating pleiotropic associations for five cancer sites (lung, breast, colorectal, ovary and prostate) including histology and subtypes. We identified three new pleiotropic associations that were supported by results in GAME-ON/GECCO data and our independent replication data sets. We identified a novel pleiotropic association at the 1q22 region involving breast cancer and lung SqCC, neither of which was previously known to be associated with genetic variation in this region. The association with lung SqCC was further supported by its functional significance as demonstrated by eQTL analysis. Further novel results include a locus at CASP8/ALS2CR12, known to be associated with breast cancer and melanoma, that was found to be associated with prostate cancer; while genetic variation at the 9p21.3 region, known to be associated with breast cancer and lung SqCC, appears to be associated with lung adenocarcinoma and prostate cancer. We also confirmed an association between a known lung and breast cancer locus at BRCA2 with serous ovarian cancer risk.
The locus at 1q22 represented by rs1057941, rs3814316 and rs4072037 lies in a region of LD that includes KRTCAP2, MTX1, TRIM46, MUC1, GBA, THBS3, ADAM15 and ASH1L. Of these variants, rs4072037 at MUC1 might be functional as it was shown to regulate alternative splicing of the second exon in MUC1 and modifies the gene’s transcriptional activity (33). Aberrantly glycosylated MUC1 is overexpressed in most epithelial cancers and is known to have an oncogenic effect. It mediates the production of growth factors such as connective tissue growth factor (CTGF), and platelet driven growth factor A and B (PDGF-A and PDGF-B) that promote activation of the MAPK and PI3k/Akt pathways potentiating proliferation and survival of tumor cells (34). It also plays a critical role in EGFR signalling, promoting survival of NSCLC cells (35).
Our lung eQTL investigation found no association with MUC1 expression but the risk allele, A, of rs4072037 was associated with increased expression of two other genes in the region (ADAM15 and THBS3). This result suggests other mechanisms by which this variant could influence cancer risk. ADAM15 is of particular interest since it was shown to be overexpressed in both lung and breast cancer (27), consistent with our finding of pleiotropic associations of these two cancer sites. Overexpression in breast cancer is associated with Her2/neu expression and evidence from breast cancer cell lines indicates that ADAM15 catalyzes the cleavage of E-cadherin which in turn binds to and enhances ErbB receptor signalling (36). A recent meta-analyses provided support for an association of this variant with gastric cancer in Asian populations (37), and a recent GWAS in the Icelandic population also suggested an association between this region and gastric cancer in European populations. These results lend further support for the etiological role of this region on cancer susceptibility in general (38).
For 2q33.1, we found evidence for a pleiotropic effect for rs13016963 (at ALS2CR12) on breast and prostate cancer. This region is known to harbor breast cancer susceptibility loci: rs1045485, encoding the missense alteration D302H in CASP8 (adjacent to ALS2CR12)(29), rs1830298 (mentioned above) at ALS2CR12 and rs1045494 at CASP8 (30,31). Of these three variants, rs1830298 was found to have the most significant association with breast cancer (P = 1.02 × 10−7) in our study, and was associated with prostate cancer risk (P = 5.2 × 10−4); whereas rs1045485 and rs1045494 were not. For prostate cancer, rs13016963 represented the peak association, although we point out strong LD between rs13016963 and rs1830298 (R2 = 0.74). Interestingly, rs13016963 was also found to be associated with risk of melanoma in a previous GWAS in subjects of European descent (28) indicating this variant may be associated with both prostate cancer and melanoma in Caucasian populations. It was also found to be associated with esophageal squamous cell carcinoma in Han Chinese (39).
Previous research has examined associations between other genetic variants in this region and prostate cancer risk. A possible association between the CASP8 histidine variant D302H and the more aggressive form of prostate cancer in European populations was reported (40). This variant was however not associated with aggressive prostate cancer (P = 0.11) or overall prostate cancer (P = 0.14) in GAME-ON/GECCO.
Our prostate eQTL analysis suggested that rs13016963 influences the expression of BZW1. Previous studies indicate a role for BZW1 in carcinogenesis. BZW1 can activate histone H4 gene transcription and serves as a co-regulator of other transcription factors involved in cell cycling. It has been implicated in promoting mucoepidermoid carcinoma tumor growth (41). We also found two potential functional variants in the region (rs700636 and rs1035142). These variants are in strong LD with rs13016963 (R2≥0.93), and have associations with prostate cancer similar to rs13016963 in strength and are predicted to sit in miRNA binding sites.
The 9q21.3 region encoding CDKN2B-AS1 has been much studied in cancer research. We observed a pleiotropic association of rs62560775 (located in the intronic region of CDKN2B-AS1) involving lung adenocarcinoma and breast cancer. The association with breast cancer might to be due to LD (R2 = 0.38) with a previously identified breast cancer susceptibility variant, rs1011970. Interestingly, we did replicate an association between rs1011970 and prostate cancer, suggesting this specific variant, or variants in LD with it contribute to risk for both breast and prostate cancer. Timofeeva et al, found an association between this region and lung SqCC, represented by rs1333040 (42), but this variant was not associated with adenocarcinoma in our data set (P = 0.62). Previous GWAS also report associations between this region and risk of glioma (43), melanoma (28) and basal cell carcinoma (44) and a recent pleiotropy study indicated an association with esophageal SqCC (45). LD between variants reported in these studies and either rs62560775 or rs1011970 range from R2 = 0.21 to R2 = 0.60, indicating that multiple variants in this region contribute to cancer risk. Despite that associations for different cancers were exerted from multiple variants, our results represent an important pleiotropic finding, as all variants are located in the same gene, CDKN2B-AS1.
Modification of CDKN2B-AS1 activity could be the mechanism through which this locus influences cancer risk. CDKN2B-AS1, also known as ANRIL (antisense non-coding RNA in the INK4 locus) is known to recruit a polycomb repression complex (PRC2) that silences CDKN2B but not CDKN2A (46). Although there is no known function for rs62660775 or rs1011970, a variant with which rs62660775 is in strong LD, rs3217986 (at R2 = 0.75), was identified to be located in a miRNA binding site (47) and classified as likely to affect binding by Regulome.
Our pleiotropy Q-Q plots (supplementary Fig. S4A–C) suggest pleiotropy between some sites: breast and ovarian cancer, breast and prostate cancer and prostate and lung cancer, with some evidence provided for pleiotropic associations involving prostate and lung and colorectal and ovarian cancer. There was little evidence for pleiotropic associations involving other site combinations using this approach.
Although we have built in studies from non-European descendants, including Asians, African Americans and Hispanics, we did not observe an association with any of the loci in other ethnic groups. It is likely that the smaller sample sizes for the non-European descendants restricted our power to detect associations. Variation in LD across ethnicities could also have reduced our power to detect associations. More research is needed to determine if associations in these regions can be generalized to different ethnic groups based on larger sample size. These efforts could make meaningful contributions to addressing disparities in health between populations.
Power to detect associations is also relevant when considering our primary analyses involving subjects of European ancestry. Our initial investigation using the GAME-ON and GECCO data set identified 33 regions and 190 variants that we further examined in replication data sets. We were able to replicate the associations for four of these regions. Since our replication datasets sample sizes were often smaller than that of our discovery (GAME-ON/GECCO) set (depending on cancer site), we may have insufficient power to replicate true associations particularly for less common variants, underlining the importance of sample size for investigations of pleiotropy.
In summary, using data from the GAME-ON initiative and GECCO, we have found four regions that show associations with multiple cancers, including a novel association between genetic variation at 1q22 and breast cancer and lung squamous cell carcinoma. This is the largest study to date examining pleiotropy across multiple cancer sites. There are likely more loci that are associated with multiple cancers but these will require additional efforts with larger data series for detection. Our results provide important insights into common carcinogenesis across multiple major cancers and highlight the value of pleiotropy analysis.
Supplementary Material
Acknowledgments
This study was supported by the grant from the National Institute of Health (NIH) (U19CA148127). Additional funding was obtained from NIH grants (5R01CA055769, 5R01CA127219, 5R01CA133996, and 5R01CA121197). See supplementary materials for additional acknowledgements.
Footnotes
Disclosure of Potential Conflicts of Interest: Dr. David Nickle is a full time employee at Merck & Co. No potential conflicts of interest were disclosed by other authors.
References
- 1.Hindorff L, MacArthur J, Morales J, Junkins H, Hall P, Klemm A, et al. [Accessed 2014 13/10/2014];A Catalog of Published Genome-Wide Association Studies. < www.genome.gov/gwastudies>.
- 2.Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. Journal of the National Cancer Institute. 2008;100:962–6. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rafnar T, Sulem P, Stacey SN, Geller F, Gudmundsson J, Sigurdsson A, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nature genetics. 2009;41:221–7. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharjee S, Rajaraman P, Jacobs KB, Wheeler WA, Melin BS, Hartge P, et al. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. American journal of human genetics. 2012;90:821–35. doi: 10.1016/j.ajhg.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peters U, Jiao S, Schumacher FR, Hutter CM, Aragaki AK, Baron JA, et al. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology. 2013;144:799–807. e24. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nature genetics. 2013;45:392–8. 98e1–2. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nature genetics. 2013;45:353–61. 61e1–2. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nature genetics. 2013;45:362–70. 70e1–2. doi: 10.1038/ng.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siddiq A, Couch FJ, Chen GK, Lindstrom S, Eccles D, Millikan RC, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Human molecular genetics. 2012;21:5373–84. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, McKay JD, Rafnar T, Wang Z, Timofeeva MN, Broderick P, et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nature genetics. 2014;46:736–41. doi: 10.1038/ng.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Burnett T, Kono S, Haiman CA, Iwasaki M, Wilkens LR, et al. Trans-ethnic genome-wide association study of colorectal cancer identifies a new susceptibility locus in VTI1A. Nature communications. 2014;5:4613. doi: 10.1038/ncomms5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung RJ, Ulrich CM, Goode EL, Brhane Y, Muir K, Chan AT, et al. Cross Cancer Genomic Investigation of Inflammation Pathway for Five Common Cancers: Lung, Ovary, Prostate, Breast, and Colorectal Cancer. Journal of the National Cancer Institute. 2015;107:djv246. doi: 10.1093/jnci/djv246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gudbjartsson DF, Helgason H, Gudjonsson SA, Zink F, Oddson A, Gylfason A, et al. Large-scale whole-genome sequencing of the Icelandic population. Nature genetics. 2015;47:435–44. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- 14.Asomaning K, Miller DP, Liu G, Wain JC, Lynch TJ, Su L, et al. Second hand smoke, age of exposure and lung cancer risk. Lung Cancer. 2008;61:13–20. doi: 10.1016/j.lungcan.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu G, Wheatley-Price P, Zhou W, Park S, Heist RS, Asomaning K, et al. Genetic polymorphisms of MDM2, cumulative cigarette smoking and nonsmall cell lung cancer risk. International journal of cancer Journal international du cancer. 2008;122:915–8. doi: 10.1002/ijc.23178. [DOI] [PubMed] [Google Scholar]
- 16.Al Olama AA, Kote-Jarai Z, Berndt SI, Conti DV, Schumacher F, Han Y, et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nature genetics. 2014;46:1103–9. doi: 10.1038/ng.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiraishi K, Kunitoh H, Daigo Y, Takahashi A, Goto K, Sakamoto H, et al. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nature genetics. 2012;44:900–3. doi: 10.1038/ng.2353. [DOI] [PubMed] [Google Scholar]
- 18.Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z, et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12. 2 in Han Chinese. Nature genetics. 2011;43:792–6. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- 19.Cai Q, Zhang B, Sung H, Low SK, Kweon SS, Lu W, et al. Genome-wide association analysis in East Asians identifies breast cancer susceptibility loci at 1q32.1, 5q14.3 and 15q26. 1. Nature genetics. 2014;46:886–90. doi: 10.1038/ng.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng I, Chen GK, Nakagawa H, He J, Wan P, Laurie CC, et al. Evaluating genetic risk for prostate cancer among Japanese and Latinos. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21:2048–58. doi: 10.1158/1055-9965.EPI-12-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fejerman L, Ahmadiyeh N, Hu D, Huntsman S, Beckman KB, Caswell JL, et al. Genome-wide association study of breast cancer in Latinas identifies novel protective variants on 6q25. Nature communications. 2014;5:5260. doi: 10.1038/ncomms6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen F, Chen GK, Stram DO, Millikan RC, Ambrosone CB, John EM, et al. A genome-wide association study of breast cancer in women of African ancestry. Human genetics. 2013;132:39–48. doi: 10.1007/s00439-012-1214-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haiman CA, Chen GK, Blot WJ, Strom SS, Berndt SI, Kittles RA, et al. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nature genetics. 2011;43:570–3. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andreassen OA, Zuber V, Thompson WK, Schork AJ, Bettella F, Djurovic S, et al. Shared common variants in prostate cancer and blood lipids. International journal of epidemiology. 2014;43:1205–14. doi: 10.1093/ije/dyu090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obeidat M, Miller S, Probert K, Billington CK, Henry AP, Hodge E, et al. GSTCD and INTS12 regulation and expression in the human lung. PloS one. 2013;8:e74630. doi: 10.1371/journal.pone.0074630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Seo JH, Stranger B, McKenna A, Pe’er I, Laframboise T, et al. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell. 2013;152:633–41. doi: 10.1016/j.cell.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuefer R, Day KC, Kleer CG, Sabel MS, Hofer MD, Varambally S, et al. ADAM15 disintegrin is associated with aggressive prostate and breast cancer disease. Neoplasia. 2006;8:319–29. doi: 10.1593/neo.05682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett JH, Iles MM, Harland M, Taylor JC, Aitken JF, Andresen PA, et al. Genome-wide association study identifies three new melanoma susceptibility loci. Nature genetics. 2011;43:1108–13. doi: 10.1038/ng.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nature genetics. 2007;39:352–8. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 30.Khan S, Greco D, Michailidou K, Milne RL, Muranen TA, Heikkinen T, et al. MicroRNA related polymorphisms and breast cancer risk. PloS one. 2014;9:e109973. doi: 10.1371/journal.pone.0109973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin WY, Camp NJ, Ghoussaini M, Beesley J, Michailidou K, Hopper JL, et al. Identification and characterization of novel associations in the CASP8/ALS2CR12 region on chromosome 2 with breast cancer risk. Human molecular genetics. 2014;24:285–98. doi: 10.1093/hmg/ddu431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nature genetics. 2010;42:504–7. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saeki N, Saito A, Choi IJ, Matsuo K, Ohnami S, Totsuka H, et al. A functional single nucleotide polymorphism in mucin 1, at chromosome 1q22, determines susceptibility to diffuse-type gastric cancer. Gastroenterology. 2011;140:892–902. doi: 10.1053/j.gastro.2010.10.058. [DOI] [PubMed] [Google Scholar]
- 34.Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends in molecular medicine. 2014;20(6):332–42. doi: 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kharbanda A, Rajabi H, Jin C, Tchaicha J, Kikuchi E, Wong K-K, et al. Targeting the Oncogenic MUC1-C Protein Inhibits Mutant EGFR-Mediated Signaling and Survival in Non–Small Cell Lung Cancer Cells. Clinical Cancer Research. 2014;20:5423–34. doi: 10.1158/1078-0432.CCR-13-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Najy AJ, Day KC, Day ML. The ectodomain shedding of E-cadherin by ADAM15 supports ErbB receptor activation. The Journal of biological chemistry. 2008;283(26):18393–401. doi: 10.1074/jbc.M801329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X, Wang Z, Zhang X, Chang J, Tang W, Gan L, et al. MUC1 gene polymorphism rs4072037 and susceptibility to gastric cancer: a meta-analysis. SpringerPlus. 2014;3:599. doi: 10.1186/2193-1801-3-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helgason H, Rafnar T, Olafsdottir HS, Jonasson JG, Sigurdsson A, Stacey SN, et al. Loss-of-function variants in ATM confer risk of gastric cancer. Nature genetics. 2015;47(8):906–10. doi: 10.1038/ng.3342. [DOI] [PubMed] [Google Scholar]
- 39.Abnet CC, Wang Z, Song X, Hu N, Zhou FY, Freedman ND, et al. Genotypic variants at 2q33 and risk of esophageal squamous cell carcinoma in China: a meta-analysis of genome-wide association studies. Human molecular genetics. 2012;21(9):2132–41. doi: 10.1093/hmg/dds029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lubahn J, Berndt SI, Jin CH, Klim A, Luly J, Wu WS, et al. Association of CASP8 D302H polymorphism with reduced risk of aggressive prostate carcinoma. The Prostate. 2010;70(6):646–53. doi: 10.1002/pros.21098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S, Chai Z, Li Y, Liu D, Bai Z, Situ Z. BZW1, a novel proliferation regulator that promotes growth of salivary muocepodermoid carcinoma. Cancer letters. 2009;284(1):86–94. doi: 10.1016/j.canlet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 42.Timofeeva MN, Hung RJ, Rafnar T, Christiani DC, Field JK, Bickeboller H, et al. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Human molecular genetics. 2012;21(22):4980–95. doi: 10.1093/hmg/dds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nature genetics. 2009;41(8):899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stacey SN, Sulem P, Masson G, Gudjonsson SA, Thorleifsson G, Jakobsdottir M, et al. New common variants affecting susceptibility to basal cell carcinoma. Nature genetics. 2009;41(8):909–14. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li WQ, Pfeiffer RM, Hyland PL, Shi J, Gu F, Wang Z, et al. Genetic polymorphisms in the 9p21 region associated with risk of multiple cancers. Carcinogenesis. 2014;35:2698–705. doi: 10.1093/carcin/bgu203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30(16):1956–62. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic acids research. 2009;37(Web Server issue):W600–5. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.